Abstract
Apolipoprotein E (apoE), one of the major plasma lipoproteins, also is expressed in a variety of cell types, including the glial cells of the nervous system. apoE is involved in processes of degeneration and regeneration after nerve lesions as well as in the pathogenesis of Alzheimer’s disease (AD). Glial synthesis of apoE is activated in response to injury both in the peripheral and central nervous system. We now report that the activity of the proximal apoE promoter in astrocytes is upregulated by cAMP and retinoic acid, which act synergistically. Sequence analysis of the apoE promoter indicated the presence of several AP-2 consensus sequences that could mediate the stimulatory effect of cAMP and retinoic acid. The possible functional role of AP-2 was examined by cotransfection of AP-2-deficient HepG2 cells with an apoE promoter construct and a human AP-2 expression construct. Cotransfection with AP-2 significantly elevated apoE promoter activity. DNase I footprinting technique revealed the existence of two binding sites for recombinant AP-2 in regions from −48 to −74 and from −107 to −135 of the apoE promoter. Mutations in these regions markedly impaired the trans-stimulatory effect of AP-2. These results indicate the existence of functional AP-2 sites in the promoter region of apoE that could contribute to the complex regulation of this gene in developmental, degenerative, and regenerative processes of the nervous system.
Keywords: apolipoprotein E, astrocytes, AP-2, cAMP, retinoic acid, promoter
Apolipoprotein E (apoE) is a major component of various classes of plasma lipoproteins. It is a single-chain polypeptide of 299 amino acids that plays a prominent role in transport and metabolism of plasma cholesterol and triglycerides as a result of its ability to interact with lipoprotein receptors (Mahley, 1988). The major site of synthesis is the liver, but the protein also is produced in extrahepatic tissues such as adrenals and nervous system. In brain, the synthesis take place in astrocytic cells (Boyles et al., 1985;Pitas et al., 1987), whereas in the peripheral nervous system apoE is synthesized by nonmyelinating glial cells and resident macrophages (Boyles et al., 1985). The protein has been implicated both in peripheral and central nerve regeneration. Synthesis of apoE is increased dramatically after injury to the sciatic nerve (Müller et al., 1985; Ignatius et al., 1986; Snipes et al., 1986) as well as after lesions of the optic nerve or the spinal cord (Boyles et al., 1989). More recently, a link between apoE and Alzheimer’s disease (AD) has been found. The amino acid sequence of the human protein presents two polymorphic sites that generate three alleles (apoE2, apoE3, and apoE4). The isoforms differ in arginine or cysteine content at positions 112 and 158 (Rall et al., 1982). Recent genetic studies have identified the apoE4 allele as a major risk factor for developing AD, both in sporadic and in familial late onset AD (Corder et al., 1993;Strittmatter et al., 1993).
ApoE synthesis is regulated in hepatic and steroidogenic cells by a complex interaction of developmental, hormonal, and dietary factors (Basu et al., 1981; Lin-Lee et al., 1981; Reue et al., 1984;Elshourbagy et al., 1985; Kayden et al., 1985). The regulatory complexity emerges from interactions of a number of proteins that bind to the proximal promoter region as well as to far downstream elements involved in its tissue-specific expression (Paik et al., 1988; Smith et al., 1988; Simonet et al., 1990, 1993; Berg et al., 1995). The regulation in brain cells, however, remains unexplored despite the importance of this protein in processes of degeneration and regeneration of the nervous system. In this report we have studied the regulation of the expression of the proximal apoE promoter in astrocytic cells. We show the presence of two functional binding sites for the transcription factor AP-2 in this region of the promoter that mediate the stimulatory effect of the differentiating agents cAMP and retinoic acid on the activity of the promoter.
MATERIALS AND METHODS
Materials
Taq polymerase was obtained from Perkin-Elmer (Norwalk, CT). DNase I, DOTAP, ligase, and restriction enzymes were obtained from Boehringer Mannheim (Mannheim, Germany). Recombinant AP-2 was obtained from Promega (Madison, WI) and poly(dI-dC) from Pharmacia (Uppsala, Sweden). Oligonucleotides were obtained from Isogen (Maarseen, The Netherlands). All other reagents were obtained in the purest form available.
Methods
Plasmid constructions. The 5′ region between positions −1011 and +400 of the apoE gene was amplified by PCR using human genomic DNA as a template and the following primers: CAAGGTCACACAGCTGGCAACT and TCCAATCGACGGCTAGCTACC. The amplified fragment was ligated to the pCRII vector (Invitrogen, San Diego, CA) (apoE–pCRII construct), and its identity was confirmed by sequencing with the Fentomol kit (Promega). The 1.4 kb-cloned DNA fragment was subcloned in front of the luciferase reporter gene in theXhoI/HindIII sites of the pXP2 vector (Nordeen, 1988). Different deletions were generated by using a similar PCR strategy: oligonucleotides were designed at the desired positions and used as primers with the apoE–pCRII construct as a template. Amplified fragments were cloned in the pCRII plasmid, sequenced, and subcloned in the XhoI/HindIII sites of the luciferase expression vector pXP2.
Site-directed mutagenesis. The PCR-based site-directed mutagenesis strategy followed to destroy the AP-2 binding sites of the apoE promoter was a modification of the method of Higuchi (1990), as described by Olivares et al. (1995). The deleted construct −227 was used as a template, and the following oligonucleotides were used as primers: GTCCCGCCCCCT GTCGACCGGATAGGGCGGGC, which exchanged the −60 AP-2 element for the underlined SalI restriction site, and CCCTCTGCCCTGCT GAATTCGGAGAACAGCCCA, which exchanged the −117 AP-2 element for the underlined EcoRI restriction site. The mutated PCR fragments were introduced into pCRII, identified by restriction analysis, sequenced, and transferred to theXhoI/HindIII sites of the luciferase expression vector pXP2.
Cell culture and transfections. U87 and HepG2 cells were grown in DMEM containing 10% fetal bovine serum. The day before transfection, confluent cells were subcultured by trypsinization, and 1–3 × 104 cells per well were plated in 24 well tissue culture plates. HepG2 cells were transfected by the calcium phosphate method. Briefly, calcium phosphate–DNA coprecipitates were prepared by adding, dropwise, a 220 mm calcium chloride solution to an equal volume of vortexing HEPES-buffered saline solution [(in mm) 275 NaCl, 40 HEPES, 10 KCl, 1.4 sodium phosphate, pH 7.05, and 12 glucose] containing plasmid. Then 0.5 ml of the DNA precipitates (0.8 μg each of test and reference plasmid constructions) was added to each well and allowed to incubate with the cells for 14 hr. HepG2 cells were shocked by adding a 10% DMSO solution in PBS for 2.5 min at room temperature. Cells were washed once with serum-free DMEM and replenished with growth medium. Then cells were harvested on day 2 after transfection. U87 cells were transfected with 0.5 μg of DNA per well by lipofection withN-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate (DOTAP; Boehringer Mannheim), according to the manufacturer’s instructions. The different drug treatments were initiated immediately after the transfection procedure and kept until cells were harvested.
Luciferase and β-galactosidase assays. Cells were harvested on day 2 after transfection with 150 μl of a lysis buffer containing 25 mm Tris-phosphate, 2 mm DTT, 2 mm EDTA, 10% glycerol, and 1% Triton X-100. Unsolubilized material was removed by 2 min of centrifugation, and the luciferase and β-galactosidase activities of the extracts were determined. Luciferase was measured by the Luciferase Assay System (Promega) in a Monolight 2010 luminometer (Analytical Luminescence Laboratory, San Diego, CA) by incubation of 10 μl of cell extract with 90 μl of luciferase assay reagent, as recommended by the manufacturer. β-Galactosidase was determined in a 96 well microtiter plate by incubating 20 μl of cellular extract with 80 μl of a solution containing 3 mg/ml of o-nitrophenyl-β-d-galactopyranoside, as described (Sambrook et al., 1989). Absorbance at 405 nm was determined in an MR 5000 microplate reader (Dynatech, West Sussex, UK).
DNase I protection assay. Oligonucleotides were end-labeled by T4 polynucleotide kinase in the presence of [γ-32P]ATP. These oligonucleotides were used as PCR primers as described (Krummel, 1990). The amplification products were purified via a DNA Purification System (Promega). The footprinting reactions were performed in 20 μl containing 100,000 cpm of labeled probe and 1–4 footprinting units of AP-2 protein (Promega), containing (in mm): 20 HEPES, pH 7.9, 100 KCl, 0.2 EDTA, 12 MgCl2, 0.5 DTT, and 0.5 phenylmethylsulfonyl fluoride with 20% glycerol, 0.3 μg/ml leupeptin, and 4 μg of poly(dI-dC). The mixture was incubated for 10 min on ice and for 20 min at room temperature and treated with DNase I for 30 sec by the addition of 50 μl of a solution containing 1 unit DNase I in 10 mmMgCl2. The digestion was terminated by adding 150 μl of 8m urea, 0.5% sodium dodecyl sulfate, and 5 mmEDTA. The DNA was phenol-extracted and ethanol-precipitated before being subjected to electrophoresis in a gel containing 6% polyacrylamide and 6 m urea. The position of the protected regions was determined by comparison with a G+A sequence ladder, as described (Maxam and Gilbert, 1980).
RESULTS
Positive and negative regulatory regions within apoE promoter mediate expression in astrocytoma cells
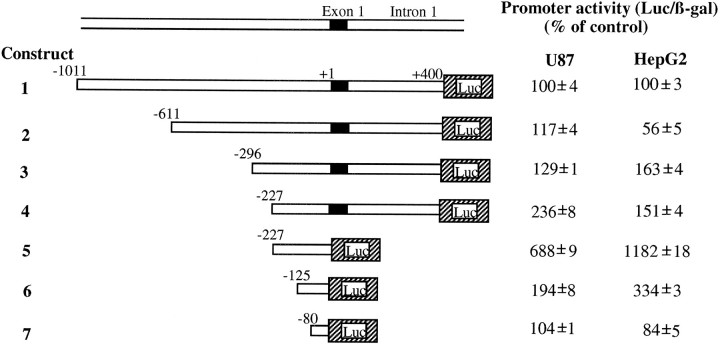
The expression of the apoE proximal promoter in astrocytic cells was studied in a transient expression system that allowed us to map which sequences were necessary to yield maximal expression of apoE in these cells. Fragments containing different sequences comprising between −1011 and +400 of the apoE gene 5′region were generated by PCR and fused to the coding sequence of the luciferase gene in the pXP2 vector. A series of constructions with various deletions (Fig.1) were transfected into U87 astrocytoma cells, and the luciferase activity was determined 48 hr later. Along with the test constructions, each plate was cotransfected with a β-galactosidase expression vector that served as an internal reference for transfection efficiency. For comparison we transfected the hepatoma cell line HepG2 with the same constructions. As shown, a stepwise increase in the luciferase activity was observed in U87 cells by successive deletions in the 5′ end of the promoter region up to the nucleotide −227. The activity increased from 100% in the longest construct assayed (construct 1) to 117% in construct 2, 129% in construct 3, and 236% in construct 4. These data indicated the presence of negative regulatory elements in this region, especially between nucleotides −227 and −296. The promoter activity pattern was clearly different in HepG2, where a positive element was defined in the region between nucleotides −1011 and −611, because the luciferase activity decreased from 100% in the long construction to 56% in construct 2 (Fig. 1). The increased luciferase activity obtained with construct 3 (160% of longer construct), as compared with construct 2, defined a negative element between nucleotides −611 and −296. In addition, a negative element was defined in both cell types in the first intron, because deletion of the region +1 to +400 produced a strong increase in the activity of the reporter, especially in HepG2. Further deletions to nucleotides −125 and −80 defined a further positive element as the reporter activity decreased stepwise in both cell types.
Fig. 1.
Promoter activity of different constructs containing truncated forms of apoE promoter fused to luciferase reporter gene. The structure of the constructs is shown at theleft. The genomic structure of the apoE gene 5′ region is indicated at the top. Luc, Luciferase reporter gene. Luciferase/β-galactosidase ratios are expressed at theright, as percentages of activity of construct 1, for U87 and HepG2 cells. Values are expressed as the mean ± SEM of two triplicate determinations and are corrected for the activities measured by using the promoterless pXP2 plasmid.
cAMP and retinoic acid differentially regulate apoE promoter in astrocytoma and hepatoma cell lines
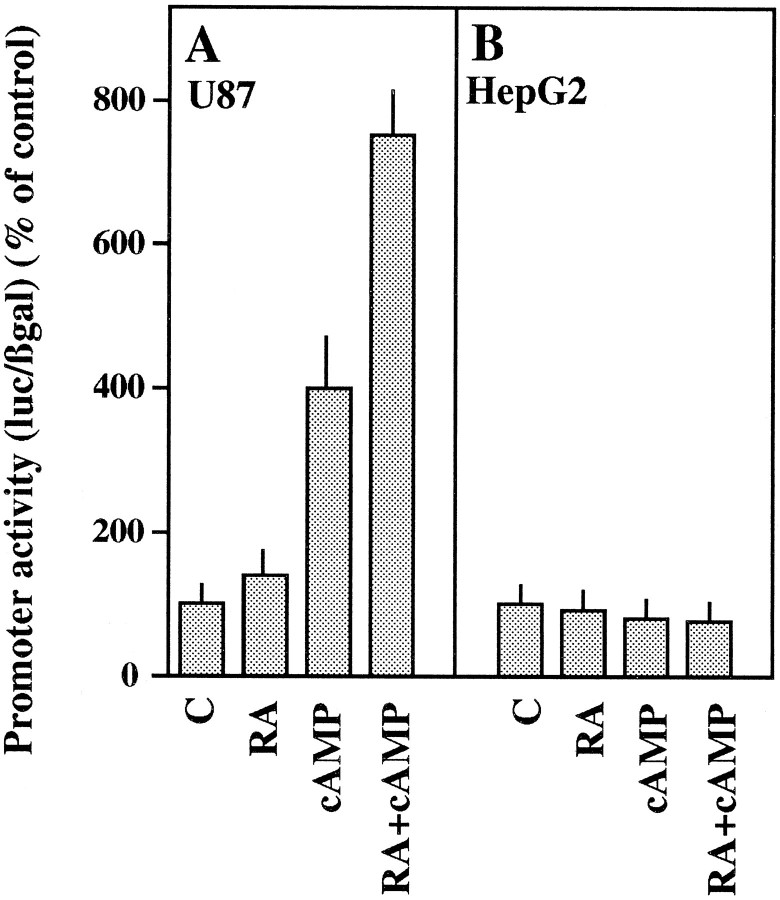
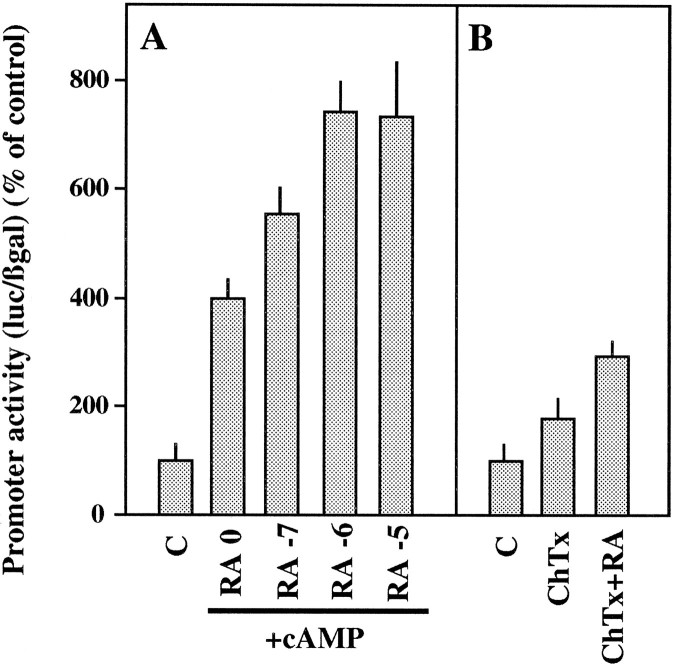
Transformation of normal astrocytes to reactive astrocytes, which occurs in brain after injury, is accompanied by a dramatic increase in the synthesis of apoE in these cells. Many of the changes that occur in reactive gliosis can be mimicked in vitro by treatment with cAMP (Fedoroff et al., 1984; Sharma and Raj, 1987). To study whether cAMP had any effect on the activity of the apoE promoter, we examined reporter expression in U87 astrocytoma cells transiently transfected with construct 4 and treated with dibutyryl-cAMP (dBcAMP). For comparison, the same treatment was performed in transfected HepG2 cells. Figure 2 shows that dBcAMP treatment produced a differential effect in U87 and HepG2 cells, stimulating approximately fourfold over the basal level in astrocytoma but being ineffective in hepatoma. Interestingly, the effect of dBcAMP in U87 cells was potentiated by retinoic acid (RA), a potent morphogenetic and teratogenic agent. Although the effect of RA alone was rather weak (140% over the nontreated control), it acted synergistically with dBcAMP in U87 cells, obtaining a stimulation of 750% when both substances were added together to the culture medium (Fig.2A). None of these treatments affected the promoter activity in HepG2 cells (Fig. 2B). Quantitatively similar results were obtained by transfecting both cell lines with construct 1 (data not shown). The synergistic effect of RA and dBcAMP was observable at RA concentrations of 10−7mbut was maximal at 10−6m (Fig.3A). Other treatments, such as addition of cholera toxin, which is known to increase the intracellular cAMP content, were also stimulatory of the apoE promoter activity (Fig.3B), although the observed effect was less remarkable.
Fig. 2.
Effect of cAMP and RA on the apoE promoter activity. U87 (A) or HepG2 (B) cells transiently transfected with construct 4 and a β-galactosidase expression construct were incubated for 48 hr in the absence (C) or the presence of 1 μm RA (RA), 1 mm dBcAMP (cAMP), or both agents simultaneously (RA+cAMP). Luciferase and β-galactosidase activities were determined as indicated in Materials and Methods. Results are expressed as a percentage of activities of untreated control cells (C). Values are the mean ± SEM of two triplicate determinations and are corrected for the activities measured by using the promoterless pXP2 plasmid.
Fig. 3.
Effect of RA at different concentrations and cholera toxin on the apoE promoter activity. U87 cells were transiently transfected with construct 4 and a β-galactosidase expression construct. A, Cells were incubated for 48 hr in the absence (C) or the presence of 1 mm dBcAMP (+cAMP) plus either 0 (RA 0), 10−7(RA-7), 10−6(RA-6), or 10−5m RA (RA-5). B, Cells were incubated in the absence (C) or the presence of 1.25 μg/ml cholera toxin (ChTx), or 1.25 μg/ml cholera toxin plus 1 μm RA (ChTx+RA). Luciferase and β-galactosidase activities were determined as indicated in Materials and Methods. Results are expressed as a percentage of activities of untreated controls. Values are the mean ± SEM of two triplicate determinations and are corrected for the activities measured by using the promoterless pXP2 plasmid.
AP-2 upregulates apoE by binding to −117 and −60 AP-2 consensus sequences
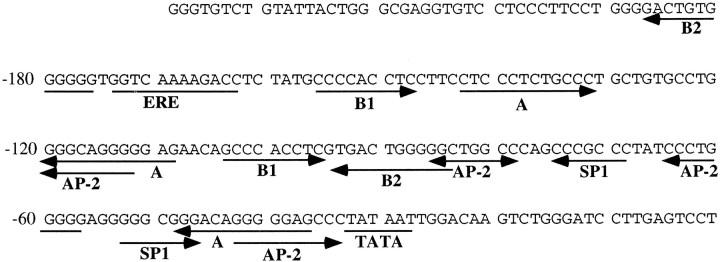
It is well established for a number of genes that regulatory responses to cAMP can be transduced by CRE, AP-2, or NFκB binding sites located in their promoter regions. A search of consensus recognition patterns in the apoE promoter revealed the presence of several putative regulatory sequences, among them four AP-2 binding sites (Fig. 4). AP-2 is known to be upregulated by RA; moreover, HepG2 is an AP-2 deficient cell line. Therefore, our data suggested the possibility that cAMP and RA could upregulate the apoE gene by a mechanism mediated by AP-2. To investigate this hypothesis, we studied the ability of AP-2 to stimulate transcription from the apoE promoter by cotransfecting the apoE promoter–luciferase construct 4 with an expression vector of AP-2. The experiments were performed in HepG2, which, as mentioned above, is deficient in AP-2. Cotransfection of construct 4 and AP-2 produced a stimulation of 1040 ± 60% (mean ± SEM of two triplicate determinations) of the reporter activity, as compared with control cells transfected with construct 4 (100 ± 5%), thus suggesting a regulatory role of AP-2 over apoE promoter.
Fig. 4.
Nucleotide sequence of the upstream region of the human apoE gene. The sequence is numbered relative to the transcription start site (+1). Localization of some of the putative regulatory elements is indicated below the sequence.TATA, The TATA box element; SP1, the GC box element; AP-2, AP-2-like binding sequence;ERE, element with homology to the estrogen-responsive elements; A, B1, and B2 are the apoE A, B1, and B2 elements, respectively, as defined by Smith et al. (1988).
To analyze whether the observed effect of AP-2 was attributable to direct binding of this transcription factor to apoE promoter, we first studied the ability of recombinant AP-2 to bind the end-labeled DNA fragment −1 to −227. As shown in Figure 5, DNase I footprinting experiments identified two protected areas in this DNA fragment. The boundaries of the binding sites were located in nucleotides −48 to −74 (footprint −60) and −104 to −135 (footprint −117). The two footprints were observed in both the noncoding (Fig.5A) and the coding strands (Fig. 5B). Second, the functional importance of these regions in AP-2 stimulatory effect was confirmed by site-directed mutagenesis of the two footprints. Mutants in the −117 and in the −60 AP-2 binding sites of construct 4 were generated and cotransfected with the AP-2 expression vector in HepG2 cells. As shown in Figure 6, mutations at both the −117 and the −60 sites markedly reduced the trans-stimulatory effect of AP-2 (52 and 66% inhibition, respectively; Fig. 6A), although the simultaneous mutation of both −117 and −60 footprints did not produce a higher level of inhibition. These experiments clearly established that these two binding sites are used functionally by AP-2. A weaker protection by recombinant AP-2 also was observed in the −85 to −100 region (Fig. 5). However, site-directed mutagenesis of this area did not affect the trans-stimulatory effect of AP-2 (data not shown). Mutations in the other putative AP-2 site located around nucleotide −40 were also ineffective (data not shown).
Fig. 5.
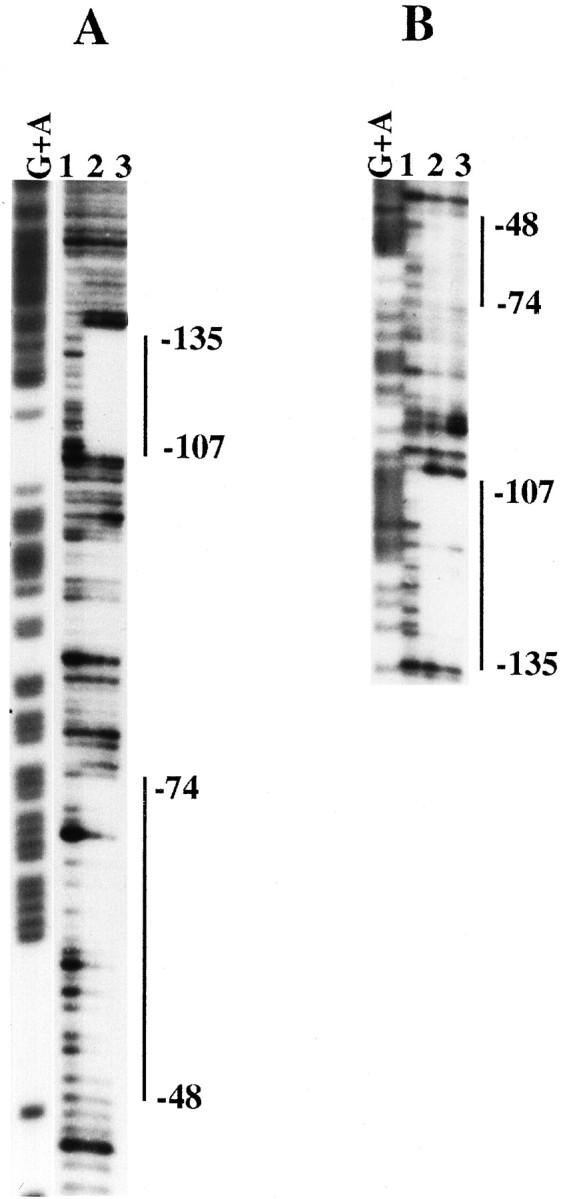
DNase I footprinting analysis of the proximal apoE promoter. A DNA fragment containing −1 to −227 bp of the apoE promoter was end-labeled with [32P]ATP on the antisense (A) or sense (B) strands. Lane G+A in A and B represents the Maxam and Gilbert sequencing ladder of the apoE promoter. Lane 1 in A and B represents reactions performed in the absence of recombinant AP-2. Lanes 2and 3 in A and B represent reactions performed in the presence of 1 or 4 footprinting units of recombinant AP-2, respectively. The protected sequence boundaries around the −60 and the −117 footprints are indicated.
Fig. 6.
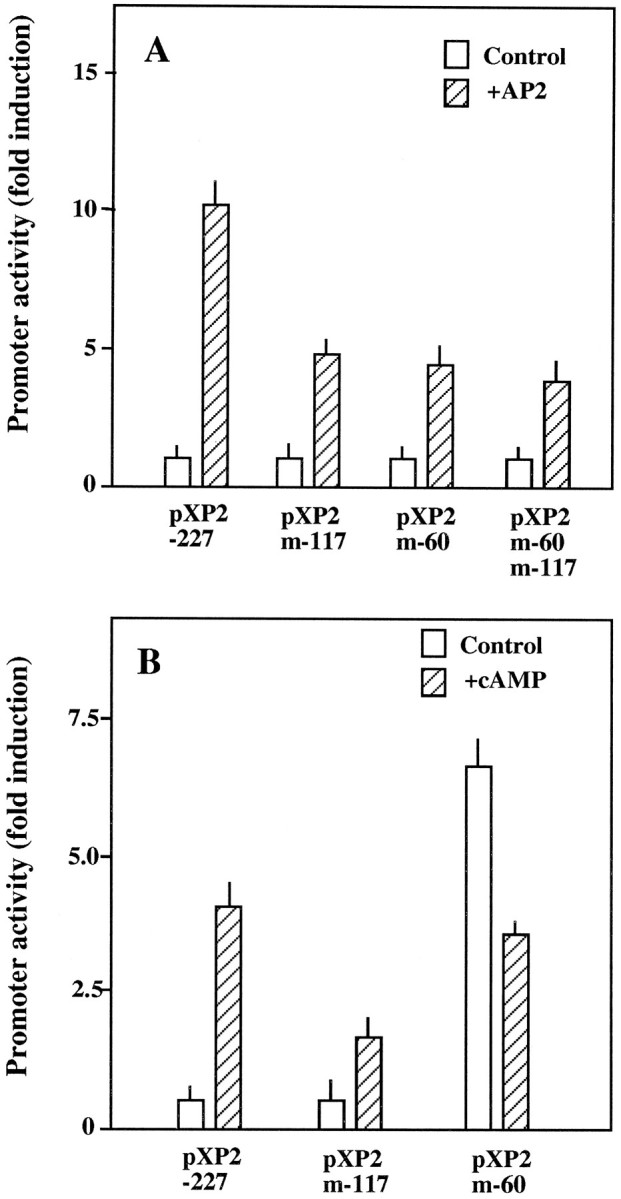
Effect of site-directed mutagenesis of footprints −117 and −60 on apoE promoter activity. A, HepG2 cells were transiently transfected with pXP2 (pXP2), construct 4 (pXP2-227), or construct 4 mutated at the −117 (pXP2-m117), −60 (pXP2m-60), or both footprints (pXP2m-60m-117). Cells were cotransfected with an AP-2 expression construct (hatched bars) or with pBluescript (empty bars). B, U87 cells were transfected with the indicated construct and incubated in the absence (empty bars) or presence (hatched bars) of 1 mm dBcAMP. Luciferase activity was determined 48 hr later. Fold induction values are relative to those obtained with untreated construct 4. Values are the mean ± SEM of two triplicate determinations and are corrected for the activities measured by using the promoterless pXP2 plasmid.
Finally, to confirm that the stimulatory effect of cAMP in astrocytic cells was mediated by interaction with these two sites, we transfected the mutant constructs into U87 cells and measured the luciferase activity after cAMP treatment (Fig. 6B). Mutation in the −117 region clearly reduced the stimulatory effect of cAMP by 69%, strongly suggesting that cAMP effect in astrocytes was mediated via the AP-2 binding site in the −117 region. However, mutation of the −60 region yielded more complex results, because basal activity of this mutant was greatly augmented (625% of the wild type) in untreated cells. This effect was cell-specific, because it was not observed in HepG2. Moreover, treatment with cAMP of U87 cells transfected with this mutant did not further increase the activity of the promoter but was inhibitory (56% of the corresponding basal level) (Fig.6B).
DISCUSSION
In the present report we describe the upregulation of the apoE promoter by cAMP and RA in astrocytic cells. This regulatory process is mediated by two AP-2 binding sites located in the proximal region of the promoter. The determination of the molecular mechanism involved in the regulation of apoE synthesis in brain is currently a matter of the greatest importance, considering the possible roles of this protein in processes of repair after traumatic injury (Müller et al., 1985;Ignatius et al., 1986; Boyles et al., 1989) or in the pathogenesis of AD (Corder et al., 1993; Strittmatter et al., 1993). The reactive gliosis accompanying these pathologies involves a series of morphological and biochemical changes in the astrocytes. One of the more abundant proteins produced by activated astrocytes is apoE (Poirier et al., 1991). The exact physiological role of this increased expression remains primarily speculative, but experiments in vitro have shown an effect of apoE on neurite morphogenesis by cultured neurons. Thus, apoE reduces the amount of neurite branching and promotes neurite extension (Handelmann et al., 1992; Nathan et al., 1994). Interestingly, this effect was observed only with the addition of the apoE3 isoform, whereas addition of the apoE4 isoform, which is associated to a high risk of late onset AD, resulted in a reduction of both branching and extension of the neurites (Nathan et al., 1994).
Regulation of the apoE promoter has been investigated thoroughly in a variety of cell types, mainly hepatoma cells, where it has been shown to contain an array of tissue-specific cis-acting regulatory elements that are distributed along a 20 kb region spanning the apoE gene (Smith et al., 1988; Simonet et al., 1990, 1993). The regulatory elements required for an efficient expression in HepG2 cells are located in the proximal 5′-flanking region and in the first intron (Paik et al., 1988;Smith et al., 1988; Berg et al., 1995). Data in the present report also indicate that this region of the apoE promoter drives efficient expression in astrocytoma cells, with the maximal activity of the promoter being obtained with the fragment located between nucleotides −1 and −227. Our truncation and site-directed mutagenesis studies show that the pattern of positive and negative regulatory elements is different in astrocytoma and hepatoma cells, indicating that, as in other cell types, synthesis of apoE in astrocytes is regulated by tissue-specific factors.
In a variety of in vitro experimental systems, including primary astrocytic cultures or glioblastoma cultures, it has been shown that many of the changes occurring in reactive gliosis can be mimicked by treatment with cAMP (Fedoroff et al., 1984; Sharma and Raj, 1987). In our experimental system, cAMP stimulated the activity of the apoE promoter by a molecular mechanism that involves the transcription factor AP-2. This factor is a 52 kDa protein that binds as a dimer to the palindromic recognition sequence 5′-GCCNNNGGC-3′ (Mitchell et al., 1987; Williams et al., 1988), although many AP-2-binding sites deviate from this consensus (Williams and Tjian, 1991). Cell culture andin vitro experiments have demonstrated a role for AP-2 in regulating a variety of target genes, including human metallothionein-IIA (Imagawa et al., 1987), human growth hormone (Courtois et al., 1990), human T-cell leukemia virus type I (Muchardt et al., 1992), human proenkephalin (Hyman et al., 1989), acetylcholinesterase (Ekström et al., 1993), or Na+/H+ exchanger (Dyck et al., 1995). Sequence analysis of the apoE promoter indicated the presence of a consensus sequence around position −117. The DNase I footprinting experiments described herein indeed showed that purified AP-2 protects the region located between nucleotides −105 to −134, and the site-directed mutagenesis experiments also supported the functionality of this AP-2 binding site. A second binding site also was identified by the same approaches and was targeted to the region located between nucleotides −48 and −74. These findings are consistent with the presence of another putative AP-2-binding site in the same region. The sequence analysis also indicates that this region may be the target of a complex regulation by a number of other transcription factors; specifically, two putative SP1 binding sites exist that flank the AP-2 core sequence. These predictions are also consistent with our observation that mutations in this AP-2 site dramatically increase the apoE promoter expression basal level in astrocytoma cells, but not in HepG2 cells, strongly suggesting the existence of an astrocyte-specific inhibitory factor that interacts with the same region of the promoter. The proximal region of the apoE promoter has been shown to be extremely rich in regulatory sequences in cells of diverse tissular origin (Paik et al., 1988; Smith et al., 1988; Berg et al., 1995), indicating that this region could be involved in some aspects of the complex tissue-specific regulation of the apolipoprotein E gene.
AP-2 is expressed by neurons and astrocytes, and it has been suggested to play a role in the development of the neuronal and astrocytic cell linages as well as in the cAMP-dependent activation of astrocytes (Lüscher et al., 1989; Mitchell et al., 1991; Philipp et al., 1994). Moreover, the expression of AP-2, which is low in primary brain astrocytes, is induced rapidly and strongly by cAMP stimulation of these cells (Philipp et al., 1994). On the other hand, RA, a potent morphogenetic and teratogenic agent on the developing nervous system, also is known to regulate at the transcriptional level the expression of AP-2 (Lüscher et al., 1989). Therefore, it is reasonable to presume that the observed synergistic effect of RA and cAMP on the apoE promoter described in the present work is probably attributable to a RA-promoted increase in the cellular content of AP-2, followed by a post-transcriptional increase of the AP-2 activity mediated by cAMP (Imagawa et al., 1987; Lüscher et al., 1989).
Analysis of AP-2 expression during embryogenesis has suggested that AP-2 may be required during differentiation of some neural cell types (Lüscher et al., 1989; Mitchell et al., 1991). In the mouse embryo, AP-2 is expressed in neural crest cells and in specific portions of the nervous system, where it may play a role during establishment of the peripheral nervous system and its connection with the central nervous system (Mitchell et al., 1991). More recently, multiple congenital defects and perinatal death have been reported in mice containing a homozygous disruption of the AP-2 gene (Schorle et al., 1996; Zhang et al., 1996). Nevertheless, it remains unexplored whether apoE is involved in some of the AP-2-dependent processes during embryogenesis and whether AP-2 is induced in the brain after diverse types of brain injury. In this context, it is suggestive of the increase in the synthesis of AP-2 in primary afferents that have been described to take place during acute inflammation (Donaldson et al., 1995). Our finding that AP-2 could participate in the regulation of apoE expression in astrocytes provides a common background to improve our understanding of the action mechanisms by which these proteins play their roles in the development and repairing of the nervous system and in several pathogenic conditions, such as Alzheimer’s disease.
In summary, the results reported herein indicate the existence of functional AP-2 sites in the promoter region of apoE that could contribute to the complex regulation of this gene in developmental, degenerative, and regenerative processes of the nervous system .
Footnotes
This work was supported by Boehringer Ingelheim España, S.A., Spanish Dirección General de Investigación Científica y Técnica (Grant PB93-0182), and an institutional grant from the Fundación Ramón Areces. M.A.G. is a recipient of a fellowship from the Spanish Ministerio de Educación y Ciencia. We thank Dr. Buettner for providing us with the AP-2 expression vector.
Correspondence should be addressed to Dr. Francisco Zafra, Centro de Biología Molecular “Severo Ochoa,” Facultad de Ciencias, Universidad Autónoma de Madrid, E-28049 Madrid, Spain.
REFERENCES
- 1.Basu SK, Brown MS, Ho YK, Havel RJ, Goldstein JL. Mouse macrophages synthesize and secrete a protein resembling apolipoprotein E. Proc Natl Acad Sci USA. 1981;78:7545–7549. doi: 10.1073/pnas.78.12.7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg DT, Calnek DS, Grinnell BW. The human apolipoprotein E gene is negatively regulated in human liver HepG2 cells by the transcription factor BEF-1. J Biol Chem. 1995;270:15447–15450. doi: 10.1074/jbc.270.26.15447. [DOI] [PubMed] [Google Scholar]
- 3.Boyles JK, Pitas RE, Wilson E, Mahley RW, Taylor JM. Apolipoprotein E associated with astrocytic glia of the central nervous system and with nonmyelinating glia of the peripheral nervous system. J Clin Invest. 1985;76:1501–1513. doi: 10.1172/JCI112130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyles JK, Zoellner CD, Anderson LJ, Kosik LM, Pitas RE, Weisgraber KH, Hui DY, Mahley RW, Gebicke-Haerter PJ, Ignatius MJ, Shooter EM. A role for apolipoprotein E, apolipoprotein A-I, and low density lipoprotein receptors in cholesterol transport during regeneration and remyelination of the rat sciatic nerve. J Clin Invest. 1989;83:1015–1031. doi: 10.1172/JCI113943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corder EH, Saunders AM, Strittmatter WJ, Schmechel D, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of AD in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 6.Courtois SJ, Lafontaine DA, Lemaigre FP, Durviaux SM, Rousseau GG. Nuclear factor-I and activator protein-2 bind in a mutually exclusive way to overlapping promoter sequences and transactivate the human growth hormone gene. Nucleic Acids Res. 1990;18:57–64. doi: 10.1093/nar/18.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donaldson LF, McQueen DS, Seckl JR. Induction of transcription factor AP-2 mRNA expression in rat primary afferent neurons during acute inflammation. Neurosci Lett. 1995;196:181–184. doi: 10.1016/0304-3940(95)11870-3. [DOI] [PubMed] [Google Scholar]
- 8.Dyck JRB, Silva NLCL, Fliegel L. Activation of the Na+/H+ exchanger gene by the transcription factor AP-2. J Biol Chem. 1995;270:1375–1381. doi: 10.1074/jbc.270.3.1375. [DOI] [PubMed] [Google Scholar]
- 9.Ekström TJ, Klump WM, Getman D, Karin M, Taylor P. Promoter elements and transcriptional regulation of the acetylcholinesterase gene. DNA Cell Biol. 1993;12:63–72. doi: 10.1089/dna.1993.12.63. [DOI] [PubMed] [Google Scholar]
- 10.Elshourbagy NA, Liao WS, Mahley RW, Taylor JM. Apolipoprotein E mRNA is abundant in the brain and adrenals, as well as in the liver, and is present in other peripheral tissues of rats and marmosets. Proc Natl Acad Sci USA. 1985;82:203–207. doi: 10.1073/pnas.82.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fedoroff S, McAuley WAJ, Houle JD, Devon RM. Astrocytic cell linage. V. Similarity of astrocytes that form in the presence of dBcAMP in culture to reactive glia. J Neurosci Res. 1984;30:359–371. doi: 10.1002/jnr.490120103. [DOI] [PubMed] [Google Scholar]
- 12.Handelmann GE, Boyles JK, Weisgraber KH, Mahley RW, Pitas RE. Effects of apolipoprotein E, β-very low density lipoproteins, and cholesterol on the extension of neurites by rabbit dorsal root ganglion neurons in vitro. J Lipid Res. 1992;33:1677–1688. [PubMed] [Google Scholar]
- 13.Higuchi R. Recombinant PCR. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols. Academic; New York: 1990. pp. 177–183. [Google Scholar]
- 14.Hyman SE, Comb M, Pearlberg J, Goodman HM. An AP-2 element acts synergistically with the cyclic AMP- and phorbol ester-inducible enhancer of the human proenkephalin gene. Mol Cell Biol. 1989;9:321–324. doi: 10.1128/mcb.9.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ignatius MJ, Gebicke-Haerter PJ, Skene JHP, Schilling JW, Weisgraber KH, Mahley RW, Shooter EM. Expression of apolipoprotein E during nerve degeneration and regeneration. Proc Natl Acad Sci USA. 1986;83:1125–1129. doi: 10.1073/pnas.83.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imagawa M, Chiu R, Karin M. Transcription factor AP-2 mediates induction by two different signal-transduction pathways: protein kinase C and cAMP. Cell. 1987;51:251–260. doi: 10.1016/0092-8674(87)90152-8. [DOI] [PubMed] [Google Scholar]
- 17.Kayden HJ, Maschio F, Traber MG. The secretion of apolipoprotein E by human monocyte-derived macrophages. Arch Biochem Biophys. 1985;239:388–395. doi: 10.1016/0003-9861(85)90704-0. [DOI] [PubMed] [Google Scholar]
- 18.Krummel B. DNase I footprinting. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols. Academic; New York: 1990. pp. 184–188. [Google Scholar]
- 19.Lin-Lee YC, Tanaka Y, Lin CT, Chan L. Effects of an atherogenic diet on apolipoprotein E biosynthesis in the rat. Biochemistry. 1981;20:6474–6480. doi: 10.1021/bi00525a028. [DOI] [PubMed] [Google Scholar]
- 20.Lüscher B, Mitchell PJ, Williams T, Tjian R. Regulation of transcription factor AP-2 by the morphogen retinoic acid and second messengers. Genes Dev. 1989;3:1507–1517. doi: 10.1101/gad.3.10.1507. [DOI] [PubMed] [Google Scholar]
- 21.Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 22.Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell PJ, Wang C, Tjian R. Positive and negative regulation of transcription in vitro: enhancer-binding protein AP-2 is inhibited by SV-40 T antigen. Cell. 1987;50:847–861. doi: 10.1016/0092-8674(87)90512-5. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell PJ, Timmons PM, Hebert JM, Rigby PW, Tjian R. Transcription factor AP-2 is expressed in neural crest cell lineages during mouse embryogenesis. Genes Dev. 1991;5:105–119. doi: 10.1101/gad.5.1.105. [DOI] [PubMed] [Google Scholar]
- 25.Muchardt C, Seeler JS, Nirula A, Gong S, Gaynor R. Interleukin-1β regulates proenkephalin gene expression in astrocytes cultured from rat cortex. Glia. 1992;6:206–212. doi: 10.1002/glia.440060308. [DOI] [PubMed] [Google Scholar]
- 26.Müller HW, Gebicke-Härter PJ, Hangen DH, Shooter EM. A specific 37,000-dalton protein that accumulates in regenerating but not in nonregenerating mammalian nerves. Science. 1985;228:499–501. doi: 10.1126/science.3983637. [DOI] [PubMed] [Google Scholar]
- 27.Nathan BP, Bellosta S, Sanan DA, Weisgraber KH, Mahley RW, Pitas RE. Differential effects of apolipoprotein E3 and E4 on neuronal growth in vitro. Science. 1994;264:850–852. doi: 10.1126/science.8171342. [DOI] [PubMed] [Google Scholar]
- 28.Nordeen SK. Luciferase reporter gene vector for analysis of promoters and enhancers. Biotechniques. 1988;6:454–457. [PubMed] [Google Scholar]
- 29.Olivares L, Aragón C, Giménez C, Zafra F. The role of N-glycosylation in the targeting and activity of the GLYT1 glycine transporter. J Biol Chem. 1995;270:9437–9442. doi: 10.1074/jbc.270.16.9437. [DOI] [PubMed] [Google Scholar]
- 30.Paik YK, Chang DJ, Reardon CA, Walker MD, Taxman E, Taylor JM. Identification and characterization of transcriptional regulatory regions associated with expression of the human apolipoprotein E gene. J Biol Chem. 1988;263:13340–13349. [PubMed] [Google Scholar]
- 31.Philipp J, Mitchell PJ, Malipiero U, Fontana A. Cell type-specific regulation of expression of transcription factor AP-2 in neuroectodermal cells. Dev Biol. 1994;165:602–614. doi: 10.1006/dbio.1994.1279. [DOI] [PubMed] [Google Scholar]
- 32.Pitas RE, Boyles JK, Lee SH, Foss D, Mahley RW. Astrocytes synthesize apolipoprotein E and metabolize apolipoprotein E-containing lipoproteins. Biochim Biophys Acta. 1987;917:148–161. doi: 10.1016/0005-2760(87)90295-5. [DOI] [PubMed] [Google Scholar]
- 33.Poirier J, Hess M, May PC, Finch CE. Astrocytic apolipoprotein E mRNA and GFAP mRNA in hippocampus after entorhinal cortex lesioning. Mol Brain Res. 1991;11:97–106. doi: 10.1016/0169-328x(91)90111-a. [DOI] [PubMed] [Google Scholar]
- 34.Rall SC, Jr, Weisgraber KH, Mahley RW. Human apolipoprotein E. The complete amino acid sequence. J Biol Chem. 1982;257:4171–4178. [PubMed] [Google Scholar]
- 35.Reue KL, Quon DH, O’Donnell KA, Dizikes GJ, Fareed GC, Lusis AJ. Cloning and regulation of messenger RNA for mouse apolipoprotein E. J Biol Chem. 1984;259:2100–2107. [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual, 2nd Ed. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 37.Schorle H, Meier P, Buchert M, Jaenisch R, Mitchell PJ. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature. 1996;381:235–238. doi: 10.1038/381235a0. [DOI] [PubMed] [Google Scholar]
- 38.Sharma SK, Raj AB. Transient increase in intracellular concentrations of adenosine 3′:5′-cyclic monophosphate results in morphological and biochemical differentiation of C6 glioma cells in culture. J Neurosci Res. 1987;17:137–141. doi: 10.1002/jnr.490170207. [DOI] [PubMed] [Google Scholar]
- 39.Simonet WS, Bucay N, Lauer SJ, Wirak DO, Stevens ME, Weisgraber KH, Pitas RE, Taylor JM. In the absence of downstream element, the apolipoprotein E gene is expressed at high level in kidneys of transgenic mice. J Biol Chem. 1990;265:10809–10812. [PubMed] [Google Scholar]
- 40.Simonet WS, Bucay N, Lauer SJ, Taylor JM. A far-downstream hepatocyte-specific control region directs expression of the linked human apolipoprotein E and C-1 genes in transgenic mice. J Biol Chem. 1993;268:8221–8229. [PubMed] [Google Scholar]
- 41.Smith JD, Melián A, Leff T, Breslow JL. Expression of the human apolipoprotein E gene is regulated by multiple positive and negative elements. J Biol Chem. 1988;263:8300–8308. [PubMed] [Google Scholar]
- 42.Snipes GJ, McGuire CB, Norden JJ, Freeman JA. Nerve injury stimulates the secretion of apolipoprotein E by non-neuronal cells. Proc Natl Acad Sci USA. 1986;83:1130–1134. doi: 10.1073/pnas.83.4.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: high avidity binding to β-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci USA. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams T, Tjian R. Analysis of the DNA-binding and activation properties of the human transcription factor AP-2. Genes Dev. 1991;5:670–682. doi: 10.1101/gad.5.4.670. [DOI] [PubMed] [Google Scholar]
- 45.Williams T, Admon A, Lüscher B, Tjian R. Cloning and expression of AP-2, a cell type-specific transcription factor that activates inducible enhancer elements. Genes Dev. 1988;2:1557–1569. doi: 10.1101/gad.2.12a.1557. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J, Hagopian-Donaldson S, Serbedzija G, Elsemore J, Plehn-Dujowich D, McMahon AP, Flavell RA, Williams T. Neural tube, skeletal, and body wall defects in mice lacking transcription factor AP-2. Nature. 1996;381:238–241. doi: 10.1038/381238a0. [DOI] [PubMed] [Google Scholar]