Abstract
Time-lapse confocal microscopy has been used to image cells in mitosis at the apical surface of neuroepithelium from the rat cerebral cortex during the period of neurogenesis. Staining with vital chromatin dyes reveals that mitotic spindles that are aligned parallel to the surface of the tissue are highly motile, rotating within the plane of the epithelium throughout metaphase, and come to rest only as anaphase begins. Spindles may make several complete turns, parallel to the epithelium, but only rarely tumble into an orientation perpendicular to the epithelial sheet. Analysis shows that spindles do not rotate randomly; rather, they spend most of their time aligned parallel or antiparallel to the direction in which they will later enter anaphase and undergo cell division. This conclusion is strongly supported by statistical analyses of the data. Stereotyped movements of this kind show that the direction of division is determined early in mitosis. This suggests the existence of intracellular and perhaps intercellular signals that define the polarity of the cell both in the apico–basal direction and within the plane of the epithelium. Such mechanisms may be important for maintaining the structure of the epithelium and cell–cell communication during development and may also provide a mechanism for the precise distribution of cytoplasmic determinants that might influence the fate of the daughter cells at a time when neuronal fate is being determined.
Keywords: mitosis, neurogenesis, neuroepithelium, confocal microscopy, time-lapse, development, cerebral cortex, cell polarity, epithelium
The cerebral cortex, like other regions of the mammalian CNS, develops from a simple, pseudostratified neuroepithelium. The adult structure contains many specialized cell types within distinct architectonic areas, making specific connections both within the cortex and to the rest of the brain to perform the highest of mental activities. How these specialized populations of cells are generated from a simple, proliferating epithelium remains a mystery, but it is now becoming clear that important decisions in the developmental process are made at the time of neurogenesis (for review, see Barbe, 1996). The work of Sauer and others has shown that the neuroepithelium is a pseudostratified epithelium in which the location of each nucleus follows an interkinetic motion in the depth of the epithelium over the period of the cell cycle (Sauer, 1935, 1936;Fujita, 1960). Mitosis takes place at the apical or ventricular surface of the tissue. Through G1, nuclei descend to the basal-most extent of the proliferative zone, where DNA is replicated, and then rise again to complete the cycle adjacent to the ventricular wall in G2 and early prophase. Each cell maintains both an apical and a basal process that together span the extent of the neuroepithelium at all stages except mitosis, when the basal process is retracted and the cell rounds at the apical surface (Hinds and Ruffet, 1971; Seymour and Berry, 1975). In most cases, the cell then divides with its mitotic axis aligned parallel to the apical surface of the epithelium to produce two radially symmetrical cells (Hinds and Ruffet, 1971; Smart, 1973).
It is known that the location of the mitotic spindle will direct the plane of cell division to be perpendicular to the axis of the spindle (Rappaport, 1961). However, we know very little of how the orientation of mitosis and cell division is spatially controlled within the three-dimensional structure of tissues (Reinsch and Karsenti, 1994). Studies within the developing nervous system (Langman et al., 1966; Smart, 1973; Zamenhof, 1978;Landrieu and Goffinet, 1979; Zieba et al., 1986; Chenn and McConnell, 1995) and in other tissues (Smart, 1970; Fristrom, 1988; Lamprecht, 1990) show a tissue-specific, stereotypic arrangement of mitotic figures, implying that there are control mechanisms that determine their organization. Cells within many epithelia divide within the plane of that sheet, maintaining cell contacts during the process (Hinds and Ruffet, 1971; Sandig and Kalnins, 1990; Reinsch and Karsenti, 1994), but the mechanisms and dynamics of these events are largely unexplored. Despite the importance of cell division to development of the nervous system, very few studies have investigated this process in situ, especially during the later stages of organogenesis (Chenn and McConnell, 1995). In this study, I have used time-lapse confocal microscopy to follow mitosis at the apical surface of neuroepithelium of the rat cerebral cortex. I show that mitotic spindles rotate extensively during metaphase but that these motions respect the direction in which the cell will later divide, showing that this orientation is specified early in mitosis.
MATERIALS AND METHODS
All chemicals were purchased from Sigma (Poole, UK) unless otherwise stated.
Tissue culture. Patches of cortex were dissected from the lateral cerebral wall of Sprague Dawley rat embryos between embryonic day 12 (E12) and E20 (the day of conception is E0). Patches were cultured in Petriperm dishes (petri dishes with a base constructed from a thin, gas-permeable membrane; Bachofer GmbH) in serum-free DMEM plus N2 culture medium supplement (Life Technologies) (Bottenstein and Sato, 1979). Cultures were maintained at 37°C and in an atmosphere of 95% air/5% CO2. Proliferation, as seen by cells entering mitosis, continues for 24–48 hr under these conditions, but all observations in this study were made within 36 hr of tissue explantation. To visualize mitosis, dishes were kept at 37°C in a purpose-built stage heater mounted on an inverted, laser-scanning confocal microscope (Leica Lasertechnik GmbH). The surface of the medium was covered with a thin layer of silicone oil (BDH) to prevent evaporation and changes in pH of the culture medium during the experiment. Slices were stained immediately before imaging by addition of 3.3 μm acridine orange (Molecular Probes, Eugene, OR) into the culture medium for 10 min, replaced with fresh culture medium, and subsequently imaged for no more than 2 hr on the microscope. The 488 nm line of an argon–krypton laser was used to excite acridine orange; emitted light was passed through an 80% transmission mirror and a 515 nm long-pass filter. Images were collected in a single scan or line 2× or 4× line averaging. Excitation was kept to a minimum by using the lowest possible laser intensities, minimal averaging, and collecting images in a single plane in most instances. The process of mitosis is highly temperature-sensitive; for this reason, either long-working-distance objectives [10×, 0.3 numerical aperture (NA) and 20× 0.4 NA (Olympus)] or a 25×, 0.75 NA, water-immersion lens (Leica) with custom-made lens heater was used for imaging living preparations.
Analysis. Mitosis was followed in living preparations using time-lapse confocal microscopy. Sequences of images of dividing cells were taken at 30 or 60 sec intervals. In most instances, images at a single image plane were collected, but some data were collected at multiple planes at each time point. A custom computer program was written using LabView (National Instruments) running on a Macintosh computer to control, via a serial connection, the collection and storage of images by the confocal microscope system. Sequences of images were analyzed on a Macintosh computer using the public domain National Institutes of Health Image program (developed at National Institutes of Health and available from the Internet by anonymous FTP from zippy.nimh.nih.gov or on floppy disk from the National Technical Information Service, Springfield, VA, part number PB95-500195GEI). Quantitative analysis of the orientation of the fluorescently stained chromatin of 24 cells dividing within the plane of E19 tissue was calculated for each frame of the movies collected at 30 sec intervals. The orientations of mitotic spindles before the onset of anaphase were calculated as the perpendicular to a line drawn through the bar of stained chromatin at the center of the spindle (i.e., a line drawn perpendicular to the axis of the spindle). The orientation of the spindle from anaphase through cytokinesis was measured by drawing a line between the two parting clusters of chromatin, parallel to the mitotic spindle. Individual measurements were reproducible to within 4°. An average of all data points after the clear onset of anaphase through to cytokinesis was calculated for each cell. All measured angles for the entire sequence from each cell were then normalized relative to this mean angle. 0° corresponds to parallel and 180° antiparallel to the mean direction of anaphase and cytokinesis. Time sequences are presented as polar plots of orientation versus time. Orientation distributions grouped from a population of cells were binned at 10° intervals and plotted as rose diagrams for which the area of each sector is proportional to its relative frequency (radius is proportional to the square root of frequency) such that the total area of each diagram is equal. Customized plotting procedures were written to present to data.
Statistical tests of the data against models of circular distributions were programmed within the numerical analysis package HiQ (National Instruments) using the methods described in Mardia (1972) and Fisher (1993). Briefly, a goodness-of-fit measure for a uniform (random) distribution of angles was established by calculating the WatsonU2 statistic (Mardia, 1972). AU2 fit-statistic for a unimodal von Mises distribution (a circular analog of the linear Normal distribution) was calculated using the cumulative frequency values of the data against the maximum-likelihood estimates of the data mean direction and κ parameters (the latter found by solving the first- to zero-order modified Bessel function ratio equal to the mean resultant vector of the data) (Fisher, 1993). Parameter estimation for a bimodal mixture of two von Mises distributions was found by the method of moments (using Nelder–Mead minimization) to find the five parameters: mean of distribution 1, κ of distribution 1, mean of distribution 2, κ of distribution 2, and p, the proportion of the first distribution to the total (Fisher, 1993). AU2 fit-statistic was calculated, as above, using the cumulative frequency values of the weighted mixture of the two distributions.
RESULTS
Mitotic cells in the ventricular zone of the cerebral cortex
Sheets of developing neuroepithelium from the lateral wall of the cerebral cortex of rat embryos were cultured for up to 36 hr. Tissue was taken between the ages of E12 and E20, spanning the entire period of neurogenesis (Bayer and Altman, 1991; Ignacio et al., 1995). Proliferation in the ventricular zone is much diminished by E20; only about half of the preparations from this age showed cells in mitosis at the ventricular surface, when imaged as described below. The nuclei of cells located at the ventricular (apical) surface of the tissue were visualized by staining with the vital dye acridine orange and imaging the living preparation by confocal microscopy. In this preparation, acridine orange stained chromatin almost exclusively, with very little cytoplasmic staining, making mitotic figures at the apical surface of the intact, living tissue very clear (Fig.1a). Nuclei of cells in prophase rise to the apical surface where their chromatin condenses as the chromosomes collect at the center of the cell to form the metaphase plate. In the experiments described here, the tissue was viewed from the ventricular surface in a plane parallel to the plane of the epithelial sheet. The mitotic spindles formed parallel to the plane of the tissue, and the plane of division was radial, parallel to the apico–basal axis of the cell. These divisions gave rise to two daughter cells that were adjacent within the sheet, each presumably maintaining apical contacts (Hinds and Ruffet, 1971). All of the mitotic figures directly visible by this technique and described in this paper were oriented in this way. We would expect, however, that at least 10% of cells will be dividing perpendicular to this, with their mitotic spindle parallel to the apico–basal axis of the cell and cleavage plane horizontal within the epithelium (Smart, 1973; Chenn and McConnell, 1995) (R. Adams and J. Nangla, unpublished observations). In some instances, a small number of nuclei, which may have been dividing in a perpendicular direction, could be distinguished from adjacent interphase nuclei by using image-processing methods, on time-lapse movies, to detect the arrival or loss of nuclei with no clear metaphase bar. It is likely that they are rarely seen because the perpendicular disk of chromatin is not readily distinguishable from adjacent interphase cells. Because they could not be unambiguously detected in this preparation, they will not be described further.
Fig. 1.
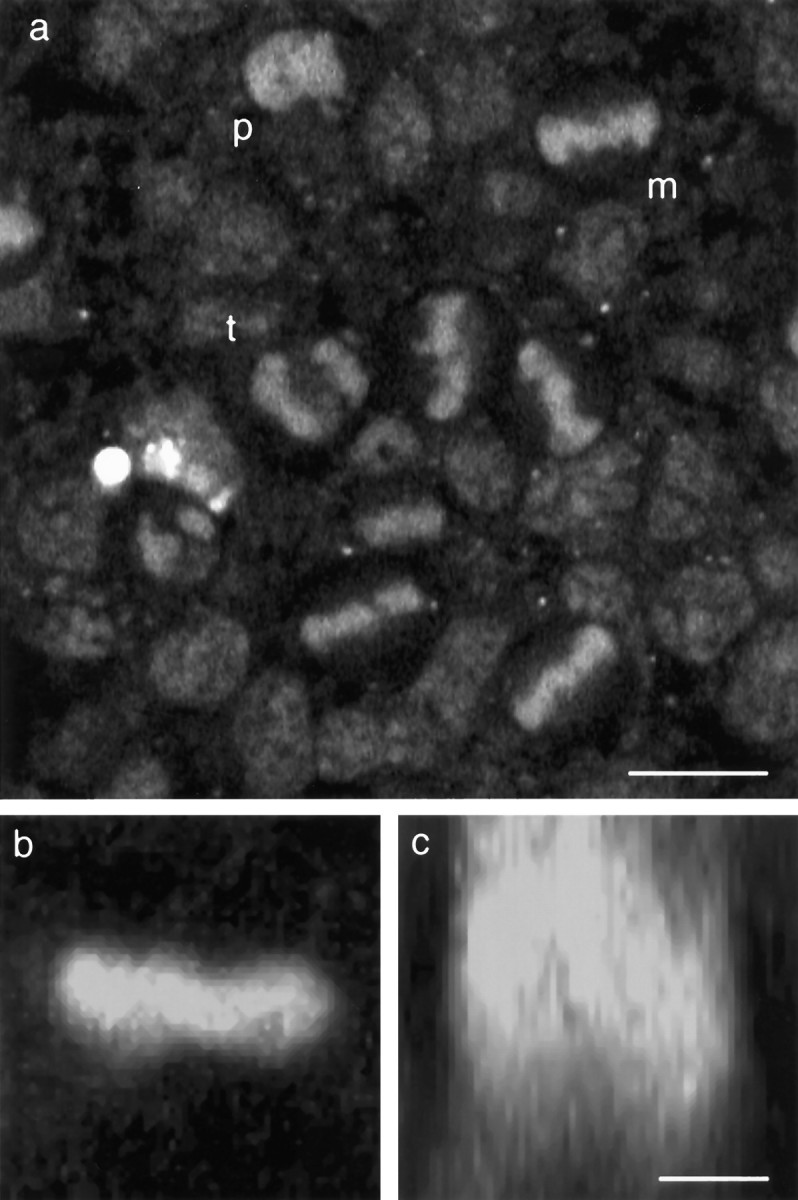
Confocal microscopy of mitotic cells at the ventricular surface of living neuroepithelium from the rat cerebral cortex. Explants from the lateral wall of the neocortical neuroepithelium of embryonic rat between the ages of E12 and E20 were cultured in vitro. The chromatin dye acridine orange (3.3 μm) was used to visualize mitotic figures at the ventricular surface of the tissue imaged by confocal microscopy.a, Single optical section taken parallel to the surface of a sheet of tissue from an E14 rat embryo. Mitotic nuclei in prophase (p), metaphase (m), and anaphase/telophase (t) can be seen. b,c, Two-dimensional extended-focus images generated from thirty-two 1 μm optical sections through a single metaphase cell dividing parallel to the surface of the epithelium. Projected along the apico–basal axis, the image appears as a bright bar (b) but rotated 90° to project along the axis of the spindle shows this to be a disk of chromatin within the depth of the tissue (c). Scale bars: a, 10 μm;b, c, 5 μm.
Viewed from the apical surface, the discoid metaphase plate appeared as a bar of chromatin in the center of the cell. The alignment of the bar indicating the orientation of the mitotic spindle within the plane of the epithelium. Three-dimensional reconstruction of optical sections through cells in metaphase confirmed that these bars were disk-shaped in the z-direction (Fig. 1b,c). Cells in anaphase had two smaller, parallel bars of chromatin (Fig.1a) that will have become more rounded by telophase and cytokinesis. It should be noted that individual chromosomes could not be resolved, so the stages of prometaphase and metaphase are not easily distinguishable (Rieder et al., 1994); for the sake of brevity, therefore, I shall refer to this entire period as metaphase.
Time-lapse imaging of mitosis
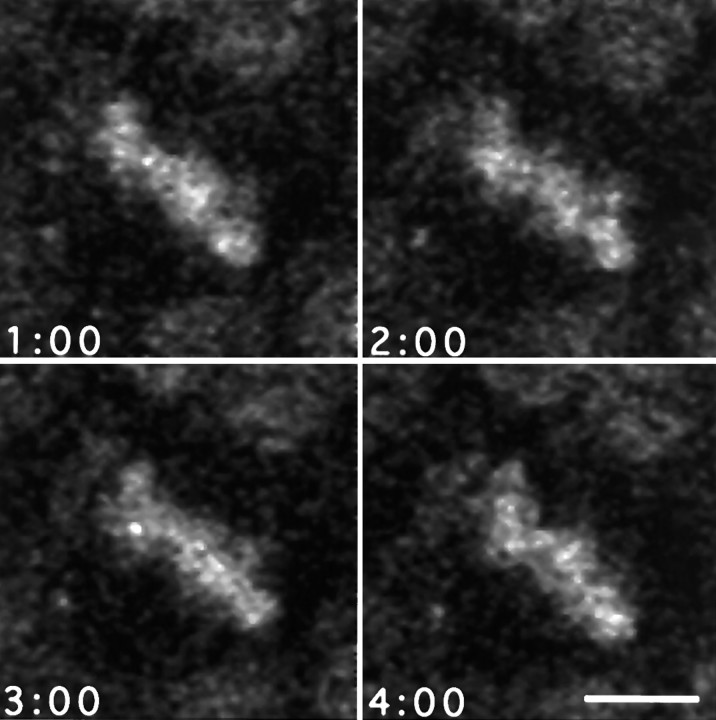
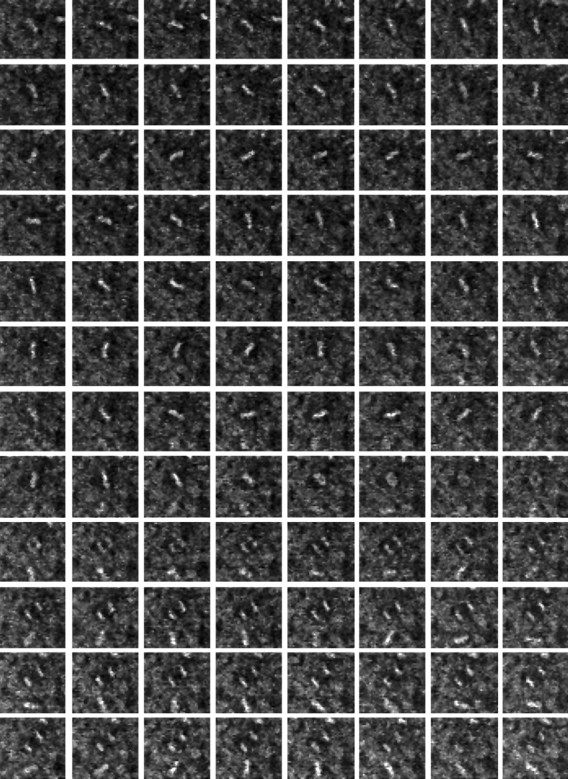
Time-lapse confocal microscopy of the ventricular surface of sheets of neuroepithelium shows that the process of mitosis is highly dynamic. No qualitative difference has been found in the behavior of mitoses in tissue taken over the entire period of E12–E20. Nuclei in prophase became visible as they rose to the apical surface. Their chromatin was in constant rotational motion, probably caused by both chromosomal condensation and alignment and to rotation of the entire nucleus. The nuclei of adjacent cells in interphase did not rotate. Within a short time, the chromosomes collected to the bar-like metaphase plate at the center of the cell. In many cases, the metaphase plate continued to display persistent and substantial rotational motion within a plane parallel with the plane of the tissue (Fig.2). The extent of rotation may range from a few degrees about a mean position to large excursions encompassing complete 360° rotations. Many rotations were of 180° or more. The total angular displacement, within the plane, may encompass one or more complete rotations; this suggests that this was a movement of the entire mitotic spindle rather than a realignment of the chromatin within the spindle complex. Importantly, the metaphase plate is recognizable as a bar very early in mitosis, indicating that the alignment of the spindle perpendicular to the apico–basal axis was relatively fast (and presumable the same for cells dividing perpendicular to the sheet) but that rotation within that plane continues for much longer. On occasions, however, the bar-like appearance of the metaphase plate may transform briefly into a disk, interpreted as a tumbling motion of the spindle into an orientation perpendicular to the plane of the epithelium. This orientation is short-lived, and the spindle rapidly returns to a parallel attitude. Smaller changes in shape of the metaphase plate could also be detected while the spindle is rotating (Fig. 3). These probably reflect the movements of individual chromosomes as they align to complete bipolar connections within the spindle, equivalent to the oscillations of chromosomes seen in some cultured cells (Rieder et al., 1994).
Fig. 2.
Time-lapse sequence of a single mitotic cell in the neuroepithelium of an E19 rat embryo. Neuroepithelium from an E19 embryo was stained with acridine orange and then imaged at 30 sec intervals for 1 hr by time-lapse confocal microscopy. Frames proceed by rows from left to right starting at thetop left. The metaphase plate of the cell seen in the center of frame 1 is in constant motion until frame 61 (row 8, column 5), 30 min into the sequence, when it enters anaphase. The two sets of daughter chromatids separate over the remainder of the frames with little change in the orientation of the spindle. Each frame is 40 μm wide.
Fig. 3.
Chromosomal movements within the metaphase plate during rotation. Four frames from a sequence of high-magnification images taken at 60 sec intervals during metaphase showing the changes in shape of the metaphase plate, consistent with the oscillatory behaviors of individual chromosomes described by others (Rieder et al., 1994). These movements are superimposed on the larger scale rotation of the entire spindle within the dividing cell. Scale bar, 5 μm.
Movement was continuous through metaphase but abruptly ceased as cells entered anaphase. Cells then proceeded through chromatin separation (anaphase and telophase) and cytokinesis with relatively little change in orientation of the spindle relative to the tissue. On passage from metaphase into anaphase, the central disk of chromatin split into two smaller masses that slowly moved to opposite poles of the cell. After the completion of anaphase, and presumably cytokinesis, some daughter nuclei descended again into the depth of the epithelial sheet and were lost from the narrow depth of field of the confocal microscope, but others remained longer at the apical surface. The cessation of movement at the onset of anaphase suggests a change in the activity of force-generating elements, possibly involving interactions of astral microtubules with the cell cortex (Hyman and White, 1987).
Quantitative analysis of movement
Quantitative analysis of the movements of mitotic spindles revealed more of the underlying behavior. The orientation of the spindle was constantly changing throughout metaphase, but this movement was not random. When the orientation over time of individual spindles was plotted as a polar graph, with angle depicting orientation relative to the mean anaphase direction and radius proportional to time, it was evident that for many cells angular excursions were large, encompassing rotations of 180° or even 360° (Fig. 4). Polar plots of movements and rose diagrams of the distributions of angles occupied during metaphase (Fig. 4) showed that not all orientations were equivalent; rather, the spindle, for many cells, spent most of its time restricted to one or both of two diagonally opposed quadrants parallel and antiparallel to the direction it will later enter anaphase, stop rotating, and divide (Fig. 4). For these cells, the time spent perpendicular to the orientation of entry into anaphase was always short. Cells with this behavior constituted about half of those studied. Most of the remaining cells showed a predominantly unimodal preference of orientation, close to the direction of anaphase. A smaller population of cells showed more extensive movements, in which their axial preferences were less clear.
Fig. 4.
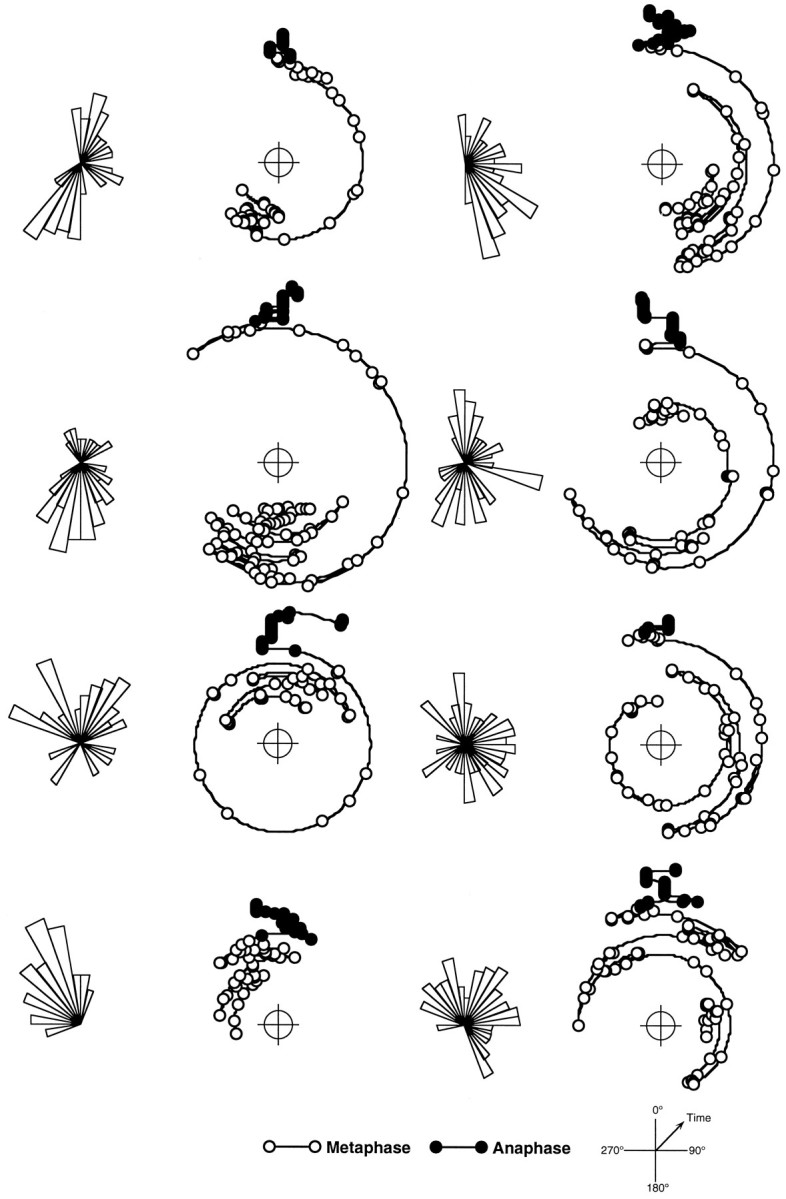
Paths of spindle rotation during mitosis in the ventricular zone. Time course of orientation of eight representative mitotic spindles measured at 30 sec intervals through metaphase (open symbols) and anaphase through cytokinesis (filled symbols). Cells in movies of the kind shown in Figure 3 were analyzed frame by frame to measure the orientation of mitotic spindle over time. Results are presented as polar plots with increasing radius representing the progression of time and orientation normalized to the mean angle of all anaphase measurements for each cell (0°). To the left of each polar plot is a rose diagram showing the relative frequency distribution of orientations during metaphase. The top four examples show distinct bipolar preferences relative the direction of anaphase. The bottom four show either a unimodal preference or more extensive movements with no clear preference. This selection is representative of the total population measured.
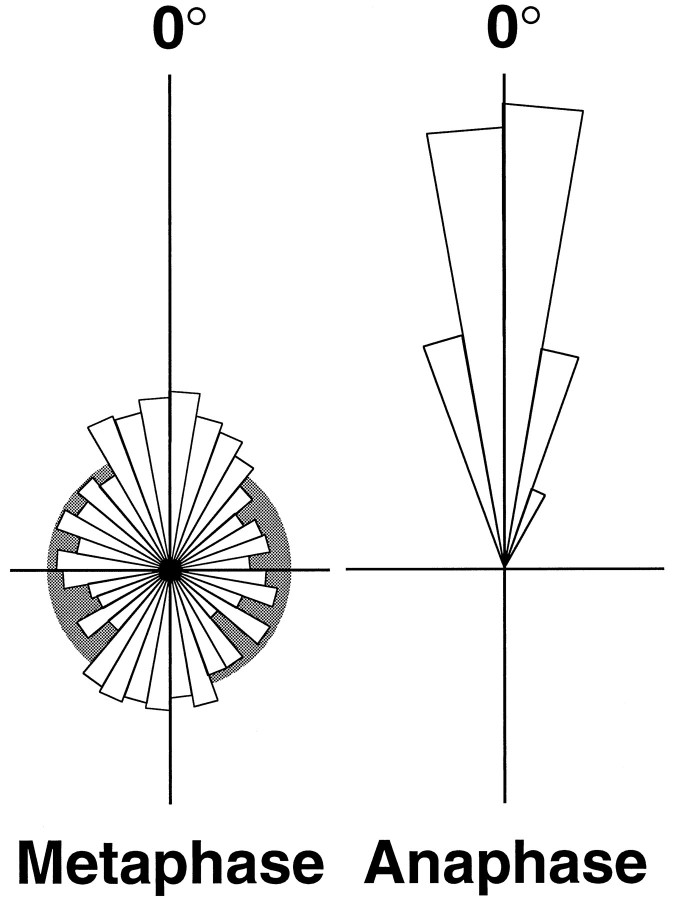
To investigate quantitatively the behavior of mitotic spindles for a population of cells, the combined frequency distribution of orientation for those cells that could be followed from metaphase through to cytokinesis was calculated. Measurements, taken from movies collected at 30 sec intervals, were separated into two groups, before and after the detectable onset of anaphase. Each group was normalized by angle, on a cell-by-cell basis, so that the mean orientation of the spindle through anaphase is 0° and then plotted as a rose diagram in which the area of each 10° bin is proportional to its relative frequency (Fig. 5). The distribution of orientations measured after the onset of anaphase showed a very restricted variation about the mean angle (0° ± 0.6°, mean ± 95% confidence limits). This clearly confirmed the impression that there was very little net rotation once the sister chromatids had begun their separation. However, the distribution of orientations of spindles during metaphase was very much broader with prominent peaks at 0° and 180°, parallel and antiparallel to the angle of karyokinesis and cytokinesis. In Figure 5, the metaphase distribution is compared to a graphical representation of an uniform (random) distribution, which clearly shows that the metaphase data are concentrated in the regions of 0° and 180° relative to 90° and 270°. If the data are grouped about the four cardinal directions, they spend 39 and 27% of their time in the quadrants 0° ± 45° and 180° ± 45°, respectively (together comprising two-thirds of the total time), compared to an average of 17% of the time in each of the perpendicular quadrants.
Fig. 5.
Frequency distributions of orientations of spindles in anaphase and metaphase relative to the orientation of division. The orientations measured over time for a population of 24 dividing cells were individually normalized to their mean orientation during anaphase. Measurements were separated into two groups corresponding to those after the onset of anaphase and those preceding them, during metaphase. Each data set was binned at 10° intervals and presented as a rose diagram. The area of each sector is proportional to its frequency (radius proportional to the square root of frequency) such that the total areas of the two plots are equal. The distribution of 591 measurements of spindles in anaphase shows very little rotational movement. The distribution of orientations during metaphase (1051 measurements) is much broader, reflecting the extensive motion of the metaphase spindle. There are two major peaks in this distribution, one at 2.7°, parallel to the mean angle after the cessation of rotation, and the other at 179.6°. This shows that the spindles spend significantly more time aligned in this axis within the plane of the tissue than at angles perpendicular to it. The metaphase distribution is superimposed on the distribution (gray) that would be expected for a uniform (random) distribution of orientation.
The data for the distribution of orientations of metaphase spindles relative to their anaphase direction have been analyzed more rigorously by comparing them to statistical models of circular data. Calculation of the Watson U2 statistic allows us to reject confidently a random (uniform) model (U2 = 1.54,p < 0.005) in which a spindle spends equal time in all orientations irrespective of the direction of division. A unimodal von Mises model of the metaphase data, a circular analog of a normal distribution, in which the data are clustered about a single mean direction, can also be strongly rejected (U2 = 0.84, p < 0.005). A plausible fit of the metaphase data may be found with a bimodal mixture of two von Mises distributions (U2 = 0.038, p > 0.25) with peaks centered at 2.7° ± 4.1° and 179.6° ± 5.5° (mean ± approximate 95% confidence limits). The contributions of the two distributions are weighted at 0.55 and 0.45, with κ coefficients of 1.9 and 1.5, respectively. This finding strongly supports the hypothesis that there is an axial preference for alignment of the metaphase spindle parallel with the axis of anaphase and division, about which it will tend to align, parallel or antiparallel, before its arrest at anaphase.
An alternative way to analyze these data is to ask to what extent the mean axial direction for each cell will predict the direction in which it will enter anaphase. The direction data of each metaphase measurement were converted to axial data by doubling the angle modulo 360, the mean direction determined and divided by two. For each cell, the mean axial direction was found and divided by two. The axial predictions for all cells analyzed have a mean orientation of 359.2° ± 13.7° (mean ± 95% confidence of the mean).
This behavior shows that there exists a physical basis for the determination of direction of cell division in both the apico–basolateral axis, which determines whether the cell will divide parallel or perpendicular to the tissue, and within the plane of the neuroepithelium for those cells dividing parallel to the surface. This polarity is defined at or before the time that the spindle forms, and the motion of the highly motile metaphase spindle is strongly biased to keep it aligned to this orientation.
DISCUSSION
Cells of the neuroepithelium divide at the ventricular surface of the tissue. This has permitted the use of time-lapse confocal microscopy to image mitosis during the period of neurogenesis in the cerebral cortex of the rodent. The major finding of this study is that metaphase mitotic spindles are in constant rotational motion within a plane parallel with that of the epithelium. Rotation arrests at the onset of anaphase, and the orientation of the spindle then remains essentially constant through cytokinesis. Analysis of the rotation of the metaphase spindle shows that although it can make large angular displacements encompassing complete 360° rotations, it will spend significantly more of its time aligned to the direction in which it will enter anaphase or 180° opposed to it. This behavior indicates that the direction in which a cell will divide must be determined early in mitosis and that the motion of the spindle during metaphase is to ensure that mitosis (and therefore cytokinesis) becomes aligned correctly in this direction.
In studies of the orientation of cell division within neuroepithelia, most cells are reported to divide within the plane of the tissue (Hinds and Ruffet, 1971; Smart, 1973), although there may be regional or developmental variation in this pattern (Zamenhof, 1978; Chenn and McConnell, 1995). It follows, therefore, that there must be a mechanism within the cell to orient the mitotic spindle relative to its apico–basal axis, determining whether a cell will divide parallel or perpendicular to the epithelium. We do not know whether the mechanism that aligns the mitotic spindle relative to the apico–basal axis is the same as that responsible for orientation within the plane. It is entirely possible that a single cortical cytoplasmic element acts to direct the spindle in both of these axes. We do know, however, that alignment must take place on different time scales in neuroepithelial cells (although specification may not); only occasionally do we see the “bar” of chromatin of the metaphase plate transform into a disk as the spindle flips into a vertical position and then rapidly revert back to a bar. Therefore, although the spindle spends almost all of metaphase correctly aligned in the apico–basal axis, it spends longer searching for its orientation within the plane of the tissue. (Presumably, any cells dividing horizontally must also spend most of their time in their preferred orientation because they are rarely as transient bars in this study.) Rotation stops as cells enter anaphase, implying that there is a change in the location of net force generation, as there is for movements within the spindle itself to begin the separation of sister chromatids and spindle poles. In a cultured epithelial cell line (Reinsch and Karsenti, 1994), the position of the centrioles has been described to align during prometaphase as the spindle becomes horizontal within the cell.
There are examples in which the direction the spindle is specifically controlled to produce an asymmetric division (Strome, 1993; Rhyu and Knoblich, 1995; White and Strome, 1996). Astral microtubules may interact with elements of the cortical actin cytoskeleton to instigate this positioning (Hyman and White, 1987; Waddle et al., 1994), and a number of the other genes involved, in Caenorhabditis elegans, are being found (White and Strome, 1996). Cells dividing within epithelia may also need to coordinate their plane of division with neighboring cells so as to maintain the integrity of cell–cell contacts during cytokinesis (Hinds and Ruffet, 1971; Sandig and Kalnins, 1990; Schoenwolf and Alvarez, 1992; Reinsch and Karsenti, 1994). Polarized cell divisions referenced to a neighboring cell have also been seen in early C. elegans development (Goldstein, 1995). Studies on living material will be needed to show whether the behavior reported here is common to all epithelia.
No pattern is seen to the directions with which neighboring cells divide within the explant. The orientations of divisions appear to be random, but we do not yet know whether they are aligned locally to neighboring cells; alignment to radial glial cell processes, for instance, might guide daughter cells from the ventricular zone after division. There are instances in the literature in which significant alignments have been found within the neural plate (Tuckett and Morriss-Kay, 1985). These have been correlated with a polarized growth of the epithelium, but whether cell division is responding to or contributing to mechanical or directional forces is still unknown. That no pattern has yet been discerned in the preparation reported here may be, in part, because the tissue is removed from the mechanical context of the rapidly expanding lateral ventricles. The sheets themselves are highly contractile and change shape while in culture.
There are several published examples in which the alignment of the mitotic spindle has been associated with a subsequent determination of cell fate as a consequence of the segregation of cytoplasmic factors from the mother cell (Horvitz and Herskowitz, 1992; Strome, 1993; White and Strome, 1996). In these examples, the mitotic spindle is rotated to locate cortical cytoplasmic factors at one pole so that just one daughter cell will inherit them. This behavior is of particular interest given the nature of the developmental decisions that are taking place in the neuroepithelium at this time (McConnell and Kaznowski, 1991; Barbe, 1996). Throughout the developmental period of this study, divisions are giving rise to daughters that include further progenitor cells—perhaps with diminishing potential—and postmitotic neurons. Significantly, in the development of the central and peripheral nervous systems of Drosophila, the determination of cell fate depends on a combination of intrinsic and extrinsic signals (Posakony, 1994; Rhyu et al., 1994; Doe and Spana, 1995; Jan and Jan, 1995). The asymmetric inheritance of two intracellular proteins, a membrane-associated protein, Numb, and a transcription factor, Prospero, is necessary for the correct specification of sibling cell fate (Rhyu et al., 1994; Hirata et al., 1995; Knoblich et al., 1995; Spana and Doe, 1995). Early in mitosis, these proteins are segregated to one side of the dividing precursor cell, and the mitotic spindle then aligns such that only one daughter cell will inherit them. The behavior reported here for the rat neuroepithelium is entirely consistent with (but not proof of) an asymmetric division of this kind. Active spindle alignment relative to a predefined cellular axis, as reported here, is an absolute prerequisite for mechanisms involving the segregation of cytoplasmic determinants and is similar to other reported cases (Strome, 1993; White and Strome, 1996). Intriguingly, Numb and Prospero have both been reported to be expressed in vertebrates (Oliver et al., 1993; Zhong et al., 1996). Mammalian Numb protein is localized to the apical or apico–lateral domains of cells dividing in the neuroepithelium. The relationship between the movements reported here and the inheritance of this protein will be important to establish.
It has been reported recently that there is an asymmetric distribution of Notch1 protein between daughters of those neuroepithelial cells that divide horizontally within in vitro preparations of ferret cerebral cortex (Chenn and McConnell, 1995) (perpendicular to those described in this report). The basal-most daughter of this division also displays a different behavioral phenotype, migrating more rapidly from the ventricular surface of the tissue than its apical sister. Whether the divisions that I describe here produce cells with asymmetric fates is unknown. Certainly not all asymmetric divisions necessarily produce postmitotic cells, as in the cascade of stem cells in the hematopoietic system; neither is this inconsistent with the observations of differing fate for cells of horizontal divisions (Chenn and McConnell, 1995). If it is argued that the cell must already have a mechanism to align divisions parallel or perpendicular to the apico–basal axis and within the plane of the tissue, it is quite possible that this same mechanism may be used to segregate factors asymmetrically, in either direction.
The relative contributions of perpendicular and horizontal divisions to the neuronal complement of the cerebral cortex are still not known; the final fates of the cells produced by divisions in either plane must still be established. We do know that the biology of the mammalian neuroepithelium is complex. It is likely that cell divisions within the neuroepithelium are producing not only proliferative progenitor cells and postmitotic neurons but that some must generate the cells of the subventricular zone and others may involve radial glial cells (Misson et al., 1988; Bayer and Altman, 1991; Takahashi et al., 1995). A number of proliferative progenitor cells may leave the neuroepithelium to migrate tangentially (Fishell et al., 1993), possibly to divide again elsewhere in the cortex (Kornack and Rakic, 1995; Reid et al., 1995). How they leave or reestablish connections at the ventricular wall is unknown, but this may resemble behavior seen in the chick otic anlage, where proliferative cells may delaminate by a horizontal division from the epithelium (Alvarez et al., 1989). The events taking place at the time of neurogenesis are clearly of great importance, making these and other studies on the behaviors of cells within the neuroepithelium vital for our elucidation of this fascinating process.
Footnotes
This work was supported by The Medical Research Council Research Centre for Brain and Behavior, Oxford University. I thank Mr. William Hinkes for his expert technical assistance with this project. I am also indebted to Professor Colin Blakemore, Iain Smart, John Scholes, Aurora Lombardo, Miguel Concha, Zoltán Molnár, and Jyoti Nangla for critically reading versions of this manuscript, and to Dr. Mario Cortina-Borja.
Correspondence should be addressed to Richard J. Adams, University Laboratory of Physiology, Oxford University, Parks Road, Oxford OX1 3PT, UK.
REFERENCES
- 1.Alvarez IS, Martín-Patido G, Rodríguez-Gallardo L, González-Ramos C, Navasués J. Cell proliferation during early development of the chick otic anlage: quantitative comparison of migratory and nonmigratory regions of the otic epithelium. J Comp Neurol. 1989;290:278–288. doi: 10.1002/cne.902900208. [DOI] [PubMed] [Google Scholar]
- 2.Barbe MF. Tempting fate and commitment in the developing forebrain. Neuron. 1996;16:1–16. doi: 10.1016/s0896-6273(00)80015-x. [DOI] [PubMed] [Google Scholar]
- 3.Bayer SA, Altman J. Neocortical development. Raven; New York: 1991. [Google Scholar]
- 4.Bottenstein JE, Sato GH. Growth of a rat neuroblastoma cell line in serum-free supplemented medium. Proc Natl Acad Sci USA. 1979;83:514–517. doi: 10.1073/pnas.76.1.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chenn A, McConnell SK. Cleavage orientation and the asymmetric inheritance of notch1 immunoreactivity in mammalian neurogenesis. Cell. 1995;82:631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- 6.Doe CQ, Spana EP. A collection of cortical crescents: asymmetric protein localization in CNS precursor cells. Neuron. 1995;15:991–995. doi: 10.1016/0896-6273(95)90088-8. [DOI] [PubMed] [Google Scholar]
- 7.Fishell G, Mason CA, Hatten ME. Dispersion of neural progenitors within the germinal zones of the forebrain. Nature. 1993;362:636–638. doi: 10.1038/362636a0. [DOI] [PubMed] [Google Scholar]
- 8.Fisher NI. Statistics of directional data. Cambridge UP; Cambridge, UK: 1993. [Google Scholar]
- 9.Fristrom D. The cellular basis of epithelial morphogenesis: a review. Tissue Cell. 1988;20:645–690. doi: 10.1016/0040-8166(88)90015-8. [DOI] [PubMed] [Google Scholar]
- 10.Fujita S. Mitotic pattern and histogenesis of the central nervous system. Nature. 1960;185:702–703. doi: 10.1038/185702a0. [DOI] [PubMed] [Google Scholar]
- 11.Goldstein B. Cell contacts orient some cell-division axes in the Caenorhabditis elegans embryo. J Cell Biol. 1995;129:1071–1080. doi: 10.1083/jcb.129.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hinds JW, Ruffet TL. Cell proliferation in the neural tube: an electron microscopic and Golgi analysis in the mouse cerebral vesicle. Z Zellforsch. 1971;115:226–264. doi: 10.1007/BF00391127. [DOI] [PubMed] [Google Scholar]
- 13.Hirata J, Nakagoshi H, Nabeshima Y-I, Matsuzaki F. Asymmetric segregation of the homeodomain protein Prospero during Drosophila development. Nature. 1995;377:627–630. doi: 10.1038/377627a0. [DOI] [PubMed] [Google Scholar]
- 14.Horvitz HR, Herskowitz I. Mechanisms of asymmetric cell division: two Bs or not two Bs, that is the question. Cell. 1992;68:237–255. doi: 10.1016/0092-8674(92)90468-r. [DOI] [PubMed] [Google Scholar]
- 15.Hyman AA. Centrosome movement in the early divisions of Caenorhabditis elegans: a cortical site determining centrosome position. J Cell Biol. 1989;109:1185–1193. doi: 10.1083/jcb.109.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyman AA, White JG. Determination of cell-division axes in the early embryogenesis of Caenorhabditis elegans. J Cell Biol. 1987;105:2123–2135. doi: 10.1083/jcb.105.5.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ignacio MPD, Kimm EJ, Kageyama GH, Yu J, Robertson RT. Postnatal migration of neurons and formation of laminae in rat cerebral cortex. Anat Embryol. 1995;191:89–100. doi: 10.1007/BF00186782. [DOI] [PubMed] [Google Scholar]
- 18.Knoblich JA, Jan LY, Jan YN (1995) Asymmetric segregation of numb and prospero during cell-division. 377:624–627. [DOI] [PubMed]
- 19.Kornack DR, Rakic P. Radial and horizontal deployment of clonally related cells in the primate neocortex: relationship to distinct mitotic lineages. Neuron. 1995;15:311–321. doi: 10.1016/0896-6273(95)90036-5. [DOI] [PubMed] [Google Scholar]
- 20.Lamprecht J. Symmetric and asymmetric cell division in rat corneal epithelium. Cell Tissue Kinet. 1990;23:203–216. doi: 10.1111/j.1365-2184.1990.tb01116.x. [DOI] [PubMed] [Google Scholar]
- 21.Landrieu P, Goffinet A. Mitotic spindle orientation in relation to cell migration in the neo-cortex of normal and reeler mouse. Neurosci Lett. 1979;13:69–72. doi: 10.1016/0304-3940(79)90077-6. [DOI] [PubMed] [Google Scholar]
- 22.Langman J, Guerrant RL, Freeman BG. Behavior of neuro-epithelial cells during closure of the neural tube. J Comp Neurol. 1966;127:399–412. doi: 10.1002/cne.901270308. [DOI] [PubMed] [Google Scholar]
- 23.Mardia KV. Statistics of directional data. Academic; London: 1972. [Google Scholar]
- 24.McConnell SK, Kaznowski CE. Cell-cycle dependence of laminar determination in developing neocortex. Science. 1991;254:282–285. doi: 10.1126/science.254.5029.282. [DOI] [PubMed] [Google Scholar]
- 25.Misson JP, Edwards MA, Yamamoto M, Caviness VS. Mitotic cycling of radial glial-cells of the fetal murine cerebral wall: a combined autoradiographic and immunohistochemical study. Dev Brain Res. 1988;38:183–190. doi: 10.1016/0165-3806(88)90043-0. [DOI] [PubMed] [Google Scholar]
- 26.Oliver G, Sosapineda B, Geisendorf S, Spana EP, Doe CQ, Gruss P. Prox-1, a prospero-related homeobox gene expressed during mouse development. Mech Dev. 1993;44:3–16. doi: 10.1016/0925-4773(93)90012-m. [DOI] [PubMed] [Google Scholar]
- 27.Posakony JW. Nature versus nurture: asymmetric cell divisions in Drosophila bristle development. Cell. 1994;76:415–418. doi: 10.1016/0092-8674(94)90105-8. [DOI] [PubMed] [Google Scholar]
- 28.Rappaport R. Experiments concerning cleavage stimulus in sand dollar eggs. J Exp Zool. 1961;148:81–89. doi: 10.1002/jez.1401480107. [DOI] [PubMed] [Google Scholar]
- 29.Reid CB, Liang I, Walsh C. Systematic widespread clonal organization in cerebral-cortex. Neuron. 1995;15:299–310. doi: 10.1016/0896-6273(95)90035-7. [DOI] [PubMed] [Google Scholar]
- 30.Reinsch S, Karsenti E. Orientation of spindle axis and distribution of plasma membrane proteins during cell division in polarised MDCKII cells. J Cell Biol. 1994;126:1509–1526. doi: 10.1083/jcb.126.6.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhyu MS, Knoblich JA. Spindle orientation and asymmetric cell fate. Cell. 1995;82:523–526. doi: 10.1016/0092-8674(95)90022-5. [DOI] [PubMed] [Google Scholar]
- 32.Rhyu MS, Jan LY, Jan YN. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 33.Rieder CL, Schultz A, Cole R, Sluder G. Anaphase onset in vertebrate somatic-cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J Cell Biol. 1994;127:1301–1310. doi: 10.1083/jcb.127.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandig M, Kalnins VI. Reorganization of circumferential microfilament bundles in retinal epithelial-cells during mitosis. Cell Motil Cytoskeleton. 1990;17:133–141. doi: 10.1002/cm.970170208. [DOI] [PubMed] [Google Scholar]
- 35.Sauer FC. Mitosis in the neural tube. J Comp Neurol. 1935;62:377–405. [Google Scholar]
- 36.Sauer FC. The interkinetic migration of embryonic epithelial nuclei. J Morphol. 1936;60:1–11. [Google Scholar]
- 37.Schliwa M, Vanblerkom J. Structural interaction of cytoskeletal components. J Cell Biol. 1981;90:222–235. doi: 10.1083/jcb.90.1.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schoenwolf GC, Alvarez IS. Role of cell rearrangement in axial morphogenesis. Curr Top Dev Biol. 1992;27:129–173. doi: 10.1016/s0070-2153(08)60534-7. [DOI] [PubMed] [Google Scholar]
- 39.Seymour RM, Berry M. Scanning and transmission electron microscope studies of interkinetic nuclear migration in the cerebral vesicles of the rat. J Comp Neurol. 1975;160:105–126. doi: 10.1002/cne.901600107. [DOI] [PubMed] [Google Scholar]
- 40.Smart IHM. Variation in the plane of cell cleavage during the process of stratification in the mouse epidermis. Br J Dermatol. 1970;82:276–282. doi: 10.1111/j.1365-2133.1970.tb12437.x. [DOI] [PubMed] [Google Scholar]
- 41.Smart IHM. Proliferative characteristics of the ependymal layer during the early development of the mouse neocortex: a pilot study based on the number, location and plane of cleavage of mitotic figures. J Anat. 1973;116:67–91. [PMC free article] [PubMed] [Google Scholar]
- 42.Spana EP, Doe CQ (1995) The prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. 121:3187–3195. [DOI] [PubMed]
- 43.Strome S. Determination of cleavage planes. Cell. 1993;72:3–6. doi: 10.1016/0092-8674(93)90041-n. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi T, Nowakowski RS, Caviness VS. Early ontogeny of the secondary proliferative population of the embryonic murine cerebral wall. J Neurosci. 1995;15:6058–6068. doi: 10.1523/JNEUROSCI.15-09-06058.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tuckett F, Morriss-Kay GM. The kinetic behavior of the cranial neural epithelium during neurulation in the rat. J Embryol Exp Morphol. 1985;85:111–119. [PubMed] [Google Scholar]
- 46.Waddle JA, Cooper JA, Waterston RH. Transient localized accumulation of actin in Caenorhabditis elegans blastomeres with oriented asymmetric divisions. Development. 1994;120:2317–2328. doi: 10.1242/dev.120.8.2317. [DOI] [PubMed] [Google Scholar]
- 47.White J, Strome S. Cleavage plane specification in C. elegans: how to divide the spoils. Cell. 1996;84:195–198. doi: 10.1016/s0092-8674(00)80974-5. [DOI] [PubMed] [Google Scholar]
- 48.Zamenhof S. Quantitative studies of mitoses in fetal rat brain: orientations of the spindles. Dev Brain Res. 1978;31:143–146. doi: 10.1016/0165-3806(87)90094-0. [DOI] [PubMed] [Google Scholar]
- 49.Zhong W, Feder JN, Jiang M-M, Jan LY, Jan YN. Asymmetric localisation of a mammalian numb homolog during mouse cortical neurogenesis. Neuron. 1996;17:43–53. doi: 10.1016/s0896-6273(00)80279-2. [DOI] [PubMed] [Google Scholar]
- 50.Zieba P, Strojny P, Lamprecht J. Positioning and stability of mitotic spindle orientation in the neuroepithelial cell. Cell Biol Int. 1986;10:91–100. doi: 10.1016/0309-1651(86)90092-5. [DOI] [PubMed] [Google Scholar]