SUMMARY
Moderate mitochondrial stress can lead to persistent activation of cytoprotective mechanisms – a phenomenon termed mitohormesis. Here, we show that mitohormesis primes a subpopulation of cancer cells to basally upregulate mitochondrial stress responses, such as the mitochondrial unfolded protein response (UPRmt) providing an adaptive metastatic advantage. In this subpopulation, UPRmt activation persists in the absence of stress, resulting in reduced oxidative stress indicative of mitohormesis. Mechanistically, we showed that the SIRT3 axis of UPRmt is necessary for invasion and metastasis. In breast cancer patients, a 7-gene UPRmt signature demonstrated that UPRmt-HIGH patients have significantly worse clinical outcomes, including metastasis. Transcriptomic analyses revealed that UPRmt-HIGH patients have expression profiles characterized by metastatic programs and the cytoprotective outcomes of mitohormesis. While mitohormesis is associated with health and longevity in non-pathological settings, these results indicate that it is perniciously used by cancer cells to promote tumor progression.
Graphical Abstract

In Brief
Mitohormesis, a phenomenon resulting in persistent activation of cytoprotective mechanisms, has been studied in the context of aging and longevity. Kenny et al. demonstrate that breast cancer cells co-opt mitohormesis to promote their invasion and metastasis via activation of the mitochondrial unfolded protein response (UPRmt).
INTRODUCTION
The Warburg effect has led to the misconception that the mitochondria of cancer cells are non-functional (Vander Heiden et al., 2009). It is now recognized that they are, in fact, critical for tumor growth and exhibit drastic reprogramming to support the unique metabolic and biosynthetic needs of a cancer cell (Pavlova and Thompson, 2016; Vyas et al., 2016; Wallace, 2012). Mitochondrial reprogramming is characterized by elevated oxidative stress via reactive oxygen species (ROS). As the majority of ROS are produced at the mitochondrial inner membrane, the mitochondria of cancer cells are especially prone to their effects. Increased ROS can serve as signaling molecules important for oncogenesis and tumor formation (D’Autréaux and Toledano, 2007; Schieber and Chandel, 2014). On the other hand, ROS can damage lipids, DNA, and proteins by oxidation–causing protein misfolding. In metastatic cells, the effects of ROS may be exacerbated due to anchorage-independent growth and the foreign microenvironments experienced during dissemination. Excessive ROS poses a severe risk to mitochondrial network integrity and, in turn, cancer cell viability. These observations emphasize the need for cancer cells to maintain mitochondrial fitness through adaptive mechanisms, including proteostasis, antioxidant machinery, mitochondrial biogenesis, and mitophagy.
Hormesis describes the biphasic response of a cell or organism to increasing amounts of a substance or condition. At high doses, these toxins or stressors are determinantal, but at low level exposure within the “hormetic zone,” favorable biological responses are produced (Mattson, 2008).
Mitochondrial ROS within the hormetic zone activates mitoprotective mechanisms, which paradoxically persist when the acute stress subsides–termed mitohormesis (Ristow and Zarse, 2010; Yun and Finkel, 2014). Mitohormesis elicited by mitochondrial ROS extends lifespan in C. elegans and yeast (Bonawitz et al., 2007; Lee et al., 2010; Yang and Hekimi, 2010; Feng et al., 2001; Van Raamsdonk and Hekimi, 2009;Schulz et al., 2007). In flies and worms, the mitochondrial unfolded protein response (UPRmt) was identified as a pathway essential for mitohormesis-induced longevity (Dillin et al., 2002; Feng et al., 2001; Merkwirth et al., 2016; Tian et al., 2016; Owusu-Ansah et al., 2013; Nargund et al., 2015; Durieux et al., 2011). The UPRmt was originally discovered by the Hoogenraad group, who described retrograde signaling mediated by C/EBP homologous protein (CHOP) leading to the induction of mitochondrial proteases, such as HSP60 (Zhao et al., 2002). Using HSP60 as a UPRmt reporter in C. elegans, subsequent work has identified ATFS-1 and DVE-1/UBL5 as important transcriptional activators of the UPRmt (Benedetti et al., 2006; Durieux et al., 2011; Fiorese et al., 2016; Gitschlag et al., 2016; Haynes et al., 2007, 2010; Lin et al., 2016; Lisanti et al., 2016; Merkwirth et al., 2016; Nargund et al., 2015, 2012; Pellegrino et al., 2014; Rauthan et al., 2013; Tian et al., 2016).
In mammalian cells, we reported a SIRT3-dependent axis of the UPRmt that activates antioxidant genes, mitochondrial biogenesis, and mitophagy (Kenny and Germain, 2017a, 2017b; Kenny et al., 2017a, 2017b; Papa and Germain, 2014). A strikingly similar sirtuin UPRmt axis exists in C. elegans (Mouchiroud et al., 2013). Recently, mitohormesis was described in mammalian cells using an inducible and reversible SOD2 knockdown mouse (iSOD2-KD) (Cox et al., 2018). In this study, knockdown of SOD2 in development caused increased ROS and mitohormesis, resulting in mitochondrial biogenesis and antioxidant programs (Cox et al., 2018).
In the context of development and aging, mitohormesis in the absence of underlying pathology is beneficial. In the context of cancer, however, the persistent activation of cytoprotective mechanisms by mitohormesis may favor tumor growth and progression. We initiated the current study to determine if mitohormesis occurs under endogenous oxidative stress in breast cancer and to assess its potential impact on disease progression.
Our data suggest the existence of two subpopulations of cancer cells characterized by mitochondrial ROS levels. We found that the ROS-positive subpopulation upregulates mitoprotective pathways, such as the UPRmt, which maintain ROS in the hormetic zone and result in increased mitochondrial fitness in a manner consistent with mitohormesis. Consequently, these cells are more resistant to oxidative stress and have increased metastatic capacity. Our data suggest that cancer cells exploit mitohormesis to enhance metastatic disease progression.
RESULTS
Mitohormesis in Primary Tumors Identifies a Metastatic Subset of Cancer Cells
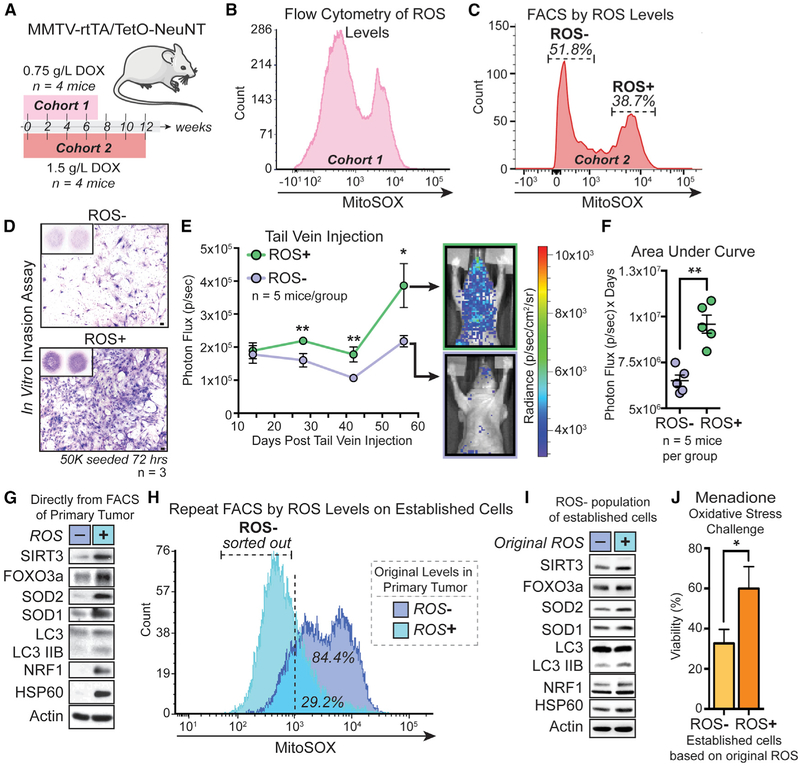
Two cohorts of 3-month-old female MTTV-rtTA/TetO-NeuNT mice (Moody et al., 2002) were given doxycycline supplemented drinking water to generate primary mammary tumors at different rates (Figure 1A). Using MitoSOX, flow cytometry was performed on primary tumors from all mice, and a distinct biphasic distribution of mitochondrial ROS was observed (Figures 1B and 1C). Low (ROS−) and high ROS (ROS+) subpopulations were isolated by fluorescence-associated cell sorting (FACS) and expanded ex vivo (Figure 1C). By transwell invasion assay, we found the ROS+ subpopulation significantly more invasive than its matched ROS− counterpart (Figure 1D). Further, we found that this difference is maintained over continued passage, indicating a stable phenotype (Figure 1D). This was not due to different proliferation rates (Figure S1A). After confirming no intrinsic differences in bioluminescence (Figure S1B), we performed tail vein injections to test metastasis differences in vivo. We found that ROS+ cells were significantly more metastatic than the ROS− subpopulation (Figures 1E and 1F). As ROS activates the UPRmt (Papa and Germain, 2014) and correlates with an invasive phenotype (Kenny et al., 2017a), we probed both subpopulations directly after FACS (Figure 1C) for markers of the UPRmt (Figure 1G). We found the ROS+ subpopulation upregulates UPRmt markers, including the SIRT3 axis (SIRT3, FOXO3a, LC3, NRF1, SOD2) and the CHOP/ATF5 axis (HSP60) (Figure 1D). Using an unbiased multi-omic approach, SOD1 was identified as a part of the UPRmt (Münch and Harper, 2016). We therefore included SOD1 and also found it upregulated in ROS+ cells (Figure 1G).
Figure 1. ROS Identifies Invasive Tumor Cells Primed to Upregulate the UPRmt by Mitohormesis.
(A) Female MMTV-rtTA/TetO-NeuNT mice given doxycycline water at 0.75 g/L for 7 weeks or 1.5 g/L for 12 weeks.
(B) Flow cytometry of mitochondrial ROS levels (MitoSOX) in cohort 1 primary tumor (n = 4 mice).
(C) Flow cytometry of cohort 2 as in (B) (n = 4). ROS− and ROS+ populations isolated by FACS and sub-cultured.
(D) Representative transwell invasion (scale, 100 μm) n = 3 with ≥ 2 technical replicates.
(E) ROS− or ROS + cells tail vein injected and metastasis assessed by bioluminescence. Mann-Whitney, two-tailed, *p < 0.05, **p < 0.01, ns = not significant; mean ± SEM; n = 5 mice/group.
(F) Area under curve of each mouse. Mann-Whitney, two-tailed, **p < 0.01; mean ± SEM; n = 5 mice/group.
(G) Western for UPRmt in isolated ROS− and ROS+ subpopulations from (C) before ex vivo expansion.
(H) Flow cytometry of mitochondrial ROS levels on ROS− and + cells cultured ex vivo. FACS isolation of ROS− populations of original ROS− and + cells.
(I) Western for UPRmt in ROS- from (H) after brief ex vivo expansion.
(J) Percent viability of attached cells after 24 h 50 μM menadione. Multiple t tests, Holm-Sidak method, *p < 0.05; Mean ± SEM; n = 3 biological replicates with 3 technical replicates.
See also Figure S1.
To test if mitohormesis was occurring, we assessed mitochondrial ROS levels following ex vivo expansion (Figure 1H). Consistent with persistent activation of cytoprotective mechanisms by mitohormesis, we found the original ROS+ subpopulation had significantly lower levels of ROS (Figure 1H), suggesting that this subpopulation is resistant to oxidative stress from ex vivo growth. We then isolated the ROS− portion of the original subpopulations by FACS to query activation of the UPRmt under lower levels of oxidative stress (Figure 1H). We found that the original ROS+ subpopulation still had enhanced activation of the UPRmt (Figure 1I). This observation supports the conclusion that oxidative stress in primary tumors induces mitohormesis in a population of cells. To functionally demonstrate the consequence of mitohormesis, we subjected the original ROS− and ROS+ subpopulations to an oxidative stress challenge using menadione–a redox cycling compound that promotes mitochondrial ROS production and mitochondrial stress (Cox et al., 2018). We found that the ROS+ subpopulation was significantly more resistant to cell death by menadione than its ROS− counterpart (Figure 1J). Taken together, these results suggest that cells primed by mitohormesis maintain activation of the UPRmt and are resistant to subsequent stress.
After finding different UPRmt subpopulations within the primary tumor, we sought to assess UPRmt activation in situ and ask if metastatic lesions are enriched for the UPRmt. Using MMTV-rtTA/TetO-NeuNT mice that develop spontaneous lung metastases (Figure S1C), we first established that doxycycline alone does not activate the UPRmt (Figures S1D–S1F; see STAR Methods). We then used immunohistochemistry (IHC) to monitor UPRmt activation in primary tumors and lung metastases of these mice. In primary tumors, we found heterogeneous staining of all UPRmt markers (Figure 2A). This suggests that the UPRmt is focally activated in primary tumors under endogenous stress. Further, we found that all markers of the UPRmt except FOXO3a were increased in lung metastases compared with primary tumors (Figure 2B), indicating in vivo that metastatic cells activate the UPRmt. Using overlaid serial sections stained by IHC, we found that all markers of the UPRmt co-localize in primary tumors (Figure 2C) and metastatic lesions (Figure 2D). Collectively, these results demonstrate that mitohormesis is observed in cells under endogenous stress, leading to UPRmt activation and a selective advantage to metastasize.
Figure 2. Focal Activation of UPRmt in Primary Murine Tumors and Enrichment of UPRmt in Metastases.
Female MMTV-rtTA/TetO-NeuNT mice given doxycycline in water at 1.5 g/L for 12 weeks.
(A) Primary tumors stained by IHC with UPRmt markers. n = 7 mice; scale, 100 μm.
(B) Primary tumors and lung metastases from serial sections stained by IHC with UPRmt markers. 1+, 2+ or 3+ scoring system (scale, 10 μm). Samples scored by intensity or intensity × extent. Intensity distribution with representative images (scale, 50 μm). Intensity × extent compared in primary and metastatic lesions. n = 7 mice; 8 primary tumors with 8 regions scored; all lung metastases (14-23/stain) scored. Unpaired t test, two-sided, *p < 0.05, **p < 0.01, ****p < 0.0001; mean (red line) ± SEM.
(C) 5 μm serial sections of primary tumors stained with UPRmt markers, pseudocolored, and overlaid (scale, 100 μm). ROI highlighted and enlarged below. SIRT3 (green); FOXO3a (pink); LC3B (purple); NRF1 (red); SOD2 (yellow); HSP60 (blue); SOD1 (orange). n = 7 mice.
(D) 5 μm serial sections of lung metastases as in (C). n = 7 mice.
Endogenous Mitochondrial Stress in Human Breast Cancer Cell Lines Activates the UPRmt and Increases Invasion
To determine if mitohormesis and its contribution to metastasis occurred in human breast cancer, we used MCF7 and MDA-MB-231 cells in which we previously characterized the UPRmt (Kenny et al., 2017a). Having established that endogenous mitohormesis primed a subpopulation of cancer cells in vivo to become more invasive, we hypothesized that the invasive subpopulation of a given cell line should have increased UPRmt activation. To test this possibility, we performed a transwell invasion assay with MCF7 cells and isolated the invasive subpopulation, MCF7 iteration 1 (I1) (Figure 3A). As expected, MCF7 I1 cells were more invasive than the parental (Figure 3B). When mitochondrial ROS levels were compared, MCF7 I1 cells had less than the parental, consistent with mitohormesis (Figure 3C). Importantly, MCF7 I1 cells had more UPRmt activation (Figure 3D). We also analyzed levels of NRF2, as it is a master transcription factor for antioxidant genes and binds to the promoter of SIRT3 (DeNicola et al., 2011; Hayes and McMahon, 2006; Padmanabhan et al., 2006; Satterstrom et al., 2015). Compared with parental, MCF7 I1 had more NRF2 (Figure S2A).
Figure 3. Identification of Invasive, UPRmt-HIGH Subpopulation of Human Cell Lines by Endogenous Mitohormesis.
(A) Invasive subpopulation of MCF7 cells isolated by transwell invasion and sub-cultured to generate MCF7 iteration 1 (I1).
(B) Representative transwell invasion (scale, 100 μm).
(C) Flow cytometry of endogenous mitochondrial ROS levels in MCF7 parental (P) and I1.
(D) Western for UPRmt in MCF7 P and I1.
(E) ROS+ population of MCF7 isolated by FACS and sub-cultured to generate MCF7 ROS+ cells.
(F) Representative images of transwell invasion assays (scale, 100 μm).
(G) Western for UPRmt in MCF7 P and ROS+.
(H) Invasive subpopulation of MDA-MB-231 P cells isolated by transwell invasion and sub-cultured to generate I1 and repeated to generate I2.
(I) Representative transwell invasion (scale, 100 μm).
(J) Flow cytometry of endogenous mitochondrial ROS levels in MDA-MB-231 P, I1, and I2.
(K) Western for UPRmt markers in MDA-MB-231 P, I1, and I2.
(L) ROS+ population of MDA-MB-231 cells isolated by FACS and sub-cultured to generate MDA-MB-231 ROS+ cells.
(M) Representative images of transwell invasion (scale, 100 μm).
(N) Western for UPRmt in MDA-MB-231 P and ROS+.
(O) MDA-MB-231 treated with non-targeting siNC1 or siSIRT3. Representative western of UPRmt.
(P) Quantification of UPRmt after siRNA. n = 3 experiments. Unpaired t test, two-sided, *p < 0.05, **p < 0.01, ns = not significant; mean ± SEM.
(Q) Representative transwell invasion of MDA-MB-231 siNC1 or siSIRT3 (scale, 100 μm). n = 3 experiments with R 3 technical replicates.
(R) Western for indicated markers in MDA-MB-231 generated with stable shRNA control (shScrambled) or targeting (shSIRT3-345 and shSIRT3-752) constructs in pLV-H1-CMV-green vector. Tail vein injection and collected after 4 weeks.
(S) Metastases visualized in freshly excised lungs using multiphoton microscopy. GFP, green. Collagen, purple (scale, 100 μm). shScrambled n = 5; shSIRT3-345 n = 4; shSIRT3-752 n = 5 mice.
(T) Whole lung sections from S stained by IF for GFP. Lesions per section counted and size noted as single cells, clusters, or micro-metastases. Total lung lesions compared. Mann-Whitney, two-tailed, *p < 0.05, **p < 0.01; mean ± SEM.
See also Figure S2.
Having demonstrated low and high ROS subpopulations in primary tumors, we asked if this was true in MCF7 cells. We identified and isolated a small ROS+ subpopulation of MCF7 by FACS (Figure 3E). We found that the ROS+ subpopulation was significantly more invasive than the parental (Figure 3F) and had increased activation of the UPRmt-SIRT3 but not HSP60 or SOD1 (Figure 3G).
To further test these observations, we repeated the same analyses in invasive, UPRmt-SIRT3 positive MDA-MB-231 cells. Two consecutive rounds of invasion were used to generate MDA-MB-231 I1 and I2 cells (Figure 3H). We found that MDA-MB-231 I1 and I2 were progressively more invasive than the parental (Figure 3I) and had decreased ROS (Figure 3J). Consistent with mitohormesis, MDA-MB-231 I1 and I2 showed successive increases in the UPRmt (Figure 3K). We also queried NRF2 levels and observed cumulative increases in the invasive sub-populations (Figure S2B). When we isolated the ROS+ of MDA-MB-231 cells by FACS (Figure 3L), we again found that the ROS+ subpopulation was more invasive than the parental (Figure3M). Additionally, ROS+ MDA-MB-231 had enhanced UPRmt activation compared with the parental, except for HSP60 (Figure 3N). To test if mitohormetically primed cells are resistant to oxidative stress, we again used menadione. As before (Figure 1J), we found that MDA-MB-231 I1, I2, and ROS+ cells were more resistant to cell death than the parental (Figure S2C).
To address causality between ROS reduction by UPRmt activation and the invasive phenotype, we treated parental MDA-MB-231 with the antioxidant N-acetyl-l-cysteine (NAC) and compared invasion with parental and I2 cells without NAC (Figure S2D). We found that treatment with NAC significantly increased invasion of MDA-MB-231 parental cells in comparison to the control but not to the same level as I2 cells (Figure S2D). We conclude that reduction of ROS by UPRmt activation promotes invasion but cannot alone explain the phenotype. We therefore used gamitrinib-triphenylphosphonium (G-TPP), a compound shown to activate the UPRmt (Kang et al., 2009; Münch and Harper, 2016; Papa and Germain, 2014) to exogenously activate the UPRmt and analyze invasion capacity. When treated with 5 μM G-TPP, MDA-MB-231 showed robust activation of all UPRmt markers (Figure S2E) and enhanced invasion capacity (Figure S2F) compared with DMSO control. These results demonstrate that UPRmt activation promotes invasion.
To address causality between UPRmt activation and invasion mechanistically, we performed small interfering RNA (siRNA) knockdown of SIRT3 in MDA-MB-231. SIRT3 knockdown leads to a subsequent decrease of UPRmt-SIRT3 markers (FOXO3a, SOD2, NRF1) but not SOD1 or HSP60 (Figures 3O and 3P). Interestingly, SIRT3 knockdown significantly increased LC3, suggesting a potential non-SIRT3-mediated mechanism of upregulation to maintain viability after UPRmt-SIRT3 knockdown (Figures 3O and 3P). Importantly, transwell assays of MDA-MB-231 treated with siSIRT3 or non-targeting control revealed that SIRT3 knockdown significantly reduced invasion (Figure 3Q). The inhibitory effect of SIRT3 knockdown on invasion was validated using a second siRNA (Figures S2A and S2B). Additionally, SIRT3 knockdown did not affect proliferation (Figure S2C) or viability (Figure S2D), demonstrating a specific effect on invasion. To assess the role of SIRT3 in metastasis in vivo, we generated stable control (shScrambled) and SIRT3 knockout (shSIRT3-345 and shSIRT3-752) MDA-MB-231 cells (Figure 3R). Western blotting of these cells confirmed near complete loss of SIRT3 in shSIRT3-345 and shSIRT3-752 compared to shScrambled and ubiquitous GFP expression in all three lines (Figure 3R). We queried in vitro invasion capacity of these cells and found that SIRT3 loss reduced invasion capacity (Figure S2K) without impacting in vitro proliferation (Figure S2L), as in our siRNA experiments (Figures 3Q and S2I). We then performed tail vein injections with these cells and harvested 4 weeks later. Two-photon microscopy was used on freshly excised lungs to visualize GFP+ metastases. While disseminated cells could be identified in all three experimental groups, markedly larger metastatic lesions were detected in shScrambled compared with either shSIRT3 group (Figures 3S and S2M). These results indicate that SIRT3 is necessary for overt lung metastasis formation. Whole lung sections were stained by immunofluorescence (IF) for GFP to quantify single disseminated cells, clusters, and micro-metastases. As observed by two-photon microscopy, the shScrambled group had appreciably more multi-cellular lesions than either shSIRT3 group by IF (Figures 3T and S2N). Importantly, we found significantly more metastatic lesions irrespective of size in shScrambled mice compared with shSIRT3-345 or shSIRT3-752 (Figures 3T and S2N). This suggests that SIRT3 promotes the invasion and extravasation capacity of metastatic breast cancer cells at secondary organs. Taken together, these results demonstrate that SIRT3 is necessary for multiple steps of the metastatic cascade.
UPRmt Gene Signature Identifies Breast Cancer Patients with Worse Prognoses and Mitohormetic Transcriptional Profiles
To address the translational relevance of our findings to patients, we took advantage of publicly available primary breast cancer expression datasets. In a cohort of 1809 patients (Györffy et al., 2010), we extracted expression levels of SIRT3, FOXO3a, SOD2, SOD1, LC3B, NRF1, and HSP60 and computed an average UPRmt expression score per patient (Figure 4A). We found a clear subset of patients with higher expression of all markers and called them UPRmt-HIGH (Figure 4A). Treating URPmt score as a categorical value, we found a significant association between UPRmt-HIGH and estrogen receptor (ER)-negative disease (Figure 4A). This association was also significant when UPRmt score was treated as a continuous variable (Figure S3A). UPRmt score was also calculated in the The Cancer Genome Atlas (TCGA) breast cancer cohort and again showed a significant association between ER-negative disease and elevated UPRmt (Figure S3B). The molecular classifications in the TCGA cohort enabled comparison of UPRmt among PAM50 intrinsic subtypes (Figure S3C). We found that the UPRmt scores were significantly different between breast cancer subtypes, with basal-like and HER2-Enriched having significantly higher levels than luminal A and luminal B (Figure S3C).
Figure 4. Increased Activation of UPRmt in Patients Correlates with Worse Clinical Outcomes and Is Associated with a Distinct Transcriptional Signature.
(A) Heatmap of UPRmt expression in 1809 breast cancer patients. Top ranked tertile (n = 603) called UPRmt-HIGH. Estrogen receptor (ER) status displayed. Negative, white; positive, black. Significant correlation: ER negative and UPRmt-HIGH. Pearson’s chi-square, Yates’ continuity correction, p = 2.45e-20, n = 1231.
(B) Kaplan-Meier (KM) analysis of overall survival (OS) with UPRmt classifier (n = 1402). HR = hazard ratio.
(C) KM of relapse-free survival (RFS) with UPRmt classifier (n = 3951).
(D) KM of distant metastasis-free survival (DMFS) with UPRmt classifier (n = 1764).
(E) KM of RFS in lymph node positive patients with UPRmt classifier (n = 1133).
(F) Volcano plot UPRmt-HIGH versus UPRmt-LOW patients (n = 1809). Log2 fold change versus false discovery rate (FDR). Significant genes, red (FDR < 0.05 & log2 fold change R ≥ ± 0.6)
(G) Gene set enrichment analysis (GSEA) of UPRmt-HIGH versus UPRmt-LOW (n = 1809). Negatively (left) and positively (right) enriched gene ontology terms. Bar length, normalized enrichment score (NES). Bar color, FDR. Gene set source, color code.
We then assessed the impact of UPRmt expression on outcomes using the larger Kaplan-Meier (KM) plotter cohort (Lánczky et al., 2016). Alone, the individual UPRmt genes had mixed prognostic value (Figures S3D–S3J). A multigene classifier of these 7 genes was used to compare clinical outcomes between UPRmt-HIGH and UPRmt-LOW patients. We found that UPRmt-HIGH patients had significantly worse overall survival (Figure 4B), relapse-free survival (Figure 4C), and distant metastasis–free survival (Figure 4D) compared with UPRmt-LOW patients. Even in lymph node-positive patients, a population which is at significant increased risk of metastatic relapse, UPRmt-HIGH patients had significantly worse distant metastasis-free survival (Figure 4E).
To further understand differences between UPRmt-HIGH and UPRmt-LOW patients, we compared global gene expression between the groups. Differential gene expression analysis between UPRmt-HIGH and UPRmt-LOW patients identified 141 genes with a log2 fold change ≥ ±0.6 and a false discovery rate <0.05 (Figure 4F; Tables S1 and S2).
Transcripts significantly upregulated in UPRmt-HIGH patient tumors included metalloproteases (MMP1, ADAM15, MM7, MMP12) and immune-related genes, such as MARCO (Figure 4F; Tables S1 and S2). MARCO identifies a subset of tumor-associated macrophages that promotes tumor growth and metastatic dissemination (Georgoudaki et al., 2016). Of interest, PDK1 was significantly increased in UPRmt-HIGH patients (Figure 4F). PDK1 was also upregulated in the livers of mitohormetically primed iSOD2-KD mice (Cox et al., 2018). Transcripts significantly downregulated in UPRmt-HIGH patient tumors include the metabolic enzyme MGAM, and PTHLH, which is implicated in epithelial-mesenychmal interactions in the developing mammary gland (Figure 4F; Tables S1 and S2).
To understand the gene expression programs differing between UPRmt-HIGH and UPRmt-LOW patients, we performed pre-ranked gene set enrichment analysis (GSEA) (Table S3; Subramanian et al., 2005). We found that cholesterol homeostasis, cell cycle, and peroxisome proliferator-activated receptor (PPAR) signaling were positively enriched in UPRmt-HIGH patients (Figure 4G; Table S4). PPAR signaling was identified as the defining transcriptional signature of mitohormetically primed iSOD2-KD mice (Cox et al., 2018). In addition, mTORC1 signaling and glycine, serine, and threonine metabolism were positively enriched in UPRmt-HIGH patients, in agreement with data that these pathways are activated by mitochondrial stress (Quirós et al., 2017). Interestingly, it has been shown that mTORC1 regulates SOD1 activity (Tsang et al., 2018) and HSP60 as part of the mitochondrial integrated stress response (ISRmt) (Khan et al., 2017). This highlights the integrated nature of mitochondrial stress responses and substantiates the UPRmt as a complex network of signaling axes. Importantly, metastasis, angiogenesis, and cell migration gene sets were positively enriched in UPRmt-HIGH patients (Figure 4G; Table S4). Innate immunity and inflammation-related gene sets were also positively enriched, in agreement with observations in C. elegans that linked the UPRmt to innate immunity (Pellegrino et al., 2014) (Figure 4G; Table S4).
Conversely, we found processes related to protein production, such as translational elongation and the ribosome, negatively enriched in UPRmt-HIGH patients (Figure 4G; Table S5). This is consistent with work showing translational inhibition in response to mitochondrial stress (Münch and Harper, 2016; Ruan et al., 2017; Wrobel et al., 2015).
Taken together, these results demonstrate that UPRmt activation is observed in patient samples and is significantly associated with worse clinical outcomes including metastasis. Further, we found that the global gene expression of UPRmt-HIGH patients is indicative of mitohormetic priming and persistent cytoprotective mechanisms.
DISCUSSION
The mitochondria of cancer cells show profound reprogramming and elevated ROS. ROS levels in cancer must be maintained to ensure viability. The ability of ROS manipulation to impact tumor biology is supported by studies demonstrating that antioxidants promote cancer growth and metastasis (Bjelakovic and Gluud, 2007; Le Gal et al., 2015; Sayin et al., 2014).
Here, we provide evidence that mitohormesis, a phenomenon normally associated with health and longevity, is perniciously used by cancer cells to promote tumor progression. Mitohormesis in cancer cells results in persistent activation of the UPRmt and reduction in oxidative stress. Resultantly, mitohormetically primed cancer cells are more metastatic. The molecular mechanisms underlying mitohormetic activation and UPRmt persistence in cancer remains elusive. A likely possibility is that epigenetic regulation is involved, as shown in C. elegans (Ma et al., 2019; Merkwirth et al., 2016; Tian et al., 2016). Future work will address such outstanding questions.
Our findings indicate that elevated UPRmt in patients results in significantly worse survival. Analysis of the transcriptomes of UPRmt-HIGH patients shows simultaneous activation of prometastatic programs and the global cytoprotective effects of mitohormesis. Our patient-derived UPRmt signature showed significant overlap with mito-protective pathways obtained from in vitro or genetic manipulations of mitochondrial stress (Cardamone et al., 2018; Münch and Harper, 2016; Pellegrino et al., 2014; Quirós et al., 2017; Tian et al., 2016). Of note, translational repression was strongly affected by the UPRmt classifer, consistent with reports characterizing exogenous UPRmt induction in vitro (Münch and Harper, 2016; Quirós et al., 2017). Mitochondria can influence innate immunity through the cytoplasmic release of mtDNA (West and Shadel, 2017; West et al., 2015). Interestingly, we see significant enrichment of innate immunity gene sets in UPRmt-HIGH patients suggesting a possible role for mitohormesis in shaping the tumor microenvironment. Several neurodegenerative gene sets were negatively enriched in UPRmt-HIGH patients. This is consistent with our work showing that UPRmt activation delayed symptom onset in a familial amyotrophic lateral sclerosis (ALS) mouse model (Riar et al., 2017). This observation suggests that mitohormesis, which we show promotes tumor progression, is predicted to oppose neurodegeneration.
In the context of cancer biology, our results are also in agreement with numerous studies reporting mitochondrial changes in recurrent tumors and circulating cancer cells. These changes included oxidative phosphorylation, mitochondrial biogenesis, and SOD2 and PGC-1α upregulation, although a unifying mechanism was not identified (Hu et al., 2012; LeBleu et al., 2014; Viale et al., 2014). The results of our study suggest that activation of the UPRmt-SIRT3 mechanistically underlies these observations.
This possibility is further supported by the fact that SIRT3 is a common denominator linking mitochondrial biogenesis, PGC1α, and SOD2. SIRT3 directly controls the activity of SOD2 through deacetylation (Lombard et al., 2007; Tao et al., 2010). PGC1-α, in complex with ERRα regulates several genes involved in mitochondrial biogenesis as well as SOD2 and SIRT3 (Kong et al., 2010; Giralt et al., 2011). SIRT3 indirectly regulates PGC1-α through AMPK signaling, which promotes CREB phosphorylation and PGC1α expression (Shi et al., 2005). Therefore, SIRT3 and PGC1α crosstalk establishes a positive feedback loop to regulate mitochondrial biogenesis and antioxidant defenses. In this study, the transcriptional signature of UPRmtHIGH patients was significantly enriched for PPAR signaling, of which PGC1α is a key component. Notably, the PPAR signaling pathway and PGC1α were key differences seen in mitohormetically primed mice (Cox et al., 2018).
In cancer biology, SIRT3 has been reported to act as an oncogene (Alhazzazi et al., 2011; Cheng et al., 2013; Choi et al., 2016; Cui et al., 2015; George et al., 2016; Li et al., 2010; Papa and Germain, 2014; Wang et al., 2015; Wei et al., 2013) and a tumor suppressor (Allison and Milner, 2007; Desouki et al., 2014; Dong et al., 2016; Finley et al., 2011; Kim et al., 2010; Li et al., 2010; Wang et al., 2014; Wei et al., 2013; Yu et al., 2016; Zhang and Zhou, 2012). This discrepancy suggests that the function of SIRT3 in cancer is context dependent. Loss of SIRT3 during tumor formation leads to increased ROS levels and promotes mitochondrial reprogramming (Finley et al., 2011). Importantly, SIRT3 loss is heterozygous, suggesting selective pressure to maintain an intact copy of the gene (Finley et al., 2011). When the prognostic value of individual UPRmt genes was analyzed, elevated SIRT3 was associated with better relapse-free survival (Figure S3F), in agreement with a recent report (Lee et al., 2018). This observation is consistent with a study demonstrating that SIRT3 overexpression in MDA-MB-231 cells delays tumor growth in xenografts (Gonzalez Herrera et al., 2018). It is important to note that this overexpression far exceeds the endogenous levels found in invasive cells, such as MDA-MB-231. The same group also demonstrated that SIRT3 is downregulated in the invasive front of MCF10A cells (Lee et al., 2018). To reconcile these observations, we propose the following model (Figure S4). In normal cells, SIRT3 levels are highest. This is supported by the observation that MCF10A cells have significantly higher SIRT3 levels than breast cancer cells lines (Papa and Germain, 2014). During transformation, heterozygous SIRT3 loss increases ROS, which stabilizes HIF1α and produces a glycolytic switch (Brunelle et al., 2005; Chandel et al., 1998, 2000; Finley et al., 2011; Haigis et al., 2012). During tumor progression, however, the retained SIRT3 allele is used to promote invasion and metastasis.
In conclusion, we demonstrate that endogenous mitohormesis leads to persistent activation of the UPRmt in a subset of cancer cells. Consistent with the pro-longevity effect of mitohormesis, its activation leads to global changes in cytoprotective mechanisms, which results in resistance to subsequent stress. In the context of cancer, however, mitohormesis promotes metastatic disease progression.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by Doris Germain, DG (doris.germain@mssm.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Culture
MDA-MB-231 and MCF7 cells, and their respective sub-lines were cultured in DMEM media supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (P/S). LentiX-293T cells (Takara Clonetech) were cultured in DMEM media supplemented with 10% FBS and 1% P/S. Cells derived from MMTV-rtTA/TetO-NeuNT mammary tumors were grown in DMEM / F12 (50/50) supplemented with 10% FBS, 1% P/S, 25 ng/mL Hydrocortisone, 0.25 μg/mL Amphotericin B, and 2 μg/mL Doxycycline and cultured on collagen coated plates (50 μg/mL Collagen 1 rat tail in 0.01M HCL).
Animals
All animal experiments were performed under an IUCAC approved protocol. MMTV-rtTA/TetO-NeuNT mice were originally generated and kindly provided by Lewis Chodosh. Genotypes of mice were confirmed using the following primers: rtTA (5′-TGCCGCCAT TATTACGACAAGC-3′; 5′-ACCGTACTCGTCAATTCCAAGGG-3′) and Neu: (5′-TTTCCTGCAGCAGCCTACGC-3′, 5′-CGGAACCCA CATCAGGCC-3′). Experimental MMTV-rtTA/TetO-NeuNT were 12-week-old females. Non-transgenic FVBN and C57BL/6N were also used in this study. Experimental FVBN mice were 8-week-old females. Experimental C57BL/6N mice were 8-week-old males. Female Athymic Nude-Foxn1nu mice 9 or 12 weeks of age were used for tail vein injection experiments. All mice were housed in vivariums at Mount Sinai with ad libitium access to food and water.
METHOD DETAILS
siRNA Transfection
Transfections were performed for 72 hours on cells seeded in antibiotic-free medium using Lipofectamine RNAi Max (Invitrogen 13778-100) and Opti-MEM (GIBCO) following manufacturer recommendations. Transfections were performed with non-targeting DsiNC1 (IDT #51-01-14-03) or SIRT3 targeting siRNA: siSIRT3 #1 (5’-GCCCAACGUCACUCACUACTT –3′, 5′-GUAGUGAGUGAC GUUGGGCTT-’3; GeneLink) and siSIRT3 #2 (5’-ACUCCCAUUCUUCUUUCACTT-3′, 5′- GUGAAAGAAGAAUGGGAGUTT-3′; GeneLink). Percent of cells alive and total cell number were determined using the Countess automated cell counter and tryphan blue (Invitrogen) on MDA-MB-231 cells treated with siNC1, siSIRT3 #1, or siSIRT3 #2 after 72 hours transfection to determine viability and in vitro proliferation (Figure S2).
Generation of stable shRNA cell lines
A pre-designed shRNA vector set was purchased from Biosettia with shRNAs designed against SIRT3 (Gene ID 23410; Accession NM_001017524.2) in the pLV-H1-CMV-Green plasmid (Biosettia #SORT-B01). NEB stable competent E. Coli cells (New England BioLabs #C3040) were transformed by heat shock with plasmids obtained from Biosettia. Transformed colonies were picked and expanded under antibiotic selection. Plasmids were prepared from bacterial cultures by Maxiprep (QIAGEN #12165). OptiMEM (GIBCO) and Lipofectamine 2000 (Invitrogen #11668019) were used to transfect 9 μg of pLV-H1-CMV-Green plasmid, 6 μg psPAX2 plasmid (Addgene #12260), and 3 μg pMD2.G plasmid (Addgene #12259) into LentiX 293T packaging cells in antibiotic free medium. pMD2.G and psPAX2 plasmids were generous gifts from Didier Trono. Lentivirus containing media was then collected, filtered and used with polybrene to infect target MDA-MB-231 Parental cells. Transduced cells were selected for using fluorescence associated cell sorting (FACS) to generate stable shRNA cell lines.
Transwell Invasion Assays
Cells were trypsinized and collected in respective serum-free media (see Cell Culture) and pelleted at 2500 rpm for 5 minutes. Cells were resuspended in respective serum-free media and counted using the Countess automated cell counter and tryphan blue (Invitrogen). Equal cell numbers were seeded in respective serum-free media on 8 μm pore size cell culture inserts for 24-well plates (Corning). Prior to seeding cell seeding, cell culture inserts were coated with growth factor reduced matrigel (GIBCO) diluted 1:100 in PBS that was incubated at room temperature for 2 hours and removed. Respective media with 10% FBS was used in the lower chamber and assays were performed for time frames indicated (see Figures) in a tissue culture incubator. Following incubation, invaded cells were fixed, stained using the Hema 3 Manual Staining Stat Pack according to manufacturer’s guidelines (Thermo Fisher Scientific), and placed on glass slides with mounting media (Permount). To generate invasive clones of MCF7 and MDA-MB-231 cells, the protocol outlined above was followed except 8 μm pore size cell culture inserts for 6-well plates (Corning) were used and invaded cells were detached by trypsin and subsequently cultured.
Western Blotting
Cells, washed with PBS, or pulverized flash frozen tissue was lysed in cold NP-40 lysis buffer with protease inhibitors (50 mM Tris, pH 7.5, 250 mM NaCl, 5 mM EDTA, 0.5% Nonidet P-40, 50 mM NaF, 0.2 mM Na3 VO4, 1 g/ml leupeptin, 1 g/ml pepstatin, 100 g/ml phenylmethylsulfonyl fluoride), sonicated for 5 s at 20% amplitude, and centrifuged at 14,000 rpm for 20 minutes at 4°C. Protein concentrations of lysates were assayed using the Bradford method (Bio-Rad Protein Assay). Equal amounts of protein were separated by SDS-PAGE electrophoresis and transferred to nitrocellulose membrane (GE Healthcare). Following blocking, membranes were probed with the following primary antibodies overnight at 4°C: SIRT3 (Cell Signaling 2627S), SIRT3 (EMD Millipore 07-1596), HSP60 (BD Transduction 611563), ATF5 (abcam ab184923), NRF1 (abcam ab55744), ClpP (abcam ab 124822), SOD1 (Santa Cruz sc-11407), FOXO3a (Cell Signaling 2497S), Actin (EMB Millipore MAB1501R), SOD2 (EMB Millipore 06-984), LC3 (MBL International PM036), SDHA (abcam ab14715), MTCO1 (abcam ab45918), GFP (Santa Cruz sc-9996), NRF2 (cell Signaling 12721). Blots were then probed with horseradish peroxidase conjugated anti-mouse (Jackson ImmunoResearch or KwikQuant) or anti-rabbit secondary antibodies (Thermo Fisher Scientific or KwikQuant) and detected using enhanced chemiluminescence (GE Healthcare or KwikQuant).
Flow Cytometry and Fluorescence Associated Cell Sorting (FACS)
Cells growing in culture were detached with trypsin, collected in respective media, and centrifuged to pellet cells. Cells were then washed in PBS and finally resuspended in 0.5% Bovine Serum Albumin (BSA) in Hank’s Balanced Salt Solution (HBSS). Appropriate volume of freshly made 5 mM stock of MitoSOX Red (Thermo Fisher Scientific M36008) was added to cells in 0.5% BSA HBSS to yield final staining concentration of 5 μM. Staining was perfomed for 30 minutes at 37°C in the dark. Following staining, cells were pelleted by centrifugation, washed in 0.5% BSA HBSS, and resuspended in 0.5% BSA HBSS and passed through a single cell strainer before flow cytometry or FACS. Using the BD FACSAria II and BD FACSDiva software, samples were excited and captured at wavelengths consentient with the fluorescent spectrum of MitoSox. When sorting was performed, noted populations were selected and sorted for subsequent analyses or culture.
Primary mammary tumors from MMTV-rtTA/TetO-NeuNT mice were extracted from euthanized animals and placed in DMEM / F12 (50/50) supplemented with 25 ng/mL Hydrocortisone, 0.25 μg/mL Amphotericin B, and 2 μg/mL Doxycycline and brought into a tissue culture hood. Tumors were mechanically cut and digested using scalpels and the homogenized tumor was placed in a 50 mL falcon tube and filled with supplemented DMEM/F12 (50/50) media. Tumor homogenate and media was mixed by repeated inversion and centrifuged at 900 rpm to separate pelleted cells from fat and debris. Pelleted cells were washed in PBS and then resuspended in red blood cell (RBC) lysis buffer and mixed gently for 1-2 minutes. Supplemented media was then added to cells in RBC lysis buffer and centrifuged at 200-500 g for 7 minutes. Pellet was resuspended in supplemented media plus 1 mg/mL collagenase (Sigma-Aldrich C9891). This was incubated at 37°C for 30 minutes with pipetting performed twice during the incubation to aid in digestion. Following incubation, the solution was briefly vortexed and stood upright for 1-2 minutes to allow large chunks to settle. The supernatant was collected and centrifuged at 900 rpm for 5 minutes. The pellet was then resuspended in supplemented media and passed through a 70 μm nylon strainer to achieve single cell suspension. Cells were then pelleted, washed with PBS, and resuspended in 0.5% BSA HBSS. Staining and flow cytometry/FACS was then performed as above.
Animal Experiments: MMTV-rtTA/TetO-NeuNT mice
12-week-old female MMTV-rtTA/TetO-NeuNT were given doxycycline in drinking water (Sigma Aldrich) at a concentration of 0.75 g/L for 7 weeks or 1.5 g/L for 12 weeks. Doxycycline water was changed biweekly. At euthanasia, primary tumors and lungs were collected and formalin fixed, flash frozen, and/or processed for FACS.
Animal Experiments: Doxycycline treatment, analysis of UPRmt
As doxycycline has been reported to inhibit mitochondrial translation and induce mitochondrial stress (Moullan et al., 2015), we first tested whether, doxycycline alone induces the UPRmt in non-transgenic mice treated with doxycycline. We found no changes in markers of either the UPRmt-SIRT3 or URPmt-CHOP (Figure S1D). As the ratio of MTCO1 to SDHA has been used previously as a marker of mitochondrial stress and imbalance between the mitochondria and nuclear genome (Moullan et al., 2015), this ratio was also tested. No difference was observed in either the mammary gland (Figure S1D) or the liver (Figure S1E). As this result is in contrast to the reported effect of doxycycline (Moullan et al., 2015), we repeated the experiment using the same gender, strain, and experimental conditions as the published study (Moullan et al., 2015). Using these conditions, we found that doxycycline promotes mitonuclear protein imbalance as measured by the ratio of MTCO1 to SDHA, (Figure S1E) but did not lead to a concomitant activation of either the UPRmt-SIRT3 or UPRmt-CHOP (Figure S1E). This observation is consistent with a more recent report from the Auwerx group showing that doxycycline does not activate canonical UPRmt pathways(Quirós et al., 2017). It also suggests that the effect of doxycycline varies between mouse strains and/or gender.
8-week-old female FVBN mice were given doxycycline (Sigma Aldrich) at a concentration of 1.5 g/L in drinking water for 30 days. Doxycycline water was changed biweekly. At euthanasia, livers and mammary glands were flash frozen for subsequent analysis. 8-week-old male C57BL/6N mice were given amoxicillin (Sigma Aldrich) at 50/mg/kg/day for 15 days or 500mg/kg/day doxycycline for 15 or 30 days in 50 g/L sucrose water. Both amoxicillin and doxycycline sucrose water was changed every 48 hours. At euthanasia, livers were flash frozen for subsequent analysis.
Animal Experiments: Tail Vein Injections
For experiments in Figure 1, 12-week-old female Athymic Nude-Foxn1nu mice were given doxycycline at a concentration of 1.5 g/L in drinking water 2 weeks prior to tail vein injection and this was continued throughout the experiment. Doxycycline water was changed biweekly. FACS sorted primary tumor subpopulations (ROS- or ROS+) expanded ex vivo were used for tail vein injection. Prior to tail vein injection, the bioluminescence of ROS− and ROS+ subpopulations was confirmed in vitro and was found to be insignificantly different between the two. 5×105 cells in 100 μL PBS was injected into the tail vein of each mouse. Five mice per group were injected.
For experiments in Figure 3, 9-week-old female Athymic Nude-Foxn1nu mice were used for tail injections. 1×106 cells in 100 uL PBS supplemented with 1% FBS were injected into the tail vein of each mouse. Five mice per group were injected.
Histology and Immunohistochemistry (IHC)
Tissue was fixed in 10% formalin (Thermo Fisher Scientific) and then processed and paraffin embedded for sectioning by the Biorepository Core Facility at Mount Sinai. Hematoxylin and eosin (H&E) or unstained paraffin embedded slides were obtained the from the core facility. Serial 5 μm sections were used for all IHC staining. Briefly, sections were deparrafinized and rehydrated in xylene followed by decreasing alcohol gradients. Antigen retrieval was performed for 30 minutes at 95°C in either citrate buffer pH 6 or 10 mM TRIS 1 mM EDTA pH 9. Following antigen retrieval, endogenous peroxidase activity was quenched using Dako Dual Endogenous Enzyme Block (#S20003) followed by serum blocking (MP Biomedicals Normal Antibody Diluent #98063) at room temperature. Primary antibody staining was performed at room temperature or 4°C for an optimized length of time. Secondary antibody staining with either Biotynlated Anti-Rabbit (Jackson ImmunoResearch #711-065-152) or Biotynlated Anti-Mouse (Jackson ImmunoResearch #115-065-205) diluted in TBS was incubated at room temperature for an optimized length of time. Streptavidin-HRP (Vector Labs #SA-5004) was diluted in TBS and incubated for 30 minutes at room temperature. All washes were performed with TBS with 0.04% Tween 20. Antigen detection was performed with freshly prepared AEC Solution (Vector Labs #SK-4200) according to manufacturer guidelines for an optimized length of time. Sections were counterstained with hematoxylin, sufficiently rinsed in water, and mounted in Vectamount aqueous mounting media (Vector Labs #H-5501). For specific conditions used for each antigen staining, see Table S6.
Sequentially sectioned (5 μm apart) primary tumors and lungs were stained with all indicated antibodies and matching regions of interest were imaged at the same magnification using a Zeiss AX10 light microscope. These images were imported into Adobe Photoshop individually and a layer encompassing all positive staining created. This layer was pseduocolored and saved alone. Multiple psuedocolored layers from different stainings were superimposed on one another after proper alignment. These overlays were saved and displayed on a black background to generate the images seen in Figure 2.
Immunofluorescence
Sections were deparaffinized and rehydrated in xylene followed by decreasing alcohol gradients. Antigen retrieval was performed in citrate buffer pH 6 for 20 minutes in the microwave. Blocking was performed with 10% serum in 0.5% Tween-20 PBS for 2 hours at room temperature. Primary antibody (GFP; Santa Cruz sc-9996) staining was performed overnight at 4°C at a concentration of 1:100 in 1% serum 0.5% Tween-20 PBS. Secondary antibody (Alexa Flour 488 goat anti-mouse; Invitrogen #A11001) staining was performed for 2 hours at roomperature at a concentration of 1:250 in 1% serum 0.5% Tween-20 PBS. Finally, sections were stained with DAPi for 20 minutes at room temperature prior to mounting with Flouromount-G (Southern Biotech). All washes were performed in 0.5% Tween-20 PBS. Sections stained following the protocol above with the exception of no primary antibody were used as negative controls. Metastatic lesions were visualized, counted, and imaged using a Leica DM5500 B upright automated microscope.
Multiphoton Microscopy
Freshly excised lungs from mice used in Figure 3 were imaged using an Olympus FV1000 multiphoton microscope with an Olympus 25X 1.05 NA water immersion objective. As previously described (Olmeda et al., 2017), a Coherent Chameleon Vision II laser was used at 880 nm to excite GFP. Collagen was visualized by second harmonic generation. 512 × 512 images were taken at 12.5 μs/pixel with a Z step-size of 5 μm using a Kalman filter. For all images, the RFP channel was also captured to rule out artifactual autofluorescence.
Bioluminescence Imaging
Given that the MMTV-rtTA/TetO-NeuNT mice have an IRES-Firefly Luciferase sequence downstream of the Tet-inducible NeuNT, we performed whole body bioluminescence imaging on anthymic nude mice that were tail vein injected with isolated MMTV-rtTA/TetO-NeuNT primary tumor populations expanded ex vivo. Bioluminescence imaging was conducted using the IVIS Spectrum In Vivo Imaging System (Perkin Elmer) in accordance with manufacturer protocols. Mice anesthetized with isoflurane were imaged before and after D-Luciferin (dissolved in PBS) injection intra peritoneally (150mg/kg body weight) at equal exposure times to determine bioluminescence signal. A kinetic study of bioluminescent signal after Luciferin injection was performed to determine peak signal time.
Bioinformatic Analyses
For Kaplan Meier analysis, KM Plotter was used(Györffy et al., 2010; Lánczky et al., 2016). The optimal probes to be used for each UPRmt marker were identified using JetSet(Li et al., 2011) and were as follows: SIRT3 (221913_at), FOXO3a (217399_s_at), SOD2 (215223_s_at), SOD1 (200642_at), NRF1 (204651_at), LC3B (208786_s_at), and HSP60 (200807_s_at). Publicly available expression data for primary breast cancers (GEO: GSE11121, GSE12093, GSE12276, GSE1456, GSE16391, GSE2034, GSE2990, GSE3494, GSE5327, GSE6532, GSE7390, and GSE9195) was downloaded and compiled. Normalized expression of UPRmt markers (SIRT3, FOXO3a, SOD2, SOD1, NRF1, LC3, HSP60) were averaged for each patient to calculate a UPRmt Expression Score. Patients were ranked from lowest to highest UPRmt Expression Score and the highest third of patients (33.3%, n = 603) were designated UPRmt-HIGH and the bottom two thirds of patients (66.6%, n = 1206) were designated UPRmt-LOW. The designations were used as classifiers for Comparative Marker Selection available through GenePattern(Reich et al., 2006). All genes with a false discovery rate (FDR) less than 0.05 were ranked by Log2 Fold Change and this was used for pre-ranked gene set enrichment analysis (GSEA) available through GenePattern (Mootha et al., 2003; Reich et al., 2006; Subramanian et al., 2005). Enrichment scores were determined as a running sum statistic at maximum deviation from zero.
QUANTIFICATION AND STATISTICAL ANLYSIS
Statistical tests used are described in figure legends with additional detail. Statistical significance was defined as a p value below 0.05. ns = not significant, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. Data displayed as mean ± standard error of the mean (SEM) or mean ± standard deviation (SD) and noted in figure legends. Statistical analyses were performed using GraphPad Prism Software or R.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-SIRT3 (C73E3) | Cell Signaling Technology | Cat#2627; RRID:AB_2188622 |
| Rabbit polyclonal anti-SIRT3 | Millipore | Cat#07-1596; RRID:AB_1977497 |
| Mouse monoclonal anti-HSP60 | BD Biosciences | Cat#611563; RRID:AB_399009 |
| Rabbit monoclonal anti-ATF5 [EPR18286] | Abcam | Cat#ab184923; RRID:AB_2800462 |
| Mouse monoclonal anti-NRF1 | Abcam | Cat#ab55744; RRID:AB_2154534 |
| Rabbit monoclonal anti-CLPP [EPR7133] | Abcam | Cat#ab124822; RRID:AB_10975619 |
| Rabbit polyclonal anti-SOD-1 (FL-154) | Santa Cruz Biotechnology | Cat#sc-11407; RRID:AB_2193779 |
| Rabbit monoclonal anti-FOXO3a (75D8) | Cell Signaling Technology | Cat#2497; RRID:AB_836876 |
| Mouse monoclonal anti-Actin, clone C4 | Millipore | Cat#MAB1501; RRID:AB_2223041 |
| Rabbit polyclonal anti-Mn-SOD | Millipore | Cat#06-984; RRID:AB_310325 |
| Rabbit polyclonal anti-LC3 | MBL International | Cat#PM036; RRID:AB_2274121 |
| Mouse monoclonal anti-SDHA [2E3GC12FB2AE2] | Abcam | Cat#ab14715; RRID:AB_301433 |
| Rabbit polyclonal anti-MTCO1 | Abcam | Cat#ab45918; RRID:AB_944283 |
| Mouse monoclonal anti-GFP (B-2) | Santa Cruz Biotechnology | Cat#sc-9996; RRID:AB_627695 |
| Rabbit monoclonal anti-NRF2 (D1Z9C) | Cell Signaling Technology | Cat#12721; RRID:AB_2715528 |
| Mouse monoclonal anti-ErbB2 (3B5) | Abcam | Cat#ab16901; RRID:AB_443537 |
| Rabbit polyclonal anti-SIRT3 | Abcam | Cat#ab86671; RRID:AB_10861832 |
| Rabbit monoclonal anti-FOXO3a (D19A7) | Cell Signaling Technology | Cat#12829; RRID:AB_2636990 |
| HRP Goat anti-Mouse IgG | Jackson ImmunoResearch Labs | Cat#115-035-003; RRID:AB_10015289 |
| HRP Goat anti-Rabbit IgG | Thermo Fisher Scientific | Cat#65-6120; RRID:AB_2533967 |
| HRP Digital anti-Mouse IgG | Kindle Biosciences | Cat#R1005; RRID:AB_2800463 |
| HRP Digital anti-Rabbit IgG | Kindle Biosciences | Cat#R1006; RRID:AB_2800464 |
| Biotin Goat anti-Mouse IgG | Jackson ImmunoResearch Labs | Cat#115-065-205; RRID:AB_2338571 |
| Biotin Donkey anti-Rabbit IgG | Jackson ImmunoResearch Labs | Cat#711-065-152; RRID:AB_2340593 |
| Alexa Flour 488 Goat anti-Mouse IgG | Thermo Fisher Scientific | Cat#A-11001; RRID:AB_2534069 |
| Bacterial and Virus Strains | ||
| NEB Stable Competent E. coli (High Efficiency) | New England BioLabs | Cat#3040 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Antibody Diluent, Normal | MP Biomedicals | Cat#08980641; RRID:AB_2335238 |
| Streptavidin, Horseradish Peroxidase concentrate for IHC | Vector Laboratories | Cat#SA-5004; RRID:AB_2336509 |
| Lipofectamine 2000 Transfection Reagent | Thermo Fisher Scientific | Cat#11668019 |
| Lipofectamine RNAiMAX Transfection Reagent | Thermo Fisher Scientific | Cat#13778100 |
| Collagen I, Rat Tail | Corning | Cat#354236 |
| Polybrene | Santa Cruz Biotechnology | Cat#sc-134220 |
| Matrigel Growth Factor Reducted (GFR) Basement Membrane Matrix | Corning | Cat#354230 |
| MitoSOX Red Mitochondrail Superoxide Indicator | Thermo Fisher Scientific | Cat#M36008 |
| gamitrinib-triphenylphosphonium (G-TPP) | Kang et al., 2009 | N/A |
| Doxycycline hyclate | Sigma-Aldrich | Cat#D9891 |
| Amoxicillin trihydrate | Sigma-Aldrich | Cat#PHR1127 |
| D-Luciferin | Sigma-Aldrich | Cat#L9504 |
| Collagenase | Sigma-Aldrich | Cat#C9891 |
| Red Blood Cell Lysing Buffer Hybri-Max | Sigma-Aldrich | Cat#R7757 |
| Critical Commercial Assays | ||
| AEC Peroxidase Substrate Kit | Vector Laboratories | Cat#SK-4200; RRID:AB_2336076 |
| Plasmid Maxi Kit | QIAGEN | Cat#12165 |
| Cell Culture Insert, Transparent PET Membrane, 8 μm pore size | Corning | Cat#353097; Cat#353093 |
| Hema 3 Manual Staining System and Stat Pack | Fisher Scientific | Cat#23-123869 |
| Deposited Data | ||
| Primary breast cancer gene expression data | Gene Expression Omnibus | GEO: GSE11121, GSE12093, GSE12276, GSE1456, GSE16391, GSE2034, GSE2990, GSE3494, GSE5327, GSE6532, GSE7390, and GSE9195 |
| TCGA Breast Cancer Dataset | National Cancer Institute GDC Data Portal | TGCA-BRCA, DbGAP Study Accession #phs000178 |
| Experimental Models: Cell Lines | ||
| MCF7 | ATCC | Cat#HTB-22 |
| MDA-MB-231 | ATCC | Cat#HTB-26 |
| Lenti-X 293T | Takara Clontech | Cat#632180 |
| MMTV-rtTA/TetO-NeuNT mammary tumor cell lines | This Paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: MMTV-rtTA/TetO-NeuNT: FVB Tg(MMTV-rtTA) 1Lach Tg(TetO-Erbb2)1Lach | Moody et al., 2002 | Mouse Genome Informatics: 5506798 |
| Mouse: FVBN: FVB/NCrl | Charles River | Strain Code: 207 |
| Mouse: C57BL/6: C57BL/6NCrl | Charles River | Strain Code: 027 |
| Mouse: Nude: Hsd:Athymic Nude-Foxn1nu | Envigo | Model Code: 069 |
| Oligonucleotides | ||
| rtTA Primer Forward 5′-TGCCGCCATTATTACGACAAGC-3′ | This Paper | N/A |
| rtTA Primer Reverse 5′-ACCGTACTCGTCAATTCCAAGGG-3′ | This Paper | N/A |
| Neu Primer Forward 5′-TTTCCTGCAGCAGCCTACGC-3′ | This Paper | N/A |
| Neu Primer Reverse 5′-CGGAACCCACATCAGGCC-3′ | This Paper | N/A |
| siSIRT3 #1 Forward 5’-GCCCAACGUCACUCACUACTT –3′ | This Paper | N/A |
| siSIRT3 #1 Reverse 5′-GUAGUGAGUGACGUUGGGCTT-3′ | This Paper | N/A |
| siSIRT3 #2 Forward 5’-ACUCCCAUUCUUCUUUCACTT-3′ | This Paper | N/A |
| siSIRT3 #2 Reverse 5′- GUGAAAGAAGAAUGGGAGUTT-3′ | This Paper | N/A |
| DsiNC1 | Integrated DNA Technologies | Cat#51-01-14-03 |
| Recombinant DNA | ||
| shRNA vector set against SIRT3 in pLV-H1-CMV-Green plasmid | Biosettia | Cat#SORT-B01, Gene ID:23410; Accession NM_001017524.2 |
| Software and Algorithms | ||
| KM Plotter | Györffy et al., 2010; Lánczky et al., 2016 | N/A |
| JetSet | Li et al., 2011 | N/A |
| GraphPad Prism v6.0c | https://www.graphpad.com/ | N/A |
| FIJI | https://fiji.sc/ | N/A |
| FCS Express Flow 6 | http://www.denovosoftware.com/site/downloadresearch.shtml | N/A |
| GenePattern | Reich et al., 2006 | N/A |
Highlights.
Evidence of endogenous mitohormesis in breast cancer cell lines and primary tumors
Mitohormetic priming of tumor cells leads to increased basal activation of the UPRmt
Mitohormesis and UPRmt activation makes cancer cells more invasive and metastatic
UPRmt enriched breast cancer patients have worse clinical outcomes
ACKNOWLEDGMENTS
Some figures use modified illustrations from Servier Medical Art licensed under a Creative Commons Attribution 3.0 Unported License. T.C.K. and A.J.C. received support from NIH T32 CA078207 Training Program in Cancer Biology. T.C.K. is currently funded by NIH F31 FCA228259A. The Tisch Cancer Institute NIH P30 CA196521 resources grant to the Flow Cytometry and Microscopy CoRE Facilities supported this work. Microscopy was performed at the Microscopy CoRE Facility with funding from NIH Shared Instrumentation Grant 1S10RR026639. A.V. is supported by the US Department of Defense grant CA150272P3. This work was funded by NIH RO1 CA172046 to D.G.
Footnotes
DECLARATION OF INTERESTS
A.V. has received consulting fees from Guidepoint and Fujifilm; advisory board fees from Exact Sciences, Nucleix, and NGM; and lecture fees from Exelixis.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.04.095.
REFERENCES
- Alhazzazi TY, Kamarajan P, Joo N, Huang J-Y, Verdin E, D’Silva NJ, and Kapila YL (2011). Sirtuin-3 (SIRT3), a novel potential therapeutic target for oral cancer. Cancer 117, 1670–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison SJ, and Milner J (2007). SIRT3 is pro-apoptotic and participates in distinct basal apoptotic pathways. Cell Cycle 6, 2669–2677. [DOI] [PubMed] [Google Scholar]
- Benedetti C, Haynes CM, Yang Y, Harding HP, and Ron D (2006). Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics 174, 229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjelakovic G, and Gluud C (2007). Surviving antioxidant supplements. J. Natl. Cancer Inst 99, 742–743. [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Chatenay-Lapointe M, Pan Y, and Shadel GS (2007). Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 5, 265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelle JK, Bell EL, Quesada NM, Vercauteren K, Tiranti V, Zeviani M, Scarpulla RC, and Chandel NS (2005). Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab. 1, 409–414. [DOI] [PubMed] [Google Scholar]
- Cardamone MD, Tanasa B, Cederquist CT, Huang J, Mahdaviani K, Li W, Rosenfeld MG, Liesa M, and Perissi V (2018). Mitochondrial Retrograde Signaling in Mammals Is Mediated by the Transcriptional Cofactor GPS2 via Direct Mitochondria-to-Nucleus Translocation. Mol. Cell 69, 757–772.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel NS, Maltepe E, Goldwasser E, Mathieu CE, Simon MC, and Schumacker PT (1998). Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc. Natl. Acad. Sci. USA 95, 11715–11720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel NS, McClintock DS, Feliciano CE, Wood TM, Melendez JA, Rodriguez AM, and Schumacker PT (2000). Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. J. Biol. Chem 275, 25130–25138. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Ren X, Gowda AS, Shan Y, Zhang L, Yuan YS, Patel R, Wu H, Huber-Keener K, Yang JW, et al. (2013). Interaction of Sirt3 with OGG1 contributes to repair of mitochondrial DNA and protects from apoptotic cell death under oxidative stress. Cell Death Dis. 4, e731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Koh E, Lee YS, Lee HW, Kang HG, Yoon YE, Han WK, Choi KH, and Kim KS (2016). Mitochondrial Sirt3 supports cell proliferation by regulating glutamine-dependent oxidation in renal cell carcinoma. Biochem. Biophys. Res. Commun 474, 547–553. [DOI] [PubMed] [Google Scholar]
- Cox CS, McKay SE, Holmbeck MA, Christian BE, Scortea AC, Tsay AJ, Newman LE, and Shadel GS (2018). Mitohormesis in Mice via Sustained Basal Activation of Mitochondrial and Antioxidant Signaling. Cell Metab. 28, 776–786.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Qin L, Wu J, Qu X, Hou C, Sun W, Li S, Vaughan ATM, Li JJ, and Liu J (2015). SIRT3 Enhances Glycolysis and Proliferation in SIRT3-Expressing Gastric Cancer Cells. PLoS ONE 10, e0129834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Autréaux B, and Toledano MB (2007). ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol 8, 813–824. [DOI] [PubMed] [Google Scholar]
- DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, et al. (2011). Oncogeneinduced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 475, 106–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desouki MM, Doubinskaia I, Gius D, and Abdulkadir SA (2014). Decreased mitochondrial SIRT3 expression is a potential molecular biomarker associated with poor outcome in breast cancer. Hum. Pathol 45, 1071–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Hsu A-L, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, and Kenyon C (2002). Rates of behavior and aging specified by mitochondrial function during development. Science 298, 2398–2401. [DOI] [PubMed] [Google Scholar]
- Dong XC, Jing LM, Wang WX, and Gao YX (2016). Down-regulation of SIRT3 promotes ovarian carcinoma metastasis. Biochem. Biophys. Res. Commun 475, 245–250. [DOI] [PubMed] [Google Scholar]
- Durieux J, Wolff S, and Dillin A (2011). The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell 144, 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bussière F, and Hekimi S (2001). Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev. Cell 1, 633–644. [DOI] [PubMed] [Google Scholar]
- Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, et al. (2011). SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell 19, 416–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorese CJ, Schulz AM, Lin Y-F, Rosin N, Pellegrino MW, and Haynes CM (2016). The Transcription Factor ATF5 Mediates a Mammalian Mitochondrial UPR. Curr. Biol 26, 2037–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J, Nihal M, Singh CK, Zhong W, Liu X, and Ahmad N (2016). Pro-Proliferative Function of Mitochondrial Sirtuin Deacetylase SIRT3 in Human Melanoma. J. Invest. Dermatol 136, 809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgoudaki A-M, Prokopec KE, Boura VF, Hellqvist E, Sohn S, Östling J, Dahan R, Harris RA, Rantalainen M, Klevebring D, et al. (2016). Reprogramming Tumor-Associated Macrophages by Antibody Targeting Inhibits Cancer Progression and Metastasis. Cell Rep. 15, 2000–2011. [DOI] [PubMed] [Google Scholar]
- Giralt A, Hondares E, Villena JA, Ribas F, Díaz-Delfín J, Giralt M, Iglesias R, and Villarroya F (2011). Peroxisome proliferator-activated receptor-γ coactivator-1α controls transcription of the Sirt3 gene, an essential component of the thermogenic brown adipocyte phenotype. J. Biol. Chem 286, 16958–16966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitschlag BL, Kirby CS, Samuels DC, Gangula RD, Mallal SA, and Patel MR (2016). Homeostatic Responses Regulate Selfish Mitochondrial Genome Dynamics in C. elegans. Cell Metab. 24, 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez Herrera KN, Zaganjor E, Ishikawa Y, Spinelli JB, Yoon H, Lin J-R, Satterstrom FK, Ringel A, Mulei S, Souza A, et al. (2018). Small-Molecule Screen Identifies De Novo Nucleotide Synthesis as a Vulnerability of Cells Lacking SIRT3. Cell Rep. 22, 1945–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, and Szallasi Z (2010). An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat 123, 725–731. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Deng CX, Finley LWS, Kim HS, and Gius D (2012). SIRT3 is a mitochondrial tumor suppressor: a scientific tale that connects aberrant cellular ROS, the Warburg effect, and carcinogenesis. Cancer Res. 72, 2468–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, and McMahon M (2006). The double-edged sword of Nrf2: subversion of redox homeostasis during the evolution of cancer. Mol. Cell 21, 732–734. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Petrova K, Benedetti C, Yang Y, and Ron D (2007). ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev. Cell 13, 467–480. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Yang Y, Blais SP, Neubert TA, and Ron D (2010). The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol. Cell 37, 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Hwang SS, Liesa M, Gan B, Sahin E, Jaskelioff M, Ding Z, Ying H, Boutin AT, Zhang H, et al. (2012). Antitelomerase therapy provokes ALT and mitochondrial adaptive mechanisms in cancer. Cell 148, 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang BH, Plescia J, Song HY, Meli M, Colombo G, Beebe K, Scroggins B, Neckers L, and Altieri DC (2009). Combinatorial drug design targeting multiple cancer signaling networks controlled by mitochondrial Hsp90. J. Clin. Invest 119, 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny TC, and Germain D (2017a). mtDNA, Metastasis, and the Mitochondrial Unfolded Protein Response (UPRmt). Front. Cell Dev. Biol 5, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny TC, and Germain D (2017b). From discovery of the CHOP axis and targeting ClpP to the identification of additional axes of the UPRmt driven by the estrogen receptor and SIRT3. J. Bioenerg. Biomembr 49, 297–305. [DOI] [PubMed] [Google Scholar]
- Kenny TC, Hart P, Ragazzi M, Sersinghe M, Chipuk J, Sagar MAK, Eliceiri KW, LaFramboise T, Grandhi S, Santos J, et al. (2017a). Selected mitochondrial DNA landscapes activate the SIRT3 axis of the UPRmt to promote metastasis. Oncogene 36, 4393–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny TC, Manfredi G, and Germain D (2017b). The Mitochondrial Unfolded Protein Response as a Non-Oncogene Addiction to Support Adap-tation to Stress during Transformation in Cancer and Beyond. Front. Oncol 7, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan NA, Nikkanen J, Yatsuga S, Jackson C, Wang L, Pradhan S, Kivelä R, Pessia A, Velagapudi V, and Suomalainen A (2017). mTORC1 Regulates Mitochondrial Integrated Stress Response and Mitochondrial Myopathy Progression. Cell Metab. 26, 419–428.e5. [DOI] [PubMed] [Google Scholar]
- Kim H-S, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, et al. (2010). SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 17, 41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Wang R, Xue Y, Liu X, Zhang H, Chen Y, Fang F, and Chang Y (2010). Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS ONE 5, e11707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lánczky A, Nagy Á, Bottai G, Munkácsy G, Szabó A, Santarpia L, and Győrffy B (2016). miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res. Treat 160, 439–446. [DOI] [PubMed] [Google Scholar]
- Le Gal K, Ibrahim MX, Wiel C, Sayin VI, Akula MK, Karlsson C, Dalin MG, Akyürek LM, Lindahl P, Nilsson J, and Bergo MO (2015). Antioxidants can increase melanoma metastasis in mice. Sci. Transl. Med 7, 308re8. [DOI] [PubMed] [Google Scholar]
- LeBleu VS, O’Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC, de Carvalho FM, Damascena A, Domingos Chinen LT, Rocha RM, et al. (2014). PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat. Cell Biol 16, 992–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-J, Hwang AB, and Kenyon C (2010). Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr. Biol 20, 2131–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, van de Ven RAH, Zaganjor E, Ng MR, Barakat A, Demmers JJPG, Finley LWS, Gonzalez Herrera KN, Hung YP, Harris IS, et al. (2018). Inhibition of epithelial cell migration and Src/FAK signaling by SIRT3. Proc. Natl. Acad. Sci. USA 115, 7057–7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Banck M, Mujtaba S, Zhou M-M, Sugrue MM, and Walsh MJ (2010). p53-induced growth arrest is regulated by the mitochondrial SirT3 deacetylase. PLoS ONE 5, e10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Birkbak NJ, Gyorffy B, Szallasi Z, and Eklund AC (2011). Jetset: selecting the optimal microarray probe set to represent a gene. BMC Bioinformatics 12, 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-F, Schulz AM, Pellegrino MW, Lu Y, Shaham S, and Haynes CM (2016). Maintenance and propagation of a deleterious mitochondrial genome by the mitochondrial unfolded protein response. Nature 533, 416–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisanti S, Garlick DS, Bryant KG, Tavecchio M, Mills GB, Lu Y, Kossenkov AV, Showe LC, Languino LR, and Altieri DC (2016). Transgenic Expression of the Mitochondrial Chaperone TNFR-associated Protein 1 (TRAP1) Accelerates Prostate Cancer Development. J. Biol. Chem 291, 25247–25254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB, Alt FW, Cheng H-L, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, et al. (2007). Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol. Cell. Biol 27, 8807–8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Niu R, Huang T, Shao L-W, Peng Y, Ding W, Wang Y, Jia G, He C, Li C-Y, et al. (2019). N6-methyldeoxyadenine is a transgenerational epigenetic signal for mitochondrial stress adaptation. Nat. Cell Biol 21, 319–327. [DOI] [PubMed] [Google Scholar]
- Mattson MP (2008). Hormesis defined. Ageing Res. Rev 7, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkwirth C, Jovaisaite V, Durieux J, Matilainen O, Jordan SD, Quiros PM, Steffen KK, Williams EG, Mouchiroud L, Tronnes SU, et al. (2016). Two Conserved Histone Demethylases Regulate Mitochondrial Stress-Induced Longevity. Cell 165, 1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody SE, Sarkisian CJ, Hahn KT, Gunther EJ, Pickup S, Dugan KD, Innocent N, Cardiff RD, Schnall MD, and Chodosh LA (2002). Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell 2, 451–461. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. (2003). PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet 34, 267–273. [DOI] [PubMed] [Google Scholar]
- Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Cantó C, Mottis A, Jo Y-S, Viswanathan M, Schoonjans K, et al. (2013). The NAD(+)/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell 154, 430–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moullan N, Mouchiroud L, Wang X, Ryu D, Williams EG, Mottis A, Jovaisaite V, Frochaux MV, Quiros PM, Deplancke B, et al. (2015). Tetracyclines Disturb Mitochondrial Function across Eukaryotic Models: A Call for Caution in Biomedical Research. Cell Rep. 10, 1681–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch C, and Harper JW (2016). Mitochondrial unfolded protein response controls matrix pre-RNA processing and translation. Nature 534, 710–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, and Haynes CM (2012). Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 337, 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Fiorese CJ, Pellegrino MW, Deng P, and Haynes CM (2015). Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPR(mt). Mol. Cell 58, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmeda D, Cerezo-Wallis D, Riveiro-Falkenbach E, Pennacchi PC, Contreras-Alcalde M, Ibarz N, Cifdaloz M, Catena X, Calvo TG, Cañón E, et al. (2017). Whole-body imaging of lymphovascular niches identifies pre-metastatic roles of midkine. Nature 546, 676–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E, Song W, and Perrimon N (2013). Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell 155, 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, Kang M-I, Kobayashi A, Yokoyama S, and Yamamoto M (2006). Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell 21, 689–700. [DOI] [PubMed] [Google Scholar]
- Papa L, and Germain D (2014). SirT3 regulates the mitochondrial unfolded protein response. Mol. Cell. Biol 34, 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova NN, and Thompson CB (2016). The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 23, 27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino MW, Nargund AM, Kirienko NV, Gillis R, Fiorese CJ, and Haynes CM (2014). Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature 516, 414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirós PM, Prado MA, Zamboni N, D’Amico D, Williams RW, Finley D, Gygi SP, and Auwerx J (2017). Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol 216, 2027–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauthan M, Ranji P, Aguilera Pradenas N, Pitot C, and Pilon M (2013). The mitochondrial unfolded protein response activator ATFS-1 protects cells from inhibition of the mevalonate pathway. Proc. Natl. Acad. Sci. USA 110, 5981–5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, and Mesirov JP (2006). GenePattern 2.0. Nat. Genet 38, 500–501. [DOI] [PubMed] [Google Scholar]
- Riar AK, Burstein SR, Palomo GM, Arreguin A, Manfredi G, and Germain D (2017). Sex specific activation of the ERα axis of the mitochondrial UPR (UPRmt) in the G93A-SOD1 mouse model of familial ALS. Hum. Mol. Genet 26, 1318–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow M, and Zarse K (2010). How increased oxidative stress promotes longevity and metabolic health: The concept of mitochondrial hormesis (mitohormesis). Exp. Gerontol 45, 410–418. [DOI] [PubMed] [Google Scholar]
- Ruan L, Zhou C, Jin E, Kucharavy A, Zhang Y, Wen Z, Florens L, and Li R (2017). Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature 543, 443–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterstrom FK, Swindell WR, Laurent G, Vyas S, Bulyk ML, and Haigis MC (2015). Nuclear respiratory factor 2 induces SIRT3 expression. Aging Cell 14, 818–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayin VI, Ibrahim MX, Larsson E, Nilsson JA, Lindahl P, and Bergo MO (2014). Antioxidants accelerate lung cancer progression in mice. Sci. Transl. Med 6, 221ra15. [DOI] [PubMed] [Google Scholar]
- Schieber M, and Chandel NS (2014). ROS function in redox signaling and oxidative stress. Curr. Biol 24, R453–R462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, and Ristow M (2007). Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 6, 280–293. [DOI] [PubMed] [Google Scholar]
- Shi T, Wang F, Stieren E, and Tong Q (2005). SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J. Biol. Chem 280, 13560–13567. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, Coleman MC, Pennington JD, Ozden O, Park S-H, Jiang H, Kim H-S, Flynn CR, Hill S, Hayes McDonald W, et al. (2010). Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol. Cell 40, 893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Garcia G, Bian Q, Steffen KK, Joe L, Wolff S, Meyer BJ, and Dillin A (2016). Mitochondrial Stress Induces Chromatin Reorganization to Promote Longevity and UPR(mt). Cell 165, 1197–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang CK, Chen M, Cheng X, Qi Y, Chen Y, Das I, Li X, Vallat B, Fu L-W, Qian C-N, et al. (2018). SOD1 Phosphorylation by mTORC1 Couples Nutrient Sensing and Redox Regulation. Mol. Cell 70, 502–515.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk JM, and Hekimi S (2009). Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. PLoS Genet. 5,e1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, and Thompson CB (2009). Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324,1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sánchez N, Marchesini M, Carugo A, Green T, Seth S, Giuliani V, et al. (2014). Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 514, 628–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas S, Zaganjor E, and Haigis MC (2016). Mitochondria and Cancer. Cell 166, 555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC (2012). Mitochondria and cancer. Nat. Rev. Cancer 12,685–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J-X, Yi Y, Li Y-W, Cai X-Y, He H-W, Ni X-C, Zhou J, Cheng Y-F, Jin J-J, Fan J, and Qiu SJ (2014). Down-regulation of sirtuin 3 is associated with poor prognosis in hepatocellular carcinoma after resection. BMC Cancer 14, 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Li L, Li CY, Pei Z, Zhou M, and Li N (2015). SIRT3 protects cells from hypoxia via PGC-1α- and MnSOD-dependent pathways. Neuroscience 286, 109–121. [DOI] [PubMed] [Google Scholar]
- Wei L, Zhou Y, Dai Q, Qiao C, Zhao L, Hui H, Lu N, and Guo Q-L (2013). Oroxylin A induces dissociation of hexokinase II from the mitochondria and inhibits glycolysis by SIRT3-mediated deacetylation of cyclophilin D in breast carcinoma. Cell Death Dis. 4, e601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, and Shadel GS (2017). Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat. Rev. Immunol 17, 363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, et al. (2015). Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520, 553–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel L, Topf U, Bragoszewski P, Wiese S, Sztolsztener ME, Oeljeklaus S, Varabyova A, Lirski M, Chroscicki P, Mroczek S, et al. (2015). Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature 524, 485–488. [DOI] [PubMed] [Google Scholar]
- Yang W, and Hekimi S (2010). A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS Biol. 8, e1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Denu RA, Krautkramer KA, Grindle KM, Yang DT, Asimakopoulos F, Hematti P, and Denu JM (2016). Loss of SIRT3 provides growth advantage for B cell malignancies. J. Biol. Chem 291, 3268–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J, and Finkel T (2014). Mitohormesis. Cell Metab. 19, 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YY, and Zhou LM (2012). Sirt3 inhibits hepatocellular carcinoma cell growth through reducing Mdm2-mediated p53 degradation. Biochem. Biophys. Res. Commun 423, 26–31. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, and Hoogenraad NJ (2002). A mitochondrial specific stress response in mammalian cells. EMBO J. 21, 4411–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.