Abstract
A developmental increase in density of delayed rectifier potassium current (IKv) in embryonicXenopus spinal neurons shortens action potential durations and limits calcium influx governing neuronal differentiation. Although previous work demonstrates that maturation ofIKv depends on general mRNA synthesis, it is not known whether increases in K+ channel gene transcripts direct maturation of the current. Accordingly, the developmental appearance of specific Kv potassium channel genes was determined using single-cell reverse transcription-PCR techniques after whole-cell recording of IKv during the period of its development. Detection of a coexpressed housekeeping gene along with the potassium channel gene controlled for successful aspiration of cellular mRNA and allowed scoring of cells in which Kv gene transcripts were not detected. Diverse types of Xenopusspinal neurons exhibit homogenous development ofIKv both in vivo and in culture. In contrast, transcripts of two genes encoding delayed rectifier current, Kv1.1 (Shaker) and Kv2.2 (Shab), are expressed heterogeneously during the period in which the current develops. Kv1.1 mRNA achieves maximal appearance in ∼30% of cells, while IKv is immature; Kv2.2 mRNA appears later in ∼60% of mature neurons. Kv1.1 and 2.2 are thus candidates for generation of IKv, and spinal neurons are a heterogeneous population with respect to potassium channel gene expression. Moreover, correlation of gene expression with current properties shows that neurons lacking Kv2.2 have a characteristic voltage dependence of activation ofIKv.
Keywords: potassium current, RT-PCR, single cell gene expression, Kv genes, spinal neurons, Xenopus embryo
Interest in molecular mechanisms of development of delayed rectifier potassium current (IKv) in embryonic spinal neurons arises from the essential role that it plays in neuronal development. Differentiation ofIKv is the major determinant in the conversion of prolonged calcium-dependent action potentials to short sodium-dependent impulses, as increases in current density and kinetics of activation decrease their duration (Barish, 1986; O’Dowd et al., 1988; Lockery and Spitzer, 1992). Calcium influx is diminished (Gu et al., 1994), probably terminating a period of calcium-dependent differentiation (Gu and Spitzer, 1995). Development ofIKv occurs during a period of 1 d in vivo (Desarmenien et al., 1993) and during the same periodin vitro (Barish, 1986; O’Dowd et al., 1988).
A developmental increase in the number of functionally available potassium channels, rather than an increase in single-channel conductance, accounts for increased current density (Harris et al., 1988). An increase in the number of functional channels could occur in several ways, including activation of preexisting channels or new channel synthesis initiated at the transcriptional level. The increase in density of IKv depends on new RNA synthesis during the period in which current increases (Ribera and Spitzer, 1989), consistent with the possibility that elevated levels of potassium channel gene transcripts initiate the increase. To examine this hypothesis, we identified potassium channel genes expressed during the period of current development.
We investigated expression of Xenopus Shaker Kv1.1 andShab Kv2.2 (Ribera and Nguyen, 1993; Burger and Ribera, 1996), because they encode delayed rectifier potassium currents when expressed in Xenopus oocytes. Moreover, in situhybridization studies localize their transcripts to the developing spinal cord. If potassium channel mRNAs initiate development ofIKv, the frequency of expression of Kv1.1 and Kv2.2 genes in spinal neurons should increase during current development.
Single-cell resolution of gene expression is critical because the neurons constitute a mixed population, although they are homogeneous with respect to development of IKv (O’Dowd et al., 1988; Desarmenien and Spitzer, 1991). The single-cell reverse transcription-PCR (RT-PCR) method permits correlation of electrophysiological and molecular profiles of individual neurons (Lambolez et al., 1992; Sucher and Deitcher, 1995). Codetection of a ubiquitously expressed gene (EF-1α; Krieg et al., 1989) controls for successful aspiration of mRNA, allowing scoring of cells in which Kv gene transcripts are not detected. Potassium channel gene expression was examined during the period of differentiation ofIKv and correlated with biophysical properties of mature current.
Kv2.2 transcripts are present in ∼60% of mature neurons, whereas Kv1.1 transcripts are expressed in only ∼30%. Thus, spinal neurons are a heterogeneous population with respect to potassium channel gene expression, although they exhibit uniform development ofIKv. Furthermore, neurons that lack Kv2.2 transcripts are distinguished by currents that have a more positive voltage-dependence of activation. Moreover, the frequency of appearance of these transcripts is developmentally regulated. These results identify potassium channel genes whose expression at early stages of differentiation may contribute to functional maturation ofIKv.
MATERIALS AND METHODS
Cell culture. Cultures were prepared from the neural plate of Xenopus embryos at stage 15, as described previously (Spitzer and Lamborghini, 1976; Blair, 1983). Cells from a single embryo were plated onto 35 mm tissue culture dishes and maintained in fully defined medium (in mm): 116 NaCl, 0.67 KCl, 1.31 MgSO4, 10 CaCl2, and 4.6 Tris buffer, pH 7.8, adjusted with HCl. Recordings were made at 6–7 hr, 8–10 hr (young neurons), or 18–22 hr (mature neurons) in culture. Cultures contained a mixed population of sensory, motor, and interneurons (Spitzer and Lamborghini, 1976; Lamborghini, 1980; Bixby and Spitzer, 1984;Lamborghini and Iles, 1985), as well as morphologically undifferentiated cells. Because the somitic mesoderm was not removed from the neural plate, cultures contained myocytes as well.
Electrophysiological recording and data analysis.Whole-cell delayed rectifier potassium currents were recorded from young and mature neurons, using patch-clamp techniques (Hamill et al., 1981). Neurons were identified morphologically by the presence of neurites with growth cones, but different neuronal types were not distinguishable anatomically. Isolated neurons that did not make contacts with other cells were selected for recording. They typically had soma diameters of 20–30 μm and between one and four neurites, none of which were longer than 60 μm, to assure adequate space clamp. Previous studies demonstrated that such selection of cells with short neurites does not bias the sample toward currents with a particular amplitude or kinetics (O’Dowd et al., 1988; Desarmenien and Spitzer, 1991). Neurons were examined at room temperature using Nomarski optics at 400× magnification. The bath solution contained (in mm): 80 NaCl, 3 KCl, 5 MgCl2, 10 CoCl2, 5 HEPES, pH 7.4, adjusted with NaOH, and 0.25 μg/ml tetrodotoxin (TTX). TTX and CoCl2 were added to block voltage-gated Na+, Ca2+, and Ca2+-activated currents. The recording pipette contained 7 μl of internal recording solution (in mm): 104–105 KCl, 3 MgCl2, and 10 HEPES, pH 7.4, adjusted with KOH.
Potassium currents were recorded using an Axopatch 1D amplifier and the pCLAMP suite of computer programs (Axon Instruments, Burlingame, CA). Whole-cell currents were induced by applying voltage steps to potentials ranging between −30 and +120 mV from a holding potential of −40 mV. Currents were filtered at 10 kHz and digitized at 50 μsec.
Steady-state amplitudes of whole-cell delayed-rectifier potassium currents were averaged over a 3 msec interval after achievement of the steady state. Conductance (G) was calculated from current asG = I/(Vc − IRs − Vr), where I is the steady-state current,Vc is the command potential,Rs is the series resistance, andVr is the calculated reversal potential for potassium current based on composition of extracellular and intracellular recording solutions (−86 mV).Rs was calculated as the ratio of the applied voltage step to the peak capacitative current (Marty and Neher, 1995). Conductance at various membrane potentials was normalized to maximal conductance, Gmax, and plots ofG/Gmax versus membrane potential were fitted with Boltzmann sigmoids to determineV1/2, the voltage of half-maximal activation.
Both current and conductance were expressed per μm2 membrane surface. Surface area was derived from membrane capacitance (1 μF/cm2), which was calculated by dividing the whole-cell capacitative charge by the applied voltage. Capacitative charge was calculated as the integral of the whole-cell capacitative current transient. Individual values of currents, conductances, V1/2, and times to half-maximal activation, t1/2, were presented in scatter plots to emphasize the distribution within neuronal populations. Because it was not apparent whether the distributions of values for current properties were Gaussian, the significance of differences between properties of various neuronal groups was tested by both the two-tailed nonparametric Mann–Whitney test and by the two-tailed Student’s t test. In all cases, the conclusions from both tests were the same. Mean values are presented ±SEM.
Harvest of cell contents. After recording of the current, the intracellular contents of a cell were aspirated by applying negative pressure until the pipette was removed from the bath. Aspiration was confirmed visually by shrinkage of the neuron around the tip of the recording pipette. Cell contents and recording solution contained in the pipette were transferred to individual test tubes containing reaction mixture for subsequent DNase treatment and RT reaction, assuming a transferred volume of 5.5 μl (Lambolez et al., 1992).
Amplification of mRNA from single neurons using RT-PCR. The cell aspirate was treated with DNase (Deoxyribonuclease I, Amplification Grade DNase, RNase free, Gibco, Gaithersburg, MD) to prevent genomic DNA from serving as template in subsequent PCR reactions (Dilworth and McCarrey, 1992). The DNase step is essential, because elimination of DNase digestion and RT steps resulted in amplification of gene fragments from genomic DNA (see Fig.4). RT of mRNA into cDNA was then followed by two PCR amplification steps. DNase treatment and RT were performed in a 10 μl solution containing 90 mm KCl, 3 mmMgCl2, 20 mm Tris-HCl, 5 mm HEPES (acid), 10 mm DTT, 1 mm of each of four deoxynucleotides (dNTP mixture; Boehringer Mannheim, Indianapolis, IN), 2 μm oligo-dT (Gibco), 2 μm potassium channel gene-specific primer (Operon Technologies, Alameda, CA) (Table 1), and 20 U of ribonuclease inhibitor (rRNasin; Promega, Madison, WI). DNA was degraded by addition of 100 U of DNase followed by 75 min at room temperature. DNase was subsequently inactivated by a 10 min incubation at 95°C. Moloney murine leukemia virus RT (100–150 U; Gibco) was then added and incubated for 1 hr at 35°C. Inactivation of the enzyme and denaturation of the RNA-DNA complexes were achieved by incubating at 95°C for 10 min. The RT-reaction mixture was placed immediately on ice.
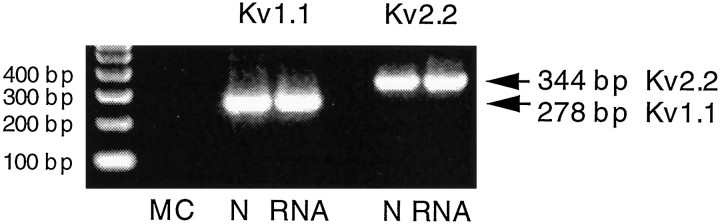
Fig. 4.
Amplification of Kv1.1 and Kv2.2 PCR fragments from genomic DNA of single neurons (N) and contaminant DNA in spinal RNA preparations (RNA). Both DNase and RT treatments were eliminated from the procedure. Fragments are 278 bp and 344 bp, as expected for Kv1.1 and Kv2.2, respectively.
Table 1.
Oligonucleotide primers used for RT-PCR assay
| Gene | Sequence | Nucleotides | Reference |
|---|---|---|---|
| Kv1.1 (nested) | F1 GCCATCGCTGGTGTGCTGACA | 1159-1179 | Ribera and Nguyen (1993) |
| F2 CTAACAGTGATCTGAGTCGACG | 1292-1313 | ||
| R1 CCCTAATTTGTACTAGGCCTC | 1571-1551 | ||
| Kv2.2 (nested) | F1 GGATTTACCCTTAGACGGAGT | 973-993 | Burger and Ribera (1996) |
| R2 GCTTCTCTTCTCTTTATTGCC | 1316-1296 | ||
| R1 CCATTTTATTAGACCCATCTTCTG | 1438-1415 | ||
| EF-1α (nested) | F1 CTTCTCAGACTACCCTCCTCT | 1292-1312 | Krieg et al. (1989) |
| R2 GCCACAAACAAATGTGTCCAC | 1610-1590 | ||
| R1 TGGTCTCAAATTTGGTGACAG | 1693-1673 |
F and R refer to forward and reverse primers. Nucleotide numbers refer to positions within published sequences, and bold numbers border nested fragments that were amplified in the second PCR. The published length of the nested sequence of Kv1.1 is 280 bases; however, sequencing the Kv1.1 PCR product revealed that this sequence was two bases shorter because of variations in the 3′ untranslated region.
PCR products of two genes were amplified for each experiment. Expression of the ubiquitously expressed gene EF-1α (Krieg et al., 1989) was analyzed in 2 μl of RT-reaction mixture to ascertain successful aspiration of cell contents, whereas the remaining 8 μl was used to amplify mRNA of a specific potassium channel gene. Aliquots were brought to 10 μl by addition of pipette recording solution.
The first PCR was performed in a volume of 100 μl of reaction mixture containing 10 mm Tris-HCl, 3 mm MgCl2, 100 mm KCl, 200 μm dNTP, 500 nm of each primer, and 2.5 U Taqpolymerase (Boehringer Mannheim), pH 8.3. For second PCR amplification of a smaller nested fragment, 1 μl of the first reaction mixture was added to a reaction mixture in which either the reverse or the forward primer was substituted by a nested primer with a sequence expected to be present within the sequence of the first PCR product (Table 1). Nested PCR of Kv2.2 was carried out with the same reaction condition as the first PCR. Nested PCR amplification of EF-1α was performed in 50 μl of reaction mixture containing 60 mmTris-HCl, 15 mm(NH4)SO4, 2 mm MgCl2, 250 μm dNTP, pH 9.5 (Buffer J, PCR Optimizer kit; Invitrogen, San Diego, CA), 1 μm of each of the primers, and 1.25 U Taq polymerase (Boehringer Mannheim). Nested amplification of Kv1.1 was accomplished similarly except that the MgCl2 concentration was 1.5 mm and pH 8.5 (Buffer A, PCR Optimizer kit, Invitrogen). Ten microliters of the second PCR mixture were separated on 3% agarose gels and visualized with ethidium bromide. Transcripts of Kv2.2, Kv1.1, and EF-1α were not detected in single mature cells by a single round of PCR using the primer sets of the first or second PCR alone, indicating that the two-step reaction was necessary. For each experiment, positive controls consisted of RNA (Chomczynski and Sacchi, 1987) isolated either from brains (brain RNA) of 1-month-old tadpoles or from the posterior neural region (spinal RNA) of stage 32–34 tadpole embryos. Omission of RT provided a negative control. The average frequency of gene expression was expressed as the mean ± SEM of individual experiments. The significance of differences between various neuronal groups was the same when evaluated by both the two-tailed nonparametric Mann–Whitney test and the two-tailed Student’s t test.
Analysis of RT-PCR products. For restriction digests, the second PCR was repeated to obtain a large quantity of PCR product which was then purified using DNA binding resin (Magic PCR Preps Purification System, Promega, Madison WI). Restriction analysis with individual enzymes was performed using the buffers and protocols provided with them. Selected PCR products were cloned using TA cloning (Invitrogen) and sequenced using Sequenase 2.0 (USB, Cleveland, OH).
Primer design. Table 1 summarizes the primers used. For each gene, F1 and R1 were used for the first PCR and F1 and R2 (nested) or F2 (nested) and R1 were used for the second. A nested primer refers to a primer sequence present within the sequence of the first PCR fragment. Thus, the sequence that was amplified in the second PCR was smaller than that amplified in the first. The most 3′ reverse primer was used to prime RT reactions for Kv genes, whereas oligo-dT was used for EF-1α.
RESULTS
Embryonic Xenopus spinal neurons constitute a heterogeneous population with respect to potassium channel gene expression
The expression of Kv1.1 and Kv2.2 mRNAs was first examined in individual neurons at 1 d in culture, whenIKv has achieved its mature density and kinetics of activation (O’Dowd et al., 1988). After recording of potassium current in the whole-cell configuration, the contents of an individual cell were collected for analysis of gene expression using RT-PCR (Lambolez et al., 1992; Sucher and Deitcher, 1995). Specific RT and PCR primers were designed (Table 1) for detection of the potassium channel mRNA of interest, Kv1.1 or Kv2.2 (Ribera and Nguyen, 1993; Burger and Ribera, 1996), and a control, ubiquitously expressed mRNA species, EF-1α (Krieg et al., 1989). Detection of EF-1α ascertained that the cell contents had been harvested successfully, and only cells positive for EF-1α were included in the results.
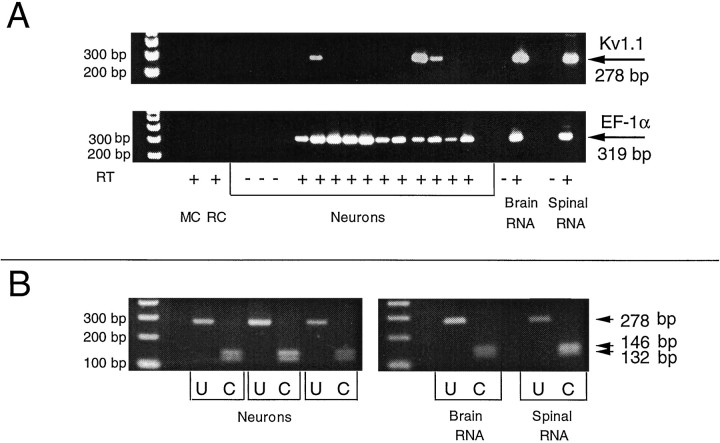
Single-cell RT-PCR analysis of Kv1.1 indicates that 35 ± 7% of mature neurons express this transcript (51 neurons examined in 10 experiments) (Figs. 1A, 3). The identity of PCR products was determined initially by the size of the PCR product and the specificity of the three primer sequences used for the two-step PCR protocol (see Materials and Methods). Restriction analysis provided further confirmation of the identity of the Kv1.1 PCR product (Fig.1B). Digestion with HpaI produced 132 and 146 basepair (bp) fragments as expected. The HpaI site was present in all Kv1.1 products amplified from single spinal neurons or total RNA isolated from either the brains of 1-month-old tadpoles or the spinal cord region of tailbud embryos. Similarly, expected restriction digest patterns were obtained by digestion withStuI (fragment sizes of 258 and 20 bp) and AluI (159 and 119 bp). Samples from three of six cells and from spinal RNA tested for AluI had an additional restriction site. Sequencing of samples from a neuron and from RNA that had only the predicted restriction sites, and from a neuron that had an additionalAluI site, confirmed the identity of PCR products. The first two samples differed from the published sequence by the same single base that did not change the encoded amino acid. In the third sample, the sequence varied by 12 bases, but the predicted additionalAluI site was not found. Six of these bases were in the coding region, although only one created a change in the encoded amino acid.
Fig. 1.
Kv1.1 transcripts expressed in mature neurons.A, In three experiments, 11 neurons were sampled for Kv1.1 and EF-1α mRNA. Although all are positive for EF-1α (319 bp,bottom), only three are positive for Kv1.1 transcripts (278 bp, top). + and − indicate reactions in which reverse transcriptase (RT) was present or absent, respectively. MC, Reaction mixture control; RC, recording solution control. Brain and spinal RNA serve as positive controls. Reactions in which RT was absent constitute negative controls. B, Restriction digestion with HpaI produces the predicted two fragments, 146 bp and 132 bp in length. This site is present in Kv1.1 products amplified from single spinal neurons, from brain, and from spinal RNA. U, Uncut fragments;C, cut fragments.
Fig. 3.
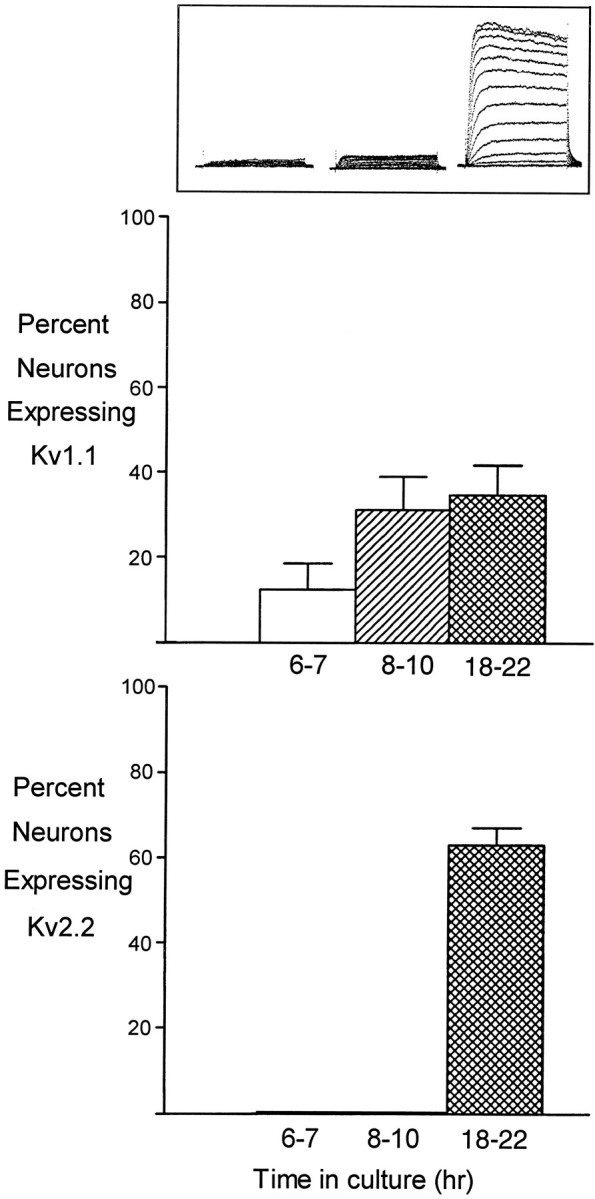
Expression of Kv1.1 precedes that of Kv2.2 during development of IKv. Neurons were examined for either Kv1.1 or Kv2.2 transcripts at times in culture when potassium current (top) is still small (6–7 and 8–10 hr) and when the current is fully mature (18–22 hr). Frequency of transcript expression of both Kv1.1 and Kv2.2 is developmentally regulated. Kv1.1 transcripts are detected in 12 ± 6% of cells by the onset of morphological differentiation, whereas Kv2.2 transcripts are below the level of detection as late as 8–10 hr in culture. For these three time points, the numbers of cells examined (number of experiments) were 29 (5), 29 (4), and 51 (10) for Kv1.1, and 12 (2), 12 (3), and 32 (6) for Kv2.2. Expression of Kv1.1 is significantly different between 6–7 hr and later times (p < 0.05).
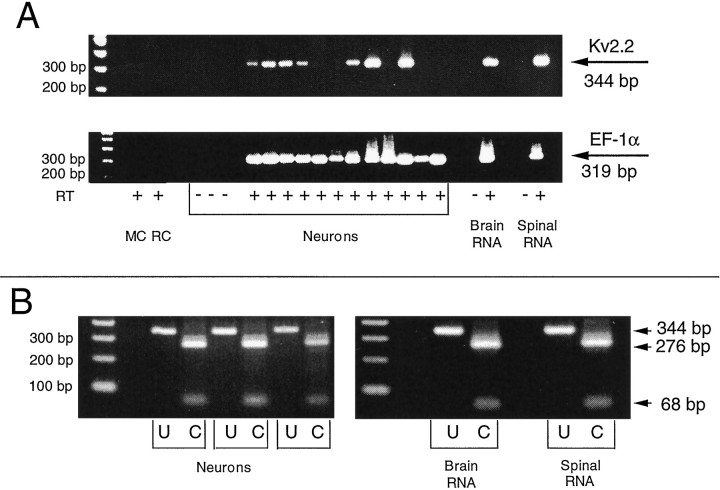
Kv2.2 mRNA was detected in 63 ± 10% of mature neurons (32 neurons examined in six experiments) (Figs.2A, 3), significantly higher than the expression frequency of 35% found for Kv1.1 (p < 0.01). As for Kv1.1, product size, specificity of primers, restriction analysis of PCR products, and sequencing provided verification of their identity. For example, Sau3AI digestion was expected to yield 68 and 276 bp fragments (Fig. 2B). This pattern was observed for Kv2.2 sequences amplified from single spinal neurons, as well as from total RNA isolated from either the brains of 1-month-old tadpoles or the spinal cord region of tailbud embryos. The expected restriction digest patterns were also obtained using Mae I (124 and 220 bp); however, restriction sites for Fok I (107 and 237 bp) and EcoO109 I (220 and 124 bp) were missing in some PCR products. The absence or addition of restriction sites occurred in one case for some neurons from the same embryo and in one case in all neurons from the same embryo. Sequencing of one sample from a neuron that had all the tested restriction sites, and two samples from RNA lacking two sites, confirmed the identity of the PCR products. The first one was identical to the published sequence, whereas the other two varied in the same eight bases. These substitutions were in the third position of amino acid codons, however, and none resulted in alteration of the predicted amino acid sequence. Two base substitutions were at the Fok I and Eco0109 I restriction sites, and thus explain the incomplete digests with these enzymes of the samples derived from RNA.
Fig. 2.
Kv2.2 transcripts expressed in mature neurons.A, In two experiments, 12 neurons were sampled for Kv2.2 and EF-1α mRNA. All 12 are positive for EF-1α (319 bp,bottom), and 7 contain Kv2.2 mRNA (344 bp, top). + and − indicate reactions in which reverse transcriptase (RT) was present or absent, respectively. MC, Reaction mixture control; RC, recording solution control. Brain and spinal RNA serve as positive controls. Reactions in which RT was absent constitute negative controls. B, Restriction digestion with Sau3AI produces the predicted two fragments, 68 bp and 276 bp in length. This site is present in Kv2.2 products amplified from single spinal neurons, from brain, and from spinal RNA.U, Uncut fragments; C, cut fragments.
The identity of EF-1α PCR fragments was ascertained by use of three specific PCR primers, generation of a PCR product of expected size (319 bp), and restriction analysis using restriction enzymesSau3AI, SspI, and HaeIII. Samples were tested from experiments in which Kv1.1 or Kv2.2 was amplified.
The appearance of Kv2.2 and Kv1.1 mRNAs is developmentally regulated
The developmental appearance of potassium channel gene transcripts is hypothesized to initiate potassium channel synthesis and ultimately increase the current. Accordingly, we determined the time of appearance of Kv1.1 and Kv2.2 transcripts during the period in which the current develops. Kv1.1 transcripts were detected in 12 ± 6% of the cells (29 neurons examined in five experiments) as early as neurons could be identified in culture (6–7 hr). By 8–10 hr in culture, one third of the neurons expressed Kv1.1, similar to the frequency of expression in mature neurons (Fig. 3), although the current has not yet achieved its mature amplitude. The expression frequency of Kv1.1 was significantly lower at 6–7 hr than at later developmental times (p < 0.05). In contrast, Kv2.2 transcripts were not detected at either 6–7 or 8–10 hr in culture (12 neurons examined at both times, in two and three experiments), although it was expressed in ∼60% of mature cells (Fig. 3). These results suggest that the Kv2.2 gene does not encode the current in young neurons; its later appearance may drive the synthesis of more channels, resulting in an increased current density as cells mature. These observations demonstrate that expression of both genes is regulated developmentally and that increase in RNA synthesis or stability is a likely first step in generating more channels encoding potassium current. The presence of current in young neurons in which neither Kv1.1 nor Kv2.2 was detected (88% of the neurons at 6–7 hr and 69% of the neurons at 8–10 hr in culture) suggests that expression of other genes accounts forIKv at early times.
It is unlikely that mRNA went undetected early in development. Two PCR steps were sufficient to detect Kv1.1 and Kv2.2 genes from genomic DNA of a single cell, when DNase treatment was eliminated (Fig.4). Bands of the size expected for Kv1.1 and Kv2.2 were amplified from cell aspirates from two of four and from four of four neurons, respectively. To determine whether lack of Kv1.1 amplification in two cells resulted from failure to aspirate the nucleus, 15 neurons were assayed after clearly visible nuclear aspiration; Kv1.1 was detected in all of these cells. When the nucleus was not unequivocally visibly aspirated, Kv1.1 was detected in only five of an additional eight neurons. Because the Xenopus genome is tetraploid (Kobel and DuPasquier, 1986), as few as four copies of these genes were detectable. Thus, if RT generates only a few copies of cDNA, subsequent PCR amplification is expected to ensure their detection. In situ hybridization indicates that Kv1.1 and Kv2.2 transcripts are present in the cell soma (Ribera and Nguyen, 1993; Burger and Ribera, 1996), suggesting that mRNA was not undetected because it was present in neurites and not aspirated.
Lack of detection of Kv2.2 transcripts is correlated with a characteristic property of mature potassium current
The biophysical properties of developingIKv have been investigated previously in several studies, but subpopulations of mature neurons with different current properties were not distinguished (Barish, 1986; O’Dowd et al., 1988; Desarmenien and Spitzer, 1991). We compared the biophysical properties of IKv from Kv1.1 or Kv2.2 mRNA-positive and -negative mature neurons to determine whether molecular heterogeneity is correlated with functional heterogeneity. The range of voltages used to activate the current was expanded relative to previous studies (O’Dowd et al., 1988; Desarmenien and Spitzer, 1991) to obtain maximal conductance values, and the rapidly inactivating A-current was removed by setting the holding potential at −40 mV (Ribera and Spitzer, 1990).
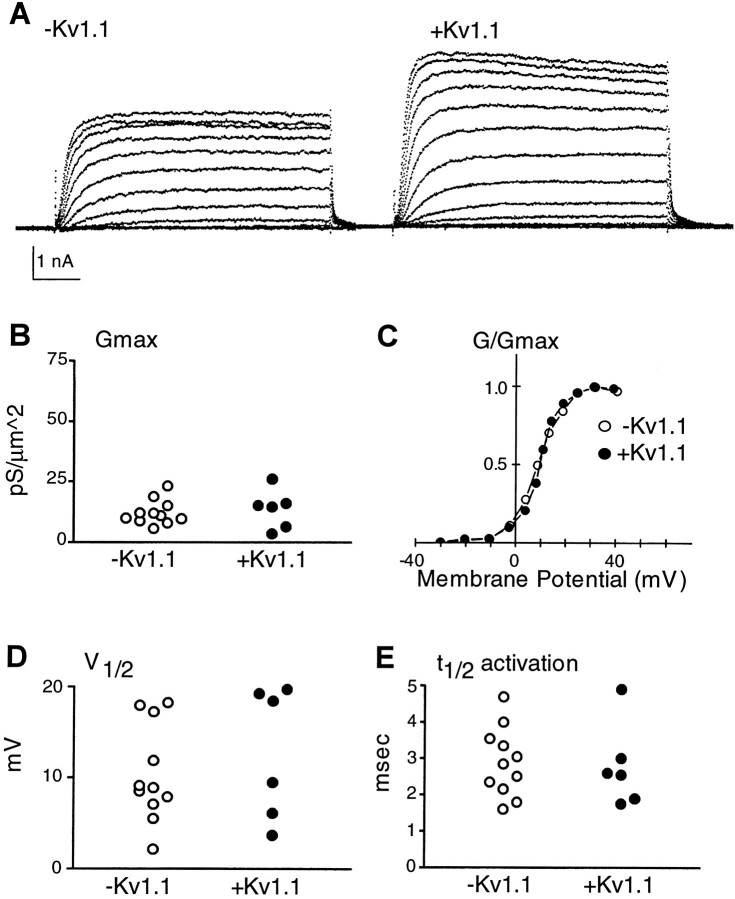
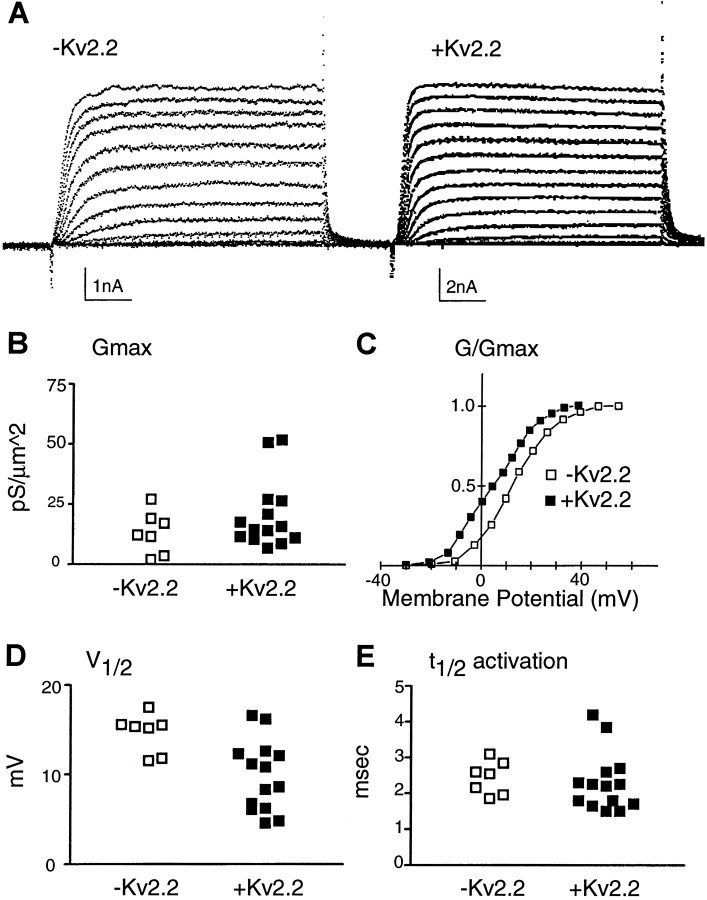
Kv1.1 mRNA-positive mature neurons could not be distinguished from the rest of the neuronal population on the basis of their maximal conductance (Gmax), voltage for half-maximal conductance (V1/2), or time to half-maximal (t1/2) activation. Both the mean values and the distribution for these current characteristics were similar (Fig. 5). Furthermore, no differences in current properties were observed among young neurons that do or do not express Kv1.1. In contrast, current in mature neurons lacking Kv2.2 transcripts was characterized by a significantly more positive meanV1/2 of steady-state activation (p < 0.01) and a tighter distribution than that of current in neurons expressing Kv2.2 transcripts. Maximal conductance (Gmax) and t1/2of activation were similar in Kv2.2-positive and -negative neurons (Fig. 6).
Fig. 5.
Comparison of biophysical and molecular profiles of individual mature neurons. A, Potassium currents from neurons that either do not (left) or do (right) express Kv1.1 mRNA. Calibration: 5 msec. B, Neurons that are + or −Kv1.1 have maximal conductance density (Gmax) within the same range; average values are (in pS/μ2) −Kv1.1, 12.2 ± 1.5, n = 11; +Kv1.1, 13.6 ± 3.2, n = 6. C, Normalized conductances (G/Gmax) plotted versus membrane potential for the currents recorded from two neurons shown in A are not different. D, Neurons that are + or −Kv1.1 exhibit V1/2 within the same range; average values are (in mV): −Kv1.1, 10.4 ± 1.6, n = 11; +Kv1.1, 12.8 ± 2.9, n = 6. E, Neurons that are + or −Kv1.1 have t1/2 within the same range; average values for t1/2 at 30 mV are (in msec): −Kv1.1, 2.9 ± 0.3, n = 11; +Kv1.1, 2.8 ± 0.5, n = 6.
Fig. 6.
Potassium current from mature neurons expressing Kv2.2 transcripts activate at a more negative membrane potential.A, Potassium currents from neurons that either do not (left) or do (right) express Kv2.2 mRNA. Calibration: 5 msec. B, Neurons that are + or −Kv2.2 have maximal conductance density (Gmax) within the same range; average values are (in pS/μ2): −Kv2.2, 13.2 ± 3.3, n = 7; +Kv2.2, 20.5 ± 3.8, n = 14. C, The normalized conductance (G/Gmax) plotted versus membrane potential for the currents recorded from two neurons shown in Aindicates that neurons that lack Kv2.2 have currents that activate at more depolarized potentials. D, MeanV1/2 for neurons that are negative for Kv2.2 (14.6 ± 0.8 mV, n = 7) is significantly higher than that for neurons positive for Kv2.2 (9.8 ± 1.0 mV, n = 14, p < 0.01). E, Neurons that are + or −Kv2.2 have t1/2 within the same range; average values for t1/2 at 30 mV are (in msec): −Kv2.2, 2.4 ± 0.2, n = 7; +Kv2.2, 2.3 ± 0.2, n = 14.
DISCUSSION
Heterogeneous gene expression
Xenopus embryonic spinal neurons are a heterogeneous population with respect to potassium channel gene expression. Xenopus Kv1.1 and Kv2.2 genes are expressed in subpopulations of mature spinal neurons, whenIKv is maximally developed. In retrospect, these molecular phenotypic variations of spinal neurons perhaps should have been expected, because the cultured neuron population consists of sensory, motor, and interneurons. However, neuronal morphology in these cultures is not a predictor of either neuronal type or subtype of gene expressed. Molecular markers for different classes of embryonic neurons will be needed to identify neuronal types in these cultures; Kv genes are candidates for such markers.
Variations in the sequence of PCR-amplified fragments of Kv1.1 and Kv2.2 were revealed by restriction digests and sequencing analysis. It is unlikely that they are attributable to random PCR error, because their presence in coding regions generally did not result in a change of the encoded amino acid and they occurred in the same bases in different samples. Although only cDNA clones for these genes are available, variations in Kv1.1 are unlikely to represent splice variants, because the coding region of mammalian Kv1.1 lies within a single exon (Chandy et al., 1990; Chandy and Gutman, 1994). TheXenopus Kv1.2 gene, for which a genomic clone has been identified, also has its coding region within a single exon (Ribera, 1990). There is no information about possible splice variants ofXenopus Kv2.2. The pseudotetraploidy of theXenopus genome implies that a given gene can have four alleles. It is most likely that allelic differences between cDNA clones and PCR fragments amplified from total RNA account for this variability, which has been observed previously in clones of Kv2.2 and Kv2.1 (Burger and Ribera, 1996).
Heterogeneous current properties
Lack of detection of Kv2.2 transcripts identifies neurons in which the current is activated at a more positive voltage. This observation does not necessarily imply that Kv2.2 encodes a channel that activates at a more negative potential; however, it enables recognition of variations in properties of whole-cell potassium current among these spinal neurons. Although shifts in V1/2 can affect the duration of the action potential (Lockery and Spitzer, 1992), the 5 mV shift associated with lack of expression of Kv2.2 produces a negligible effect, given the similarity of slope factors of the Boltzmann fits to conductance versus voltage for neurons in which Kv2.2 transcripts were or were not detected. Other current properties were distributed similarly despite differential gene expression. The developmental increase in kinetics of activation of the current is similar in all neurons even though expression of Kv1.1 and Kv2.2 is heterogeneous, suggesting that activation kinetics are not correlated with expression of these genes unless both genes encode kinetically similar currents.
Present evidence does not allow association of the steady-state voltage of activation with either the 15 or the 30 pS channels that underlie the whole-cell current (Harris et al., 1988).Popen as a function of voltage was examined only to +40 mV, at which the open probability is not maximal. Furthermore, single-channel recordings of potassium currents have not been made from neurons expressing or apparently lacking Kv1.1 or Kv2.2 transcripts, nor have they been made from oocytes expressing Kv1.1 or Kv2.2 homomultimers.
The steady-state voltage of activation (V1/2) of IKvof spinal neurons is more similar to that of current in oocytes expressing homomultimers of Kv1.1 (Ribera and Nguyen, 1993) than to the current in oocytes expressing homomultimers of Kv2.2 (Burger and Ribera, 1996). The V1/2 of current in oocytes expressing homomultimers of Kv2.2 is more positive. There are limits to the extent that such oocyte data can be used to predict the voltage-dependent properties of endogenous channels that contain either Kv1.1 or Kv2.2 subunits, because currents in spinal neurons are likely to be encoded by heteromultimers. Kv1.1 or Kv2.2 may coassemble with other Kv1 or Kv2 gene products, respectively, to form heteromeric channels with novel properties. Additionally, post-translational modification of subunits may alter channel properties (Desarmenien and Spitzer, 1991).
Coexpression versus segregated expression of Kv1.1 and Kv2.2
Expression of Kv1.1 and Kv2.2 in 35% and 63% of mature neurons could indicate either that neurons express exclusively one of the two genes or that there is full or partial overlap in gene expression. Although we did not test for coexpression of both genes in the same cells, analysis of current properties suggests that they are coexpressed in some neurons. The population of Kv2.2(−) neurons lacked negatively shifted V1/2s. If all Kv1.1(+) neurons were part of this Kv2.2(−) population, then no Kv1.1(+) neurons should have had negatively shiftedV1/2s. This was not the case, however, implying that some Kv1.1(+) neurons were also part of the Kv2.2(+) population, and that Kv1.1 and Kv2.2 are coexpressed. Because potassium channel subunits from different subfamilies do not coassemble to form functional channels (Covarrubias et al., 1991; Sheng et al., 1993), coexpression of Kv1.1 and Kv2.2 would indicate that channels of different subfamilies (i.e., Shaker and Shab) contribute to the whole-cell potassium current in a single neuron. Given that coassembly of potassium channel gene products from the same subfamily can form functional heteromultimeric channels and are normally coexpressed (Christie et al., 1990; Isacoff et al., 1990;Ruppersberg et al., 1990; Sheng et al., 1993; Wang et al., 1993), the presence of Kv1.1 and Kv2.2 mRNAs may indicate the expression of other Kv1 or Kv2 genes, respectively. The extent of coexpression in mature neurons is not known and will determine the extent to which neurons express neither of these transcripts. If all neurons express one or the other, only 2% of neurons express neither; however, if all neurons expressing Kv1.1 also express Kv2.2, as many as 37% of neurons may express neither transcript.
Developmental regulation
Our observation that Kv1.1 is expressed in ∼30% of cultured neurons, equivalent to stages 24–32 in vivo, complements the later detection of Kv1.1 by in situ hybridization at stages 33–42 in a subpopulation of spinal neurons developing in vivo, the Rohon-Beard cells (Ribera and Nguyen, 1993). Moreover, one third of neurons in culture have a neurotransmitter sensitivity profile similar to that of Rohon-Beard neurons in situ(Bixby and Spitzer, 1982, 1984; Rohrbough and Spitzer, 1996), consistent with the observations that one third of neurons in culture express Kv1.1 mRNA and that these transcripts are localized to Rohon-Beard neurons in situ (Ribera and Nguyen, 1993). In contrast, Kv2.2 expression in cultured neurons seems delayed with respect to its expression in the embryo. Kv2.2 mRNA is detected in the ventrolateral spinal cord at stages 23–26, and only faintly by stage 35 (Burger and Ribera, 1996). Expression is not observed in neurons at 6–10 hr in culture (equivalent to stage 24), but is observed in a large fraction of neurons at 1 d in vitro (equivalent to stage 32–34). The delayed appearance of Kv2.2 relative to Kv1.1 in cultured neurons may result from the later development of motor neurons relative to Rohon-Beard neurons (Hartenstein, 1993).
How can homogeneous current development be reconciled with temporally and anatomically selective gene expression? Temporal regulation in response to developmental cues may require genes with different regulatory regions but similar functional properties. The significance of anatomically selective expression may lie in other processes, such as selective neurotransmitter modulation of current in different cell types.
Developmental expression of delayed rectifier potassium current in the GH3 pituitary cell line after hormonal stimulation has been demonstrated by following levels of Kv1.5 mRNA, protein, andIKv (Takimoto et al., 1993, 1995) and demonstrating that this current is encoded by Kv1.5 (Chung et al., 1995). The developmental regulation of K+ channel mRNAs (Drewe et al., 1992; Perney et al., 1992; Weiser et al., 1994) and of K+ channel proteins (Maletic-Savatic et al., 1995) most often have been studied separately in neurons; however, the presence of particular mRNA species has been correlated with the proteins that they may encode (Trimmer, 1993). Here we have correlated the pattern of appearance of mRNA with the developing current.
The development of potassium current in spinal neurons was shown previously to be dependent on RNA synthesis during the period in which the current matures (Ribera and Spitzer, 1989), consistent with the idea that synthesis of new channels initiated by transcription of potassium channel genes is the rate-limiting step in development of the current. This hypothesis is supported by the finding that transcripts for two potassium channel genes appear during this interval. The earlier appearance of Kv1.1 transcripts may indicate sequential synthesis of specific channel types in specific types of neurons. Although the current present in spinal neurons is likely to result from expression of several potassium channel genes of different subfamilies, common elements in Kv1.1 or Kv2.2 promoter regions may point to cues that control expression of these and other channel subunits that coassemble with them during development. Future work will characterize the appearance of channel proteins in the surface membrane and identify the functional role of specific channel genes in the development of delayed rectifier current.
Footnotes
This work was supported by National Institutes of Health Grants NS25217 (A.B.R.) and NS25916 (N.C.S.). We thank the members of our laboratories for discussions, Drs. E. Gleason and M. Ferrari for comments on this manuscript, and S. D. Watt for technical assistance. We are grateful to Dr. Corinna Burger for sharing the Kv2.2 sequence and information regarding in situ localization before publication.
Correspondence should be addressed to Devorah Gurantz, Department of Biology 0357, University of California at San Diego, 9500 Gilman Drive, La Jolla, CA 92093-0357.
REFERENCES
- 1.Barish ME. Differentiation of voltage-gated potassium current and modulation of excitability in cultured amphibian spinal neurones. J Physiol (Lond) 1986;375:229–250. doi: 10.1113/jphysiol.1986.sp016114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bixby JL, Spitzer NC. The appearance and development of chemosensitivity in Rohon-Beard neurones of the Xenopus spinal cord. J Physiol (Lond) 1982;330:513–536. doi: 10.1113/jphysiol.1982.sp014356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bixby JL, Spitzer NC. The appearance and development of neurotransmitter sensitivity in Xenopus embryonic spinal neurones in vitro . J Physiol (Lond) 1984;353:143–155. doi: 10.1113/jphysiol.1984.sp015328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blair LAC. The timing of protein synthesis required for the development of the sodium action potential in embryonic spinal neurons. J Neurosci. 1983;3:1430–1436. doi: 10.1523/JNEUROSCI.03-07-01430.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burger C, Ribera AB. Xenopus spinal neurons express Kv2 potassium channel transcripts during embryonic development. J Neurosci. 1996;16:1412–1421. doi: 10.1523/JNEUROSCI.16-04-01412.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandy KG, Gutman GA. CRC handbook of receptors and channels, CRC; Boca Raton: 1994. Voltage-gated K+ channel genes. pp. 1–71. [Google Scholar]
- 7.Chandy KG, Williams CB, Spencer RH, Aguilar BA, Ghanshani S, Tempel BL, Gutman GA. A family of three mouse potassium channel genes with intronless coding regions. Science. 1990;247:973–975. doi: 10.1126/science.2305265. [DOI] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Christie MJ, North RA, Osborne PB, Douglass J, Adelman JP. Heteropolymeric potassium channels expressed in Xenopus oocytes from cloned subunits. Neuron. 1990;4:405–411. doi: 10.1016/0896-6273(90)90052-h. [DOI] [PubMed] [Google Scholar]
- 10.Covarrubias M, Wei AA, Salkoff L. Shaker , Shal , Shab , and Shaw express independent K+ current systems. Neuron. 1991;7:763–773. doi: 10.1016/0896-6273(91)90279-9. [DOI] [PubMed] [Google Scholar]
- 11.Chung S, Saal DB, Kaczmarek LK. Elimination of potassium channel expression by antisense oligonucleotides in a pituitary cell line. Proc Natl Acad Sci USA. 1995;92:5955–5959. doi: 10.1073/pnas.92.13.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desarmenien MG, Spitzer NC. Role of calcium and protein kinase C in development of the delayed rectifier potassium current in Xenopus spinal neurons. Neuron. 1991;7:797–805. doi: 10.1016/0896-6273(91)90282-5. [DOI] [PubMed] [Google Scholar]
- 13.Desarmenien MG, Clendening B, Spitzer NC. In vivo development of voltage-dependent ionic currents in embryonic Xenopus spinal neurons. J Neurosci. 1993;13:2575–2581. doi: 10.1523/JNEUROSCI.13-06-02575.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dilworth DD, McCarrey JR. Single-step elimination of contaminating DNA prior to reverse transcriptase PCR. PCR Methods Appl. 1992;1:279–282. doi: 10.1101/gr.1.4.279. [DOI] [PubMed] [Google Scholar]
- 15.Drewe JA, Verma S, Frech G, Joho RH. Distinct spatial and temporal expression patterns of K+ channel mRNAs from different subfamilies. J Neurosci. 1992;12:538–548. doi: 10.1523/JNEUROSCI.12-02-00538.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu X, Spitzer NC. Distinct aspects of neuronal differentiation encoded by frequency of spontaneous Ca2+ transients. Nature. 1995;375:784–787. doi: 10.1038/375784a0. [DOI] [PubMed] [Google Scholar]
- 17.Gu X, Olson EC, Spitzer NC. Spontaneous neuronal calcium spikes and waves during early differentiation. J Neurosci. 1994;14:6325–6335. doi: 10.1523/JNEUROSCI.14-11-06325.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamill OP, Marty A, Neher E, Sakmann B, Sigworth FJ. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflügers Arch. 1981;391:85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- 19.Harris GL, Henderson LP, Spitzer NC. Changes in densities and kinetics of delayed rectifier potassium channels during neuronal differentiation. Neuron. 1988;1:739–750. doi: 10.1016/0896-6273(88)90172-9. [DOI] [PubMed] [Google Scholar]
- 20.Hartenstein V. Early pattern of neuronal differentiation of the Xenopus embryonic brain stem and spinal cord. J Comp Neurol. 1993;328:213–231. doi: 10.1002/cne.903280205. [DOI] [PubMed] [Google Scholar]
- 21.Isacoff EY, Jan YN, Jan LY. Evidence for the formation of heteromultimeric potassium channels in Xenopus oocytes. Nature. 1990;345:530–534. doi: 10.1038/345530a0. [DOI] [PubMed] [Google Scholar]
- 22.Kobel HR, DuPasquier L. Genetics of polyploid Xenopus . Trends Genet. 1986;2:310–315. [Google Scholar]
- 23.Krieg PA, Varnum SM, Wormington WM, Melton DA. The mRNA encoding elongation factor-1α (EF-1α) is a major transcript at the midblastula transition in Xenopus . Dev Biol. 1989;133:93–100. doi: 10.1016/0012-1606(89)90300-x. [DOI] [PubMed] [Google Scholar]
- 24.Lambolez B, Audinat E, Bochet P, Crepel F, Rossier J. AMPA receptor subunits expressed by single Purkinje cells. Neuron. 1992;9:247–258. doi: 10.1016/0896-6273(92)90164-9. [DOI] [PubMed] [Google Scholar]
- 25.Lamborghini JE. Rohon-Beard cells and other large neurons in Xenopus embryos originate during gastrulation. J Comp Neurol. 1980;189:323–333. doi: 10.1002/cne.901890208. [DOI] [PubMed] [Google Scholar]
- 26.Lamborghini JE, Iles A. Development of a high-affinity GABA uptake system in embryonic amphibian spinal neurons. Dev Biol. 1985;112:167–176. doi: 10.1016/0012-1606(85)90130-7. [DOI] [PubMed] [Google Scholar]
- 27.Lockery SR, Spitzer NC. Reconstruction of action potential development from whole-cell currents of differentiating spinal neurons. J Neurosci. 1992;12:2268–2287. doi: 10.1523/JNEUROSCI.12-06-02268.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maletic-Savatic M, Lenn NJ, Trimmer JS. Differential spatiotemporal expression of K+ channel polypeptides in rat hippocampal neurons developing in situ and in vitro . J Neurosci. 1995;15:3840–3851. doi: 10.1523/JNEUROSCI.15-05-03840.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marty A, Neher E. Tight-seal whole-cell recording. In: Sakmann B, Neher E, editors. Single channel recording. Plenum; New York: 1995. pp. 31–52. [Google Scholar]
- 30.O’Dowd DK, Ribera AB, Spitzer NC. Development of voltage-dependent calcium, sodium, and potassium currents in Xenopus spinal neurons. J Neurosci. 1988;8:792–805. doi: 10.1523/JNEUROSCI.08-03-00792.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perney TM, Marshall J, Martin KA, Hockfield S, Kaczmarek LK. Expression of the mRNAs for the Kv3.1 potassium channel gene in the adult and developing rat brain. J Neurophysiol. 1992;68:756–766. doi: 10.1152/jn.1992.68.3.756. [DOI] [PubMed] [Google Scholar]
- 32.Ribera AB. A potassium channel gene is expressed at neural induction. Neuron. 1990;5:691–701. doi: 10.1016/0896-6273(90)90223-3. [DOI] [PubMed] [Google Scholar]
- 33.Ribera AB, Nguyen DA. Primary sensory neurons express a Shaker -like potassium channel gene. J Neurosci. 1993;13:4988–4996. doi: 10.1523/JNEUROSCI.13-11-04988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ribera AB, Spitzer NC. A critical period of transcription required for differentiation of the action potential of spinal neurons. Neuron. 1989;2:1055–1062. doi: 10.1016/0896-6273(89)90229-8. [DOI] [PubMed] [Google Scholar]
- 35.Ribera AB, Spitzer NC. Differentiation of IKA in amphibian spinal neurons. J Neurosci. 1990;10:1886–1891. doi: 10.1523/JNEUROSCI.10-06-01886.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohrbough J, Spitzer NC. Regulation of intracellular Cl− levels by Na+-dependent Cl−cotransport distinguishes depolarizing from hyperpolarizing GABAA receptor-mediated responses in spinal neurons. J Neurosci. 1996;16:82–91. doi: 10.1523/JNEUROSCI.16-01-00082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruppersberg JP, Schroter KH, Sakmann B, Stocker M, Sewing S, Pongs O. Heteromultimeric channels formed by rat brain potassium-channel proteins. Nature. 1990;345:535–537. doi: 10.1038/345535a0. [DOI] [PubMed] [Google Scholar]
- 38.Sheng M, Liao YJ, Jan YN, Jan LY. Presynaptic A-current based on heteromultimeric K+ channels detected in vivo . Nature. 1993;365:72–75. doi: 10.1038/365072a0. [DOI] [PubMed] [Google Scholar]
- 39.Spitzer NC, Lamborghini JE. The development of the action potential mechanism of amphibian neurons isolated in culture. Proc Natl Acad Sci USA. 1976;73:1641–1645. doi: 10.1073/pnas.73.5.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sucher NJ, Deitcher DL. PCR and patch-clamp analysis of single neurons. Neuron. 1995;14:1095–1100. doi: 10.1016/0896-6273(95)90257-0. [DOI] [PubMed] [Google Scholar]
- 41.Takimoto K, Fomina AF, Gealy R, Trimmer JS, Levitan ES. Dexamethasone rapidly induces Kv1.5 K+ channel gene transcription and expression in clonal pituitary cells. Neuron. 1993;11:359–369. doi: 10.1016/0896-6273(93)90191-s. [DOI] [PubMed] [Google Scholar]
- 42.Takimoto K, Gealy R, Fomina AF, Trimmer JS, Levitan ES. Inhibition of voltage-gated K+ channel gene expression by the neuropeptide thyrotropin-releasing hormone. J Neurosci. 1995;15:449–457. doi: 10.1523/JNEUROSCI.15-01-00449.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trimmer JS. Expression of Kv2.1 delayed rectifier K+ channel isoforms in the developing rat brain. FEBS Lett. 1993;324:205–210. doi: 10.1016/0014-5793(93)81394-f. [DOI] [PubMed] [Google Scholar]
- 44.Wang H, Kunkel DD, Martin TM, Schwartzkroin PA, Tempel BL. Heteromultimeric K+ channels in terminal and juxtaparanodal regions of neurons. Nature. 1993;365:75–79. doi: 10.1038/365075a0. [DOI] [PubMed] [Google Scholar]
- 45.Weiser M, Vega-Saenz de Miera E, Kentros C, Moreno H, Franzen L, Hillman D, Baker H, Rudy B. Differential expression of Shaw -related K+ channels in the rat central nervous system. J Neurosci. 1994;14:949–972. doi: 10.1523/JNEUROSCI.14-03-00949.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]