Abstract
The D2-class of dopamine receptors (D2, D3, and D4) is a target for typical and atypical neuroleptic drugs. They have been considered, therefore, as factors that may contribute to the pathophysiology of psychotic disorders. Interestingly, in cortical brain tissues obtained postmortem form patients with chronic schizophrenia D3 mRNA was found to be significantly lower than in the corresponding anatomic regions of controls. Because the expression of a truncated D3-like mRNA (named D3nf) appeared to be unaffected in schizophrenic brains, these findings suggest the possibility that the loss of D3 mRNA results from an abnormal splicing of D3 pre-mRNA in schizophrenia that is accompanied by an increased accumulation of the truncated D3nf mRNA. To test this, three approaches were taken. (1) Substrate D3 pre-mRNA was spliced in vitroin HeLa nuclear extracts. Results from these experiments show that D3nf mRNA results from the alternative removal of a short spliceosomal intron in D3 pre-mRNA that has a noncanonical 3′ splice site. (2) Substrate D3 pre-mRNA was splicedin vivo in stably transfected rat GH3 cells. Despite the atypical 3′ cleavage that is necessary to generate D3nfmRNA, D3 and D3nf mRNA were found to be processed at similar amounts. (3) The relative D3/D3nf splicing efficiencies were then determined in the anterior cingulate cortex of postmortem brains obtained from controls and from patients with chronic schizophrenia. Significant differences were found between the relative levels of D3 and D3nf mRNA, suggesting that an enhanced D3nf-specific splicing of D3 pre-mRNA in schizophrenia leads to a decreased expression of D3mRNA.
Keywords: D3 pre-mRNA, in vitro splicing, in vivo splicing, primer extension, S1 nuclease protection, postmortem brain RNA
The cloned dopamine receptor subtypes, named D2 (Bunzow et al., 1988; Dal Toso et al., 1989; Grandy et al., 1989; Giros et al., 1989; Monsma et al., 1989), D3(Sokoloff et al., 1990), and D4 (Van Tol et al., 1991), are targets for drugs with antipsychotic efficacy and have been repeatedly suggested as factors in the pathophysiology of schizophrenia. Although there is at present no evidence of linkage between the D3gene and schizophrenia, several studies suggest that a distinct polymorphism in the first coding exon of the D3 gene increases the susceptibility to schizophrenia (Crocq et al., 1992; Mant et al., 1994; Asherson et al., 1996; Shaikh et al., 1996). Although the full significance of this finding is not yet understood (see Macciardi et al., 1994), it might suggest a contributory etiological role of the D3 receptor in schizophrenia. Furthermore, a postmortem study showed that D3 mRNA was lost in certain cortical regions of brains obtained from patients with chronic schizophrenia, whereas a truncated D3-like mRNA (named D3nf) could readily be detected in the same anatomic regions (Schmauss et al., 1993).
Several different truncated dopamine D3-like mRNAs that do not encode G-protein-coupled receptors have been identified (Giros et al., 1991; Snyder et al., 1991; Nagai et al., 1993; Schmauss et al., 1993). Their function is unknown. The longest of these mRNAs, named D3nf, results from a deletion of 98 nt that constitute the C-terminal region of the putative third cytoplasmic domain of the D3 receptor and, therefore, encodes a D3-like protein with a different C terminus (Schmauss et al., 1993; Liu et al., 1994). In human brain, D3nf mRNA was shown to be abundantly expressed and also translated into protein (Liu et al., 1994), suggesting that D3nf mRNA does not result from an RNA processing error. Another interesting observation is that D3nf mRNA was expressed in certain neocortical regions of brains obtained from patients with chronic schizophrenia, regions in which the expression of D3 mRNA was found to be lost (Schmauss et al., 1993) .
The human genome contains a single, intron-containing D3-encoded gene in which the 98 nt that are deleted in D3nf are embedded within a large continuous exon (Liu et al., 1994). Thus, to generate D3nf mRNA via alternative splicing, these 98 nt must be recognized by the splicing machinery as an alternative intron. Removal of this intron, however, would require cleavage of a rare (and nonconforming) 3′ splice site sequence that lacks the penultimate AG dinucleotides. However, these dinucleotides are known to be the most highly conserved dinucleotides of 3′ splice sites (Reed, 1989) and have been found to be essential for both lariat formation (Reed and Maniatis, 1985) and snRNP binding (Charbot and Steitz, 1987).
The present study shows that D3nf mRNA is indeed generated via atypical alternative splicing and, therefore, suggests a mechanism by which our previous results in schizophrenic brains would be explained: that an increased splicing of D3 pre-mRNA leads to a decrease of D3 mRNA and an increased accumulation of D3nf mRNA.
MATERIALS AND METHODS
Splicing of substrate D3 pre-mRNA in vitro. The sequence schematically diagrammed in Figure 1(top), which codes for the 3′ part of the putative third cytoplasmic domain of the D3 receptor and the 5′ region of the putative 6th transmembrane-spanning domain, was cloned into the plasmid vector pCR II (Invitrogen, San Diego, CA). A 5′-capped 2.3 kb pre-mRNA transcript (see Fig. 1, top) of this recombinant plasmid was synthesized in vitro using 10 U of Sp6 RNA polymerase (Stratagene, La Jolla, CA). This D3-encoded substrate pre-mRNA (20 fmol) was added to 50 μl of HeLa nuclear extract [prepared as described by Dignam et al. (1983) and supplemented with MgCl2, ATP, and creatine phosphate as described by Krainer et al. (1984)]. Aliquots (10.5 μl) were incubated at 30°C for 1–4 hr. At the end of each incubation, the splicing reaction was treated with proteinase K, extracted with phenol/chloroform, and precipitated. First-strand cDNA was synthesized with 200 U of Moloney murine leukemia virus (M-MLV) reverse transcriptase (United States Biochemical, Cleveland, OH) and the primer sequence—5′-GTATTGAGAACATGGGT-3′—which is complementary to the most 3′ sequences of exon II (see Fig. 1). The same primer, and a 5′ primer—5′-GAGGAGAAGACTCGGAATTC-3′ (identical to the most 5′ sequences of exon I; see Fig. 1)—were used for the PCR amplification to selectively amplify the ligated exons of the in vitrosplicing reaction. PCR products were separated on a 1.5% agarose/TBE gel, transferred to membrane, and subjected to Southern blot analysis using a 32P-radiolabeled random-primed cDNA encoding the human D3 receptor as a probe. D3- and D3nf-specific PCR products were cloned into the plasmid pCR II (Invitrogen) and subjected to nucleotide sequence analysis of both strands.
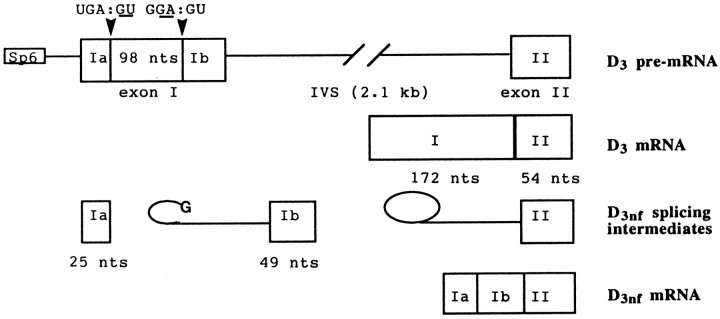
Fig. 1.
Splicing of D3 pre-mRNA in vitro. Schematic diagram of the structure of the substrate pre-mRNA, the products, and D3nf-specific intermediates ofin vitro splicing. The D3-encoded genomic locus (cloned into the plasmid pCR II) contains exon sequences that code for the 3′ part of the putative third cytoplasmic domain and the 5′ part of the putative transmembrane spanning domain 6 of the human D3 receptor.
Primer extension analysis of intermediates and products ofin vitro-spliced wild-type and mutant substrate D3 pre-mRNA. For these experiments, a 5′ capped 223 nt T7 RNA polymerase (Stratagene) transcript of the wild-type or mutant exon I sequences (see Fig. 1) was spliced in vitro in HeLa nuclear extracts as described above. 3′ splice site mutants were generated in vitro using the Chameleon Double-Stranded, Site-Directed Mutagenesis kit (Stratagene). Briefly, exon I was cloned into the plasmid pRC II, which served as the target plasmid DNA to which two oligonucleotide primers were simultaneously annealed to one of the strands. The selection primer was identical for all mutations. To introduce mutations into the D3nf-specific alternative 3′ splice site, the following oligonucleotide primers were used:
Wild-type sequence: 5′-CCCCTGCAACCTCGGG GAGTGCCACTTC- GGGAGAAG-3′
Mutant 1: 5′-CTGCAACCTCGGG AGGTGCCACTTCGGGAG-3′
Mutant 2: 5′-CTGCAACCTCGGG TAGTGCCACTTCGGGAGA-3′
Mutant 3: 5′-CCCCTGCAACCTCGGG - - - - GCCACTTCGG- GAGAAG-3′.
Extended and digested plasmid DNA was then transformed into the repair-deficient Escherichia coli strain XLmutS, and plasmid DNA extracted from these colonies was transformed into Epicuran Coli XL1-Blue competent cells. The correctness of each mutation was verified by nucleotide sequencing of the entire exon I.
In vitro-spliced wild-type or mutant RNA was precipitated and annealed to 5 × 105 cpm of a32P-end-labeled oligonucleotide sequence (5′-CAAGCACAATGGCCACC-3′) that is complementary to the most 3′ sequence of exons I and Ib (see Fig. 1) at 52°C for 10 min and then incubated with 2 U/μl AMV reverse transcriptase (Boehringer Mannheim, Indianapolis, IN) for 1 hr at 42°C. The primer extension products were separated on a 7 m urea/6% acrylamide gel.
Splicing of substrate D3 pre-mRNA in vivo. A D3 minigene schematically diagrammed in Figure 1(top) was cloned into the expression vector plasmid pCEP4 (Invitrogen), which also encodes the hygromycin-resistance gene. The recombinant plasmid (pCEP4/D3mg) was transfected into rat GH3 cells via lipofection as described previously (Liu et al., 1994). Hygromycin-resistant cells were isolated by ring cloning and expanded to cell lines. The cytoplasmic abundance of spliced transcripts of the minigene was analyzed by S1 nuclease protection analysis as described below using 10 μg of total cytoplasmic RNA extracted from each hygromycin-resistant clone.
RT-PCR analysis of D3 and D3nf mRNA expression in D3 minigene-expressing GH3 cells and in human cingulate cortex. First-strand cDNA was generated from 10 μg of total RNA extracted from transfected GH3 cells using an oligo-dT15 primer in conjunction with 200 U of M-MLV reverse transcriptase. The cDNA was then amplified by PCR using a 3′ primer (5′-GTATTGAGAACATGGGT-3′) that is complementary to the most 3′ sequence of exon II (see Fig. 1) and a 5′ primer (5′- ATTCGGCTTGAGGAGAA-3′) that is identical to the 5′ sequence of the transcript derived from pCEP4/D3mg (the underlined sequence is derived from the transcribed vector sequence immediately upstream of the 5′ sequence of exon I; see Fig. 1). Thus, this primer pair specifically enables the amplification of cRNA that is derived from the primary transcript of the transfected plasmid pCEP4/D3mg. PCR products were separated on 1.5% agarose/TBE gels, transferred to membrane, and subjected to Southern blotting using a γ-32P-radiolabeled oligonucleotide probe (5′-CAAGCACAATGGCCACC-3′) that is complementary to the most 3′ sequence of exon I (see Fig. 1).
For amplification of D3 and D3nf mRNAs expressed in the anterior cingulate cortex of control and schizophrenic brains, first-strand cDNA (generated as described above) was amplified by PCR using the primer pair D3S5′: 5′-TACCTGCCCTTTGGAGT-3′ and D33′: 5′-CTCCCTC- AGCAAGACAG-3′. This pair of primers allows the simultaneous amplification of the C-terminal halves of both D3 and D3nf cDNA. Details of this PCR amplification were described previously (Schmauss et al., 1993). PCR products were separated on ethidium bromide-stained 1% agarose gels. The relative quantities of D3- and D3nf-specific amplification products were compared to the quantities of ethidium bromide-stained amplification products that resulted from parallel experiments with plasmid DNA templates consisting of a mixture of equal amounts of the plasmids pRC/CMV/D3 and pRC/CMV/D3nf (see Schmauss et al., 1993) ranging from 0.5 fg to 0.5 ng.
RNA extraction and S1 nuclease protection assays. RNA was extracted from stably transfected GH3 cells (107 cells per clone) or from 0.5 gm of each postmortem tissue using the guanidine isothiocyanate/CsCl ultracentrifugation methods as described previously (Schmauss et al., 1993). To protect D3- and D3nf-specific mRNAs from ribonuclease digestion, 10 μg (cell lines) or 20 μg (brain tissues) of total cytoplasmic RNA was hybridized to 5 × 105 cpm of32P-end-labeled 39-mer antisense oligonucleotide at 50°C overnight. The nucleotide sequences of these oligonucleotides are either complementary to sequences found in both D3 and D3nf mRNAs (D3ALL: 5′-TCCACCCAAGGCAGTGTCCTGGCAGAT- GCTGTAGTAACG-3′) or they are complementary to sequences found only in D3 mRNA (D3-AS1: 5′-GAGCTTAGGCGCTATGGTGGGAC- TCAGGGAATTCCGAGT-3′) or in D3nf mRNA (D3nf-AS1: 5′-GCCTTC- TTCTCCCGAAGTGGCACTCAGGGAATTCCGAGT-3′). In parallel, sense oligonucleotides were incubated with RNA to control for the specificity of the RNA protection from ribonuclease digestion. Furthermore, 10 μg of total cytoplasmic RNA extracted from each brain tissue was also hybridized to a 215-nt-long [32P]UTP-labeled antisense RNA encoding the 5′-untranslated sequence of the human snRNP-associated protein N (Schmauss et al., 1989) before digestion with S1 nuclease. The N-encoded antisense riboprobe is an Sp6 RNA-polymerase transcript of the EcoRI-linearized plasmid pCRII-N. Sense-N-encoded mRNA (a T7 RNA-polymerase transcript of the same plasmid linearized with HindIII) was used in parallel experiments to control for the specificity of the RNase protection experiments. Hybridized RNA was digested with 500 U of S1 nuclease (Life Technologies, Gaithersburg, MD) at 37°C for 30 min. Protected fragments were precipitated and separated on 7 m urea/6% acrylamide gels. Triplicate experiments were performed for each sample.
Tissues sources. Normal control tissues [n= 8; age (mean ± SD) 60 ± 11.4 years; postmortem interval [PMI (mean ± SD) 11.7 ± 3.6 hr] were obtained from the National Neurological Research Specimen Bank (VAMC, Los Angeles, CA; sponsored by NINDS/National Institutes of Health, National Multiple Sclerosis Society, Hereditary Disease Foundation, Comprehensive Epilepsy Program, Tourette Syndrome Association, Dystonia Medical Research Foundation, and Veterans Health Services and Research Administration, Department of Veterans Affairs). Age-matched tissues from long-term hospitalized patients with chronic schizophrenia were obtained from the Schizophrenia Brain Bank of the Department of Psychiatry at Mount Sinai School of Medicine. Details of the recruitment and assessment of these patients and their brains were reported previously (Schmauss et al., 1993).
RESULTS
To clarify whether D3nf mRNA is indeed generated via atypical splicing of D3 pre-mRNA, in vitrosplicing experiments were done. A 2.3 kb in vitro-synthesized RNA transcript of the D3-encoded genomic locus (shown in Fig. 1) was incubated with HeLa nuclear extracts, and the products and intermediates of this substrate pre-mRNA-spliced in vitro were analyzed.
In the first set of experiments, the spliced RNA was precipitated and used as a template for first-strand cDNA synthesis that was primed with a 17-mer oligonucleotide sequence complementary to the most 3′ sequence of the distal exon of the substrate pre-mRNA (exon II; see Fig. 1). The cDNA was then amplified by PCR using the same 3′ primer in conjunction with a 5′ primer whose sequence is identical to the most 5′ sequence of the proximal exon of the pre-mRNA (exon I; Fig. 1). A Southern blot of the PCR products is shown in Figure2A. The lengths of the three hybridizing products correspond to the lengths of the unreacted pre-mRNA (2.3 kb), the D3-specific ligated exons (226 nt), and the D3nf-specific ligated exons (128 nt). The nucleotide sequences of the latter two products were found to be identical to D3- and D3nf-specific sequences expressed in vivo (10) (Fig.2B). Thus, in vitro splicing of the substrate D3 pre-mRNA resulted in the removal of the 2.1 kb intron to allow religation of the proximal exon (I) and distal exon (II) to generate D3 mRNA (constitutive splicing; see Figs.1, 2). In addition, the proximal exon (exon I) of some of the pre-mRNA is further cleaved to remove the 98-nt-long spliceosomal intron [resulting in the consecutive religation of the two short proximal exons (exons Ia and Ib) and the distal exon II] to yield D3nf mRNA (alternative splicing; see Figs. 1, 2). These results suggest that the sequence UGA: GU in exon I is recognized as a (typical) 5′ splice site, whereas the sequence G GA:GU serves as an AG-independent 3′ splice site. Because such a 3′ splice site sequence is indeed atypical, it should be stressed that the possibility of a sequencing compression at the boldfaced G of the 3′ splice site (GA:GT), which would make canonical splicing quite feasible ( AG:GT), has been excluded by analyzing multiple sequencing reactions of both strands using 7-deazaGTP and dITP.
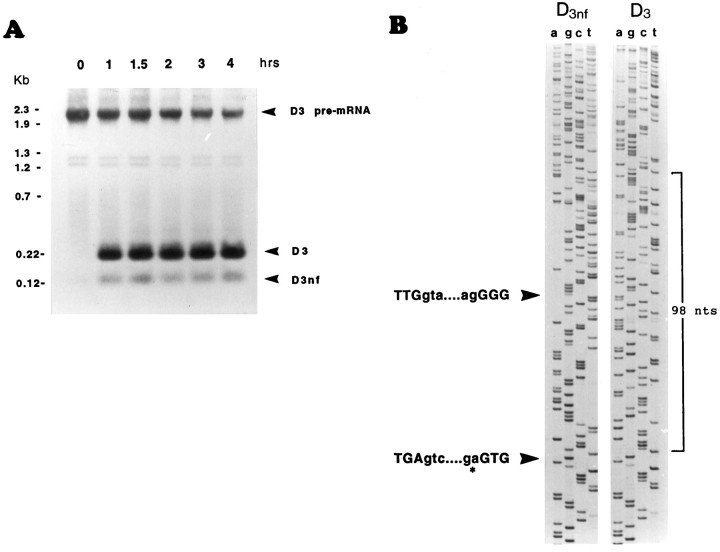
Fig. 2.
Analysis of the products and intermediates ofin vitro-spliced D3 pre-mRNA.A, Southern blot of PCR-amplified products of thein vitro-spliced substrate pre-mRNA. The time points shown on top of each lane indicate the length of the incubation of the substrate pre-mRNA with HeLa nuclear extracts at 30°C. Time point 0 corresponds to a 30 min incubation at 4°C. The blot was exposed to film for 10 min. The PCR amplification allowed the detection of ligated D3- and D3nf-specific exons after a 1 hr incubation of the pre-mRNA with nuclear extracts. B, Nucleotide sequence of ligated D3- and D3nf-specific exons. The sequences of D3- and D3nf-specific PCR amplification products shown in A precisely match the sequence of D3 and D3nf mRNA expressed in human brain (see Schmauss et al., 1993). The splice-junctional sequences resulting from both D3nf-specific splicing events are indicated by arrowheads. The lower arrowhead indicates the position of the splice junction resulting from cleavage of an atypical 3′ splice site (*) in the alternative intron.
For mammalian introns, AG-independent 3′ splice sites are extremely rare. Thus, to verify further the cleavage of the noncanonical 3′ splice site GA:GU, primer extension experiments were performed with wild-type substrate pre-mRNA corresponding to exon I (see Fig. 1) and various mutants thereof with altered or lacking 3′ splice site sequences. These substrate pre-mRNAs were spliced in vitroin HeLa nuclear extracts at 30°C for 2 hr, precipitated, and subjected to primer extension analysis using a primer sequence that is complementary to the most 3′ 17 nt of exon I (see Fig. 1). Primer extension products were expected to correspond in size to the unspliced substrate (D3) RNA (238 nt; the 5′ ends of each extension product are determined by the transcription start site of T7 RNA polymerase in pCR II/exon I, which is located 66 nt upstream of the first nucleotide in exon I), the religated exons Ia and I b (140 nt), and a splicing intermediate resulting from cleavage of the 5′ splice site and release of the 3′ exon attached to the branched 98-nt-long intron. As shown in Figure 3, when wild-type exon I is spliced in vitro, prominent extension products of 238 nt (corresponding in size to unspliced D3 RNA) and of 140 nt (corresponding in size to the religated exons Ia and Ib after splicing of the alternative 98 nt intron) are indeed obtained. This result is is consistent with the results shown in Figure 2.
Fig. 3.
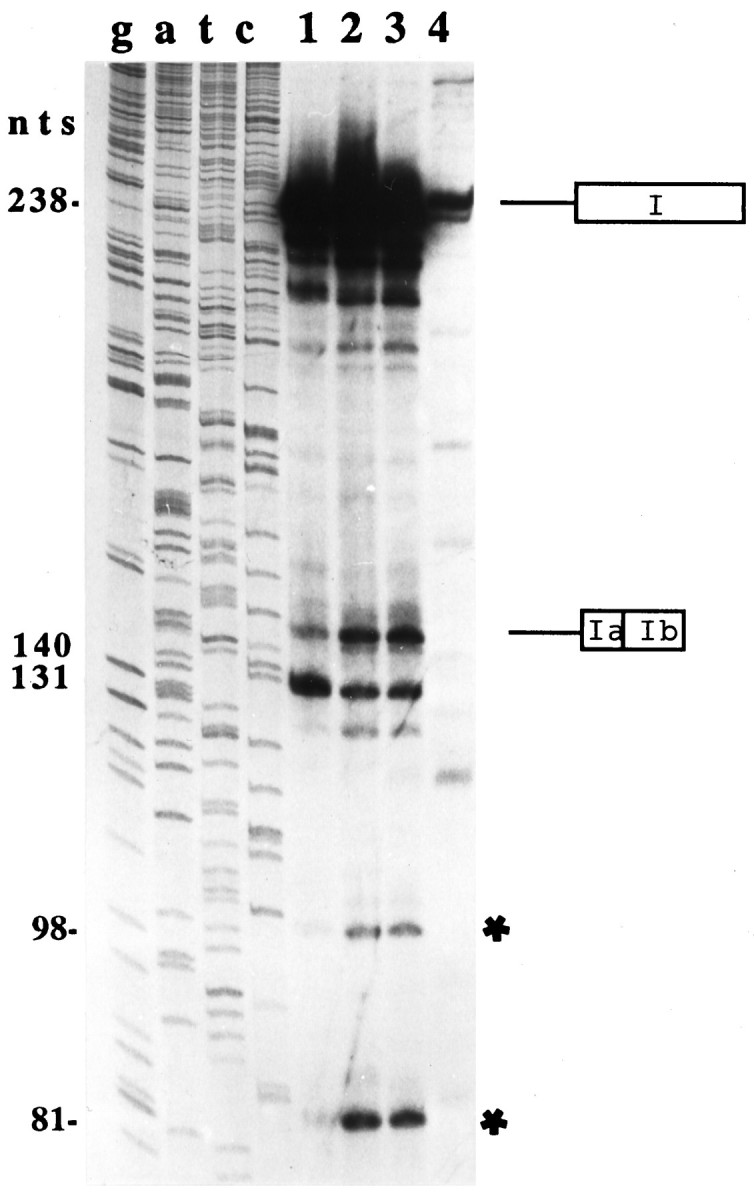
Determination of the length of the splicing intermediates and products of in vitro-spliced wild-type and mutant substrate pre-mRNAs by primer extension. A T7 RNA polymerase transcript of exon I (see Fig. 1) was spliced in vitroin HeLa nuclear extracts for 2 hr at 30°C. The 238 nt extension product corresponds in size to the unspliced D3 mRNA, and the 140 nt extension product corresponds to the spliced and religated D3nf-specific exons Ia and Ib (see Fig. 1). In contrast to wild-type pre-mRNA (lane 2), the D3nf-specific splicing of mutant 1 ( AG:GU) substrate pre-mRNA appears to be less efficient (lane 1). D3nf-specific alternative splicing of mutant 2 ( UG:GU) pre-mRNA appears to be indistinguishable from that of wild-type pre-mRNA (lane 3). In mutant 3, the 3′ splice site sequence GA:GU is deleted, and no D3nf-specific splicing is observed (lane 4). Although the precise nature of the 131 and 81 nt extension products is presently unknown, it is likely that the 98 nt extension product resulted from the extension of the splicing intermediate exon Ib/branched intron (see Results). Primer–extension products were separated on 7 m urea/6% polyacrylamide gels. Gels were exposed to film for 15 hr.
In addition, three mutant substrate pre-mRNAs were spliced in vitro and subsequently analyzed by primer extension. Mutant 1 contains the consensus AG ( AG:GU) dinucleotides, rather than the wild-type GA ( GA:GU) dinucleotides, penultimate to the 3′ cleavage site. Interestingly, although alternative splicing of the 98 nt intron is still obtained, the efficiency of this alternative splicing appears to be reduced (rather than enhanced) as indicated by the decreased signal of the primer extension product that corresponds in length to the D3nf-specific religated exons Ia and Ib (Fig. 3, lanes 1, 2). Furthermore, when mutant 2, which carries a single nucleotide change from GA:GU (wild-type) to UA:GU, is spliced in vitro, the D3nf-specific alternative splicing appears to be unaffected, i.e., the signal intensity of the extension product that corresponds in length to the D3nf-specific religated exons Ia and Ib is similar to those obtained for the spliced wild-type RNA (Fig. 3, lanes 2, 3). In another mutant substrate RNA (mutant 3), all of the four nucleotides (GA:GU) that constitute the 3′ splice site are deleted. As expected, no D3nf-specific splicing could be detected (Fig. 3, lane 4). Compared to experiments with wild-type and mutant 1 or 2 substrate RNA, less RNA was recovered after splicing of mutant 3 in vitro. However, it is apparent that the smaller amount of the mutant 3 RNA cannot account for the lack of visible D3nf-specific primer extension products, because other extension products are visible that do not correspond in size to any of the specific extension products seen in lanes 1–3 of Figure 3.
Additional extension products of 81, 98, and 131 nt were also detected after splicing of wild-type and mutant 1 or 2 substrate RNAs. The 131 nt extension product could perhaps have resulted from a strong impediment for reverse transcriptase that is provided by the 5′ sequence (derived from the plasmid pRC II) of the extended RNA template. Alternatively, T7 RNA polymerase may have used two different transcription start sites, one located 9 nt downstream of the other. If the latter scenario applies, however, one has to assume that the splicing of mutant 1 is as efficient as the splicing of the wild-type RNA sequence (see Fig. 3, lanes 1, 2).
Because the branchpoint of an intron is known to be an impediment for reverse transcriptase, either the 98 or the 81 nt extension products could have resulted from the extension of exon Ib/branched intron that results after cleavage of the 5′ splice site. Because the 81 nt extension product terminates at a cytosine nucleotide (which would be atypical for a branchpoint), it is more likely that the 98 nt extension product indicates the position of the branchpoint. This would be an adenosine located 49 nt upstream of the 3′ cleavage site. This putative branchpoint adenosine is preceded by a short pyrimidine-rich sequence.
In conclusion, the analysis of the intermediates and products of D3 pre-mRNA spliced in vitro revealed clearly that D3nf mRNA is derived from D3 pre-mRNA via removal of a short alternative spliceosomal intron that has a noncanonical 3′ splice site. However, these experiments did not directly address the relative D3/D3nfsplicing efficiencies. Therefore, additional in vivosplicing experiments were performed with transfected rat GH3 cells that stably express the D3 minigene that is schematically diagrammed in Figure 1 (top). To test whether transcripts of the transfected D3 minigene are processed in GH3 cells to yield cytoplasmic D3- and D3nf-specific mRNA sequences, RNA was extracted from stable transfectants and analyzed by RT-PCR analysis. PCR primers were used that specifically enable the amplification of the expression vector pCEP4/D3mg-derived transcripts (see Materials and Methods). These amplification products are expected to be 235 nt (D3 mRNA) and 137 nt (D3nf mRNA) in length. Figure4A shows a Southern blot of the PCR-amplified products. In 2 of 4 hygromycin-resistant GH3 cell clones, D3- and D3nf-specific amplification products were obtained; the other two clones did not express the minigene. Nucleotide sequence analysis (data not shown) revealed that the sequences of these two PCR products are identical to those shown in Figure 2B. Thus, as in HeLa nuclear extracts, in the neuroendocrine cell-derived GH3 cells D3 pre-mRNA is also processed constitutively and alternatively to yield D3 and D3nf mRNA.
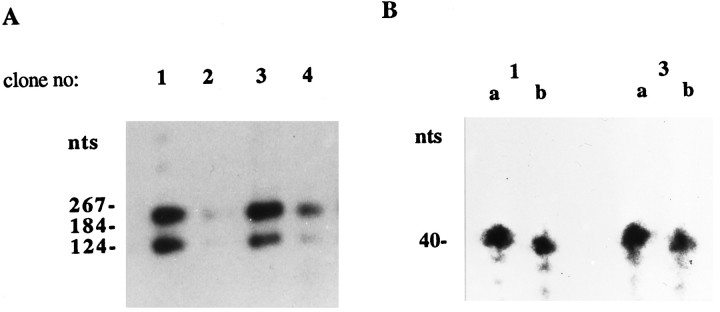
Fig. 4.
In vivo splicing of D3pre-mRNA transcripts derived from a D3 minigene stably inserted into the genome of GH3 cells. A, Southern blot of RT-PCR-amplified cytoplasmic D3- and D3nf-specific mRNA sequences. PCR products were separated on 1.5% agarose/TBE gels. The Southern blot was probed with a γ-32P-radiolabeled oligonucleotide probe (5′-CAAGCA- CAATGGCCACC-3′) that is complementary to the most 3′ sequence of exon I (see Fig. 1). Two of four randomly selected stable GH3 transfectants express both D3 and D3nf mRNA sequences that are derived from the transfected plasmid pCEP4/D3 mg (lanes 1, 3). B, Determination of the relative abundance of cytoplasmic D3and D3nf mRNA sequences in stable GH3 transfectants by S1 nuclease protection assays. Ten micrograms of total cytoplasmic RNA extracted from GH3 clones 1 and 3 were hybridized to either D3 (a)- or D3nf(b)-specific antisense oligonucleotides (see Materials and Methods) and subsequently digested with S1 nuclease. Protected fragments were separated on 7 m urea/6% polyacrylamide gels. The gel was exposed to film for 1 hr.
To analyze further the relative D3/D3nf splicing efficiencies, S1 nuclease protection assays were performed with RNAs extracted from the two D3 minigene-expressing cell clones (see Fig.4A). For these experiments, antisense oligonucleotides that are either complementary to a region of the sequence of D3 mRNA (D3-AS1) that is missing in D3nf or complementary to the splice-junctional sequence of D3nf mRNA (D3nf-AS1), which is not present in D3, were used (see Materials and Methods). Thus, D3 and D3nf mRNAs are separately protected from S1 nuclease protection, allowing their relative abundance to be compared. As shown in Figure 4B, in both clones that were shown to express transcripts of the D3 minigene (see Fig. 4A) the relative abundance of cytoplasmic D3/D3nf mRNA sequences is approximately equal. The sense sequence of both oligonucleotides did not specifically protect the respective RNAs from ribonuclease digestion (data not shown).
As shown previously, D3nf mRNA is abundantly expressed in human brain (Liu et al., 1994), indicating that in vivo the atypical alternative intron described here is spliced efficiently. Interestingly, whereas both D3 and D3nf mRNAs are expressed in normal neocortical tissues, only D3nf mRNA was detected by RT-PCR in motor and parietal cortices of chronic schizophrenics (Schmauss et al., 1993). One possible explanation for these results is that D3 pre-mRNA is generally decreased in chronic schizophrenics. Another possibility, especially given the above findings, is that the loss of D3mRNA in chronic schizophrenia results from enhanced D3nf-specific splicing of the D3-encoded primary transcript. The first possibility would predict a decreased abundance (relative to controls) of both D3 and D3nf mRNAs, whereas the abundance of D3nf mRNA should be unaltered or increased if the second possibility is obtained. To test these two possibilities, S1 nuclease protection assays were performed on total cytoplasmic RNA extracted from the anterior cingulate cortex of postmortem brains derived from the same population of patients that were examined previously (Schmauss et al., 1993), as well as from matched controls (see Materials and Methods). This particular anatomic region was chosen because a nonquantitative RT-PCR analysis demonstrated coexpression of D3 and D3nf in both diagnostic groups (see below).
In a first series of experiments, RNA extracted from 8 control and 8 schizophrenic tissues was subjected to S1 nuclease protection assays using a 215-nt-long antisense riboprobe that codes for the 5′-untranslated region of mRNA encoding the snRNP-associated protein N (Schmauss et al., 1989). This initial control experiment was performed to test for the integrity of each RNA preparation. N-encoded mRNA was chosen for this control experiment because it is exclusively expressed in neurons, and it is found in all major neuronal populations throughout the brain (Schmauss et al., 1992). As shown in Figure 5A, the expression of the abundant N-encoded mRNA (protected from S1 nuclease digestion by an Sp6 RNA polymerase transcript of the EcoRI-linearized plasmid pCRII-N; see Materials and Methods) could readily be detected in all schizophrenic and control brains. Although the levels of N mRNA vary between samples, no significant differences were found in the range of N mRNA levels between both diagnostic groups. A sense N-encoded riboprobe (a T7 RNA polymerase transcript of theHindIII-linearized plasmid pCRII-N) did not protect RNA from S1 nuclease digestion.
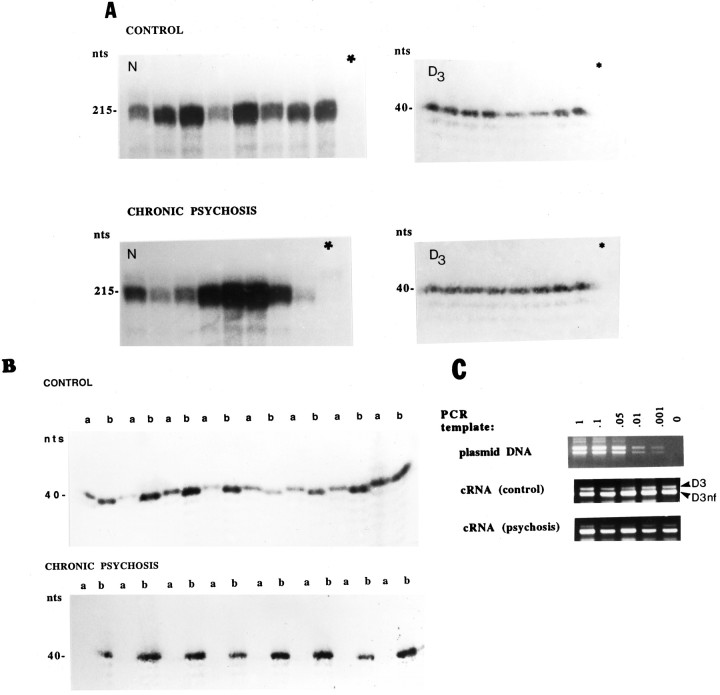
Fig. 5.
Expression of D3 and D3nfmRNAs in the cingulate cortex (Brodmann areas 24/25) of control brains and brains from patients with chronic schizophrenia. A, Determination of the expression of N-encoded mRNA (left) and of the sum of D3 and D3nf mRNA (right). Ten micrograms of total cytoplasmic RNA were hybridized to a 215-nt-long antisense riboprobe encoding the human snRNA N mRNA, which is known to be expressed exclusively and at high levels in all major neuronal populations of the brain. The gel was exposed to film for 5 hr. Twenty micrograms of the same RNA samples were hybridized to an antisense oligonucleotide (D3ALL) that is complementary to a sequence common to both D3 and D3nf mRNA. The gel was exposed to film for 12 hr. Sense N-encoded riboprobe and the sense sequence of the oligonucleotide D3ALL did not specifically protect the respective mRNAs from ribonuclease digestion (lanes marked asterisks).B, Separate detection of cytoplasmic D3 and D3nf mRNA sequences. S1 nuclease protection analysis of 20 μg of the same total cytoplasmic RNA samples hybridized to either a D3-specific (a) or a D3nf-specific (b) 39-mer antisense oligonucleotide (see Materials and Methods). The gels were exposed to film for 24 hr and developed simultaneously. The individual samples were loaded in the same order on all gels. C, Amplification of D3 and D3nf cDNAs by PCR. D3 and D3nf cDNAs, derived either from a mixture of equal amounts of D3- and D3nf-encoded plasmid DNA templates at concentrations ranging from 0.5 fg to 0.5 ng or from RNA extracted from the anterior cingulate cortex of brains from five controls and five schizophrenics, were simultaneously amplified by PCR. A single pair of PCR primers (see Materials and Methods) results in the coamplification of the C-terminal halves of D3 (619 nt) and D3nf (521 nt) cDNAs. PCR products were separated on ethidium bromide-stained agarose gels. To be able to visualize the small amount of D3-specific amplification products of the schizophrenic samples on such gels, products of a 35 cycle PCR amplification are shown here.
A second series of experiments, performed on the same RNAs, involved the simultaneous protection of D3 and D3nf mRNA via hybridization of a 39-mer antisense oligonucleotide (D3ALL; see Materials and Methods) that is complementary to a sequence that codes for the N terminus of the putative third cytoplasmic domain of D3. This probe, therefore, protects both D3 and D3nf mRNAs from S1 nuclease digestion. As shown in Figure 5A, the signals obtained for the sum of D3 and D3nf mRNAs were also found to be similar in the control and schizophrenic tissues. The sense sequence of the oligo-D3ALL did not protect these mRNAs from S1 nuclease digestion.
It is noted that the variability of the sum of D3 mRNA expression between samples is substantially less than that of N-encoded mRNA. This is perhaps because of differences in the turnover rates (stabilities) between both mRNAs. The abundance of the 1.6 kb N-encoded mRNA has been shown to be similar to that of the very abundant actin-encoded mRNA (Schmauss and Lerner, 1990). In contrast, D3 mRNA (8.3 kb) is known to be expressed at very low levels (Sokoloff et al., 1990). It is possible that an mRNA with a high transcription rate also has a relatively short turnover rate and that differences in the postmortem intervals (PMIs) between samples (the SDs of the PMIs of the samples analyzed here are ∼3 hr), therefore, result in the detection of different levels of N mRNA. This is supported by the observation that higher levels of N-encoded mRNA were detected in samples with shorter PMIs. Thus, if D3 mRNA transcripts are substantially more stable than N mRNA, the magnitude of the differences between the individual PMIs of the samples analyzed here may not be large enough to result in the detection of similarly different levels of D3 mRNA.
A third series of experiments targeted D3 and D3nf sequences separately, using D3- and D3nf-specific antisense 39-mer oligonucleotides. As shown in Figure 5B, in 5 of 8 cingulate cortices obtained postmortem from normal controls D3nf mRNA was more abundant than D3 mRNA, although D3 mRNA was clearly detected in all 8 individuals. In contrast, no D3 mRNA could be detected in cingulate cortical tissues obtained from 8 long-term hospitalized patients with chronic schizophrenia, whereas in all 8 tissues D3nf mRNA was found to be expressed at levels that, in most cases, are higher than the corresponding levels found in control tissues.
Differences in the expression of D3 mRNA in control and schizophrenic brains are also apparent in RT-PCR experiments in which D3 and D3nf cDNAs are simultaneously amplified. This is demonstrated in Figure 5C. PCR experiments on the five control samples that have a higher ratio of D3nf/D3 mRNA (see Fig. 5B) also indicate that the amplification of D3 cDNA is 5- to 10-fold less than that of D3nf if one compares ethidium bromide-stained signals of these PCR products to the corresponding ones obtained from PCR amplifications, performed in parallel, of a mixture of equal amounts of D3- and D3nf-encoded plasmid DNA templates that range from 0.5 fg to 0.5 ng (Fig.5C; see Materials and Methods). Both D3- and D3nf-specific amplifications are also obtained from five randomly selected schizophrenic samples, indicating that not all of the D3 pre-mRNA is spliced in a D3nf-specific manner. However, for the schizophrenic samples the difference in the amounts of D3nf and D3 amplification products is at least 100-fold (Fig.5C).
In summary, the analysis of the relative abundance of D3 and D3nf mRNA by RNase protection as well as by RT-PCR revealed a significant decrease of D3 mRNA in the cingulate cortex of brains of patients with chronic psychosis. In the same anatomical region, however, D3nf mRNA is abundantly expressed and this can explain why the sum of D3 and D3nf mRNAs (see Fig. 3A) is similar between control and schizophrenic brains. Altogether, these data support the suggestion that the loss of D3 mRNA in some neocortical regions obtained postmortem from patients with chronic schizophrenia (Schmauss et al., 1993), and the decrease of D3 mRNA in the cingulate cortex found in this study, results from an enhanced D3nf-specific splicing of the D3-encoded primary transcript.
DISCUSSION
The results of in vitro and in vivo splicing experiments shown here demonstrate that the truncated D3-like mRNA (D3nf) results from D3pre-mRNA via removal of a short alternative spliceosomal intron that is retained in D3 mRNA. This intron is flanked by a noncanonical 3′ splice site sequence ( GA:GU).
Although in general one would predict that the in vivosplicing efficiency of such a rare and nonconforming alternative 3′ splice site is significantly lower than the cleavage of constitutive and/or canonical 3′ splice sites, results from in vivosplicing experiments with GH3 cells that stably express RNA transcripts of a transfected D3 minigene revealed that both D3 (constitutive)- and D3nf(alternative)-specific splicing events occur with similar frequency. Furthermore, in human brain the expression levels of D3nfmRNA are either equal to or higher than the corresponding levels of the constitutively spliced D3 mRNA (Liu et al., 1994) (this study). Interestingly, substitution of the penultimate GA ( GA:GU) dinucleotides of the D3nf-specific 3′ splice site of substrate D3 pre-mRNA with the consensus AG ( AG:GU) dinucleotides does not increase the D3nf-specific alternative splicing efficiency. Somewhat unexpectedly, substitution of the boldfaced G (GA:GU) of the wild-type 3′ splice site with a uridine (UA:GU) also does not affect the 3′ cleavage (see Fig. 3). Thus, the recognition signals for cleavage of the noncanonical 3′ splice site of the D3nf-specific intron are obviously not determined by the sequence of the 3′ splice site ( GA:GU) alone.
Because the frequency of D3nf-specific splicing of D3 pre-mRNA ultimately determines the level of D3 mRNA, this alternative splicing could function as an important regulator of D3-receptor expression. It is of interest, therefore, to note that different relative ratios of D3/D3nf mRNA were observed in the anterior cingulate cortex of controls and chronic schizophrenics. Whereas D3 mRNA levels are lower in schizophrenics compared to controls, D3nf mRNA levels are increased. This suggests that the D3nf-specific splicing activity is higher in brains of the chronic schizophrenic population studied here (and inSchmauss et al., 1993).
It is unlikely that the results of the S1 nuclease protection assays and RT-PCR experiments shown here reflect different stabilities of D3 and D3nf mRNA in schizophrenic and normal brains (which could obscure estimates of the relative D3/D3nf-specific splicing activities). First, in schizophrenia, D3 and D3nf mRNAs change in opposite directions and the level of the sum of both mRNAs is not different from the corresponding level found in control brains (see Fig. 5). Second, in stably and tetracycline-regulated D3- and D3nf-expressing GH3 cells (see Howe et al., 1995) the stabilities of both cDNA-derived RNAs (determined with transcription inhibition experiments) were identical (t1/2 = 80 min; C. Schmauss, unpublished observation). Although this result does not exclude the possibility that other destabilizing or stabilizing factors operate on full-length D3 and D3nf mRNAs, it clearly shows that the absence or presence of the alternative 98 nt intron has no significant effect on the stability of either RNA.
In summary, the results reported here demonstrate that it is possible to reconstitute both in vitro and in vivo an alternative splicing pathway of D3 pre-mRNA that involves cleavage of a noncanonical 3′ splice site to generate the truncated D3nf mRNA. A comparison between the relative levels of the products of this splicing (D3 and D3nf mRNA) in the anterior cingulate cortex of schizophrenics and controls suggests an abnormal post-transcriptional processing of D3 pre-mRNA in schizophrenia. This result may implicate altered activities of post-transcriptional regulators of gene expression in this disease. Clearly, it will now be of interest to search for other introns with a 3′ splice site sequence specificity similar to the one described here and to analyze the corresponding splicing pattern in schizophrenics and controls. It is possible that the D3nf-specific alternative intron is a member of a novel class of minor introns. In this respect, it is of interest to note that recent studies on another novel group of minor introns, called the AU–AC introns (which have atypical, but evolutionarily highly conserved, 5′ and 3′ splice sites), have shown that a deviation of consensus-sequence splice sites of major-class introns is accompanied by the assembly of different snRNPs (Hall and Padgett, 1996; Tarn and Steitz, 1996). Four pre-mRNAs are presently known that have such AU–AC introns. Importantly, these introns are spliced by a U11/U12 and U5 snRNP-containing spliceosome that is completely different from the U1, U2, and U6 snRNP-containing spliceosome known to be essential for cleavage of all major-class introns (GU–AG introns). It is possible, therefore, that the cleavage of the alternative D3nf-specific intron requires the presence of unique splicing factors and that an altered expression of such splicing factors may underlie the observed abnormality in the D3 pre-mRNA processing. If unique splicing factors indeed mediate the cleavage of atypical introns like the one described here, they are likely to be found in the common nuclear repertoire of snRNPs. This assumption is supported by the observation that, in addition to neuronal cells, D3 pre-mRNA is also spliced both constitutively and alternatively in two completely different cell types, human HeLa cells and rat GH3 cells.
It is also important to note that the patient population studied here and previously (Schmauss et al., 1993) represents a rather unique group of schizophrenic patients that were chronically and severely ill, therapy-resistant, and hospitalized long-term. Future studies will show whether a similar alteration of D3 and D3nf mRNA expression can be detected in brains of schizophrenic patients with a less severe course of the disease. Another unresolved issue is whether the altered splicing of D3 pre-mRNA is an outcome of neuroleptic treatment, which is the most common therapeutic strategy for schizophrenia. It is impossible, at present, to address this issue with studies on tissues obtained from drug-naive schizophrenic patients because of an extremely limited availability of such brains. However, the reconstitution of D3 pre-mRNA splicing in vivo in transfected cells that express D2 and/or D2-like receptors (D3, D4) will now allow us to test directly whether different pharmacological manipulations of these receptors have effects on the splicing of D3 pre-mRNA. If neuroleptic drugs can alter the splicing pattern of D3 pre-mRNA, this would not necessarily imply that another side effect of such drugs has been discovered. It could, in fact, be a mechanism by which neuroleptics mediate their antipsychotic effect.
Footnotes
This work was supported by a National Alliance for Research on Schizophrenia and Depression Young Investigator Award. I thank the staff members of the National Neurological Research Specimen Bank (Veterans’ Administration Medical Center, Los Angeles, CA) and of the Schizophrenia Brain Bank of the Department of Psychiatry at Mount Sinai School of Medicine (New York, NY) for providing the brain tissues and the corresponding diagnostic and neuropathological evaluations for this study. Dr. Boris Skryabin and Julie Yoon assisted in the generation of mutant plasmids and the preparation of nuclear extracts, respectively.
Correspondence should be addressed to Dr. Claudia Schmauss, Mount Sinai School of Medicine, Box 1229, One Gustave L. Levy Place, New York, NY 10029.
REFERENCES
- 1.Asherson P, Mant R, Holmans P, Williams J, Cardno A, Murphy K, Jones L, Collier D, McGuffin P, Owen MJ. Linkage, association and mutational analysis of the dopamine D3 receptor gene in schizophrenia. Mol Psychiatry. 1996;1:125–132. [PubMed] [Google Scholar]
- 2.Bunzow JR, Van Tool HM, Grandy DK, Albert P, Salon J, Christie M, Machida CA, Neve KA, Civelli O. Cloning and expression of a rat D2 dopamine receptor cDNA. Nature. 1988;336:783–787. doi: 10.1038/336783a0. [DOI] [PubMed] [Google Scholar]
- 3.Charbot B, Steitz JA. Multiple interactions between the splicing substrate and small nuclear ribonucleoproteins in spliceosomes. Mol Cell Biol. 1987;7:281–293. doi: 10.1128/mcb.7.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crocq MA, Mant R, Asherson P, Williams J, Hode Y, Mayerova A, Collier D, Lannfelt L, Sokoloff P, Schwartz JC, Gill M, Macher JP, McGuffin P, Owen MJ. Association between schizophrenia and homozygosity at the dopamine D3 receptor gene. J Med Genet. 1992;29:858–860. doi: 10.1136/jmg.29.12.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dal Toso R, Sommer B, Ewert M, Herb A, Pritchett DB, Bach A, Shivers B, Seeburg PH. The dopamine D2 receptor: two molecular forms generated by alternative splicing. EMBO J. 1989;8:4025–4034. doi: 10.1002/j.1460-2075.1989.tb08585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dignam JD, Lebovitz RM, Roeder RG. Acurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giros B, Sokoloff P, Martres MP, Riou JF, Emorine LJ, Schwartz JC. Alternative splicing directs the expression of two D2 dopamine receptor isoforms. Nature. 1989;342:923–926. doi: 10.1038/342923a0. [DOI] [PubMed] [Google Scholar]
- 8.Giros B, Martres MP, Pilon C, Sokoloff P, Schwartz JC. Shorter variants of the D3 dopamine receptor produced through various patterns of alternative splicing. Biochem Biophys Res Commun. 1991;176:1584–1592. doi: 10.1016/0006-291x(91)90469-n. [DOI] [PubMed] [Google Scholar]
- 9.Grandy DK, Marchionni MA, Makam H, Stofko RE, Alfano M, Frothingham L, Fischer JB, Burke-Howie KJ, Bunzow JR, Server A, Civelli O. Cloning of the cDNA and gene for a human D2 dopamine receptor. Proc Natl Acad Sci USA. 1989;86:9762–9766. doi: 10.1073/pnas.86.24.9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall SL, Padgett RA. Requirement of U12 snRNA for in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science. 1996;271:1716–1718. doi: 10.1126/science.271.5256.1716. [DOI] [PubMed] [Google Scholar]
- 11.Howe JR, Skryabin BV, Belcher SM, Zerillo CA, Schmauss C. The responsiveness of tetracycline-sensitive expression system differs in different cell lines. J Biol Chem. 1995;270:14168–14174. doi: 10.1074/jbc.270.23.14168. [DOI] [PubMed] [Google Scholar]
- 12.Krainer AR, Maniatis T, Ruskin B, Green M. Normal and mutant human β-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984;36:993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- 13.Liu K, Bergson C, Levenson R, Schmauss C. On the origin of mRNA encoding the truncated dopamine D3-type receptor D3nf and detection of D3nf-like immunoreactivity in human brain. J Biol Chem. 1994;269:29220–29226. [PubMed] [Google Scholar]
- 14.Macciardi F, Verga M, Kennedy JL, Petronis A, Bersani G, Pancheri P, Smeraldi E. An association study between schizophrenia and the dopamine receptor genes DRD3 and DRD4 using haplotype relative risk. Hum Hered. 1994;44:328–336. doi: 10.1159/000154240. [DOI] [PubMed] [Google Scholar]
- 15.Mant R, Williams J, Asherson P, Parfitt E, McGuffin P, Owen MJ. Relationship between homozygosity at the dopamine D3 receptor gene and schizophrenia. Am J Med Genet. 1994;54:21–26. doi: 10.1002/ajmg.1320540106. [DOI] [PubMed] [Google Scholar]
- 16.Monsma F, McVittie LD, Gerfen CR, Mahan LC, Sibley DR. Multiple D2 dopamine receptors produced by alternative RNA splicing. Nature. 1989;342:926–929. doi: 10.1038/342926a0. [DOI] [PubMed] [Google Scholar]
- 17.Nagai Y, Ueno S, Saeki Y, Soga F, Yanagihara T. Expression of the D3 dopamine receptor gene and a novel variant transcript generated by alternative splicing in human peripheral blood lymphocytes. Biochem Biophys Res Commun. 1993;194:368–374. doi: 10.1006/bbrc.1993.1829. [DOI] [PubMed] [Google Scholar]
- 18.Reed R. The organization of 3′ splice-site sequences in mammalian introns. Genes Dev. 1989;3:2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- 19.Reed R, Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell. 1985;41:95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- 20.Schmauss C, Lerner MR. The closely related snRNP polypeptides N and B/B′ are distinguishable by antibodies as well as by differences in their mRNA and gene structures. J Biol Chem. 1990;265:10733–10739. [PubMed] [Google Scholar]
- 21.Schmauss C, McAllister G, Ohosone Y, Hardin JA, Lerner M R. A comparison of the snRNP-associated Sm-autoantigens human N, rat N and human B/B′. Nucleic Acids Res. 1989;17:1733–1744. doi: 10.1093/nar/17.4.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmauss C, Brines ML, Lerner MR. The gene that encodes the snRNP-associated protein N is expressed at high levels in neurons. J Biol Chem. 1992;267:8521–8529. [PubMed] [Google Scholar]
- 23.Schmauss C, Haroutunian V, Davis KL, Davidson M. Selective loss of dopamine D3 receptor mRNA expression in the parietal and motor cortex in patients with chronic schizophrenia. Proc Natl Acad Sci USA. 1993;90:8942–8946. doi: 10.1073/pnas.90.19.8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaikh S, Collier DA, Sham PC, Ball D, Aitchison K, Vallada H, Smith I, Gill M, Kerwin RW. Allelic association between a Ser-9-Gly polymorphism in the dopamine D3 receptor gene and schizophrenia. Hum Genet. 1996;97:714–719. doi: 10.1007/BF02346178. [DOI] [PubMed] [Google Scholar]
- 25.Snyder LA, Roberts JL, Sealfon SC. Alternative transcripts of the rat and human dopamine D3 receptor. Biochem Biophys Res Commun. 1991;180:1031–1035. doi: 10.1016/s0006-291x(05)81169-6. [DOI] [PubMed] [Google Scholar]
- 26.Sokoloff P, Giros B, Martres MP, Bouthenet ML, Schwartz JC. Molecular cloning and characterization of a novel dopamine receptor (D3) as a targets for neuroleptics. Nature. 1990;347:146–151. doi: 10.1038/347146a0. [DOI] [PubMed] [Google Scholar]
- 27.Tarn WY, Steitz JA. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell. 1996;84:801–811. doi: 10.1016/s0092-8674(00)81057-0. [DOI] [PubMed] [Google Scholar]
- 28.Van Tol HHM, Bunzow JR, Guan HC, Sunahara RK, Seeman P, Niznik HB, Civelli O. Cloning of the gene for a human dopamine D4 receptor with high affinity for the antipsychotic clozapine. Nature. 1991;350:610–614. doi: 10.1038/350610a0. [DOI] [PubMed] [Google Scholar]