Abstract
Mutations in the gene for proteolipid protein (PLP) have been associated with CNS dysmyelination and abnormal oligodendrocyte death in spontaneous mouse mutants and in Pelizaeus-Merzbacher disease; however, the effect of mutations on PLP structure and function are little understood. We have identified a monoclonal antibody directed against a novel cell surface epitope of PLP, termed O10. By immunofluorescence analysis, COS-7 cells transiently transfected to express PLP (or its isoform DM20) can be stained with antibody O10 and another antibody (A431) directed against the C terminus of PLP/DM20. The subcellular distribution of immunofluorescence labels for the two antibodies is not identical, suggesting that the O10 epitope is acquired post-translationally. When PLP/DM20 from jimpy, jimpymsd, and rumpshaker mutant mice is expressed in COS-7 cells and compared with wild-type PLP/DM20, none of the mutant isoforms displays the O10 epitope, whereas the C-terminal epitope is detected. Because the O10 but not the A431 epitope is also sensitive to SDS and reducing agents, this strongly suggests abnormal protein folding in the PLP mutants. PLP fromjimpymsd mice is obviously misfolded, because the amino acid substitution (Ala242 → Val) is located within a transmembrane domain to which the O10 antibody does not bind. We propose that the O10 epitope emerges as the full length protein reaches a functional tertiary structure and that the absence of this epitope marks a structural defect of PLP that leads to dysmyelination.
Keywords: oligodendrocyte differentiation, myelin assembly, proteolipid protein, protein misfolding, apoptosis, jimpymouse, rumpshaker mouse, Pelizaeus-Merzbacher disease
In the rodent CNS, oligodendrocytes differentiate and elaborate myelin during postnatal life, concluding one of the final steps in neural development. The requirement of myelin for normal brain function is dramatically illustrated by naturally occurring mutations and dysmyelinating diseases. Several mutations that have been identified at the genomic level have been associated with a defect of oligodendrocyte differentiation, but the underlying mechanisms remain obscure (Mikoshiba et al., 1991; Lemke, 1993; Snipes et al., 1993;Nave, 1995).
Oligodendrocyte development from glial precursor cells follows an intrinsic genetic program, and the progression along this pathway has been described morphologically and with the help of stage-specific antigenic markers. In one earlier study, the immunization of mice with corpus callosal membranes (Schachner et al., 1981; Sommer and Schachner, 1981; Kuhlmann-Krieg et al., 1988) has yielded a series of monoclonal antibodies (O1–O11) that recognize developmentally regulated cell surface antigens and define subsequent stages of differentiating oligodendrocytes. Although most O-antigens have not been defined biochemically, at least two antibodies that recognize predominantly galactocerebroside (O1) and sulfatide (O4) are widely used as references to monitor oligodendrocyte development (Schachner et al., 1981; Sommer and Schachner, 1981, 1984; Pfeiffer et al., 1993).
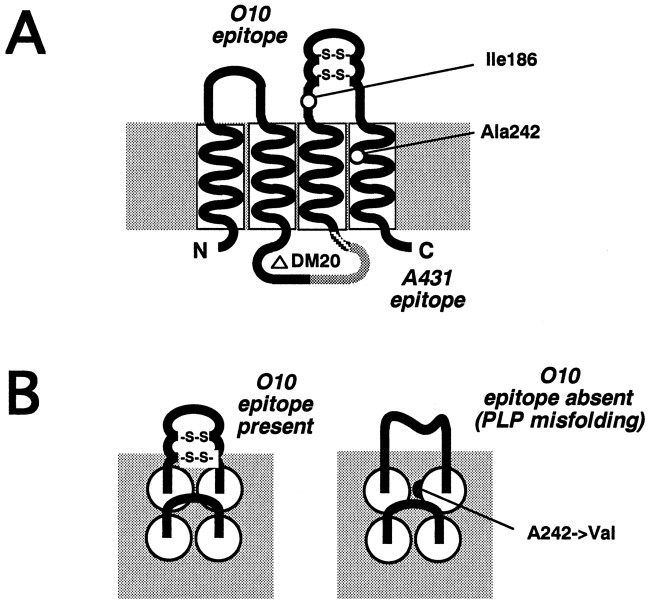
Among the most terminal differentiation markers of oligodendrocytes are myelin-associated proteins, such as myelin basic protein (MBP) and proteolipid protein (PLP). PLP is the most abundant integral membrane protein and is specific to CNS myelin (Lees and Brostoff, 1984; Stoffel et al., 1984; Nave and Milner, 1989). Four hydrophobic regions of this molecule constitute transmembrane domains (Popot et al., 1991; Weimbs and Stoffel, 1992) (see Fig. 10A). DM20, a smaller PLP isoform that is generated by alternative mRNA splicing (Nave et al., 1987), lacks residues 116–150 but shares the overall “four-helix-span” topology.
Fig. 10.
Hypothetical model of wild-type PLP and misfolded PLP in the dysmyelinated mouse mutantjimpymsd. A, Based on the position of four highly hydrophobic stretches (transmembrane domains TM1-4), PLP is depicted as a four-helix-bundle protein within the membrane, exposing one intracellular and two extracellular loop regions (Popot et al., 1991). Two intramolecular disulfide bonds link Cys200–Cys219 and Cys183–Cys227 within the second extracellular loop (Shaw et al., 1989; Weimbs and Stoffel, 1992). In DM20, residues 116–150 are absent from the intracellular loop (shaded). The relative positions of two amino acid substitutions that cause dysmyelination in vivo are shown (Ile186 → Thr in rumpshaker; Ala242 → Val in jimpymsd). Injimpy, the last 42 amino acids, which include TM 4, are replaced by an aberrant C terminus. Also indicated are the intra- and extracellular localization of epitopes A431 and O10, respectively (the exact binding site for antibody O10 is not known). It is suggested that the O10 epitope emerges as PLP reaches a functional three-dimensional conformation, possibly after formation of disulfide bridges and/or oligomerization. B, Hypothetical model of the PLP four-helix-bundle from jimpymsd mice (top-down view on the membrane). It is suggested that the spatial demand of one extra methyl group in TM4 (Val242) prevents the necessary tight alignment of hydrophobic domains, which allows PLP to obtain its final three-dimensional conformation. Val242may also interfere with the alignment of two PLP molecules during oligomerization.
PLP and DM20 are highly conserved in evolution (for review, see Yoshida and Coleman, 1996). The identification of numerous point mutations in the PLP gene, initially in rodents and subsequently in patients with Pelizaeus-Merzbacher disease, has demonstrated that even conservative single amino acid substitutions are not tolerated (Hodes et al., 1994; Nave and Boespflug-Tanguy, 1996).
The best known mutation in the mouse PLP gene, jimpy, is predicted to alter the structure of PLP/DM20 as a result of abnormal mRNA splicing (Nave et al., 1986; Hudson et al., 1987). By cDNA analysis, jimpy-PLP lacks the C-terminal transmembrane domain TM4, and dysmyelination in jimpy mice is largely caused by oligodendrocyte death (Knapp et al., 1986; Vermeesch et al., 1990). The allelic mutation jimpymsd (Gencic and Hudson, 1990) is defined by a single conservative substitution (Ala242 → Val) located within TM4. The phenotype is similar to jimpy and includes cell death. A third mutation, rumpshaker, is defined by the single substitution Ile186 → Thr in the second extracellular loop. Inrumpshaker mice, however, CNS dysmyelination is not associated with increased oligodendrocyte death (Schneider et al., 1992).
In the present study we provide evidence that the different mutant alleles have one common effect at the protein level. By monitoring a novel PLP epitope, which is defined by the monoclonal antibody O10 and localized on the cell surface, we demonstrate that even single amino acid substitutions can lead to a conformational change of PLP. Our data provide evidence that protein misfolding causes the intracellular retention of mutant PLP and DM20, which in vivo interferes with oligodendrocyte differentiation and survival.
MATERIALS AND METHODS
Antibodies. Hybridomas secreting monoclonal antibodies of the “O-series” were generated by fusion of a nonsecreting myeloma line with splenocytes of mice immunized with homogenates from bovine corpus callosum (described in detail by Sommer and Schachner, 1981). Hybridoma cells were maintained in RPMI 1640, supplemented with 10% fetal calf serum, and subcloned twice by limiting dilution. Ig subclasses were determined by Ochterlony immunodiffusion using subclass-specific anti-mouse antibodies (Miles, Elkhart, IN). Originally, two clones of different subclasses were obtained that had the same specificity (termed O10), as evidenced by identical immunostaining pattern in tissue sections and on cultured cells and by the competition for the same cell surface epitope in double-labeling experiments (data not shown). One clone was found to belong to the IgG subclass and was used for a preliminary report (Sommer and Schachner, 1984) but is no longer available. The second clone was an IgM and was used for the experiments described in this paper.
A polyclonal antibody (A431), directed against both PLP and DM20, was generated by coupling the C-terminal hexapeptide (GRGTKF) to a carrier protein (KLH) and immunizing New Zealand rabbits. The IgG fraction was enriched by affinity chromatography using protein A-Sepharose. A rabbit polyclonal anti-peptide antibody (8410) specific for the C terminus ofjimpy-PLP (Nave et al., 1986) was kindly provided by R. Skoff (Wayne State University). For immunofluorescence analysis, fluorochrome-conjugated secondary antibodies (DTA-fluorescein-goat-anti-mouse/rat-IgM, Cy3-goat-anti-rabbit-IgG) were purchased from Dianova (Hamburg, Germany).
Western blot analysis. Myelin was prepared by standard procedures from adult mouse brains (Norton, 1974). Myelin proteins (1–10 μg) were separated by SDS-PAGE (12%) under reducing conditions and transferred to supported nitrocellulose membranes (BA-S 85, Schleicher & Schuell, Dassel, Germany) by semi-dry electroblotting. Membranes were blocked for at least 2 hr at room temperature with 5% nonfat dry milk, 0.01% gelatin, 1% BSA, and 0.02% Tween 20 in Tris-buffered saline (TBS; 50 mm Tris base, 150 mm NaCl, pH 7.4). Incubation with antibodies (1:40 for A431 and 1:10 for O10) in the same buffer was for 2 hr at room temperature. After two washes (0.1% Tween 20 in TBS), secondary horseradish peroxidase-conjugated antibodies (Amersham, Braunschweig, Germany, and Dianova, diluted 1:1000) were applied for 2 hr. Immunoreactive proteins were detected with an enhanced chemiluminescence kit (ECL, Amersham) according to the manufacturers instructions, using ECL-Hyperfilm.
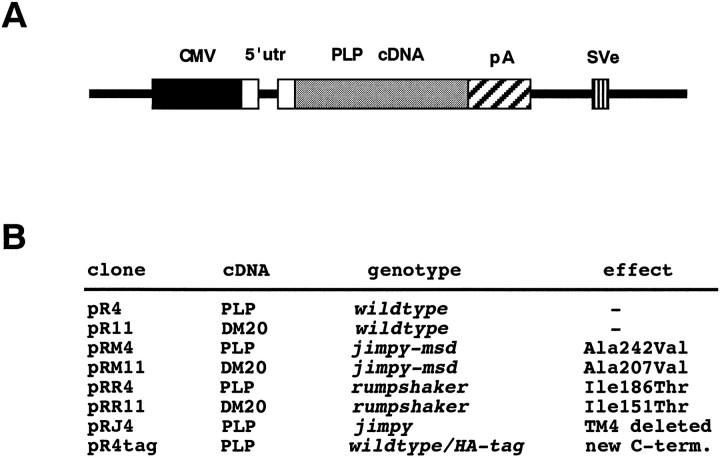
Molecular cloning. All cDNA expression constructs were derivatives of clones pC4 and pC11, which encode wild-type mouse PLP and DM20, respectively (Nave et al., 1987). cDNA inserts were excised with BamHI and PstI and ligated into the expression vector pCMV, generating clones pR4 and pR11. Expression of PLP (in pR4) and DM20 (in pR11) was under the control of the human CMV promoter. A β-globin gene-derived intron was located in the 5′ untranslated region and the SV40 polyadenylation signal in the 3′ untranslated region of each construct. The presence of the SV40 early region allowed maintenance of each plasmid at a high copy number in transfected COS-7 cells. Mutant derivatives of pR4 (listed in Fig.2B) were generated by substituting internal restriction fragments against homologous fragments from mutant cDNAs. For jimpy PLP this was a BglII–PstI fragment excised from clone pJ31 (Nave et al., 1986). pRJ4 contained PLP exons 1–3 from clone p27 of Milner et al. (1985).Jimpymsd derivatives were generated by PCR amplification of PLP and DM20 cDNA fragments from total brain cDNA of hemizygous mutant mice (kindly provided by S. Billings-Gagliardi, Worcester, MA). By using primers flanking exon 6 (primer A: 5′-CATCACCTATGCCCTGA-3′ and primer B: 5′-ATGGATCCAC-AAAGGGGAGTTTCTATGGGA-GCTC-3′), a 400 bp fragment with the mutation was obtained, digested with NcoI andSpeI, and ligated into the homologous position of pR4 and pR11. A rumpshaker clone was constructed by amplification of mutant brain cDNA, as described by Schneider et al. (1992), and ligation of mutant exon 4 into the BsrGI/SpeI sites of clone pR4 and pR11. A hemagglutinin (HA) epitope tag replacing the last six residues of wild-type PLP with the 12′ mer RGPYPYDVPDYA was generated by PCR using primer C (5′-GCGATCCTGCA-GTCAGGCGTAGTCGGGAACGTCGTAGGGGTAAGGACCTCGTC-GGCCCATGAGTTT-3′) instead of primer B. All PCR-generated fragments were confirmed by DNA sequence analysis.
Fig. 2.
PLP expression constructs. A, Schematic drawing of expression constructs in which a mouse PLP (or DM20) cDNAs are transcribed under control of the humanCMV promoter (not to scale). For efficient expression, a β-globin intron has been introduced into the 5′ untranslated region (5′utr), and the cDNA is flanked by a polyadenylation signal (pA). The SV40 early region (SVe) yields high plasmid copy number in cells that express large T antigen. B, Listing of cDNA constructs that have been generated to express PLP/DM20 wild-type and mutant isoforms in transfected COS-7 cells (for references of PLP coding sequences, see Materials and Methods).
Cell culture. COS-7 cells were maintained on untreated tissue-culture dishes (Falcon) in DMEM containing 10% fetal bovine serum. Cells were grown at 37°C in a 5% CO2 atmosphere, and medium was changed every third day. For passaging cells, confluent plates were washed once with PBS, followed by a short trypsination with 0.05% trypsin-EDTA (Sigma, St. Louis, MO). Cultures with purified rat oligodendrocytes (McCarthy and DeVellis, 1980) were kindly provided by J. Trotter (University of Heidelberg).
Immunohistology. Animals were anesthetized deeply, their brains and optic nerves were quickly removed, and cryosections (10 μm) from freshly frozen tissues were thawed onto glass coverslips. Indirect immunofluorescence was carried out as described by Schnitzer and Schachner (1981), using O10 as primary antibody and an FITC-conjugated anti-mouse IgM as secondary antibody.
DNA transfection. The day before transfection, 3 × 105 COS-7 cells were plated on a 6-cm-diameter culture dish, and medium was changed 5 hr before DNA precipitation. For transfection, 25 μl CaCl2 (2.5 m) was mixed with 10 μg DNA, and ddH2O was added to a 250 μl final volume. The calcium/DNA suspension was dispersed under standard conditions with 250 μl HEPES-buffered saline (HBS) (2× HBS: 280 mm NaCl, 50 mm HEPES, 2.8 mmNa2HPO4, pH 7.1). Precipitates were formed at room temperature (10 min) before transfection. After 5 hr, excess precipitates were removed with a PBS wash, and fresh medium was added. Approximately 24 hr after transfection, cells were plated either on untreated glass coverslips (for permeabilization) or in chamber slides (Nunc, Naperville, IL) for live staining. Chamber slides were coated with 0.01% poly-l-lysine (2 hr at 37°C) and washed twice with ddH20.
Immunostaining of transfected cells. Immunostaining was carried out 48 hr after transfection. The following steps were performed at room temperature, except for the incubation of primary antibodies (4°C). Cells grown on coverslips were washed twice in DMEM with 10 mm HEPES and were fixed for 30 min in 2% paraformaldehyde/DMEM/10 mm HEPES. After they were washed twice for 20 min in TBS (25 mm Tris, 136 mmNaCl, 2.6 mm KCl, pH 7.5), cells were permeabilized with 0.1% saponin in TBS (for 30 min). Coverslips were placed into a humidified chamber and incubated in blocking buffer (TBS containing 2% goat serum, 2% BSA, 0.02% biotin, 0.1% porcine skin gelatin) for at least 30 min. Primary antibodies were incubated overnight at 4°C. After three washes in TBS (20 min), fluorochrome-conjugated secondary antibodies were applied for at least 1 hr, followed in some experiments by nuclear counterstaining with DAPI (50 ng/ml in TBS). Finally, cells were washed in TBS (3× 20 min), rinsed in distilled water, and mounted in Aqua-Poly/Mount (Polysciences, Warrington, PA) on glass slides. Antibodies were diluted as follows: O10 hybridoma supernatant, 1:20; A431, 1:1000; dichlorotriazinylamino fluorescein-goat-anti-mouse/rat-IgM, 1:100; and Cy3-goat-anti-rabbit-IgG, 1:1000.
Cells grown in chamber slides for live staining were washed with Dulbecco’s PBS (DPBS; 0.7 mm CaCl2, 2.6 mm KCl, 1.5 mm KH2PO4, 0.5 mm MgCl2, 136 mm NaCl, 8.1 mm Na2HPO4), before nonspecific binding sites were blocked with DPBS containing 3% goat serum. Cells were incubated with primary antibodies (diluted in the same buffer) for 30–60 min. After two washes in DPBS, cells were fixed in 2% paraformaldehyde (in DMEM) for 10 min. After two additional washes in DPBS, fluorochrome-conjugated secondary antibodies (diluted in DPBS with 3% goat serum) were applied for at least 30 min. Finally, cells were washed twice in DPBS, rinsed in ddH2O, and mounted.
Immunofluorescence analysis was carried out with a Zeiss Axiophot epifluorescence microscope. Results were documented on Kodak Ektachrome film (400 ASA) and by processing of digitalized images (Nikon/Macintosh).
Immunoprecipitation. Twenty-four hours after transfection, culture medium of COS-7 cells was exchanged for a medium containing 25 μCi/ml 35S-methionine and 35S-cysteine (Amersham), followed by a 4 hr incubation. Cells were washed with PBS, and a crude membrane preparation was obtained by cell lysis with 1% NP-40 and 0.5% sodium desoxycholate (in 150 mm NaCl, 50 mm Tris-HCl, pH 8.0) for 30 min on ice. Cellular debris was removed by centrifugation at 100,000 × g for 30 min (4°C). For immunoprecipitation, antibody A431 was added (1:100) to the supernatant and incubated under agitation at 4°C for 2 hr. Immunoprecipitates were obtained with Staphylococcus aureusprotein A coupled to Sepharose CL-4B (Sigma), and size-separated by SDS-PAGE (12%). Gels were dried, and labeled proteins were visualized with a phosphoimager (Molecular Dynamics, Sunnyvale, CA).
RESULTS
Immunolabeling of oligodendrocytes and CNS white matter
A panel of monoclonal antibodies has been obtained by the immunization of mice with purified membranes of CNS white matter. This O-series of hybridomas has been used to define successive stages of oligodendrocyte development in vitro. Studies on the expression of O-antigens have been reported elsewhere (Sommer and Schachner, 1981; Kuhlmann-Krieg et al., 1984).
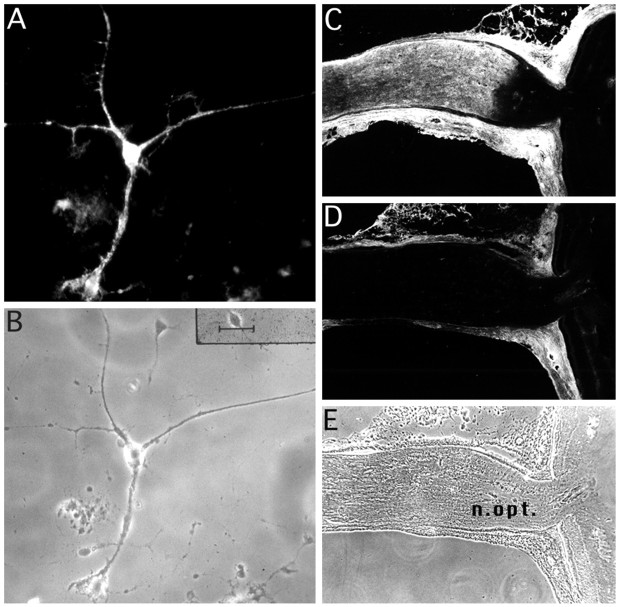
Antibody O10 belongs to the IgM subclass and marks a cell-specific epitope that appears late in the oligodendrocyte lineage (for a preliminary report, see Sommer and Schachner, 1984). Purified oligodendrocytes in culture (Fig.1A,B) were strongly immunostained with O10 at a developmental stage when MBP and other myelin-specific markers were also expressed (data not shown). Cells could be stained live and after permeabilization, indicating that the antigen is localized on the oligodendrocyte cell surface and resistant to treatment with 0.1% saponin. The O10 epitope was oligodendrocyte-specific, because no other neural cell type could be labeled, and cells of the peripheral nervous system and other cell types tested were O10-negative (Sommer and Schachner, 1984; Kuhlmann-Krieg et al., 1988). In fresh-frozen sections of the adult rodent CNS, O10 stained white matter areas, as illustrated for the mouse optic nerve (Fig. 1C–E).
Fig. 1.
Monoclonal antibody O10 labels oligodendrocytes and CNS white matter. O10, a mouse IgM, defines a developmentally regulated protein epitope that is expressed on cultured oligodendrocytes and in CNS white matter regions. A, Immunofluorescence staining of a rat oligodendrocyte cell body and its processes. B, Phase contrast. Scale bar, 40 μm.C, In adult mice, O10 stains the highly myelinated optic nerve. Note that the secondary anti-mouse IgM antibody stains non-neural mouse tissue unspecifically, which is demonstrated in the absence of the primary antibody (D). A phase-contrast image of the sectioned optic nerve is shown in E.n.opt., Optic nerve.
A cell surface epitope of PLP
The O10 epitope is highly sensitive to detergent and protease digestion (Sommer and Schachner, 1984), suggesting that the antigenic site belongs to a protein. The CNS-specific immunolabeling of O10 and the inability to identify the antigen in jimpy mice led us to consider that the antibody is associated with PLP and/or DM20.
To determine the relationship between PLP, DM20, and the O10 antigen, we sought to overexpress both myelin proteins by transfection of cells. We constructed a panel of cDNA expression vectors, including several mutant derivatives, as summarized in Figure 2. Using CaPO4-mediated DNA transfection and indirect immunofluorescence (Gow et al., 1994a; Sinoway et al., 1994), we monitored the ectopic expression of PLP. For these and subsequent experiments, we also generated a polyclonal rabbit antibody, termed A431, directed against the C-terminal hexapeptide common to both PLP and DM20. Positive A431 staining thus indicated the synthesis of full-length polypeptides.
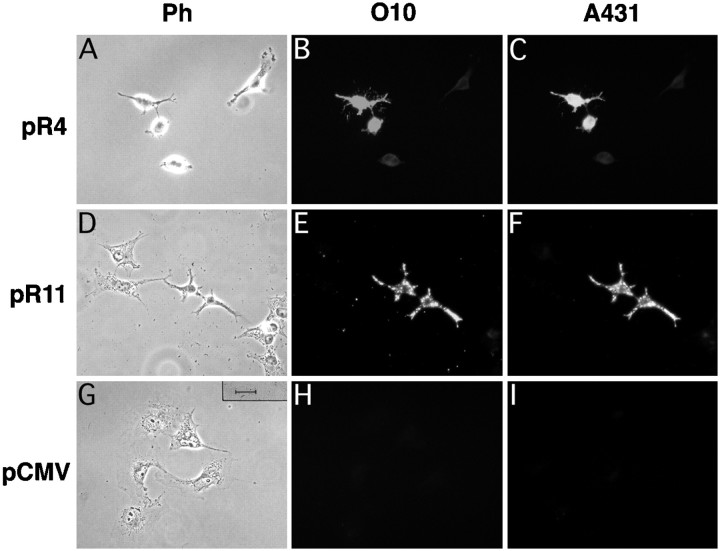
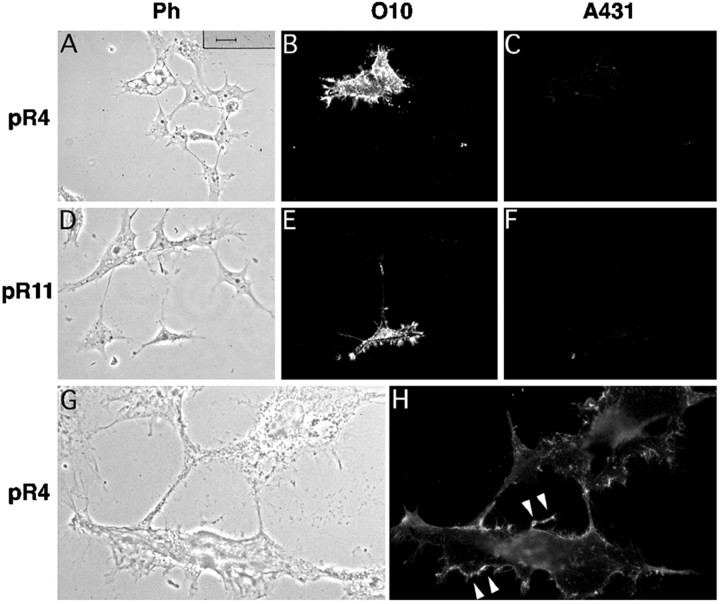
COS-7 cells, transiently transfected with vector pR4 (encoding wild-type PLP) or pR11 (encoding wild-type DM20), were fixed 48 hr after transfection, permeabilized with 0.1% saponin, and immunostained (Fig. 3). Transfected cells were strongly positive with either antibody O10 or A431. By double-immunostaining, we could show that cells expressing the O10 epitope are also positive for A431. These results provided first evidence that the O10 epitope is part of PLP and displayed by both protein isoforms.
Fig. 3.
O10 defines an epitope of myelin PLP. Cultured COS-7 cells, shown in phase contrast (A, D, G), were transiently transfected with expression constructs encoding wild-type mouse PLP (pR4), wild-type mouse DM20 (pR11), or a vector control (pCMV) and analyzed by double-immunofluorescence 48 hr later. Cells expressing PLP or DM20 were detected with a polyclonal antibody (A431) directed against the C terminus common to both isoforms (C, F). PLP/DM20-positive cells express the O10 epitope (B, E), which is absent from cells transfected with the vector control (H, I). Scale bar, 25 μm.
When antibodies O10 and A431 were applied to live cells, only the O10 epitope was labeled but not the C terminus (Fig. 4). Thus, the monoclonal antibody recognizes an extracellular domain of PLP/DM20, whereas the C terminus resides inside the cell, in agreement with staining of live oligodendrocytes and with the current topological model of PLP (Popot et al.,1991; Weimbs and Stoffel, 1992) (see Fig.10A).
Fig. 4.
O10 marks a PLP ectodomain. COS-7 cells, as shown in phase contrast (A, D, G), were transiently transfected to express PLP (A–C, G, H) and DM20 (D–F). Staining of live cells with O10 (B, E, H) demonstrates that the O10 epitope is localized on the cell surface (arrowheads), whereas they are A431-negative (C, F), in agreement with the topological model of PLP (see Fig. 10A). Note O10 staining of the plasma membrane and filopodia at higher magnification (H). Scale bars: A–F, 25 μm;G, H, 10 μm.
The O10 epitope emerges post-translationally
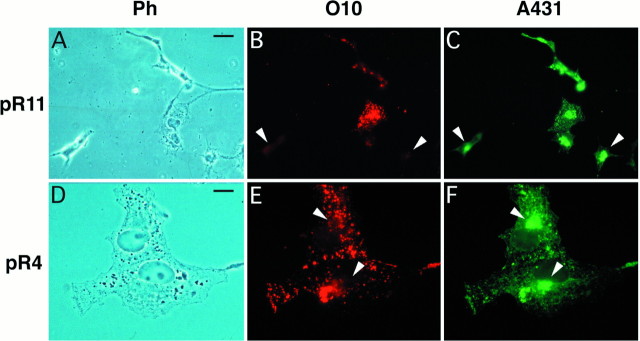
By light-microscopic inspection at higher magnification (Fig.5), COS-7 cells expressing wild-type PLP (pR4) or DM20 (pR11) displayed a noticeably different intracellular distribution of PLP/DM20 immunoreactivity, depending on which antibody was used. In general, permeabilized cells immunostained with O10 alone displayed a more vesicular pattern, whereas cells stained only with A431 showed a reticular, endoplasmic reticulum (ER)-like distribution of the antigen. Neither subcellular staining pattern, however, was exclusive for one antibody. By double-immunolabeling, the subcellular antigen distribution was generally overlapping, but in many cells the two epitopes seemed to be localized differentially, with antibody A431 staining the ER more prominently (compare Fig. 5E and 5F). In some double-labeling experiments, up to twice as many cells were scored as PLP-positive by A431 staining (as compared with O10 staining). In contrast, we never observed O10-positive cells in the absence of A431 staining. Taken together, this indicated that antibody O10 binds to a subset of PLP molecules, most likely because the O10 epitope is generated after the A431 epitope. Because antibody A431 recognizes the C terminus of full-length PLP, the O10 epitope may be specific to a sufficiently folded protein and is referred to as post-translational.
Fig. 5.
Differential localization of the O10 and A431 epitope of PLP/DM20. COS-7 cells, shown in phase contrast (A, D), were tranfected to express wild-type DM20 (A–C) or PLP (D–F) and after fixation and permeabilization were double-stained with O10 (B, E) and A431 (C, F). Immunoreactivity was strongest inside the cells and largely overlapping for both primary antibodies, but the intracellular antigen distribution was not identical. In general, more areas could be stained for the PLP/DM20 C terminus (A431) than for the O10 epitope, suggesting that the O10 antibody recognizes a subset of all full-length PLP/DM20 polypeptides. Some A431-positive cells are O10 negative (B, C). Note, at higher magnification (D–F), the absence of O10 staining in a perinuclear region, which is A431-positive (arrowheads). Scale bars:A–C, 25 μm; D–F, 10 μm.
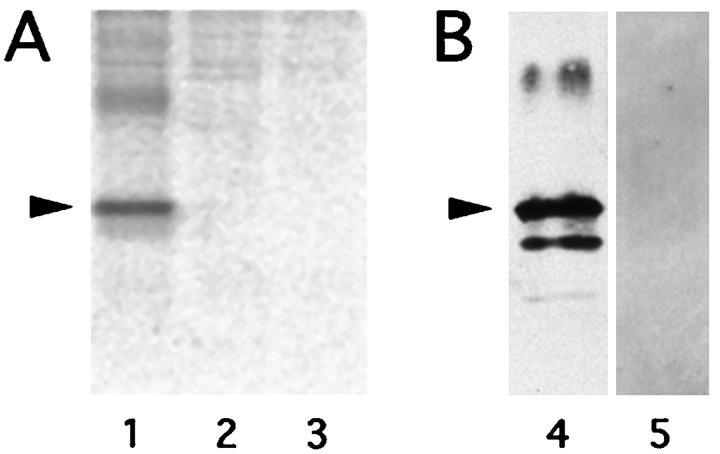
To confirm that A431, our positive control antibody, binds specifically to PLP in COS-7 cells, we performed an immunoprecipitation of35S-labeled proteins solubilized 24 hr after transfection (Fig. 6A). In these experiments, we never observed A431 binding to proteins other than PLP (or DM20; not shown), which suggests strongly that the colocalization of O10 with A431 in COS-7 cells marks PLP/DM20-specific epitopes that are distinct from each other but associated with the same molecule.
Fig. 6.
The O10 but not the A431 epitope of PLP/DM20 is denaturation-sensitive. A, The specificity of antibody A431 for PLP was demonstrated by immunoprecipitation of S35-labeled proteins from COS-7 cells transiently transfected with pR4 (encoding wild-type PLP, lane 1), or with pCMV as a vector control (lane 3). The preimmune serum does not precipitate PLP from cells transfected with pR4 (lane 2). The position of PLP with a relative mobility of 24 kDa is indicated (arrowhead). Unspecific aggregates of PLP with a lower mobility can also be seen.B, By Western blot analysis of denaturing acrylamide gels, antibody A431 (directed against the C-terminal hexapeptide of PLP and DM20) detects both PLP isoforms in 1 μg of purified CNS myelin (lane 4). The position of PLP is indicated (arrowhead). DM20 has a relative mobility of 20 kDa and is visible below. Under these conditions, PLP and DM20 cannot be detected with the antibody O10 in 10 μg of protein (lane 5), suggesting that the O10 epitope is denaturation-sensitive.
PLP is an acylated membrane protein but is not known to be glycosylated (Weimbs and Stoffel, 1992; Lees and Brostoff, 1984). We investigated the possibility that monoclonal antibody O10 binds to a short, previously unrecognized glycosyl chain, reasoning that carbohydrate epitopes are generally stable in denaturing gels. By Western blot analysis of detergent-solubilized, purified myelin (in which PLP is abundant), antibody A431 detected both PLP and DM20 as expected; however, we never observed any PLP or DM20 staining with antibody O10 (Fig. 6B). This suggests that epitope O10, which appears post-translational in COS-7 cells, is indeed linked to the three-dimensional structure of the polypeptide chain itself, i.e., to a protein conformation that is lost in denaturing gels.
The O10 epitope is not detectable in PLP/DM20 fromjimpymsd mice
Dysmyelination in the jimpymsd mouse has been associated with a single amino acid substitution in PLP (Ala242 → Val). It remains unclear, however, why this conservative change, localized within TM 4, interferes with PLP function. We have hypothesized that a substitution at this position interferes with the alignment of helices in the four-helix-bundle structure, causing PLP misfolding that can be monitored with the help of O10.
PLP and DM20 cDNA vectors were constructed that harbor thejimpymsd substitution (Fig.2B) and were expressed individually in COS-7 cells. By staining of live cells, neither antibody O10 nor A431 was able to detect jimpymsd PLP and DM20. Lack of A431 staining was in agreement with the topological model of PLP (Popot et al., 1991) (see Fig. 10A). To determine whether the absence of O10 from live cells was attributable to an impairment of intracellular transport or to the loss of the O10 epitope itself, cells were permeabilized before immunostaining. All mutant PLP and DM20 transfectants were strongly labeled with antibody A431, but immunoreactivity was largely restricted to the ER and was highest in the perinuclear region (in a pattern rarely seen when wild-type PLP was expressed). None of these A431-positive cells could be stained with O10 (Fig. 7). Thus, the jimpymsdmutation alters the overall structure of PLP/DM20, which can be monitored by the loss of the O10 epitope at the luminal side of the ER membrane. This abnormal three-dimensional structure of mutant PLP (referred to as misfolding) is the most likely cause of its retention inside the transfected cell (Fig. 7C,F).
Fig. 7.
PLP and DM20 fromjimpymsd mice lack the O10 epitope. COS-7 cells, shown in phase contrast (A, D), were transiently tranfected to express DM20 (A–C) or PLP (D–F) from jimpymsdmice and double-stained after permeabilization with O10 (B, E) and A431 (C, F). In these experiments, O10 immunoreactivity was never observed, although the mutant proteins were well detectable. Absence of the O10 epitope provides evidence that the substitution Ala242 → Val alters the three-dimensional structure of PLP and DM20. Note that both PLP isoforms are largely retained in the ER (C, F). Scale bars: A–C, 25 μm: D–F, 10 μm.
The O10 epitope is not detectable in rumpshaker andjimpy mice
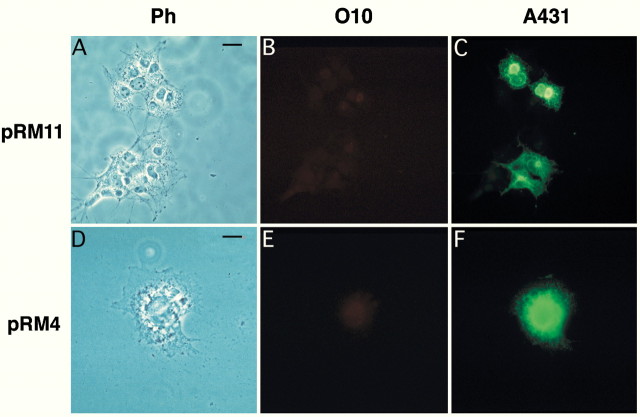
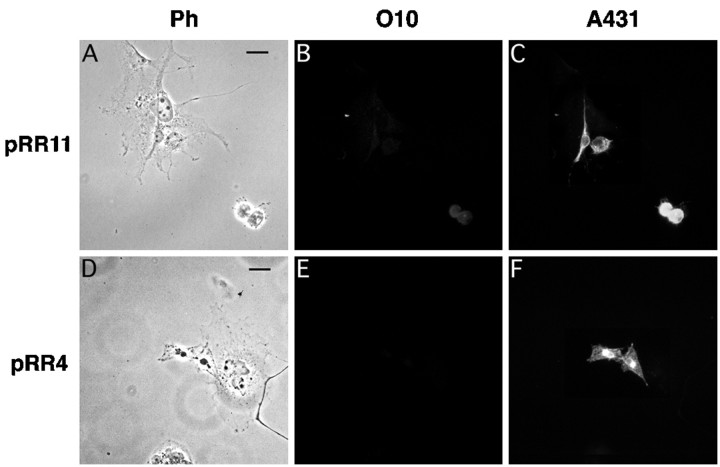
To investigate whether the effect of thejimpymsd point mutation on the formation of the O10 epitope is shared by other mutant alleles, we performed the same analysis with PLP and DM20 derived from rumpshaker andjimpy mice. PLP and DM20 cDNAs harboring therumpshaker mutation (Ile186 → Thr) were expressed in COS-7 cells, but the encoded proteins could not be stained with O10 (Fig. 8). Again, positive staining with A431 demonstrated that the full-length rumpshaker proteins were present in transfected cells (Fig. 8C,F). Also by live staining, rumpshaker PLP or DM20 expressing COS-7 cells was O10-negative (data not shown). It is of interest thatrumpshaker DM20 stains weakly O10-positive when expressed in BHK cells (C. Thompson and I. Griffiths, unpublished observations), but we note that rumpshaker oligodendrocytes in primary culture are almost all O10-negative (Fanarraga et al., 1993).
Fig. 8.
PLP and DM20 from rumpshaker mice lack the O10 epitope. Clones pRR11 (encoding rumpshakerDM20) and pRR4 (rumpshaker PLP) were overexpressed in COS-7 cells, as shown by phase contrast (A, D). When double-stained 48 hr after transfection, permeabilized cells were A431-positive (C, F) but O10-negative (B, E). A faint signal in B stems from the brightly stained cell in C and was not observed when only antibody O10 was used (not shown). Scale bar, 25 μm.
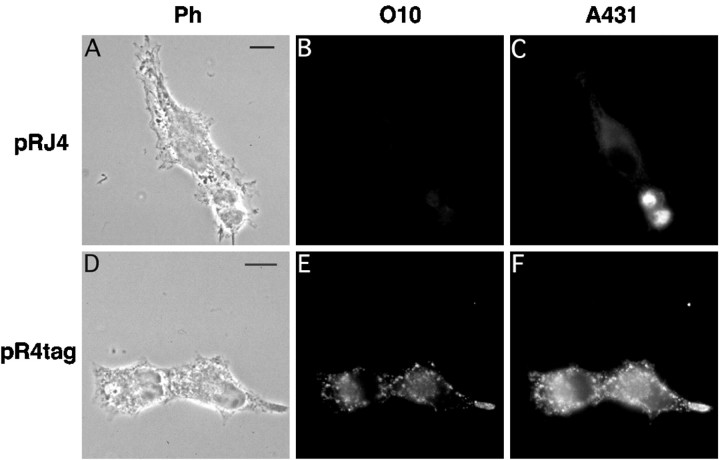
As expected, the severely truncated jimpy PLP was also O10-negative (Fig. 9A–C). In this experiment, A431 could not serve to control for protein synthesis because jimpy PLP contains an aberrant C terminus. Instead, we used a polyclonal antiserum (kindly provided by R. Skoff) directed against the jimpy-specific C terminus. Jimpy PLP protein was clearly expressed and retained inside the cell.
Fig. 9.
PLP with a C-terminal HA epitope tag, but notjimpy-PLP, displays the O10 epitope. COS-7 cells, as shown by phase contrast (A, D), were transiently transfected to express either wild-type PLP modified in the last six positions with the HA epitope tag (D–F) orjimpy PLP with an aberrant C terminus that deletes TM 4 (A–C). The epitope-tagged PLP was stained with both O10 (E) and a specific anti-HA antibody (F) and was therefore presumably in a wild-type conformation. In contrast, jimpy PLP was O10-negative (B) but was visualized with the jimpyPLP-specific antibody (C). Scale bar, 10 μm.
In contrast, when wild-type PLP was modified by replacing the normal C-terminal hexapeptide with the HA epitope tag (mimicking a “mutation” that has not been associated with a disease), the expressed protein could be stained in COS-7 cells with antibody O10, and it showed the same subcellular distribution as wild-type PLP (Fig.9D–F). Epitope-tagged PLP was also detected on the cell surface after live staining with O10 (data not shown).
Taken together, mutations of the mouse PLP/DM20 gene that cause dysmyelination in vivo also cause a defect in the three-dimensional structure of PLP, which can be visualized directly by a disappearance of the O10 epitope. Thus, O10 serves as a genetic marker for the functional expression of PLP and DM20 at the cell surface.
DISCUSSION
To elucidate the role of cell surface molecules in oligodendrocyte differentiation, monoclonal antibodies were produced that recognize antigens on oligodendrocytes and in myelin (Schachner et al., 1981;Sommer and Schachner, 1981; Kuhlmann-Krieg et al., 1988). One antigen, O10, had the characteristics of a protein, although it was not biochemically identified at the time. In the present study, we have provided molecular–genetic evidence that the antibody O10, which stains CNS white matter and live oligodendrocytes in culture, is directed against an ectodomain of myelin PLP. O10 recognizes both alternatively spliced isoforms PLP and DM20, and it binds a protein epitope that seems to emerge post-translationally.
The only known post-translational modification of PLP/DM20 (in addition to the formation of two disulfide bridges) is the intracellular acylation of cysteine residues (Bizzozero et al., 1990;Weimbs and Stoffel, 1992). Thus, the O10 epitope on the cell surface is most likely part of the protein backbone itself. We like to speculate that the O10 epitope emerges after proper folding, i.e., when the correct disulfide bridges have formed (schematically depicted in Fig.10A) or an oligomeric structure has formed.
PLP/DM20 constitutes the major protein of CNS myelin, and mutations of its gene underlie dysmyelination in mouse and man. The molecular–genetic basis and the clinical features have been documented (for review, see Duncan, 1995; Nave, 1995; Nave and Boespflug-Tanguy, 1996); however, little has been learned at the level of the affected protein. In particular, it has been puzzling that seemingly conservative point mutations, predicted to cause subtle structural changes, are phenotypically indistinguishable from mutations that severely alter the topology of this protein.
We have used O10 as a tool to address the question of a mutation-induced misfolding of PLP and DM20 and have shown that the antibody O10 distinguishes between wild-type PLP/DM20 and mutant isoforms. Two amino acid substitutions, Ile186 → Thr inrumpshaker and Ala242 → Val injimpymsd, are separated by 56 residues in the primary sequence, and both affect formation of the O10 epitope, even in heterologous cells. Moreover, one of these residues (Val242in jimpymsd mice) is localized within a membrane-spanning domain to which the antibody cannot bind. Staining of live cells has demonstrated that in wild-type PLP the O10 epitope is exposed at the cell surface, which is topologically equivalent to the luminal surface of the ER and Golgi where the mutant PLP isoforms are localized. Thus, a complete loss of the O10 epitope from cells expressing jimpymsd PLP demonstrates abnormal folding of PLP, and this conformational change can be picked up at some distance from Val242. Becausejimpymsd is the most conservative of more than 30 identified amino acid substitutions in mouse and man (for review, see Hodes et al., 1994; Nave and Boespflug-Tanguy, 1996), this suggests that the three-dimensional structure of PLP/DM20 is easily perturbed. O10 may provide a first tool to differentiate between functional and nonfunctional PLP at the protein level.
The finding of protein misfolding provides a rationale for the intracellular transportation arrest (Hurtley and Helenius, 1989;Hammond and Helenius, 1994). PLP from jimpymsdmice fails to exit the ER at the normal rate and is not expressed at the cell surface when visualized with A431. A colocalization of PLP with the ER marker BiP has been documented by Gow et al. (1994b) and was confirmed in the present study (data not shown). Injimpy oligodendrocytes, a similar mechanism may explain why PLP fails to reach the myelin compartment and is then difficult to detect biochemically (Kerner and Carson, 1984; Yanasigawa and Quarles, 1986; Benjamins et al., 1994; Fannon et al., 1994).
When expressed in COS-7 cells, PLP forms biochemically stable complexes (M. Jung and K.-A. Nave, unpublished observations). It is possible that homophilic protein interactions are required for PLP to exit the ER and that misfolding prevents this oligomerization step. In fact, we cannot formally exclude the possibility that the O10 epitope itself is generated as PLP (or DM20) forms oligomers.
We were unable to detect the O10 epitope on rumpshakerPLP/DM20, expressed in COS-7 cells under conditions that allowed us to label the wild-type proteins. Interestingly, rumpshaker mice have a low but significant amount of PLP and DM20 incorporated into CNS myelin (Mitchell et al., 1992; Schneider et al., 1992), in marked contrast to jimpy and jimpymsd mice in which PLP is barely detectable (Kerner and Carson, 1984; Gardinier and Macklin, 1988). Fanarraga et al. (1993) used O10 as a biochemically undefined differentiation marker and found that rumpshakeroligodendrocytes in culture are nearly all O10-negative, which is in agreement with our data on O10 expression in transfected COS-7 cells. The developmental defect of rumpshaker oligodendrocytes (Schneider et al., 1992) thus correlates with a near-complete loss of the O10 epitope and a reduced incorporation of PLP/DM20 into myelin. Taken together, this suggests that the O10 epitope provides a more stringent assay for functional PLP/DM20 expression than the immunolocalization of these proteins in myelin.
Theoretically, misfolding can occur when PLP is arrested in an “immature” conformation. For example, one or both disulfide bonds, which bridge Cys200–Cys219 and Cys183–Cys227 in the second extracellular loop (Shaw et al., 1989; Weimbs and Stoffel, 1992), may simply fail to form. Such a model assumes that four transmembrane domains of PLP must tightly associate in the plane of the membrane before the correct disulfide bonds can be formed. Amino acid substitutions that affect the width of any one transmembrane helix (as injimpymsd and related mutations) can potentially prevent the necessary close alignment of helices, as shown schematically in Figure 10B.
PLP misfolding may also occur if alternative disulfide bonds form, a situation that could be favored by an incorrect orientation of the transmembrane helices or the extracellular loop regions. Theoretically, aberrant disulfide bonds could be intra- or intermolecular, and they could involve other membrane proteins synthesized in the ER. The latter is speculative but intriguing, because it offers an explanation of why misfolded PLP is “toxic” in vivo. The survival of newly generated oligodendrocytes depends, for example, on the receptors for survival factors, which are made in the ER. Misfolded PLP could negatively affect the rate at which these receptors are transported to the cell surface. One would predict that oligodendrocytes that compete for survival factors (Barres et al., 1992) are more susceptible to a negative effect of misfolded PLP than immortalized cells. Indeed, transfected COS-7 cells are remarkably resistant to the intracellular accumulation of misfolded proteins, in contrast tojimpy oligodendrocytes, which die even before PLP becomes immunodetectable (Vermeesch et al., 1990).
Transgenic experiments have shown that the X chromosome-linked PLP mutations act in a “dominant–negative” fashion, because the naturally occurring mutant mice could not be fully rescued with a wild-type PLP transgene (Nadon et al., 1994; Schneider et al., 1995). Moreover, the wild-type PLP gene itself turns into a bona fide disease gene when expressed above a critical threshold level in transgenic mice (Kagawa et al., 1994; Readhead et al., 1994). Thus, the dysmyelinated phenotype results from a combination of loss- and gain-of-function effects. More recently, gene targeting experiments have shown that the dysmyelination is moderate when expression of a mutant allele is downregulated (Boison and Stoffel, 1994; Boison et al., 1995) and that functional myelin is made in a PLP null allele (M. Klugmann, M. Schwab, I. Griffiths, and K. A. Nave, unpublished observations). Taken together, the PLP/DM20 protein product contributes to the development of a dysmyelinated phenotype. The finding that mutant PLP/DM20 genes, which cause dysmyelination in vivo, encode polypeptides that misfold demonstrates directly the effect of genomic changes at the protein level. They are in good agreement with an earlier observation of Roussel et al. (1987) that PLP is mislocalized in the CNS ofjimpy mice. PLP misfolding and the subcellular consequences of impaired protein trafficking may provide an explanation for the fact, discussed above, that dysmyelination requires PLP gene expression.
In transiently transfected COS-7 cells, mutant and wild-type PLP cDNAs have been “overexpressed” (with the help of a strong viral promoter and a high plasmid copy number), but the degree of overexpression, e.g., relative to PLP in oligodendrocytes, is difficult to estimate. It is possible that chaperones or other ER proteins that assist in normal membrane protein folding become rate-limiting, leaving some wild-type PLP misfolded and retained inside the cell. It has been calculated that polytopic membrane proteins, e.g., the cystic fibrosis transmembrane conductance regulator, are misfolded (up to 80%) in the course of their normal biosynthesis (Ward and Kopito, 1994). It is likewise possible that some PLP misfolding occurs in normal oligodendrocytes, which is tolerated at the normal expression level; however, under conditions of overexpression, e.g., when the PLP/DM20 gene dosage is increased in transgenic mice (Kagawa et al., 1994; Readhead et al., 1994) or in patients with a PLP gene duplication, the degree of PLP misfolding may reach a critical threshold level and affect oligodendrocyte function. Again, immortalized cells, such as COS-7, may be more resistant to the adverse side effects of misfolded PLP. Although still speculative, such mechanisms may provide an explanation of why point mutations and increased gene dosage can result in a similar cellular pathology. New molecular probes such as the O10 antibody for PLP/DM20 will allow these questions to be answered.
Footnotes
This work was funded by a grant from the Deutsche Forschungsgemeinschaft (SFB317). We thank U. Bartsch and S. Druffel-Augustin for help with immunocytochemistry, H. Krischke for DNA sequencing, and A. Schneider for molecular cloning. Purified oligodendrocytes were kindly provided by J. Trotter. We also thank S. Billings-Gagliardi for jimpymsd mice and R. Skoff for a jimpy PLP-specific antibody.
Correspondence should be addressed to Dr. Klaus-Armin Nave, Zentrum für Molekulare Biologie (ZMBH), Universität Heidelberg, Im Neuenheimer Feld 282, D-69120 Heidelberg, Germany.
REFERENCES
- 1.Barres BA, Hart IK, Coles HS, Burne JF, Voyvodic JT, Richardson WD, Raff MC. Cell death and control of cell survival in the oligodendrocyte lineage. Cell. 1992;70:31–46. doi: 10.1016/0092-8674(92)90531-g. [DOI] [PubMed] [Google Scholar]
- 2.Benjamins JA, Studzinski DM, Skoff RP. Analysis of myelin proteolipid protein and F0 ATPase subunit 9 in normal and jimpy CNS. Neurochem Res. 1994;19:1013–1022. doi: 10.1007/BF00968711. [DOI] [PubMed] [Google Scholar]
- 3.Bizzozero OA, Good LK, Evans JH. Cysteine-108 is an acylation site in myelin proteolipid protein. Biochem Biophys Res Commun. 1990;170:375–382. doi: 10.1016/0006-291x(90)91284-y. [DOI] [PubMed] [Google Scholar]
- 4.Boison D, Stoffel W. Disruption of the compacted myelin sheath of axons of the central nervous system in proteolipid protein-deficient mice. Proc Natl Acad Sci USA. 1994;91:11709–11713. doi: 10.1073/pnas.91.24.11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boison D, Büssow H, D’Urso D, Müller HW, Stoffel W. Adhesive properties of proteolipid protein are responsible for the compaction of CNS myelin sheaths. J Neurosci. 1995;15:5502–5513. doi: 10.1523/JNEUROSCI.15-08-05502.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duncan ID. Inherited disorders of myelination of the central nervous system. In: Kettenmann H, Ransom B, editors. Neuroglial cells. Oxford UP; New York: 1995. pp. 990–1009. [Google Scholar]
- 7.Fanarraga ML, Sommer IU, Griffiths IR, Montague P, Groome NP, Nave KA, Schneider A, Brophy PJ, Kennedy PGE. Oligodendrocyte development and differentiation in the rumpshaker mutation. Glia. 1993;9:146–156. doi: 10.1002/glia.440090208. [DOI] [PubMed] [Google Scholar]
- 8.Fannon AM, Mastronardi FG, Moscarello MA. Isolation and identification of proteolipid proteins in jimpy mouse brains. Neurochem Res. 1994;19:1005–1012. doi: 10.1007/BF00968710. [DOI] [PubMed] [Google Scholar]
- 9.Gardinier M, Macklin WB. Myelin proteolipid protein gene expression in jimpy and jimpymsd mice. J Neurochem. 1988;51:360–369. doi: 10.1111/j.1471-4159.1988.tb01047.x. [DOI] [PubMed] [Google Scholar]
- 10.Gencic S, Hudson L. Conservative amino acid substitution in the myelin proteolipid protein of jimpymsd mice. J Neurosci. 1990;10:117–124. doi: 10.1523/JNEUROSCI.10-01-00117.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gow A, Friedrich VL, Lazzarini RA. Intracellular transport and sorting of the oligodendrocyte transmembrane proteolipid protein. J Neurosci Res. 1994a;37:563–573. doi: 10.1002/jnr.490370503. [DOI] [PubMed] [Google Scholar]
- 12.Gow A, Friedrich VL, Lazzarini RA. Many naturally occurring mutations of myelin proteolipid protein impair its intracellular transport. J Neurosci Res. 1994b;37:574–583. doi: 10.1002/jnr.490370504. [DOI] [PubMed] [Google Scholar]
- 13.Hammond C, Helenius A. Quality control in the secretory pathway: retention of a misfolded viral membrane glycoprotein involves cycling between the ER, intermediate compartment, and Golgi apparatus. J Cell Biol. 1994;126:41–52. doi: 10.1083/jcb.126.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodes ME, Pratt VM, Dlouhy SR. Genetics of Pelizaeus-Merzbacher disease. Dev Neurosci. 1994;15:383–394. doi: 10.1159/000111361. [DOI] [PubMed] [Google Scholar]
- 15.Hudson LD, Berndt JA, Puckett C, Kozak CA, Lazzarini RA. Aberrant splicing of proteolipid protein mRNA in the dysmyelinating jimpy mutant mouse. Proc Natl Acad Sci USA. 1987;84:1454–1458. doi: 10.1073/pnas.84.5.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurtley SM, Helenius A. Protein oligomerization in the endoplasmic reticulum. Annu Rev Cell Biol. 1989;5:277–307. doi: 10.1146/annurev.cb.05.110189.001425. [DOI] [PubMed] [Google Scholar]
- 17.Kagawa T, Ikenaka K, Inoue Y, Kuriyama S, Tsujii T, Nakajima J, Nakao J, Aruga J, Okano H, Mikoshiba K. Glial cell degeneration and hypomyelination caused by overexpression of the myelin proteolipid protein gene. Neuron. 1994;13:427–442. doi: 10.1016/0896-6273(94)90358-1. [DOI] [PubMed] [Google Scholar]
- 18.Kerner AL, Carson JH. Effect of the jimpy mutation on expression of myelin proteins in heterozygous and hemizygous mouse brain. J Neurochem. 1984;43:1706–1715. doi: 10.1111/j.1471-4159.1984.tb06099.x. [DOI] [PubMed] [Google Scholar]
- 19.Knapp PE, Skoff RP, Redstone DW. Oligodendroglial cell death in jimpy mice: an explanation for the myelin deficit. J Neurosci. 1986;6:2813–2822. doi: 10.1523/JNEUROSCI.06-10-02813.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhlmann-Krieg S, Sommer I, Schachner M. Ultrastructural features of cultured oligodendrocytes expressing stage-specific cell-surface antigens. Brain Res. 1988;467:269–280. doi: 10.1016/0165-3806(88)90031-4. [DOI] [PubMed] [Google Scholar]
- 21.Lees M, Brostoff SL. Proteins of myelin. In: Morell P, editor. Myelin, Vol 2. Plenum; New York: 1984. pp. 197–224. [Google Scholar]
- 22.Lemke G. The molecular genetics of myelination: an update. Glia. 1993;7:263–271. doi: 10.1002/glia.440070402. [DOI] [PubMed] [Google Scholar]
- 23.McCarthy KD, DeVellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mikoshiba K, Okano H, Tamura T, Ikenaka K. Structure and function of myelin protein genes. Annu Rev Neurosci. 1991;14:201–217. doi: 10.1146/annurev.ne.14.030191.001221. [DOI] [PubMed] [Google Scholar]
- 25.Milner RJ, Lai C, Nave KA, Lenoir D, Ogata J, Sutcliffe JG. Nucleotide sequences of two mRNAs for rat brain myelin proteolipid protein. Cell. 1985;42:931–939. doi: 10.1016/0092-8674(85)90289-2. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell LS, Gillespie SC, McAllister F, Fanarraga ML, Kirkham D, Kelly B, Brophy PJ, Griffiths IR, Montague P, Kennedy P. Developmental expression of major myelin protein genes in the CNS of X-linked hypomyelinating mutant rumpshaker. J Neurosci Res. 1992;33:205–217. doi: 10.1002/jnr.490330204. [DOI] [PubMed] [Google Scholar]
- 27.Nadon N, Arnheiter H, Hudson LD. A combination of PLP and DM20 transgenes promotes partial myelination in the jimpy mouse. J Neurochem. 1994;63:822–833. doi: 10.1046/j.1471-4159.1994.63030822.x. [DOI] [PubMed] [Google Scholar]
- 28.Nave KA. Neurological mouse mutants: a molecular genetic analysis of myelin proteins. In: Kettenmann H, Ransom B, editors. Neuroglial cells. Oxford UP; New York: 1995. pp. 571–586. [Google Scholar]
- 29.Nave KA, Boespflug-Tanguy O. X-linked developmental defects of myelination: from mouse mutants to human genetic diseases. Neuroscientist. 1996;2:33–43. [Google Scholar]
- 30.Nave KA, Milner RJ. Proteolipid proteins: structure and genetic expression in normal and myelin-deficient mutant mice. CRC Crit Rev Neurobiol. 1989;5:65–91. [PubMed] [Google Scholar]
- 31.Nave KA, Lai C, Bloom FE, Milner RJ. Jimpy mutant mouse: a 74-base deletion in the mRNA for myelin proteolipid protein and evidence for a primary defect in RNA splicing. Proc Natl Acad Sci USA. 1986;83:9264–9268. doi: 10.1073/pnas.83.23.9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nave KA, Lai C, Bloom FE, Milner RJ. Splice site selection in the proteolipid protein (PLP) gene transcript and primary structure of the DM-20 protein of central nervous system myelin. Proc Natl Acad Sci USA. 1987;84:5665–5669. doi: 10.1073/pnas.84.16.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Norton WT. Isolation of myelin from nerve tissue. In: Fleischer S, Packer L, editors. Methods in enzymology, Vol 31. Academic; New York: 1974. pp. 435–444. [DOI] [PubMed] [Google Scholar]
- 34.Pfeiffer SE, Warrington A, Bansal R. The oligodendrocyte and its many cellular processes. Trends Cell Biol. 1993;3:191–197. doi: 10.1016/0962-8924(93)90213-k. [DOI] [PubMed] [Google Scholar]
- 35.Popot JL, Pham-Dinh D, Dautigny A. Major myelin proteolipid: the 4-α-helix topology. J Membr Biol. 1991;120:233–246. doi: 10.1007/BF01868534. [DOI] [PubMed] [Google Scholar]
- 36.Readhead C, Schneider A, Griffiths I, Nave KA. Premature arrest of myelination in transgenic mice with increased proteolipid protein gene dosage. Neuron. 1994;12:583–595. doi: 10.1016/0896-6273(94)90214-3. [DOI] [PubMed] [Google Scholar]
- 37.Roussell G, Neskovic NM, Trifilieff E, Artault JC, Nussbaum JC. Arrest of proteolipid transport through the Golgi apparatus of jimpy brain. J Neurocytol. 1987;16:195–204. doi: 10.1007/BF01795303. [DOI] [PubMed] [Google Scholar]
- 38.Schachner M, Kim SU, Zehnle R. Developmental expression in central and peripheral nervous system of oligodendrocyte cell surface antigens (O-antigens) recognized by monoclonal antibodies. Dev Biol. 1981;83:328–338. doi: 10.1016/0012-1606(81)90478-4. [DOI] [PubMed] [Google Scholar]
- 39.Schneider A, Montague P, Griffiths IR, Fanarraga M, Kennedy P, Brophy P, Nave KA. Uncoupling of hypomyelination and glial cell death by a mutation in the proteolipid protein gene. Nature. 1992;358:758–761. doi: 10.1038/358758a0. [DOI] [PubMed] [Google Scholar]
- 40.Schneider A, Readhead C, Griffiths I, Nave KA. Dominant-negative action of the jimpy mutation in mice complemented with an autosomal transgene for myelin proteolipid protein. Proc Natl Acad Sci USA. 1995;92:4447–4451. doi: 10.1073/pnas.92.10.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schnitzer J, Schachner M. Expression of Thy -1, H-2, and NS-4 cell surface antigens and tetanus toxin receptors in early postnatal and adult mouse cerebellum. J Neuroimmunol. 1981;1:429–456. doi: 10.1016/0165-5728(81)90022-9. [DOI] [PubMed] [Google Scholar]
- 42.Shaw SY, Laursen RA, Lees MB. Identification of thiol groups and a disulfide crosslink site in bovine myelin proteolipid protein. FEBS Lett. 1989;250:306–310. doi: 10.1016/0014-5793(89)80744-6. [DOI] [PubMed] [Google Scholar]
- 43.Sinoway MP, Kitagawa K, Timsit S, Hashim GA, Colman DR. Proteolipid protein interactions in transfectants: implications for myelin assembly. J Neurosci Res. 1994;37:551–562. doi: 10.1002/jnr.490370502. [DOI] [PubMed] [Google Scholar]
- 44.Snipes GJ, Suter U, Shooter E. The genetics of myelin. Curr Opin Neurobiol. 1993;3:694–702. doi: 10.1016/0959-4388(93)90140-t. [DOI] [PubMed] [Google Scholar]
- 45.Sommer I, Schachner M. Monoclonal antibodies (O1 to O4) to oligodendrocyte surfaces: an immunological study in the central nervous system. Dev Biol. 1981;83:311–327. doi: 10.1016/0012-1606(81)90477-2. [DOI] [PubMed] [Google Scholar]
- 46.Sommer I, Schachner M. Stage-specific antigens on oligodendrocyte cell surfaces. In: Dupret AM, Kato AC, Weber M, editors. The role of cell interactions in early neurogenesis. Plenum; New York: 1984. pp. 201–205. [Google Scholar]
- 47.Stoffel W, Hillen H, Gierseifen H. Structure and molecular ar- rangement of proteolipid protein of central nervous system myelin. Proc Natl Acad Sci USA. 1984;81:5012–5016. doi: 10.1073/pnas.81.16.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vermeesch MK, Knapp PE, Skoff RP, Studzinski DM, Benjamins JA. Death of individual oligodendrocytes in jimpy brain precedes expression of proteolipid protein. Dev Neurosci. 1990;12:303–315. doi: 10.1159/000111859. [DOI] [PubMed] [Google Scholar]
- 49.Ward CL, Kopito RR. Intracellular turnover of cystic fibrosis transmembrane conductance regulator: inefficient processing and rapid degradation of wild-type and mutant proteins. J Biol Chem. 1994;269:25710–25718. [PubMed] [Google Scholar]
- 50.Weimbs T, Stoffel W. Proteolipid protein (PLP) of CNS myelin: positions of free, disulfide-bonded, and fatty acid thioester-linked cysteine residues and implications for the membrane topology of PLP. Biochemistry. 1992;31:12289–12296. doi: 10.1021/bi00164a002. [DOI] [PubMed] [Google Scholar]
- 51.Yanagisawa K, Quarles RH. Jimpy mice: quantitation of myelin-associated glycoprotein and other proteins. J Neurochem. 1986;47:322–325. doi: 10.1111/j.1471-4159.1986.tb02866.x. [DOI] [PubMed] [Google Scholar]
- 52.Yoshida M, Colman DR. Parallel evolution and coexpression of the proteolipid proteins and protein zero in vertebrate myelin. Neuron. 1996;16:1115–1126. doi: 10.1016/s0896-6273(00)80138-5. [DOI] [PubMed] [Google Scholar]