Abstract
Sorting of RNAs to specific subcellular loci occurs in diverse settings from fly oocytes to mammalian neurons. Using the membrane-permeable nucleic acid stain SYTO 14, we directly visualized the translocation of endogenous RNA in living cells. Labeled RNA was distributed nonrandomly as discrete granules in neuronal processes. The labeled granules colocalized with poly(A+) mRNA, with the 60S ribosomal subunit, and with elongation factor 1α, suggesting that granules represent a translational unit. A subset of labeled granules colocalized with β-actin mRNA. Correlative light and electron microscopy indicated that the fluorescent granules corresponded to clusters of ribosomes at the ultrastructural level. Poststaining of sections with heavy metals confirmed the presence of ribosomes within these granules. In living neurons, a subpopulation of RNA granules was motile during the observation period. They moved at an average rate of 0.1 μm/sec. In young cultures their movements were exclusively anterograde, but after 7 d in culture, one-half of the motile granules moved in the retrograde direction. Granules in neurites were delocalized after treatment with microtubule-disrupting drugs. These results raise the possibility of a cellular trafficking system for the targeting of RNA in neurons.
Keywords: RNA granules, cytoskeleton, ribosomes, β-actin mRNA, elongation factor 1α, photo-oxidation, RNA translocation
Translocation of endogenous mRNAs to specific destinations in the cytoplasm has been increasingly suspected, but not observed directly. The deduction that RNAs translocate is based on the clear segregation of specific RNAs to discrete regions in many cell types. For instance, in Drosophila oocytes, where the segregation of mRNAs is a key factor in development, localized maternal mRNAs control anterior–posterior axis formation. The bicoidmRNA, localized to the anterior pole, encodes a protein that establishes a morphogenic gradient defining the pattern of the head and thorax (Driever, 1993). At the posterior pole oskar mRNA specifies the site of formation of pole plasm, which contains the posterior determinant encoded by the nanos mRNA, required for abdominal development (Ephrussi et al., 1991; Kim-Ha et al., 1991;Lehmann and Nusslein-Volhard, 1991; Wang and Lehmann, 1991; Ephrussi and Lehmann, 1992).
Several experiments have suggested that RNAs translocate in mammalian neurons. Using high-resolution electron microscopic in situ hybridization, we found the chaoptic RNA to be distributed nonrandomly in the cytoplasm of photoreceptor neurons and to be associated with discrete patches of the rough endoplasmic reticulum (ER; Pollack et al., 1990). Cultured hippocampal neurons labeled with 3H-uridine and analyzed by autoradiography transported newly synthesized RNA into dendrites at the rate of ∼0.5 mm/d (Davis et al., 1987). Candidate-transported RNAs are the subset of RNAs found in dendrites, including microtubule-associated protein 2 (Garner et al., 1988; Bruckenstein et al., 1990; Kleiman et al., 1990), the α subunit of calcium/calmodulin-dependent protein kinase II (Burgin et al., 1990), the polymerase III transcript BC1 (Tiedge et al., 1991), and probably many others (Chicurel et al., 1993; Miyashiro et al., 1994). The delivery of membrane proteins via localized mRNAs, such as the glutamate receptor (Miyashiro et al., 1994), represents a distinct problem that requires passage through the rough ER complex. The discovery of localized RNAs fits well with the earlier observation of polyribosomes beneath postsynaptic sites at the base of dendritic spines (Steward and Levy, 1982). Translational “machinery” stationed near the synapse could engage specific mRNAs to effect local synthesis and perhaps influence synaptic function.
Recent observations suggest that mRNA is transported as part of a larger structure. Myelin basic protein mRNA, when exogenously labeled and microinjected into cultured oligodendrocytes, was observed directly to translocate (Ainger et al., 1993). The unit of translocation was a granule, a structure also observed in fibroblasts (Taneja et al., 1992). In Drosophila, microinjection of the 3′ UTR ofbicoid into the embryo resulted in the recruitment of staufen protein into large nucleoprotein particles that were translocated in a microtubule-dependent manner (Ferrandon et al., 1994). The existence of similar granules in neurons has not yet been determined.
Here we examined the spatial localization and movement of endogenous RNA in living neurons. To visualize RNA, we used the membrane-permeable dye SYTO 14, which fluoresces on binding to nucleic acids. The motility characteristics of RNA granules, their components, and ultrastructural correlates in primary neuronal cultures are shown.
MATERIALS AND METHODS
Cell culture. The method of cell culture has been described in detail by Banker (for review, see Goslin and Banker, 1991) and modified for use with cortical neurons in our laboratory (Kosik and Finch, 1987). Cerebral cortex was dissected from embryonic day 18 rats and digested with 0.25% trypsin in HBSS. Tissue was washed twice in HBSS, placed in modified Eagle’s medium with 10% fetal calf serum, and mechanically dissociated by pipetting. Neurons were plated at low density (1000 cells/cm2) on poly-l-lysine-coated (1.0 mg/ml; overnight) coverslips or glass-bottom microwells (MatTek, Ashland, MA). After 2 hr when neurons had attached to the substrate, coverslips were inverted onto a monolayer of astrocytes. Astrocytes were prepared from postnatal day 1 rat cortex by culturing dissociated cortex in modified Eagle’s medium with 10% fetal calf serum on untreated tissue culture plates. The coculture was maintained in glutamate-free modified Eagle’s medium with N2 supplements, which include transferrin (100 μg/ml), insulin (5 μg/ml), progesterone (20 nm), putrescine (100 μm), and selenium dioxide (30 nm). In addition, extra glucose (600 mg/l) and sodium pyruvate (1 mm) were used.
SYTO 14 labeling of cultures. For live-cell analysis, cells were incubated with 50 nm SYTO 14 for 10 min in modified Eagle’s medium with N2 supplements plus 10 mm HEPES at 37°C and then washed twice in media before evaluation. Under these conditions, neurons remained healthy for the 2 hr period of observation as determined by neuronal morphology, viability, and organelle movement. For cells to be fixed for immunocytochemistry, cells were incubated with 500 nm SYTO 14 for 10 min in modified Eagle’s medium with N2 supplements at 37°C and then washed twice in media before fixation.
Mitotracker labeling of cultures. To subtract out mitochondria that were labeled with SYTO 14, we colabeled cells with a mitochondrion-selective dye Mitotracker CMXROS (Molecular Probes, Eugene, OR), with an excitation and emission maximum of 579 and 599 nm at 50 nm for 15 min at 37°C.
Video microscopy. An image was taken with a Cy5 filter that detected Mitotracker-labeled objects and did not detect SYTO 14; this image was compared with one taken with a Resorufin filter (p/n 41010 with excitation and emission spectra of 570 ± 5 and 600 ± 25 nm; Chroma Technology, Brattleboro, VT) that detected both SYTO 14- and Mitotracker-labeled objects. Signals between the two images were compared with a line scan that generated signal intensity profiles for each image. Then average pixel intensities of labeled structures were compared between each image. Signal that colocalized in both the Cy5 and resorufin images were considered mitochondria. Signals that only appeared in the resorufin image were considered nonmitochondrion SYTO 14-labeled objects. A series of images was taken every 20 sec for 5 min, and movement was visualized as an animated loop with time compression. Cells were viewed with a Zeiss Axiovert 135TV microscope and 63×/1.4 objective on a stage kept at 37°C by a Thermomix BU water bath (B. Braun, Melsungen, Germany). Fluorescent light was generated by a 100 W mercury arc lamp and filtered with Chroma HiQ bandpass filters. The images were captured with a cooled CCD camera (Photometrics, Tucson, AZ) that used a 35 mm shutter and driver (Uniblitz, Rochester, NY) for 10–50 msec exposure times through a 3% neutrodensity filter. Images were processed by Metamorph 2.0 (Universal Imaging, West Chester, PA) running on an Intel 486DX2 processor and viewed on a Viewsonic 17 monitor. Images were recorded on a Panasonic Optical Disk drive.
RNase Treatment. After incubation with SYTO 14 that followed the above protocol, the cells were fixed on the microscope stage with 4% paraformaldehyde (in PBS with 5 mm MgCl2) for 15 min. After fixation, cells were permeabilized with 0.5% Triton X-100 for 10 min. An image was taken with a fluorescein filter. Cells were incubated with RNase Plus (5 Prime–3 Prime, Boulder, CO) 1:250 in Tris-buffered solution and ribonuclease T2 (Sigma, St. Louis, MO) at 1000 U/ml for 1 hr at 37°C. A second image was taken of the labeled cell, and the intensity of the two signals was compared by Metamorph 2.0. Some cells were incubated in DNase 1 (Sigma), 10 μg/ml, for 1 hr after fixation and permeabilization. Control cells incubated in buffer for 1 hr after fixation and permeabilization did not have a decrease in signal intensity.
Hybridization. Neurons were labeled with SYTO 14 that followed the above protocol and then hybridized to poly(A+) or β-actin mRNA probes, as described previously (Bassell et al., 1994b). Synthetic oligo-dT (55 nucleotides) and five oligonucleotide sequences (50 bases each) complementary to β-actin 3′ untranslated sequences were 3′ end-labeled with digoxigenin-11-dUTP (Boehringer Mannheim, Indianapolis, IN) by using terminal transferase. Oligonucleotide probes that were made complementary to β-galactosidase mRNA and oligo(dA) were used as controls. Column-purified probes then were mixed with hybridization buffer, and cells were hybridized for 2 hr at 37°C. The probes were detected with affinity-purified sheep antibody to digoxigenin conjugated to horseradish peroxidase (Boehringer Mannheim) and affinity-purified goat antibody to horseradish peroxidase conjugated to Cy5 (Jackson ImmunoResearch, West Grove, PA). Images of SYTO 14 labeling were taken with a fluorescein filter and of poly(A+) with a Cy5 filter. Colocalization was determined by Metamorph 2.0, which provides precise x and y coordinates from the signal obtained with each filter.
Immunocytochemistry. Neurons were labeled with SYTO 14 that followed the above protocol and then were fixed and permeabilized as described above. Cells were incubated with either a rabbit polyclonal antibody to Artemia EF1α provided by Wim Moller (Rijks University, Leiden, The Netherlands) or a rabbit polyclonal antibody to rat 60S subunit provided by John Hesketh (Rowett Research Institute, Aberdeen, UK) and detected with a secondary antibody conjugated to Cy5. Immunoblot analysis showed the antibodies to recognize five major proteins of the large subunit; these were identified as L4, L6, L7, L15, and L17 by two-dimensional electrophoresis (Horne and Hesketh, 1990). Images of SYTO 14 labeling were taken with a fluorescein filter and of EF1α and 60S subunit with a Cy5 filter. Colocalization was determined by aligning the signal from precise coordinates obtained with each filter.
Photo-oxidation and electron microscopy. Fluorescent and transmitted light images of cultures labeled with SYTO 14 and Mitotracker were recorded with a laser-scanning confocal attachment (MRC-1000; Bio-Rad, Cambridge, MA) that used a krypton/argon laser and then was photo-oxidized, as described previously (Deerinck et al., 1994). After fluorescence images were recorded, the cultured cells were fixed for 15 min with 2% glutaraldehyde, 2% paraformaldehyde, and 5 mm MgCl2 in PBS and then rinsed in 0.1m sodium cacodylate buffer for 10 min. Then the samples were immersed in a 4°C chilled solution of 2.8 mmdiaminobenzidine (DAB) in 0.1 m sodium cacodylate bubbled with pure 02, final pH 7.4, and irradiated by using 515 ± 10 nm excitation from a 75 W mercury light source. Continuous observations were made during the photo-oxidation procedure by using transmitted light. The fluorescence faded within a few minutes; between 15–25 min a brownish reaction product appeared in place of the fluorescence. The process was stopped by halting the excitation. Then the samples were rinsed several times in 0.1m sodium cacodylate and treated with 2% osmium tetroxide in 0.1 m sodium cacodylate for 30 min. Samples that were not reacted were stained with 1% uranyl acetate for 30 min and Satoh lead for 10 min. After being washed in H2O several times, the cells were dehydrated in an ethanol series, infiltrated with Durcupan ACM resin, and polymerized for 24 hr at 60°C.
Sections were cut with a Reichert Ultracut E at a thickness of 90 nm by using a diamond knife and examined with a JEOL 100CX at 80 keV. Low power electron microscopic images were scanned into PhotoShop and rotated to align the image precisely with the fluorescent and phase images of the same cell.
Drug treatments. To disrupt microfilaments, we treated cells with 10 μg/ml cytochalasin-D (Sigma) in culture media at 37°C for 2 hr before fixation. To depolymerize microtubules, we treated cells with 20 μg/ml colchicine (Sigma) in culture media for 2 hr before labeling with SYTO 14 and Mitotracker, as described above. Then cells were evaluated for live-cell microscopy or fixed as described above. Stock solutions of cytochalasin-D and colchicine were made up in DMSO and ethanol, respectively, and the concentration of these solvents was diluted below 0.1% in culture media so as not to be toxic to neurons.
RESULTS
SYTO 14 staining in cortical neurons
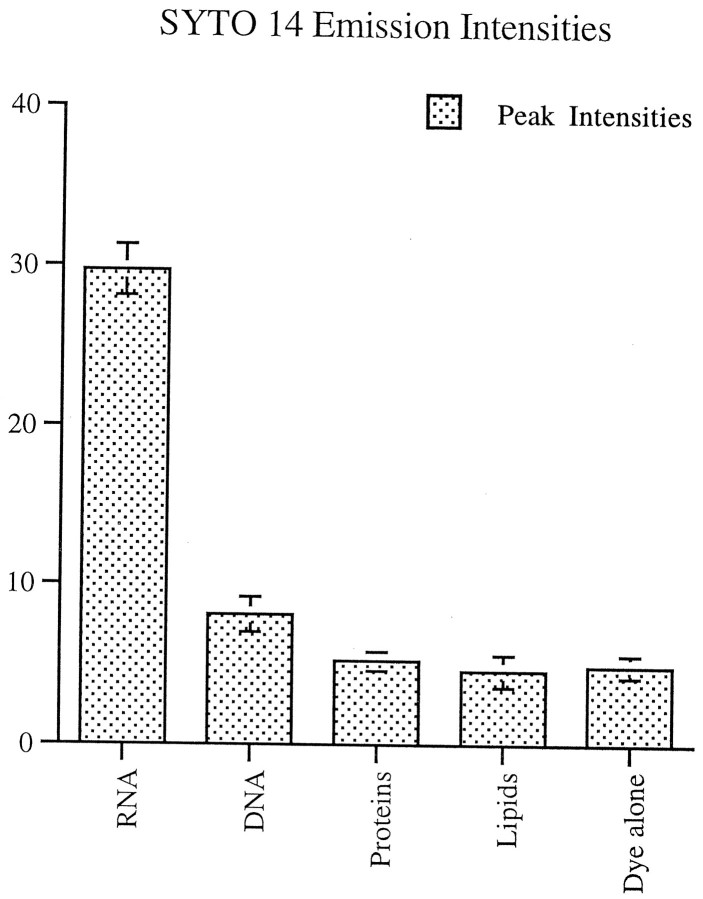
SYTO 14 (Molecular Probes) is a cell-permeant dye with fluorescent enhancement on binding to nucleic acids and at least a 50% greater quantum yield for RNA versus DNA. The spectral characteristics of this dye in the presence of RNA are an absorption maximum of 521 nm and a fluorescence emission maximum of 547 nm. Emission intensities of SYTO 14 in the presence of RNA, DNA proteins, and lipids were compared. Samples of total RNA purified from yeast (Boehringer Mannheim) 1 mg/ml, salmon sperm DNA 1 mg/ml, albumin from bovine serum (Boehringer Mannheim) 10 mg/ml, and lipid standard (Sigma) 10 mg/ml were incubated with 1 μm SYTO 14 for 10 min in HEPES-buffered water (10 mm HEPES in DEPC-treated water, pH 7.3). Then emission profiles were generated with a luminescence spectrometer (Perkin-Elmer, Norwalk, CT) at an absorbance of 500 nm, and peak intensities were calculated (Fig. 1). Peak emission intensity for RNA was sixfold greater than that for either protein or lipids.
Fig. 1.
Emission intensity profiles of RNA, DNA, proteins, and lipids incubated with SYTO 14 and scanned at an absorbance of 500 nm. Peak emission intensity for RNA was sixfold greater than that for either proteins or lipids. It differed from proteins, lipids, and DNA with a significance of p < 0.001. The emission intensity for DNA differed from proteins and lipids with a significance of p < 0.05.
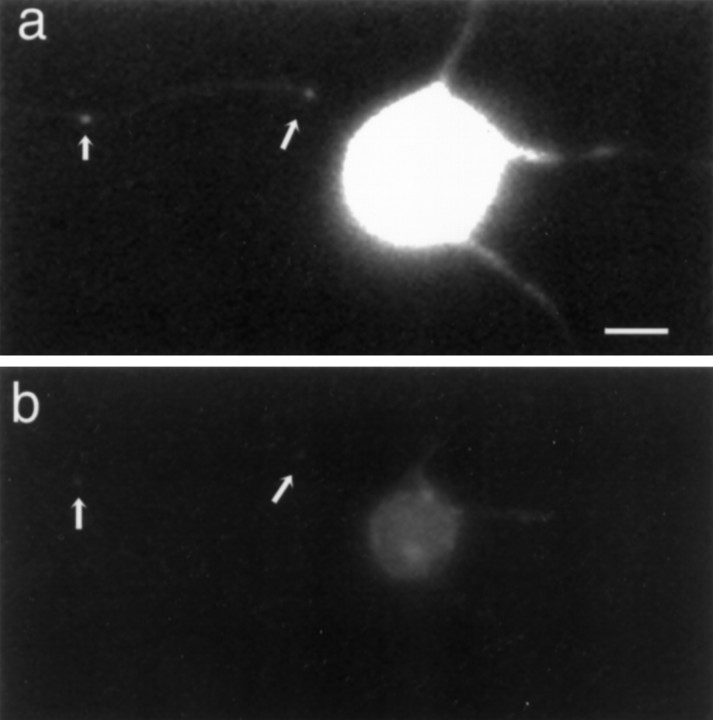
Cultures of rat cortical neurons incubated with SYTO 14 were brightly labeled. Within the cell body the nucleus stained intensely, and the cytoplasm was labeled in a dense granular pattern (Fig.2a). In neurites the dye stained two classes of structures: elongated, cigar-shaped organelles and small granules. The more elongated structures were mitochondria, as shown by double-labeling with the mitochondrial marker Mitotracker CMXROS (Fig.2b). The signal intensity of each labeled structure was evaluated through a resorufin filter and a Cy5 filter (Table1). For evaluation and quantification, SYTO 14-labeled nonmitochondrial granules were defined as any distinct structure that had an average pixel intensity of >50 in the resorufin channel and <40 in the Cy5 channel. The absence of small SYTO 14-labeled granules in the Cy5 channel was not attributable to threshold effects of less intensely labeled small granules, because mitochondria with the minimum intensity of 50 in the resorufin channel were detected in the Cy5 channel (Fig. 2, arrowhead). In 4-d-old cortical cultures, 21 of 100 SYTO 14-labeled structures were nonmitochondrial granules.
Fig. 2.
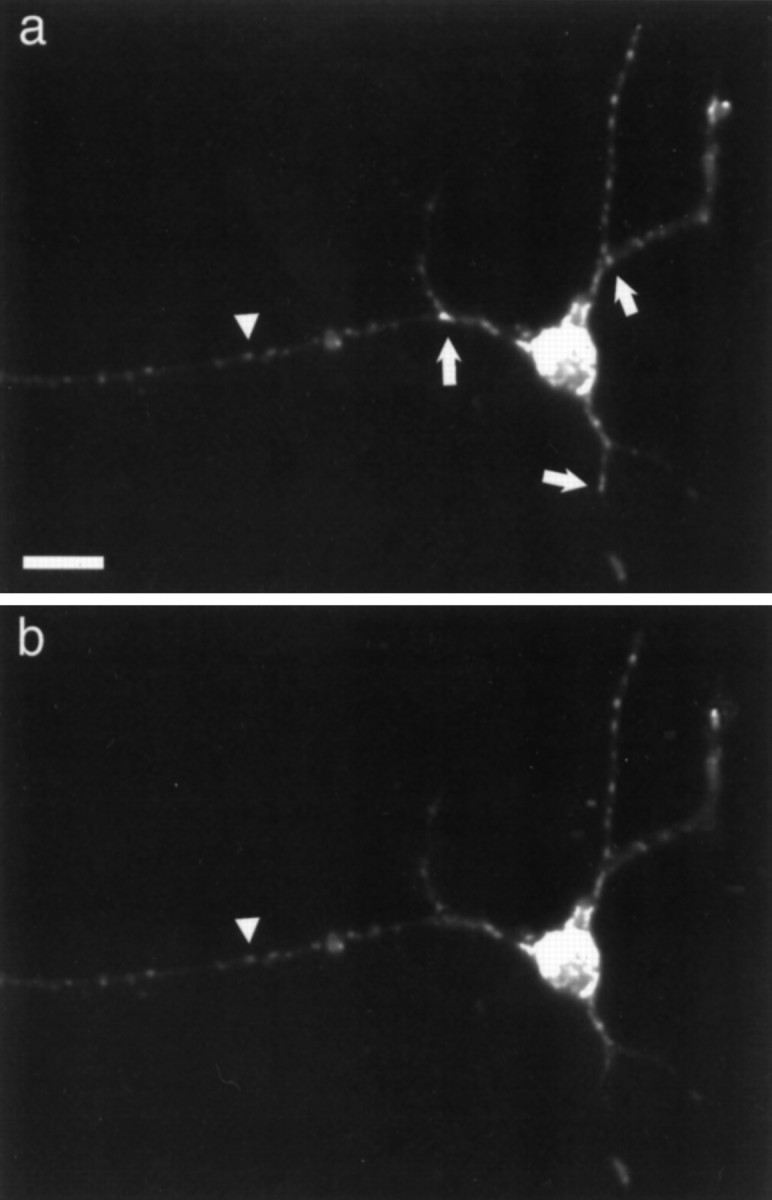
Cortical neurons colabeled with SYTO 14 (a) and Mitotracker (b).Arrowhead indicates mitochondria that colocalize with both markers. Small arrows indicate granules only observed with SYTO 14 labeling. Scale bar, 20 μm.
Table 1.
Average signal intensities
| Filler | Mitochondria | Granules | Background |
|---|---|---|---|
| Resorufin | 90.2 ± 1.7 | 65.4 ± 0.7 | 22.3 ± 0.4 |
| Cy5 | 89.7 ± 1.6 | 31.5 ± 0.5 | 31.3 ± 0.6 |
Average signal intensities of SYTO 14-labeled mitochondria (n = 100) and nonmitochondria granules (n = 100) detected with a Resorufin filter and the corresponding signal intensities in the identical regions caused by Mitotracker labeling detected with the Cy5 filter. Background intensities were generated by examining the average pixel intensity in random 1 × 1 μm regions in the neurite shaft that did not contain any detectable mitochondria or nonmitochondria granules (n = 100). Signal intensities of mitochondria and nonmitochondria granules are significantly above background detected with the Resorufin filter (p < 0.001). Mitochondria intensities are significantly above background with the cy5 filter (p < 0.001), whereas the signal from granules is not significantly above background.
Distribution of RNA granules
Beyond the cell body and proximal shaft of neurites, SYTO 14-labeled granules that did not colocalize with Mitotracker signal were distributed uniformly with 6.7 ± 0.4 granules per 40 μm. The average distance of a granule from the cell body was 58 ± 17 μm. The only exception to this distribution occurred in those cultures that had differentiated one neurite into an axon-like process. After 3 d in culture, very few nonmitochondrial granules could be detected in axons. In minor neurites, granules could be detected throughout the shaft and into the distal tips where granules occasionally extended into growth cones. Staining of nonmitochondrial granules was intense in the proximal shaft of all neurites; therefore, individual granules could not be resolved. This pattern of intense labeling in proximal neurites and near-elimination of label from axons is reminiscent of the distribution of poly(A+) mRNA (Bassell, 1994b) and ribosomes in neurons (Bartlett and Banker, 1984). One region of nonhomogeneity was branch points where RNA granules tended to cluster proximal to the junction. Fifty-six percent of neurite branch points had granules detected within 3 μm of the branch point. Similar measurements taken in nonbranch point segments yielded only 14% with granules. Although spreading of the fluorescent signal does not permit us to gauge an accurate size for these granules, their apparent size was remarkably uniform.
Identification of SYTO 14-labeled granules
SYTO 14-labeled granules were sensitive to treatment with RNase. When cultured neurons were fixed, permeabilized, and treated with RNase for 1 hr, all SYTO 14-labeled structures showed a reduction in signal intensity to <9.8% ± 0.7% (n = 10) of pre-RNase-treated levels in neurites and 19.2% ± 1.5% (n = 10) in the cell body (Fig. 3). The reduction of signal in mitochondria is most likely attributable to SYTO 14 labeling of mitochondrial RNA. The greater resistance of the cell body to RNase is attributable, in part, to SYTO 14 labeling of DNA in the nucleus. There was a modest decrement in fluorescence intensity in the cell body and no detectable decrement in the processes at sites of SYTO 14-labeled granules after DNase treatment.
Fig. 3.
Cortical neurons labeled with SYTO 14 before (a) and after (b) RNase treatment.Arrows indicate SYTO 14-labeled granules. Scale bar, 10 μm.
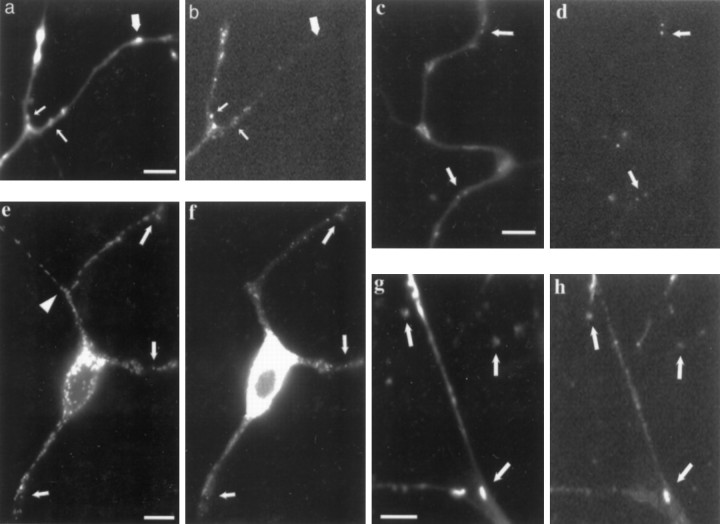
Components of protein synthesis are spatially organized together. In oligodendrocyte processes, granules containing myelin basic protein mRNA colocalize with ribosomal RNA, arginyl-tRNA, and elongation factor 1α (Barbarese et al., 1995). The constituents of the SYTO 14-labeled granules were assessed by colocalization studies with several different markers. A digoxigenin-labeled oligo-dT probe used for fluorescencein situ hybridization colocalized with SYTO14-labeled granules. In many cells one of the neurites was considerably longer than the others, suggesting features of an axon. Just as with the nonmitochondrial SYTO 14 labeling, there was minimal labeling of poly(A+) probe in these axon-like processes beyond an intensely labeled region in the initial segment. The majority of the poly(A+) RNA hybridization colocalized with SYTO 14-labeled small granules (Fig. 4a,b, small arrows); however, the converse was not true. In 4-d-old cultures, 32 of 100 SYTO 14-labeled structures colocalized with the poly(A+) probe. Most of the noncolocalized SYTO 14-labeled structures were elongated and mitochondria-like (Fig. 4b, large arrow). Some regions of the neurite shaft seem to have both mitochondria and granules densely packed together (Fig. 4a,b, top leftneurite branch). Previous observations with the poly(A+) probe have shown hybridization to mitochondrial poly(A+) was infrequent with normal hybridization protocols, perhaps because of reduced penetrability of the probe (Somasundaran et al., 1994). Therefore, on the basis of RNase sensitivity and poly(A+) colocalization, the small granules represent nonmitochondrial mRNA-containing structures.
Fig. 4.
Cortical neurons labeled with SYTO 14 and translational components. a, SYTO 14-labeled granules (small arrows) and mitochondria (large arrow) staining in neurites. b, Poly(A+) mRNA-containing granules detected in neurites (small arrows) and no signal at location of mitochondria (large arrow). Scale bar, 5 μm. c, SYTO 14-labeled granules in neurites. d, β-Actin mRNA-containing granules in neurites. Examples of colocalization are indicated with arrows. Scale bar, 5 μm.e, SYTO 14-labeled granule staining in neurites.f, Ribosomal 60S subunit labeling in neurites. Examples of colocalization are indicated with arrows.Arrowhead in e points to start of axon-like neurite. Scale bar, 10 μm. g, SYTO 14-labeled granule staining in neurites. h, EF1α labeling in neurites. Examples of colocalization are indicated witharrows. Phase contrast showed that indicated granules were located on neurite branches (data not shown). Scale bar, 5 μm.
To determine whether specific messages were present in the SYTO 14-labeled granules, we hybridized a probe to β-actin mRNA to neurites that previously were labeled with SYTO 14. In 4-d-old cultures, 14 of 100 SYTO 14-labeled structures colocalized with the β-actin probe, and 43% of the nonmitochondrial granules contained β-actin mRNA. These β-actin mRNA-containing SYTO 14 structures all seemed to be small granules (Fig. 4c,d, arrows). None of the mitochondria-like structures were labeled with the β-actin probe.
Proteins involved in translation may be associated with these RNA granules. One structure expected to be present in functional polyribosomes is the 60S ribosomal subunit (Horne and Hesketh, 1990). In 4-d-old cultures, 23 of 100 SYTO 14-labeled structures colocalized with anti-60S subunit antibodies (Fig. 4e,f, arrows). The staining pattern with this antibody, like the poly(A+) mRNA signal, tended to be excluded from axon-like processes that were significantly longer than the other neurites, were not tapered, and had few branches. SYTO 14 mitochondrial staining, as demonstrated in the axon-like process (Fig. 4e, arrowhead), did not colocalize with the anti-60S ribosomal subunit antibodies. Another protein involved with translation, elongation factor 1α (EF1α), binds to GTP and aminoacyl-tRNA, leading to the codon-dependent placement of the aminoacyl-tRNA at the A site of the ribosome (Riis et al., 1990). EF1α has been shown to colocalize with poly(A+) hybridization (Bassell et al., 1994a). In 4-d-old cultures, 29 of 100 SYTO 14-labeled structures colocalized with antibodies against EF1α (Fig. 4g,h).
Motility characteristics of SYTO 14-labeled granules
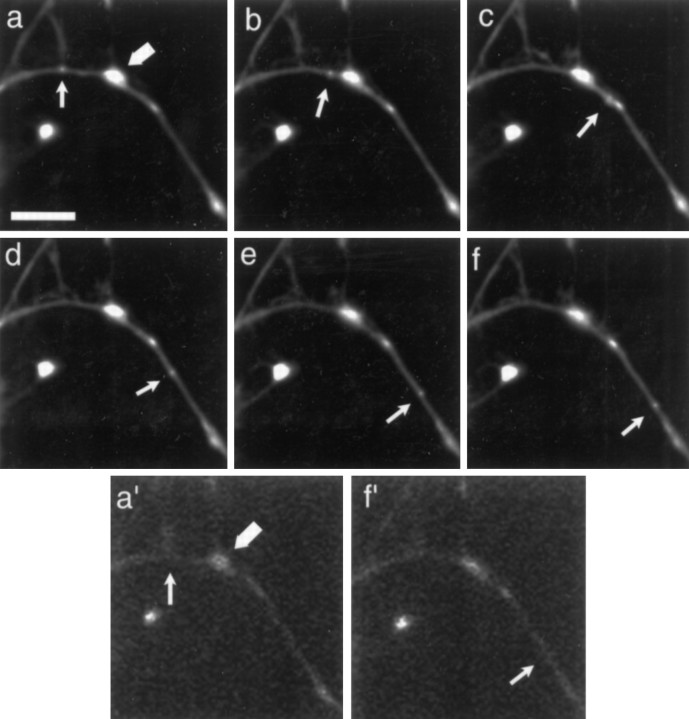
Subtraction of the Mitotracker image from that obtained with SYTO 14 allowed unambiguous identification of RNA granules in living neurons (Fig. 5). Both mitochondria and RNA granules were motile, each with distinct characteristics. Three percent of the granules moved over a 5 min time period, suggesting that RNA granules exist principally in a stationary phase. Nevertheless, in a 24-d-old cultured hippocampal neuron the total distance traveled by all of the moving granules in the cell would be over 52 mm/24 hr, assuming a total dendritic length of 2800 μm (Banker and Waxman, 1988). In the presence of dye, 20% of mitochondria moved, similar to the reported value in chick sympathetic neurons by phase-contrast microscopy (Morris and Hollenbeck, 1993). The motile granules had an average velocity of 0.10 ± 0.01 μm/sec (n = 19), which is slower than mitochondria that move with an average velocity of 0.31 ± 0.05 μm/sec (n = 38; p < 0.001).
Fig. 5.
Time-lapse analysis of SYTO 14-labeled small granule movement in neurites. a–f, Images after granule (small arrow) and mitochondria (large arrow) at times 0, 1, 2, 3, 4, 5 min. a′, Image taken of mitochondria (large arrow) just before time-lapse. f′, Image taken of mitochondria just after time-lapse. Scale bar, 10 μm.
In 2- to 4-d-old cultures, only anterograde movement was observed in granules, whereas 57% of the mitochondria had at least one retrograde movement and 39% had a net retrograde displacement. However, in cultures older than 7 d, one-half of the granules moved in the anterograde direction and one-half in the retrograde direction. Of 12 observations in 2- to 4-d-old cultures, 100% were anterograde; of 6 observations in 5- to 7-d-old cultures, 67% were anterograde and 33% were retrograde; of 6 observations in 8- to 10-d-old cultures, 50% were anterograde and 50% were retrograde. The rates of transport in the retrograde direction were identical to those moving in the anterograde direction. In addition to observations of individual motile granules, a dense array of punctate signal seemed to move bidirectionally in the proximal shaft of some neurites. Individual movements of granules within this bulk flow could not be tracked with any precision.
Cytoskeletal-disrupting drugs
The most likely track for the observed movements is the cytoskeleton. Cytochalasin-D, which binds to the barbed ends of actin filaments and induces their disorganization (Cooper, 1987), had no effect on the distribution of SYTO 14-labeled RNA granules in 4-d-old neuronal cultures. In contrast, colchicine caused a decline in the density of granules from 6.7 ± 0.4 per 40 μm to 2.6 ± 0.3 per 40 μm (n = 60; p < 0.001; Fig.6). This 62% drop in granular density after colchicine treatment suggested that microtubules are involved in the anchoring of SYTO 14-labeled granules. Movements of SYTO 14-labeled granules were evaluated in the presence of colchicine and cytochalasin-D. No movements were seen in the presence of colchicine, whereas 2.5% of the granules moved in the presence of cytochalasin-D (n = 40). Granules moving in cytochalasin-D-treated cells had similar average velocities to those of control cells. This value does not differ significantly from untreated cultures. Together, these observations suggest that intact microtubules are necessary for RNA granule translocation in the neurite shaft.
Fig. 6.
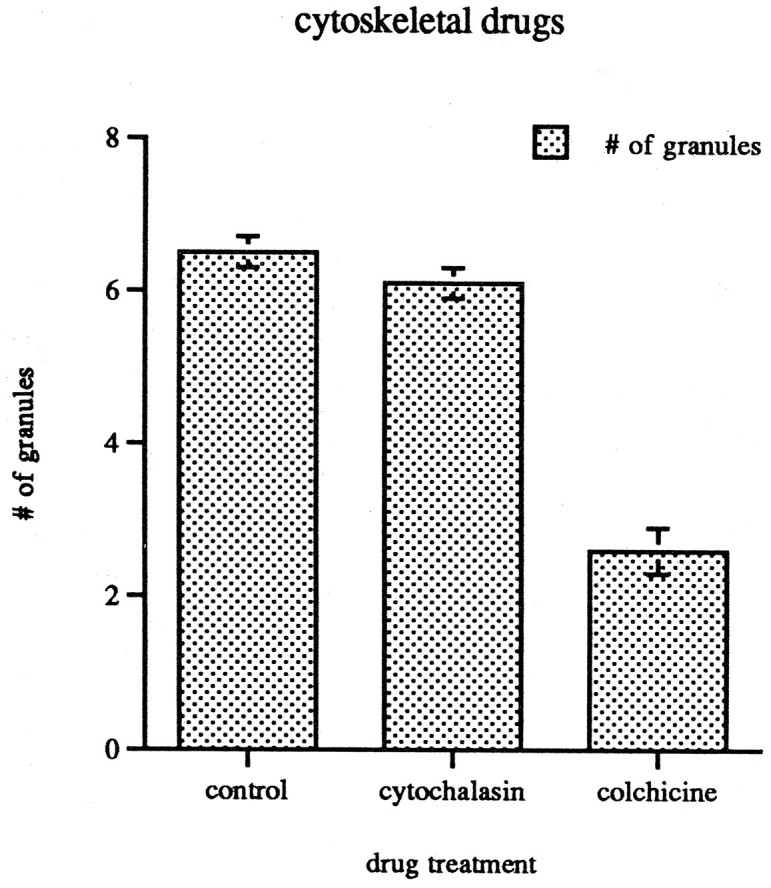
Distribution of SYTO 14-labeled granules in 40 μm segments in the neurite shafts of 4-d-old cortical neuronal cultures. Treatment with cytochalasin-D does not significantly alter the distribution of RNA granules, whereas treatment with colchicine decreases the number by 62% (p < 0.001).
Ultrastructural identification of SYTO 14-labeled granules
To determine the ultrastructural correlates of RNA granules labeled with SYTO14, we performed fluorescence photo-oxidation of DAB with SYTO 14 to provide localization of granules at the light and electron microscopic levels. Granules appeared as circular clusters of ribosomes (Fig. 7A, arrow). Within these clusters, some ribosomes were grouped together with an interparticle spacing typical of polyribosomes (Steward and Falk, 1986). No membrane could be detected enclosing the ribosomal cluster. The diameter of these labeled granules was relatively uniform, ranging from 175 to 600 nm. Sometimes a slight clearing was observed in the center of granules. Only a subset of the total cellular ribosome population formed discrete granules. Mitochondria also photo-oxidized DAB under these conditions but were easily distinguished from granules (Fig. 7A, arrowhead).
Fig. 7.
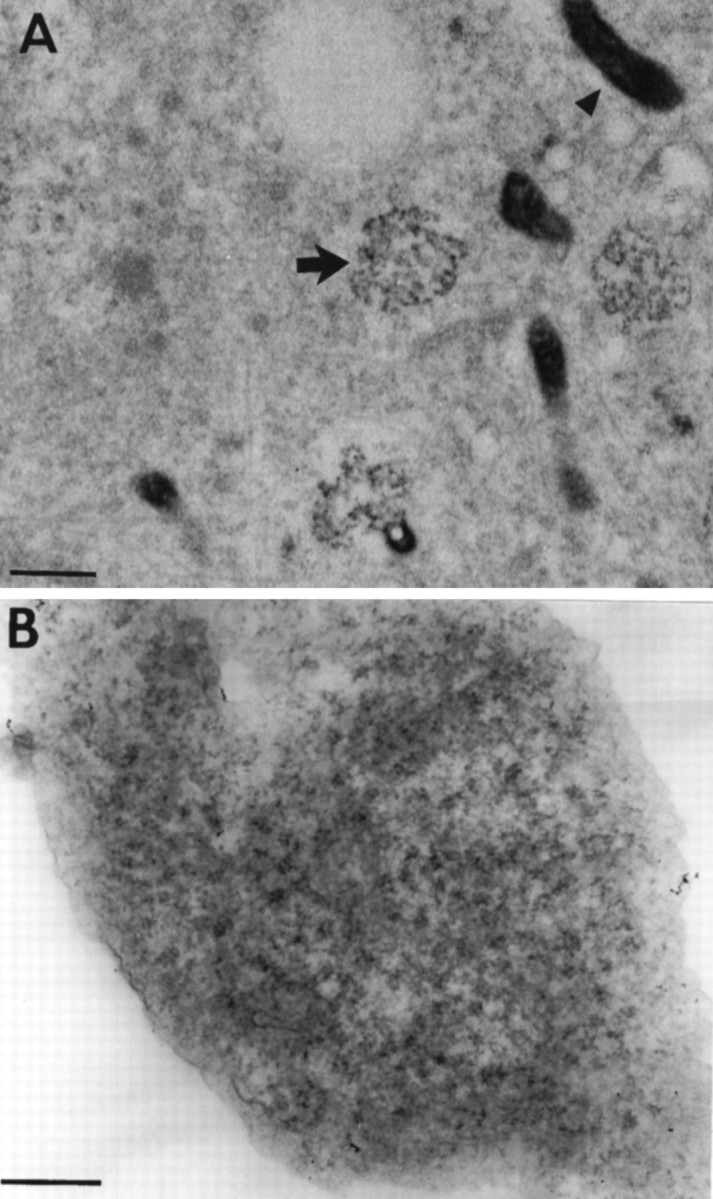
A, Cortical neurons labeled with SYTO 14, photoconverted in the presence of DAB, and viewed with an electron microscope. Arrows point to electron-dense DAB reaction product that corresponds to the precise location of the SYTO 14 dye. The labeled structures appear as clusters of ribosomes (arrow) and organelles that seem to be mitochondria (arrowhead). Sections are 90 μm thick. Scale bar, 500 nm. B, Electron micrograph of a single RNA granule. The site shown here precisely corresponds to a SYTO 14-labeled granule in a culture of cortical neurons. Scale bar, 100 nm.
Because it was possible that the photo-oxidation technique artifactually might create the appearance of a ribosomal cluster or obscure fine structural detail, SYTO14-labeled granules were located ultrastructurally without using photo-oxidation for their identification by digitally correlating the light and electron microscopic images. At the electron microscopic level, clusters of ribosomes, as detected by poststaining with heavy metals, were observed in regions exhibiting fluorescent granules at the light microscopic level (Fig. 7B).
DISCUSSION
RNA granules are actively transported
The dye SYTO 14 makes it possible to visualize a motile pool of RNA granules in living cells. From the extensive data on the localization of RNA, one would presume that RNAs are motile; however, very few studies address RNA translocation, and none has observed directly the translocation of endogenous RNA. RNA granules traveled at an average rate of 0.1 μm/sec, a rate that is within the range of fast transport (Brady et al., 1982; Brady and Lasek, 1982). This rate is ∼20-fold faster than the rate calculated for RNA transport with autoradiographic imaging of [3H]uridine label at timed intervals (Davis et al., 1987). This discrepancy may be explained if, over the 5 min period used here, the vast majority of RNA granules were stationary. Rates more consistent with those observed here were obtained by microinjecting labeled myelin basic protein into oligodendrocytes (Ainger et al., 1993).
Active transport of RNA was suggested in fibroblasts in which the localization of the β-actin mRNA was regulated by GTPase signal transduction cascades (Hill et al., 1994; Latham et al., 1994). In these cells platelet-derived growth factor can induce a translocation of β-actin mRNA to the leading lamella within 2–5 min (Latham et al., 1994). The RNA granule movement observed here is also likely to represent active transport and not simple diffusion. When individual granules were observed to translocate in regions where there was no concentration gradient, 12 of 12 granules moved solely in the anterograde direction in young cultures. Even in older cultures, in which one-half of the granules moved in the retrograde direction, each individual granule over a 5 min period (15 images/granule) moved in only one direction. This strongly suggests that the granules are transported by an active mechanism and not by passive diffusion. Because SYTO 14-labeled granules represent only a subset of the total ribosomal population of the cell, diffusion may explain the transport of some RNA, as suggested by Kleiman et al. (1993).
The direction of RNA granule movement was dependent on the age of the neuronal culture. In the midportion of developing dendrites, microtubule orientation went from near 100% plus-end distal to 50% plus-end distal/50% minus-end distal during the time period (Baas et al., 1989) when we observed the onset of bidirectional RNA granule movement. These developmental changes in directionality are consistent with a plus-end-directed microtubule motor powering the granules. One candidate motor is the kinesin-like molecule KIF-4, which is a plus-end-directed motor protein with transport rates similar to the rates observed for RNA granules (Sekine et al., 1994).
Localization of RNA granules
The conclusion that RNA granules have the motility characteristics of an actively transported organelle leads to the possibility that their translocation is along cytoskeletal tracks. During the 5 min observation period we observed a large stationary pool of mRNA granules that became delocalized with colchicine. Although these RNA granules either have paused or remained stationary, there must exist a means to anchor them to the cytoskeleton. Previous pharmacological manipulations with colchicine support the finding that mRNAs are anchored to microtubules. In Xenopus oocytes, the translocation of veg-1 RNA to the vegetal pole required intact microtubules (Yisraeli et al., 1990), and in Drosophila, bicoid mRNA transport depended on microtubules (Pokrywka and Stephenson, 1991). In somatic cells, mRNA anchored to actin filaments in fibroblasts (Taneja et al., 1992; Bassell et al., 1994a) and to microtubules in neurons (Bassell et al., 1994b) have both been demonstrated. That a single organelle can translocate from one cytoskeletal system to another in the squid giant axon (Kuznetsov et al., 1992) may account for these apparently contradictory observations. Perhaps RNA granules use both systems—microtubules for the relatively long distances in neurons and oocytes and actin filaments for the relatively short distances in fibroblasts (Atkinson et al., 1992).
As observed for both poly(A+) mRNA and 60S ribosomal antibody labeling, SYTO 14-labeled granules are nearly excluded from the axon, a structure known to have considerably fewer ribosomes than dendrites (Deitch and Banker, 1993). SYTO 14-labeled granules are punctate, an appearance consistent with the punctate labeling of poly(A+) mRNA (Taneja et al., 1992; Bassell et al., 1994b) of the 60S ribosomal antibody (Fig. 4f) and the punctate appearance of specific mRNAs such as myelin basic protein in oligodendrocytes (Ainger et al., 1993), Xlsirt in frog oocytes (Kloc and Etkin, 1994), staufen/bicoid RNP particles in fly oocytes (Ferrandon et al., 1994), and actin mRNA in chick fibroblasts (Sundell and Singer, 1991).
Electron microscopic images of neurons often reveal clusters of polyribosomes (Deitch and Banker, 1993). These clusters may concentrate RNA and thereby reach a signal threshold that generates a discrete signal. Fluorescent photo-oxidation of DAB has been shown to provide ultrastructural resolution of specific markers (Deerinck et al., 1994). Photo-oxidation with SYTO 14 revealed ribosome clusters that might not be apparent against a background of ribosomes that fail to generate a discrete signal. Unequivocal electron microscope detection of granules without photo-oxidation required a precise alignment with the light microscopic SYTO 14 signal (Fig. 7B). These data suggest that populations of ribosomes coalesce to form a granule capable of moving cohesively through the cytoplasm. By analogy with other nonmembrane-bound bodies in cells, these structures might be called cytosomes (Chicurel et al., 1995). Whether these structures are translation-competent is unknown; they do, however, have many of the necessary translational components—polyribosomes, EF1α, and poly(A+) mRNA.
The function of RNA transport
The relatively large amount of RNA transported into dendrites raises the question of why neurons need an ever-constant supply of protein synthetic machinery delivered to their neurites (for review, see St. Johnston, 1995). Possibly the transport of mRNA to a site where its translation products will be used represents an efficient means of sorting and delivering a specific number and array of mRNAs. Site-specific translation at the neuronal synapse, for example, probably requires the stoichiometric delivery of component proteins for the assembly of synaptic structures and for the creation of the correct synaptic milieu. Furthermore, mRNAs may require discrete localizations to avoid inappropriate interactions of their translation products. Myelin basic protein has a strong propensity to bind tightly to membranes in vitro, a property that could disrupt intracellular membranes unless the protein were targeted to the plasma membrane before its synthesis (for review, see Brophy et al., 1993). Likewise, MAP2 and tau, which have the ability to interact with any microtubule, may require segregation of their mRNAs to discrete cellular domains so they interact only with appropriate microtubules destined for the axon or dendrite. It is likely that translocating granules carry many or all of the components observed under fixation conditions. If so, why does the neuron translocate its synthetic machinery in addition to mRNAs? Protein synthetic machinery is located at the base of dendritic spines (Steward, 1983; Steward and Fass, 1983), and this apparatus may engage the RNA granule to initiate protein synthesis. In this way the local synthesis of proteins may be responsible for local spine morphologies, of which there is considerable diversity (Harris and Kater, 1994), or serve as a local determinant of synapse identity.
Footnotes
This work was supported by National Institutes of Health Grant NS29031 and the American Health Assistance Foundation to K.S.K. and National Institutes of Health, National Center for Research Resources, P41 04050 to M.H.E. We thank Marc Kirschner for generously allowing the use of microscopy equipment for some of these experiments. Antibody to EF1α was kindly provided by Wim Moller (Rijks University, Leiden, The Netherlands), and antibody to 60S ribosomal subunit was generously provided by John Hesketh (Rowett Research Institute, Aberdeen, UK). We thank Ying Zhang for her technical support.
Correspondence should be addressed to Dr. Kenneth S. Kosik, Brigham and Women’s Hospital, 221 Longwood Avenue, Boston, MA 02115.
REFERENCES
- 1.Ainger K, Avossa D, Morgan F, Hill SJ, Barry C, Barbarese E, Carson JH. Transport and localization of exogenous MBP mRNA microinjected into oligodendrocytes. J Cell Biol. 1993;123:431–441. doi: 10.1083/jcb.123.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson S, Doberstein SK, Pollard TD. Moving off the beaten track. Curr Biol. 1992;2:326–328. doi: 10.1016/0960-9822(92)90896-i. [DOI] [PubMed] [Google Scholar]
- 3.Baas PW, Black MW, Banker GA. Changes in microtubule polarity orientation during the development of hippocampal neurons in culture. J Cell Biol. 1989;109:3085–3094. doi: 10.1083/jcb.109.6.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banker GA, Waxman AB. Hippocampal neurons generate natural shapes in cell culture. In: Lasek RJ, Black MM, editors. Studies of the intrinsic determinants of neuronal form and function. Liss; New York: 1988. pp. 61–82. [Google Scholar]
- 5.Barbarese E, Koppel DE, Deutscher MP, Smith CL, Ainger K, Morgan F, Carson JH. Protein translation components are colocalized in granules in oligodendrocytes. J Cell Sci. 1995;108:2781–2790. doi: 10.1242/jcs.108.8.2781. [DOI] [PubMed] [Google Scholar]
- 6.Bartlett WP, Banker GA. An electron microscopic study of the development of axons and dendrites by hippocampal neurons in culture. J Neurosci. 1984;4:1944–1965. doi: 10.1523/JNEUROSCI.04-08-01944.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bassell GJ, Powers CM, Taneja KL, Singer RH. Single mRNAs visualized by ultrastructural in situ hybridization are principally localized at actin filament intersections in fibroblasts. J Cell Biol. 1994a;126:863–876. doi: 10.1083/jcb.126.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassell GJ, Singer RH, Kosik KS. Association of poly(A+) mRNA with microtubules in cultured neurons. Neuron. 1994b;12:571–582. doi: 10.1016/0896-6273(94)90213-5. [DOI] [PubMed] [Google Scholar]
- 9.Brady ST, Lasek RJ. Axonal transport: a cell biological method for studying proteins that associate with the cytoskeleton. Methods Cell Biol. 1982;25:326–398. doi: 10.1016/s0091-679x(08)61434-x. [DOI] [PubMed] [Google Scholar]
- 10.Brady ST, Lasek RJ, Allen RD. Fast axonal transport in extruded axoplasm from giant squid axon. Science. 1982;218:1129–1131. doi: 10.1126/science.6183745. [DOI] [PubMed] [Google Scholar]
- 11.Brophy PJ, Boccaccio GL, Colman DR. The distribution of myelin basic protein mRNAs within myelinating oligodendrocytes. Trends Neurosci. 1993;16:515–521. doi: 10.1016/0166-2236(93)90196-s. [DOI] [PubMed] [Google Scholar]
- 12.Bruckenstein DA, Lein PJ, Higgins D, Fremeau RT. Distinct spatial localization of specific mRNAs in cultured sympathetic neurons. Neuron. 1990;5:809–819. doi: 10.1016/0896-6273(90)90340-l. [DOI] [PubMed] [Google Scholar]
- 13.Burgin KE, Waxham MN, Rickling S, Westgate SA, Mobley WC, Kelly PT. In situ hybridization histochemistry of Ca2+/calmodulin-dependent protein kinase in developing rat brain. J Neurosci. 1990;10:1788–1798. doi: 10.1523/JNEUROSCI.10-06-01788.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chicurel ME, Terrian DM, Potter H. mRNA at the synapse: analysis of a synaptosomal preparation enriched in hippocampal dendritic spines. J Neurosci. 1993;13:4054–4063. doi: 10.1523/JNEUROSCI.13-09-04054.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chicurel ME, DeFranco C, Potter H. RNA localization and RNA-cytoskeletal interactions in neurons. Lipshitz H. Localized RNAs. 1996. Landes; Austin, TX: Biomedical, in press. [Google Scholar]
- 16.Cooper JA. Effects of cytochalasin and phalloidin on actin. J Cell Biol. 1987;105:1473–1478. doi: 10.1083/jcb.105.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis L, Banker GA, Steward O. Selective transport of RNA in hippocampal neurons in culture. Nature. 1987;330:477–479. doi: 10.1038/330477a0. [DOI] [PubMed] [Google Scholar]
- 18.Deerinck TJ, Martone ME, Lev-Ram V, Green DPL, Tsien RY, Spector DL, Huang S, Ellisman MH. Fluorescence photo-oxidation with eosin: a method for high resolution immunolocalization and in situ hybridization detection for light and electron microscopy. J Cell Biol. 1994;126:901–910. doi: 10.1083/jcb.126.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deitch JS, Banker GA. An electron microscopic analysis of hippocampal neurons developing in culture: early stages in the emergence of polarity. J Neurosci. 1993;13:4301–4315. doi: 10.1523/JNEUROSCI.13-10-04301.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Driever W. The development of Drosophila melanogaster (Bate M, Martinez-Arias A, eds), pp 301–324. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1993. [Google Scholar]
- 21.Ephrussi AL, Lehmann R. Induction of germ cell formation by oskar. Nature. 1992;358:387–392. doi: 10.1038/358387a0. [DOI] [PubMed] [Google Scholar]
- 22.Ephrussi AL, Dickinson LK, Lehmann R. Oskar organizes the germ plasm and direct localization of the posterior determinant nanos. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- 23.Ferrandon D, Elphick L, Nusslein-Volhard C, St. Johnston D. Staufen protein associates with the 3′ UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell. 1994;79:1221–1232. doi: 10.1016/0092-8674(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 24.Garner CC, Tucker RP, Matus A. Selective localization of mRNA for cytoskeletal protein MAP2 in dendrites. Nature. 1988;336:674–679. doi: 10.1038/336674a0. [DOI] [PubMed] [Google Scholar]
- 25.Goslin K, Banker GA. Culturing nerve cells. MIT; London: 1991. [Google Scholar]
- 26.Harris KH, Kater SB. Dendritic spines: cellular specializations imparting both stability and flexibility to synaptic function. Annu Rev Neurosci. 1994;17:341–371. doi: 10.1146/annurev.ne.17.030194.002013. [DOI] [PubMed] [Google Scholar]
- 27.Hill MA, Schedlich L, Gunning P. Serum-induced signal transduction determines the peripheral location of beta-actin mRNA within the cell. J Cell Biol. 1994;126:1221–1230. doi: 10.1083/jcb.126.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horne Z, Hesketh J. Immunological localization of ribosomes in striated rat muscle. J Biochem. 1990;268:231–236. doi: 10.1042/bj2680231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim-Ha J, Smith JL, Macdonald PM. Oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell. 1991;66:23–55. doi: 10.1016/0092-8674(91)90136-m. [DOI] [PubMed] [Google Scholar]
- 30.Kleiman R, Banker GA, Steward O. Differential subcellular localization of particular mRNAs in hippocampal neurons in culture. Neuron. 1990;5:821–830. doi: 10.1016/0896-6273(90)90341-c. [DOI] [PubMed] [Google Scholar]
- 31.Kleiman R, Banker G, Steward O. Inhibition of protein synthesis alters the subcellular distribution of mRNA in neurons but does not prevent dendritic transport of RNA. Proc Natl Acad Sci USA. 1993;90:11192–11196. doi: 10.1073/pnas.90.23.11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kloc M, Etkin LD. Delocalization of Vg1 mRNA from the vegetal cortex in Xenopus oocytes after destruction of Xlsirt RNA. Science. 1994;265:1101–1103. doi: 10.1126/science.7520603. [DOI] [PubMed] [Google Scholar]
- 33.Kosik KS, Finch EA. MAP2 and tau segregate into dendritic and axonal domains after the elaboration of morphologically distinct neurites: an immunocytochemical study of cultured rat cerebrum. J Neurosci. 1987;7:3142–3153. doi: 10.1523/JNEUROSCI.07-10-03142.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuznetsov SA, Langford GM, Weiss DG. Actin-dependent organelle movement in squid axoplasm. Nature. 1992;356:722–725. doi: 10.1038/356722a0. [DOI] [PubMed] [Google Scholar]
- 35.Latham VM, Kislauskis EH, Singer RH, Ross AF. Beta-actin mRNA localization is regulated by signal transduction mechanisms. J Cell Biol. 1994;126:1211–1219. doi: 10.1083/jcb.126.5.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehmann R, Nusslein-Volhard C. The maternal gene nanos has a central role in pattern formation of the Drosophila embryo. Development (Camb) 1991;112:679–691. doi: 10.1242/dev.112.3.679. [DOI] [PubMed] [Google Scholar]
- 37.Miyashiro K, Dichter M, Eberwine J. On the nature and differential distribution of mRNAs in hippocampal neurites: implications for neuronal functioning. Proc Natl Acad Sci USA. 1994;91:10800–10804. doi: 10.1073/pnas.91.23.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morris RL, Hollenbeck PJ. The regulation of bidirectional mitochondrial transport is coordinated with axonal outgrowth. J Cell Sci. 1993;104:917–927. doi: 10.1242/jcs.104.3.917. [DOI] [PubMed] [Google Scholar]
- 39.Pokrywka NJ, Stephenson EC. Microtubules mediate the localization of bicoid RNA during Drosophila oogenesis. Development (Camb) 1991;113:55–66. doi: 10.1242/dev.113.1.55. [DOI] [PubMed] [Google Scholar]
- 40.Pollock JA, Ellisman MH, Benzer S. Subcellular localization of transcripts in Drosophila photoreceptor neurons: chaoptic mutants have an aberrant distribution. Genes Dev. 1990;4:806–821. doi: 10.1101/gad.4.5.806. [DOI] [PubMed] [Google Scholar]
- 41.Riis B, Ruttan SIS, Clark BFC, Merrick WC. Eukaryotic protein elongation factors. Trends Biochem Sci. 1990;15:98–102. doi: 10.1016/0968-0004(90)90279-k. [DOI] [PubMed] [Google Scholar]
- 42.Sekine Y, Okada Y, Noda Y, Kondo S, Aizawa H, Takemura R, Hirokawa N. A novel microtubule-based motor protein (KIF4) for organelle transports, whose expression is regulated developmentally. J Cell Biol. 1994;127:187–201. doi: 10.1083/jcb.127.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Somasundaran MA, Zapp ML, Beattie LK, Pang L, Byron KS, Bassell GJ, Sullivan JL, Singer RH. Localization of HIV RNA in mitochondria of infected cells: potential role in cytopathogenicity. J Cell Biol. 1994;126:1353–1360. doi: 10.1083/jcb.126.6.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steward O. Polyribosomes at the base of dendritic spines of central nervous system neurons: the possible role in synapse construction and modification. Cold Spring Harbor Symp Quant Biol. 1983;48:745–759. doi: 10.1101/sqb.1983.048.01.077. [DOI] [PubMed] [Google Scholar]
- 45.Steward O, Falk PM. Protein synthetic machinery at postsynaptic sites during synaptogenesis: a quantitative study of the association between polyribosomes and developing synapses. J Neurosci. 1986;6:412–423. doi: 10.1523/JNEUROSCI.06-02-00412.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steward O, Fass B. Polyribosomes associated with dendritic spines in the denervated dendrite gyrus: evidence for local regulation of protein synthesis during reinnervation. Prog Brain Res. 1983;58:131–136. doi: 10.1016/S0079-6123(08)60013-8. [DOI] [PubMed] [Google Scholar]
- 47.Steward O, Levy WB. Preferential localization of polysomes under the base of dendritic spines in granule cells of the dentate gyrus. J Neurosci. 1982;2:248–291. doi: 10.1523/JNEUROSCI.02-03-00284.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.St. Johnston D. The intracellular localization of messenger RNAs. Cell. 1995;81:161–170. doi: 10.1016/0092-8674(95)90324-0. [DOI] [PubMed] [Google Scholar]
- 49.Sundell CL, Singer RH. Requirement of microfilaments in sorting of actin messenger RNA. Science. 1991;253:1275–1277. doi: 10.1126/science.1891715. [DOI] [PubMed] [Google Scholar]
- 50.Taneja KL, Lifshitz LM, Fay FS, Singer RH. Poly(A+) RNA codistribution with microfilaments: evaluation by in situ hybridization and quantitative digital imaging microscopy. J Cell Biol. 1992;119:1245–1260. doi: 10.1083/jcb.119.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tiedge H, Fremeau RT, Weinstock PH, Arancio O, Brosius J. Dendritic location of neural BCI RNA. Proc Natl Acad Sci USA. 1991;88:2093–2097. doi: 10.1073/pnas.88.6.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang C, Lehmann R. Nanos is the localized posterior determinant in Drosophila. Cell. 1991;66:637–647. doi: 10.1016/0092-8674(91)90110-k. [DOI] [PubMed] [Google Scholar]
- 53.Yisraeli JK, Sokol S, Melton DA. A two step model for the localization of maternal mRNA in Xenopus oocytes: involvement of microtubules and microfilaments in the translocation and anchoring of Vg1 mRNA. Development (Camb) 1990;108:289–298. doi: 10.1242/dev.108.2.289. [DOI] [PubMed] [Google Scholar]