Abstract
The STEP family of protein tyrosine phosphatases is highly enriched within the CNS. Members of this family are alternatively spliced to produce both transmembrane and cytosolic variants. This manuscript describes the distinctive intracellular distribution and enzymatic activity of the membrane-associated isoform STEP61. Transfection experiments in fibroblasts, as well as subcellular fractionations, sucrose density gradients, immunocytochemical labeling, and electron microscopy in brain tissue, show that STEP61is an intrinsic membrane protein of striatal neurons and is associated with the endoplasmic reticulum. In addition, structural analysis of the novel N-terminal region of STEP61 reveals several motifs not present in the cytosolic variant STEP46. These include two putative transmembrane domains, two sequences rich in Pro, Glu, Asp, Ser, and Thr (PEST sequences), and two polyproline-rich domains. Like STEP46, STEP61 is enriched in the brain, but the recombinant protein has less enzymatic activity than STEP46. Because STEP46 is contained in its entirety within STEP61 and differs only in the extended N terminus of STEP61, this amino acid sequence is responsible for the association of STEP61 with membrane compartments and may also regulate its enzymatic activity.
Keywords: protein tyrosine phosphatase, intracellular PTP, basal ganglia, alternative splicing, SH3 domain, polyproline domain, PEST sequence, signal transduction, endoplasmic reticulum
Protein tyrosine phosphorylation plays a central role in neuronal development and function. Tyrosine phosphorylation has been implicated in axonal navigation (Winslow et al., 1995; Desai et al., 1996; Krueger et al., 1996), growth cone elongation (Maness et al., 1988), synapse formation (Qu et al., 1990; Cudmore and Gurd, 1991), cell–cell or cell–extracellular matrix interactions (Atashi et al., 1992; Doherty and Walsh, 1992), and differentiation (Girault et al., 1992; Sahin and Hockfield, 1993; Walton et al., 1993; Zhang and Longo, 1995). It is also clear that the biological effects of neurotrophic factors on neuronal survival and differentiation are partly attributable to regulating tyrosine phosphorylation (Cordon-Cardo et al., 1991; Kaplan et al., 1991; Klein et al., 1991;Schlessinger and Ullrich, 1992). These and other observations have stimulated efforts to identify novel neuronal protein tyrosine phosphatases (PTPs) and protein tyrosine kinases (PTKs) and to characterize their functions within the CNS (for review, see Walton and Dixon, 1993; Naegele and Lombroso, 1994; Bult et al., 1995).
The PTPs are classified on the basis of their structural organization and are broadly divided into receptor-like or intracellular PTPs (for review, see Fischer et al., 1991; Tonks et al., 1991; Charbonneau and Tonks, 1992). The STEP family comprises intracellular PTPs enriched within the basal ganglia and related structures (Lombroso et al., 1991,1993; Boulanger et al., 1995; Sharma et al., 1995). Immunocytochemical and biochemical studies have demonstrated that the STEP family of polypeptides includes a group of lower molecular weight (MW) proteins enriched in cytoplasm and a group of higher MW proteins associated with particulate fractions (Lombroso et al., 1993; Boulanger et al., 1995). We reported previously the isolation of several STEP-related cDNAs (Li et al., 1995; Sharma et al., 1995). The present study was undertaken to better understand the structural and functional characteristics of one of these clones, STEP61.
A major finding of this work is that STEP61 is targeted to the endoplasmic reticulum (ER) of neurons. Outside of the nervous system, PTPs have been identified that are targeted to the cytoskeleton (Gu et al., 1991; Yang and Tonks, 1991; Sawada et al., 1994), the perinuclear region (Cool et al., 1990; Faure and Posner, 1993), the plasma membrane of neurosecretory granules (Solimena et al., 1996), and the nucleus (McLaughlin and Dixon, 1993; Flores et al., 1994). To date, the subcellular localization to the ER of two intracellular PTPs has been reported (Frangioni et al., 1992; Woodford-Thomas et al., 1992;Lorenzen et al., 1995). The present work is notable in that it is the first demonstration of such a localization within the CNS and includes electron microscopic, biochemical, and immunocytochemical data.
MATERIALS AND METHODS
Reagents. All reagents and chemicals were obtained from Sigma (St. Louis, MO) unless otherwise indicated.
Sequence analysis. Sequence analysis of STEP61was performed using the MacVector sequence analysis software (Eastman Kodak, New Haven, CT), and homologies with other sequences were determined using the GCG software package (University of Wisconsin). To determine PEST sequence scores, the PEST-FIND program (Rogers et al., 1986) was obtained through the generosity of Dr. M. Rechsteiner (University of Utah).
Northern analysis. Poly(A+) RNA was obtained from different mouse tissues, and ∼2 μg was electrophoresed on a 1.2% agarose–formaldehyde gel, transferred to nylon membrane, and fixed by UV irradiation (blot obtained from Clontech). A STEP61-specific probe was generated using PCR amplification and was 32P-randomly primed. Primers used to generate this 327 bp probe were 5′-AGCTCG GATCCA CTAGTA ACGGCC-3′ (sense oligomer, nucleotides 1–24) and 5′-ACATTT CTTTGT CGACGT CCACCG-3′ (antisense oligomer, nucleotides 304–327). Hybridization was performed under stringent conditions as described (Lombroso et al., 1991). Films were exposed overnight at −80°C with intensifier screens.
Filters were stripped and reprobed using a cDNA for a 28 S ribosomal protein to compare amounts of RNA loaded per lane. Equivalent amounts of mRNA were loaded in all lanes, except for a slight underloading of spleen. However, prolonged exposure (1 week) did not reveal STEP mRNA transcripts in this tissue. This is consistent with previous Northern analyses using the full-length STEP46 cDNA in which no STEP transcripts were detected in spleen after 2 week exposures (Lombroso et al., 1991).
Immunoblotting. Ten percent SDS–polyacrylamide gels were used according to the method of Laemmli (1970) and Towbin et al. (1979). Adult female Long Evans rats were killed and decapitated and their brains rapidly removed. For total brain homogenates, CNS tissue was homogenized with a Teflon homogenizer in glass at a speed of 2000 rpm for 10 strokes in homogenization buffer (0.32 msucrose, 4 mm HEPES, pH 7.3, 1 mmphenylmethylsulfonyl fluoride (PMSF), 10 mm EDTA, 1 mm benzamidine, 0.2 mg/ml aprotinin), and protein concentrations were determined using the method of Bradford (1976). Protein samples were aliquoted and stored at −80°C until further processed.
Antibodies included a monoclonal antibody generated against STEP isoforms (23E5), diluted 1:2000 (Boulanger et al., 1995); a rabbit polyclonal antibody generated against the synaptic vesicle protein synaptophysin (p38), diluted 1:250 (kindly provided by Dr. R. Jahn, Yale University School of Medicine); and a rabbit polyclonal antibody generated against the ER protein α-calnexin, diluted 1:250 (kindly provided by Dr. A. Helenius, Yale University School of Medicine). The specificity of the STEP monoclonal antibody (23E5) has been established previously by the loss of immunoreactivity on both Western blots and immunohistochemistry after preabsorption with STEP fusion protein or the peptide used as the immunogen (Boulanger et al., 1995).
Brain cell fractionation was adapted from Huttner et al. (1983) to obtain P1, P2, P3, S3, LP1, LP2, and LS2. Continuous sucrose gradients were performed following the protocol of Walch-Solimena et al. (1993). In brief, P3 and LP2 pellets were resuspended in 0.32 msucrose buffer and layered on top of a continuous sucrose gradient (0.4–2 m). Gradients were ultracentrifuged for 5 hr at 65,000 × g at 4°C. Aliquots were removed and analyzed for refractive index calculation for verification of the density gradient. Samples were stored at −80°C until further analysis.
To determine the nature of the association of STEP61 with membrane fractions, P3 pellets were washed in different buffers and ultracentrifuged, and the resulting pellet and supernatants were analyzed by immunoblotting. Approximately 1 mg of the P3 pellet was resuspended in 0.32 m sucrose, 10 mm HEPES and either 1 m NaCl, 0.1 mNa2CO3, pH 11.5, 2% Triton X-100, or 1% SDS was added to each tube. Samples were then incubated on ice for 30 min and ultracentrifuged at 200,000 × g for 1 hr (Fujiki et al., 1982). Equivalent amounts of supernatant and pellets were analyzed for STEP61 by immunoblot.
Deglycosylation. Deglycosylation of brain membrane fractions were essentially as described previously (Naegele and Barnstable, 1991). P3 and LP2 fractions were diluted to a final concentration of 1 mg/ml in incubation buffer (100 mm potassium phosphate, pH 7.9, 25 mm EDTA, 1% Triton X-100, 0.2% SDS, 1% β-mercaptoethanol, 0.5 mm PMSF), boiled for 3 min, and cooled on ice. After the addition of 25 U of N-glycosidase F (Boehringer Mannheim, Indianapolis, IN), the samples were incubated overnight at 37°C with shaking. Control samples were treated identically except that the enzyme was omitted. Samples (50 μg) were loaded onto 10% SDS–polyacrylamide gels, transferred to nitrocellulose, and immunoblotted with anti-STEP antibody (23E5) or anti-synaptophysin antibodies.
Transfections. The open reading frames (ORFs) of STEP61 and STEP46 were amplified using PCR with clone-specific primers containing an XbaI restriction site. Amplified fragments were placed in both sense and antisense orientation at the unique XbaI site of the eukaryotic expression vector pRc/CMV (In vitrogen, San Diego, CA) and checked for proper orientation by restriction enzyme digestions. DNA was purified by two cesium chloride ultracentrifugations before use in transient transfections of Chinese Hamster Ovary (CHO) cells using 10 μg of plasmid DNA with 30 μl of lipofectin (1 mg/ml, Life Technologies, Grand Island, NY) as described previously (Solimena et al., 1993).
Immunohistochemistry. A STEP61-specific antibody was generated by immunizing rabbits with a 20 amino acid synthetic peptide present in STEP61 and absent in STEP46(amino acids 36–55) (Fig. 1A). Crude antiserum from rabbit “Nod” was affinity-purified as follows: antiserum was diluted twofold in 0.1 mNa2CO3, pH 7.2 and centrifuged at 10,000 × g for 20 min, and total IgG was isolated on a G-protein column (BioRad, Melville, NY). The eluant was subsequently affinity-purified on a STEP61 fusion protein column and dialyzed extensively before use. CHO cells were transiently transfected with either STEP61 or STEP46 cDNA and were fixed and processed for immunocytochemistry as described previously (Cameron et al., 1991). For immunohistochemical analyses, dilutions for Nod ranged from 1:10 to 1:400, and for the monoclonal antibody 23E5, which recognizes all isoforms isolated to date, the dilution was 1:80. Secondary antibodies were rhodamine-conjugated goat anti-rabbit or anti-mouse IgG at 1:50 dilution. Because the Nod antiserum does not stain STEP isoforms in Western blot experiments, the monoclonal was used for those experiments (Boulanger et al., 1995).
Fig. 1.
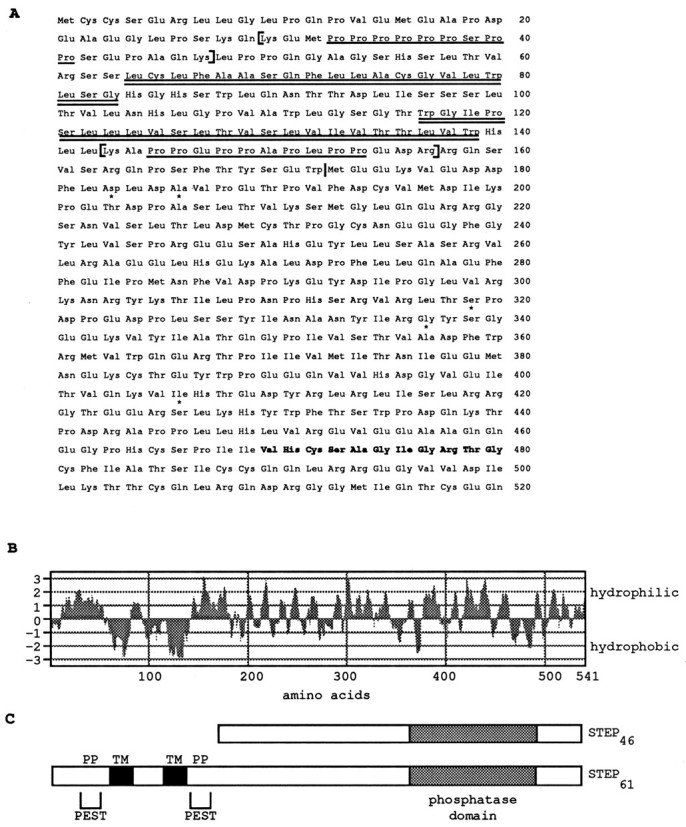
STEP61 encodes a PTP with two transmembrane, two PEST sequences, and two potential SH3 binding sites.A, The two hydrophobic domains are indicated by thedouble underline, and the two polyproline-rich domains are indicated by a single underline. Two PEST sequences are enclosed in brackets, and the phosphatase domain is shown in bold. Five conservative amino acid changes from the original rat STEP46 sequence are indicated byasterisks and reflect the expected variations of sequence between mouse and rat. STEP46 sequence begins at methionine residue 173 (indicated by a vertical bar).B, Hydrophilicity analysis of the STEP61-predicted amino acid sequence indicates two stretches of hydrophobic amino acids of 20 and 23 amino acids are present at the N terminus. The plot was obtained using the MacVector sequence analysis software and a Kyte–Doolittle algorithm with a window of seven amino acids. C, Schematic representation of STEP61 and comparison with the cytosolic STEP variant STEP46. The transmembrane domains (TM), PEST sequences, and polyproline domains (PP) are shown.
A marker for the ER, protein disulfide isomerase (PDI) (mouse monoclonal anti-PDI, clone 1D3, StressGen, Sidney, Canada) was colocalized with STEP61 by two-color immunofluorescent staining in two adult rats. The CNS was fixed by transcardiac perfusion, and staining was performed as described previously, with slight modifications (Dunn et al., 1995; Raghunathan et al., 1996). Sections were incubated in a cocktail of Nod (1:200) and anti-PDI (1:600) overnight at RT. After extensive washing, sections were labeled with a cocktail of secondary antibodies; PDI immunoreactivity was detected with a horse anti-mouse IgG-Texas Red (1:500, Vector, Burlingame, CA), and STEP61 was detected with a biotinylated goat anti-rabbit IgG (1:300, Vector), followed by streptavidin-FITC (1:1000, Vector). After extensive washes, sections were coverslipped in Vectashield (Vector) and photographed on a Zeiss Axiophot fluorescence photomicroscope.
Immunoelectron microscopy. Deeply anesthetized Long Evans rats were perfused with 0.1 m sodium phosphate buffer, pH 7.4, containing 0.1 U/ml sodium heparin, followed by perfusion with ∼250 ml of a fixative containing 4% paraformaldehyde and 0.2% glutaraldehyde in 0.1 m phosphate buffer, pH 7.4. The brains were left in the skull for 2 hr at 4°C, removed, blocked, and post-fixed in the perfusion solution for 4 hr (Naegele et al., 1988). Vibratome sections (100 μm thick) were cut in coronal plane and collected in cold 0.1 m phosphate buffer. For EM embedding, sections were post-fixed in 1% osmium tetroxide for 30 min on ice. Sections were dehydrated in ethanol and propylene oxide (EM Sciences, Gibbstown, NJ). Tissue strips of striatum were embedded in LR White (Ted Pella, Redding, CA) and cured for 48 hr at 56°C in a vacuum oven. Semithin sections (1 μm thick) were cut with an LKB ultramicrotome and stained with 1% toluidine blue and basic fuchsin. Ultrathin sections (90 nm) were cut with a diamond knife and collected on 200 mesh Formvar-coated nickel grids (EM Sciences).
The procedure for EM immunolabeling is essentially as described byGriffiths et al. (1984). Ultrathin sections were incubated on the grids in a blocking solution of 1% BSA (type V, Sigma) in PBS, then in Nod sera (1:10) or anti-synaptophysin (1:50). After extensive washing, primary antibodies were detected by incubating grids in A-protein–gold probes (15 nm, Drs. Posthuma and Slot, Utrecht University, The Netherlands) diluted 1:45 in 0.1% BSA in PBS for 1 hr at room temperature. Control sections were incubated in protein A-gold alone or in Nod antibody that was preabsorbed with 20 μg of STEP61peptide. All incubations were performed overnight at 4°C. Stained grids were washed in PBS, stabilized in 1% glutaraldehyde, washed, then stained with 2.5% uranyl acetate for 10 min and 2% lead citrate for 5 min. Sections were examined in a Zeiss transmission electron microscope with an acceleration voltage of 80 kV.
Phosphatase assays. Construction of glutathione S-transferase fusion proteins in the bacterial expression vector pGEX-2T (Smith and Johnson, 1988) was performed as described previously (Lombroso et al., 1993). The entire ORF STEP61 was used for these constructs. Controls for these experiments included a previously constructed STEP46-GST and GST-alone fusion protein (Lombroso et al., 1993). Transformed cells were induced with isopropyl-β-d-thiogalactopyranoside (IPTG), and fusion proteins were affinity-purified using glutathione-agarose beads (Sigma). For some experiments, digestion of recombinant fusion proteins was performed by the addition of excess thrombin (2 μg) to 150 μl of buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 2.5 mm CaCl2, 0.1% 2-mercaptoethanol) at room temperature for 1 hr with shaking. GST and uncut fusion protein were removed by affinity purification against glutathione–agarose beads. Enzymatic assays were conducted in the presence of 1% Triton X-100 and used paranitrophenyl phosphate (pNPP) as substrate. Assays were performed in imidazole buffer, pH 6.2, with 1.9 mmpNPP for 30 min at 30°C, and the reactions were terminated by adding 900 μl of 0.2N NaOH. The production ofparanitrophenolate ion was expressed as a concentration using a molar extinction coefficient of 1.78 × 104m−1 cm−1 at 410 nm.
RESULTS
Structural analysis of STEP61
The full-length cDNA clone encodes a peptide of 541 amino acids with a predicted MW of 61 kDa (Fig. 1A). A single PTP catalytic sequence VHCSAGIGRTG (residues 470–480) is present and conforms to the consensus sequence (I/V)HCXAGXXR(S/T)G found in members of this family (Charbonneau et al., 1989; Fischer et al., 1991). STEP61 contains the entire STEP46 amino acid sequence at its COOH end and 172 novel amino acids at its N terminus. Five amino acids differ between the shared sequences. The original STEP46 clone was isolated from a rat brain cDNA library, and these changes represent conservative amino acid changes found among related species.
The 172 amino acids at the N terminus of STEP61 contain several motifs that may have functional significance. First, there are two potential transmembrane domains of 20 and 23 hydrophobic amino acids, respectively (Fig. 1A,B). Second, two PEST sequences are present (A). These sequences are defined as stretches of amino acids that begin and end with a positively charged residue (H, K, or R) and contain at least eight internal residues. The internal sequences are enriched for P, E, D, S, and T and are identified by calculating a PEST score of between −50 and +35. To qualify as a PEST sequence, the PEST score value must be >−5.0. When the stretch lacks E/D or S/T, then its PEST score must be >0 (Wang et al., 1989). The two PEST sequences of STEP61have highly positive scores of 18 and 16, respectively. Third, two polyproline-rich domains are present and match the consensus sequence for the binding site for proteins containing SH3 domains (Fig.1A) (Ren et al., 1993; Yu et al., 1994; Cohen et al., 1995). Finally, a putative site for N-glycosylation is present at the asparagine at amino acid position 91.
Northern analyses
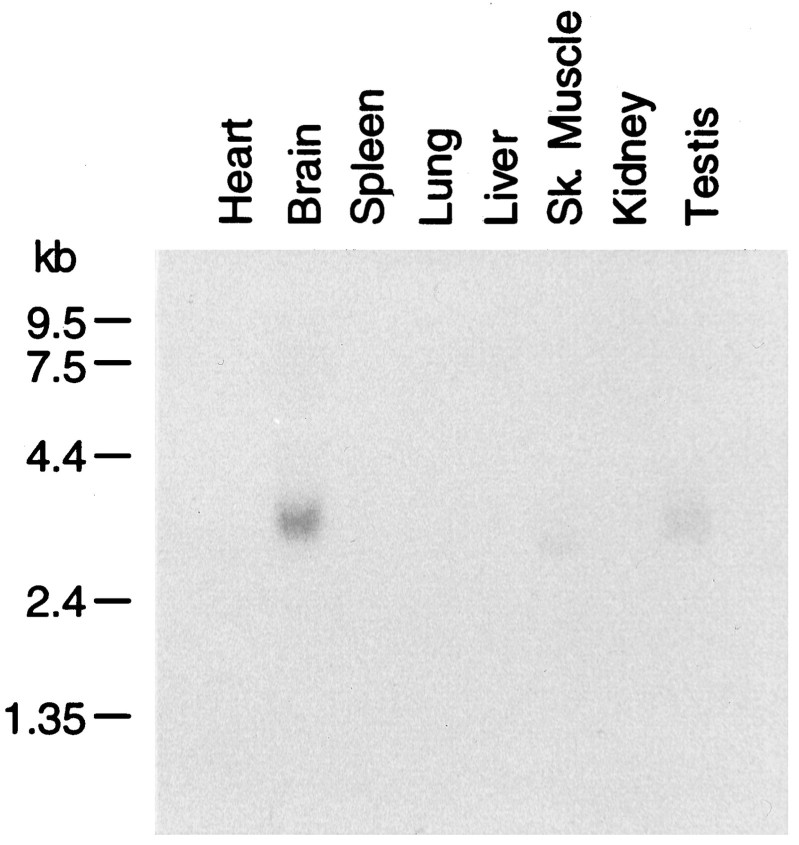
Northern analyses were performed on various mouse organs to determine the expression patterns of STEP61 transcripts (Fig. 2). An ∼3 kb transcript is detected in brain using a STEP61-specific probe. These results are similar to what has been found with all previously identified members of the STEP family, which have been detected in brain only and not in the peripheral tissues tested (Lombroso et al., 1991, 1993; Sharma et al., 1995; Raghunathan et al., 1996). In the present study, we also probed against mRNA obtained from skeletal muscle and testes. A transcript of ∼3 kb is present in testes and a smaller transcript (2.8 kb) in skeletal muscle. It is not yet known whether these additional mRNA transcripts encode STEP-related peptides or are the result of cross-hybridization with unrelated transcripts. Prolonged exposure (1 week) did not reveal hybridization signals from the other tissues tested (data not shown). The similarity in size between the cDNA (3158 bp) and the signal seen on Northern analysis suggests that the full-length, or near full-length, cDNA was obtained.
Fig. 2.
STEP61 mRNA is enriched in the CNS. Poly(A+)-selected mRNA (2 μg) from the indicated mouse organs was loaded onto a denaturing gel, electrophoresed, and transferred to nylon membrane. A STEP61-specific probe was generated by PCR and randomly primed with 32P. After 18 hr of hybridization, the blot was washed and exposed with intensifying screens overnight at −80°C.
Biochemical analyses
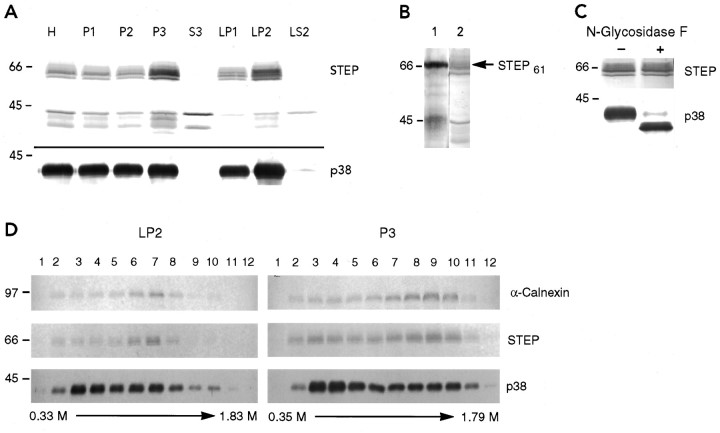
The distribution of STEP61 in brain was determined by immunoblotting after subcellular fractionations. As shown in Figure3A, a group of STEP-immunoreactive bands with apparent MWs between 60 and 66 kDa were enriched in particulate fractions (P3 and LP2) and were not detected in soluble fractions (S3 and LS2). P3 and LP2 fractions were enriched with small organelles and membranes from either neuronal cell bodies or synaptosomes, respectively. These results confirm earlier findings showing enrichment of higher MW STEP-immunoreactive polypeptides in particulate fractions, whereas lower MW STEP immunoreactive proteins are detected in soluble as well as in particulate fractions (Fig. 3A) (Boulanger et al., 1995). As expected, the membrane marker of synaptic vesicles, synaptophysin (p38) (Jahn et al., 1985), was detected in particulate fractions only.
Fig. 3.
STEP61 in brain: biochemical analysis.A, Immunoblots of subcellular fractionation of rat brain. Equivalent amounts of protein from each fraction (50 μg) were subjected to SDS-PAGE, transferred to nitrocellulose, and probed with the STEP61 monoclonal antibody 23E5 and with a polyclonal antibody against synaptophysin (p38). H, Total homogenate; P1, consisting primarily of unbroken cells and nuclei; P2, plasma membrane and larger organelles;P3, membranes of smaller organelles; S3, cytosol; LP1, pellet after hypotonic lysis of synaptosomes and contains larger organelles within synaptosomes;LP2, high-spin pellet of LS1 supernatant and containing microsomal fraction within synaptosomes; LS2, supernatant of LP2 spin. MW standards are shown on theleft. B, Thrombin digestion of STEP61-fusion protein. The released recombinant STEP polypeptide (lane 1) was compared with the mobilities of endogenous STEP proteins enriched in P3 fraction (lane 2). C, N-glycosidase F treatment of P3 membranes. Samples were processed at 37°C overnight in the presence (+) or absence (−) of enzyme. Blots were processed with antibodies against STEP (top) or against the control synaptophysin (p38, bottom). D, Distribution of STEP61 after sucrose density gradient fractionation and comparison with ER and non-ER-associated proteins. Membrane fractions enriched for the higher MW STEP-immunoreactive peptides (P3 and LP2) were applied to continuous sucrose gradients (0.4–2 m). After ultracentrifugation, equal volumes from each of 12 aliquots were loaded onto 10% SDS polyacrylamide gels and processed in parallel with antibodies against calnexin, STEP, and synaptophysin (p38), as indicated on the right. MW markers are shown on the left.
To establish the relationship between the higher MW STEP-immunoreactive bands and the product of the STEP61 cDNA, we compared the mobility of the recombinant STEP61 protein with endogenous STEP isoforms. For this purpose, the STEP61-GST fusion protein was digested with thrombin before immunoblotting (Fig.3B). The recombinant protein had a very similar mobility to the higher MW membrane-enriched isoforms present in P3 (Fig.3B, lane 2), suggesting that the isolated cDNA encodes one of these higher MW STEP variants. However, this experiment did not allow us to identify the specific polypeptide that corresponds to STEP61.
Next, we assessed whether varying levels of glycosylation might generate the multiple STEP-immunoreactive bands observed in particulate fractions. N-glycosidase F selectively removes asparagine-linked oligosaccharides from glycoproteins and should alter the electrophoretic mobility of STEP61 if it were glycosylated. Treated samples were separated by SDS-PAGE, and immunoreactivity for STEP61 and the glycoprotein synaptophysin was detected on Western blots (Fig. 3C). As reported previously (Rehm et al., 1986), digesting membrane fractions with N-glycosidase F resulted in a faster electrophoretic mobility of synaptophysin (p38), whereas none of the STEP-immunoreactive bands had altered mobilities. These results indicate that the multiple higher MW STEP-immunoreactive bands do not correspond to different N-glycosylated forms of a single STEP polypeptide.
Because STEP61 immunoreactivity is enriched in P3 and LP2 fractions, these fractions were processed further through continuous sucrose density gradients. The pattern of STEP immunoreactivity was compared with that of calnexin and synaptophysin in Figure3D. Calnexin is a resident protein of the ER (Wada et al., 1991), whereas synaptophysin is an intrinsic membrane protein of synaptic vesicles (Jahn et al., 1985). In LP2, the peak of STEP-immunoreactivity for the higher MW STEP isoforms colocalized with the peak of calnexin immunoreactivity (fraction 7; 1.04 msucrose) but not with the peak of synaptophysin (fractions 3–4; 0.61–0.69 m sucrose). In P3 fractions, the peak of STEP-immunoreactivity also overlapped with the peak of calnexin (fraction 9; 1.35 m sucrose), although a second less intense STEP-immunoreactivity peak was detected in fraction 3 (0.60m sucrose). These results suggest that the higher MW STEP isoforms are enriched in the ER.
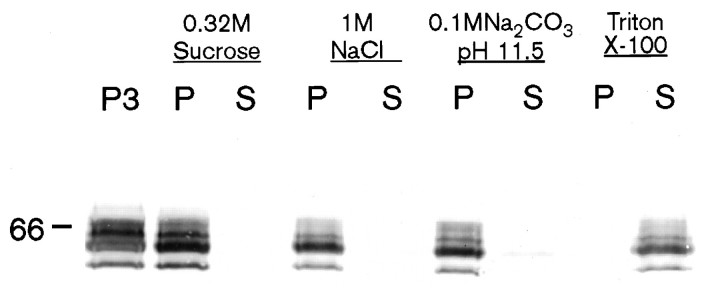
The primary amino acid sequence of STEP61 predicts for an intrinsic membrane protein with two transmembrane spanning domains. Consistent with that sequence, the higher MW STEP-immunoreactive bands were efficiently released from particulate fractions only after washes with detergents, including 2% Triton X-100 (Fig. 4,lanes 6, 7) and SDS (data not shown).
Fig. 4.
STEP61 is an integral membrane protein. Membrane fractions (P3) were washed in different buffer conditions as indicated and spun, and pellets (P) and supernatants (S) were collected. Protein (∼50 μg) from each fraction was subjected to SDS-PAGE, transferred to nitrocellulose, and probed with the monoclonal antibody 23E5. The majority of STEP isoforms remain membrane-associated until washed in 2% Triton X-100.
Immunocytochemical analyses
Peroxidase staining or two-color immunofluorescent double labeling was carried out to compare the subcellular distribution of STEP61 with PDI, a resident protein of the ER, that acts as a catalyst for rearrangement of disulfide bonds (Vaux et al., 1990). Figure 5A shows punctate immunoperoxidase staining of STEP61 in pyramidal neurons in the cerebral cortex. The staining in the perinuclear region extended into proximal dendrites. Immunofluorescent double labeling is shown in the remaining panels of Figure 5. Pyramidal neurons of the cerebral cortex contained both STEP61 (C) and PDI (D) immunoreactivity in the perinuclear region. Similarly, in pyramidal neurons of the hippocampus (Fig.5E,F), these two markers colocalized in the perinuclear region. PDI immunoreactivity did not extend far into dendrites or axons. Additionally, in the striatum, most neurons exhibited colocalization of both markers in the perinuclear region (data not shown). In contrast to the punctate perinuclear labeling by STEP61 antisera, a more diffuse pattern of staining was observed with either monoclonal or polyclonal antisera that recognized both cytosolic and membrane-associated forms of STEP (Lombroso et al., 1993; Boulanger et al., 1995). In control sections, STEP immunostaining was abolished by preabsorbing antiserum with STEP61 fusion protein (Fig. 5B).
Fig. 5.
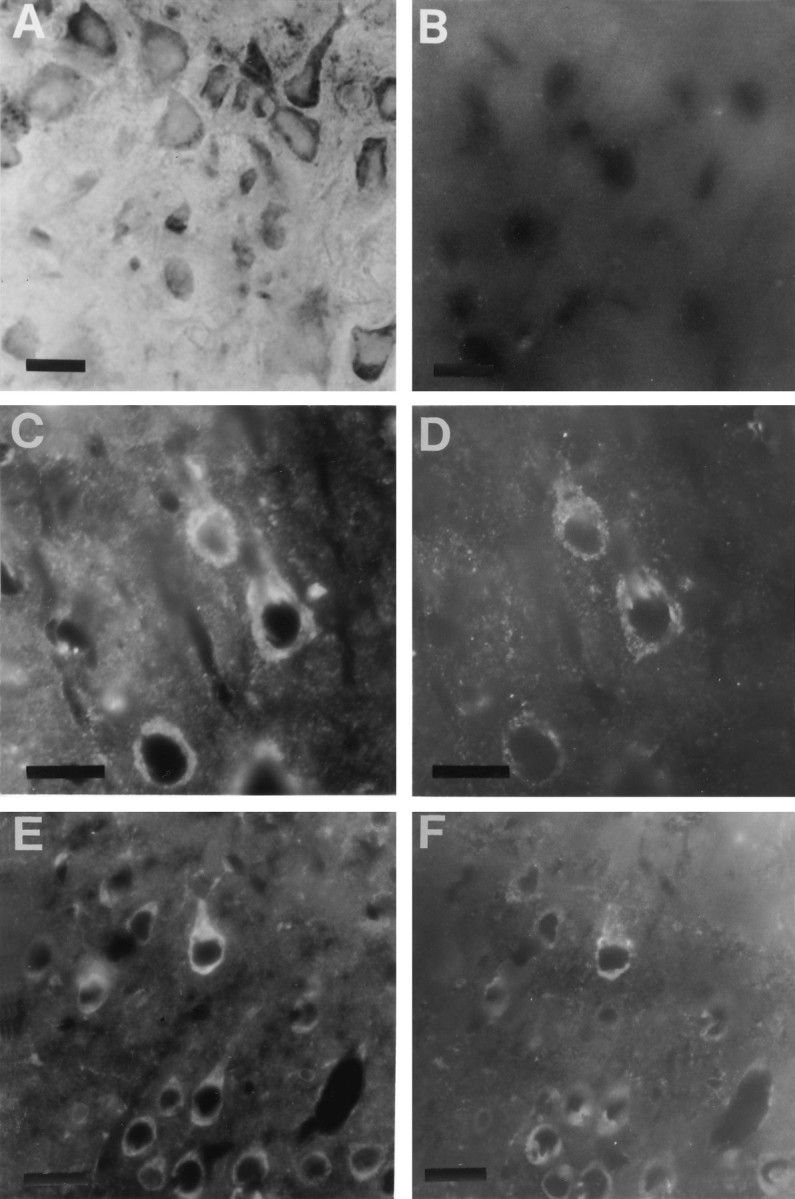
STEP61 immunoreactivity is localized in the perinuclear region. A, Immunoperoxidase labeling of cortical neurons with Nod antisera showed that the perinuclear region of pyramidal neurons was labeled. B, Preabsorption of Nod antisera with STEP61 fusion protein eliminated all immunofluorescent staining in control sections.C–F, To determine whether STEP immunoreactivity was associated with the ER, immunofluorescent double-labeling studies were performed with the Nod antisera (C, E) and an antibody specific for PDI, a resident protein of the ER (D,F). In cortical neurons (C,D), STEP colocalized with PDI in proximal dendrites and surrounding the nucleus. A similar pattern was also seen in hippocampal neurons (E, F). Scale bar, 10 μm.
The perinuclear staining for STEP isoforms was confirmed at the ultrastructural level with postembedding immunogold labeling. In the perinuclear region, punctate gold labeling was observed over free ribosomes (Fig. 6A) and rough ER (B). In control sections, immunogold labeling was abolished by preabsorption of antiserum with STEP61 peptide (C) or by omission of primary antibodies (data not shown). The Nod antiserum also failed to stain distal processes of neurons, including synaptic boutons (E). The specificity of the staining was verified by comparing STEP labeling with that of an antibody against synaptophysin (Fig.6D,F). In contrast to the STEP antiserum, synaptophysin antiserum gave little or no background labeling of rough ER in the perinuclear region (D), whereas synaptic terminals were heavily labeled (F). Omission of synaptophysin antibody in the incubation steps eliminated immunogold labeling of synaptic terminals (data not shown).
Fig. 6.
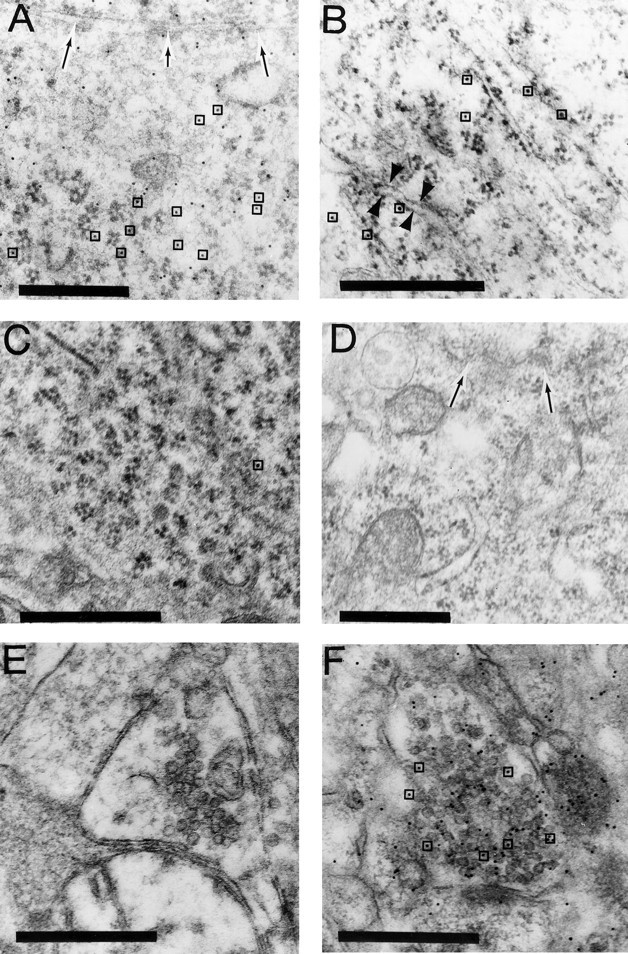
Electron microscopy demonstrates localization of STEP61 to ER. In sections incubated in Nod antibody against STEP61, a high density of gold particles was evident over free ribosomes (A) and the rough ER (B). Preabsorption of STEP antisera with STEP peptide-abolished staining (C). Specificity of staining was also shown by labeling sections with synaptophysin antibodies, which failed to label rough ER (D) but gave strong labeling of synaptic vesicles in terminal boutons (F). By contrast, STEP immunogold staining was not observed over more distal processes, including axons and synaptic boutons (E). InA and D, arrows indicate nuclear envelope. In A and C,boxes highlight some gold particles to distinguish them from ribosomes. In B, arrowheads show labeled rough ER. Scale bar, 0.5 μm.
Transfection experiments
The immunohistochemical and electron microscopic experiments presented above demonstrate that members of the STEP family are present in the ER. An independent line of investigation supports these findings. The full ORFs for STEP61 and the cytosolic variant STEP46 were transiently transfected into CHO cells, a fibroblast cell line that does not normally express STEP gene products. STEP proteins were localized using immunocytochemical staining with the monoclonal antibody 23E5, which recognizes both STEP isoforms. A reticular pattern of staining was seen after STEP61 transfection (Fig. 7,top). STEP46, instead, was evenly distributed in the cytoplasm, as expected for a soluble cytosolic protein (bottom). The polyclonal antibody Nod, generated to recognize membrane and not cytosolic variants, produced a similar staining pattern in STEP61-transfected cells but, as expected, did not stain STEP46-transfected cells (data not shown).
Fig. 7.
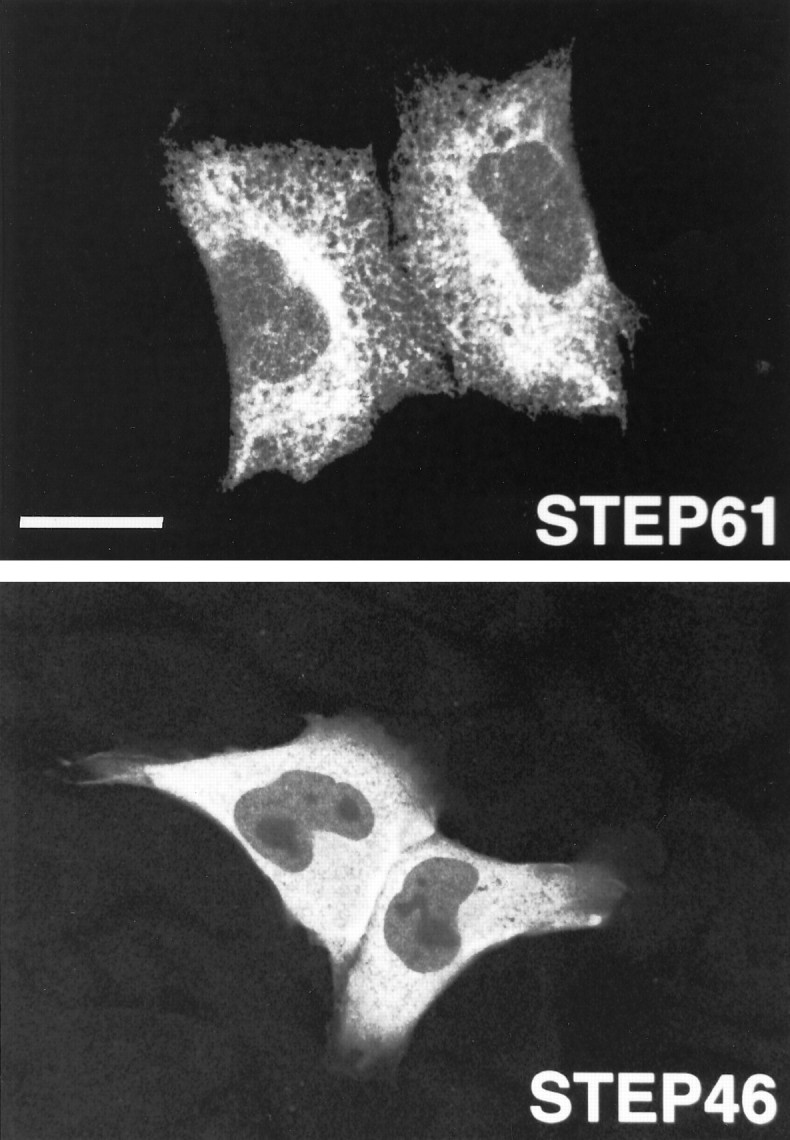
Transfection of STEP61 into fibroblasts shows a reticular staining pattern relative to the cytosolic STEP46 variant. STEP61 and STEP46 cDNAs were transiently transfected into CHO cells and detected by immunohistochemistry using the monoclonal antibody 23E5 followed by rhodamine-conjugated goat anti-mouse IgG. In transfected cells, STEP61 presented the characteristic reticular distribution of proteins associated with the ER (top). Note the perinuclear accumulation of STEP61. In contrast, STEP46 immunoreactivity was evenly distributed in the cytoplasm of transfected cells, consistent with STEP46being a soluble cytosolic protein (bottom). Scale bar, 22 μm.
Tyrosine phosphatase activity
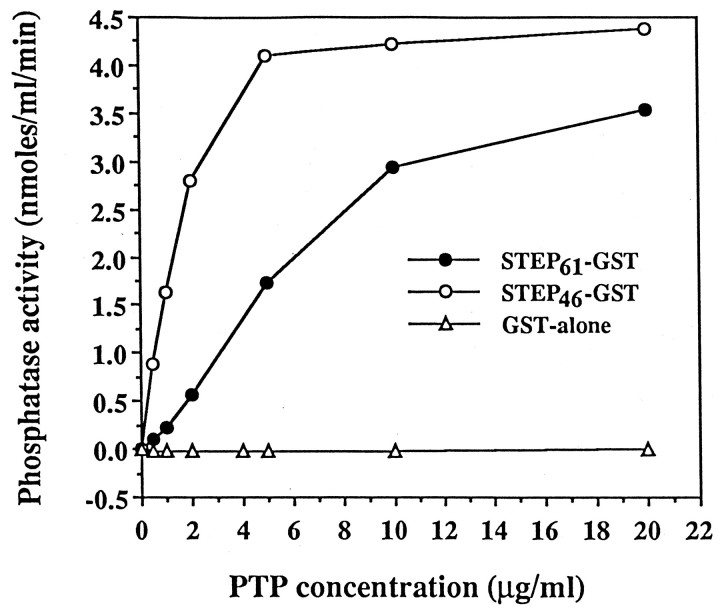
The phosphatase activity of STEP61 was compared with the activity of the cytosolic variant STEP46. Because it was possible that the STEP61 recombinant protein might form protein aggregates through its two hydrophobic domains, all phosphatase assays were performed in the presence of Triton X-100. STEP61-GST fusion protein showed phosphatase activity against pNPP substrate, but the level of activity was approximately sixfold lower than that of STEP46-GST fusion protein (Fig. 8). Phosphatase assays were also repeated after thrombin cleavage and affinity purification of STEP polypeptides, and STEP61 again had approximately 10-fold less activity than STEP46. The decrease in STEP61 activity thus was not likely to be attributable to the interference of GST. As expected, GST fusion protein alone did not have phosphatase activity.
Fig. 8.
STEP61 is less active than the cytosolic variant STEP46. The phosphatase activity of the recombinant proteins STEP61 and STEP46 was compared. STEP61 had approximately sixfold less phosphatase activity than STEP46. STEP61-GST and STEP46-GST fusion proteins were affinity-purified on glutathione–agarose beads after induction by IPTG and assayed for phosphatase activity in the presence of 1% Triton-X 100 using the substrate pNPP. The values of STEP61 and STEP46 represent the mean ± SE of two separate assays. GST fusion protein alone (GST-alone) is the negative control.
Both STEP61-GST and STEP46-GST were inhibited by the tyrosine phosphatase inhibitors sodium vanadate and ammonium molybdate (data not shown). The IC50 values for sodium vanadate were 200 nm for STEP61-GST and 1 mm for STEP46-GST. The IC50 values for ammonium molybdate were 400 nm for STEP61-GST and 200 nm for STEP46-GST. These values are similar to those obtained previously for STEP46 (Lombroso et al., 1993).
DISCUSSION
A key point emerging from the present study is that the intracellular distribution of STEP61 is markedly different from previously characterized STEP family members. The distinctive localization of STEP61 to membrane compartments has been shown by several types of experiments. Taken together, the subcellular fractionations, detergent extractions, and sucrose density gradients clearly indicate that STEP61 is an integral membrane protein and is enriched in fractions in which the endoplasmic reticular protein calnexin is also enriched. Immunofluorescent double labeling for PDI and STEP61, as well as electron microscopic localization, demonstrates that STEP61 is most enriched in the perinuclear ER. This is in contrast to the more diffuse pattern of neuronal staining that was seen with antibodies generated to recognize all STEP isoforms (Lombroso et al., 1993; Boulanger et al., 1995), suggesting that the polyclonal antibody (Nod) recognizes a subgroup of STEP isoforms.
The transfection experiments provide additional support for these findings. In isolation, transfection experiments must be viewed with caution, because it is known that some proteins are not targeted to their proper final destination when transfected into cells that do not normally process them. However, the purpose of these experiments was to determine the targeting pattern of recombinant STEP61 in cells that do not have additional STEP isoforms present and compare it to the pattern seen with the cytosolic variant STEP46. The results lend support to the hypothesis that the N-terminal region of STEP61 confers to this protein an intracellular localization pattern that is distinct from the cytosolic pattern seen with STEP46.
Although these experiments do not rule out the possibility that a pool of STEP61 is associated with other membrane compartments, they clearly show that STEP61 is enriched in the ER of neurons. The presence of two hydrophobic domains in STEP61that are not present in other known STEP variants suggests that these domains provide the necessary information for the selective compartmentalization of STEP61 to neuronal ER. The differential centrifugation results indicate that several other higher MW membrane-associated STEP isoforms exist, as well as a pool of lower MW STEP protein that are present in both particulate and soluble fractions (Fig. 3A). It is possible that these STEP isoforms will also be found to localize to the ER or to additional intracellular membranes. However, the antiserum used in this study failed to detect SDS-denatured proteins on Western blots and could not be used to test this hypothesis. To identify the subcellular targets of these other variants, specific antibodies will need to be generated.
The present study supports recent findings of important amino acid domains outside the phosphatase domain. For example, PTPs have been identified containing Src homology 2 domains (Shen et al., 1991; Matthews et al., 1992; Plutzky et al., 1992; Yi et al., 1992;Pawson, 1995), and polyproline-rich sequences that match the consensus sequence for the binding site of Src homology 3 domains have also been identified (Sawada et al., 1994). These domains are thought to provide a mechanism by which PTPs associate with downstream effector molecules.
A number of studies have demonstrated that alternative splicing is responsible for targeting PTPs to distinct intracellular regions and compartments (Matthews et al., 1990; Price, 1992; McLaughlin and Dixon, 1993; Oon et al., 1993; Mauro and Dixon, 1994; Elson and Leder, 1995). This has the effect of compartmentalizing PTPs in the vicinity of their substrates or anchoring them to membrane storage sites until released or activated by appropriate intracellular signals. The present study on STEP61 extends these findings to the CNS. We have shown that alternative splicing within the STEP family leads to the production of either cytosolic polypeptides or proteins targeted to the ER.
Although the functional significance of having STEP61associated with the ER is not yet known, recent studies on a sterol regulatory element-binding protein 1 (SREBP-1) are relevant. SREBP-1 is a transcription factor that is synthesized as a 125 kDa precursor attached to the nuclear envelope and ER (Wang et al., 1994). Under the appropriate cellular signal (low intracellular concentration of cholesterol), the membrane-bound precursor is cleaved to generate a smaller cytosolic fragment that rapidly translocates to the nucleus, where it activates transcription of proteins involved in sterol pathways (Wang et al., 1994).
Several observations suggest that an analogous mechanism might be at work with STEP61. We have shown that STEP61 is bound to the ER membrane. In addition, PEST sequences are present, and these sequences are thought to signal proteolytic cleavage of the proteins in which they are found. They have now been identified in several additional PTPs (Matthews et al., 1992; Takekawa et al., 1992;Yang et al., 1993; Garton and Tonks, 1994), although it has not yet been determined in these proteins whether the PEST sequences are functional. If proteolysis of STEP61 were to occur, then the predicted MW of the largest released fragment would be ∼44 kDa. This size is close to the observed mobilities of some members of the cytosolic group of STEP-immunoreactive proteins. In future studies, it may be possible to determine whether higher MW STEP proteins are cleaved to release the cytosolic isoforms by employing [35S]methionine pulse chase experiments. Alternatively, careful peptide mapping or amino acid sequencing of each of the STEP immunoreactive bands will be necessary to determine the relationship of the different variants to each other.
Similarly to STEP, two other intracellular PTPs that have been localized to the ER (Frangioni et al., 1993; Lorenzen et al., 1995). It is interesting to note that proteolysis has been suggested as a mechanism that modulates the enzymatic activity for both of these PTPs. Cleavage of the C-terminal sequence of T-cell PTP stimulates its phosphatase activity in vitro (Cool et al., 1990; Zander et al., 1991), and PTP1B shows a twofold increase of phosphatase activity after limited proteolysis by calpain (Frangioni et al., 1993).
In this study, we have shown that the recombinant membrane-associated isoform STEP61 has significantly less phosphatase activity than the cytosolic variant STEP46. This was a surprising observation, because STEP46 is contained entirely within STEP61, and the only difference in their sequences are the novel 172 amino acids at the N terminus of STEP61. There are several possible explanations for the observed decrease in enzymatic activity. The N-terminal extension of the recombinant STEP61 protein may directly interfere by limiting the accessibility of pNPP to the catalytic domain. In addition, aggregation of recombinant protein is likely to occur through the two hydrophobic domains. We attempted to address these issues by performing all enzymatic assays in the presence of detergent as well as digesting with thrombin before phosphatase assays. The difference in activity remained, suggesting that neither protein aggregation nor interference by GST accounts for the decrease of phosphatase activity seen with STEP61. Nonetheless, the assays were conducted against an artificial substrate, and additional work is required to demonstrate whether this decrease in activity occurs in vivo.
In addition to the PEST sequences, STEP61 has two polyproline motifs that match the consensus sequence for the binding site of SH3 domains (Ren et al., 1993; Cohen et al., 1995). Each of the PEST sequences contains one of the polyproline-rich domains and suggests a possible functional relationship. Protein–protein interactions are capable of masking underlying PEST signals, thereby preventing proteolysis of these proteins and prolonging their half-life (Shanklin et al., 1987; Rechsteiner, 1988).
Based on these considerations, a model for how STEP61functions within neurons can now be proposed. STEP61 is normally attached to membrane compartments. In this form, STEP61 interacts with other protein(s), and these protein–protein interactions mask underlying PEST sequences. Only after neuronal stimulation (e.g., growth factor or neurotransmitter binding that leads to phosphorylation or Ca2+ influx) are the protein–protein complexes disrupted and the proteolytic sites exposed. STEP61 is then cleaved, and smaller isoforms are released into the cytosol.
In conclusion, the present study has characterized a new member of the STEP family of brain-enriched PTPs. We have shown that this family consists of cytosolic and transmembrane isoforms produced by alternative splicing mechanisms. The biological effects of alternative splicing include changing the subcellular localization of protein isoforms as well as modulating their enzymatic activity. The significance of these mechanisms is especially evident when the proteins are regulatory molecules, such as PTPs, in which subtle changes in their structure may effect their localization pattern, enzymatic activity, or potential access to substrate molecules.
Footnotes
This work was supported by the National Alliance for Research on Schizophrenia and Depression, NATO, and National Institutes of Health Grants P0149351, MH52711, NS35989 (P.J.L.), MH18268 (A.B.), and EY09749 (J.R.N.), and by a Juvenile Diabetes Foundation Career Development Award (M.S.). The sequence reported in this article was submitted to GenBank under accession number U28217. A.B. and F.Z. contributed equally to this work. We thank P. DeCamilli, R. Jahn, J. Leckman, M. Ogren, and F. Vaccarino for critical review of this manuscript; and Reinhard Jahn and Ari Helenius, Yale University School of Medicine, for antibodies against synaptophysin and calnexin.
Correspondence should be addressed to Dr. Paul Lombroso, Child Study Center, 230 South Frontage Road, New Haven, CT 06520-0900.
Dr Bult’s present address: Michigan State University, Department of Psychology, Psychology Research Building, East Lansing, MI 48824.
REFERENCES
- 1.Atashi JR, Klinz SG, Ingraham CA, Matten WT, Schachner M, Maness PF. Neural cell adhesion molecules modulate tyrosine phosphorylation of tubulin in nerve growth cone membranes. Neuron. 1992;8:831–842. doi: 10.1016/0896-6273(92)90197-l. [DOI] [PubMed] [Google Scholar]
- 2.Boulanger LM, Lombroso PJ, Raghunathan A, During MJ, Wahle P, Naegele JR. Cellular and molecular characterization of a brain-enriched protein tyrosine phosphatase. J Neurosci. 1995;15:1532–1544. doi: 10.1523/JNEUROSCI.15-02-01532.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Bult A, Naegele JR, Sharma E, Lombroso PJ. Isolation and characterization of brain enriched protein tyrosine phosphatases. Neuroprotocols. 1995;6:91–104. [Google Scholar]
- 5.Cameron PL, Südhof TC, Jahn R, De Camilli P. Colocalization of synaptophysin with transferrin receptors: implications for synaptic vesicle biogenesis. J Cell Biol. 1991;115:151–164. doi: 10.1083/jcb.115.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charbonneau H, Tonks NK. 1002 protein phosphatases? Annu Rev Cell Biol. 1992;8:463–493. doi: 10.1146/annurev.cb.08.110192.002335. [DOI] [PubMed] [Google Scholar]
- 7.Charbonneau H, Tonks NK, Dumar S, Diltz C, Harrylock M, Cool DE, Krebs EG, Fischer EJ, Walsh KA. Human placenta protein-tyrosine-phosphatase: amino acid sequence and relationship to a family of receptor-like proteins. Proc Natl Acad Sci USA. 1989;89:5252–5256. doi: 10.1073/pnas.86.14.5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen GB, Ren R, Baltimore D. Modular binding domains in signal transduction proteins. Cell. 1995;80:237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- 9.Cool D, Tonks N, Charbonneau H, Walsh K, Fischer E, Krebs E. Expression of a human T-cell protein-tyrosine-phosphatase in baby hamster kidney cells. Proc Natl Acad Sci USA. 1990;87:7280–7284. doi: 10.1073/pnas.87.18.7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cordon-Cardo C, Tapley P, Jing S, Nanduri V, O’Rourke E, Lamballe F, Kovary K, Klein R, Jones KR, Reichardt LF, Barbacid M. The trk tyrosine protein kinase mediates the mitogenic properties of nerve growth factor and neurotrophin-3. Cell. 1991;66:1–20. doi: 10.1016/0092-8674(91)90149-s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cudmore SB, Gurd JW. Postnatal age and protein tyrosine phosphorylation at synapses in the developing rat brain. J Neurochem. 1991;57:1240–1248. doi: 10.1111/j.1471-4159.1991.tb08285.x. [DOI] [PubMed] [Google Scholar]
- 12.Desai C, Gindhart J, Goldstein L, Zinn K. Receptor tyrosine phosphatases are required for motor axon guidance in the Drosophila embryo. Cell. 1996;84:599–609. doi: 10.1016/s0092-8674(00)81035-1. [DOI] [PubMed] [Google Scholar]
- 13.Doherty P, Walsh FS. Cell adhesion molecules, second messengers and axonal growth. Curr Opin Neurobiol. 1992;2:595–601. doi: 10.1016/0959-4388(92)90024-f. [DOI] [PubMed] [Google Scholar]
- 14.Dunn JA, Kirsch JD, Naegele JR. Transient immunoglobulin-like molecules are present in the subplate zone and cerebral cortex during postnatal development. Cereb Cortex. 1995;5:494–505. doi: 10.1093/cercor/5.6.494. [DOI] [PubMed] [Google Scholar]
- 15.Elson A, Leder P. Identification of a cytoplasmic, phorbol ester-inducible isoform of protein tyrosine phosphatase epsilon. Cell Biol. 1995;92:12235–12239. doi: 10.1073/pnas.92.26.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faure R, Posner B. Differential intracellular compartmentalization of phosphotyrosine phosphatases in a glial cell line: TC-PTP versus PTP-1B. Glia. 1993;9:311–314. doi: 10.1002/glia.440090409. [DOI] [PubMed] [Google Scholar]
- 17.Fischer EH, Charbonneau H, Tonks NK. Protein tyrosine phosphatases: a diverse family of intracellular and transmembrane enzymes. Science. 1991;253:401–406. doi: 10.1126/science.1650499. [DOI] [PubMed] [Google Scholar]
- 18.Flores E, Roy G, Patel D, Shaw A, Thomas M. Nuclear localization of the PEP protein tyrosine phosphatase. Mol Cell Bio. 1994;14:4938–4946. doi: 10.1128/mcb.14.7.4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frangioni JV, Beahm PH, Shifrin V, Jost CA, Neel BG. The nontransmembrane tyrosine phosphatase PTP1B localizes to the endoplasmic reticulum via its 35 amino acid C-terminal sequence. Cell. 1992;68:545–560. doi: 10.1016/0092-8674(92)90190-n. [DOI] [PubMed] [Google Scholar]
- 20.Frangioni JV, Oda A, Smith M, Salzman EW, Neel BG. Calpain-catalyzed cleavage and subcellular relocation of protein phosphotyrosine phosphatase 1B (PTP-1B) in human platelets. EMBO J. 1993;12:4843–4856. doi: 10.1002/j.1460-2075.1993.tb06174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujiki Y, Hubard A, Fowler S, Lazarow P. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garton A, Tonks N. PTP-PEST: a protein tyrosine phosphatase regulated by serine phosphorylation. EMBO J. 1994;13:3763–71. doi: 10.1002/j.1460-2075.1994.tb06687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Girault J-A, Chamak B, Bertuzzi G, Tixier H, Wang JK, Pang DT, Greengard P. Protein phosphotyrosine in mouse brain: developmental changes and regulation by epidermal growth factor, type I insulin-like growth factor, and insulin. J Neurochem. 1992;58:518–528. doi: 10.1111/j.1471-4159.1992.tb09751.x. [DOI] [PubMed] [Google Scholar]
- 24.Griffiths G, McDowall A, Back R, Dubocher J. On the preparation of cryosections for immunocytochemistry. J Ultrastruct Res. 1984;89:65–78. doi: 10.1016/s0022-5320(84)80024-6. [DOI] [PubMed] [Google Scholar]
- 25.Gu MX, York JD, Warshawsky I, Majerus PW. Identification, cloning, and expression of a cytosolic megakaryocyte protein-tyrosine-phosphatase with sequence homology to cytoskeletal protein 4.1. Proc Natl Acad Sci USA. 1991;88:5867–5871. doi: 10.1073/pnas.88.13.5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huttner WB, Schiebler W, Greengard P, DeCamilli P. Synapsin I (protein I), a nerve terminal specific phosphoprotein. III. Its association with synaptic vesicles studied in a highly purified synaptic vesicle preparation. J Cell Biol. 1983;96:1374–1388. doi: 10.1083/jcb.96.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jahn R, Schiebler W, Ouimet C, Greengard P. A 38,000-dalton membrane protein (p38) present in synaptic vesicles. Proc Natl Acad Sci USA. 1985;82:4137–4141. doi: 10.1073/pnas.82.12.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaplan DR, Hempstead BL, Martin-Zanca D, Chao MV, Parada LF. The trk proto-oncogene product: a signal transducing receptor for nerve growth factor. Science. 1991;252:554–557. doi: 10.1126/science.1850549. [DOI] [PubMed] [Google Scholar]
- 29.Klein R, Jing S, Nanduri V, O’Rourke E, Barbacid M. The trk proto-oncogene encodes a receptor for nerve growth factor. Cell. 1991;65:189–197. doi: 10.1016/0092-8674(91)90419-y. [DOI] [PubMed] [Google Scholar]
- 30.Krueger N, Van Vactor D, Wan H, Gelbart W, Goodman C, Saito H. The transmembrane tyrosine phosphatase DLAR controls motor axon guidance in Drosophila. Cell. 1996;84:611–622. doi: 10.1016/s0092-8674(00)81036-3. [DOI] [PubMed] [Google Scholar]
- 31.Laemmli UK. Cleavage of structural proteins during the preassembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 32.Li X, Luna J, Lombroso P, Francke U. Molecular cloning of the human homolog of a striatum-enriched phosphatase (STEP) gene and chromosomal mapping of the human and murine loci. Genomics. 1995;28:442–449. doi: 10.1006/geno.1995.1173. [DOI] [PubMed] [Google Scholar]
- 33.Lombroso PJ, Murdoch G, Lerner M. Molecular characterization of a protein-tyrosine phosphatase enriched in striatum. Proc Natl Acad Sci USA. 1991;88:7242–7246. doi: 10.1073/pnas.88.16.7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lombroso PJ, Naegele JR, Sharma E, Lerner M. A protein tyrosine phosphatase expressed within dopaminoceptive neurons of the basal ganglia and related structures. J Neurosci. 1993;13:3064–3074. doi: 10.1523/JNEUROSCI.13-07-03064.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lorenzen JA, Dadabay CY, Fischer EH. COOH-terminal sequence motifs target the T cell protein phosphatase to the ER and nucleus. J Cell Biol. 1995;131:631–643. doi: 10.1083/jcb.131.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maness PF, Aubury M, Shores CG, Frame L, Pfenniger KH. c-src gene product in developing rat brain is enriched in nerve growth cone membranes. Proc Natl Acad Sci USA. 1988;85:5001–5005. doi: 10.1073/pnas.85.14.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matthews R, Cahir E, Thomas M. Identification of an additional member of the protein tyrosine phosphatase family: evidence for alternative splicing in the PTP domain. Proc Natl Acad Sci USA. 1990;87:4444–4448. doi: 10.1073/pnas.87.12.4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matthews R, Bowne D, Flores E, Thomas M. Characterization of hematopoietic intracellular protein tyrosine phosphatases: description of a phosphatase containing an SH2 domain and another enriched in proline-, glutamic acid-, serine-, and threonine-rich sequences. Mol Cell Biol. 1992;12:2396–2405. doi: 10.1128/mcb.12.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mauro LJ, Dixon JE. “Zip codes” direct intracellular protein tyrosine phosphatases to the correct cellular “address”. Trends Biochem Sci. 1994;19:151–155. doi: 10.1016/0968-0004(94)90274-7. [DOI] [PubMed] [Google Scholar]
- 40.McLaughlin S, Dixon JE. Alternative splicing gives rise to a nuclear protein tyrosine phosphatase in Drosophila. J Biol Chem. 1993;268:6839–6842. [PubMed] [Google Scholar]
- 41.Naegele J, Barnstable C. A carbohydrate epitope defined by monoclonal antibody VC1.1 is found on N-CAM and other cell adhesion molecules. Brain Res. 1991;559:118–129. doi: 10.1016/0006-8993(91)90294-6. [DOI] [PubMed] [Google Scholar]
- 42.Naegele J, Lombroso P. Protein tyrosine phosphatases in the central nervous system. Crit Rev Neurobiol. 1994;9:115–124. [PubMed] [Google Scholar]
- 43.Naegele J, Arimatsu Y, Schwartz P, Barnstable C. Selective staining of a subset of GABAergic neurons in cat visual cortex by monoclonal antibody VC1.1. J Neurosci. 1988;8:79–89. doi: 10.1523/JNEUROSCI.08-01-00079.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oon S, Hong A, Yang X, Chia W. Alternative splicing in a novel tyrosine phosphatase gene (DPTP4E) of Drosophila melanogaster generates two large receptor-like proteins which differ in their carboxyl termini. J Biol Chem. 1993;268:23964–23971. [PubMed] [Google Scholar]
- 45.Pawson T. Protein modules and signalling networks. Nature. 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 46.Plutzky J, Neel B, Rosenberg R. Isolation of a src homology 2-containing tyrosine phosphatase. Proc Natl Acad Sci USA. 1992;89:1123–1127. doi: 10.1073/pnas.89.3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Price BD. Signalling across the endoplasmic reticulum membrane: potential mechanisms. Cell Signal. 1992;4:465–470. doi: 10.1016/0898-6568(92)90015-z. [DOI] [PubMed] [Google Scholar]
- 48.Qu Z, Moritz E, Huganir RL. Regulation of tyrosine phosphorylation of the nicotinic acetylcholine receptor at the rat neuromuscular junction. Neuron. 1990;2:367–378. doi: 10.1016/0896-6273(90)90049-l. [DOI] [PubMed] [Google Scholar]
- 49.Raghunathan A, Matthews G, Lombroso P, Naegele J. Transient compartmental expression of a family of protein tyrosine phosphatases in the developing striatum. Dev Brain Res. 1996;91:190–199. doi: 10.1016/0165-3806(95)00176-x. [DOI] [PubMed] [Google Scholar]
- 50.Rechsteiner M. Regulation of enzyme levels by proteolysis: the role of pest regions. Adv Enzyme Regul. 1988;27:135–151. doi: 10.1016/0065-2571(88)90014-3. [DOI] [PubMed] [Google Scholar]
- 51.Rehm H, Wiedenmann B, Betz H. Molecular characterization of synaptophysin, a major calcium-binding protein of the synaptic vesicle membrane. EMBO J. 1986;5:535–541. doi: 10.1002/j.1460-2075.1986.tb04243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ren R, Mayer BJ, Cicchetti P, Baltimore D. Identification of a 10-amino acid proline-rich SH3 binding site. Science. 1993;259:1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- 53.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 54.Sahin M, Hockfield S. Protein tyrosine phosphatases expressed in the developing rat brain. J Neurosci. 1993;13:4968–4978. doi: 10.1523/JNEUROSCI.13-11-04968.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sawada M, Ogata M, Fujino Y, Hamaoka T. cDNA cloning of a novel protein tyrosine phosphatase with homology to cytoskeletal protein 4.1 and its expression in T-lineage cells. Biochem Biophys Res Commun. 1994;203:479–484. doi: 10.1006/bbrc.1994.2207. [DOI] [PubMed] [Google Scholar]
- 56.Schlessinger J, Ullrich A. Growth factor signaling by receptor tyrosine kinases. Neuron. 1992;9:383–391. doi: 10.1016/0896-6273(92)90177-f. [DOI] [PubMed] [Google Scholar]
- 57.Shanklin J, Jabben M, Vierstra R. Red light-induced formation of ubiquitin-phytochrome conjugates: identification of possible intermediates of phytochrome degradation. Proc Natl Acad Sci USA. 1987;84:359–363. doi: 10.1073/pnas.84.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharma E, Zhao F, Bult A, Lombroso PJ. Isolation and characterization of STEP20: an isoform of a family of brain-enriched protein tyrosine phosphatases. Mol Brain Res. 1995;32:87–93. doi: 10.1016/0169-328x(95)00066-2. [DOI] [PubMed] [Google Scholar]
- 59.Shen S, Bastien L, Posner B, Chretien P. A protein-tyrosine phosphatase with sequence similarity to the SH2 domain of the protein-tyrosine kinases. Nature. 1991;352:736–739. doi: 10.1038/352736a0. [DOI] [PubMed] [Google Scholar]
- 60.Smith DB, Johnson KS. Single step purification of polypeptides expressed in E. coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 61.Solimena M, Aggujaro D, Muntzel C, Dirkx R, Butler M, De Camilli P, Hayday A. Association of GAD65, but not of GAD67, with the Golgi complex is mediated by the NH2 terminal region. Proc Natl Acad Sci USA. 1993;90:3073–3077. doi: 10.1073/pnas.90.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Solimena M, Dirkx R, Hermel J-M, Pleasic-Williams S, Shapiro JA, Rabin DU. ICA 512, an autoantigen of insulin-dependent diabetes mellitus is an ubiquitous intrinsic membrane protein of neurosecretory granules. EMBO J. 1996;15:2102–2114. [PMC free article] [PubMed] [Google Scholar]
- 63.Takekawa M, Itoh F, Hinoda Y, Arimura Y, Toyota M, Sekiya M, Adachi M, Imai K, Yachi A. Cloning and characterization of a human cDNA encoding a novel putative cytoplasmic protein-tyrosine-phosphatase. Biochem Biophys Res Commun. 1992;189:1223–1230. doi: 10.1016/0006-291x(92)92335-u. [DOI] [PubMed] [Google Scholar]
- 64.Tonks NK, Yang Q, Guida P. Structure, regulation, and function of protein tyrosine phosphatases. Cold Spring Harb Symp Quant Biol. 1991;56:265–273. doi: 10.1101/sqb.1991.056.01.032. [DOI] [PubMed] [Google Scholar]
- 65.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vaux D, Tooze J, Fuller S. Identification by anti-idiotype antibodies of an intracellular membrane protein that recognizes a mammalian endoplasmic reticulum retention signal. Nature. 1990;345:495–502. doi: 10.1038/345495a0. [DOI] [PubMed] [Google Scholar]
- 67.Wada I, Rindress D, Cameron P, Ou W, Doherty J, Louvard D, Bell A, Dignard D, Thomas D, Bergeron J. SSRα and associated calnexin are major calcium binding proteins of the endoplasmic reticulum membrane. J Biol Chem. 1991;266:19599–19610. [PubMed] [Google Scholar]
- 68.Walch-Solimena C, Takei K, Marek K, Midyett K, Sudhof T, De Camilli P, Jahn R. Synaptotagmin: a membrane constituent of neuropeptide-containing large dense-core vesicles. J Neurosci. 1993;13:3895–3903. doi: 10.1523/JNEUROSCI.13-09-03895.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walton DM, Dixon JE. Protein tyrosine phosphatase. Annu Rev Biochem. 1993;62:101–120. doi: 10.1146/annurev.bi.62.070193.000533. [DOI] [PubMed] [Google Scholar]
- 70.Walton KM, Martell KJ, Kwak SP, Dixon JE, Largent BL. A novel receptor-type protein tyrosine phosphatase is expressed during neurogenesis in the olfactory neuroepithelium. Neuron. 1993;11:387–400. doi: 10.1016/0896-6273(93)90193-u. [DOI] [PubMed] [Google Scholar]
- 71.Wang K, Villalobo A, Roufogalis B. Calmodulin-binding proteins as calpain substrates. Biochem J. 1989;262:693–706. doi: 10.1042/bj2620693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang X, Sato R, Brown M, Hua X, Goldstein J. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 73.Winslow J, Moran P, Valverde J, Shih A, Yuan J, Wong S, Tsai S, Goddard A, Henzel W, Hefti F, Beck K, Caras I. Cloning of AL-1, a ligand for an Eph-related tyrosine kinase receptor involved in axon bundle formation. Neuron. 1995;14:973–981. doi: 10.1016/0896-6273(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 74.Woodford-Thomas TA, Rhodes JD, Dixon JE. Expression of a protein tyrosine phosphatase in normal and v-src-transformed mouse 3T3 fibroblasts. J Cell Biol. 1992;117:401–414. doi: 10.1083/jcb.117.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang Q, Tonks NK. Isolation of a cDNA clone encoding a human protein-tyrosine phosphatase with homology to the cytoskeletal-associated proteins band 4.1, ezrin, and talin. Proc Natl Acad Sci USA. 1991;88:5949–5953. doi: 10.1073/pnas.88.14.5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang Q, Co D, Sommercorn J, Tonks NK. Cloning and expression of PTP-PEST. A novel, human, nontransmembrane protein tyrosine phosphatase. J Biol Chem. 1993;268:6622–6628. [PubMed] [Google Scholar]
- 77.Yi T, Cleveland J, Ihle J. Protein tyrosine phosphatase containing SH2 domains: characterization, preferential expression in hematopoietic cells, and localization to human chromosome 12p12–p13. Mol Cell Biol. 1992;12:836–846. doi: 10.1128/mcb.12.2.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu H, Chen JK, Feng S, Dalgarno DC, Brauer AW, Schreiber SL. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- 79.Zander NF, Lorenzen J, Cool D, Tonks N, Daum G, Krebs E, Fischer E. Purification and characterization of a human recombinant T-cell protein-tyrosine-phosphatase from a baculovirus expression system. Biochemistry. 1991;30:6964–6970. doi: 10.1021/bi00242a022. [DOI] [PubMed] [Google Scholar]
- 80.Zhang JS, Longo FM. LAR tyrosine phosphatase receptor: alternative splicing is preferential to the nervous system, coordinated with cell growth and generates novel isoforms containing extensive CAG repeats. J Cell Biol. 1995;128:415–431. doi: 10.1083/jcb.128.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]