Abstract
cDNAs encoding two novel 25 kDa Ras-like proteins, Rit and Rin, were isolated from mouse retina using a degenerate PCR-based cloning strategy. Using the expressed sequence tag database, human orthologs were also obtained and sequenced. The protein sequences of Rit and Rin, which are 64% identical, are more similar to each other than to any known Ras protein. Their closest homologs in the databases areMucor racemosus Ras2 and Ras3, to which they show ∼48% identity. Rit and Rin both bind GTP in vitro. An unusual feature of their structure is that they lack a known recognition signal for C-terminal lipidation, a modification that is generally necessary for plasma membrane association among the Ras subfamily of proteins. Nonetheless, transiently expressed Rit and Rin are plasma membrane-localized. Both proteins contain a C-terminal cluster of basic amino acids, which could provide a mechanism for membrane association. Deletion analysis suggested that this region is important for Rit membrane binding but is not necessary for Rin. Rit, like most Ras-related proteins, is ubiquitously expressed. Rin, however, is unusual in that it is expressed only in neurons. In addition, Rin binds calmodulin through a C-terminal binding motif. These results suggest that Rit and Rin define a novel subfamily of Ras-related proteins, perhaps using a new mechanism of membrane association, and that Rin may be involved in calcium-mediated signaling within neurons.
Keywords: small G-protein, Ras-like protein, CAAX box, calcium signaling, signal transduction, neuron-specific, calmodulin
The Ras superfamily of small GTP-binding (G) proteins comprises a group of structurally related proteins of low molecular weight (20–30 kDa). They are involved in signal transduction and the regulation of a wide variety of cellular processes, including cell growth, transformation, differentiation and morphogenesis, vesicle trafficking and secretion, nucleocytoplasmic transport, and apoptosis (Hall, 1990; Bischoff and Ponstingl, 1991; Pryer et al., 1992; Fischer et al., 1994; Hall, 1994). Based on structural and functional similarities, five broad subgroups of small G-proteins have been defined: Ras, Rho, Rab, Ran, and ADP-ribosylation factor. Despite the differences between the subfamilies, all small G-proteins contain five highly conserved domains (G1–G5) and act as molecular switches by alternating between an active GTP-bound form and an inactive GDP-bound form (Hall, 1990; Bourne et al., 1991; Lowy and Willumsen, 1993). The relative proportion of molecules in the active versus the inactive configuration is influenced by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) (Bollag and McCormick, 1991). GEFs favor the active form by inducing the release of GDP, whereas GAPs favor the inactive form by stimulating the intrinsic GTPase activity of the G-protein.
Most members of the Ras subfamily are plasma membrane-associated (Casey, 1994, 1995). Membrane binding generally requires a C-terminal isoprenyl group, which is added post-translationally by a mechanism that involves recognition of a terminal cysteine-aliphatic amino acid-aliphatic amino acid-any amino acid (CAAX) motif. [In some Ras superfamily members, CXC or CC motifs are present instead of CAAX (Hancock et al., 1989).] A second component of binding energy is often provided by either internal palmitoylation or a C-terminal cluster of basic amino acids (Hancock et al., 1990; Cadwallader et al., 1994). Some unusual Ras-related proteins such as Rad, Kir, and Gem lack a CAAX or similar box (Reynet and Kahn, 1993; Cohen et al., 1994: Maguire et al., 1994); however, they do contain a cysteine residue at the seventh position from the C terminus, which provides a putative site for isoprenylation.
The membrane association of Ras-related proteins is both essential and central to their biological activity. Mutations that interfere with membrane association, as well as inhibitors of isoprenylation, block the transforming activity of Ras (Kohl et al., 1993, 1994; Manne et al., 1995). In addition, modification of Raf, which acts downstream of Ras, to achieve Ras-independent membrane association of Raf leads to constitutively active signal transduction, which is independent of Ras (Leevers et al., 1994).
A number of studies have implicated Ras-mediated signal transduction in retinal development and function. Cell fate determination of theDrosophila R7 photoreceptor progenitor cell requires activation of the sevenless tyrosine kinase receptor by the boss ligand (Simon et al., 1992). Sevenless signaling, in turn, requires Ras1 activity (Simon et al., 1991; Wassarman et al., 1995), and expression of a constitutively activated mutant of Ras1 (val12) in photoreceptor precursor cells replaces the need for activation of the sevenless receptor and causes the formation of supernumerary R7 cells (Fortini et al., 1992). Sevenless signaling activity is upregulated by the GEF Son of sevenless (Sos) (Bonfini et al., 1992), and it is downregulated by Gap1 (Gaul et al., 1992). The farnesyl transferase inhibitor FTI-254 suppresses the ability of microinjected activated Ras1 to form supernumerary R7 cells (Kauffmann et al., 1995). On the other hand, the roughened phenotype, which often includes loss of the R7 photoreceptor, is caused by a dominant gain-of-function mutation in theDrosophila Rap1 gene, which is thought to act by inhibiting the Ras pathway (Hariharan et al., 1991). Additional gene products that act downstream of Ras1 or interact with the Ras cascade to regulate retinal cell fate determination, including photoreceptors other than R7, have also been identified, and they include Jun (Treier et al., 1995), the ets-related transcription factors pointed and yan (Lai and Rubin, 1992; O’Neill et al., 1994), phyllopod (Chang et al., 1995), the mitogen-activated protein kinase rolled (Carthew et al., 1994), downstream of receptor kinases (Simon et al., 1993), and the small subunit of TFIIA (Zeidler et al., 1996).
Although the evidence is less direct, Ras and associated pathways have also been implicated in the development and maintenance of the mammalian retina. Retrovirus-mediated alteration of the expression of epidermal growth factor (EGF) receptors, which are thought to act through a Ras-containing cascade, in retinal progenitor cells can influence the response to transforming growth factor-α (TGF-α) and affect the relative proportion of rod and Müller cells (Lillien, 1995). Inhibition of fibroblast growth factor (FGF) signal transduction, another pathway thought to involve Ras, can lead to retinal degeneration (Campochiaro et al., 1996). In addition, mutations in the Rab escort protein REPI, which is involved in Rab isoprenylation and membrane association, can cause the X-linked human retinal degeneration choroideremia (Andres et al., 1993; Seabra et al., 1993).
Based on these findings and our interest in mammalian retinal development and signal transduction, we decided to conduct a search for novel Ras family members expressed in the mouse retina. Using a PCR-based approach with degenerate primers corresponding to conserved Ras domains and mouse retinal cDNA as template, we have cloned cDNAs corresponding to two novel small G-proteins, which we have named Rin for Ras-like protein expressed in neurons and Rit for Ras-like protein expressed in many tissues. As its name implies, Rin appears to be expressed only in neurons, and it is, to our knowledge, the only Ras-subfamily member that shows such an expression pattern. In addition, Rin binds calmodulin in a Ca2+-dependent manner. In the present study, we report the cloning, sequence analysis, expression, and preliminary characterization of Rit and Rin and suggest that they constitute a new subfamily of Ras-related proteins and that Rin may be involved in mediating calcium-dependent signaling within neurons.
MATERIALS AND METHODS
Reverse transcription (RT)-PCR. Total RNA was isolated from adult mouse retina using Trizol reagent (Life Technologies, Gaithersburg, MD). Five micrograms of the extracted total retinal RNA were then used as template for oligo-dT-primed first-strand cDNA synthesis using M-MLV reverse transcriptase (Life Technologies). One tenth of the resulting cDNA was used for PCR amplification with the degenerate oligonucleotide primers R1 (5′-AT(C/T)GA(C/T)AT(C/T)CT(G/C/A)- GA(C/T)AC(C/A/T)GC-3′) and R2 (5′-GG(C/A/G)AA(C/T)AA(A/G)- TC(C/T/A)GA(C/T)CT (G/C/T)GA-3′), which correspond to the conserved Ras GTP-binding domains G3 (IDILDTA) and G4 (VGNKSDL), respectively. Thirty cycles of PCR amplification were carried out as follows: 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min. The resulting PCR product of ∼200 bp was agarose gel-purified, reamplified with the same primer pairs, and subcloned by blunt-end ligation into the SmaI site of pBluescript SK+ (Stratagene, La Jolla, CA). Inserts in individual clones were sequenced by the dideoxynucleotide-chain termination method (Sequenase, United States Biochemicals, Cleveland, OH). For analysis ofRit and Rin expression in individual RNA samples, the procedure was the same except that the cDNA was treated with RNase-free DNase (Boehringer Mannheim, Indianapolis, IN) before amplification, nondegenerate primers were used, and the amplification conditions were 94°C for 1 min, 62°C for 1 min, and 72°C for 1 min, for a total of 30 cycles.
cDNA cloning and sequencing. Oligo-dT and random-primed murine retinal cDNA libraries in Uni-ZAP XR (Stratagene) were screened using random hexamer-labeled DNA probes made from the subclonedRit and Rin PCR products. For both Ritand Rin, a total of 1.5 × 106 plaques were screened under high-stringency hybridization conditions. Positive clones were plaque-purified and in vivo-excised, and the inserts of the resulting plasmids were sequenced on both strands using both vector and internal primers. Homology comparisons were performed using the BLAST algorithm (NCBI). Sequences were aligned using ClustalW (Higgins and Sharp, 1988) and displayed using BOXSHADE.
Generation of Rit and Rin bacterial fusion proteins.The entire coding regions of Rit and Rin except the initiation ATG were subcloned into either pTrcHis (Invitrogen, San Diego, CA) or pGEX (Pharmacia, Piscataway, NJ) prokaryotic expression vectors using PCR and primers encoding unique restriction sites. The 5′ primers for Rit and Rin were GAGTCCGGAGCTCGCCCCAT and GAAGTAGAAAACGAAGCCCAC, respectively, and 3′ primers were GCAGGCACAAGGAGCACTGCA and ACCCACAAGGAGAGACAGGA, respectively. The resulting constructs were sequenced to exclude PCR-induced mutations and to confirm that the coding regions were in frame. Transformed bacteria (BL21) were induced with IPTG, and the fusion proteins were purified under denaturing condition over a nickel column (Qiagen, Hilden, Germany).
GTP binding assay. Five micrograms each of the affinity purified histidine (His)-tagged Rit and Rin fusion proteins were subjected to 12% SDS-PAGE and electroblotted onto nitrocellulose. The filter-bound fusion proteins were renatured in PBS containing 1% bovine serum albumin (BSA), 0.5 mm MgCl2, 50 mm ZnCl2, 0.1% Triton X-100, and 5 mm DTT at 4°C. The membrane was then incubated in GTP-binding buffer (50 mm TrisHCl, pH 7.5, 5 mmMgCl2, and 0.3% Tween 20) at room temperature for 15 min. [α-32P]-GTP (20 μCi; 1 Ci = 37 GBq; DuPont NEN, Wilmington, DE) was then added, and the membrane was incubated at room temperature for 90 min, washed three times in GTP-binding buffer without [α-32P]-GTP, air-dried, and exposed to film. For cold competition, unlabeled GTP, GDP, or ATP to a final concentration of 0.1 mm was added together with the labeled GTP (Ohmstede et al., 1990). GTP binding assays with the Rit and Rin del I and II deletions (see below) were performed similarly except that total bacterial lysates were used instead of purified fusion proteins.
Generation of c-myc-tagged Rit and Rin mammalian expression constructs. Constructs were generated using the same primers and PCR in a manner analogous to that described above for the generation of the His-tagged fusion constructs. The resulting PCR products were appropriately restricted and subcloned into the myc-epitope-tag-containing mammalian expression vector pRK5. The constructs were then sequenced to rule out PCR-induced mutations. For the generation of the Rit del I and del II deletions constructs, the 3′ primers were CTCCATGGCCAGTACTAGCTCCTTCTC and GACGAGGGCGTGGAAAA-CGTCGT, respectively, and for the Rindel I and del II deletions constructs, the 3′ primers were TTCCACCAAGGACAGCATG and CACTAAGCCTTGAAAAGCATCATC, respectively.
Cell culture and transfection. COS and 293 cells were grown in DMEM high-glucose medium with 10% fetal bovine serum at 37°C and 5% CO2. The day before transfection, cells were split and then seeded onto poly-l-lysine-treated coverslips in a 24-well tissue culture plate with 2 ml of medium per well. The next day the cells were transfected with purified plasmid DNA (5 μg/well) using lipofectamine (Life Technologies) according to the manufacturer’s directions.
Immunocytochemistry. Forty-eight hours after transfection, cells were fixed with 4% paraformaldehyde in PBS for 15 min at 37°C, permeabilized in PBS containing 0.25% Triton X-100 for 5 min at room temperature, and washed twice with PBS. Nonspecific sites were blocked for 1 hr with PBS containing 10% normal goat serum. Monoclonal murine anti-c-myc primary antibodies (Oncogene Science, Manhasset, NY) were applied in PBS containing 3% goat serum at a concentration of 2 ng/μl and incubated overnight at 4°C. After they were washed in PBS, the cells were incubated for 1 hr at room temperature with a 1:5000 FITC-conjugated goat anti-mouse antibody (Sigma, St. Louis, MO). Horizontal optical sections (0.75 μm thick) ∼2-3 μm above the level of cell attachment were obtained by scanning laser confocal microscopy (Bio-Rad MRC-600; Bio-Rad, Richmond, CA). Excitation wavelength was 488 nm, and emission wavelength was 510–515 nm. Images were captured and processed using COMOS version 6.03 (Bio-Rad).
Calmodulin binding assay.Escherichia coliBL21 cells were transformed with the appropriate GST-Rin fusion constructs. After induction with 0.1 mm IPTG, total bacteria lysate was prepared by solubilization with SDS sample buffer. The lysate was electrophoresed on a 12% polyacrylamide gel and then electroblotted onto a nitrocellulose membrane. The protein blot was blocked by incubation for 1 hr at room temperature with 1% BSA in 50 mm Tris-HCl, pH 7.5, 0.2 m NaCl, 0.5 mm CaCl2, and 50 mmMgCl2 (TBS/CaMg). After removal of the blocking solution, 100 ng/ml biotinylated calmodulin (Life Technologies) in the same buffer was added onto the membrane and incubated for another 2 hr. The membrane was washed twice for 10 min in TBS/CaMg containing 0.05% Tween 20. The membrane was incubated with a 1:5000 dilution of the strepavidin-alkaline phosphatase (Life Technologies) in the same buffer for 30 min, and then color development with NBT and BCIP (Life Technologies) was performed according to the manufacturer’s instructions. To assess binding in the absence of calcium, the same assay was performed with TBS/CaMg in which 5 mm EGTA was substituted for the CaCl2. To assess the amount of GST fusion protein that was present, the protein blot was incubated for 2 hr with 2.5 μl/ml primary goat anti-GST antibody (Pharmacia). Then the blot was probed with rabbit anti-goat IgG coupled to horseradish peroxidase (1:4000; Sigma) for 30 min and washed, and the peroxidase signal was detected by DAB staining.
Northern blot analysis. Total RNA was extracted from adult mouse retina, brain, liver, spleen, heart, lung, and kidney, and from brains of late gestational embryo, 1- and 3-week-old mouse pups, using Trizol reagent (Life Technologies). Twenty-five micrograms per lane of the extracted total RNA were then analyzed by Northern blot hybridization using random hexamer-labeled DNA probes and standard procedures.
In situ hybridization. BALB/cJ mice were anesthetized and perfused with 4% paraformaldehyde in PBS. The brain and other organs were dissected, post-fixed for an additional 2 hr in the same fixative, immersed in a 15% sucrose solution in PBS overnight, and then sectioned (12 μm) on a cryostat at −20°C. For retinal sections, eyes from unperfused animals were fresh-frozen, sectioned, and then post-fixed in 4% paraformaldehyde. In situhybridization was performed as described (Della et al., 1996), using a modification of published methods (Harland, 1991; Wilkinson, 1992). Sense and antisense riboprobes for Rin were generated byin vitro transcription of the full-length clone in pBluescript (Stratagene) using T3 and T7 RNA polymerase and digoxgenin-coupled UTP (Boeh-ringer Mannheim) according to the manufacturer’s protocol. Chemical hydrolysis at 60°C for 30 min was used to reduce the average length of the riboprobes to 200 bp. Pronase E (70 μg/ml for 10 min at room temperature; Sigma) was used to pretreat the sections to improve probe access to the target mRNA. Hybridization was performed overnight at 57°C using a riboprobe concentration of 0.5 μg/ml, and a posthybridization ribonuclease digestion (20 μg/ml of RNase A for 30 min at 37°C; Sigma) was included to reduce background. Sections were blocked with 40% heat-inactivated lamb serum (Sigma) in Tris-buffered saline before addition of the anti-DIG antibody (1:2000; Boehringer Mannheim). The alkaline phosphatase-mediated color reaction was performed as described (Wilkinson, 1992). Sections were mounted in an aqueous glycerol-based mounting medium. Photographs were taken with a Zeiss Axioskop photomicroscope using bright-field or Nomarski optics and Kodak T-64 35 mm film.
RESULTS
Cloning of Rit and Rin
RT-PCR was performed on mouse retinal RNA using degenerate oligonucleotide primers that correspond to the highly conserved G3 and G4 domains of Ras proteins (Fig. 1). Of the 60 PCR products that were cloned and sequenced, the vast majority were identical, or nearly identical, to already published sequences. Two sequences, however, seemed to represent novel Ras family members, which we have named Rit and Rin.
Fig. 1.
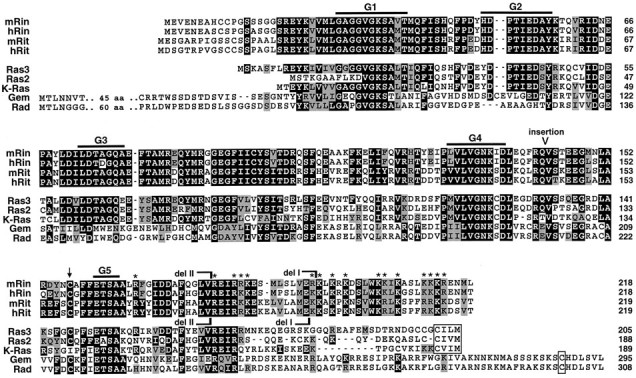
Comparison of the predicted amino acid sequences of murine Rin (GenBank accession number U71202), human Rin (U71204), murine Rit (U71205), human Rit (U71203), M. racemosusRas3 and Ras2, murine K-Ras2, murine Gem, and human Rad. The G1–G5 regions, the position of the Rit and Rin deletions (del I and del II), the conserved basic residues (asterisks), and the location of the C-terminal-most cysteine residue (arrow) in Rit and Rin are indicated. The word “insertion” indicates the position of the 64 bp insertion in the humanRin ESTs. The CAAX motifs in Ras3, Ras2, and K-Ras areboxed, as are the cysteines located seven residues from the C terminus in Gem and Rad. Black and gray shadingindicate sequence identity and conservative substitutions, respectively. Homology comparisons were performed using the BLAST algorithm (NCBI). Sequences were aligned using ClustalW (Higgins and Sharp, 1988) and displayed using BOXSHADE.
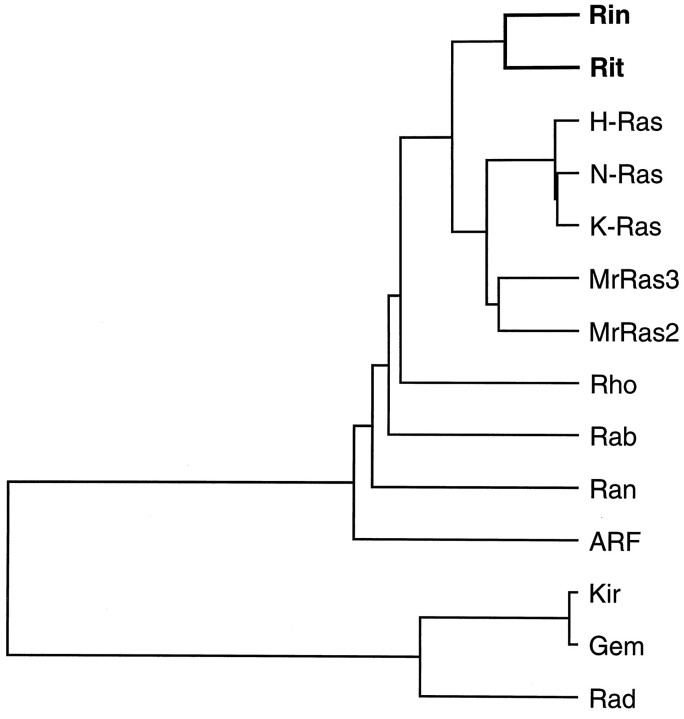
Mouse retinal cDNA libraries were screened using the clonedRit and Rin PCR fragments as probes. Sequencing of several independent Rit and Rin clones demonstrated full-length cDNAs of 1.1 and 1.8 kb, encoding proteins of 219 and 217 amino acid residues, respectively (Fig. 1). Both sequences contain good Kozak consensus sites (Kozak, 1987). The predicted molecular weights of Rit and Rin are 25.6 and 24.8 kDa, respectively, and their predicted amino acid sequences are 64% identical to each other. The highest degree of homology is in the central 167 amino acids, in which the identity is 74%. The closest homolog to Rit in the protein database is Mucor racemosus Ras3, to which it is 48% identical; the closest homolog to Rin is M. racemosusRas2, to which it is also 48% identical (Casale et al., 1990). The relationship of Rit and Rin to these and other small G-proteins is shown diagrammatically in Figure 2.
Fig. 2.
Dendrogram showing relationship of Rin and Rit to selected other small G-proteins [Rin, mouse; Rit, mouse; H-ras, rat, GenBank accession number M13011; N-ras, mouse, M12121; K-ras 2A, mouse,P32883; Ras3, M. racemosus, P22280; Ras2, M. racemosus, M55176; Rho/CDC42, mouse, U37720; Rab18, mouse,L04966; Ran, mouse, L32752; ARF 4 (ADP-ribosylation factor), rat,B54022; Kir, mouse, U10551; Gem, mouse, U10551; Rad, human, L24564]. The tree was generated using the unweighted pair group with arithmetic mean method (GeneWorks 2.3, IntelliGenetics), and it suggests that Rin and Rit constitute a subfamily within the Ras group.
Comparison of murine Rit and Rin with the human expressed sequence tag (EST) database revealed the existence of human orthologs. Given the neural specificity of Rin (see below), it is noteworthy that the Rin ESTs are derived from brain libraries. The appropriate ESTs were obtained and sequenced. Of the three available Rin ESTs, two of them (R52317 and H08460) have an additional 64 bp sequence that leads to a predicted 21 amino acid insertion with an associated frameshift and premature termination (Fig. 1). The third EST (N53351) does not contain the 64 bp insertion. Because of considerations of homology, the frameshift, and the finding that RT-PCR analysis of both mouse and human retinal RNA shows a major band corresponding to the size of the sequence without the insertion (the human retinal RT-PCR shows a barely detectable band corresponding to the sequence with the insertion; data not shown), we conclude that the major Rin transcript does not contain the 64 bp insertion. The sequences containing the insertion probably represent aberrant splicing events, although the possibility of a pseudogene cannot be ruled out. The sequence flanking the insertion shows moderate homology to the splice consensus sites (Sharp, 1994). The predicted protein sequences of murine and human Rin are 91% identical. The predicted sequences of murine and human Rit (ESTs N36448, T58089, and R81023) are 94% identical.
One of the Rit ESTs (T58089) has been mapped to chromosome 3 (62 cM offset) in the mouse (XREF database, NCBI).
Rit and Rin show specificity for GTP and GDP
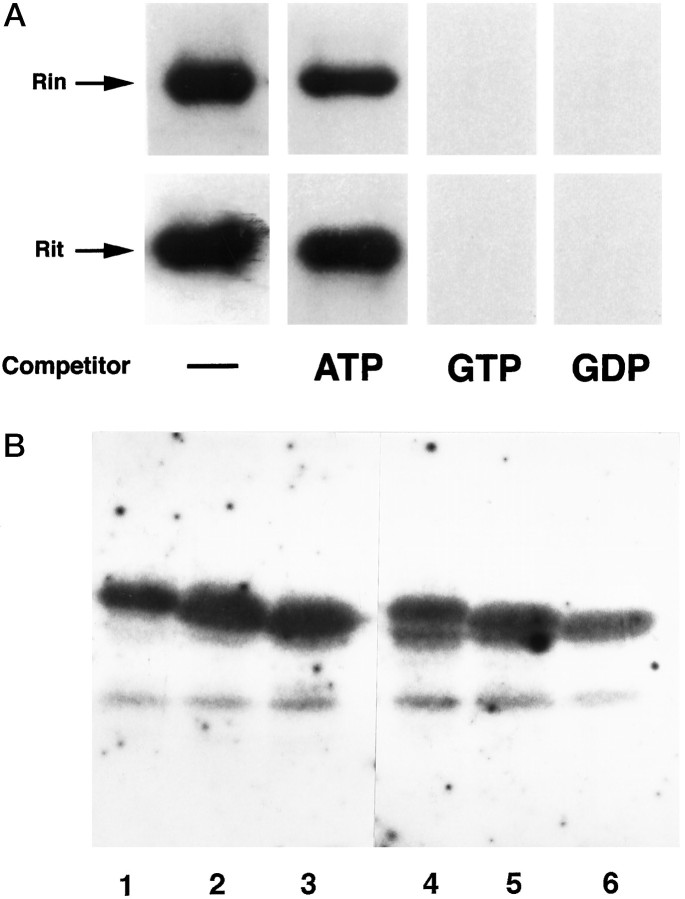
Because Rit and Rin both contain the five highly conserved domains (G1–G5) that are characteristic of small G-proteins (Fig. 1), we tested for their ability to bind GTP. Murine Rit and Rin were expressed in E. coli as His-tagged fusion proteins, affinity-purified, subjected to SDS-PAGE, transferred to a nitrocellulose membrane, and incubated with radiolabeled GTP (Fig. 3A). The Rit and Rin fusion proteins both bound [α-32P]-GTP. Binding was specific in that binding activity could be blocked by excess unlabeled GTP or GDP but not by ATP.
Fig. 3.
Nucleotide binding activity of Rin and Rit.A, His-tagged Rin and Rit fusion proteins demonstrated binding of [α-32P]-GTP. Binding was blocked by unlabeled GTP and GDP but not by ATP. For cold competition, unlabeled GTP, GDP, or ATP was added to a final concentration of 0.1 mm. B, The del I and II deletion mutants of Rin and Rit retain the ability to bind [α-32P]-GTP.Lane 1, Rin wild type; 2, Rin del I;3, Rin del II; 4, Rit wild type;5, Rit del I; 6, Rit del II.
Rit and Rin are membrane-associated despite the lack of a CAAX box
An unusual feature of both Rit and Rin is that they lack a C-terminal CAAX box (Fig. 1). Although there are other Ras-related family members that do not contain a CAAX box, such as Rad, Gem, and Kir, these proteins all have a cysteine residue at position 7 from their C terminus that provides a putative site for isoprenylation (Reynet and Kahn, 1993; Cohen et al., 1994; Maguire et al., 1994). In contrast, the closest cysteines in Rit and Rin are 62 and 61 residues, respectively, from the C terminus. Rit and Rin, however, both contain a C-terminal polybasic domain, which by analogy with other small G-proteins may provide some binding energy for membrane association. In fact, although they have minimal overall conservation in their C-terminal regions, the positions of basic amino acid residues within Rit and Rin are highly conserved in this region.
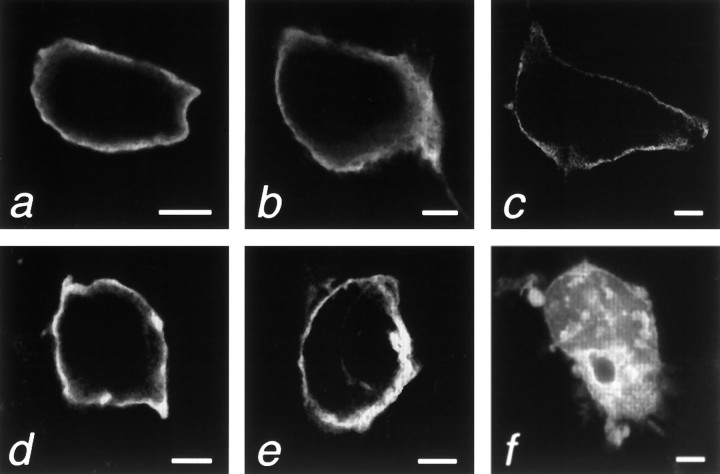
To explore whether Rit and Rin are membrane-associated, given their unusual C termini, the coding regions for the murine proteins were subcloned into a c-myc-tagged mammalian expression vector and transiently expressed in COS and 293 cells (Robertson et al., 1995). Analysis by immunofluorescent and confocal microscopy with a primary anti-myc antibody revealed that both Rit and Rin were localized to the plasma membrane in both cell types. The results with COS cells are shown (Fig. 4A,D). It is possible, although perhaps unlikely, that the localization observed for Rin with COS and 293 cells may not reflect the endogenous situation, because these cells may lack a neuron-specific interacting protein that could affect subcellular localization. Additionally, the presence of the myc tag as well as the use of transfection could potentially influence the results. Future experiments with Rin- and Rit-specific antisera will more definitively resolve this issue.
Fig. 4.
Confocal microscopy of wild-type and C-terminally deleted Rin- and Rit-c-myc-tagged fusion proteins expressed in COS cells. Rin wild-type (a), del I (b), del II (c), Rit wild- type (d), and del I (e) fusion proteins all localized to the plasma membrane, whereas Rit del II showed a vesiculated staining pattern throughout the cytoplasm (f). Scale bars, 5 μm. (The results with COS and 293 cells were identical; only the COS results are shown.) Forty-eight hours after transfection, the cells were fixed with 3% paraformaldehyde, permeabilized in 0.25% Triton X-100, blocked in 10% normal goat serum, and then incubated overnight at 4°C with a mouse anti-c-myc monoclonal antibody at a concentration of 2 ng/μl (Oncogene Science).
To examine the possible role of the C-terminal domain in membrane association, we also generated and expressed constructs coding for mutant proteins in which parts of the C-terminal polybasic region were deleted (del I and II) (Fig. 1). Rin del I lacked the C-terminal 24 amino acids, containing 11 basic amino acid residues, and del II lacked the C-terminal 39 amino acids, containing 16 basic amino acid residues (Fig. 1). Rit deletions I and II lacked the C-terminal 25 and 40 amino acids, respectively, which also contained 11 and 16 basic amino acid residues, respectively. As shown by confocal microscopy, Rin del I and II and Rit del I localized to the plasma membrane (Fig.4B,C,E). In contrast, Rit deletion II was not membrane-associated in COS and 293 cells, but rather showed a vesicular pattern localized to the cytoplasmic region (Fig.4F). The identity of the stained region is unclear, but staining with DAPI indicated that it was non-nuclear (data not shown). These results indicate that although membrane association of Rin does not require the C-terminal polybasic region, the polybasic region between residues 180 and 194 in Rit may be involved in the mechanism of membrane localization. Alternatively, we cannot rule out the possibility that overexpressed mutant Rit protein may fail to fold properly and may aggregate, thereby preventing proper intracellular sorting to the plasma membrane; however, the finding that the Rin and Rit del I and II deletions retain the ability to bind GTP (Fig.3B) suggests, at least at a first approximation, that the deleted proteins fold properly.
Rin binds to calmodulin in a calcium-dependent manner
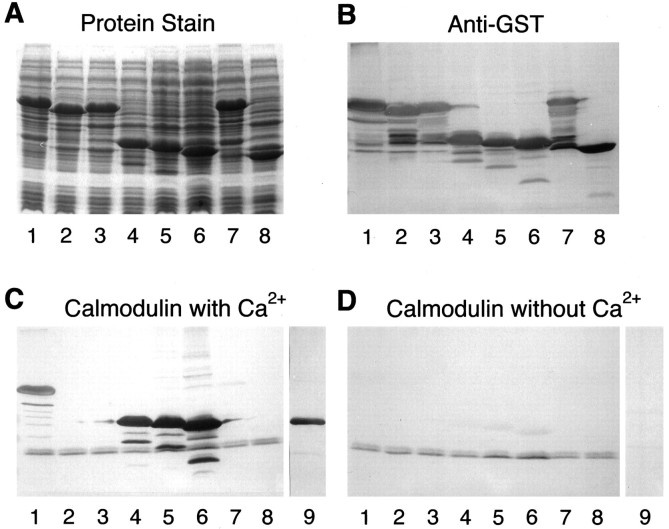
Based on the finding that a Drosophila Ras homolog that also contains a CAAX-less and polybasic C terminus binds calmodulin (Wes et al., in press), and the important but incompletely understood relationship between calcium-dependent processes and Ras signaling in neurons (Finkbeiner and Greenberg, 1996), we tested Rit and Rin for their ability to bind calmodulin. GST fusions of Rit and Rin, as well as deleted forms of the proteins, were expressed in E. coli. Total bacterial extracts or purified fusion proteins were subjected to SDS-PAGE (Fig. 5A), transferred to nitrocellulose membranes, and tested for their ability to bind biotinylated calmodulin. As shown by immunoreactivity with an anti-GST antibody, all of the fusion proteins were expressed at comparable levels (Fig. 5B). Full-length Rin fusion protein, either unpurified (Fig. 5C, lane 1) or purified (lane 9), showed strong binding to calmodulin, whereas GST itself did not demonstrate any detectable binding (lane 8). Rit fusion protein did not show significant calmodulin binding (lane 7). Although some experiments did suggest minimal binding activity associated with Rit, the level was so low that it most likely represents experimental background. Alternatively, it is possible that bacterially expressed Rit fails to bind, because it is not properly folded or post-translationally modified.
Fig. 5.
Calmodulin binding activity of Rin and Rit fusion proteins. A, Protein gel-stained with Coomassie brilliant blue. B, Western analysis using anti-GST antibody. C, Western blot probed with biotinylated calmodulin in the presence of 0.5 mm Ca2+.D, Western blot probed with biotinylated calmodulin in the absence of Ca2+ and presence of 5 mm EGTA.Lane 1, GST/Rin; 2, GST/Rin-del II;3, GST/Rin-del I; 4, 53 C-terminal amino acid residues of GST/Ren; 5, 39 C-terminal amino acid residues of GST/Ren; 6, 25 C-terminal amino acid residues of GST/Ren; 7, GST/Rit; 8, GST alone; 9, pTrcHis/Rin. Lanes 1–8 contain total bacterial extract. Lane 9 contains purified fusion protein.
To localize the calmodulin binding activity within Rin, the del I and II C-terminal deletion mutants described above were tested for binding (Fig. 5C, lanes 3 and 2). Both constructs failed to show detectable binding, demonstrating that at least part of the C-terminal 24 amino acid residue region is necessary for calmodulin binding. To determine which region is sufficient for binding, we also generated nested GST fusions containing only the 53, 39, and 25 C-terminal amino acids (lanes 4, 5, and6). All three constructs showed binding, demonstrating that the C-terminal 25 amino acid sequence in Rin is sufficient to mediate calmodulin binding.
To determine the role of calcium in the interaction between Rin and calmodulin, the same binding protocol was carried out in the absence of calcium and in the presence of the calcium chelator EGTA (Fig.5D). The binding activity of Rin and the C-terminal constructs was significantly reduced in the absence of calcium, demonstrating that, at least in this solid-phase binding assay, the interaction between Rin and calmodulin is calcium-dependent.
Rit is expressed ubiquitously, but Rin is expressed only in subsets of neurons
Northern analysis was performed to examine the expression patterns of Rit and Rin. Like most Ras-related genes, Rit is expressed ubiquitously and is present as a single 1.2 kb transcript (Fig. 6A).Rin mRNA, however, which is expressed as a ∼2.0 kb transcript, is detectable only in retina and brain. RT-PCR likewise indicated that Rin is expressed only in neural tissue (Fig.6B). Developmental Northern analysis of brain revealed that, although Rit is expressed at similar levels in embryonic and adult tissues, Rin mRNA is expressed more highly in the adult (Fig. 6C).
Fig. 6.
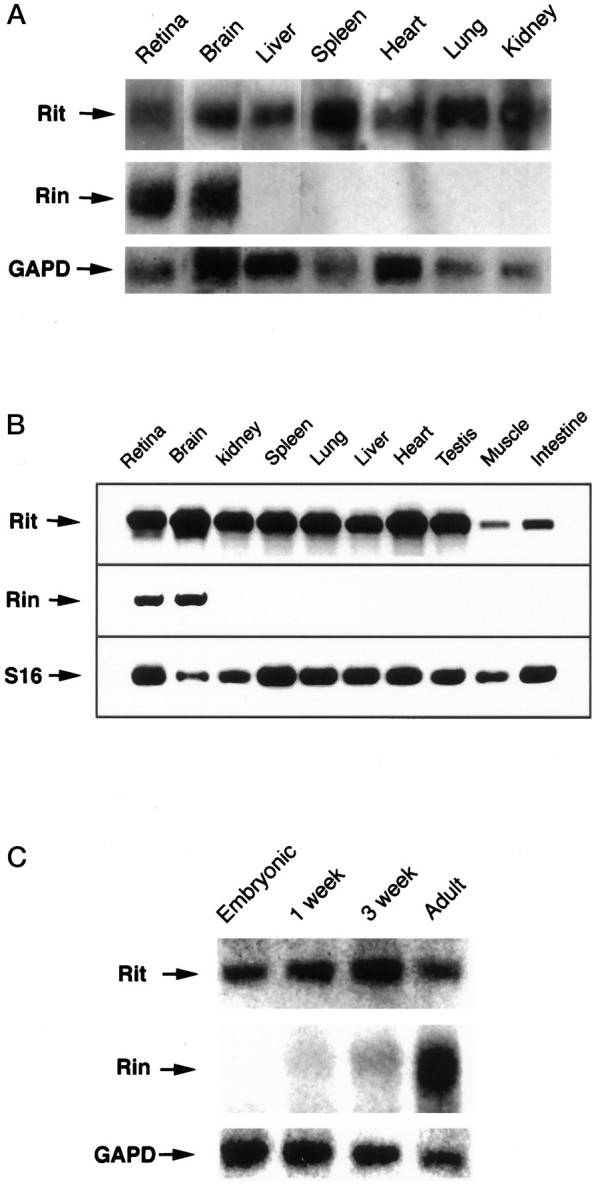
Tissue-specific and developmental analysis ofRit and Rin expression. Northern blots of total RNA (25 μg/lane) extracted from adult mouse tissues (A) and from mouse brain in late gestation embryos, 1- and 3-week-old pups, and adults (C) were hybridized withRit, Rin, and human glyceraldehyde-3-phosphate dehydrogenase (GAPD) cDNA probes. B, Similar analysis of adult tissues performed using RT-PCR to amplify Rit and Rin cDNA fragments. Amplification of riboprotein S16 was used as a control for the amount of starting RNA.
The neural-specific pattern of Rin expression was confirmed and further analyzed by in situ hybridization (Fig.7). In mouse retina, Rin mRNA was detected in the inner nuclear and ganglion cell layers, comprising the second and third order neurons, respectively (Fig. 7A). In contrast, the photoreceptor layer was negative for Rin expression. Ganglion cell expression appeared perinuclear and variable (Fig.7C). Some ganglion cells contained abundant reaction product, whereas others, even adjacent cells, revealed no detectable signal.
Fig. 7.
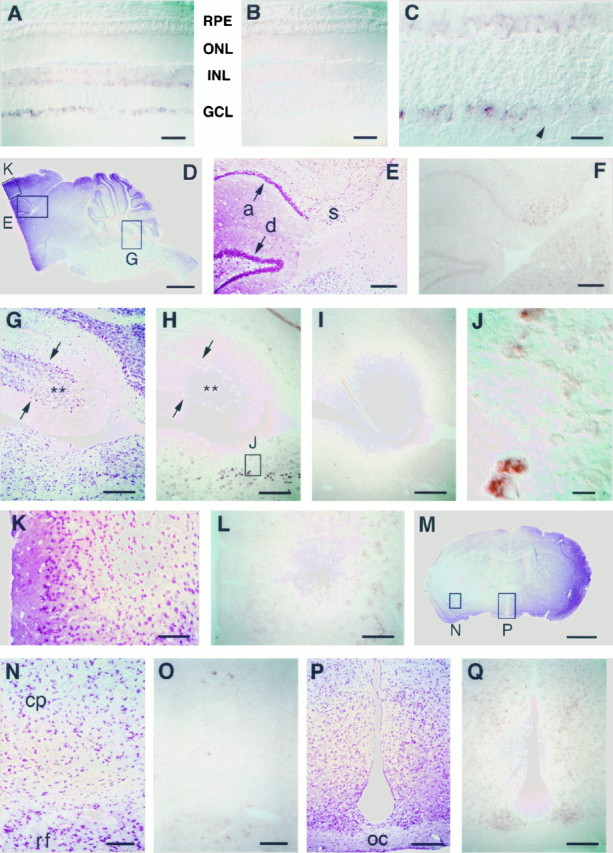
In situ hybridization ofRin in the CNS of the mouse. A, B, Sections of retina hybridized with antisense (A) and sense (B) Rin riboprobes reveal expression in the ganglion cell layer (GCL) and inner nuclear layer (INL), but not in the outer nuclear layer (ONL) or in the retinal pigment epithelium (RPE). Scale bars, 50 μm. C, High-power view showing the variability in Rin expression within the ganglion cell layer, including one cell with barely detectable expression (arrowhead). Scale bar, 20 μm.D, Low-power view of a sagittal section of brain, counterstained with 0.1% thionin, showing boxedregions corresponding to cortex (K), hippocampus (E), and dorsal brainstem and cerebellum (G). The anterior third of this brain is not shown because it was used to cut coronal sections (M). Scale bar, 1.2 mm. E, F, Sagittal sections through the hippocampus and dorsal midbrain counterstained with 0.1% thionin (E) or hybridized with the Rin antisense probe (F) reveal strong expression in the subiculum (s), lower levels of expression in the dentate gyrus (d), and Ammon’s horn (a), and little expression in neurons adjacent to the subiculum. In contrast, the majority of neurons in the dorsal midbrain express Rin. Scale bars, 200 μm. G, H, I, Sagittal sections through the cerebellum, fourth ventricle, and dorsal brainstem counterstained with 0.1% thionin (G) or hybridized with the antisense (H) or sense (I)Rin probes. The signal in the Purkinje cell layer (arrows) contrasts with its absence in the dense granular layer (asterisks) of the cerebellar cortex. Several nuclei in the dorsal brainstem show very strong expression ofRin. Scale bars, 200 μm. J, High-power detail from H shows variable levels ofRin expression in neurons and the perinuclear/cytoplasmic location of the signal. Note the marked variability in the level of expression between neighboring cells. Scale bar, 20 μm. K, L, Sagittal sections of cerebral cortex hybridized with the antisense Rin probe (K) demonstrate signal in the deeper (pyramidal) layer of neurons. The counterstain in L is 0.1% thionin. Scale bars, 100 μm. M, Low-power view of a coronal section through the anterior brain stained with 0.1% thionin, showing boxed regions corresponding to caudate putamen (N) and third ventricle region (P). Scale bar, 1.5 mm. N, O, Coronal views of the periphery of the caudate putamen (cp) and adjacent reticular formation ventrally (rf), counterstained with 0.1% thionin (N) or hybridized with the antisense Rin probe (O), demonstrate that only selected caudate neurons express Rin, but at a moderately high level. In contrast, the majority of neurons in the reticular formation appear to express Rin. Scale bars, 100 μm. P, Q, Coronal sections through the third ventricle region counterstained (P) or hybridized with the antisense Rinprobe (Q) show signal in the periventricular nuclei of the hypothalamus but not in the ventricular ependymal cells or in the axons of the optic chiasm (oc). Scale bars, 200 μm.
Sections through the mouse brain also revealed a pattern of widespread but variable expression in subsets of neurons. A moderate level ofRin expression was observed in the majority of neurons throughout the diencephalon, midbrain, and hindbrain, including neurons within named nuclei as well as neurons scattered throughout the reticular formation. In the hippocampus, strong expression was evident in the subiculum (Fig. 7F). Low to moderate expression was seen in neurons of the dentate gyrus, Ammon’s horn, and the hilus of the dentate gyrus. A high level of expression was evident in several nuclei of the dorsal medulla (Fig. 7H), including what appeared to be the dorsal motor nucleus of the vagus nerve and the perihypoglossal nucleus. Within the caudate putamen, occasional isolated neurons showed a moderate level of expression (Fig.7O). Within the cerebellum, a moderate level of expression was noted in the medial cerebellar nucleus, as well as in Purkinje cells, and in occasional neurons in the molecular layer of the cortex (Fig. 7H). No expression was evident in the granular layer. In the cerebral cortex, the deeper layers of the cortex (probably corresponding to layer V, the pyramidal layer) showed low to moderate expression, with barely detectable expression in the other layers (Fig. 7K). Glial cells, including the ventricular ependymal cells, appeared not to express Rin. Given the limits of in situ hybridization, however, it is impossible to completely exclude the possibility of glial expression ofRin in the present study. Consistent with the Northern blot and RT-PCR results, sections through mouse testis, kidney, and liver revealed no evidence of Rin expression (data not shown).
DISCUSSION
In this paper we have described the cloning and initial characterization of two novel small G-proteins, Rit and Rin. Sequence comparison suggests that the proteins are members of the Ras subfamily. Both proteins, however, demonstrate a number of unusual structural and other characteristics that differentiate them from typical Ras-related proteins and suggest that they define a new subclass.
Mechanism of membrane association
Rit and Rin are unusual in that they lack a CAAX box. The finding that Rit and Rin, when expressed as fusion proteins with a myc-epitope tag, are nonetheless membrane-localized indicates that these proteins may use a mechanism of membrane association that is distinct from the prototypic isoprenylation used by the typical Ras protein (Casey, 1995). Although a farnesyl transferase or equivalent enzyme could recognize a different consensus sequence, it is unlikely that isoprenylation of the C-terminal region is involved, because the most C-terminal cysteine residues in Rit and Rin are 62 and 61 residues from the C terminus, respectively. There are known proteins that also lack a CAAX box such as Rad, Gem, and Kir (Reynet and Kahn, 1993; Cohen et al., 1994; Maguire et al., 1994), but each of these proteins has a cysteine residue located seven residues from its C terminus, which provides a putative lipidation site. The apparent lack of a C-terminal farnesyl group in Rit and Rin may have additional implications, because isoprenyl groups have been suggested as being involved in protein–protein interactions (Marshall, 1993; Porfiri et al., 1994).
Both Rit and Rin have extended, highly conserved C-terminal polybasic regions. Because polybasic domains have been implicated in providing energy for membrane association for other Ras-related proteins (Hancock et al., 1990; Cadwallader et al., 1994), we suspected that these regions might be important for membrane association; however, deletion of the entire Rin polybasic domain had no detectable effect on membrane association under the experimental conditions used in this study. In the case of Rit, the situation appears more complicated. Deletion of the C-terminal 11 basic residues (del I) had no effect, but deletion of a larger region (16 basic residues, del II) led to a vesiculated cytoplasmic staining pattern. These results suggest that the 15 amino acid residue region between del II and del I in Rit is necessary for membrane association. Alternatively, it is possible that del II so disrupts the protein that it prevents protein folding and thereby blocks normal protein sorting in a nonspecific manner. Considering the perfect conservation between Rit and Rin of the position of the C-terminal basic residues, this difference in results for the two proteins is surprising, and it may reflect a difference in the binding energy provided by other parts of the molecules. Whether the basic mechanism of membrane localization involves direct membrane binding or interaction with an anchoring protein remains to be determined. Because both Rit and Rin have putative myristoylation sites, such a mechanism may be involved in membrane association.
Rit and Rin share a unique effector domain
A second unusual aspect of Rin and Rit is their effector (G2) domains (DPTIEDAYK). The nine amino acid residues are 100% conserved between the murine and human Rin and Rit proteins. A search of the current databases did not reveal any other proteins with an identical sequence. The closest match, seven of nine, is to the effector domain of N-Ras and other related Ras proteins (DPTIED SY R). The significance of these two amino acid residue differences is not yet clear. The overall similarity to the effector domain of Ras proteins may suggest that Rin and Rit interact with some Ras effector proteins. Alternatively, the sequence changes, although minimal, may lead to interaction with a novel set of effector molecules. It is hoped that ongoing experiments using the yeast two-hybrid system will identify effectors of Rin and Rit. Preliminary two-hybrid results suggest that Raf is not an effector for Rit (our unpublished results). In terms of which GAPs interact with Rin and Rit, it is potentially significant that the equivalent of Ras residues 12, 13, and 61, which are important for sensitivity to GAP and neurofibromin (Lowy and Willumsen, 1993), are conserved between Ras, Rin, and Rit.
Neuron-specific pattern of Rin expression provides an additional mechanism for achieving cell-type specificity of signaling
Although Ras-mediated signaling pathways are often highly cell type-specific, most Ras subfamily members are widely or ubiquitously expressed. The cellular specificity of individual pathways is generally thought to be provided by the specific expression patterns of other proteins in the cascade, such as receptors, GAPs, GEFs, and downstream effector molecules. For example, GAPIII is expressed most highly in the brain (Baba et al., 1995), and p140 Ras-GRF is reported to be neuron-specific and has been proposed to be responsible for the neuron-specific activation of Ras by calcium (see below) (Farnsworth et al., 1995). Clearly, the highly specific expression pattern of Rin within the nervous system provides another and more direct mechanism for achieving functional cell-type specificity in signaling. There are a few other small G-proteins that show limited expression patterns, such as Rab3A, which is expressed in neuroendocrine cells (Burstein and Macara, 1989; Darchen et al., 1990). Within the Ras subfamily, however, Rin is the only member that is expressed exclusively in neuronal cells.
Mechanism of interaction between Rin and calmodulin
The calcium binding protein calmodulin regulates a wide variety of cellular processes (Crivici and Ikura, 1995). In response to an increase in the intracellular concentration of free Ca2+, calmodulin undergoes a conformational change that results in its binding to a target molecule. Although calmodulin binds with high affinity and specificity to more than 20 different targets, the targets do not display a well conserved binding motif. Most target sites are small, ranging from 14 to 26 amino acid residues, and contain a high density of basic residues within an amphiphilic α-helix (O’Neil and DeGrado, 1990). Whether the calmodulin binding site of Rin fits this prototypic structure is unclear. It is small, is contained within the C-terminal 25 amino acid residues (RKLKRKDSLWKKIKASLKKKRENML), is polybasic, and includes a number of hydrophobic residues. Based on the helical wheel algorithm, however, it is not an ideal sequence for forming an amphiphilic α-helix. In addition, the location of C-terminal basic residues is identical between Rin and Rit. Rit also contains hydrophobic residues, yet Rit does not demonstrate significant calmodulin binding. Additional physical studies will be necessary to ascertain the structure of the binding site of Rin and the nature of its interaction with calmodulin.
Rin may be involved in calcium-mediated neuronal signal transduction
Intracellular calcium has been implicated in the regulation of a number of neuronal processes, including ion channel status, neurotransmitter release, neurite outgrowth, apoptosis, synaptic plasticity, and long-term potentiation (Ghosh and Greenberg, 1995). Recently, increasing evidence has accumulated to suggest that Ras proteins may be important in mediating some of these calcium-dependent processes (Rosen et al., 1994; Rusanescu et al., 1995; Finkbeiner and Greenberg, 1996). A number of mechanisms have been proposed to explain the interconnection between the calcium and Ras signaling pathways. Among them is the ability of calcium, through calmodulin, to affect the activity of Ras regulatory proteins such as p140 Ras-GRF and IQGAP1 (Weissbach et al., 1994; Farnsworth et al., 1995). The ability of Rin to interact directly with calmodulin in a Ca2+-dependent manner may provide another and more direct route for calcium signaling to act through the Ras pathway. Whether such a putative pathway acts through the MAP kinase cascade remains to be determined, but given the conservation within the Ras family, it seems likely.
The finding that Rin expression is significantly greater in the adult brain than at earlier stages suggests that it may be more important for adult neurological functions than for developmental processes. Although the physiological function of Rin remains to be determined, its membrane localization, interaction with calmodulin, and unique expression pattern within subsets of neurons are consistent with its possible involvement in functions such as learning and memory, perhaps through modulation of processes such as long-term potentiation and depression. Additional studies using dominant–negative and constitutively active forms of Rin may help to answer these questions.
Footnotes
This work was supported by grants from the National Eye Institute (EY09769 and P30 EY01765) and The Foundation Fighting Blindness, a Neil Hamilton Fairley Postdoctoral Scholarship from the National Health and Medical Research Council of Australia (N.G.D.), and unrestricted funds from Research to Prevent Blindness. We thank Drs. Robert Nickells and Wolfgang Baehr for their gifts of the oligo-dT and random-primed mouse retinal cDNA libraries, respectively; Dr. Paul Worley and his lab; Drs. Carolyn Machemer and Mark Moliver for their generous assistance; Mike Delannoy for help with the confocal microscopy; Dr. Cornelia Gorman and Genentech for the use of the expression vector pRK-5-myc; and Drs. Josh Dunaief and Marc Symons for critical reading of this manuscript.
Correspondence should be addressed to Donald J. Zack, Johns Hopkins University School of Medicine, 809 Maumenee, 600 North Wolfe Street, Baltimore, MD 21287-9289.
REFERENCES
- 1.Andres DA, Seabra MC, Brown MS, Armstrong SA, Smeland TE, Cremers FP, Goldstein JL. cDNA cloning of component A of Rab geranylgeranyl transferase and demonstration of its role as a Rab escort protein. Cell. 1993;73:1091–1099. doi: 10.1016/0092-8674(93)90639-8. [DOI] [PubMed] [Google Scholar]
- 2.Baba H, Fuss B, Urano J, Poullet P, Watson JB, Tamanoi F, Macklin WB. GapIII a new brain-enriched member of the GTPase-activating protein family. J Neurosci Res. 1995;41:846–858. doi: 10.1002/jnr.490410615. [DOI] [PubMed] [Google Scholar]
- 3.Bischoff FR, Ponstingl H. Mitotic regulator protein RCC1 is complexed with a nuclear ras-related polypeptide. Proc Natl Acad Sci USA. 1991;88:10830–10834. doi: 10.1073/pnas.88.23.10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bollag G, McCormick F. Regulators and effectors of ras proteins. Annu Rev Cell Biol. 1991;7:601–632. doi: 10.1146/annurev.cb.07.110191.003125. [DOI] [PubMed] [Google Scholar]
- 5.Bonfini L, Karlovich CA, Dasgupta C, Banerjee U. The Son of sevenless gene product: a putative activator of Ras. Science. 1992;255:603–606. doi: 10.1126/science.1736363. [DOI] [PubMed] [Google Scholar]
- 6.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 7.Burstein E, Macara IG. The ras-like protein p25 rab3A is partially cytosolic and is expressed only in neural tissue. Mol Cell Biol. 1989;9:4807–4811. doi: 10.1128/mcb.9.11.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cadwallader KA, Paterson H, Macdonald SG, Hancock JF. N-terminally myristoylated Ras proteins require palmitoylation or a polybasic domain for plasma membrane localization. Mol Cell Biol. 1994;14:4722–4730. doi: 10.1128/mcb.14.7.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campochiaro PA, Chang M, Ohsato M, Vinores SA, Nie ZQ, Hjelmeland L, Mansukhani A, Basilico C, Zack DJ. Retinal degeneration in transgenic mice with photoreceptor-specific expression of a dominant-negative fibroblast growth factor receptor. J Neurosci. 1996;16:1679–1688. doi: 10.1523/JNEUROSCI.16-05-01679.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carthew RW, Neufeld TP, Rubin GM. Identification of genes that interact with the sina gene in Drosophila eye development. Proc Natl Acad Sci USA. 1994;91:11689–11693. doi: 10.1073/pnas.91.24.11689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casale WL, McConnell DG, Wang SY, Lee YJ, Linz JE. Expression of a gene family in the dimorphic fungus Mucor racemosus which exhibits striking similarity to human ras genes. Mol Cell Biol. 1990;10:6654–6663. doi: 10.1128/mcb.10.12.6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casey PJ. Lipid modifications of G proteins. Curr Opin Cell Biol. 1994;6:219–225. doi: 10.1016/0955-0674(94)90139-2. [DOI] [PubMed] [Google Scholar]
- 13.Casey PJ. Protein lipidation in cell signaling. Science. 1995;268:221–225. doi: 10.1126/science.7716512. [DOI] [PubMed] [Google Scholar]
- 14.Chang HC, Solomon NM, Wassarman DA, Karim FD, Therrien M, Rubin GM, Wolff T. Phyllopod functions in the fate determination of a subset of photoreceptors in Drosophila. Cell. 1995;80:463–472. doi: 10.1016/0092-8674(95)90497-2. [DOI] [PubMed] [Google Scholar]
- 15.Cohen L, Mohr R, Chen YY, Huang M, Kato R, Dorin D, Tamanoi F, Goga A, Afar D, Rosenberg N, White O. Transcriptional activation of a ras-like gene (kir) by oncogenic tyrosine kinases. Proc Natl Acad Sci USA. 1994;91:12448–12452. doi: 10.1073/pnas.91.26.12448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crivici A, Ikura M. Molecular and structural basis of target recognition by calmodulin. Annu Rev Biophys Biomol Struct. 1995;24:85–116. doi: 10.1146/annurev.bb.24.060195.000505. [DOI] [PubMed] [Google Scholar]
- 17.Darchen F, Zahraoui A, Hammel F, Monteils MP, Tavitian A, Scherman D. Association of the GTP-binding protein Rab3A with bovine adrenal chromaffin granules. Proc Natl Acad Sci USA. 1990;87:5692–5696. doi: 10.1073/pnas.87.15.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Della NG, Campochiaro PA, Zack DJ. Localization of TIMP-3 mRNA expression to the retinal pigment epithelium. Invest Ophthalmol Vis Sci. 1996;37:1921–1924. [PubMed] [Google Scholar]
- 19.Farnsworth CL, Freshney NW, Rosen LB, Ghosh A, Greenberg ME, Feig LA. Calcium activation of Ras mediated by neuronal exchange factor Ras-GRF. Nature. 1995;376:524–527. doi: 10.1038/376524a0. [DOI] [PubMed] [Google Scholar]
- 20.Finkbeiner S, Greenberg ME. Ca2+-dependent routes to Ras-mechanisms for neuronal survival, differentiation, and plasticity? Neuron. 1996;16:233–236. doi: 10.1016/s0896-6273(00)80040-9. [DOI] [PubMed] [Google Scholar]
- 21.Fischer von Mollard G, Stahl B, Li C, Sudhof TC, Jahn R. Rab proteins in regulated exocytosis. Trends Biochem Sci. 1994;19:164–168. doi: 10.1016/0968-0004(94)90278-x. [DOI] [PubMed] [Google Scholar]
- 22.Fortini ME, Simon MA, Rubin GM. Signalling by the sevenless protein tyrosine kinase is mimicked by Ras1 activation. Nature. 1992;355:559–561. doi: 10.1038/355559a0. [DOI] [PubMed] [Google Scholar]
- 23.Gaul U, Mardon G, Rubin GM. A putative Ras GTPase activating protein acts as a negative regulator of signaling by the Sevenless receptor tyrosine kinase. Cell. 1992;68:1007–1019. doi: 10.1016/0092-8674(92)90073-l. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh A, Greenberg ME. Calcium signaling in neurons: molecular mechanisms and cellular consequences. Science. 1995;268:239–247. doi: 10.1126/science.7716515. [DOI] [PubMed] [Google Scholar]
- 25.Hall A. The cellular functions of small GTP-binding proteins. Science. 1990;249:635–640. doi: 10.1126/science.2116664. [DOI] [PubMed] [Google Scholar]
- 26.Hall A. Small GTP-binding proteins and the regulation of the actin cytoskeleton. Annu Rev Cell Biol. 1994;10:31–54. doi: 10.1146/annurev.cb.10.110194.000335. [DOI] [PubMed] [Google Scholar]
- 27.Hancock JF, Magee AI, Childs JE, Marshall CJ. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell. 1989;57:1167–1177. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- 28.Hancock JF, Paterson H, Marshall CJ. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- 29.Hariharan IK, Carthew RW, Rubin GM. The Drosophila roughened mutation: activation of a rap homolog disrupts eye development and interferes with cell determination. Cell. 1991;67:717–722. doi: 10.1016/0092-8674(91)90066-8. [DOI] [PubMed] [Google Scholar]
- 30.Harland R. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- 31.Higgins DG, Sharp PM. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 32.Kauffmann RC, Qian Y, Vogt A, Sebti SM, Hamilton AD, Carthew RW. Activated Drosophila ras1 is selectively suppressed by isoprenyl transferase inhibitors. Proc Natl Acad Sci USA. 1995;92:10919–10923. doi: 10.1073/pnas.92.24.10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohl NE, Mosser SD, deSolms SJ, Giuliani EA, Pompliano DL, Graham SL, Smith RL, Scolnick EM, Oliff A, Gibbs JB. Selective inhibition of ras-dependent transformation by a farnesyltransferase inhibitor. Science. 1993;260:1934–1937. doi: 10.1126/science.8316833. [DOI] [PubMed] [Google Scholar]
- 34.Kohl NE, Wilson FR, Mosser SD, Giuliani E, deSolms SJ, Conner MW, Anthony NJ, Holtz WJ, Gomez RP, Lee TJ, Smith RL, Graham SL, Hartman GD, Gibbs JB, Oliff A. Protein farnesyltransferase inhibitors block the growth of ras-dependent tumors in nude mice. Proc Natl Acad Sci USA. 1994;91:9141–9145. doi: 10.1073/pnas.91.19.9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai ZC, Rubin GM. Negative control of photoreceptor development in Drosophila by the product of the yan gene, an ETS domain protein. Cell. 1992;70:609–620. doi: 10.1016/0092-8674(92)90430-k. [DOI] [PubMed] [Google Scholar]
- 37.Leevers SJ, Paterson HF, Marshall CJ. Requirement for Ras in Raf activation is overcome by targeting Raf to the plasma membrane. Nature. 1994;369:411–414. doi: 10.1038/369411a0. [DOI] [PubMed] [Google Scholar]
- 38.Lillien L. Changes in retinal cell fate induced by overexpression of EGF receptor. Nature. 1995;377:158–162. doi: 10.1038/377158a0. [DOI] [PubMed] [Google Scholar]
- 39.Lowy DR, Willumsen BM. Function and regulation of Ras. Annu Rev Biochem. 1993;62:851–891. doi: 10.1146/annurev.bi.62.070193.004223. [DOI] [PubMed] [Google Scholar]
- 40.Maguire J, Santoro T, Jensen P, Siebenlist U, Yewdell J, Kelly K. Gem: an induced, immediate early protein belonging to the Ras family. Science. 1994;265:241–244. doi: 10.1126/science.7912851. [DOI] [PubMed] [Google Scholar]
- 41.Manne V, Yan N, Carboni JM, Tuomari AV, Ricca CS, Brown JG, Andahazy ML, Schmidt RJ, Patel D, Zahler R, Weinmann R, Channing JD, Cox AD, Hunt JT, Gordon EM, Barbacid M, Seizinger BR. Bisubstrate inhibitors of farnesyltransferase: a novel class of specific inhibitors of ras transformed cells. Oncogene. 1995;10:1763–1779. [PubMed] [Google Scholar]
- 42.Marshall CJ. Protein prenylation: a mediator of protein-protein interactions. Science. 1993;259:1865–1866. doi: 10.1126/science.8456312. [DOI] [PubMed] [Google Scholar]
- 43.Ohmstede CA, Farrell FX, Reep BR, Clemetson KJ, Lapetina EG. RAP2B: a RAS-related GTP-binding protein from platelets. Proc Natl Acad Sci USA. 1990;87:6527–6531. doi: 10.1073/pnas.87.17.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Neil KT, DeGrado WF. How calmodulin binds its targets: sequence independent recognition of amphiphilic alpha-helices. Trends Biochem Sci. 1990;15:59–64. doi: 10.1016/0968-0004(90)90177-d. [DOI] [PubMed] [Google Scholar]
- 45.O’Neill EM, Rebay I, Tjian R, Rubin GM. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- 46.Porfiri E, Evans T, Chardin P, Hancock JF. Prenylation of Ras proteins is required for efficient hSOS1-promoted guanine nucleotide exchange. J Biol Chem. 1994;269:22672–22677. [PubMed] [Google Scholar]
- 47.Pryer NK, Wuestehube LJ, Schekman R. Vesicle-mediated protein sorting. Annu Rev Biochem. 1992;61:471–516. doi: 10.1146/annurev.bi.61.070192.002351. [DOI] [PubMed] [Google Scholar]
- 48.Reynet C, Kahn CR. Rad: a member of the Ras family overexpressed in muscle of type II diabetic humans. Science. 1993;262:1441–1444. doi: 10.1126/science.8248782. [DOI] [PubMed] [Google Scholar]
- 49.Robertson D, Paterson HF, Adamson P, Hall A, Monaghan P. Ultrastructural localization of ras-related proteins using epitope-tagged plasmids. J Histochem Cytochem. 1995;43:471–480. doi: 10.1177/43.5.7537292. [DOI] [PubMed] [Google Scholar]
- 50.Rosen LB, Ginty DD, Weber MJ, Greenberg ME. Membrane depolarization and calcium influx stimulate MEK and MAP kinase via activation of Ras. Neuron. 1994;12:1207–1221. doi: 10.1016/0896-6273(94)90438-3. [DOI] [PubMed] [Google Scholar]
- 51.Rusanescu G, Qi HQ, Thomas SM, Brugge JS, Halegoua S. Calcium influx induces neurite growth through a src-ras signaling cassette. Neuron. 1995;15:1415–1425. doi: 10.1016/0896-6273(95)90019-5. [DOI] [PubMed] [Google Scholar]
- 52.Seabra MC, Brown MS, Goldstein JL. Retinal degeneration in choroideremia: deficiency of rab geranylgeranyl transferase. Science. 1993;259:377–381. doi: 10.1126/science.8380507. [DOI] [PubMed] [Google Scholar]
- 53.Sharp PA. Split genes and RNA splicing. Cell. 1994;77:805–815. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 54.Simon MA, Bowtell DD, Dodson GS, Laverty TR, Rubin GM. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell. 1991;67:701–716. doi: 10.1016/0092-8674(91)90065-7. [DOI] [PubMed] [Google Scholar]
- 55.Simon MA, Carthew RW, Fortini ME, Gaul U, Mardon G, Rubin GM. Signal transduction pathway initiated by activation of the sevenless tyrosine kinase receptor. Cold Spring Harbor Symp Quant Biol. 1992;57:375–380. doi: 10.1101/sqb.1992.057.01.042. [DOI] [PubMed] [Google Scholar]
- 56.Simon MA, Dodson GS, Rubin GM. An SH3-SH2-SH3 protein is required for p21Ras1 activation and binds to sevenless and Sos proteins in vitro. Cell. 1993;73:169–177. doi: 10.1016/0092-8674(93)90169-q. [DOI] [PubMed] [Google Scholar]
- 57.Treier M, Bohmann D, Mlodzik M. Jun cooperates with the ets domain protein pointed to induce photoreceptor R7 fate in the Drosophila eye. Cell. 1995;83:753–760. doi: 10.1016/0092-8674(95)90188-4. [DOI] [PubMed] [Google Scholar]
- 58.Wassarman DA, Therrien M, Rubin GM. The Ras signaling pathway in Drosophila. Curr Opin Genet Dev. 1995;5:44–50. doi: 10.1016/s0959-437x(95)90052-7. [DOI] [PubMed] [Google Scholar]
- 59.Weissbach L, Settleman J, Kalady MF, Snijders AJ, Murthy AE, Yan YX, Bernards A. Identification of a human rasGAP-related protein containing calmodulin-binding motifs. J Biol Chem. 1994;269:20517–20521. [PubMed] [Google Scholar]
- 60.Wes PD, Yu M, Montell RIC (1996) A calmodulin-binding Ras-like GTPase. EMBO J, in press. [PMC free article] [PubMed]
- 61.Wilkinson D. Whole mount in situ hybridization of vertebrate embryos. In: Wilkinson D, editor. In situ hybridization. A practical approach. IRL; Oxford: 1992. pp. 75–84. [Google Scholar]
- 62.Zeidler MP, Yokomori K, Tjian R, Mlodzik M. Drosophila TFIIA is up-regulated and required during ras-mediated photoreceptor determination. Genes Dev. 1996;10:50–59. doi: 10.1101/gad.10.1.50. [DOI] [PubMed] [Google Scholar]