Abstract
Serine–arginine (SR) proteins are essential splicing factors that promote numerous steps associated with mRNA processing and whose biological function is tightly regulated through multi-site phosphorylation. In the nucleus, the cdc2-like kinases (CLKs) phosphorylate SR proteins on their intrinsically disordered Arg–Ser (RS) domains, mobilizing them from storage speckles to the splicing machinery. The CLKs have disordered N termini that bind tightly to RS domains, enhancing SR protein phosphorylation. The N termini also promote nuclear localization of CLKs, but their transport mechanism is presently unknown. To explore cytoplasmic–nuclear transitions, several classical nuclear localization sequences in the N terminus of the CLK1 isoform were identified, but their mutation had no effect on subcellular localization. Rather, we found that CLK1 amplifies its presence in the nucleus by forming a stable complex with the SR protein substrate and appropriating its NLS for transport. These findings indicate that, along with their well-established roles in mRNA splicing, SR proteins use disordered protein–protein interactions to carry their kinase regulator from the cytoplasm to the nucleus.
Keywords: phosphorylation, RNA splicing, nuclear translocation, serine/threonine protein kinase, RNA processing, intrinsically disordered protein, post-transcriptional regulation, Arg–Ser repeat, cdc2-like kinase (CLK), splicing factor, SR protein
Introduction
The splicing of precursor mRNA (pre-mRNA)3 generates mature, intron-free mRNA that is exported to the cytoplasm for protein translation. Splicing occurs at the spliceosome, a macromolecular complex composed of 5 small nuclear ribonuclear proteins (U1–6 snRNPs) and over 100 auxiliary proteins (1). Spliceosome assembly and splice-site selection rely on an essential family of 12 splicing factors known as Ser–Arg (SR) proteins (SRSF1–12) (2, 3). SR proteins possess one or two folded RNA recognition motifs that facilitate pre-mRNA binding and an intrinsically disordered C terminus rich in Arg–Ser (RS) repeats that regulates this event along with other functions (3). SR-specific transportin 2 (TRN-SR2, also called TNPO3), a member of the β-karyopherin protein family, recognizes the RS domain after phosphorylation by cytoplasmic SR protein kinases (SRPKs), directing the SR protein through the nuclear pore complex (4–6). In the nucleus, SR proteins reside largely in membrane-free, storage compartments known as interchromatin granules or speckles, but additional phosphorylation mobilizes them to the nucleoplasm for splicing function (7, 8). The latter modification, catalyzed by the cdc2-like protein kinases (CLK1–4), promotes SR protein attachment to pre-mRNA and U1 snRNP, the first step in spliceosome assembly (9, 10). CLK1 phosphorylates the RS domain of the prototype SR protein SRSF1 (also called ASF/SF2) liberating it from speckles and up-regulating spliceosome assembly (9, 11, 12). Although most family members remain exclusively in the nucleus, SRSF1 is one of four SR proteins (SRSF1, SRSF3, SRSF7, and SRSF10) that transiently shuttle to the cytoplasm upon dephosphorylation, delivering spliced mRNA for protein translation (13–15).
Although CLKs share a conserved, folded kinase domain, they have N-terminal extensions of variable length (approximately 130–300 amino acids) and low residue conservation that are necessary for proper SR protein regulation. The N terminus of CLK1 both enhances overall SR protein phosphorylation rates and enables the phosphorylation of three Ser–Pro dipeptides in the RS domain of SRSF1 required for mobilization from speckles and alternative splicing control (12, 16). Although biochemical studies show that mouse CLK1 forms a highly stable complex with SRSF1 (Kd ≈10 nm) because of strong interactions between the N terminus and the RS domain (11), the mechanism underlying this interaction is still not understood. Sequence analyses suggest that the CLK N termini are intrinsically disordered, a feature that may assist binding to other disordered regions such as the RS domains. Interestingly, the N terminus of CLK1 does not readily distinguish unphosphorylated and phosphorylated SRSF1, binding with high affinity to both (16, 17). This phenomenon may be driven by charge diversity in the N terminus that sustains flexible interactions with either arginines or phosphoserines in the RS domain (see Fig. 1A). Several biophysical studies indicate that although the CLK1 kinase domain is a small monomer, the N terminus induces formation of a large oligomer in the full-length enzyme (hydrodynamic radius ≈ 100 nm) (18). This high-order CLK1 species is strongly coupled to efficient SR protein recognition and phosphorylation, although its mechanism is not understood. Irrespective of the structural basis, high-affinity interactions present a problem for conveying the phospho-SR protein from CLK1 to the spliceosome. However, this problem is circumvented in the nucleus through a secondary protein kinase (SRPK1) that peels away the CLK1 N terminus, freeing the SR protein for splicing function (17).
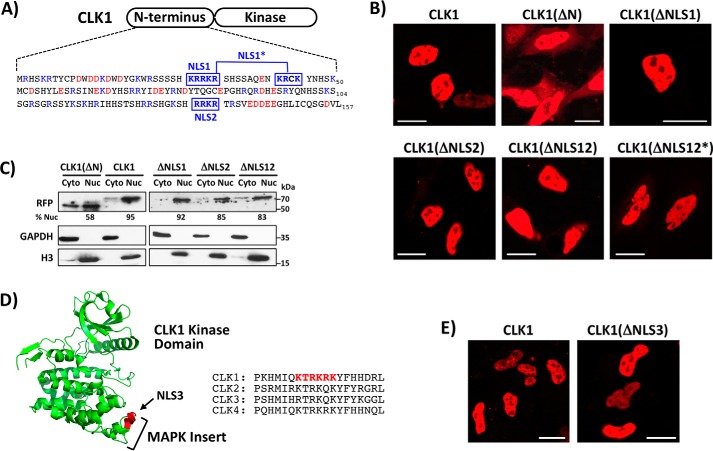
Figure 1.
CLK1 N-terminus lacks a short, positively charged NLS. A, N-terminal sequence of CLK1 and several, short potential NLSs identified by cNLS Mapper. NLS1 and NLS2 represent monopartite NLSs, whereas NLS1* represents a bipartite NLS. B, live-cell confocal imaging of HeLa cells expressing WT CLK1 with a C-terminal RFP tag along with mutant forms where the putative NLS residues are mutated to glycine. NLS regions are defined in A. ΔNLS12* represents glycine mutations in both NLS1* and NLS2. C, fractionation of HeLa cells expressing WT CLK1-RFP and several mutants. ImageJ was used to quantitate the amount of RFP-tagged proteins in the nucleus (% Nuc). D, potential NLS in the MAP kinase insert of the CLK1 kinase domain. Regional sequences of the potential NLS are compared with those in other human CLKs. E, live-cell confocal imaging of HeLa cells expressing WT CLK1-RFP and a form where Lys/Arg in NLS3 is mutated to alanine. Scale bars, 20 μm.
In addition to activity regulation and substrate specificity, the N termini of CLKs are also important for nuclear localization. Early studies showed that the mouse CLK1 (also called Clk/Sty) is exclusively expressed in the nucleus, but N-terminal deletion leads to a nuclear/cytoplasmic mixture, suggesting that this disordered sequence amplifies nuclear localization (19). Although CLK N termini share little sequence homology (Fig. S1), they may serve similar functions with regard to localization. For instance, three human (CLK1–3) and four mouse CLK isoforms (mCLK1–4) have been overexpressed and found localized exclusively to the cell nucleus, consistent with their role in mRNA splicing (12, 20, 21). Furthermore, endogenous CLK1 is expressed in the nuclei of several different cell types supporting such a general splicing function (17, 22). Overexpression of active CLK1 causes SR protein diffusion from speckles, a phenomenon important for splicing control, whereas overexpression of inactive CLK1 inhibits such movement leading to mostly speckle-localized kinase (8, 12, 23). The general importance of CLKs for splicing is now well-established, and many advances have been made in developing useful chemotherapeutic agents that inhibit these kinases for disease intervention (24–26).
Because of the ability of the N terminus to induce a large oligomeric kinase form, it is unlikely that CLK1 can passively diffuse through the nuclear pore, thus raising the question of how this important splicing kinase gains full entry to the nucleus (18). In this study we explored the mechanism underlying the subcellular localization of human CLK1. Although the CLK1 N terminus has short, positively charged regions that resemble classical NLSs, we found that their mutation had no effect on subcellular localization. Instead, we found that CLK1 possesses a diffuse NLS in the N terminus that supports exclusive nuclear localization through a “piggyback” mechanism with its substrate SR protein. Either disruption of SR protein transport by TRN-SR2 knockdown or mutation of the SRSF1 NLS impairs CLK1 nuclear localization. Binding studies reveal that CLK1 attains maximal nuclear entry by forming a complex with phosphorylated SRSF1, thereby recruiting its RS domain as an NLS. These findings suggest that CLKs use their intrinsically disordered N termini to clamp onto the disordered RS domains of SR proteins exploiting them as adaptors for efficient cytoplasmic–nuclear transport. Thus, in addition to their mRNA processing functions, SR proteins are also transport carriers for the CLK family of kinases, their principal regulators in the nucleus.
Results
CLK1 lacks a short classical NLS
To characterize residues that might constitute a classical NLS, we initially identified two monopartite sequences (NLS1 and NLS2) in the CLK1 N terminus using cNLS Mapper (Fig. 1A). To assess their role, we designed a CLK1 construct containing a C-terminal RFP (CLK1-RFP) for direct fluorescence monitoring. Using confocal microscopy, we found that this construct localized exclusively to the nucleus of live HeLa cells, whereas one lacking the N terminus (CLK1(ΔN)-RFP) was present in both the cytoplasm and nucleus (Fig. 1B). We quantified these results in fractionation experiments showing that CLK1(ΔN)-RFP partitions 58% in the nucleus, whereas the full-length CLK1 is 95% nuclear (Fig. 1C). These findings are similar to those reported for the mouse CLK1 isoform (19) and set a baseline for distribution of the RFP-tagged kinase domain in both cellular compartments. We next showed that mutation of Arg/Lys to Gly in either NLS1 or NLS2 (CLK1(ΔNLS1)-RFP or CLK1(ΔNLS2)-RFP) had no observable effect on CLK1 nuclear localization in confocal imaging and only minor effects in fractionation experiments (Fig. 1, B and C). To address whether these NLSs work cooperatively, we mutated both and found that this new construct (CLK1(ΔNLS12)) localized largely in the nucleus in confocal imaging and fractionation experiments (Fig. 1, B and C). To next address whether additional, positively charged sequences contribute to nuclear localization, we also identified a potential bipartite NLS (NLS1*), mutated these sequences along with those in NLS2 (CLK1(ΔNLS12*)-RFP), and found that the mutant was still nuclear (Fig. 1B). In addition, we identified a possible NLS in the conserved MAPK insert of the CLK1 kinase domain (NLS3) (Fig. 1D). We mutated the charged residues to alanine in the full-length kinase and found that the RFP-tagged CLK1(ΔNLS3) localized to the nucleus of live HeLa cells, similar to the WT enzyme (Fig. 1E). Overall, these findings show that the N terminus does not have a short, classical NLS for CLK1 nuclear localization.
Identifying regions important for subcellular localization
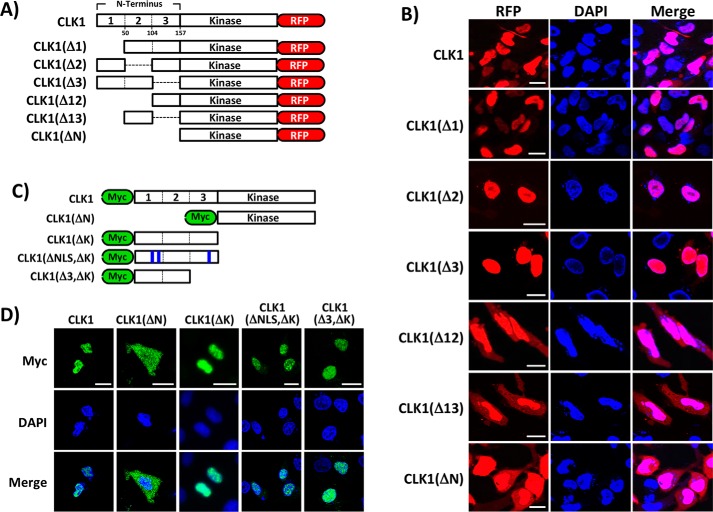
Because CLK1 lacks a short, classical NLS, we wished to determine which other N-terminal sequences control subcellular localization. Considering that the N termini of CLKs share very little sequence homology (Fig. S1), we targeted broad regions for mutagenesis. We constructed a series of deletion mutants in CLK1-RFP by dividing the N terminus into three blocks of ∼50 residues prior to the start of its canonical kinase domain (Fig. 2A). We found that deletion of any individual block did not alter the nuclear localization of CLK1 compared with the WT control in HeLa cells (Fig. 2B). These findings suggest that any two blocks of residues are sufficient for nuclear localization of the kinase. We corroborated these results by showing that deletion of other two-block combinations resulted in mixed cytoplasmic/nuclear localization similar to that for CLK1(ΔN)-RFP (Fig. 2B). Such findings suggest that the nuclear localization sequence in CLK1 is spread over a rather large region of the N terminus. We next tested whether the isolated N terminus is sufficient for nuclear entry in the absence of the kinase domain by generating Myc-tagged forms of CLK1 (Fig. 2C). We found that both full-length CLK1 and a form lacking the kinase domain (CLK1(ΔK)) localized to the nucleus in confocal imaging and fractionation experiments (Fig. 2D and Fig. S2). In control experiments, a Myc-tagged construct lacking the N terminus (CLK1(ΔN)) displayed equivalent cytoplasmic and nuclear localization (Fig. 2D), indicating that removal of the N-terminal sequences allows free diffusion of the monomeric kinase domain. We also found that an N-terminal construct with glycine mutations in the three putative NLS sequences (CLK1(ΔNLS, ΔK)) identified in Fig. 1A localized to the nucleus in both confocal imaging and fractionation experiments, confirming that the CLK1 N terminus does not require a classical NLS (Fig. 2D and Fig. S2). Furthermore, removal of box 3 in the N terminus (CLK1Δ3, ΔK) had no effect on nuclear localization, supporting the idea that two blocks are needed for nuclear localization (Fig. 2D). Together, these findings indicate that the nuclear localization sequence for CLK1 is highly delocalized along the N terminus.
Figure 2.
N-terminal regions important for CLK1 nuclear localization. A, deletions in N terminus of CLK1-RFP. B, confocal imaging of WT and mutant CLK1-RFP constructs expressed in HeLa cells. C, deletions in Myc-tagged forms of CLK1. The N terminus is divided at the same residues as shown in A for the CLK1-RFP constructs. D, confocal imaging of WT and mutant forms of Myc-CLK1 expressed in HeLa cells. Scale bars, 20 μm. DAPI, 4′,6′-diamino-2-phenylindole.
An SR-specific transportin facilitates CLK1 nuclear localization
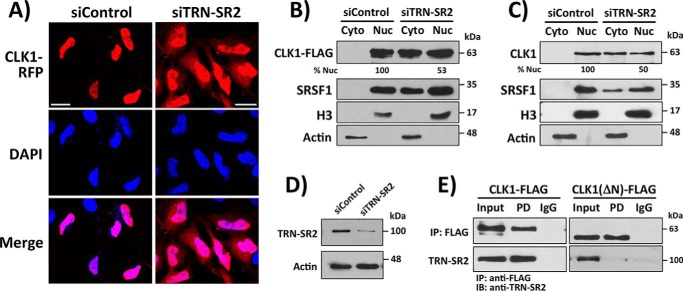
SR proteins are transported to the nucleus through interaction of their phosphorylated RS domains with TRN-SR2, an SR protein-specific transportin (4, 5, 27). Because the N terminus binds with high affinity to RS domains, we speculated that CLK1 might indirectly attain nuclear localization through this transportin. To test this idea, we monitored the subcellular localization of CLK1-RFP in HeLa cells in the absence and presence of TRN-SR2 knockdown using siRNA. We found that although CLK1-RFP localized exclusively to the nucleus, the kinase was present in both the cytoplasm and nucleus of cells treated with siRNA (Fig. 3A). We next expressed CLK1 with a C-terminal FLAG sequence (CLK1-FLAG) in HeLa cells and performed fractionation experiments. We showed that CLK1-FLAG localizes exclusively to the nucleus in the absence of siRNA but displays equivalent cytoplasmic and nuclear distribution in the presence of siRNA treatment, results consistent with the imaging data (Fig. 3B). To rule out the possibility that these effects are the result of kinase overexpression or the C-terminal tag, we showed in fractionation experiments that TRN-SR2 knockdown also results in a shift of endogenous CLK1 from the nucleus to the cytoplasm (Fig. 3C). Consistent with the SR protein transport function of TRN-SR2, knockdown impairs endogenous SRSF1 entry into the nucleus (Fig. 3, B and C). We verified by Western blotting analyses that these observed changes in subcellular localization are associated with decreases in TRN-SR2 protein levels upon siRNA treatment (Fig. 3D). Because full CLK1 nuclear entry depends on TRN-SR2, we next wished to determine whether CLK1 interacts with TRN-SR2 in whole cell lysates. To accomplish this, we expressed CLK1-FLAG and a form lacking the N terminus (CLK1(ΔN)-FLAG) in HeLa cells and monitored their interactions with endogenous TRN-SR2 using co-immunoprecipitation assays. We found that immunoprecipitated CLK1-FLAG, but not CLK1(ΔN)-FLAG, interacts with endogenous TRN-SR2 in whole cell lysates (Fig. 3E). These findings indicate that full entry of CLK1 into the nucleus is contingent on its interaction with TRN-SR2 and that this interaction depends on the N terminus, a disordered domain in CLK1 that enhances SR protein binding (16).
Figure 3.
CLK1 nuclear localization requires TRN-SR2. A, knockdown of TRN-SR2 in HeLa cells with siRNA affects subcellular localization of CLK1-RFP using confocal microscopy. B, knockdown of TRN-SR2 with siRNA in HeLa cells affects subcellular localization of CLK1-FLAG using fraction experiments. C, knockdown of TRN-SR2 with siRNA in HEK293 cells affects subcellular localization of endogenous CLK1 using fractionation experiments. ImageJ was used in B and C to quantitate the amount of CLK1-FLAG and endogenous CLK1 in the nucleus (% Nuc) in the absence and presence of siRNA treatment. D, TRN-SR2 is depleted in HeLa cells treated with siRNA. E, co-immunoprecipitation (IP) assays showing that immunoprecipitated CLK1-FLAG, but not CLK1(ΔN)-FLAG, interacts with endogenous TRN-SR2 in HeLa cell lysates. FLAG-tagged constructs are immunoprecipitated with an agarose-conjugated anti-FLAG antibody, and IgG represents the agarose resin control lacking the conjugated antibody. Scale bars, 20 μm. DAPI, 4′,6′-diamino-2-phenylindole; Cyto, cytoplasmic; Nuc, nuclear.
Phospho-SRSF1 recruits CLK1 to TRN-SR2
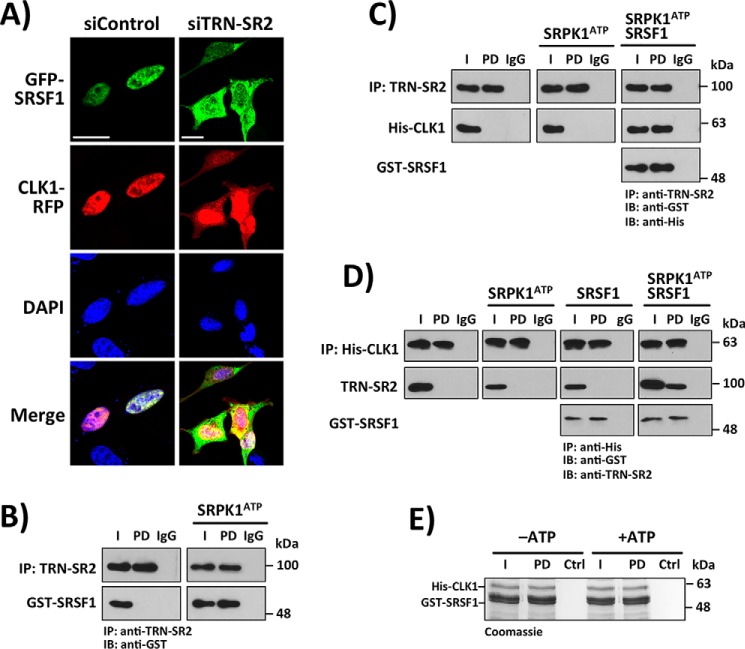
We showed previously that CLK1 forms a high-affinity complex with SRSF1, raising the possibility that the kinase may enter the nucleus indirectly through a “piggyback” mechanism with the SR protein (16). To address this hypothesis, we first showed that siRNA treatment of HeLa cells expressing CLK1-RFP and SRSF1 with an N-terminal GFP tag (GFP-SRSF1) induced both cytoplasmic CLK1-RFP and GFP-SRSF1, suggesting that both may use a common nuclear transporter (Fig. 4A). To test whether CLK1 independently associates with TRN-SR2 or instead relies upon an SR protein, we monitored the binding of recombinant His-CLK1 to endogenous TRN-SR2. We first showed that TRN-SR2 immunoprecipitated from cytoplasmic fractions lacking CLK1 and SRSF1 (Fig. S3) interacts with GST-SRSF1 only in the presence of SRPK1 and ATP, consistent with prior reports (27) (Fig. 4B). We then added recombinant His-CLK1 to immunoprecipitated TRN-SR2 and showed that they interact only in the presence of SRPK1-phosphorylated GST-SRSF1 (Fig. 4C). To corroborate these results, we also showed that immunoprecipitated His-CLK1 interacts with endogenous TRN-SR2 in cytoplasmic fractions only in the presence of SRPK1-phosphorylated GST-SRSF1 (Fig. 4D). We verified that catalytic His-SRPK1 in these experiments phosphorylates GST-SRSF1 (Fig. S4A) and also showed that although CLK1 phosphorylates SRSF1, its rate is significantly lower than that for SRPK1 at the same enzyme concentrations (Fig. S4B), supporting prior published findings that SRPK1 is the principal nuclear import driver for SRSF1 (6, 28). Finally, we demonstrated in pulldown assays that CLK1 binds to SRSF1 independent of RS domain phosphorylation, as previously reported (17), suggesting that CLK1 strongly recognizes the SR protein regardless of its phosphorylation state (Fig. 4E). Overall, these experiments indicate that the phosphorylated SR protein acts as an adaptor for CLK1 binding to TRN-SR2, consistent with a “piggyback” mechanism for CLK1 nuclear entry.
Figure 4.
Interaction of CLK1 with TRN-SR2 requires SR proteins. A, knockdown of TRN-SR2 with siRNA leads to co-localization of GFP-SRSF1 and CLK1-RFP in the cytoplasm using confocal microscopy. B, TRN-SR2 immunoprecipitated (IP) from cytoplasmic fractions interacts with SRPK1-phosphorylated recombinant GST-SRSF1. C, TRN-SR2 immunoprecipitated from cytoplasmic fractions binds to recombinant His-CLK1 only in the presence of SRPK1-phosphorylated GST-SRSF1. In B and C, TRN-SR2 is immunoprecipitated with an agarose-conjugated anti-TRN-SR2 antibody, and IgG represents the agarose resin control lacking the conjugated antibody. D, immunoprecipitated His-CLK1 binds to TRN-SR2 in cytoplasmic fractions only in the presence of SRPK1-phosphorylated GST-SRSF1. Recombinant His-CLK1 is immunoprecipitated with an agarose-conjugated anti-His antibody, and IgG represents the agarose resin control lacking the conjugated antibody. E, His-CLK1 interacts with both unphosphorylated and phosphorylated GST-SRSF1 in g-agarose pulldown assays. Ctrl refers to a control in which GST-SRSF1 is omitted. Scale bars, 20 μm. DAPI, 4′,6′-diamino-2-phenylindole; IB, immunoblot.
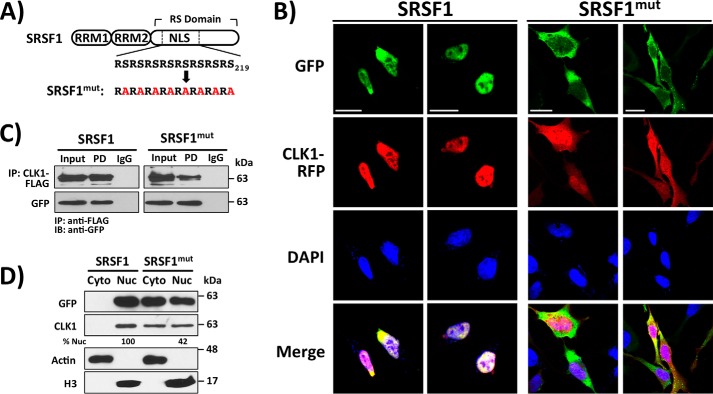
NLS in SRSF1 drives CLK1 into the nucleus
Because CLK1 binding to TRN-SR2 relies upon a phosphorylated SR protein, we tested whether selective disruption of SRSF1 nuclear entry without TRN-SR2 down-regulation also blocks CLK1 access to this compartment. To accomplish this, we used a form of SRSF1 that is defective in entering the nucleus because of mutations in its NLS. In a previous study, we demonstrated that replacing 8 serines in the N terminus of the RS domain generates a mutant SRSF1 (GFP-SRSF1mut) resistant to SRPK1-dependent nuclear transport (29) (Fig. 5A). We confirmed these results showing that GFP-SRSF1mut is indeed present in the cytoplasm of HeLa cells (Fig. 5B). Importantly, we found that GFP-SRSF1mut expression also induces cytoplasmic localization of CLK1-RFP, suggesting that CLK1 nuclear entry relies upon SR protein transport to the nucleus (Fig. 5B). We next wished to determine whether decreased CLK1 transport into the nucleus of cells expressing GFP-SRSF1mut is due to SR protein binding. We immunoprecipitated CLK1-FLAG and showed that it interacts with both GFP-SRSF1 and GFP-SRSF1mut in HeLa cell lysates, indicating that Ser-to-Ala mutation in the NLS of the RS domain does not disrupt CLK1–SRSF1 complex formation (Fig. 5C). These findings indicate that the NLS-defective SR protein retains binding to CLK1, limiting nuclear transport. Finally, we showed in fractionation experiments that overexpression of GFP-SRSF1mut results in cytoplasmic distribution of endogenous CLK1, indicating that the observed effects are not the result of kinase overexpression or the RFP tag (Fig. 5D). Overall, the combined data indicate that CLK1 gains maximal entry to the nucleus using a “piggyback” mechanism in which the N terminus of the kinase binds tightly to the SR protein whose RS domain is a phosphorylation-dependent NLS for TRN-SR2.
Figure 5.
Disruption of the NLS on SRSF1 induces cytoplasmic CLK1 localization. A, Ser-to-Ala mutations in the NLS of GFP-SRSF1. B, GFP-SRSF1mut and CLK1-RFP co-localize to the cytoplasm using confocal microscopy. C, co-immunoprecipitation (IP) assays show that CLK1-FLAG interacts with both GFP-SRSF1 and GFP-SRSF1mut in HeLa cell lysates. CLK1-FLAG is immunoprecipitated with an agarose-conjugated anti-FLAG antibody, and IgG represents the agarose resin control lacking the conjugated antibody. D, overexpression of GFP-SRSF1mut induces changes in subcellular distribution of endogenous CLK1 in fractionation experiments. ImageJ was used to quantitate the amount of endogenous CLK1 in the nucleus (% Nuc) upon the expression of GFP-SRSF1 and GFP-SRSF1mut. Scale bars, 20 μm. Cyto, cytoplasmic; Nuc, nuclear.
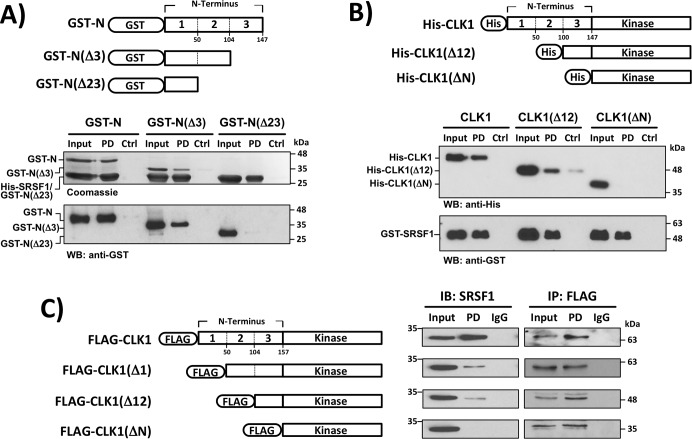
SRSF1 binding correlates with CLK1 nuclear transitions
We showed previously that the CLK1 N terminus binds with high affinity to the RS domain of SRSF1 (11) and wanted to determine whether this stable interaction correlates with CLK1 nuclear entry. We first made a soluble, GST-tagged form of the N terminus (GST-N) that reconstituted stable binding to His-SRSF1 in pulldown assays (Fig. 6A). We then made deletions in this construct and found that the apparent affinity of the N terminus for His-SRSF1 decreased as a function of serial deletion. In these experiments, we used Western blotting to monitor relative binding because one of the constructs, GST-N(Δ23), co-migrated with His-SRSF1 (Fig. 6A). These studies indicate not only that SRSF1 interaction with the CLK1 N terminus is length-dependent but also that the N terminus does not require phosphorylation for SR protein interactions because the GST-N constructs are bacterially expressed and unphosphorylated. We next wished to determine whether similar N-terminal deletions in His-CLK1 also cause a proportional decrease in SRSF1 binding. In pulldown assays, we found that serial deletion of the N terminus in His-CLK1 caused progressive reductions in GST-SRSF1 binding compared with WT His-CLK1 based on comparisons of input and pulldown lanes (Fig. 6B). To correlate these observed length-dependent changes in substrate binding with CLK1 nuclear localization, we expressed several CLK1 deletions with an N-terminal FLAG tag in HeLa cells that mirror those made in the CLK1-RFP constructs (Fig. 2A) and monitored their binding to endogenous SRSF1 in co-immunoprecipitation assays (Fig. 6C). We found that sequential deletion diminished interaction with endogenous SRSF1, suggesting that SR protein affinity depends on CLK1 N-terminal length. Thus, the removal of two or three blocks in the N terminus reduces SRSF1 interactions (Fig. 6) in correspondence with reductions in nuclear localization in the imaging data (Figs. 1 and 2). Overall, these binding experiments suggest that the N terminus drives high-affinity SRSF1 interactions propelling full localization of CLK1 in the nucleus.
Figure 6.
N terminus of CLK1 supports binding to SRSF1. A, C-terminal deletions of the CLK1 N terminus reduce binding affinity to His-SRSF1 in pulldown assays. His-SRSF1 is immobilized on a nickel resin and incubated with GST-N constructs before washing. Ctrl refers to a control in which His-SRSF1 is omitted. B, N-terminal deletions in His-CLK1 reduce binding affinity to GST-SRSF1 in pulldown assays. GST-SRSF1 is immobilized on GSH–agarose resin and incubated with His-CLK1 constructs before washing. Ctrl refers to a control in which GST-SRSF1 is omitted. C, N-terminal deletions in FLAG-CLK1 reduces binding to endogenous SRSF1 in HeLa cell lysates. FLAG-CLK1 is immunoprecipitated (IP) with an agarose-conjugated anti-FLAG antibody, and IgG represents the agarose resin control lacking the conjugated antibody. IB, immunoblot; WB, Western blot.
Discussion
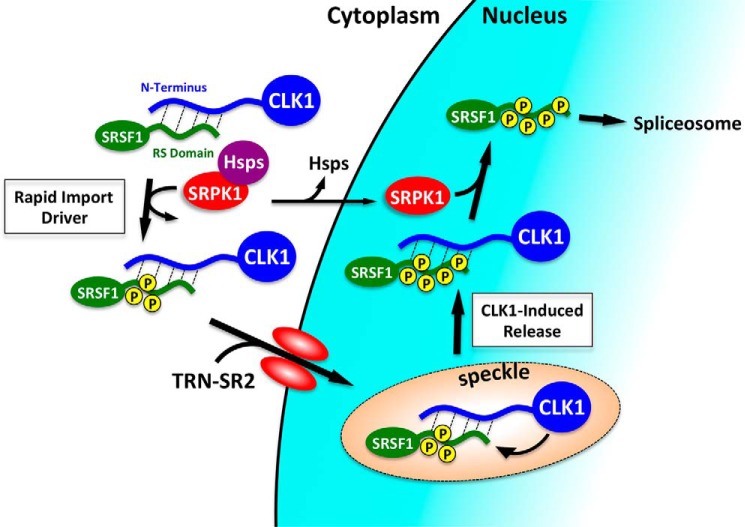
Phosphorylation plays a highly specialized role in the cytoplasmic–nuclear trafficking and splicing function of SR proteins. Although the two key enzymes that catalyze these important post-translational modifications, the SRPKs and CLKs, target RS domains, they incorporate very distinctive catalytic mechanisms that have significant implications for the cellular processing of these essential splicing factors. Both enzymes bind with high affinity to SR proteins but possess divergent mechanisms for substrate recognition and product dissociation. SRPKs use a conserved docking groove in a structured kinase domain that binds Arg–Ser repeats in RS domains, but upon phosphorylation by SRPKs, electrostatic clashes prompt rapid phospho-SR protein release (30). The latter is likely to be critical for the subsequent association of TRN-SR2 and passage of the SR protein through the nuclear pore (Fig. 7). In comparison, CLKs lack a docking groove and instead incorporate a disordered N terminus that binds the disordered RS domain of the SR protein. This provides a sticky platform for SR protein recognition but does not offer a means for facile dissociation of the phospho-SR protein. We showed previously that this dilemma can be overcome by recruiting SRPK1 to the nucleus in a signal-dependent manner. SRPK1 is typically maintained in the cytoplasm through the association of chaperone complexes, but upon EGF stimulation and release of these cytoplasmic linkages (31, 32), free SRPK1 enters the nucleus and expedites CLK1 release by peeling away the N terminus from SRSF1. This structural change frees the splicing factor for spliceosomal functions averting a nonproductive enzyme complex (17). In this new study, we now show that this high-affinity product complex also provides a means for nuclear transport of CLK1 (Fig. 7).
Figure 7.
Model showing CLK1 entry into the nucleus via a piggyback mechanism with an SR protein. CLK1 forms a high-affinity complex with SRSF1 using interactions between the N terminus and RS domain. SRPK1 phosphorylation of several serines in the RS domain drives binding to TRN-SR2 and nuclear import of the SR protein. The contacts between SRSF1 and CLK1 can be broken in the nucleus for splicing function by SRPK1.
Large proteins (>40 kDa) destined for the nucleus often contain a short, positively charged NLS that binds to adaptor proteins from the β-karyopherin family, thereby assisting passage through the nuclear pore complex (33). Because CLK1 is a large oligomeric protein and unlikely to diffuse freely through the nuclear pore (18), we explored the possibility that nuclear localization of the kinase depends on a classical NLS. CLK1 contains sequences resembling such classical NLSs in both the N terminus and kinase domain but does not appear to utilize them for nuclear localization. Instead, we showed that CLK1 strongly interacts with an SR protein appropriating its NLS for maximal nuclear entry (Fig. 7). Although the disordered N terminus is clearly an important facilitator of this transition, other regions may participate because we observed that CLK1 lacking its N terminus is not excluded from the nucleus. The latter observation could be the result of some basic affinity of the kinase domain for its substrate in the absence of N-terminal residues. Indeed, prior studies showed that the kinase domain is still active in the absence of the N terminus, although its binding affinity for SRSF1 is diminished by 15-fold (16). Thus, without its N terminus, CLK1 can still form transient interactions with SRSF1, not readily detected in a stringent immunoprecipitation assay, that may facilitate the observed basal level of nuclear localization. It is also possible that upon deletion of the N terminus, a latent signal in the kinase domain may be uncovered that drives some nuclear localization. Finally, removal of the CLK1 N terminus has been shown previously to reduce the kinase from a large oligomer that cannot passively transit through the nuclear pore to a small monomer that is expected to diffuse freely between the cytoplasmic and nuclear compartments (18). This change in quaternary structure may account for a large amount of nuclear CLK1(ΔN) in our experiments. Regardless, the data indicate that the N terminus is a strong modifier of CLK1 subcellular localization and that a significant pathway for such nuclear–cytoplasmic transition involves a ternary complex composed of CLK1, phospho-SRSF1, and TRN-SR2.
An interesting question arising from our present study is why the N terminus of CLK1 appears to contain short, positively charged sequences that resemble classical NLSs but does not utilize them in the typical mechanism for nuclear localization with the importin α/β heterodimer (34). We speculate upon two possible reasons for this result. First, we have shown that CLK1 and its substrate form a high-affinity complex (Km = ≈100 nm) that is far more stable than most kinase–substrate pairs (11, 35). It is possible that the high affinity of CLK1 for SRSF1 may diminish any free CLK1 that could be used in a classic transport mechanism. Second, we have shown previously that CLK1 forms oligomers through its disordered N terminus (18). Interestingly, this oligomer formation is vital for both the high-affinity recognition and specific phosphorylation of the RS domains in SR proteins. It is conceivable that such higher-order species, although vital for biological function, could also mask classical NLSs in CLK1. Although our data indicate that CLK1 nuclear entry can be induced by its substrate SR protein, there may be conditions where its classical NLS subsumes this function if the kinase oligomerization state is reduced because of cytoplasmic factors or variances in the CLK1-SR protein stoichiometry.
CLKs are vital catalysts for SR protein phosphorylation and subsequent incorporation into the developing spliceosome (9). Not surprisingly, they are largely found in the nucleus of interphase cells either concentrated in speckles or diffusely spread in the nucleoplasm to carry out their splicing function. However, our new mechanism for CLK1 transport to the nucleus via a “piggyback” mechanism with an SR protein raises the possibility that CLKs could serve additional functions outside the nucleus (Fig. 7). Several SR proteins including SRSF1 shuttle between the nucleus and cytoplasm escorting processed mRNA (6, 27, 36). SRSF1 has been shown to interact with ribosomal proteins in the cytoplasm activating the translation of a reporter mRNA (13, 37). Furthermore, it has been shown that environmental factors can induce the translocation of SRSF1 from the nucleus to cytoplasmic stress granules where it can down-regulate mRNA translation (38). Stress granules are highly dynamic structures whose assembly/disassembly is regulated, in part, by protein phosphorylation (39). SR protein-rich granules have also been observed in the cytoplasm of some mouse tissues, particularly testes (40). Interestingly, CLK1 along with SRPK1 have been detected in such granules, where they are thought to induce SR protein phosphorylation and regulate alternative splicing. Because of the high affinity of CLKs for SR proteins, it is possible that these kinases may influence such cytoplasmic events in a phosphorylation-dependent manner. In summary, this new mechanism not only explains how CLKs can maximize their presence in the nucleus using disordered protein–protein interactions but also offers a possible means for signal-dependent transport to the cytoplasm for additional functions.
Experimental procedures
Materials
ATP, Mops, HEPES, Tris, MgCl2, MnCl2, NaCl, KCl, DTT, EDTA, Brij 35, Nonidet P-40, glycerol, acetic acid, lysozyme, DNase, RNase, Phenix imaging film, BSA, nickel resin, GSH, and liquid scintillant were obtained from Fisher Scientific. [32P]ATP was obtained from NEN Products, FuGENE reagent was obtained from Promega, Lipofectamine 2000 was obtained from Thermo Fisher, and protease inhibitor mixture was obtained from Roche. Anti–TRN-SR2 (anti-TNPO3) and anti-RFP antibodies were obtained from Abcam. His tag and TNPO3 agarose–conjugated, anti–histone H3, and anti-FLAG antibodies were obtained from Cell Signaling. Anti-GAPDH antibodies were purchased from R&D Systems. Anti-GST antibodies were obtained from BioLegend. Anti-actin antibodies were obtained from Sigma. InstantBlue was purchased from Expedeon. siRNA was obtained from Bioneer.
Expression and purification of recombinant proteins
Recombinant SRPK1 and SRSF1 were expressed and purified from pET19b vectors containing an N-terminal His tag as previously described (16). CLK1 virus was transfected and expressed in Hi5 insect cells, and His-tagged CLK1 was purified using a nickel resin and a previously described procedure (12). GFP-SRSF1 constructs were expressed from pcDNA3.1+N-eGFP vectors, and CLK constructs were expressed from pcDNA3-mRFP vectors. All proteins containing a GST tag were expressed and purified from a pGEX vector as previously described unless otherwise stated (16).
Phosphorylation reactions
Phosphorylation of GST-SRSF1 and His-SRSF1 by His-SRPK1 or His-CLK1 were carried out in the presence of 100 mm Mops (pH 7.4), 10 mm Mg2+ at 37 °C, using 100 μm [32P]ATP with a specific activity of 4000–8000 cpm/pmol. All reactions were carried out in a total volume of 10 μl and quenched with 10 μl of SDS/PAGE loading buffer. Phosphorylated SR proteins were separated from unreacted [32P]ATP by SDS-PAGE (12–16%), cut from the dried gel, and quantified on the 32P channel in liquid scintillant.
Pulldown assays
GST-tagged proteins (10 μm) were incubated with His-tagged proteins (2–10 μm) in binding buffer (0.1% Nonidet P-40, 20 mm Tris/HCl (pH 7.5), and 75 mm NaCl) in a total volume of 40 μl for 30 min before incubating with 25 μl of either GSH–agarose or nickel–agarose resin for 30 min at room temperature. Where phosphorylation is performed, GST-tagged proteins were incubated with His-tagged enzyme (100 nm SRPK1 or 2 μm CLK1) in the absence and presence of 100 μm ATP with 10 mm Mg2+, 20 mm Tris/HCl (pH 7.5), and 75 mm NaCl at 37 °C for 60 min, followed by incubating with 25 μl of either GSH–agarose or nickel–agarose resin for 30 min at room temperature. In all cases, the resin was washed four times with 200 μl of binding buffer, and the bound proteins were eluted with SDS quench buffer and boiled for 5 min. Retained protein was resolved by SDS-PAGE (12% or 18% gel) and visualized by Instant Blue Coomassie stain.
Immunoprecipitation and confocal imaging experiments
HeLa or HEK293 cell lysates (200 μl at 1 mg/ml) were incubated with gentle rocking overnight at 4 °C with 10 μl immobilized antibody-bead conjugate, followed by five 500 μl of 1× PBS washes at 4 °C with centrifugation at 1000 × g. The bands were visualized by Western immunoblotting. Cell fractionations were performed using a fractionation kit purchased from Cell Signaling. Whole cell lysates were generated by incubating cells with 1× radioimmune precipitation assay buffer on ice followed by sonication. For ImageJ analyses of Western blots, a rectangular frame was drawn around each protein band in a lane, and a profile plot was generated displaying the relative density of the contents of the rectangle. For live-cell confocal imaging, HeLa cells were plated on 2.5-cm2 MatTek poly-d-lysine plates and transfected with 2 μg of construct for 24 h and washed with PBS prior for imaging. For analyzing intrinsic fluorescence of GFP- and RFP-tagged proteins, HeLa cells were fixed to coverslips using 1% paraformaldehyde and directly imaged. For immunofluorescence, HeLa cells were fixed to coverslips using 1% paraformaldehyde, permeabilized using Triton X-100, blocked with 20% goat serum, and incubated with anti-Myc antibody and 4′,6′-diamino-2-phenylindole–stained. All transfected cells were analyzed using an Olympus FV1000 as described previously (12). For knockdown experiments, Lipofectamine 2000 was used to transfect 50 pmol of siRNA in a 12-well poly-d-lysine plate.
Author contributions
A. G. and B. E. A. data curation; A. G., B. E. A., and L. F. formal analysis; A. G., B. E. A., and L. F. methodology; J. A. A. conceptualization; J. A. A. supervision; J. A. A. writing-original draft; J. A. A. project administration; J. A. A. writing-review and editing.
Supplementary Material
Acknowledgment
We thank Dr. Nicolas Villanueva for help with purifying GST-N constructs.
This work was supported by National Institutes of Health Grants GM67969 and GM98528. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S4.
- mRNA
- precursor mRNA
- CLK
- cdc2-like kinase
- RS domain
- domain rich in arginine–serine dipeptide repeats
- SR protein
- splicing factor containing arginine-serine dipeptide repeats
- SRPK
- serine-arginine–specific protein kinase
- SRSF1
- SR protein splicing factor 1 (also called ASF/SF2)
- TRN-SR2
- SR-specific transportin 2
- NLS
- nuclear localization sequence
- RFP
- red fluorescent protein
- GST-N
- GST-tagged form of the N terminus.
References
- 1. Müller S., Wolpensinger B., Angenitzki M., Engel A., Sperling J., and Sperling R. (1998) A supraspliceosome model for large nuclear ribonucleoprotein particles based on mass determinations by scanning transmission electron microscopy. J. Mol. Biol. 283, 383–394 10.1006/jmbi.1998.2078 [DOI] [PubMed] [Google Scholar]
- 2. Stojdl D. F., and Bell J. C. (1999) SR protein kinases: the splice of life. Biochem. Cell Biol. 77, 293–298 10.1139/o99-046 [DOI] [PubMed] [Google Scholar]
- 3. Jeong S. (2017) SR proteins: binders, regulators, and connectors of RNA. Mol. Cells 40, 1–9 10.14348/molcells.2017.2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maertens G. N., Cook N. J., Wang W., Hare S., Gupta S. S., Öztop I., Lee K., Pye V. E., Cosnefroy O., Snijders A. P., KewalRamani V. N., Fassati A., Engelman A., and Cherepanov P. (2014) Structural basis for nuclear import of splicing factors by human transportin 3. Proc. Natl. Acad. Sci. U.S.A. 111, 2728–2733 10.1073/pnas.1320755111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kataoka N., Bachorik J. L., and Dreyfuss G. (1999) Transportin-SR, a nuclear import receptor for SR proteins. J. Cell Biol. 145, 1145–1152 10.1083/jcb.145.6.1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lai M. C., Lin R. I., and Tarn W. Y. (2001) Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Natl. Acad. Sci. U.S.A. 98, 10154–10159 10.1073/pnas.181354098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aubol B. E., Keshwani M. M., Fattet L., and Adams J. A. (2018) Mobilization of a splicing factor through a nuclear kinase-kinase complex. Biochem. J. 475, 677–690 10.1042/BCJ20170672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sacco-Bubulya P., and Spector D. L. (2002) Disassembly of interchromatin granule clusters alters the coordination of transcription and pre-mRNA splicing. J. Cell Biol. 156, 425–436 10.1083/jcb.200107017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cho S., Hoang A., Sinha R., Zhong X. Y., Fu X. D., Krainer A. R., and Ghosh G. (2011) Interaction between the RNA binding domains of Ser–Arg splicing factor 1 and U1–70K snRNP protein determines early spliceosome assembly. Proc. Natl. Acad. Sci. U.S.A. 108, 8233–8238 10.1073/pnas.1017700108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kohtz J. D., Jamison S. F., Will C. L., Zuo P., Lührmann R., Garcia-Blanco M. A., and Manley J. L. (1994) Protein–protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature 368, 119–124 10.1038/368119a0 [DOI] [PubMed] [Google Scholar]
- 11. Aubol B. E., Plocinik R. M., Hagopian J. C., Ma C. T., McGlone M. L., Bandyopadhyay R., Fu X. D., and Adams J. A. (2013) Partitioning RS domain phosphorylation in an SR protein through the CLK and SRPK protein kinases. J. Mol. Biol. 425, 2894–2909 10.1016/j.jmb.2013.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Keshwani M. M., Aubol B. E., Fattet L., Ma C. T., Qiu J., Jennings P. A., Fu X. D., and Adams J. A. (2015) Conserved proline-directed phosphorylation regulates SR protein conformation and splicing function. Biochem. J. 466, 311–322 10.1042/BJ20141373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sanford J. R., Gray N. K., Beckmann K., and Cáceres J. F. (2004) A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 18, 755–768 10.1101/gad.286404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang Y., Yario T. A., and Steitz J. A. (2004) A molecular link between SR protein dephosphorylation and mRNA export. Proc. Natl. Acad. Sci. U.S.A. 101, 9666–9670 10.1073/pnas.0403533101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Twyffels L., Gueydan C., and Kruys V. (2011) Shuttling SR proteins: more than splicing factors. FEBS J. 278, 3246–3255 10.1111/j.1742-4658.2011.08274.x [DOI] [PubMed] [Google Scholar]
- 16. Aubol B. E., Plocinik R. M., Keshwani M. M., McGlone M. L., Hagopian J. C., Ghosh G., Fu X. D., and Adams J. A. (2014) N-terminus of the protein kinase CLK1 induces SR protein hyperphosphorylation. Biochem. J. 462, 143–152 10.1042/BJ20140494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aubol B. E., Wu G., Keshwani M. M., Movassat M., Fattet L., Hertel K. J., Fu X. D., and Adams J. A. (2016) Release of SR proteins from CLK1 by SRPK1: a symbiotic kinase system for phosphorylation control of pre-mRNA splicing. Mol. Cell 63, 218–228 10.1016/j.molcel.2016.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Keshwani M. M., Hailey K. L., Aubol B. E., Fattet L., McGlone M. L., Jennings P. A., and Adams J. A. (2015) Nuclear protein kinase CLK1 uses a non-traditional docking mechanism to select physiological substrates. Biochem. J. 472, 329–338 10.1042/BJ20150903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Duncan P. I., Howell B. W., Marius R. M., Drmanic S., Douville E. M., and Bell J. C. (1995) Alternative splicing of STY, a nuclear dual specificity kinase. J. Biol. Chem. 270, 21524–21531 10.1074/jbc.270.37.21524 [DOI] [PubMed] [Google Scholar]
- 20. Duncan P. I., Stojdl D. F., Marius R. M., Scheit K. H., and Bell J. C. (1998) The Clk2 and Clk3 dual-specificity protein kinases regulate the intranuclear distribution of SR proteins and influence pre-mRNA splicing. Exp. Cell Res. 241, 300–308 10.1006/excr.1998.4083 [DOI] [PubMed] [Google Scholar]
- 21. Nayler O., Stamm S., and Ullrich A. (1997) Characterization and comparison of four serine- and arginine-rich (SR) protein kinases. Biochem. J. 326, 693–700 10.1042/bj3260693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Prasad J., and Manley J. L. (2003) Regulation and substrate specificity of the SR protein kinase Clk/Sty. Mol. Cell Biol. 23, 4139–4149 10.1128/MCB.23.12.4139-4149.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Colwill K., Pawson T., Andrews B., Prasad J., Manley J. L., Bell J. C., and Duncan P. I. (1996) The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 15, 265–275 10.1002/j.1460-2075.1996.tb00357.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sako Y., Ninomiya K., Okuno Y., Toyomoto M., Nishida A., Koike Y., Ohe K., Kii I., Yoshida S., Hashimoto N., Hosoya T., Matsuo M., and Hagiwara M. (2017) Development of an orally available inhibitor of CLK1 for skipping a mutated dystrophin exon in Duchenne muscular dystrophy. Sci. Rep. 7, 46126 10.1038/srep46126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zu M., Li C., Fang J. S., Lian W. W., Liu A. L., Zheng L. S., and Du G. H. (2015) Drug discovery of host CLK1 inhibitors for influenza treatment. Molecules 20, 19735–19747 10.3390/molecules201119653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jain P., Karthikeyan C., Moorthy N. S., Waiker D. K., Jain A. K., and Trivedi P. (2014) Human CDC2-like kinase 1 (CLK1): a novel target for Alzheimer's disease. Curr. Drug Targets 15, 539–550 10.2174/1389450115666140226112321 [DOI] [PubMed] [Google Scholar]
- 27. Lai M. C., Lin R. I., Huang S. Y., Tsai C. W., and Tarn W. Y. (2000) A human importin-beta family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J. Biol. Chem. 275, 7950–7957 10.1074/jbc.275.11.7950 [DOI] [PubMed] [Google Scholar]
- 28. Koizumi J., Okamoto Y., Onogi H., Mayeda A., Krainer A. R., and Hagiwara M. (1999) The subcellular localization of SF2/ASF is regulated by direct interaction with SR protein kinases (SRPKs). J. Biol. Chem. 274, 11125–11131 10.1074/jbc.274.16.11125 [DOI] [PubMed] [Google Scholar]
- 29. Aubol B. E., Serrano P., Fattet L., Wüthrich K., and Adams J. A. (2018) Molecular interactions connecting the function of the serine-arginine-rich protein SRSF1 to protein phosphatase 1. J. Biol. Chem. 293, 16751–16760 10.1074/jbc.RA118.004587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ngo J. C., Giang K., Chakrabarti S., Ma C. T., Huynh N., Hagopian J. C., Dorrestein P. C., Fu X. D., Adams J. A., and Ghosh G. (2008) A sliding docking interaction is essential for sequential and processive phosphorylation of an SR protein by SRPK1. Mol. Cell 29, 563–576 10.1016/j.molcel.2007.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou Z., Qiu J., Liu W., Zhou Y., Plocinik R. M., Li H., Hu Q., Ghosh G., Adams J. A., Rosenfeld M. G., and Fu X. D. (2012) The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol. Cell 47, 422–433 10.1016/j.molcel.2012.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhong X. Y., Ding J. H., Adams J. A., Ghosh G., and Fu X. D. (2009) Regulation of SR protein phosphorylation and alternative splicing by modulating kinetic interactions of SRPK1 with molecular chaperones. Genes Dev. 23, 482–495 10.1101/gad.1752109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim Y. H., Han M. E., and Oh S. O. (2017) The molecular mechanism for nuclear transport and its application. Anat. Cell Biol. 50, 77–85 10.5115/acb.2017.50.2.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Soniat M., and Chook Y. M. (2015) Nuclear localization signals for four distinct karyopherin-β nuclear import systems. Biochem. J. 468, 353–362 10.1042/BJ20150368 [DOI] [PubMed] [Google Scholar]
- 35. Adams J. A. (2001) Kinetic and catalytic mechanisms of protein kinases. Chem. Rev. 101, 2271–2290 10.1021/cr000230w [DOI] [PubMed] [Google Scholar]
- 36. Lai M. C., and Tarn W. Y. (2004) Hypophosphorylated ASF/SF2 binds TAP and is present in messenger ribonucleoproteins. J. Biol. Chem. 279, 31745–31749 10.1074/jbc.C400173200 [DOI] [PubMed] [Google Scholar]
- 37. Sanford J. R., Ellis J. D., Cazalla D., and Cáceres J. F. (2005) Reversible phosphorylation differentially affects nuclear and cytoplasmic functions of splicing factor 2/alternative splicing factor. Proc. Natl. Acad. Sci. U.S.A. 102, 15042–15047 10.1073/pnas.0507827102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Delestienne N., Wauquier C., Soin R., Dierick J. F., Gueydan C., and Kruys V. (2010) The splicing factor ASF/SF2 is associated with TIA-1–related/TIA-1–containing ribonucleoproteic complexes and contributes to post-transcriptional repression of gene expression. FEBS J. 277, 2496–2514 10.1111/j.1742-4658.2010.07664.x [DOI] [PubMed] [Google Scholar]
- 39. Wippich F., Bodenmiller B., Trajkovska M. G., Wanka S., Aebersold R., and Pelkmans L. (2013) Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 152, 791–805 10.1016/j.cell.2013.01.033 [DOI] [PubMed] [Google Scholar]
- 40. Saitoh N., Sakamoto C., Hagiwara M., Agredano-Moreno L. T., Jiménez-García L. F., and Nakao M. (2012) The distribution of phosphorylated SR proteins and alternative splicing are regulated by RANBP2. Mol. Biol. Cell 23, 1115–1128 10.1091/mbc.e11-09-0783 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.