Abstract
Heartland virus (HRTV) is a pathogenic phlebovirus recently identified in the United States and related to severe fever with thrombocytopenia syndrome virus (SFTSV) emerging in Asia. We previously reported that SFTSV disrupts host antiviral responses directed by interferons (IFNs) and their downstream regulators, signal transducer and activator of transcription (STAT) proteins. However, whether HRTV infection antagonizes the IFN–STAT signaling axis remains unclear. Here, we show that, similar to SFTSV, HRTV also inhibits IFN-α– and IFN-λ–mediated antiviral responses. As expected, the nonstructural protein (NSs) of HRTV (HNSs) robustly antagonized both type I and III IFN signaling. Protein interaction analyses revealed that a common component downstream of type I and III IFN signaling, STAT2, is the target of HNSs. Of note, the DNA-binding and linker domains of STAT2 were required for an efficient HNSs–STAT2 interaction. Unlike the NSs of SFTSV (SNSs), which blocks both STAT2 and STAT1 nuclear accumulation, HNSs specifically blocked IFN-triggered nuclear translocation only of STAT2. However, upon HRTV infection, IFN-induced nuclear translocation of both STAT2 and STAT1 was suppressed, suggesting that STAT1 is an additional HRTV target for IFN antagonism. Consistently, despite HNSs inhibiting phosphorylation only of STAT2 and not STAT1, HRTV infection diminished both STAT2 and STAT1 phosphorylation. These results suggest that HRTV antagonizes IFN antiviral signaling by dampening both STAT2 and STAT1 activities. We propose that HNSs-specific targeting of STAT2 likely plays an important role but is not all of the “tactics” of HRTV in its immune evasion.
Keywords: immunology, infectious disease, innate immunity, interferon, virus, signal transducers and activators of transcription 1 (STAT1), Heartland virus, nonstructural protein, phlebovirus, STAT2, phosphorylation, nuclear translocation
Introduction
Heartland virus (HRTV)2 is an emerging pathogenic phlebovirus (Phlebovirus genus, Phenuiviridae family, Bunyavirales order) first isolated from two Missouri farmers hospitalized with severe fever, leucopenia, and thrombocytopenia in 2009 (1). HRTV is the first known autochthonous phlebovirus pathogenic to humans in North America (1, 2). As of September 2018, sporadic human cases of HRTV infection have been identified from 10 states in the Midwestern and southern United States (https://www.cdc.gov/heartland-virus/statistics/index.html, accessed October 2, 2018). Based on the virus RNA detection in arthropods, the Lone Star tick (Amblyomma americanum) has been implicated as a vector of HRTV (4), whereas serological assessment of HRTV exposure showed that many domestic and wild animals likely are potential amplification hosts of the virus (2). HRTV is genetically closely related to the severe fever with thrombocytopenia syndrome virus (SFTSV), another highly pathogenic tick-borne phlebovirus emerging in China (5–7) and neighboring countries (8, 9). SFTSV has been associated with thousands of cases of symptomatic human disease characterized by similar hemorrhagic fever–like clinical manifestations with HRTV infection (6, 10). With the emergence of SFTSV and HRTV, more SFTSV/HRTV-related phleboviruses of potential high pathogenicity have been isolated in recent years around the world (11). It is apparent that SFTSV, HRTV, and other related emerging phleboviruses have posed a severe threat to worldwide human health (12, 13). However, there is currently no vaccine or medication available to prevent or treat these virus infections (12). Moreover, it is still poorly understood in terms of the phlebovirus–host interactions (13).
Type I and III interferons (IFNs), as “antiviral IFNs,” are the key components of innate immunity in host defense against invading viral pathogens (14–17). IFN responses upon virus infection comprise two phases, IFN induction and IFN signaling (14, 15). In the induction phase, viral infection promptly stimulates expression and secretion of type I IFNs (especially IFN-α and IFN-β) and type III IFNs (IFN-λ) (14, 15, 18, 19). In the IFN signaling phase, although type I and III IFNs employ different cell surface receptors, they share a similar downstream Janus kinase (JAK)-signal transducer and activator of transcription (STAT) signaling (14, 15, 18, 19). The binding of type I and III IFNs to their receptors initiates activation of several JAKs, including tyrosine kinase 2 (TYK2), JAK1, and JAK2, leading to the subsequent activation of STAT1 and STAT2 by phosphorylation (14, 20). Phosphorylated STAT1 and STAT2 heterodimerize and interact with IFN regulatory factor 9 (IRF9) to form an IFN-stimulated gene factor 3 (ISGF3) transcription complex (14). ISGF3 translocates to the nucleus, where it binds to the IFN-stimulated response element (ISRE) of the interferon-stimulated gene (ISG) promoters, resulting in the expression of these antiviral genes and the establishment of host antiviral state (14, 21, 22).
Although the pathogenesis of the emerging phleboviruses remains elusive, most (if not all) of the pathogenic viruses have evolved various countermeasures against host innate immunity and particularly the IFN system (e.g. encoding a robust IFN-antagonizing protein) (23, 24). Like all members of the genus Phlebovirus, HRTV and SFTSV have a genome organization that consists of three single-stranded negative-sense RNA segments, designated L, M, and S (13). The L segment encodes the RNA-dependent RNA polymerase, the M segment encodes the glycoproteins, and the S segment encodes the nucleocapsid protein (NP) and a nonstructural protein (NSs) by ambisense strategy (13). The NSs proteins of some phleboviruses likely function as the crucial virulence factor and contribute to viral pathogenesis by subverting host interferon responses through diverse strategies (24). For instance, our previous studies showed that SFTSV NSs (SNSs) irreversibly hijacks host kinases TBK1/IKKϵ (25) and transcriptional activators STAT2/STAT1 (26) into SNSs-induced inclusion bodies (IBs), leading to the blockade of IFN induction and downstream signaling, respectively. Furthermore, we found that HRTV NSs (HNSs) interacts with TBK1 and blocks the association of TBK1 with its substrate IRF3, thus inhibiting IFN induction (27). However, it is unclear whether IFN signaling can be counteracted by HRTV infection, and the potential role of HNSs therein needs further detailed elucidation as well.
In this study, we demonstrate that HRTV infection can block both type I and III IFN antiviral signaling. Furthermore, the mechanism underlying HRTV manipulation of type I and III IFN signaling was unraveled by detailed comparative studies in the context of HRTV infection and transient expression of potential IFN-inhibiting protein, HNSs. Finally, a function and mechanism model for IFN antagonism by HRTV as well as HNSs was proposed compared with SFTSV and SNSs.
Results
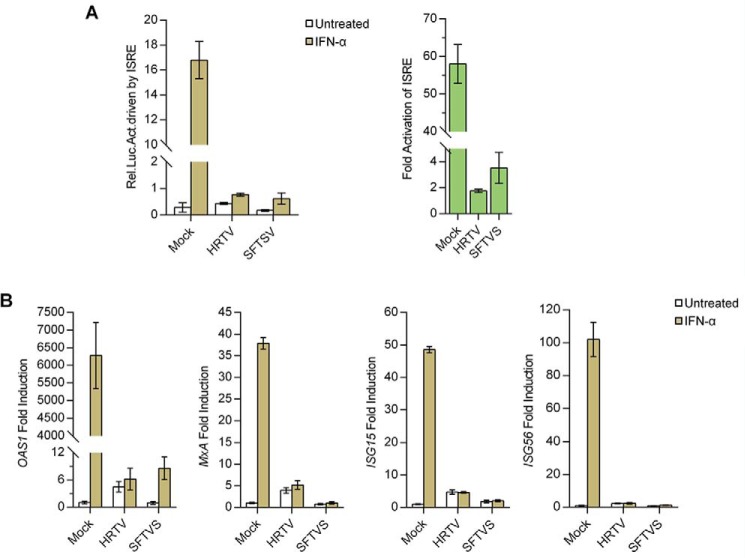
Similar to SFTSV, HRTV potently suppresses type I IFN–directed antiviral signaling and ISG expression
To determine whether HRTV infection interferes with type I IFN signaling, we first examined the impact of HRTV infection (with SFTSV infection serving as a positive control) on IFN-α–triggered ISRE promoter activation by a dual-luciferase reporter (DLR) assay. As shown in Fig. 1A, in mock-infected cells, ISRE promoter was expectably activated by the addition of IFN-α; however, following HRTV infection, the activation of ISRE promoter was evidently abolished, indicating the robust type I IFN–inhibiting capacity of HRTV. Meanwhile, in accordance with the previous report (26), a similar inhibitory effect on ISRE promoter activation was observed in the context of SFTSV infection (Fig. 1A). Furthermore, we detected IFN-α–driven expression of several representative ISGs, including oligoadenylate synthetase 1 (OAS1), myxovirus-resistance A (MxA), ISG15, and ISG56, by real-time quantitative PCR (qPCR). Consistently, the infection with HRTV as well as SFTSV greatly blocked the ISG induction by IFN-α (Fig. 1B), confirming the type I IFN–antagonistic activity of HRTV and meanwhile suggesting the functional conservativeness of these emerging phleboviruses in type I IFN antagonism.
Figure 1.
Suppression of type I IFN signaling by HRTV infection. A, HRTV infection inhibits IFN-α–driven activation of the ISRE promoter. HEK293 cells were cotransfected with an ISRE reporter plasmid and an internal control plasmid (pRL-TK). At 12 h posttransfection, cells were mock-infected or infected with HRTV (MOI = 5) or SFTSV (MOI = 5), respectively. At 24 hpi, cells were left untreated or treated with IFN-α (1000 units/ml) for 18 h, followed by the measurement of luciferase activities. Relative luciferase activity (Rel. Luc. Act.) and -fold activation of ISRE are shown, respectively. B, HRTV infection blocks IFN-α–induced ISG expression. HEK293 cells were mock-infected or infected with HRTV (MOI = 5) or SFTSV (MOI = 5). At 24 hpi, cells were left untreated or treated with IFN-α (1000 units/ml) for 10 h, followed by the detection of ISG mRNA expression by real-time qPCR. Graphs show mean ± S.D. (error bars) (n = 3).
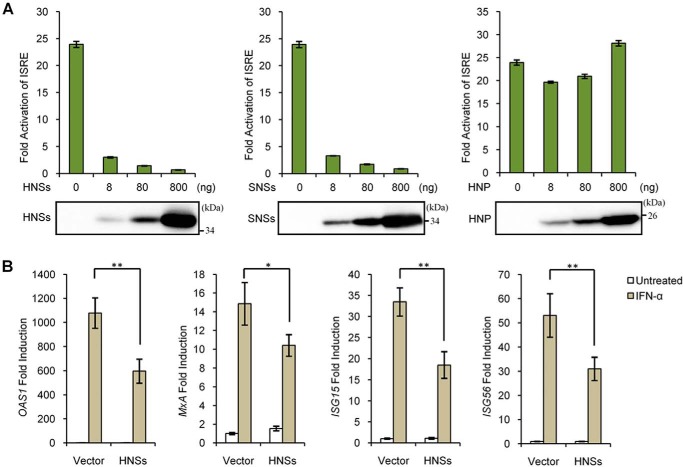
HNSs, but not HNP, antagonizes IFN-α–elicited antiviral signaling and ISG expression
It has been established that SNSs can disable both IFN induction and signaling (25, 26, 28–30). In addition, a previous study by us has shown that HNSs suppresses the production of type I IFNs (27). Considering the homology of HNSs with SNSs (∼60% identity in amino acid sequence) (1), we hypothesized that HNSs may also perturb type I IFN signaling like SNSs. To test this hypothesis, we assessed the effect of HNSs on IFN-α–induced ISRE promoter activation by a DLR assay. As shown in Fig. 2A, similar to SNSs, HNSs efficiently inhibited the activation of ISRE promoter by IFN-α in a dosage-dependent manner; however, by contrast, HNP expression did not exhibit such an inhibitory effect, suggesting the specific inhibition of IFN-α-mediated ISRE activation by HNSs. Next, we examined the influence of HNSs expression on IFN-α–induced expression of ISGs. Real-time qPCR analyses showed that HNSs expression by transient transfection significantly weakens the induction of ISGs by IFN-α (Fig. 2B), confirming the suppression of type I IFN signaling pathway by HNSs. Together, these results validate the role of HNSs as an antagonist of type I IFN antiviral signaling.
Figure 2.
HNSs, but not HNP, is an antagonist of type I IFN signaling. A, HNSs, but not HNP, inhibits IFN-α–stimulated activation of the ISRE promoter. HEK293 cells were transfected with the indicated amounts of viral protein expression plasmids, together with the reporter plasmids. At 24 h posttransfection, cells were left untreated or treated with IFN-α for 18 h before the measurement of luciferase activities. Meanwhile, viral protein expression levels were monitored by WB. B, HNSs suppresses IFN-α–triggered ISG expression. HEK293 cells transfected with the HNSs expression plasmid or the empty control plasmid (vector) were left untreated or treated with IFN-α for 10 h, followed by the analyses of the indicated ISG mRNA expression through real-time qPCR. Graphs show mean ± S.D. (error bars) (n = 3). *, p < 0.05; **, p < 0.01.
HRTV infection and HNSs transient expression both can inhibit type III IFN signaling
Type III IFNs are the most recently described “antiviral IFNs,” which are structurally similar to members of the interleukin-10 (IL-10) family but functionally similar to type I IFNs (19). Type III and I IFNs signal through distinct receptor complexes, but downstream they drive the similar JAK-STAT signaling and largely overlapping ISG expression (14). Thus, we next investigated whether HRTV and its HNSs perturb type III IFN signaling. As presented in Fig. 3A, IFN-λ–induced activation of the ISRE promoter was reduced by the infection of HRTV as well as SFTSV. It should be noted here that viral infections themselves appear to trigger a slight activation of the ISRE promoter (Fig. 3A, left), and the viral inhibitory effects on IFN-λ signaling were manifested more evidently by calculating the -fold activation of ISRE over the corresponding untreated groups (Fig. 3A, right). Furthermore, ISRE promoter activation by IFN-λ was likewise impaired in cells transfected with the plasmids expressing HNSs or SNSs (Fig. 3B), suggesting that HNSs, like SNSs, is also an antagonist of type III IFN signaling.
Figure 3.
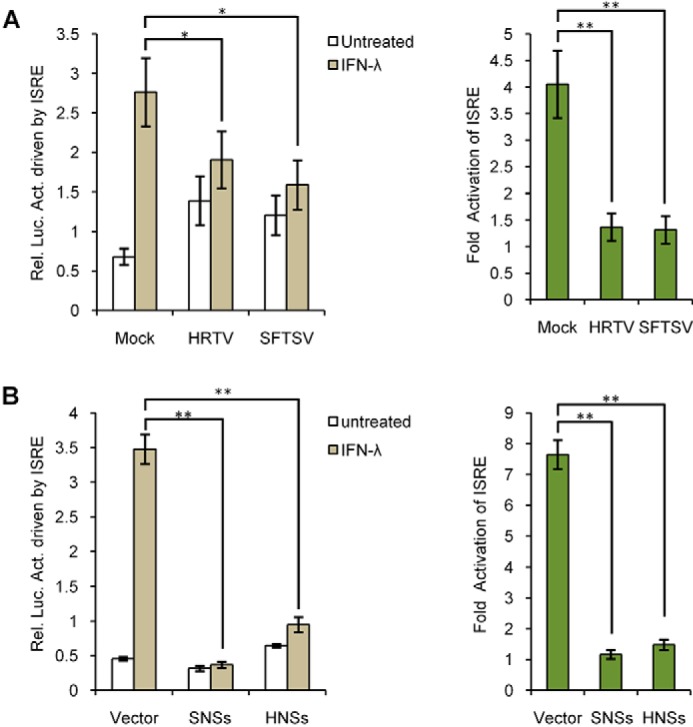
Suppression of type III IFN signaling by HRTV infection and HNSs expression. A, HRTV infection interferes with IFN-λ–stimulated ISRE activation. Huh7 cells transfected with the reporter plasmids were mock-infected or infected with HRTV or SFTSV for 24 h. Cells were then stimulated with IFN-λ (100 ng/ml) for 18 h or left unstimulated, before the measurement of luciferase activities. B, HNSs functions as an antagonist of type III IFN signaling. Huh7 cells were transfected with the reporter plasmids, along with the vector plasmid or the plasmids encoding SNSs or HNSs. At 24 h posttransfection, cells were left untreated or treated with IFN-λ (100 ng/ml) for 18 h, followed by the detection of luciferase activities. Graphs show mean ± S.D. (error bars) (n = 3). *, p < 0.05; **, p < 0.01.
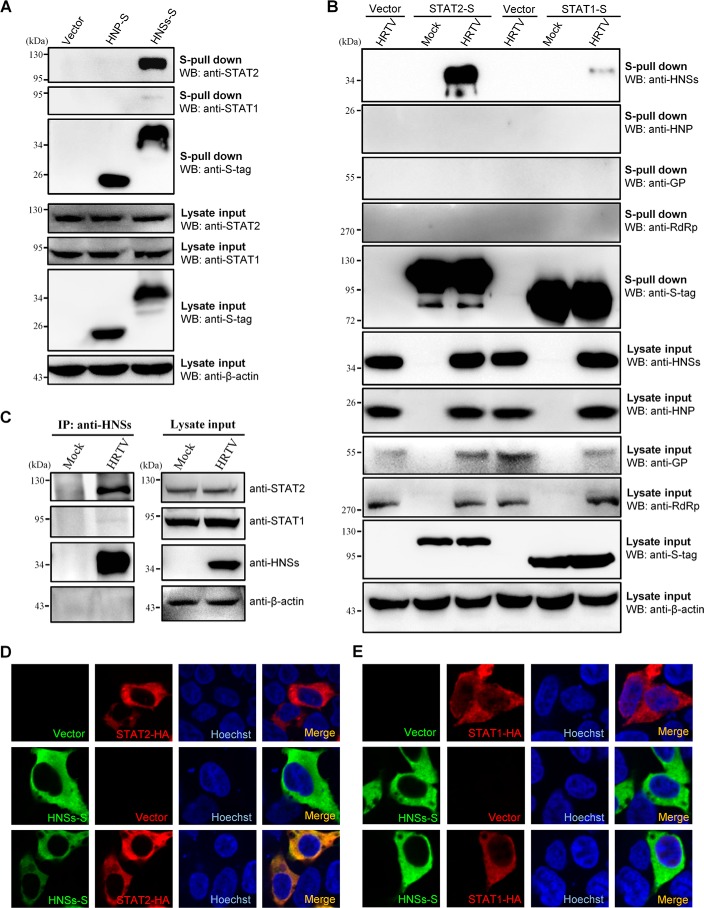
HNSs targeting of STAT2
The suppression of both type I and type III IFN signaling by the viral NSs proteins suggests that their cellular target(s) is likely shared by the two IFN signaling cascades. Indeed, our previous studies showed that STAT2 and STAT1, the common transcription activators downstream of type I and III IFN signaling, are the targets of SNSs (26). Thus, we examined whether HRTV and HNSs also target the STAT proteins. First, the interaction of S-tagged HNSs (HNSs-S) with STAT2 and STAT1 was evaluated by S-tag pulldown (S-pulldown) assays. As indicated in Fig. 4A, STAT2 was strongly co-precipitated by HNSs (but not HNP), whereas only a faint signal of STAT1 could be detected in the HNSs co-precipitates, even with a long-term exposure. Then, a reciprocal protein interaction S-pulldown assay was performed in the context of HRTV infection. Consistently, HNSs (but not the other viral proteins) could be high efficiently enriched in the S-tagged STAT2 (but not STAT1) co-precipitates in HRTV-infected cells (Fig. 4B), indicating the strong and specific interaction between HNSs and STAT2. Furthermore, the interaction of endogenous STAT proteins with HNSs was determined in the context of HRTV infection as well. As shown in the co-immunoprecipitation (co-IP) assay, endogenous STAT2 but not STAT1 was co-precipitated with HNSs (Fig. 4C), further confirming the specific HNSs–STAT2 interaction. Next, the subcellular colocalization of HNSs and STAT proteins was analyzed by an immunofluorescence assay (IFA). In line with the findings of the protein interaction assays, HNSs was obviously colocalized with STAT2 in the cytoplasm (Fig. 4D), whereas no comparable colocalization between HNSs and STAT1 was observed (Fig. 4E). These results validated the specific targeting of STAT2 by HNSs.
Figure 4.
Interaction and colocalization of HNSs with STAT2. A, HEK293 cells were transfected with the plasmids encoding S-tagged HNP (HNP-S) or HNSs (HNSs-S) or the vector plasmid. At 36 h posttransfection, cells were harvested for the S-pulldown assay. S-pulldown products and cell lysates were subjected to WB analyses with the indicated antibodies. B, HEK293 cells were transfected with the plasmids expressing S-tagged STAT2 (STAT2-S) or STAT1 (STAT1-S) or the control vector. At 12 h posttransfection, cells were mock-infected or infected with HRTV (MOI = 5) for 36 h and then lysed for the S-pulldown assay and WB analyses. Therein, GP expression was monitored with the detection of GN (the N-terminal region of GP) using anti-GN antibody. C, mock- or HRTV-infected HEK293 cells were lysed for a co-IP assay at 36 hpi. The lysate supernatants were first pretreated with preimmune serum and protein A/G–agarose and then used for co-IP with the HNSs-specific antiserum. Immunoprecipitates (IP) and cell lysates were delivered to WB analysis using the indicated antibodies. D and E, HEK293 cells were transfected with the plasmids expressing the indicated protein or the control vector. At 36 h posttransfection, cells were fixed for IFA to visualize the indicated proteins (HNSs in green and STAT2/STAT1 in red) by confocal microscopy. Nuclei were stained with Hoechst, as shown in blue.
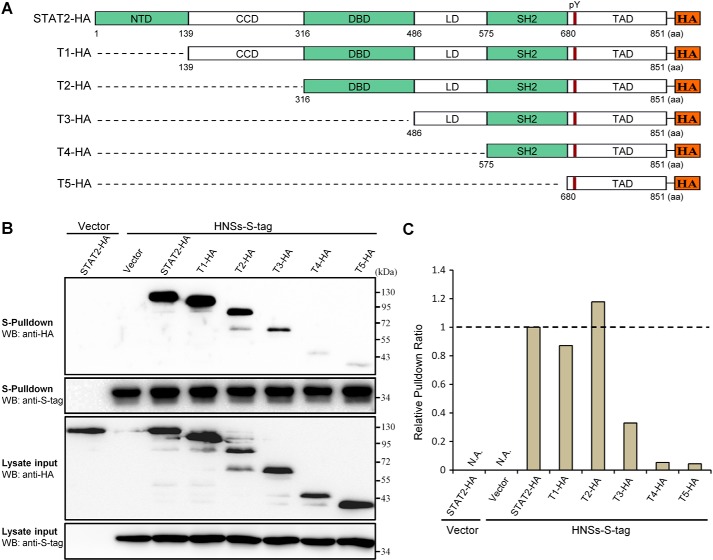
The DNA-binding domain (DBD) and linker domain (LD) of STAT2 are required for efficient HNSs–STAT2 interaction
To gain insight into the region(s) within STAT2 required for the HNSs–STAT2 interaction, we assayed the ability of a series of N-terminally truncated STAT2 mutants (Fig. 5A) to co-precipitate with HNSs by an S-pulldown assay. Expression of all of the STAT2 mutants by transfection was readily detected, whereas some differences in the expression levels were still observed (Fig. 5B). To better assess the protein interactions, band intensities of full-length or truncated STAT2 in Western blotting (WB) analyses of the lysate inputs and HNSs-S pulldown products (Fig. 5B) were respectively measured, and the pulldown ratios of STAT2 and its mutants (pulldown over input) were then calculated and compared. As shown in Fig. 5 (B and C), STAT2 mutants lacking the N-terminal domain (NTD) and coiled-coil domain (CCD) retained the capability of strongly interacting with HNSs; however, the deletion of DBD resulted in a substantial reduction of the pulldown ratio, and further deletion of LD almost abolished the interaction with HNSs (Fig. 5C). Taken together, these data suggest that the middle regions, DBD and LD, within STAT2 are likely important for efficient HNSs–STAT2 interaction.
Figure 5.
Mapping of the STAT2 domains required for efficient HNSs–STAT2 interaction. A, domain organization of full-length or N-terminal truncated STAT2 C-terminally fused with HA tag. NTD, N-terminal domain; CCD, coiled-coil domain; SH2, src homology domain-2; TAD, transactivation domain; pY, tyrosine (Tyr-690) phosphorylation site. The HA-tagged truncated STAT2 proteins were named T1-HA, T2-HA, T3-HA, T4-HA, and T5-HA, respectively. B, HEK293 cells were transfected with the HNSs-S expression plasmid and the plasmids encoding full-length or truncated STAT2 proteins or the corresponding control vectors, as indicated. At 48 h posttransfection, interactions of HNSs with the full-length or truncated STAT2 were analyzed with the S-pulldown assay, followed by WB with the indicated antibodies. C, band intensities of full-length or truncated STAT2 proteins in B were respectively measured by ImageJ software. To calculate the pulldown ratio of full-length or truncated STAT2, band intensities of the proteins co-precipitated with HNSs were then normalized to the corresponding band intensities in lysate input. The relative pulldown ratio of full-length STAT2 was set to 1, for reference. N.A., not analyzed.
Different from SNSs disruption of both STAT2 and STAT1 nuclear accumulation, HNSs specifically blocks IFN-induced nuclear translocation of STAT2, but not STAT1
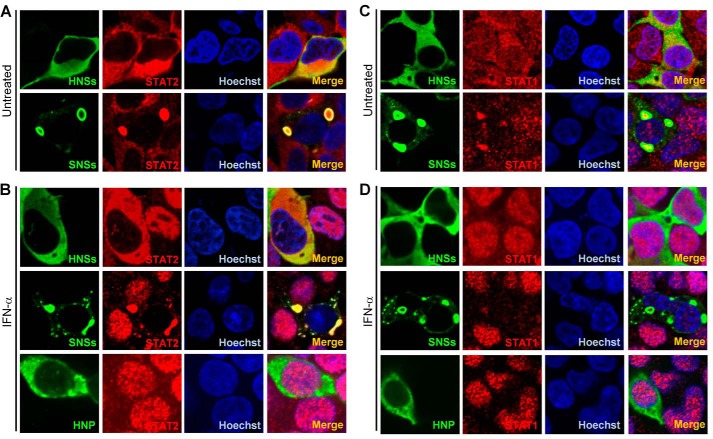
Given the potent interaction of HNSs with STAT2, we investigated whether HNSs can affect IFN-induced STAT2 nuclear translocation, which is the essential step for the antiviral signal transduction. HEK293 cells transfected with viral protein expression plasmids were left untreated (Fig. 6, A and C) or treated with IFN-α (Fig. 6, B and D) and then fixed to visualize the expression and localization of the viral proteins and endogenous STAT1 and STAT2 by IFA. In accordance with the previous reports (25, 27), SNSs was located in cytoplasmic SNSs IBs, whereas HNSs was distributed diffusely in cytoplasm (Fig. 6). Furthermore, in SNSs-expressing cells, both of STAT2 and STAT1, were relocated into SNSs IBs, and their nuclear accumulation induced by IFN-α was obviously suppressed; however, HNSs expression could specifically block the nuclear translocation of STAT2, but not STAT1 (Fig. 6), reflecting a difference between the two viral NSs proteins. Meanwhile, HNP did not notably influence the nuclear translocation of either STAT2 or STAT1 (Fig. 6, B and D), in line with the findings that HNP cannot inhibit IFN-α-triggered ISRE promoter activation (Fig. 2A) or co-precipitate the STAT proteins (Fig. 4A). Additionally, similar results regarding the effects of the viral proteins on STAT nuclear translocation were obtained from experiments under the treatment of IFN-λ (data not shown). Collectively, these data indicate that HNSs only can block STAT2 nuclear translocation induced by type I and III IFNs, different from SNSs-mediated inhibition of both STAT1 and STAT2 nuclear accumulation.
Figure 6.
HNSs specifically inhibits nuclear translocation of STAT2 (but not STAT1), in contrast to SNSs inhibition of both STAT2 and STAT1 nuclear accumulation. HEK293 cells were transfected with the HNSs, SNSs, or HNP expression plasmids. At 24 h posttransfection, cells were left untreated (A and C) or treated with IFN-α (1000 units/ml) (B and D) for 30 min. After fixation, subcellular localization of the indicated viral proteins and STAT2 (A and B) or STAT1 (C and D) was visualized by IFA and confocal microscopy. Nuclei were stained with Hoechst.
HRTV infection interferes with IFN-induced nuclear translocation of both STAT2 and STAT1
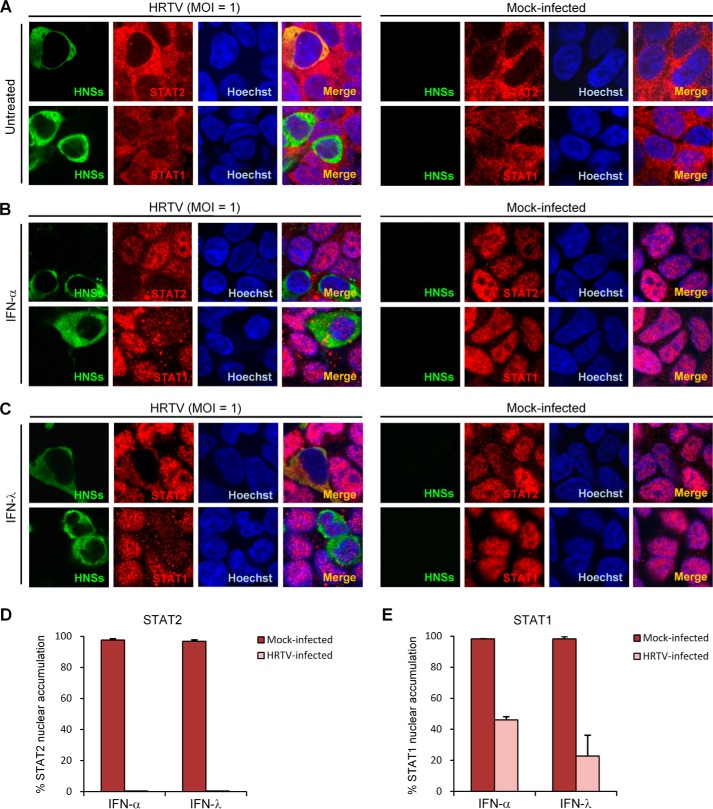
We next investigated whether IFN-triggered STAT nuclear translocation can be affected in the context of HRTV infection exactly as that mediated by HNSs expression alone with transient transfection. HEK293 cells infected with HRTV were left untreated or treated with IFN-α or IFN-λ for 30 min, followed by fixation for IFA. As shown in Fig. 7 (A–C), IFN-α or IFN-λ treatment resulted in STAT2 and STAT1 accumulation into the nucleus in uninfected cells, whereas in the HRTV-infected cells, the nuclear translocation of STAT2 was nearly abolished (Fig. 7, B–D), consistent with the observation in the cells transiently expressing HNSs. However, to our surprise, the nuclear translocation of STAT1 appeared to be also diminished by HRTV infection (Fig. 7, B, C, and E), different from the inhibitory ability of HNSs confined to STAT2 nuclear translocation (Fig. 6). To further confirm HRTV inhibition of STAT1 nuclear translocation, nuclear proteins were extracted by cellular fractionation. WB analyses of the subcellular fractions further showed that nuclear translocation of both STAT1 and STAT2 induced by IFN-α or IFN-λ was inhibited by HRTV infection (Fig. S1). These findings suggest that HRTV infection represses the nuclear translocation of not only STAT2 but also STAT1, likely together leading to the viral antagonism of type I and III IFN signaling.
Figure 7.
HRTV infection interferes with type I and III IFN-triggered nuclear translocation of both STAT2 and STAT1. HEK293 cells infected with HRTV (MOI = 1) or mock-infected were left untreated (A) or treated with IFN-α (1000 units/ml) (B) or IFN-λ (100 ng/ml) (C) for 30 min at 24 hpi. After fixation, IFA was performed to visualize the subcellular localization of HNSs and STAT2 or STAT1 under a confocal microscope. Nuclei were stained with Hoechst. D and E, cells mock-infected or infected with HRTV (HNSs-positive) from the experiments of B and C were scored for STAT2 (D) or STAT1 (E) nuclear accumulation, respectively. For each group, ∼100 cells were counted. Percentages of the cells with noticeable STAT2 or STAT1 nuclear accumulation were calculated, respectively. Data are presented as mean ± S.D. (error bars) (n = 3). Also see Fig. S1.
Differential inhibition of STAT2 and STAT1 phosphorylation by HNSs transient expression and HRTV infection
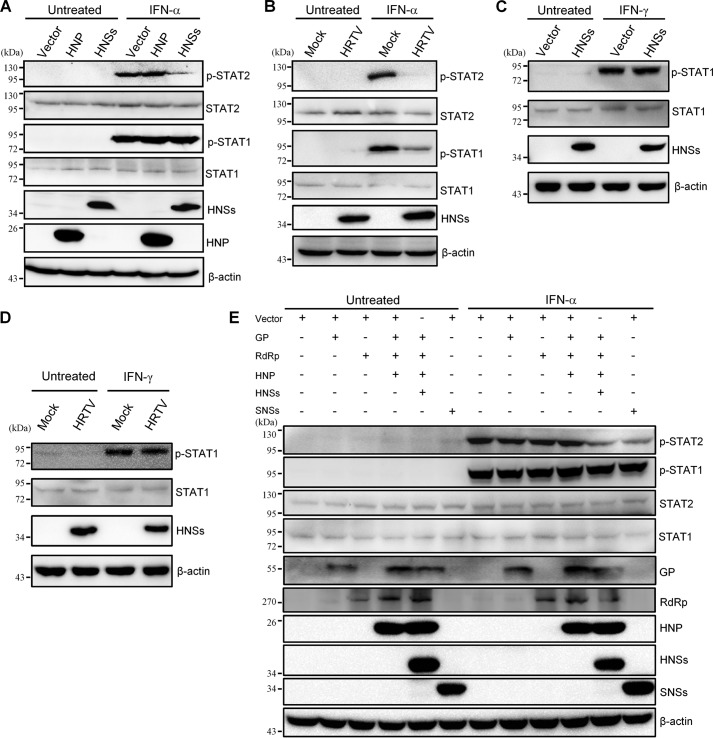
Tyrosine phosphorylation of STAT2 and STAT1 represents the activation of the transcription factors and is the prerequisite for their nuclear accumulation to stimulate ISG transcription (14, 15). Thus, we tested the effects of HNSs transient expression and HRTV infection on IFN-induced STAT phosphorylation, respectively. In a transient transfection–based experiment, HNSs expression resulted in an evident inhibition of IFN-α–elicited STAT2 phosphorylation but in contrast did not exhibit any noticeable influence on the phosphorylation of STAT1 (Fig. 8A), confirming the confined targeting of STAT2 by HNSs. Meanwhile, as expected, HNP had no significant effect on the activation of either STAT (Fig. 8A). Importantly, in the context of HRTV infection, the phosphorylation of STAT2 stimulated by IFN-α was nearly undetectable (Fig. 8B), in accordance with the robust inhibitory activity of HNSs to STAT2 activation. Further, intriguingly, a substantial decrease of IFN-α–triggered STAT1 phosphorylation was also resulted from HRTV infection (Fig. 8B), consistent with the suppression of STAT1 nuclear translocation by HRTV infection. Additionally, HRTV infection, similarly, could disable both STAT2 and STAT1 phosphorylation induced by IFN-λ as well (data not shown). Type II IFN, IFN-γ, can also trigger the phosphorylation of STAT1 that further forms an activated homodimer (i.e. IFN-γ activation factor, GAF) rather than ISGF3 via different receptors and JAK-STAT signaling (20). To gain further insights into HRTV inhibition of STAT1 activation, we also tested the effect of HRTV infection and HNSs transient expression on IFN-γ–induced STAT1 phosphorylation. Interestingly, neither HNSs transient expression (Fig. 8C) nor HRTV infection (Fig. 8D) detectably impaired IFN-γ–elicited STAT1 phosphorylation, revealing that HRTV infection specifically inhibits STAT1 phosphorylation and activation directed by the antiviral type I and III IFNs.
Figure 8.
Differential inhibition of STAT2 and STAT1 phosphorylation by HNSs transient expression and HRTV infection. A, transient expression of HNSs exclusively inhibits phosphorylation of STAT2 but not STAT1. HEK293 cells were transfected with the HNSs or HNP expression plasmids or the control vector. At 36 h posttransfection, cells were left untreated or treated with IFN-α (1000 units/ml) for 30 min. Protein levels were assessed by WB analysis with antibodies against the indicated proteins. B, HRTV infection suppresses the phosphorylation of both STAT2 and STAT1. HEK293 cells were mock-infected or infected with HRTV for 36 h and subsequently treated with IFN-α (1000 units/ml) for 30 min or left untreated. Protein levels were then analyzed by WB as in A. C and D, neither HNSs transient expression nor HRTV infection affects IFN-γ–induced STAT1 activation. Transfected (C) or infected (D) HEK293 cells were treated with IFN-γ (50 ng/ml; Peprotech) for 30 min and then subjected to WB analysis. E, effects of individual or combinatorial expression of HRTV proteins on STAT activation. HEK293 cells were transfected with the plasmids as indicated (500 ng for each viral protein expression plasmid). Total transfection amount of plasmids was kept constant by the corresponding addition of the vector. At 36 h posttransfection, cells were treated with IFN-α as in A and then delivered to WB analysis.
Furthermore, we examined the respective influence of individual expression of the other viral proteins (GP and RdRp) or combinatorial co-expression of all of the HRTV proteins on IFN-α–triggered STAT1 activation. As shown in Fig. 8E, like HNSs and HNP (Fig. 8A), HRTV GP or RdRp did not affect IFN-α–induced STAT1 phosphorylation when transiently expressed individually; moreover, combined co-expression of HRTV proteins did not detectably inhibit STAT1 activation either. These data indicate that HRTV does not encode any viral protein as the direct antagonist against STAT1. Meanwhile, it is also demonstrated that HNSs (like SNSs) is the specific STAT2 antagonist, as in the absence of HNSs, individual expression or combinatorial co-expression of the other HRTV proteins (HNP, GP, and RdRp) could not impair STAT2 activation (Fig. 8, A and E).
Altogether, these results establish that HNSs (but not the other viral proteins) exclusively abates STAT2 activation, acting as a type I and III IFN antagonist. Furthermore, although HRTV does not encode any viral protein as a direct antagonist against STAT1, in the context of infection, HRTV can abolish type I and III IFN activation of both STAT2 and STAT1 (but not IFN-γ activation of STAT1), likely leading to a specific and more rigorous disruption of the antiviral type I and III IFN signaling cascades.
Discussion
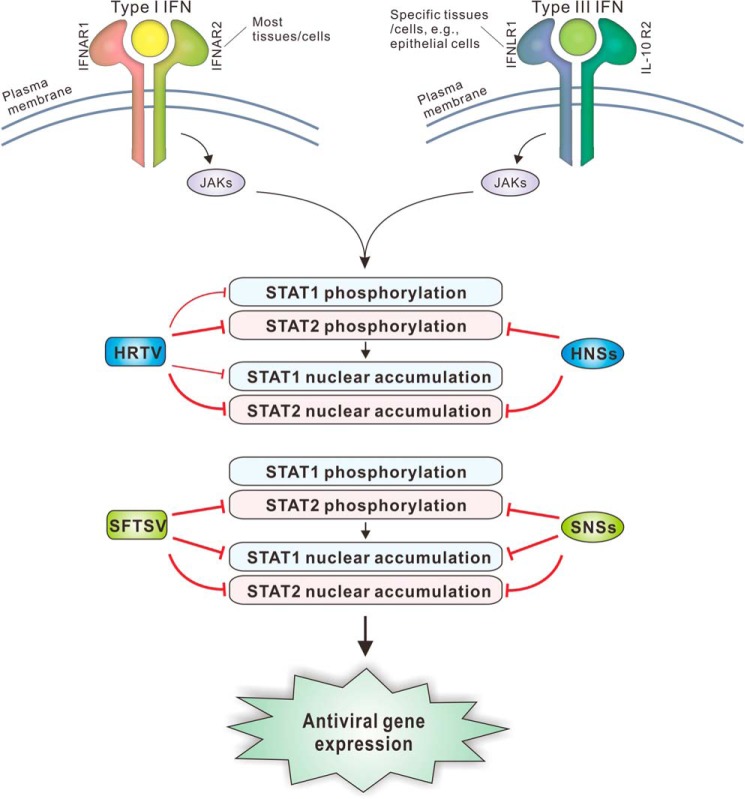
Emerging tick-borne phleboviruses, represented by SFTSV and HRTV, present a public health challenge (12, 13). There is an urgent need to understand the phleboviral pathogenesis and virus–host interactions for facilitating antiviral drug and vaccine development. Previously, several studies by us and others have investigated the complex immune evasion strategies employed by SFTSV (25, 26, 28–30). Therein, we found that SNSs induces viral IB formation and sequesters STAT2 and STAT1 in the IBs, blocking both STAT2 and STAT1 nuclear translocation (25, 26). Meanwhile, the phosphorylation of STAT2 (but not STAT1) was specifically hampered by SNSs (26). Consistent effects on STAT2 and STAT1 were observed in the context of SFTSV infection (26). Here, we demonstrated that HNSs has the conservative function in the antagonism of type I and III IFN signaling, but unlike SNSs, HNSs can only target the STAT2 actions. Furthermore, HRTV infection blocks the phosphorylation and nuclear accumulation of both STAT2 and STAT1 and hence robustly subverts type I and III IFN antiviral signaling, revealing that not only STAT2 but also STAT1 is the viral target for the immune evasion. The anti-IFN strategies of HRTV and HNSs were summarized in Fig. 9, compared with those of SFTSV and SNSs.
Figure 9.
Model for the disruption of type I and III IFN signaling by HRTV and HNSs, compared with SFTSV and SNSs. Type I and III IFNs use two different cell surface receptor complexes (broadly expressed IFNAR1/IFNAR2 for type I IFNs and tissue-specifically distributed IFNLR1/IL-10R2 for type III IFNs) and then direct the similar JAK–STAT antiviral signaling. First, binding of type I and III IFNs to their respective receptors initiates the activation of JAKs. Then the JAKs activate STAT1 and STAT2 by phosphorylation, resulting in nuclear translocation of the transcription factors and subsequent induction of hundreds of antiviral ISGs. SFTSV infection or SNSs expression by transient transfection can inhibit the phosphorylation of STAT2 and the nuclear translocation of both STAT2 and STAT1, yielding similar inhibitory effects on STAT2 and STAT1 actions. In comparison, although like SNSs, HNSs also functions as an antagonist of type I and III IFN signaling, it exclusively blocks the activities of STAT2. Consistently, STAT2 phosphorylation and nuclear accumulation can be impeded in the context of HRTV infection. However, interestingly, HRTV infection also can significantly suppress STAT1 activation and nuclear translocation, albeit to a lesser degree, as an additional strategy for the immune evasion. Together, the blockade of both STAT2 and STAT1 actions by HRTV likely leads to a more radical subversion of type I and III IFN signaling. These findings highlight the functional conservativeness and mechanism differentiation of these related emerging viruses and their NSs proteins. IFNAR, IFN-α receptor; IFNLR, IFN-λ receptor; IL-10R, IL-10 receptor.
We previously showed that HRTV can subvert IFN induction through HNSs blocking of TBK1–IRF3 interaction and signaling (27). Together with the present findings here, we demonstrate an overall view for HRTV disruption of the antiviral IFN system, including IFN induction and signaling. During the current study, based on transient transfection of HNSs expression plasmid, Rezelj et al. (31) showed that HNSs transient expression exhibited an inhibitory effect on the IFN-β (another type I IFN)–STAT2 signaling, which is in line with our parallel and expanded findings that HNSs, but not HNP, disables IFN-α–STAT2 signaling and hence antiviral ISG expression. Importantly, viral antagonism of type I IFN signaling is not only revealed here with further significant characterization and mechanistic investigation but also explored in detail in the context of HRTV infection. Intriguingly, we demonstrate that divergent with HNSs transient expression, HRTV infection impairs the phosphorylation and nuclear translocation of not only STAT2 but also STAT1, indicating that HNSs targeting of STAT2 is not the sole mechanism for HRTV antagonism of IFN signaling, and diminished STAT1 activities should contribute to the viral immune evasion as well. The most recently discovered type III IFNs engage their tissue-specifically distributed receptors, which differ from the broadly expressed type I IFN receptors (19). Thus, type III IFNs preferentially direct localized antiviral response, especially at barrier surfaces and mucosal sites (including liver and intestine epithelium), which coincidently may be involved in these phleboviral infections and the pathogenesis of the resultant hemorrhagic fever–like diseases (19, 20, 32–35). In this study, we found that HNSs can also suppress type III IFN signaling by impeding STAT2 actions. Furthermore, in the context of viral infection, HRTV disrupted type III IFN signaling by targeting the activities of both STAT2 and STAT1. These findings imply that type III IFN response may have an important role in host restriction to HRTV infection as well, further shedding light on the interactions of HRTV with innate immune system and the viral pathogenesis.
Indeed, Bosco-Lauth et al. (36) reported that Ag129 mice lacking functional IFN receptors (but not immunocompetent animals) are highly susceptible to HRTV challenge and inoculation, with even low dosages of HRTV resulting in severe illness and death, manifesting the critical role of the IFN system in host defense against HRTV infection. Additionally, Westover et al. (37) recently found that HRTV causes only moderate illness in STAT2 knockout hamsters, and most of the infected animals can recover, indicating that in addition to STAT2, another component(s) of the IFN system can also offer significant protection against HRTV infection. Here, the identification of STAT1 as an additional target of HRTV suggests the potential role of STAT1 therein. Further knockout of STAT1 may render the animals more susceptible, and this merits testing for developing better animal models of HRTV infection and for further characterizing the role of STAT1 in anti-HRTV immunity in vivo.
In the present study, we found that HNSs specifically inhibits IFN-induced phosphorylation and nuclear translocation of STAT2 but not STAT1. Interestingly, Ho et al. (38) previously reported that unphosphorylated STAT2 appeared to retain a fraction of phosphorylated STAT1 in the cytoplasm by the formation of a hemiphosphorylated STAT1–STAT2 heterodimer, whereas we observed the blockade of STAT2 nuclear translocation and simultaneously likely unaffected nuclear translocation of STAT1 triggered by IFN-α in cells transiently expressing HNSs (Fig. 6), suggesting that the unphosphorylated STAT2 here might fail to encumber STAT1 nuclear accumulation. A simple explanation is that HNSs likely hindered the hemiphosphorylated STAT1–STAT2 heterodimer formation by the robust HNSs–STAT2 interaction. Otherwise, the molecular number of STAT1 may be far more than that of STAT2 in HEK293 cells like several cell lines tested by Ho et al. (38), and thus the influence of STAT2 on STAT1 was hardly observable. Furthermore, the phenomenon (that IFN-triggered nuclear translocation of STAT2 was blocked but that of STAT1 was intact) resulting from HNSs transient expression was not similarly observed in the context of actual viral infection, as HRTV infection can inhibit the phosphorylation and nuclear accumulation of not only STAT2 but also STAT1. In contrast with the unique participation of STAT2 in type I and III IFN antiviral circuits, STAT1 is engaged in multiple signaling cascades triggered by various cellular factors, such as some interleukins and growth factors, besides the whole IFN family (39). Accordingly, the targeting of STAT1 by HRTV can not only enhance the destruction of type I and III IFN antiviral responses but may also lead to a wider perturbation of other cell signaling pathways involving STAT1 (although we excluded viral interference with IFN-γ–triggered STAT1 activation), which is worthy of future study to further expand our knowledge of HRTV–host interactions. Although we observed a weak HNSs–STAT1 co-precipitation, HNSs, unlike SNSs, cannot affect, or is insufficient to affect, STAT1 function. Additionally, we did not detect the interaction of STAT1 with the other HRTV proteins (Fig. 4B), and HRTV does not encode any viral protein as the STAT1 activation antagonist (Fig. 8E). Therefore, further details of the viral strategy underlying HRTV targeting of the STAT1 axis still remain to be clarified. In addition to encoding a direct IFN antagonist protein, some viruses can hijack the function of cellular suppressor of cytokine signaling (SOCS) proteins, which attenuate IFN–STAT signaling in a negative feedback loop (16, 40–43), leading to viral immune evasion from IFN responses. It will be interesting to address whether similar indirect strategies are employed by HRTV.
HNSs and SNSs share ∼60% identity in amino acid sequence (1). SNSs is localized in IBs induced by itself (25), whereas HNSs cannot mediate IB formation and distributes diffusely in the cytoplasm (27, 31). Although they have conservative IFN signaling–antagonizing function, in addition to the distinct subcellular localization, the two NSs proteins exhibit some other remarkable differences in their detailed activities. As mentioned previously, SNSs can interfere with the nuclear translocation of both STAT2 and STAT1 via hijacking of the two transcription factors into viral IBs (26), whereas HNSs specifically blocks STAT2 nuclear translocation through disabling STAT2 phosphorylation and activation. Additionally, by contrast with the involvement of STAT2 DBD within the SNSs–STAT2 interaction (26), both DBD and LD are likely required for the efficient targeting of STAT2 by HNSs, revealing one more difference between the two NSs. These findings highlight the function conservativeness and mechanism divergence of these homologous viral nonstructural proteins. Currently, a reverse genetic system for HRTV has not been established. Comparison of HNSs-deficient viruses yielded by reverse genetics with the WT HRTV will further unravel the significance of HNSs in viral infection and pathogenesis. Anyway, identification of these NSs proteins as robust IFN antagonists and hence the potential viral virulence factors may advance the design of attenuated vaccines and antiviral drugs by targeting NSs actions in the future.
At present, reported hospitalized and fatal cases of HRTV disease are relatively fewer (https://www.cdc.gov/heartland-virus/statistics/index.html, accessed October 2, 2018.), compared with those caused by SFTSV infection. It is sometimes considered that HRTV may be less virulent than SFTSV. However, our findings suggest that HRTV and SFTSV, as well as their NSs proteins, appear to have potent and comparable capacity to destroy type I and III IFN antiviral responses (including both IFN induction and signaling), albeit by partly divergent strategies. The pathogenic risk of HRTV may need reassessment. Recently, several other new phleboviruses genetically related to HRTV and SFTSV were successively discovered worldwide (11, 44–46). Together with HRTV and SFTSV, these clustered emerging viruses have been designated as the SFTSV/HRTV group (11). Further comparative studies on these phleboviruses and their NSs proteins will provide clues as to the molecular basis of the conservativeness and variance and promote the understanding of viral pathogenicity and virus–host interactions in evolutionary perspectives, thus likely benefiting the future development of specific or broad-spectrum antiviral therapies.
Materials and methods
Cells and viruses
HEK293 cells were cultured in minimum essential medium (Gibco) with 10% fetal bovine serum (Gibco) at 37 °C in a 5% CO2 atmosphere. Vero and Huh7 cells were maintained in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum. HRTV (strain MO-4) was obtained from the World Reference Center for Emerging Viruses and Arboviruses (University of Texas Medical Branch). The proliferation of HRTV was performed in Vero cells in a biosafety level 3 laboratory. SFTSV was propagated and handled as described previously (25–27).
Plasmids
The firefly luciferase reporter plasmid for ISRE and the Renilla luciferase control plasmid (pRL-TK) were kindly provided by Dr. Hong-Bing Shu (Wuhan University) (47–49). HA-tagged full-length or truncated STAT2 expression plasmids and the plasmids encoding HA- or S-tagged HNSs, HNP, or SNSs were described previously (25–27). Expression plasmids for S-tagged STAT1 and STAT2 were constructed by standard molecular biological approaches.
Antibodies
Primary antibodies used in this study included mouse antibodies to HA tag (H3663; Sigma-Aldrich), β-actin (60008; Proteintech), glyceraldehyde-3-phosphate dehydrogenase (60004-1-lg; Proteintech), STAT1 (sc-464; Santa Cruz Biotechnology, Inc.), or STAT2 (sc-514193; Santa Cruz Biotechnology) and rabbit antibodies to S-tag (ab18588; Abcam), histone deacetylase 1 (10197-1-AP; Proteintech), STAT1 (9172S; Cell Signaling Technology), phospho-STAT1 (9167S; Cell Signaling Technology), STAT2 (sc-22816; Santa Cruz Biotechnology), or phospho-STAT2 (sc-21689; Santa Cruz Biotechnology). Rabbit antisera against the viral proteins (NSs, NP, GP, or RdRp) were generated by serial vaccination of the corresponding viral proteins prepared in Escherichia coli. Secondary antibodies used in immunofluorescence assays included Alexa Fluor 488–conjugated goat anti-rabbit (ab150077) or anti-mouse (ab150113) IgG (H+L) and Alexa Fluor 647–conjugated goat anti-rabbit (ab150079) or anti-mouse (ab150115) IgG (H+L) (Abcam).
Reporter gene assay
Reporter gene assays were performed in HEK293 cells seeded on 24-well plates. Cells were cotransfected with 100 ng of ISRE reporter plasmid and 20 ng of pRL-TK plasmid using Lipofectamine 3000 (Invitrogen). At 12 h posttransfection, HEK293 cells were mock-infected or infected with HRTV or SFTSV at a multiplicity of infection (MOI) of 5. At 24 h postinfection (hpi), cells were stimulated with IFN-α2b (PBL Biomedical Inc.) or IFN-λ2 (Peprotech) or left unstimulated for 18 h, followed by the measurement of luciferase activities with a DLR assay kit (Promega). The firefly luciferase activities were normalized on the basis of Renilla luciferase activities to exhibit the relative luciferase activities. -Fold activation of ISRE was calculated by further normalization to the untreated controls. For the reporter gene assays with viral protein transient expression by transfection, HEK293 cells were cotransfected with 100 ng of ISRE reporter plasmid, 20 ng of pRL-TK, and the indicated amounts of HNSs, SNSs, or HNP expression plasmids. Total amounts of DNA transfected per well were constant by the addition of the empty control plasmids, correspondingly. After 24 h, cells were left untreated or treated with IFN-α or IFN-λ for 18 h before the detection of luciferase activities.
Quantitative real-time PCR
The relative mRNA levels of the indicated genes were analyzed by the 2−ΔΔCT method with quantitative real-time PCR as described previously (26).
Protein–protein interaction analysis
S-pulldown assays were used for protein interaction analysis as described previously (25–27). The principle of the S-pulldown assay is to utilize the high-specific and strong affinity of S-tag with S-protein coupled on agarose beads to precipitate the S-tagged protein and its interacting proteins. In a nutshell, transfected or infected cells were lysed in a lysis buffer (25 mm Tris, pH 7.4, 150 mm NaCl, 1 mm EDTA, and 1% Triton X-100) supplemented with a mixture of protease inhibitors (Roche Applied Science) on ice for 15 min. Following centrifugation, supernatants of the cell lysates then were mixed with the S-protein agarose slurry (Merck Novagen) by rotating incubation for 4 h at 4 °C. After extensively washing the beads, bound proteins were eluted with 1× SDS sample buffer by boiling for 5 min, followed by SDS-PAGE and WB analyses.
For the validation of endogenous STAT2–HNSs interaction in the context of HRTV infection, a co-IP assay was performed. HEK293 cells (∼5 × 107) were infected with HRTV (MOI = 5) or mock-infected and lysed in the lysis buffer at 36 hpi. Subsequently, the lysate supernatants were first pretreated with preimmune serum and protein A/G-Plus–agarose (Santa Cruz Biotechnology). After centrifugation, the pretreated supernatants were incubated with the HNSs-specific antiserum at 4 °C for 1 h and then mixed with the protein A/G–agarose at 4 °C overnight. After extensive washes, the immunoprecipitates were delivered to WB analyses.
Immunofluorescence and confocal microscopy
Immunofluorescence assays coupled with confocal microscopy were used to monitor protein expression and localization as described previously (3, 25–27). Briefly, transfected or infected cells were fixed with 4% paraformaldehyde in PBS and then permeabilized with 0.5% Triton X-100–PBS. After blocking with 2.5% BSA (Biosharp) and 2.5% normal goat serum (Jackson ImmunoResearch) in PBS, cells were stained successively with the primary antibodies for the target proteins and the corresponding fluorescently labeled secondary antibodies. For visualization of nuclei, cells were stained with Hoechst 33258 (Beyotime). Images were obtained and analyzed with a Nikon Ti confocal microscope and the matched Volocity software (PerkinElmer Life Sciences).
Cellular fractionation and WB analysis
Nuclear extracts were prepared by cellular fractionation with a nucleus/cytoplasm fractionation kit (Beyotime Biotechnology) according to the manufacturer's instructions, followed by WB analysis. WB was performed as described previously (3, 27). In brief, protein samples were subjected to SDS-PAGE and then transferred onto polyvinylidene difluoride membranes (Millipore). After blocking with 5% skim milk in TBS-Tween 20 (TBST), the polyvinylidene difluoride membranes were incubated with primary antibodies and then the corresponding secondary antibodies conjugated with horseradish peroxidase (Sigma-Aldrich) in 1% BSA-TBST. Protein bands were detected with an enhanced chemiluminescence (ECL) kit (Thermo Fisher Scientific).
Statistical analysis
Data are presented as mean ± S.D. Statistical significance was determined by Student's t test using IBM SPSS software. p values < 0.05 were considered statistically significant.
Author contributions
K. F., F. D., Z. H., H. W., and Y.-J. N. resources; K. F., F. D., Z. H., H. W., and Y.-J. N. data curation; K. F., F. D., Z. H., H. W., and Y.-J. N. formal analysis; K. F., F. D., Z. H., H. W., and Y.-J. N. validation; K. F. and Y.-J. N. investigation; K. F. and Y.-J. N. visualization; F. D., Z. H., and H. W. supervision; F. D., Z. H., H. W., and Y.-J. N. funding acquisition; F. D., Z. H., and H. W. project administration; Y.-J. N. conceptualization; Y.-J. N. methodology; Y.-J. N. writing-original draft; Y.-J. N. writing-review and editing; K. F. performance of most of the experiments.
Supplementary Material
Acknowledgments
We thank Dr. Hong-Bing Shu (Wuhan University, China) for supplying the reporter plasmids, the World Reference Center for Emerging Viruses and Arboviruses (University of Texas Medical Branch) for the HRTV strain, and the Core Facility and Technical Support of Wuhan Institute of Virology for technical assistance.
This work was supported by National Natural Science Foundation of China Grants 31621061 and 31600144; National Key Research and Development Program of China Grants 2018YFA0507202, 2016YFC1200400, and 2016YFE0113500; the European Union's Horizon 2020 project European Virus Archive Goes Global (EVAg; Grant 653316); the strategic priority research program of the Chinese Academy of Sciences (Grant XDPB0301); and the “One-Three-Five” Research Program of the Wuhan Institute of Virology. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Fig. S1.
- HRTV
- Heartland virus
- SFTSV
- severe fever with thrombocytopenia syndrome virus
- IFN
- interferon
- STAT
- signal transducer and activator of transcription
- NP
- nucleocapsid protein
- NSs
- nonstructural protein encoded by S segment
- HNSs
- HRTV NSs
- SNSs
- SFTSV NSs
- DBD
- DNA-binding domain
- LD
- linker domain
- DLR
- dual-luciferase reporter
- hpi
- hours postinfection
- IB
- inclusion body
- IFA
- immunofluorescence assay
- IL-10
- interleukin-10
- IRF
- IFN regulatory factor
- ISG
- IFN-stimulated gene
- ISGF3
- IFN-stimulated gene factor 3
- ISRE
- IFN-stimulated response element
- JAK
- Janus kinase
- MOI
- multiplicity of infection
- MxA
- myxovirus-resistance A
- OAS1
- oligoadenylate synthetase 1
- SOCS
- suppressor of cytokine signaling
- HNSs-S
- S-tagged HNSs
- S-pulldown
- S-tag pulldown
- TYK2
- tyrosine kinase 2
- WB
- Western blotting
- qPCR
- real-time quantitative PCR.
References
- 1. McMullan L. K., Folk S. M., Kelly A. J., MacNeil A., Goldsmith C. S., Metcalfe M. G., Batten B. C., Albariño C. G., Zaki S. R., Rollin P. E., Nicholson W. L., and Nichol S. T. (2012) A new phlebovirus associated with severe febrile illness in Missouri. N. Engl. J. Med. 367, 834–841 10.1056/NEJMoa1203378 [DOI] [PubMed] [Google Scholar]
- 2. Bosco-Lauth A. M., Panella N. A., Root J. J., Gidlewski T., Lash R. R., Harmon J. R., Burkhalter K. L., Godsey M. S., Savage H. M., Nicholson W. L., Komar N., and Brault A. C. (2015) Serological investigation of heartland virus (Bunyaviridae: Phlebovirus) exposure in wild and domestic animals adjacent to human case sites in Missouri 2012–2013. Am. J. Trop. Med. Hyg. 92, 1163–1167 10.4269/ajtmh.14-0702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ning Y.-J., Kang Z., Xing J., Min Y.-Q., Liu D., Feng K., Wang M., Deng F., Zhou Y., Hu Z., and Wang H. (2018) Ebola virus mucin-like glycoprotein (Emuc) induces remarkable acute inflammation and tissue injury: evidence for Emuc pathogenicity in vivo. Protein Cell 9, 389–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Savage H. M., Godsey M. S., Lambert A., Panella N. A., Burkhalter K. L., Harmon J. R., Lash R. R., Ashley D. C., and Nicholson W. L. (2013) First detection of heartland virus (Bunyaviridae: Phlebovirus) from field collected arthropods. Am. J. Trop. Med. Hyg. 89, 445–452 10.4269/ajtmh.13-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xu B., Liu L., Huang X., Ma H., Zhang Y., Du Y., Wang P., Tang X., Wang H., Kang K., Zhang S., Zhao G., Wu W., Yang Y., Chen H., et al. (2011) Metagenomic analysis of fever, thrombocytopenia and leukopenia syndrome (FTLS) in Henan Province, China: discovery of a new bunyavirus. PLoS Pathog. 7, e1002369 10.1371/journal.ppat.1002369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yu X. J., Liang M. F., Zhang S. Y., Liu Y., Li J. D., Sun Y. L., Zhang L., Zhang Q. F., Popov V. L., Li C., Qu J., Li Q., Zhang Y. P., Hai R., Wu W., et al. (2011) Fever with thrombocytopenia associated with a novel bunyavirus in China. N. Engl. J. Med. 364, 1523–1532 10.1056/NEJMoa1010095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang Y. Z., Zhou D. J., Xiong Y., Chen X. P., He Y. W., Sun Q., Yu B., Li J., Dai Y. A., Tian J. H., Qin X. C., Jin D., Cui Z., Luo X. L., Li W., et al. (2011) Hemorrhagic fever caused by a novel tick-borne Bunyavirus in Huaiyangshan, China. Zhonghua Liu Xing Bing Xue Za Zhi 32, 209–220 [PubMed] [Google Scholar]
- 8. Kim K. H., Yi J., Kim G., Choi S. J., Jun K. I., Kim N. H., Choe P. G., Kim N. J., Lee J. K., and Oh M. D. (2013) Severe fever with thrombocytopenia syndrome, South Korea, 2012. Emerg. Infect. Dis. 19, 1892–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Takahashi T., Maeda K., Suzuki T., Ishido A., Shigeoka T., Tominaga T., Kamei T., Honda M., Ninomiya D., Sakai T., Senba T., Kaneyuki S., Sakaguchi S., Satoh A., Hosokawa T., et al. (2014) The first identification and retrospective study of severe fever with thrombocytopenia syndrome in Japan. J. Infect. Dis. 209, 816–827 10.1093/infdis/jit603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li H., Lu Q.-B., Xing B., Zhang S.-F., Liu K., Du J., Li X.-K., Cui N., Yang Z.-D., Wang L.-Y., Hu J.-G., Cao W.-C., and Liu W. (2018) Epidemiological and clinical features of laboratory-diagnosed severe fever with thrombocytopenia syndrome in China, 2011–17: a prospective observational study. Lancet Infect. Dis. 18, 1127–1137 10.1016/S1473-3099(18)30293-7 [DOI] [PubMed] [Google Scholar]
- 11. Matsuno K., Weisend C., Kajihara M., Matysiak C., Williamson B. N., Simuunza M., Mweene A. S., Takada A., Tesh R. B., and Ebihara H. (2015) Comprehensive molecular detection of tick-borne phleboviruses leads to the retrospective identification of taxonomically unassigned bunyaviruses and the discovery of a novel member of the genus phlebovirus. J. Virol. 89, 594–604 10.1128/JVI.02704-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mansfield K. L., Jizhou L., Phipps L. P., and Johnson N. (2017) Emerging tick-borne viruses in the twenty-first century. Front. Cell. Infect. Microbiol. 7, 298 10.3389/fcimb.2017.00298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Elliott R. M., and Brennan B. (2014) Emerging phleboviruses. Curr. Opin. Virol. 5, 50–57 10.1016/j.coviro.2014.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoffmann H. H., Schneider W. M., and Rice C. M. (2015) Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol. 36, 124–138 10.1016/j.it.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Levy D. E., Marié I. J., and Durbin J. E. (2011) Induction and function of type I and III interferon in response to viral infection. Curr. Opin. Virol. 1, 476–486 10.1016/j.coviro.2011.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Akhtar L. N., and Benveniste E. N. (2011) Viral exploitation of host SOCS protein functions. J. Virol. 85, 1912–1921 10.1128/JVI.01857-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sun Y., Jiang J., Tien P., Liu W., and Li J. (2018) IFN-λ: a new spotlight in innate immunity against influenza virus infection. Protein Cell 9, 832–837 10.1007/s13238-017-0503-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McNab F., Mayer-Barber K., Sher A., Wack A., and O'Garra A. (2015) Type I interferons in infectious disease. Nat. Rev. Immunol. 15, 87–103 10.1038/nri3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hemann E. A., Gale M. Jr., and Savan R. (2017) Interferon λ genetics and biology in regulation of viral control. Front. Immunol. 8, 1707 10.3389/fimmu.2017.01707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chow K. T., and Gale M. Jr. (2015) SnapShot: interferon signaling. Cell 163, 1808–1808.e1 10.1016/j.cell.2015.12.008 [DOI] [PubMed] [Google Scholar]
- 21. Schneider W. M., Chevillotte M. D., and Rice C. M. (2014) Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32, 513–545 10.1146/annurev-immunol-032713-120231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun L., Liu S., and Chen Z. J. (2010) SnapShot: pathways of antiviral innate immunity. Cell 140, 436–436.e2 10.1016/j.cell.2010.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Elliott R. M., and Weber F. (2009) Bunyaviruses and the type I interferon system. Viruses 1, 1003–1021 10.3390/v1031003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wuerth J. D., and Weber F. (2016) Phleboviruses and the type I interferon response. Viruses 8, E174 10.3390/v8060174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ning Y. J., Wang M., Deng M., Shen S., Liu W., Cao W. C., Deng F., Wang Y. Y., Hu Z., and Wang H. (2014) Viral suppression of innate immunity via spatial isolation of TBK1/IKKϵ from mitochondrial antiviral platform. J. Mol. Cell. Biol. 6, 324–337 10.1093/jmcb/mju015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ning Y. J., Feng K., Min Y. Q., Cao W. C., Wang M., Deng F., Hu Z., and Wang H. (2015) Disruption of type I interferon signaling by the nonstructural protein of severe fever with thrombocytopenia syndrome virus via the hijacking of STAT2 and STAT1 into inclusion bodies. J. Virol. 89, 4227–4236 10.1128/JVI.00154-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ning Y. J., Feng K., Min Y. Q., Deng F., Hu Z., and Wang H. (2017) Heartland virus NSs protein disrupts host defenses by blocking the TBK1 kinase-IRF3 transcription factor interaction and signaling required for interferon induction. J. Biol. Chem. 292, 16722–16733 10.1074/jbc.M117.805127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Santiago F. W., Covaleda L. M., Sanchez-Aparicio M. T., Silvas J. A., Diaz-Vizarreta A. C., Patel J. R., Popov V., Yu X. J., García-Sastre A., and Aguilar P. V. (2014) Hijacking of RIG-I signaling proteins into virus-induced cytoplasmic structures correlates with the inhibition of type I interferon responses. J. Virol. 88, 4572–4585 10.1128/JVI.03021-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu X., Qi X., Qu B., Zhang Z., Liang M., Li C., Cardona C. J., Li D., and Xing Z. (2014) Evasion of antiviral immunity through sequestering of TBK1/IKKϵ/IRF3 into viral inclusion bodies. J. Virol. 88, 3067–3076 10.1128/JVI.03510-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chaudhary V., Zhang S., Yuen K. S., Li C., Lui P. Y., Fung S. Y., Wang P. H., Chan C. P., Li D., Kok K. H., Liang M., and Jin D. Y. (2015) Suppression of type I and type III IFN signalling by NSs protein of severe fever with thrombocytopenia syndrome virus through inhibition of STAT1 phosphorylation and activation. J. Gen. Virol. 96, 3204–3211 10.1099/jgv.0.000280 [DOI] [PubMed] [Google Scholar]
- 31. Rezelj V. V., Li P., Chaudhary V., Elliott R. M., Jin D.-Y., and Brennan B. (2017) Differential antagonism of human innate immune responses by tick-borne phlebovirus nonstructural proteins. mSphere 2, e00234–00217 10.1128/mSphere.00234-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu Y., Wu B., Paessler S., Walker D. H., Tesh R. B., and Yu X. J. (2014) The pathogenesis of severe fever with thrombocytopenia syndrome virus infection in α/β interferon knockout mice: insights into the pathologic mechanisms of a new viral hemorrhagic fever. J. Virol. 88, 1781–1786 10.1128/JVI.02277-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Muehlenbachs A., Fata C. R., Lambert A. J., Paddock C. D., Velez J. O., Blau D. M., Staples J. E., Karlekar M. B., Bhatnagar J., Nasci R. S., and Zaki S. R. (2014) Heartland virus-associated death in Tennessee. Clin. Infect. Dis. 59, 845–850 10.1093/cid/ciu434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lazear H. M., Nice T. J., and Diamond M. S. (2015) Interferon-λ: immune functions at barrier surfaces and beyond. Immunity 43, 15–28 10.1016/j.immuni.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Egli A., Santer D. M., O'Shea D., Tyrrell D. L., and Houghton M. (2014) The impact of the interferon-λ family on the innate and adaptive immune response to viral infections. Emerg. Microbes Infect. 3, e51 10.1038/emi.2014.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bosco-Lauth A. M., Calvert A. E., Root J. J., Gidlewski T., Bird B. H., Bowen R. A., Muehlenbachs A., Zaki S. R., and Brault A. C. (2016) Vertebrate host susceptibility to Heartland virus. Emerg. Infect. Dis. 22, 2070–2077 10.3201/eid2212.160472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Westover J. B., Rigas J. D., Van Wettere A. J., Li R., Hickerson B. T., Jung K. H., Miao J., Reynolds E. S., Conrad B. L., Nielson S., Furuta Y., Thangamani S., Wang Z., and Gowen B. B. (2017) Heartland virus infection in hamsters deficient in type I interferon signaling: protracted disease course ameliorated by favipiravir. Virology 511, 175–183 10.1016/j.virol.2017.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ho J., Pelzel C., Begitt A., Mee M., Elsheikha H. M., Scott D. J., and Vinkemeier U. (2016) STAT2 is a pervasive cytokine regulator due to its inhibition of STAT1 in multiple signaling pathways. PLoS Biol. 14, e2000117 10.1371/journal.pbio.2000117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Najjar I., and Fagard R. (2010) STAT1 and pathogens, not a friendly relationship. Biochimie 92, 425–444 10.1016/j.biochi.2010.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nan Y., Wu C., and Zhang Y. J. (2017) Interplay between Janus kinase/signal transducer and activator of transcription signaling activated by type I interferons and viral antagonism. Front. Immunol. 8, 1758 10.3389/fimmu.2017.01758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Charoenthongtrakul S., Zhou Q., Shembade N., Harhaj N. S., and Harhaj E. W. (2011) Human T cell leukemia virus type 1 Tax inhibits innate antiviral signaling via NF-κ B-dependent induction of SOCS1. J. Virol. 85, 6955–6962 10.1128/JVI.00007-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pauli E. K., Schmolke M., Wolff T., Viemann D., Roth J., Bode J. G., and Ludwig S. (2008) Influenza A virus inhibits type I IFN signaling via NF-κ B-dependent induction of SOCS-3 expression. PLoS Pathog. 4, e1000196 10.1371/journal.ppat.1000196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Song M. M., and Shuai K. (1998) The suppressor of cytokine signaling (SOCS) 1 and SOCS3 but not SOCS2 proteins inhibit interferon-mediated antiviral and antiproliferative activities. J. Biol. Chem. 273, 35056–35062 10.1074/jbc.273.52.35056 [DOI] [PubMed] [Google Scholar]
- 44. Wang J., Selleck P., Yu M., Ha W., Rootes C., Gales R., Wise T., Crameri S., Chen H., Broz I., Hyatt A., Woods R., Meehan B., McCullough S., and Wang L. F. (2014) Novel phlebovirus with zoonotic potential isolated from ticks, Australia. Emerg. Infect. Dis. 20, 1040–1043 10.3201/eid2006.140003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mourya D. T., Yadav P. D., Basu A., Shete A., Patil D. Y., Zawar D., Majumdar T. D., Kokate P., Sarkale P., Raut C. G., and Jadhav S. M. (2014) Malsoor virus, a novel bat phlebovirus, is closely related to severe fever with thrombocytopenia syndrome virus and heartland virus. J. Virol. 88, 3605–3609 10.1128/JVI.02617-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shen S., Duan X., Wang B., Zhu L., Zhang Y., Zhang J., Wang J., Luo T., Kou C., Liu D., Lv C., Zhang L., Chang C., Su Z., Tang S., et al. (2018) A novel tick-borne phlebovirus, closely related to severe fever with thrombocytopenia syndrome virus and Heartland virus, is a potential pathogen. Emerg. Microbes Infect. 7, 95 10.1038/s41426-018-0093-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen H., Li Y., Zhang J., Ran Y., Wei J., Yang Y., and Shu H. B. (2013) RAVER1 is a coactivator of MDA5-mediated cellular antiviral response. J. Mol. Cell. Biol. 5, 111–119 10.1093/jmcb/mjt006 [DOI] [PubMed] [Google Scholar]
- 48. Zhang J., Hu M.-M., Wang Y.-Y., and Shu H.-B. (2012) TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J. Biol. Chem. 287, 28646–28655 10.1074/jbc.M112.362608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xu Z. S., Zhang H. X., Zhang Y. L., Liu T. T., Ran Y., Chen L. T., Wang Y. Y., and Shu H. B. (2016) PASD1 promotes STAT3 activity and tumor growth by inhibiting TC45-mediated dephosphorylation of STAT3 in the nucleus. J. Mol. Cell. Biol. 8, 221–231 10.1093/jmcb/mjw005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.