Abstract
Tau is a microtubule-associated protein involved in the regulation of axonal microtubules in neurons. In pathological conditions, it forms fibrils that are molecular hallmarks of neurological disorders known as tauopathies. In the last 2 years, cryo-EM has given unprecedented high-resolution views of Tau in both physiological and pathological conditions. We review here these new findings and put them into the context of the knowledge about Tau before this structural breakthrough. The first structures of Tau fibrils, a molecular hallmark of Alzheimer's disease (AD), were based on fibrils from the brain of an individual with AD and, along with similar patient-derived structures, have set the gold standard for the field. Cryo-EM structures of Tau fibers in three distinct diseases, AD, Pick's disease, and chronic traumatic encephalopathy, represent the end points of Tau's molecular trajectory. We propose that the recent Tau structures may call for a re-examination of databases that link different Tau variants to various forms of dementia. We also address the question of how this structural information may link Tau's functional and pathological aspects. Because this structural information on Tau was obtained in a very short period, the new structures should be viewed in light of earlier structural observations and past and present functional data to shed additional light on Tau function and dysfunction.
Keywords: cryo-electron microscopy, tauopathy, Tau protein (Tau), amyloid, microtubule, tubulin, neurodegeneration, frontotemporal dementia, Alzheimer's disease
Introduction
If single-particle cryo-EM has taken the general field of structural biology by storm (1, 2), in its application to the subfield of neuronal physiology and pathology, it should rather be considered a tsunami. The atomic structures of the γ-secretase complex, first by itself (3, 4) and more recently in complex with a fragment of the amyloid precursor protein (5) or with Notch (6), have highlighted the mechanisms of the complex machinery that produces the amyloid-β peptide, one of the molecular hallmarks of Alzheimer's disease (AD).2 The GABAA receptor as the molecular target of the benzodiazepines used to treat depression, schizophrenia, and epilepsy, resisted structural characterization for a long time, but cryo-EM finally yielded its structural organization at atomic resolution (7, 8). Polymorphic structures of both amyloid-β (9) and α-synuclein fibrils (10, 11) were solved. Whereas all of these accomplishments have given tremendous insight into the molecular functioning of the brain, the samples that were used were invariably made from overexpressed proteins, except for one: The first structures of Tau fibrils, the other molecular hallmark of AD, were based on carefully selected fibrils from the brain of a deceased patient (12). This incredible tour de force of “structural biology on a patient”—and the subsequent structures of Tau fibrils isolated from the brain of patients suffering from Pick's disease (PiD) (13) or from chronic traumatic encephalopathy (CTE) (14)–have clearly set a gold standard for the field. Cryo-EM equally solved the structure of synthetic fibrils, widely used as a proxy for the brain-derived fibrils, and obtained by incubating Tau with a negatively charged polyanion, such as heparin (15, 16). They were found to be polymorphic, and all forms adopt a substantially different fold from the brain-derived fibrils (17). Negative-stain EM and fluorescence by dyes that more or less specifically recognize amyloid forms are hence not sufficient to distinguish different forms of the fibers, further underlining the extreme care one should take when interpreting data derived from synthetic fibrils in the framework of any amyloid disease. Before pathology, Tau as a tubulin-associated unit plays an important physiological role in the assembly and stabilization of microtubules (MTs) (18). The recent cryo-EM structure of Tau on MTs (19) has complemented our functional view of this archetypal intrinsically disordered protein (20).
Because all of this information on Tau was obtained in a very short period (the last 2 years), we propose here to analyze the novel structures in view of past structural and functional data and thereby to also define some remaining questions about its (dys)function. Starting from a comparative analysis of the global form of Tau fibers, we will zoom in on a particular peptide that has long been considered a nucleus of the aggregation process. We will illustrate how the novel structures also point to the need for a re-examination of the mutational databases that link Tau mutants to different forms of dementia. Finally, we address the question of whether and how functional and pathological aspects of Tau might be linked through the recent structural information.
Structure–(dys)function relationships of Tau before the cryo-EM data
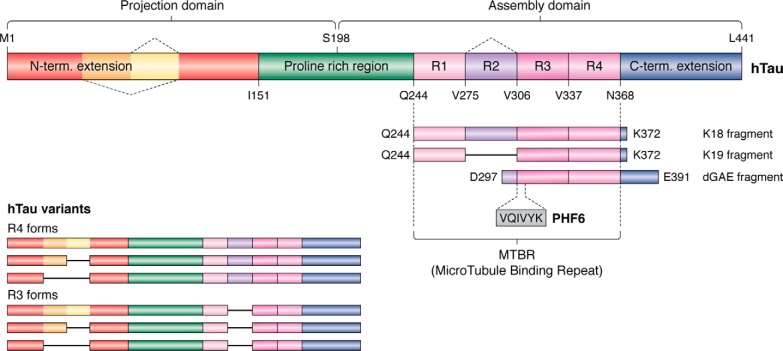
Tau was discovered as a protein involved in the assembly of tubulin into microtubules and their consequent stabilization (18). Other functions have been described (21), but as the structural information in these contexts is lacking, we will not treat them in the present review. Tau is notably characterized by the presence of three or four (according to the isoforms) imperfect repetitions of a motif of about 30 residues, known as the microtubule-binding repeats (MTBRs) (Fig. 1). Strictly speaking, each repetition is composed of an 18-amino acid imperfect repeat and a 13–14-residue interrepeat region (22), and we will make the distinction when necessary. N-terminal to the MTBR region is a proline-rich region (PRR), and these two regions are flanked by N-terminal and C-terminal extensions (Fig. 1). Early efforts to establish a structure–function relationship were based on affinity measurements of different Tau fragments to taxol-stabilized MTs. This study found (i) Tau protein binding primarily to the exterior surfaces of MTs; (ii) a negative contribution of the Tau N-terminal extension; (iii) an unstructured, noncooperative, and distributed binding by the different repeats, with the first repeat R1 binding ∼100 times more tightly than the other repeats; and (iv) within the repeats, no significant effect of the less conserved interrepeat regions (20). The last conclusion was, however, based on truncations of only the R3 interrepeat, whereas later studies showed that the R1 interrepeat does contribute to the MT binding (23). An additional fragment in the PRR (Fig. 1) was equally found to contribute significantly to the affinity (24, 25) and led to a “jaws” model, whereby this flanking PRR (and the sequence downstream of the fourth repeat) would position the repeat peptides to promote tubulin assembly (26). However, no atomic-level structural information about Tau on the MT surface was available to provide mechanistic insights into the functional aspects of Tau. One notable caveat is that most studies were done with taxol-stabilized MTs; it is not clear yet whether the same results will hold when Tau is allowed to copolymerize with tubulin as is the case in the neuron (27). From the NMR study of a functional fragment of Tau (TauF4, corresponding to the Ser208–Ser324 fragment of Tau; numbering throughout this paper is according to the longest isoform) (25) bound to a nonpolymerizable complex of two tubulin heterodimers sequestered by a stathmin-like domain protein, we proposed a model whereby the 258SKIGSTE264 peptide, embedded in the first repeat that by itself can already stimulate MT assembly (28), would bridge different tubulin dimers and change conformation in the assembly process (29). We will provide below a detailed discussion of this model in view of the recent cryo-EM data of Tau on the MT surface.
Figure 1.
Primary structure of the longest isoform of human Tau, with its different domains. Splice variants occur through the omission of one or two N-terminal inserts or of the second repeat in the MTBR.
The above-described line of research converged with that started by Alois Alzheimer when it was realized by different groups that the neuronal tangles, one of the molecular hallmarks of AD, are composed primarily of aggregates of the same Tau protein that promotes MT assembly (30–33). Further important evidence for Tau having a direct role in neurodegeneration came from the identification of distinct mutations leading to neuronal degeneration and dementia (34–36) that are, however, distinguishable from AD.
Knowledge at the structural level of Tau fibrils before the cryo-EM boom was scarce at best. Most studies have relied on synthetic fibers, commonly induced by incubating recombinant Tau forms with negatively charged polyanions, with heparin being the most popular (15, 16). Although solid-state NMR and EPR were able to assign β-strands to certain peptides (37–40) in such synthetic fibers, no atomic model was available. The crystal structure of the PHF6 peptide, thought to be a nucleus of the aggregation process (41), came out 12 years ago and described a zipper-like structure with a dry interface (42). As we will discuss below, the recent cryo-EM data on brain-derived natural fibers entirely challenges both the model of heparin-induced synthetic fibers and the biological relevance of structural data derived from the peptide crystals.
Although ill-defined, “hyperphosphorylation” is a characteristic term that is used to describe Tau in the fibrillar aggregates that characterize AD and other related dementia (commonly called “tauopathies”) (43). Accordingly, the present method of choice to clinically stage AD is by post-mortem staining of the neurons of deceased patients with the AT8 antibody (44), whose epitope was identified as the Tau peptide centered on the phosphorylated residues Ser202 and Thr205 in the PRR. Although NMR (45) and X-ray crystallography (46) gave some insights into the structural aspects of this epitope, with notably a question of whether a third phosphorylation event at Ser208 should be included in the epitope, how this epitope and, for that matter, the whole PRR connects to the fibril core is not clear at this moment. The divergence between the physiological (MT assembly) and pathological (aggregation into fibers) aspects of Tau, imposed by the loose statement “Tau gets hyperphosphorylated, detaches from the MT and becomes prone to aggregation” (47), obviously needs clarification in terms of the location and stoichiometry of the phosphorylation pattern.
In summary, the available structural data described Tau as an archetypal intrinsically disordered protein that binds to MTs in a dynamic manner and transforms into a β-sheet–rich rigid core region upon aggregation and whose function and dysfunction are regulated by numerous post-translational modifications, among which phosphorylation stands out. Detailed understanding of these different aspects was lacking because of the absence of atomic-level structures and has only become available in the last couple of years.
The global fold of Tau fibers differs substantially between ex vivo and synthetic samples
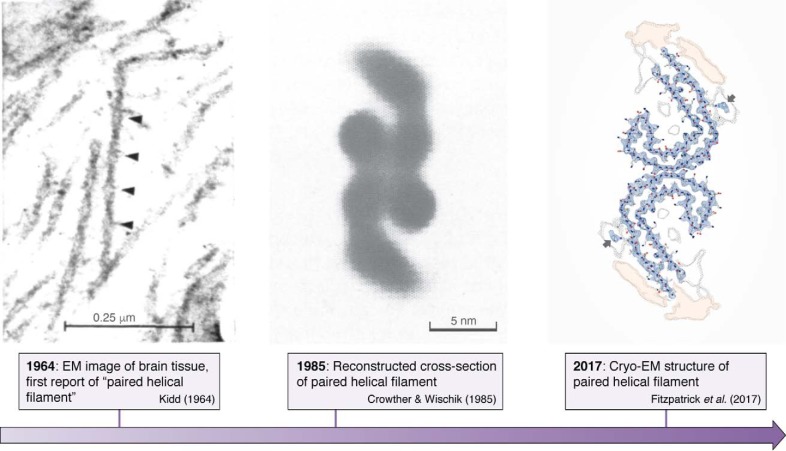
Paired helical filaments in the neurons of Alzheimer's disease patients were first described (48) and subsequently shown (49) by EM in the early 1960s. It then took some 22 years before image reconstruction by EM led to the first model of the cross-section of these fibrils (50). Finally, some 32 years elapsed before cryo-EM turned this image into an atomic-level structure of the fibrils (12) (Fig. 2). The two latter studies equally confirmed that straight filaments, a minor fraction of the Tau fibrils in AD brains, are composed of the same protofilament structure, but with a different packing. Whether the straight filaments that are dominant in progressive supranuclear palsy (51) adopt the same fold remains to be seen. In view of the extensive polymorphisms that amyloid structures can adopt (52), the currently published filament structures may represent only a small fraction of the Tau filament landscape, as there are numerous other tauopathies where atomic structures are still lacking. Nevertheless, these novel structures were eagerly awaited, as for the first time, it could be said with a high degree of confidence that they are the “real thing.” The identical structures of the fibers derived from different patients (53) furthermore underscore the idea that we are considering a disease- rather than patient-specific amyloid form of the Tau fibers.
Figure 2.
Structural detail as available over time for the Tau fibers formed in the brain of AD patients. Left, negative-stain EM image of brain tissue showed the first PHFs (49). Middle, reconstructed cross-section of the paired helical filament (50). Right, atomic model of the same cross-section obtained by cryo-EM (12).
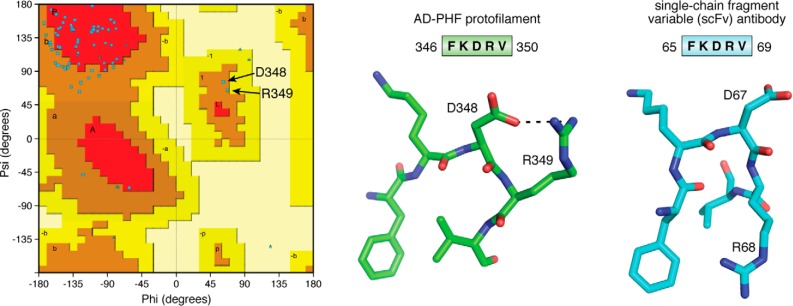
When considering the molecular arrangement of Tau in the AD fibrils, the most unexpected feature is the β-helical fold formed by the triangular arrangement of three consecutive β-sheets (12). This structure is also observed in the recent cryo-EM structure of fibrils from brains of CTE patients (professional sportsmen suffering from a specific tauopathy due to repeated head impact), but in this case, it lines a wider cavity with a presently unknown (hydrophobic) molecule (14). Composed of residues in the fourth MTBR (Fig. 1), this β-helix is rather reminiscent of a folded protein. The peculiar character of the turn is highlighted when we consider the Ramachandran plot of the structure, with Asp348 and Arg349 the only two residues characterized by positive Φ,Ψ angles (Fig. 3). When we query the Protein Data Bank (PDB) with the sequence (DFKDRV) of the peptide centered around these two residues, a unique structure of a single-chain fragment variable antibody (54) is found. In its three-dimensional structure (PDB code 6EHV), this fragment adopts a turn comparable with the one found in the AD-Tau fibrils (Fig. 3), underscoring its character of “folded protein.” Finally, this same (DFKDRV) peptide was found to enhance the affinity for the MT surface of a Tau fragment spanning the first three repeats by a factor of 2.5 (20), so it remains an open question what conformation this peptide adopts in the physiological conformation of Tau and notably at the microtubule surface (see below).
Figure 3.
The turn around Asp348 in the AD fibers. Left, Ramachandran plot of one chain of the AD PHF protofilament, showing the positive Φ,Ψ values for Asp348 and Arg349. Middle, the FKDRV peptide in the AD PHF structure (PDB code 5O3L) adopts a turn around Asp348, whose side chain also stabilizes the Arg349 side chain. Right, in the equivalent turn of the single-chain fragment variable antibody (PDB code 6EHV), Asp67 occupies the central position in the turn, but Arg68 makes a salt bridge with the Asp91 side chain on a nearby helix (not shown). In both structures, the Phe ring and Val methyls form a hydrophobic cluster.
In contrast to this peculiar conformation in the AD-Tau fibers, the same Phe346–Val350 peptide observed in the fibers isolated from the brain of a PiD patient, mainly composed of the shorter 3R isoforms, adopts a rather common extended conformation (13). Subtle forces can hence push the structure toward completely different packings. The relationship with the aggregation process and the possible intervention of cofactors and/or post-translational modifications of Tau is at this moment unresolved.
To compare the ex vivo fibers with the synthetic heparin-induced fibers, the same team used cryo-EM to solve the atomic-level structure of the latter. Despite the resemblance of the macroscopic structures as seen under negative-stain EM, differences at the atomic level are massive. The ordered core of the heparin-induced fibrils as seen by cryo-EM extends from Gly272 to His330 and thereby hardly overlaps with the core of AD paired helical filaments (PHFs) spanning the fragment from Val306 to Phe378. The structures do, however, explain why heparin readily induced fibers with our TauF4 fragment, which overlaps perfectly with the former observed core region (55). The turn region observed in AD PHFs is evidently not visible in the heparin-induced fibrils, but at least in the synthetic fibers obtained with 4R-Tau, the chain does turn on itself around a peptide centered in the R2 repeat (Lys290–Pro301) (17). In the synthetic fibers obtained with 3R-Tau, where this R2 repeat is missing, no turn is observed, but rather two molecules of Tau stacking in a parallel manner are identified. One might speculate that different cofactors or altered reaction conditions from the ones that were chosen for the in vitro experiments could produce additional types of filaments.
In conclusion, the combined cryo-EM structures clearly indicate that at the atomic level, brain-derived fibers are disease-specific and are substantially different from the heparin-derived synthetic fibers that have been used in most previous studies.
The PHF6 peptide adopts different conformations among the available fiber structures
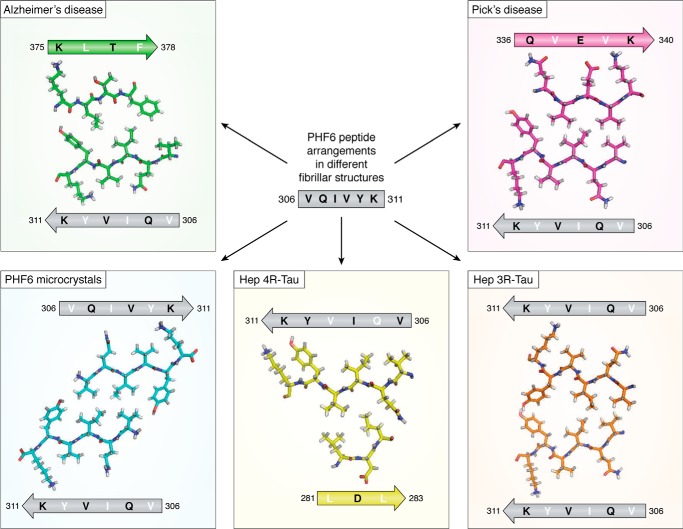
The PHF6 peptide motif, spanning the 6 residues 306VQIVYK311 in the third repeat of Tau (Fig. 1), was early on identified as one of the hot spots of the aggregation behavior (41). This same peptide motif is at the very beginning of the ordered structure of the brain-isolated AD fibrils (12). Earlier X-ray microcrystallography on crystals of the isolated peptide showed a homotypic interaction, whereby one PHF6 peptide locks into a second antiparallel one to form a steric zipper with a dry interface (42) (Fig. 4). In the AD fibrils, however, the same PHF6 motif locks into the 374HKLTF378 sequence, with Leu376 intercalating between the Ile308 and Tyr310 side chains (12) (Fig. 4). At the center of the pseudorepeat region Lys369–Thr386 directly following the MTBRs, this peptide motif also contributes largely to the MT binding of 3R- or 4R-Tau in neuronal processes through decreasing the dissociation rate (56). Ironically, whereas the first synthetic fibers without heparin were obtained with the K11 or K12 fragments running to Tyr394 in the 3R- or 4R-Tau constructs and hence spanning this Lys369–Thr386 sequence (57), the shorter K18 or K19 fragments, later extensively used as a proxy to study the aggregation of full-length Tau (58, 59), stop at Lys372 (Fig. 1) and hence are strictly unable to provide the complement of the PHF6 sequence found in AD PHFs.
Figure 4.
Different arrangements of the PHF6 peptide in the different fibrillar structures. The peptide is indicated as a gray arrow, with inward-pointing residues in white and outward-pointing residues in black. Structures are from the AD PHFs (top left, green), the PiD PHFs (top right, magenta), the PHF6 microcrystals (bottom left, blue), the heparin-induced 4R-Tau structure (bottom middle, yellow), and the heparin-induced 3R-Tau fibers (bottom right, orange). The peptide itself adopts invariably the same extended conformation but faces different peptides in every single structure. In the heparin-induced 4R-Tau fibrils, Lys311 points in the same direction as Tyr310, and both residues face the outside of the fibrillar structure.
In the PiD 3R-Tau fiber structure, the same PHF6 fragment could potentially face the same 374HKLTF378 sequence. But intriguingly, it faces another peptide from Tau, with this time the two hydrophobic side chains of Val337 and Val339 intercalating between the Val306-Ile308-Tyr310 side chains (Fig. 4) (13). And even more surprisingly, in the cryo-EM structure of heparin-induced synthetic fibers with 4R-Tau as starting material, this PHF6 sequence is literally turned inside-out, with its Val306-Ile308-Tyr310 side chains facing the outside of the fibril (Fig. 4). The side chain of Lys311 equally points outside, probably through interaction with the negatively charged heparin. Indeed, introducing a negative charge at this position was previously found to reduce aggregation in the heparin-induced aggregation assay (60). Only in the heparin-induced 3R-Tau fibrils, where two Tau molecules stack to make a fibrillary structure (rather than one single Tau molecule bending over itself to form a protofilament), do the PHF6 peptide sequences on the molecules face one another (17). However, they do so in a parallel manner, which further underscores the extreme polymorphism of which Tau is capable. One should remember that the initial study identifying this hot spot of aggregation used both a 3R-Tau construct (the K19 fragment) and heparin as inducer (41), and the latter structure is hence most relevant to interpret the results. As the peptide does adopt a β-strand conformation in all structures, the conclusion of the mutational analysis that a proline in whatever position of PHF6 abolishes fiber formation (41, 61) might, however, well hold up for every type of fibril.
The crystal structure of the PHF6 peptide, describing the steric zipper with a dry interface (42), has spurred a large research effort both to interpret at the structural level aggregation inhibitors described in the past (62–64) or to rationally develop novel inhibitors (62, 65, 66). Many of these were tested first in an in vitro assay monitoring the polyanion-induced aggregation of some Tau construct before being tested in a cell model or a transgenic organism. An important issue is that we do not know the atomic structure of Tau fibrils in the latter models, but at least in those based on overexpression of the K18/K19 fragments, the lack of the C-terminal 374HKLTF378 peptide implies that these Tau fragments cannot adopt the AD brain-derived conformation. A shift toward the dGAE fragment (67), which comprises residues from Asp297 to Glu391 (Fig. 1) and spans the sequence of Tau that is ordered in the AD PHFs, could possibly reproduce the spatial organization of the ex vivo fibrils, although this has still to be proven. Whether it is opportune to add to this fragment 16 N-terminal residues visible as an unsharpened density in the AD fiber structure (12) remains an open question.
In conclusion, the presently available structures of brain-derived fibrils and their substantial differences from the heparin-induced synthetic fibrils, in particular the differential positioning of the PHF6 peptide in these structures, invite a re-evaluation of the inhibitors in this new framework.
Revisiting Tau mutations leading to non-AD dementia in view of the novel structures
Although absent in Alzheimer's disease, these mutations can either affect the ratio of 3R/4R-Tau splicing variants, with PiD (31, 68) for example being mostly characterized by a dominant expression of 3R forms (69), or can directly introduce a point mutation without notably affecting the splicing ratio. Generally, these mutations have been first identified in a family with a history of precocious dementia, and the recombinant proteins are then evaluated in terms of their aggregation behavior and/or their capacity to assemble tubulin into microtubules. However, the aggregation assay in most cases concerns the heparin-induced fiber formation, monitored by some fluorescence method (70) and/or negative-stain EM imaging of the resulting fibers. In view of the pronounced structural differences between the ex vivo fibrils and the heparin-induced ones, we probably should reconsider these findings. One caveat hereby is that these mutations could cause still entirely different structures compared with the three available disease models. As the library of structures will increase, however, we might at some point be able to rationalize their influence.
As an example, consider the K317N mutation that was recently identified in patients suffering from globular glial tauopathy, a four-repeat tauopathy characterized by Tau-positive, globular glial inclusions (71). This mutation was found to lead to enhanced aggregation when introduced in the recombinant 4R isoform while decreasing filament formation when the mutated 3R isoform was used in the same aggregation assay. This latter assay compared by thioflavin T fluorescence the resulting fiber formation after incubation of one or the other isoform with a close-to-stoichiometric amount of heparin (71).
If we consider the most populated heparin-induced 4R-snake fiber structure, seven lysine (and two more histidine) side chains stick outward from each Tau monomer to form a charge ladder parallel with the fiber axis. Lys317 and Lys321, but also Lys274–Lys280 and Lys298–His299, form clamplike structures that require stabilization by some negative polyanion (72). In the AD-Tau fibrils, the Lys317–Lys321 tandem is organized in a similar manner, with both lysine side chains also pointing away from the fiber core. Residual electron density in the experimental cryo-EM map of these AD-Tau fibrils in the immediate vicinity of the Lys317 and Lys321 side chains (arrows in Fig. 2, right) was assigned tentatively to the 7EFE9 acidic patch that, together with at least one of the MTBRs, is thought to form the Alz50 antibody conformational epitope (73–75). NMR could also localize a transient interaction in the heparin-induced 4R-Tau fibers between the N terminus and a paramagnetic agent attached to Cys322, suggesting that this Lys317–Lys321 clamp might not be the primary target of heparin (76). Removing a single positive charge of Lys317 would hence leave more heparin available for the other sites and might thereby stimulate aggregation.
In the ordered assembly of the heparin-induced 3R-Tau fibers, two parallel chains make up the protofilament (17). In these, Lys317 and Lys321 do not provide the only positively charged outward-facing residues, but they are the only clamplike structure. The observed diminished aggregation upon mutating Lys317 might hence come from the removal of the single clamplike structure on each of the 3R-Tau monomers and suggests that the 3R-Tau fibers are truly stabilized by heparin binding to this lysine tandem.
In vivo, in the case of globular glial tauopathy, although we have as yet no structure of a 4R-Tau–only fiber and even less of a mutant Tau fiber, the K317N mutation could promote aggregation through reducing the need for charge compensation or through the lessened entropic cost of the N terminus folding back toward the repeats. Importantly, the in vitro heparin-induced aggregation assay should be taken with caution in the interpretation of in vivo data, as it might not be indicative of the same aggregation process. We thus conclude that revisiting the Tau mutation database in terms of the novel (and future) structures seems a worthwhile effort.
Tau conformations in function and dysfunction might be related
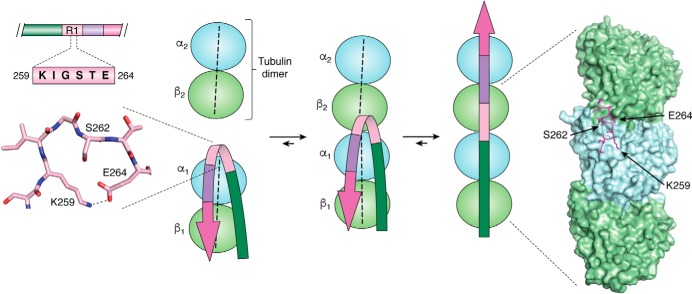
Whereas the crystal structure of tubulin in its polymerized form was determined by electron crystallography some 20 years ago (77), we had to wait until last year for cryo-EM to yield the first atomic-resolution view of Tau on the MT surface (19). Although these structures should be considered with some caution—they show an artificial construct built from a repetition of a single repeat (R1, as the largest contributor of binding energy (20), or R2, the second repeat that distinguishes 4R- from 3R-Tau), the constructs were added in excess of tubulin, and near-atomic resolution was only obtained with peluroside as a stabilizing agent and even then required extensive modeling—they do contain important novel information that completes past indirect evidence. First, the structure places the 258SKIGSTEN265 peptide in the first repeat at the interface of two tubulin dimers, where an α1 subunit contacts the β2 subunit of the next dimer (Fig. 5). NMR analysis of a Tau construct bound to soluble tubulin assemblies not only localized this peptide at exactly the same position, but suggested that it would transit from a turn toward an extended conformation when a second tubulin dimer comes in (29). The cryo-EM structure of the R1 repeat adopts a remnant of such a turn and thereby confirms the proposed mechanism (Fig. 5). As we dispose now of another structure of this R1 repeat, in the cryo-EM structure of the Pick's disease fibrils, it is interesting to link the functional and dysfunctional conformations of Tau. Indeed, in the 3R-Tau structure of PiD fibrils, the same 258SKIGSTEN265 peptide adopts a perfect turn conformation, stabilized by a salt bridge between the side chains of Lys259 and Glu264 (Fig. 5). On the tubulin surface, this bridge could break when a novel tubulin dimer comes in, with Lys259 now forming a salt bridge with α1 Asp424, whereas Glu264 stabilizes the α1/β2 interaction through a salt bridge with β2 Lys402. Beyond proving that the turn conformation is possible, the combined structures hence provide an additional link between Tau's functional and pathological conformations, in line with previous studies that hint at a role for tubulin in the aggregation process. Whether in vivo aggregation of Tau occurs at the microtubule surface (78, 79) or rather through the soluble tubulin (80) remains unclear but could be related to the recent controversy whether Tau stabilizes MTs or enables these same axonal MTs to have labile domains (81, 82).
Figure 5.
The structural switch of the 260IGSTE264 peptide. Left, turn adopted by the 260IGSTE264 peptide in the PiD 3R-Tau fibrillar structure (13); middle, NMR-based model of the transition of this peptide from a turn when bound to a single tubulin dimer to an extended conformation when anchoring a second tubulin dimer (29). Right, conformation of the same peptide in the MT cryo-EM structure with the artificial 4xR1 construct (19).
The second repeat, spanning residues Lys274–Gly304, was also resolved by cryo-EM and Rosetta modeling. Its conformation is extended and spans three tubulin units (19). Because this second repeat is invisible in the AD PHF structure and missing in the PiD 3R-Tau fibrils, we can only compare it with the same fragment in the heparin-induced 4R-Tau filaments. Although there too it adopts an extended conformation, overlap of both fragments is poor, with a general root mean square deviation of 4.5 Å. Further structural studies, notably with fragments spanning different repeats, are awaited to explain the effect of mutations and/or post-translational modifications (PTMs).
Conclusions and perspectives
As indicated before, the present cryo-EM structures of Tau fibers in three distinct diseases, AD, PiD, and CTE, represent the end points of the molecular trajectory of pathological Tau. Together with the tubulin-bound structure, however, it means that we now dispose of a structural glimpse of several stages in the lifetime of Tau, from its microtubulin bound to fiber conformation. Importantly, the slow turnover of Tau in the brain, which can span days or even weeks (83), suggests that individual molecules can take many paths. Proposed trajectories that were derived without those constraints (or with the wrong constraints, if we consider the heparin-induced fibrils), can now be re-evaluated in terms of the end points.
In all structures, large parts of Tau and notably the PRR (Fig. 1) are absent. This latter PRR contributes importantly to microtubule binding (25). It is equally one of the main regions regulated by PTMs and notably phosphorylation. The resulting heterogeneity (with many “mod-forms” (84) or “proteoforms” (85) if we include the splice variants) will necessarily reduce the constraint of homogeneity that cryo-EM can detect. Nevertheless, clinically, AD is diagnosed and staged post-mortem by the AT8 antibody raised against a phospho-epitope in this PRR (44). The definition of the latter epitope has equally evolved over the last quarter of a century, and currently, it is not clear whether the antibody “sees” two (Ser202 and Thr205) or three (with an additional Ser208) phosphorylated residues on AD-Tau (46). Importantly, the absence of these regions in the present structures therefore does not necessarily indicate their lack of importance but might rather be a consequence of their heterogeneous nature in the neuron.
A last but most important open question that is not answered (yet) by the structures remains the identification of the driving force(s) for aggregation. Is the specific phosphorylation pattern that can drive aggregation in vitro (86) also at work in vivo? What about the role of other PTMs, such as acetylation of lysines (87–89), O-GlcNacylation (90, 91), or others? But it also raises the more general (and pressing) question about the structure of Tau fibers in all models, be they at the level of molecules, cells, or organisms. Even if these models are imperfect, we need to ascertain whether they obey the structural constraint of the end point, with fibers of a comparable structure as the brain-derived ones. Negative-stain EM and thioflavin fluorescence cannot answer this question at a sufficient resolution, but we can hope that the increased access to cryo-EM platforms will provide a structural evaluation of the different models at the atomic level. With comparable filaments at the atomic level, we can hope that the trajectory of Tau mimics what is happening in the patient's brain. As such, the recently derived structures set a standard and should open a new era of increased pathological relevance of models at all length scales.
Acknowledgments
We thank Prof. M. Goedert (Cambridge, UK), who provided in advance the structures of heparin-induced filaments and CTE fibers and whose insightful comments helped to shape this review. We equally thank Dr. C. Byrne (Paris, France) for careful proofreading of the manuscript.
This work was supported in part by Agence Nationale de la Recherche (ANR) Grant CatSAmy ANR-18-CE07-0016. The authors declare that they have no conflicts of interest with the contents of this article.
- AD
- Alzheimer's disease
- PiD
- Pick's disease
- CTE
- chronic traumatic encephalopathy
- MT
- microtubule
- MTBR
- microtubule-binding repeat
- PRR
- proline-rich region
- PDB
- Protein Data Bank
- PTM
- post-translational modification
- PHF
- paired helical filament.
References
- 1. Cheng Y. (2018) Single-particle cryo-EM—how did it get here and where will it go. Science 361, 876–880 10.1126/science.aat4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Renaud J.-P., Chari A., Ciferri C., Liu W.-T., Rémigy H.-W., Stark H., and Wiesmann C. (2018) Cryo-EM in drug discovery: achievements, limitations and prospects. Nat. Rev. Drug Discov. 17, 471–492 10.1038/nrd.2018.77 [DOI] [PubMed] [Google Scholar]
- 3. Bai X.-C., Yan C., Yang G., Lu P., Ma D., Sun L., Zhou R., Scheres S. H. W., and Shi Y. (2015) An atomic structure of human γ-secretase. Nature 525, 212–217 10.1038/nature14892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lu P., Bai X.-C., Ma D., Xie T., Yan C., Sun L., Yang G., Zhao Y., Zhou R., Scheres S. H. W., and Shi Y. (2014) Three-dimensional structure of human γ-secretase. Nature 512, 166–170 10.1038/nature13567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhou R., Yang G., Guo X., Zhou Q., Lei J., and Shi Y. (2019) Recognition of the amyloid precursor protein by human γ-secretase. Science 363, eaaw0930 10.1126/science.aaw0930 [DOI] [PubMed] [Google Scholar]
- 6. Yang G., Zhou R., Zhou Q., Guo X., Yan C., Ke M., Lei J., and Shi Y. (2019) Structural basis of Notch recognition by human γ-secretase. Nature 565, 192–197 10.1038/s41586-018-0813-8 [DOI] [PubMed] [Google Scholar]
- 7. Laverty D., Desai R., Uchański T., Masiulis S., Stec W. J., Malinauskas T., Zivanov J., Pardon E., Steyaert J., Miller K. W., and Aricescu A. R. (2019) Cryo-EM structure of the human α1β3γ2 GABAA receptor in a lipid bilayer. Nature 565, 516–520 10.1038/s41586-018-0833-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Phulera S., Zhu H., Yu J., Claxton D. P., Yoder N., Yoshioka C., and Gouaux E. (2018) Cryo-EM structure of the benzodiazepine-sensitive α1β1γ2S tri-heteromeric GABAA receptor in complex with GABA. eLife 7, e39383 10.7554/eLife.39383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gremer L., Schölzel D., Schenk C., Reinartz E., Labahn J., Ravelli R. B. G., Tusche M., Lopez-Iglesias C., Hoyer W., Heise H., Willbold D., and Schröder G. F. (2017) Fibril structure of amyloid-β(1–42) by cryo-electron microscopy. Science 358, 116–119 10.1126/science.aao2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guerrero-Ferreira R., Taylor N. M., Mona D., Ringler P., Lauer M. E., Riek R., Britschgi M., and Stahlberg H. (2018) Cryo-EM structure of α-synuclein fibrils. eLife 7, e36402 10.7554/eLife.36402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li B., Ge P., Murray K. A., Sheth P., Zhang M., Nair G., Sawaya M. R., Shin W. S., Boyer D. R., Ye S., Eisenberg D. S., Zhou Z. H., and Jiang L. (2018) Cryo-EM of full-length α-synuclein reveals fibril polymorphs with a common structural kernel. Nat. Commun. 9, 3609 10.1038/s41467-018-05971-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fitzpatrick A. W. P., Falcon B., He S., Murzin A. G., Murshudov G., Garringer H. J., Crowther R. A., Ghetti B., Goedert M., and Scheres S. H. W. (2017) Cryo-EM structures of tau filaments from Alzheimer's disease. Nature 547, 185–190 10.1038/nature23002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Falcon B., Zhang W., Murzin A. G., Murshudov G., Garringer H. J., Vidal R., Crowther R. A., Ghetti B., Scheres S. H. W., and Goedert M. (2018) Structures of filaments from Pick's disease reveal a novel tau protein fold. Nature 561, 137–140 10.1038/s41586-018-0454-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Falcon B., Zivanov J., Zhang W., Murzin A. G., Garringer H. J., Vidal R., Crowther R. A., Newell K. L., Ghetti B., Goedert M., and Scheres S. H. W. (2019) Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature 568, 420–423 10.1038/s41586-019-1026-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goedert M., Jakes R., Spillantini M. G., Hasegawa M., Smith M. J., and Crowther R. A. (1996) Assembly of microtubule-associated protein tau into Alzheimer-like filaments induced by sulphated glycosaminoglycans. Nature 383, 550–553 10.1038/383550a0 [DOI] [PubMed] [Google Scholar]
- 16. Pérez M., Valpuesta J. M., Medina M., Montejo de Garcini E., and Avila J. (1996) Polymerization of tau into filaments in the presence of heparin: the minimal sequence required for tau-tau interaction. J. Neurochem. 67, 1183–1190 [DOI] [PubMed] [Google Scholar]
- 17. Zhang W., Falcon B., Murzin A. G., Fan J., Crowther R. A., Goedert M., and Scheres S. H. (2019) Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases. eLife 8, e43584 10.7554/eLife.43584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weingarten M. D., Lockwood A. H., Hwo S. Y., and Kirschner M. W. (1975) A protein factor essential for microtubule assembly. Proc. Natl. Acad. Sci. U.S.A. 72, 1858–1862 10.1073/pnas.72.5.1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kellogg E. H., Hejab N. M. A., Poepsel S., Downing K. H., DiMaio F., and Nogales E. (2018) Near-atomic model of microtubule-tau interactions. Science 360, 1242–1246 10.1126/science.aat1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Butner K. A., and Kirschner M. W. (1991) Tau protein binds to microtubules through a flexible array of distributed weak sites. J. Cell Biol. 115, 717–730 10.1083/jcb.115.3.717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guo T., Noble W., and Hanger D. P. (2017) Roles of tau protein in health and disease. Acta Neuropathol. 133, 665–704 10.1007/s00401-017-1707-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. LeBoeuf A. C., Levy S. F., Gaylord M., Bhattacharya A., Singh A. K., Jordan M. A., Wilson L., and Feinstein S. C. (2008) FTDP-17 mutations in Tau alter the regulation of microtubule dynamics: an “alternative core” model for normal and pathological Tau action. J. Biol. Chem. 283, 36406–36415 10.1074/jbc.M803519200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goode B. L., and Feinstein S. C. (1994) Identification of a novel microtubule binding and assembly domain in the developmentally regulated inter-repeat region of tau. J. Cell Biol. 124, 769–782 10.1083/jcb.124.5.769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goode B. L., Denis P. E., Panda D., Radeke M. J., Miller H. P., Wilson L., and Feinstein S. C. (1997) Functional interactions between the proline-rich and repeat regions of tau enhance microtubule binding and assembly. Mol. Biol. Cell 8, 353–365 10.1091/mbc.8.2.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fauquant C., Redeker V., Landrieu I., Wieruszeski J.-M., Verdegem D., Laprévote O., Lippens G., Gigant B., and Knossow M. (2011) Systematic identification of tubulin-interacting fragments of the microtubule-associated protein Tau leads to a highly efficient promoter of microtubule assembly. J. Biol. Chem. 286, 33358–33368 10.1074/jbc.M111.223545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gustke N., Trinczek B., Biernat J., Mandelkow E. M., and Mandelkow E. (1994) Domains of tau protein and interactions with microtubules. Biochemistry 33, 9511–9522 10.1021/bi00198a017 [DOI] [PubMed] [Google Scholar]
- 27. Makrides V., Massie M. R., Feinstein S. C., and Lew J. (2004) Evidence for two distinct binding sites for tau on microtubules. Proc. Natl. Acad. Sci. U.S.A. 101, 6746–6751 10.1073/pnas.0400992101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ennulat D. J., Liem R. K., Hashim G. A., and Shelanski M. L. (1989) Two separate 18-amino acid domains of tau promote the polymerization of tubulin. J. Biol. Chem. 264, 5327–5330 [PubMed] [Google Scholar]
- 29. Gigant B., Landrieu I., Fauquant C., Barbier P., Huvent I., Wieruszeski J.-M., Knossow M., and Lippens G. (2014) Mechanism of Tau-promoted microtubule assembly as probed by NMR spectroscopy. J. Am. Chem. Soc. 136, 12615–12623 10.1021/ja504864m [DOI] [PubMed] [Google Scholar]
- 30. Brion J. P., Flament-Durand J., and Dustin P. (1986) Alzheimer's disease and tau proteins. Lancet 2, 1098 [DOI] [PubMed] [Google Scholar]
- 31. Joachim C. L., Morris J. H., Kosik K. S., and Selkoe D. J. (1987) Tau antisera recognize neurofibrillary tangles in a range of neurodegenerative disorders. Ann. Neurol. 22, 514–520 10.1002/ana.410220411 [DOI] [PubMed] [Google Scholar]
- 32. Grundke-Iqbal I., Iqbal K., Quinlan M., Tung Y. C., Zaidi M. S., and Wisniewski H. M. (1986) Microtubule-associated protein tau: a component of Alzheimer paired helical filaments. J. Biol. Chem. 261, 6084–6089 [PubMed] [Google Scholar]
- 33. Kosik K. S., Joachim C. L., and Selkoe D. J. (1986) Microtubule-associated protein tau (tau) is a major antigenic component of paired helical filaments in Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 83, 4044–4048 10.1073/pnas.83.11.4044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Clark L. N., Poorkaj P., Wszolek Z., Geschwind D. H., Nasreddine Z. S., Miller B., Li D., Payami H., Awert F., Markopoulou K., Andreadis A., D'Souza I., Lee V. M.-Y., Reed L., Trojanowski J. Q., et al. (1998) Pathogenic implications of mutations in the tau gene in pallido-ponto-nigral degeneration and related neurodegenerative disorders linked to chromosome 17. Proc. Natl. Acad. Sci. 95, 13103–13107 10.1073/pnas.95.22.13103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hutton M., Lendon C. L., Rizzu P., Baker M., Froelich S., Houlden H., Pickering-Brown S., Chakraverty S., Isaacs A., Grover A., Hackett J., Adamson J., Lincoln S., Dickson D., Davies P., et al. (1998) Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 393, 702–705 10.1038/31508 [DOI] [PubMed] [Google Scholar]
- 36. Spillantini M. G., Goedert M., Crowther R. A., Murrell J. R., Farlow M. R., and Ghetti B. (1997) Familial multiple system tauopathy with presenile dementia: a disease with abundant neuronal and glial tau filaments. Proc. Natl. Acad. Sci. U.S.A. 94, 4113–4118 10.1073/pnas.94.8.4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Andronesi O. C., von Bergen M., Biernat J., Seidel K., Griesinger C., Mandelkow E., and Baldus M. (2008) Characterization of Alzheimer's-like paired helical filaments from the core domain of tau protein using solid-state NMR spectroscopy. J. Am. Chem. Soc. 130, 5922–5928 10.1021/ja7100517 [DOI] [PubMed] [Google Scholar]
- 38. Daebel V., Chinnathambi S., Biernat J., Schwalbe M., Habenstein B., Loquet A., Akoury E., Tepper K., Müller H., Baldus M., Griesinger C., Zweckstetter M., Mandelkow E., Vijayan V., and Lange A. (2012) β-Sheet core of tau paired helical filaments revealed by solid-state NMR. J. Am. Chem. Soc. 134, 13982–13989 10.1021/ja305470p [DOI] [PubMed] [Google Scholar]
- 39. Margittai M., and Langen R. (2004) Template-assisted filament growth by parallel stacking of tau. Proc. Natl. Acad. Sci. U.S.A. 101, 10278–10283 10.1073/pnas.0401911101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Meyer V., and Margittai M. (2016) Spin labeling and characterization of Tau fibrils using electron paramagnetic resonance (EPR). Methods Mol. Biol. 1345, 185–199 10.1007/978-1-4939-2978-8_12 [DOI] [PubMed] [Google Scholar]
- 41. von Bergen M., Friedhoff P., Biernat J., Heberle J., Mandelkow E. M., and Mandelkow E. (2000) Assembly of tau protein into Alzheimer paired helical filaments depends on a local sequence motif (306VQIVYK311) forming beta structure. Proc. Natl. Acad. Sci. U.S.A. 97, 5129–5134 10.1073/pnas.97.10.5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sawaya M. R., Sambashivan S., Nelson R., Ivanova M. I., Sievers S. A., Apostol M. I., Thompson M. J., Balbirnie M., Wiltzius J. J. W., McFarlane H. T., Madsen A. Ø., Riekel C., and Eisenberg D. (2007) Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature 447, 453–457 10.1038/nature05695 [DOI] [PubMed] [Google Scholar]
- 43. Grundke-Iqbal I., Iqbal K., Tung Y. C., Quinlan M., Wisniewski H. M., and Binder L. I. (1986) Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc. Natl. Acad. Sci. U.S.A. 83, 4913–4917 10.1073/pnas.83.13.4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Braak H., Alafuzoff I., Arzberger T., Kretzschmar H., and Del Tredici K. (2006) Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol. 112, 389–404 10.1007/s00401-006-0127-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gandhi N. S., Landrieu I., Byrne C., Kukic P., Amniai L., Cantrelle F.-X., Wieruszeski J.-M., Mancera R. L., Jacquot Y., and Lippens G. (2015) A phosphorylation-induced turn defines the Alzheimer's disease AT8 antibody epitope on the Tau protein. Angew. Chem. Int. Ed. Engl. 54, 6819–6823 10.1002/anie.201501898 [DOI] [PubMed] [Google Scholar]
- 46. Malia T. J., Teplyakov A., Ernst R., Wu S.-J., Lacy E. R., Liu X., Vandermeeren M., Mercken M., Luo J., Sweet R. W., and Gilliland G. L. (2016) Epitope mapping and structural basis for the recognition of phosphorylated tau by the anti-tau antibody AT8. Proteins 84, 427–434 10.1002/prot.24988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brandt R., and Bakota L. (2017) Microtubule dynamics and the neurodegenerative triad of Alzheimer's disease: the hidden connection. J. Neurochem. 143, 409–417 10.1111/jnc.14011 [DOI] [PubMed] [Google Scholar]
- 48. Kidd M. (1963) Paired helical filaments in electron microscopy of Alzheimer's disease. Nature 197, 192–193 10.1038/197192b0 [DOI] [PubMed] [Google Scholar]
- 49. Kidd M. (1964) Alzheimers disease—an electron microscopical study. Brain 87, 307–320 10.1093/brain/87.2.307 [DOI] [PubMed] [Google Scholar]
- 50. Crowther R. A., and Wischik C. M. (1985) Image reconstruction of the Alzheimer paired helical filament. EMBO J. 4, 3661–3665 10.1002/j.1460-2075.1985.tb04132.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tellez-Nagel I., and Winiewski H. M. (1973) Ultrastructure of neurofibrillary tangles in Steele-Richardson-Olszewski syndrome. Arch. Neurol. 29, 324–327 10.1001/archneur.1973.00490290064007 [DOI] [PubMed] [Google Scholar]
- 52. Eichner T., and Radford S. E. (2011) A diversity of assembly mechanisms of a generic amyloid fold. Mol. Cell 43, 8–18 10.1016/j.molcel.2011.05.012 [DOI] [PubMed] [Google Scholar]
- 53. Falcon B., Zhang W., Schweighauser M., Murzin A. G., Vidal R., Garringer H. J., Ghetti B., Scheres S. H. W., and Goedert M. (2018) Tau filaments from multiple cases of sporadic and inherited Alzheimer's disease adopt a common fold. Acta Neuropathol. 136, 699–708 10.1007/s00401-018-1914-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ahmad Z. A., Yeap S. K., Ali A. M., Ho W. Y., Alitheen N. B. M., and Hamid M. (2012) scFv antibody: principles and clinical application. Clin. Dev. Immunol. 2012, 980250 10.1155/2012/980250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huvent I., Kamah A., Cantrelle F.-X., Barois N., Slomianny C., Smet-Nocca C., Landrieu I., and Lippens G. (2014) A functional fragment of Tau forms fibers without the need for an intermolecular cysteine bridge. Biochem. Biophys. Res. Commun. 445, 299–303 10.1016/j.bbrc.2014.01.161 [DOI] [PubMed] [Google Scholar]
- 56. Niewidok B., Igaev M., Sündermann F., Janning D., Bakota L., and Brandt R. (2016) Presence of a carboxy-terminal pseudorepeat and disease-like pseudohyperphosphorylation critically influence tau's interaction with microtubules in axon-like processes. Mol. Biol. Cell 27, 3537–3549 10.1091/mbc.e16-06-0402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wille H., Drewes G., Biernat J., Mandelkow E. M., and Mandelkow E. (1992) Alzheimer-like paired helical filaments and antiparallel dimers formed from microtubule-associated protein tau in vitro. J. Cell Biol. 118, 573–584 10.1083/jcb.118.3.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Barghorn S., and Mandelkow E. (2002) Toward a unified scheme for the aggregation of tau into Alzheimer paired helical filaments. Biochemistry 41, 14885–14896 10.1021/bi026469j [DOI] [PubMed] [Google Scholar]
- 59. Stöhr J., Wu H., Nick M., Wu Y., Bhate M., Condello C., Johnson N., Rodgers J., Lemmin T., Acharya S., Becker J., Robinson K., Kelly M. J. S., Gai F., Stubbs G., et al. (2017) A 31-residue peptide induces aggregation of tau's microtubule-binding region in cells. Nat. Chem. 9, 874–881 10.1038/nchem.2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li W., and Lee V. M.-Y. (2006) Characterization of two VQIXXK motifs for tau fibrillization in vitro. Biochemistry 45, 15692–15701 10.1021/bi061422+ [DOI] [PubMed] [Google Scholar]
- 61. Chemerovski-Glikman M., Frenkel-Pinter M., Mdah R., Abu-Mokh A., Gazit E., and Segal D. (2017) Inhibition of the aggregation and toxicity of the minimal amyloidogenic fragment of Tau by its Pro-substituted analogues. Chem. Eur. J. 23, 9618–9624 10.1002/chem.201701218 [DOI] [PubMed] [Google Scholar]
- 62. Bulic B., Pickhardt M., Schmidt B., Mandelkow E.-M., Waldmann H., and Mandelkow E. (2009) Development of Tau aggregation inhibitors for Alzheimer's disease. Angew. Chem. Int. Ed. Engl. 48, 1740–1752 10.1002/anie.200802621 [DOI] [PubMed] [Google Scholar]
- 63. Cisek K., Cooper G. L., Huseby C. J., and Kuret J. (2014) Structure and mechanism of action of Tau aggregation inhibitors. Curr. Alzheimer Res. 11, 918–927 10.2174/1567205011666141107150331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Landau M., Sawaya M. R., Faull K. F., Laganowsky A., Jiang L., Sievers S. A., Liu J., Barrio J. R., and Eisenberg D. (2011) Towards a pharmacophore for amyloid. PLoS Biol. 9, e1001080 10.1371/journal.pbio.1001080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sievers S. A., Karanicolas J., Chang H. W., Zhao A., Jiang L., Zirafi O., Stevens J. T., Münch J., Baker D., and Eisenberg D. (2011) Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation. Nature 475, 96–100 10.1038/nature10154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang C. K., Northfield S. E., Huang Y.-H., Ramos M. C., and Craik D. J. (2016) Inhibition of tau aggregation using a naturally-occurring cyclic peptide scaffold. Eur. J. Med. Chem. 109, 342–349 10.1016/j.ejmech.2016.01.006 [DOI] [PubMed] [Google Scholar]
- 67. Al-Hilaly Y. K., Pollack S. J., Vadukul D. M., Citossi F., Rickard J. E., Simpson M., Storey J. M. D., Harrington C. R., Wischik C. M., and Serpell L. C. (2017) Alzheimer's disease-like paired helical filament assembly from truncated Tau protein is independent of disulfide crosslinking. J. Mol. Biol. 429, 3650–3665 10.1016/j.jmb.2017.09.007 [DOI] [PubMed] [Google Scholar]
- 68. Murayama S., Mori H., Ihara Y., and Tomonaga M. (1990) Immunocytochemical and ultrastructural studies of Pick's disease. Ann. Neurol. 27, 394–405 10.1002/ana.410270407 [DOI] [PubMed] [Google Scholar]
- 69. Buée L., and Delacourte A. (1999) Comparative biochemistry of tau in progressive supranuclear palsy, corticobasal degeneration, FTDP-17 and Pick's disease. Brain Pathol. 9, 681–693 10.1111/j.1750-3639.1999.tb00550.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Friedhoff P., Schneider A., Mandelkow E. M., and Mandelkow E. (1998) Rapid assembly of Alzheimer-like paired helical filaments from microtubule-associated protein tau monitored by fluorescence in solution. Biochemistry 37, 10223–10230 10.1021/bi980537d [DOI] [PubMed] [Google Scholar]
- 71. Tacik P., DeTure M., Lin W.-L., Sanchez Contreras M., Wojtas A., Hinkle K. M., Fujioka S., Baker M. C., Walton R. L., Carlomagno Y., Brown P. H., Strongosky A. J., Kouri N., Murray M. E., Petrucelli L., et al. (2015) A novel tau mutation, p.K317N, causes globular glial tauopathy. Acta Neuropathol. 130, 199–214 10.1007/s00401-015-1425-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sibille N., Sillen A., Leroy A., Wieruszeski J.-M., Mulloy B., Landrieu I., and Lippens G. (2006) Structural impact of heparin binding to full-length Tau as studied by NMR spectroscopy. Biochemistry 45, 12560–12572 10.1021/bi060964o [DOI] [PubMed] [Google Scholar]
- 73. Carmel G., Mager E. M., Binder L. I., and Kuret J. (1996) The structural basis of monoclonal antibody Alz50's selectivity for Alzheimer's disease pathology. J. Biol. Chem. 271, 32789–32795 10.1074/jbc.271.51.32789 [DOI] [PubMed] [Google Scholar]
- 74. Jicha G. A., Bowser R., Kazam I. G., and Davies P. (1997) Alz-50 and MC-1, a new monoclonal antibody raised to paired helical filaments, recognize conformational epitopes on recombinant tau. J. Neurosci. Res. 48, 128–132 [DOI] [PubMed] [Google Scholar]
- 75. Jicha G. A., Berenfeld B., and Davies P. (1999) Sequence requirements for formation of conformational variants of tau similar to those found in Alzheimer's disease. J. Neurosci. Res. 55, 713–723 [DOI] [PubMed] [Google Scholar]
- 76. Bibow S., Mukrasch M. D., Chinnathambi S., Biernat J., Griesinger C., Mandelkow E., and Zweckstetter M. (2011) The dynamic structure of filamentous tau. Angew. Chem. Int. Ed. Engl. 50, 11520–11524 10.1002/anie.201105493 [DOI] [PubMed] [Google Scholar]
- 77. Nogales E., Wolf S. G., and Downing K. H. (1998) Structure of the αβ tubulin dimer by electron crystallography. Nature 391, 199–203 10.1038/34465 [DOI] [PubMed] [Google Scholar]
- 78. Makrides V., Shen T. E., Bhatia R., Smith B. L., Thimm J., Lal R., and Feinstein S. C. (2003) Microtubule-dependent oligomerization of tau. Implications for physiological tau function and tauopathies. J. Biol. Chem. 278, 33298–33304 10.1074/jbc.M305207200 [DOI] [PubMed] [Google Scholar]
- 79. Duan A. R., and Goodson H. V. (2012) Taxol-stabilized microtubules promote the formation of filaments from unmodified full-length Tau in vitro. Mol. Biol. Cell 23, 4796–4806 10.1091/mbc.e12-05-0374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Elbaum-Garfinkle S., Cobb G., Compton J. T., Li X.-H., and Rhoades E. (2014) Tau mutants bind tubulin heterodimers with enhanced affinity. Proc. Natl. Acad. Sci. U.S.A. 111, 6311–6316 10.1073/pnas.1315983111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Qiang L., Sun X., Austin T. O., Muralidharan H., Jean D. C., Liu M., Yu W., and Baas P. W. (2018) Tau does not stabilize axonal microtubules but rather enables them to have long labile domains. Curr. Biol. 28, 2181–2189.e4 10.1016/j.cub.2018.05.045 [DOI] [PubMed] [Google Scholar]
- 82. Baas P. W., and Qiang L. (2019) Tau: it's not what you think. Trends Cell Biol. 29, 452–461 10.1016/j.tcb.2019.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sato C., Barthélemy N. R., Mawuenyega K. G., Patterson B. W., Gordon B. A., Jockel-Balsarotti J., Sullivan M., Crisp M. J., Kasten T., Kirmess K. M., Kanaan N. M., Yarasheski K. E., Baker-Nigh A., Benzinger T. L. S., Miller T. M., et al. (2018) Tau kinetics in neurons and the human central nervous system. Neuron 97, 1284–1298.e7 10.1016/j.neuron.2018.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Prabakaran S., Lippens G., Steen H., and Gunawardena J. (2012) Post-translational modification: nature's escape from genetic imprisonment and the basis for dynamic information encoding. Wiley Interdiscip. Rev. Syst. Biol. Med. 4, 565–583 10.1002/wsbm.1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Smith L. M., Kelleher N. L., and Consortium for Top Down Proteomics (2013) Proteoform: a single term describing protein complexity. Nat. Methods 10, 186–187 10.1038/nmeth.2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Despres C., Byrne C., Qi H., Cantrelle F.-X., Huvent I., Chambraud B., Baulieu E.-E., Jacquot Y., Landrieu I., Lippens G., and Smet-Nocca C. (2017) Identification of the Tau phosphorylation pattern that drives its aggregation. Proc. Natl. Acad. Sci. U.S.A. 114, 9080–9085 10.1073/pnas.1708448114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Min S.-W., Cho S.-H., Zhou Y., Schroeder S., Haroutunian V., Seeley W. W., Huang E. J., Shen Y., Masliah E., Mukherjee C., Meyers D., Cole P. A., Ott M., and Gan L. (2010) Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron 67, 953–966 10.1016/j.neuron.2010.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Cohen T. J., Guo J. L., Hurtado D. E., Kwong L. K., Mills I. P., Trojanowski J. Q., and Lee V. M. Y. (2011) The acetylation of tau inhibits its function and promotes pathological tau aggregation. Nat. Commun. 2, 252 10.1038/ncomms1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kamah A., Huvent I., Cantrelle F.-X., Qi H., Lippens G., Landrieu I., and Smet-Nocca C. (2014) Nuclear magnetic resonance analysis of the acetylation pattern of the neuronal Tau protein. Biochemistry 53, 3020–3032 10.1021/bi500006v [DOI] [PubMed] [Google Scholar]
- 90. Liu F., Iqbal K., Grundke-Iqbal I., Hart G. W., and Gong C.-X. (2004) O-GlcNAcylation regulates phosphorylation of tau: a mechanism involved in Alzheimer's disease. Proc. Natl. Acad. Sci. U.S.A. 101, 10804–10809 10.1073/pnas.0400348101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhu Y., Shan X., Yuzwa S. A., and Vocadlo D. J. (2014) The emerging link between O-GlcNAc and Alzheimer disease. J. Biol. Chem. 289, 34472–34481 10.1074/jbc.R114.601351 [DOI] [PMC free article] [PubMed] [Google Scholar]