Abstract
Under oxidative stress conditions, hydroxyl radicals can oxidize the phenyl ring of phenylalanine, producing the abnormal tyrosine isomer meta-tyrosine (m-tyrosine). m-Tyrosine levels are commonly used as a biomarker of oxidative stress, and its accumulation has recently been reported to adversely affect cells, suggesting a direct role for m-tyrosine in oxidative stress effects. We found that the Caenorhabditis elegans ortholog of tyrosine aminotransferase (TATN-1)—the first enzyme involved in the metabolic degradation of tyrosine—is up-regulated in response to oxidative stress and directly activated by the oxidative stress–responsive transcription factor SKN-1. Worms deficient in tyrosine aminotransferase activity displayed increased sensitivity to multiple sources of oxidative stress. Biochemical assays revealed that m-tyrosine is a substrate for TATN-1–mediated deamination, suggesting that TATN-1 also metabolizes m-tyrosine. Consistent with a toxic effect of m-tyrosine and a protective function of TATN-1, tatn-1 mutant worms exhibited delayed development, marked reduction in fertility, and shortened lifespan when exposed to m-tyrosine. A forward genetic screen identified a mutation in the previously uncharacterized gene F01D4.5—homologous with human transcription factor 20 (TCF20) and retinoic acid–induced 1 (RAI1)—that suppresses the adverse phenotypes observed in m-tyrosine–treated tatn-1 mutant worms. RNA-Seq analysis of F01D4.5 mutant worms disclosed a significant reduction in the expression of specific isoforms of genes encoding ribosomal proteins, suggesting that alterations in protein synthesis or ribosome structure could diminish the adverse effects of m-tyrosine. Our findings uncover a critical role for tyrosine aminotransferase in the oxidative stress response via m-tyrosine metabolism.
Keywords: oxidative stress, tyrosine, aging, amino acid, gene expression, meta-tyrosine, ribosome, tyrosine aminotransferase, metabolism, F01D4.5, reactive oxygen species, Nrf2, SKN-1, transcriptional regulation
Introduction
The damage to cellular components by reactive oxygen and nitrogen species, termed oxidative stress, has long been implicated in aging and age-related diseases (1–9). Whereas much is now known regarding the biochemical mechanisms by which oxidative stress produces damage to molecules including lipids (10), nucleic acids (11), and proteins (12), as well as the cellular responses to counteract these effects (13), recent work has suggested that our understanding of the mechanisms by which cells defend against the adverse effects of oxidative stress and the connections between oxidative stress and aging is incomplete. Specifically, work in mice and other model animals has shown that even animals lacking critical components of the oxidative stress defenses can age normally and resist the development of age-related disease (14–16). These findings could either suggest that oxidative stress is less biologically important than previously believed or that new types of antioxidant defenses still remain to be discovered.
At the cellular level, the response to oxidative stress is diverse, with the activation of multiple pathways (17, 18). One of the major defense responses is the induction of genes encoding phase II enzymes by nuclear factor E2–related factor 2 (Nrf2) (19). Nrf2 is part of the cap 'n' collar family of basic region-leucine zipper (bZip) motif transcription factors and binds to the antioxidant response element (ARE)4 within the promoter regions of various genes, thus inducing their expression (20–22). Under normal conditions, Nrf2 is bound by Kelch-like ECH-associated protein 1 (Keap1) and sequestered in the cytoplasm, where it is ubiquinated and targeted for rapid degradation; however, under conditions of oxidative stress, cysteine residues both within Nrf2 and Keap1 sense electrophilic inducers, and the interaction with Nrf2 is terminated (23–25). This allows Nrf2 to enter the nucleus and, together with Maf2, bind to the ARE to induce key detoxification genes (21).
This mechanism is conserved across species, including Caenorhabditis elegans, where the SKN-1 transcription factor is the ortholog of Nrf2 (26, 27). Whereas worms lack a direct homolog of Keap1, WDR-23—a WD40 repeat protein—serves a similar function in regulating SKN-1 nuclear accumulation and activity (28). After exposure to oxidative stress or xenobiotics, SKN-1 rapidly translocates to the nucleus, particularly within intestinal cells, where it induces the expression of GSH synthetase, NADH quinone oxidoreductase, superoxide dismutase, and catalase, among others (27).
Our interest in a possible function of tyrosine aminotransferase within the oxidative stress response stemmed from previous work that showed the garlic constituent diallyl trisulfide is able to extend the lifespan of C. elegans in a skn-1–dependent manner (29). Interestingly, when comparing microarray data from worms treated with diallyl trisulfide and worms that were subjected to oxidative stress produced by hyperbaric oxygen, the C. elegans tyrosine aminotransferase homolog tatn-1 was observed to be overexpressed in both data sets (29).
Tyrosine aminotransferase catalyzes the conversion of tyrosine to 4-hydroxyphenylpyruvate and is the rate-limiting step in the tyrosine degradation pathway—the pathway by which tyrosine is metabolized to fumarate and acetoacetate (Fig. 1A) (30, 31). In relation to oxidative stress, multiple studies have reported a potential enhancement of oxidative stress with either a reduction in tyrosine aminotransferase or administration of tyrosine (32–37). Furthermore, rats treated with mercury had greater tyrosine aminotransferase activity subsequent to higher transcriptional expression (38), and heat stress resulted in increased tyrosine aminotransferase activity in chickens (39). Together, these data suggest a role for tyrosine aminotransferase in response to stress. However, the mechanisms involved are unclear.
Figure 1.
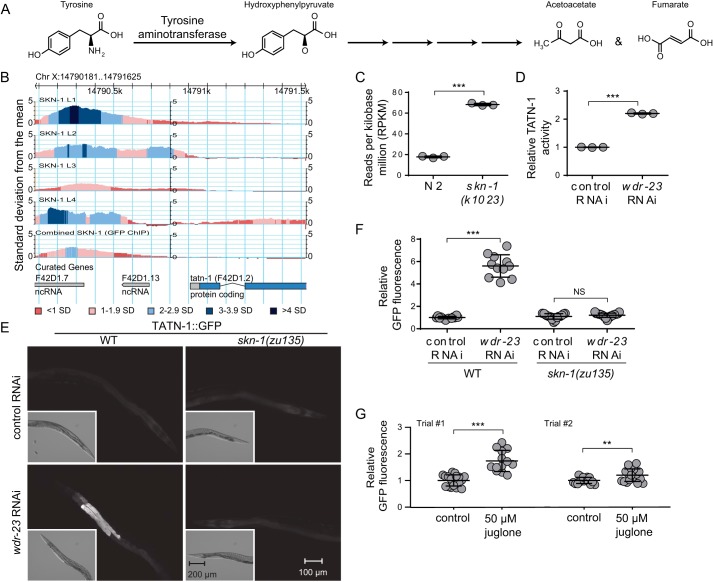
SKN-1 up-regulates TATN-1 expression and enzymatic activity. A, schematic of the tyrosine degradation pathway showing tyrosine metabolism to hydroxyphenylpyruvate by tyrosine aminotransferase as the first step. B, ChIP-Seq data reveal a significant peak of SKN-1 binding to the genomic DNA within the tatn-1 promoter during multiple stages of development. The y axis represents the level of SKN-1 binding in terms of the number of S.D. values above the mean genome-wide level of SKN-1 binding. Colors indicate genomic regions within each S.D. range. C, RNA-Seq data show a significant increase in the number of tatn-1 transcripts in a gain-of-function skn-1 mutant compared with WT C. elegans. Shown are the mean reads per kilobase million and S.D. values from each of three biological replicates. ***, p < 0.001 by t test. D, activation of SKN-1 with wdr-23 RNAi treatment increases tyrosine aminotransferase enzymatic activity. Relative TATN-1 activity was measured in protein lysates from adult N2 worms treated with control or wdr-23 RNAi. Shown are the mean enzymatic activity and S.D. of wdr-23 RNAi-treated worms relative to control RNAi-treated worms for each of three biologic replicates. ***, p < 0.001 by t test. E, activation of SKN-1 with wdr-23 RNAi treatment increases TATN-1 expression. Shown are representative photomicrographs of adult WT or skn-1 loss-of-function mutant transgenic worms expressing a TATN-1::GFP fusion protein, produced by a tatn-1p::tatn-1::GFP transgene, that were treated with control or wdr-23 RNAi. Note the increase in GFP fluorescence in the worm intestine following wdr-23 RNAi treatment. All images are captured at similar magnification. Scale bar, 100 μm (larger fluorescent images) and 200 μm (inset Nomarski images). F, quantification of relative TATN-1::GFP fluorescence in WT or skn-1 mutant worms treated with control or wdr-23 RNAi. Mean fluorescence intensity relative to control RNAi–treated worms and S.D. for each group are shown (n = 11–21). ***, p < 0.001; NS, nonsignificant by a one-way ANOVA. G, quantification of relative TATN-1::GFP fluorescence in WT worms treated with 50 μm juglone dissolved in M9 or M9 alone as a control. Mean fluorescence intensity relative to control-treated worms and S.D. for each group are shown (n = 13–19). ***, p < 0.001; **, p = 0.0031 by t test.
Here, we explore a novel role for tyrosine aminotransferase within the cellular oxidative stress response by metabolizing meta-tyrosine, thus preventing its accumulation and the subsequent consequences that are detrimental to cells and organisms. Work over the past few decades has revealed that under conditions of oxidative stress, the production of the abnormal tyrosine isomer m-tyrosine may occur when hydroxyl radicals oxidize the phenyl ring of phenylalanine (40–42). Whereas elevations in m-tyrosine concentrations were previously perceived to simply be a biological marker of oxidative stress (43), there is emerging evidence from bacterial (44, 45), yeast (46), plant (47, 48), and mammalian studies (49–55) suggesting that m-tyrosine directly produces adverse effects in cells and may contribute to oxidative stress-related pathologies (56). How m-tyrosine is harmful to cells is not entirely clear, but studies have shown that it can be erroneously charged to phenylalanyl-tRNA, which then leads to the substitution of m-tyrosine for phenylalanine during protein synthesis (46, 57–59). This could result in adverse changes to protein structure and activity, but the downstream events triggered by these substitutions remain unexplored. Nevertheless, the elimination of m-tyrosine by metabolic pathways could serve a purpose in the oxidative stress response (56).
In this work, we find that TATN-1 protein expression is controlled by the skn-1 transcription factor and increases after exposure to oxidative stress. This induction of TATN-1 plays an active role in the response to oxidative stress, as a tatn-1 mutant is sensitive to both external and internal sources of oxidative stress. The activation of TATN-1 may serve to protect animals against the accumulation of m-tyrosine, as we show that both p-tyrosine and m-tyrosine can serve as substrates for deamination by TATN-1. Consistently, only the tatn-1 mutant accumulates m-tyrosine after being fed an m-tyrosine containing diet, and this feeding of m-tyrosine to the tatn-1 mutant leads to a selective developmental delay, reduction in fertility, and lifespan shortening compared with WT animals. Together, our work suggests a novel role for tyrosine aminotransferase in the oxidative stress response through the metabolism of m-tyrosine produced by phenylalanine oxidation.
Results
tatn-1 expression is controlled by SKN-1
By re-analyzing the ChIP-Seq data of Niu et al. (60), we discovered that the SKN-1 transcription factor binds to the proximal promoter of the tatn-1 gene with peak binding that is up to 4 S.D. values greater than the local mean level (Fig. 1B). Consistent with the possible regulation of tatn-1 expression by SKN-1, the recently published RNA-Seq data of Peddibhotla et al. (61) demonstrate that tatn-1 expression is increased in worms carrying a gain-of-function skn-1 mutation, which promotes the expression of SKN-1 target genes (Fig. 1C). These results suggest that the activation of SKN-1 should lead to an increase in tyrosine aminotransferase levels in worms. To test this hypothesis, we measured tyrosine aminotransferase activity from the lysates of WT N2 worms treated with either wdr-23 or control RNAi. The WDR-23 protein binds to SKN-1 and acts to inhibit its translocation to the nucleus. Treatment with wdr-23 RNAi relieves this inhibition and has previously been shown to lead to robust expression of SKN-1 target genes (28). Treatment with wdr-23 RNAi results in a significant increase in TATN-1 enzymatic activity, which is more than double that seen in worms fed control RNAi (Fig. 1D).
To determine whether the increase in tyrosine aminotransferase activity reflected an increase in the level of TATN-1 protein, we utilized transgenic worms with a tatn-1p::tatn-1::GFP transgene that had previously been integrated into the C. elegans genome (62). We found that treatment of these worms with wdr-23 RNAi led to a nearly 5-fold increase in the expression of TATN-1::GFP compared with the level seen in worms treated with control RNAi (Fig. 1, E and F). To determine whether this increase in TATN-1::GFP with wdr-23 RNAi depends on SKN-1, we crossed the transgene into worms carrying the skn-1(zu135) loss-of-function mutation. In contrast to WT worms, we found that skn-1 homozygous mutants failed to induce the expression of TATN-1::GFP after treatment with wdr-23 RNAi (Fig. 1, E and F). Together, these results suggest tatn-1 expression is directly controlled by SKN-1.
We then tested whether exposure to oxidative stress, which is an environmental activator of SKN-1, would also lead to increases in TATN-1::GFP expression. We treated worms for 1 h with 50 μm juglone, which is a protocol known to induce the expression of the SKN-1 target gene gst-4 (28). We found that this treatment led to an increase in TATN-1::GFP expression in two separate trials, although the level of increase did differ between the experiments (Fig. 1G). Hence, the activation of SKN-1 via either genetic or environmental conditions results in the induction of TATN-1 expression.
tatn-1 mutants are sensitive to the effects of oxidative stress
Given the critical role played by SKN-1 in the oxidative stress response (63, 64), we next sought to test whether TATN-1 played a biologically significant role in the oxidative stress response by assessing the sensitivity of tatn-1 mutants to endogenous and exogenous sources of oxidative stress. Because a tatn-1 null mutation is not viable, we utilized tatn-1(qd182) mutant worms that have a hypomorphic (G171E) missense mutation, which reduces the tyrosine aminotransferase activity level in mutant worm extracts relative to WT animals by ∼90% (Fig. S1A) and leads to an increase in worm tyrosine levels as shown by MS (62). To assay the sensitivity of the tatn-1 mutant worms to exogenous stress, we used an established developmental assay, in which WT N2 and tatn-1(qd182) embryos were placed on nematode growth agar (NGA) plates supplemented with paraquat—an organic compound that produces superoxide anions in vivo—and we measured the time for each strain to develop to reproductive adults (65, 66). Whereas tatn-1 mutants are slightly delayed compared with N2 worms on control plates without paraquat (Fig. S2), they are significantly more delayed compared with N2 worms when treated with 0.4 mm paraquat, with only 4.9% developing into fertile adults by day 7 compared with 93.3% of N2 worms doing so (Fig. 2A). Similar results are seen when N2 worms are treated with paraquat and tatn-1 RNAi (Fig. 2A), which reduces TATN-1 activity by ∼75% (Fig. S1B).
Figure 2.
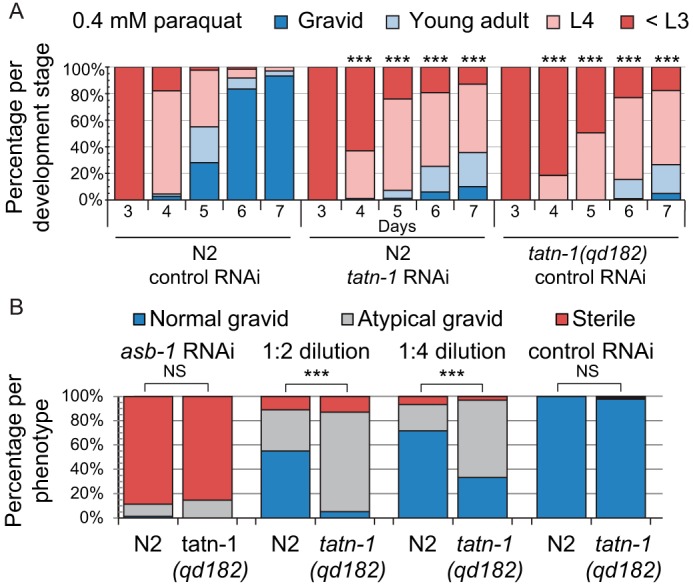
Reduction in TATN-1 activity renders worms sensitive to oxidative stress. A, the treatment of either N2 WT worms, tatn-1 mutant worms, or N2 worms treated with tatn-1 RNAi with 0.4 mm paraquat produces a developmental delay, but both the tatn-1 mutants and tatn-1 RNAi–treated animals show increased sensitivity to this source of oxidative stress. Shown are the percentage of animals in each larvae stage and reproductive stage during a developmental assay. n = ∼150 worms per genotype or RNAi treatment. ***, p < 0.001 by χ2 test for the indicated treatment and day relative to N2 animals treated with control RNAi on the same day. B, tatn-1 mutant animals are more sensitive to the adverse effects of asb-1 RNAi on fertility. Shown are the percentage of animals exhibiting each reproductive phenotype (sterile (0 eggs observed), abnormal gravid (<6 eggs), or normal gravid (≥6 eggs)) for N2 or tatn-1 mutant worms treated with the indicated dilution of asb-1 RNAi. n = ∼150 worms per genotype and treatment. ***, p < 0.001; NS, nonsignificant by χ2 test comparing tatn-1 mutant worms with N2 worms treated with the same dilution of asb-1 RNAi.
To address sensitivity to endogenous oxidative stress, tatn-1 mutants and N2 worms were treated with asb-1 RNAi. The asb-1 gene encodes a germline-specific isoform of the mitochondrial ATP synthase b subunit, and asb-1 RNAi has been shown to increase the levels of H2O2 within the cytosol of the germline and cause sterility (67, 68). Treating either N2 or tatn-1 mutant worms with undiluted asb-1 RNAi indeed resulted in sterility in both strains (Fig. 2B). However, dilution of the asb-1 RNAi with control RNAi reduced the adverse effects on the germline and revealed an increased sensitivity of tatn-1 mutants to the effects of asb-1 RNAi (Fig. 2B). Whereas asb-1 RNAi treatment has been shown to increase intracellular H2O2 levels comparable with those seen in paraquat-treated worms (68), it is unclear whether its effect on germline development and sterility is a result of increased ROS or reduced ATP production by the germline mitochondria. Thus, reductions in ATP levels could potentially contribute to the germline phenotypes observed with asb-1 RNAi treatment, and we did not conduct experiments to exclude this possibility.
Both p- and m-tyrosine are substrates for TATN-1
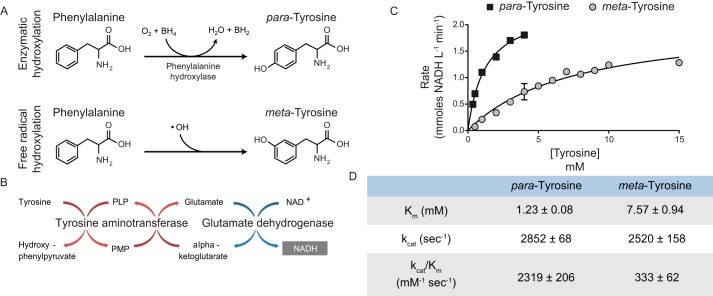
Given our findings that TATN-1 is induced by SKN-1 and contributes to oxidative stress defenses of the worm, we hypothesized that TATN-1 could act to metabolize m-tyrosine formed under conditions of oxidative stress (Fig. 3A). However, there have been no reports assessing whether m-tyrosine, in addition to p-tyrosine, is a substrate of TATN-1. To assess the ability of TATN-1 to convert m-tyrosine into the less toxic metabolite 3-hydroxyphenylpyruvic acid (47), we semi-purified C. elegans TATN-1 that had been expressed in bacteria for an in vitro assay (Fig. S3). In this assay, the tyrosine aminotransferase reaction was coupled with glutamate dehydrogenase, which allowed us to measure the formation of NADH via spectrophotometry when varying concentrations of the tyrosine isomers and phenylalanine were added to the reaction (Fig. 3, B–D). The Km for p-tyrosine (1.23 ± 0.08 mm) is similar to that reported for tyrosine aminotransferase from other species, including Mus musculus, Rattus norvegicus, Felis catus, and Trypanosoma cruzi (69–71). Despite the Km for m-tyrosine (7.57 ± 0.94 mm) being higher than p-tyrosine, TATN-1 maintains substantial activity for m-tyrosine as a substrate, particularly when considering that no detectable reaction was observed with phenylalanine at the concentrations tested.
Figure 3.
TATN-1 maintains activity with m-tyrosine as a substrate. A, schematic of the enzymatic (top) and free radical–mediated (bottom) hydroxylation of phenylalanine that results in the formation of the tyrosine isomers para- and meta-tyrosine. B, diagram of the tyrosine aminotransferase/glutamate dehydrogenase coupled reaction used to measure TATN-1 kinetics. In this reaction, glutamate, which is a product of the tyrosine aminotransferase reaction, is converted back to α-ketoglutarate by glutamate dehydrogenase. NAD+ serves as a cofactor for the glutamate dehydrogenase reaction and is reduced to NADH, which absorbs light at a wavelength of 340 nm and can be measured by spectrophotometry. C, Michaelis–Menten plots for TATN-1 activity with para- and meta-tyrosine as substrates. The initial velocities of the reaction at each of the indicated substrate concentrations was measured in triplicate. Mean rates of enzymatic activity and S.E. (error bar) are shown for both tyrosine isomers. D, the calculated kinetic parameters for TATN-1 with either p- or m-tyrosine as a substrate. Ranges represent S.E.
It should be noted that the turnover number (kcat) for TATN-1 with p-tyrosine as a substrate is considerably higher than what has been previously published for tyrosine aminotransferase in various species (69–71). However, this difference may be due to the presence of catalytically active degradation products of TATN-1 following affinity purification as a GST fusion protein (Fig. S3). Thus, the true concentration of TATN-1 catalytic sites may be underestimated for this calculation. However, as the same preparation of TATN-1 from Escherichia coli was used for all experiments, the enzyme concentration remained constant for each experiment, and the relative TATN-1 reaction rate with p-tyrosine, m-tyrosine, and phenylalanine could still be adequately assessed.
tatn-1 mutants are sensitive to the adverse effects of m-tyrosine
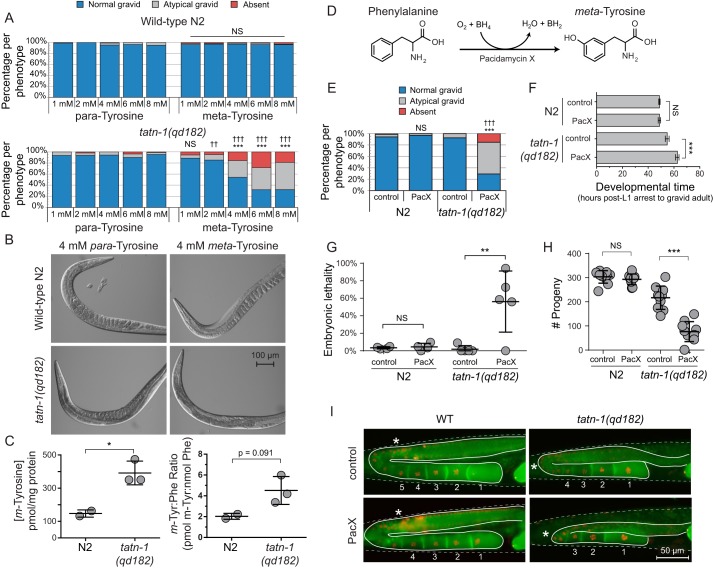
To determine whether metabolism via TATN-1 is an important mechanism that protects C. elegans from the effects of m-tyrosine, N2 WT and tatn-1 mutant worms were treated with varying concentrations of m-tyrosine that had been supplemented in the agar plates used for worm culture. Compared with WT worms, tatn-1 mutants treated with m-tyrosine show a concentration-dependent phenotype characterized by a reduction in the number of embryos present in day 1 adult hermaphrodites compared with age-matched controls (Fig. 4, A and B). This phenotype was not simply due to increased tyrosine levels in general, but is specific to m-tyrosine, as tatn-1 mutants treated with equal concentrations of p-tyrosine failed to produce this phenotype (Fig. 4, A and B).
Figure 4.
tatn-1 mutants are sensitive to the adverse effects of m-tyrosine. A, treatment of N2 and tatn-1(qd182) mutant worms with varying concentrations of either p- or m-tyrosine revealed a decrease in fertility of the tatn-1 mutants supplemented with m-tyrosine. Approximately 150 worms were scored per treatment, and the percentage of worms showing each reproductive phenotype—based on the number of eggs in the uterus of day 1 adult animals with normal gravid showing ≥6 embryos within the uterus, abnormal gravid showing <6 embryos, and absent showing zero embryos—are graphed. ***, p < 0.001 for χ2 test of the effects of equal concentrations of m- and p-tyrosine within the same strain. ††, p < 0.01; †††, p < 0.001 for χ2 test of the effects of m-tyrosine treatment of equal concentrations between strains. B, representative images of worms quantified in A. C, results of LC/MS-MRM quantification of the concentration of free m-tyrosine and the m-tyrosine/phenylalanine ratio for N2 and tatn-1(qd182) mutant worms grown on NGA plates supplemented with 4 mm m-tyrosine. Approximately 1500 worms were collected per biological replicate, and the amino acid concentrations were normalized to the concentration of soluble worm protein measured per sample. *, p < 0.05 by t test. D, diagram of the enzymatic formation of m-tyrosine from phenylalanine by the pacidamycin X enzyme produced by Streptomyces coeruleoribudus. E, treatment of N2 and tatn-1(qd182) mutant worms fed bacteria expressing PacX or an empty vector control revealed a decrease in fertility in the tatn-1 mutants treated with PacX-expressing bacteria. Scoring and statistical analysis were performed as in A. F, results of a developmental time assay in which the time required for L1 arrested N2 and tatn-1(qd182) animals treated with control or PacX-expressing bacteria to become gravid adults is measured. 4–5 worms were assayed per condition. Mean developmental time and S.D. (error bars) are shown. ***, p < 0.001; NS, nonsignificant p value by t test. G, measurement of the percentage of embryonic lethality in the worms from F treated with either control or PacX-expressing bacteria demonstrates an increase only in the tatn-1 mutants treated with the PacX-expressing bacteria. Shown are the mean percentage of embryonic lethality from each animal and the S.D. ***, p < 0.001; NS, nonsignificant p value by t test. H, measurement of the fertility of N2 and tatn-1(qd182) worms treated with control or PacX-expressing bacteria. n = 9–10 worms assayed per genotype and treatment. Shown are the mean number of progeny and S.D. for each genotype-condition pair. ***, p < 0.001; NS, nonsignificant p value by t test. I, representative images of WT and tatn-1(qd182) transgenic worms expressing fluorescent proteins that label the cell membrane (green) and nucleus (red) of the developing oocytes. The germline is outlined with a solid line, the worm body is outlined with a dashed line, mature oocytes are numbered, and a star marks the beginning of oocyte expansion. The WT worms exhibit normal gonadal architecture regardless of treatment. The control-treated tatn-1 mutant worm shows a delay in oocyte expansion. The PacX-treated tatn-1 mutant also shows a delay in oocyte expansion along with a reduction in mature oocytes and a defect where the germline fails to cross the midline.
To test whether these phenotypes could be the result of greater m-tyrosine accumulation in the tatn-1 mutants compared with the WT animals, we prepared extracts for analysis by LC/MS. This analysis demonstrated that the levels of free m-tyrosine were nearly 2-fold higher in tatn-1 mutants compared with N2 worms when treated with 4 mm m-tyrosine (Fig. 4C). Because m-tyrosine could compete with phenylalanine for charging to phenylalanyl-tRNA (46, 49, 52, 58, 59), we asked whether there may be an increase in free phenylalanine levels due to a reduction in tRNA charging after m-tyrosine supplementation. However, we observed no significant change in free phenylalanine concentrations in either strain with m-tyrosine treatment (Fig. S4). Thus, the m-tyrosine/phenylalanine ratio is elevated in tatn-1 mutants administered m-tyrosine; however, due to our low sample size and imprecise measurements, this clear trend failed to reach significance (Fig. 4C).
Besides administering m-tyrosine to the worms via supplementation in the worm medium, which is costly due to the price of the chemical isomer, we also performed experiments in which worms fed on E. coli expressing a plasmid that codes for the pacidamycin X (PacX) enzyme. Unlike the phenylalanine hydroxylase present in most other species—including humans and C. elegans—that selectively hydroxylates phenylalanine at the 4- or para position to produce p-tyrosine (76, 77), PacX is a phenylalanine 3-hydroxylase from Streptomyces coeruleoribudus that instead produces m-tyrosine (Fig. 4D) (78). Feeding tatn-1 mutant worms bacteria expressing PacX produces a phenotype similar to that resulting from m-tyrosine supplementation to NGA medium (Fig. 4E and Fig. S5). It should be noted that we were unable to detect free m-tyrosine in either N2 or tatn-1(qd182) worms via LC/MS, but this may reflect a difference in m-tyrosine uptake from the intestine and delivery to cells between m-tyrosine supplementation in the NGA medium and the consumption of PacX-expressing bacteria.
To determine whether the phenotypes produced by m-tyrosine treatment are a result of a developmental delay, a loss of fertility, or other effects on the worms, we conducted additional experiments to measure development time, percentage of embryonic lethality, and fecundity. PacX treatment significantly increased both the developmental time and percentage of embryonic lethality of tatn-1 mutant worms compared with controls (Fig. 4, F and G). In contrast, PacX treatment had no statistically significant effect on N2 development rate or the percentage of embryonic lethality. Perhaps as result of the increase in embryonic lethality, treatment with PacX also significantly reduced the total number of progeny produced by tatn-1 mutants (Fig. 4H). No difference in the number of progeny was observed in N2 worms with PacX treatment (Fig. 4H).
To determine the nature of the germline defects in tatn-1 mutant worms treated with m-tyrosine, we utilized the transgenic OD95 worm strain, which carries transgenes expressing fluorescent proteins to mark the nuclei and cell membranes of germ cells (79–82). This strain has been utilized extensively to characterize alterations in gonadal morphology that occur with RNAi treatment targeting genes linked to sterility (83). Whereas there were no obvious differences between WT worms treated with PacX or control, there were differences in the tatn-1 background. Both control and PacX-treated tatn-1 mutant worms exhibited a delay in oocyte expansion. Because delayed expansion represents a transitional state between L4 and young adults, this finding is consistent with the observed overall delay in organismal development (Fig. 4F). Further, this observation may account for the slight reduction in progeny number for the control-treated tatn-1 mutants, when compared with control-treated N2 worms (Fig. 4H). However, the PacX-treated tatn-1 mutant worms exhibited a range of additional stochastic defects in two independent experiments (Fig. 4I and Data set S1). The most prominent phenotypic defects observed were a marked reduction in the number of oocytes formed and distal tip extension and/or a germline pathfinding defect, where the germline failed to cross the midline. In an attempt to identify potential genes or proteins that may be affected by m-tyrosine treatment, we utilized the phenotypic profiling data from Green et al. (83) and sought to match the phenotypes observed in tatn-1 mutants treated with PacX with the phenotypic profiles of genes associated with sterility. The range of phenotypes observed in the tatn-1 mutants were not diagnostic enough to reveal particular genes or pathways that could be specifically linked to our observed phenotype with m-tyrosine treatment. However, oocyte expansion defects tend to be broadly observed among RNAi conditions targeting regulators of translation (see J-class (76)). However, additional work is needed to better define the nature and cause(s) of the irregular gonadal architecture.
We then examined whether the germline defects produced in this strain by PacX treatment would also be seen when m-tyrosine is delivered by direct feeding of the chemical. We treated strains carrying the reporters in both a WT and tatn-1 background, and we found that 4 mm m-tyrosine treatment produced similar phenotypes to PacX treatment but only in the tatn-1 mutant background (Data set S1). Interestingly, when 4 mm p-tyrosine treatment was used as a comparison, we did see similar germline phenotypes as with m-tyrosine treatment but of lesser severity and with lower penetrance (Data set S1). This could reflect direct actions of tyrosine, either p or m, on germline growth and development, or it could reflect an indirect increase in m-tyrosine levels as a result of the higher p-tyrosine levels outcompeting endogenously formed m-tyrosine for the available TATN-1 enzyme.
F01D4.5 mutation suppresses the adverse effects of oxidative stress and m-tyrosine treatment in tatn-1 mutant worms
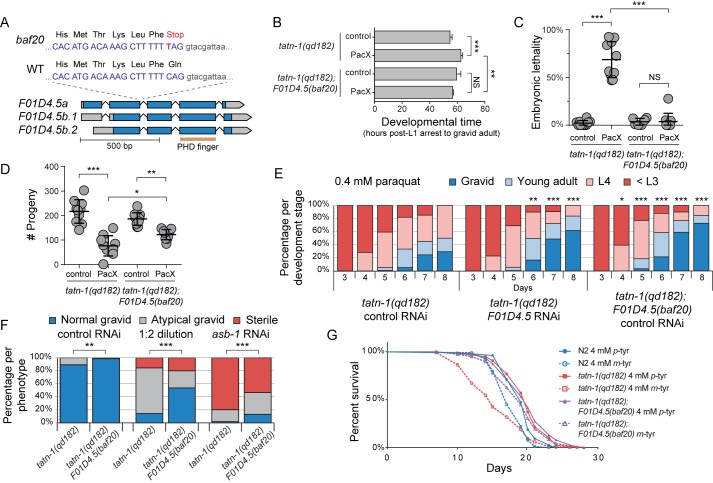
Because the mechanism(s) by which m-tyrosine is harmful to cells and tissues is incompletely understood, we used a forward genetic screen using the chemical mutagen ethyl methanesulfonate (EMS) to identify genes involved in the effects of m-tyrosine on the tatn-1 mutants (72, 84, 85). We mutagenized tatn-1(qd182) mutant worms and then screened for worms showing resistance to PacX treatment as indicated by a normal number of embryos formed within the worms on day 1 of adulthood. From this screen, we identified a strain that appeared to be highly resistant to PacX treatment and, via genomic DNA sequencing, mapped the causative mutation to the uncharacterized gene F01D4.5, which had a C to T substitution that creates a nonsense mutation early in the protein sequence—altering the 97th codon of the F01D4.5a isoform and the 56th codon of the F01D4.5b isoform (Fig. 5A and Fig. S6). The F01D4.5 protein shares homology with two human proteins—transcription factor 20 (TCF20) and retinoic acid-induced protein 1 (RAI1). This homology is principally due to a predicted plant homeodomain (PHD) finger motif within F01D4.5 that is present in both RAI1 and TCF20 and enables these proteins to bind nucleosomes (86, 87).
Figure 5.
F01D4.5 mutation suppresses the adverse effects of oxidative stress and m-tyrosine treatment in tatn-1 mutant worms. A, diagram depicting the various isoforms F01D4.5, the location of its predicted PHD finger domain, and the nature of the mutation isolated from the EMS mutagenesis screen. B, the F01D4.5 mutation reduces the development time for tatn-1(qd182) mutant worms treated with PacX-expressing bacteria to become gravid adults. Shown are the mean developmental time and S.D. (error bars) for 4–5 worms per genotype-treatment pair to develop from an arrested L1 larvae into a reproductive adult. **, p < 0.01; ***, p < 0.001; NS, nonsignificant by a one-way ANOVA. C, the F01D4.5 mutation suppresses the increased embryonic lethality of tatn-1(qd182) mutants treated with PacX-expressing bacteria. Shown are the mean percentage of embryonic lethality and S.D. from 6–10 worms assayed per genotype-treatment pair. ***, p < 0.001; NS, nonsignificant by a one-way ANOVA. D, results from a fertility assay showing an increase in the number of progeny produced by tatn-1(qd182); F01D4.5(baf20) worms treated with PacX-expressing bacteria. Shown are the mean progeny produced by 9–10 worms assayed per genotype-treatment pair along with the S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001 by one-way ANOVA. E, either the F01D4.5 mutation or F01D4.5 RNAi reduces the developmental delay produced by treatment with 0.4 mm paraquat, as shown by a developmental assay. Shown are the percentage of worms in the indicated developmental stage on each day with ∼150 worms being scored per genotype and treatment. **, p < 0.01; ***, p < 0.001 for comparison of worms deficient in F01D4.5 as a result of F01D4.5 RNAi treatment or the F01D4.5(baf20) genetic mutation to tatn-1 mutant worms treated with control RNAi of the same age by χ2 test. F, the F01D4.5(baf20) mutation attenuates the sterility resulting from the treatment of tatn-1 mutants with asb-1 RNAi. Approximately 150 worms were scored per genotype and RNAi treatment based on the number of eggs within the uterus of each animal. Shown are the percentage of worms showing each reproductive phenotype with the worms being scored as sterile (zero eggs observed), abnormal gravid (<6 eggs), or normal gravid (≥6 eggs). **, p < 0.01; ***, p < 0.001 for comparison of tatn-1(qd182); F01D4.5(baf20) mutant worms with tatn-1(qd182) worms for each dilution by χ2 test. G, treatment of WT and tatn-1 mutant animals with m-tyrosine reduces worm lifespan, and the F01D4.5 mutation reduces the adverse effect on longevity. Kaplan–Meier survival curves for N2, tatn-1(qd182), and tatn-1(qd182); F01D4.5(baf20) worms on NGA plates supplemented with either 4 mm p- or m-tyrosine. n = 108–122 worms per genotype and treatment. p < 0.0001 for N2 and tatn-1 mutants treated with m-tyrosine versus p-tyrosine, whereas p = 0.06 for tatn-1(qd182); F01D4.5(baf20) treated with m-tyrosine versus p-tyrosine determined by log-rank test. Mean lifespans were as follows: p-tyrosine–treated N2, 19.2 days; m-tyrosine–treated N2, 17.7 days; p-tyrosine–treated tatn-1(qd182), 19.7 days; m-tyrosine–treated tatn-1(qd182), 15.5 days; p-tyrosine–treated tatn-1(qd182); F01D4.5(baf20), 19.9 days; m-tyrosine–treated tatn-1(qd812); F01D4.5(baf20), 18.9 days.
RAI1 is highly expressed in neurons, where it is present both in the cytoplasm and in the nucleus and has been shown to bind to gene promoter regions and induce transcription (88–91). Haploinsufficiency of the RAI1 gene causes the rare genetic disorder Smith–Magenis syndrome, which is characterized by mild to moderate intellectual impairment, distinctive facial features, and behavioral abnormalities (92–94). Duplication of the RAI1 gene region is associated with another genetic disorder, Potocki–Lupski syndrome, which consists of intellectual disability, autistic features, and low body weight (95). In addition to these syndromes, RAI1 mutations have been associated with schizophrenia and autism (88, 96, 97).
TCF20, also known as stromelysin-1 platelet-derived growth factor–responsive element–binding protein (SPBP), is a ubiquitously expressed nuclear protein and functions as a transcriptional coactivator that enhances the activity of multiple transcription factors, including Nrf2, c-Jun, Ets1, Sp1, paired box protein Pax-6, and the androgen receptor (73–75). TCF20 also binds the phosphorylated estrogen receptor α, represses its transcriptional activity, and inhibits proliferation of the ERα-dependent breast cancer cell line MCF7 (98). Interestingly, like RAI1, mutations in TCF20 have also been linked to intellectual disability, autism spectrum disorders, and schizophrenia (99–101).
The F01D4.5 gene has two putative isoforms with the two isoforms sharing 3 exons, one of which encodes the PHD finger. F01D4.5a has an additional exon at the N terminus, and the two isoforms have distinct exons at the C terminus (Fig. 5A). The F01D4.5(baf20) mutation will affect all of the gene isoforms, and, given the location of the nonsense mutation near the N terminus of the protein, it is likely a null allele for F01D4.5.
To explore the function(s) of F01D4.5, we compared the effects of F01D4.5(baf20) on the responses to m-tyrosine treatment and oxidative stress exposure. The developmental time and percentage of embryonic lethality of PacX-treated tatn-1(qd182); F01D4.5(baf20) worms were significantly reduced compared with PacX-treated tatn-1 mutants (Fig. 5, B and C), with these measurements nearing or equaling control levels. Whereas the tatn-1(qd182); F01D4.5(baf20) mutants treated with PacX still produced fewer progeny than controls, they produced significantly more progeny than tatn-1 mutants treated with PacX (Fig. 5D). Additionally, tatn-1(qd182); F01D4.5(baf20) worms were more resistant to the developmental delay effects of paraquat and the sterility effects of asb-1 RNAi compared with tatn-1 mutants alone (Fig. 5, E and F).
Given the potential roles of oxidative stress responses in aging, we also examined the effects of m-tyrosine treatment on worm lifespan. We treated N2, tatn-1(qd182), and tatn-1(qd182); F01D4.5(baf20) worms with m-tyrosine and measured the lifespan of these animals. Whereas the mean lifespan of m-tyrosine–treated tatn-1(qd182) worms was significantly reduced compared with controls, there was not a statistically significant effect observed in tatn-1(qd82); F01D4.5(baf20) worms treated in parallel (Fig. 5G and Fig. S6). The addition of the F01D4.5 mutation did not extend the lifespan of control-treated tatn-1(qd182) animals. N2 mean lifespan was also shortened but to a lesser extent than in tatn-1(qd182) animals and only when treated with m-tyrosine supplemented in the worm media (Fig. 5G and Fig. S7).
These effects were selective to the tatn-1 background, as comparison of WT N2 and the F01D4.5(baf20) mutant showed at most minor changes in developmental time, fertility, or embryonic lethality either with or without PacX treatment (Fig. S8). Similarly, the F01D4.5(baf20) mutation alone only showed modest changes in lifespan compared with WT animals following either treatment with m-tyrosine or p-tyrosine (Fig. S8).
Previous studies have shown that chronic exposure to low levels of oxidative stress produced by paraquat treatment extends C. elegans lifespan (102), so we sought to test whether this oxidative stress treatment could interact with either the tatn-1 or F01D4.5 mutations, which might suggest a role for m-tyrosine in the effects of paraquat on lifespan. We treated tatn-1(qd182) and tatn-1(qd182); F01D4.5(baf20) worms with 0.2 mm paraquat in two independent lifespan experiments with mixed results. In the first trial, paraquat treatment extended the lifespan of WT as well as tatn-1(qd182); F01D4.5(baf20) mutants, but not tatn-1(qd182) mutants. In the second trial, paraquat treatment extended the lifespan of both WT and tatn-1(qd182) mutant worms but produced a greater lifespan extension for the tatn-1(qd182); F01D4.5(baf20) worms (Fig. S9). Although further work is required to fully understand these results, there is at least some evidence to suggest that m-tyrosine produced even by low-dose paraquat treatment could limit C. elegans lifespan, and both tatn-1 and F01D4.5 can influence these effects.
F01D4.5 is broadly expressed and may control the expression of specific isoforms of genes encoding ribosomal proteins
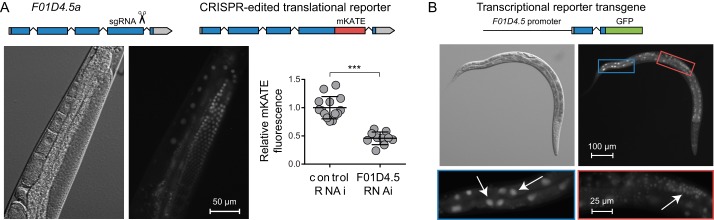
Because the F01D4.5 gene has not been well-studied in worms, we sought to determine which tissues expressed the gene as well as the subcellular localization of the translated protein. Through CRISPR, we created a translational reporter by editing the F01D4.5 gene locus to include a C-terminal tag with the red fluorescent protein mKate2. Fluorescent microscopy of the worms harboring this edited genome revealed expression within the germline with nuclear localization of the F01D4.5 protein (Fig. 6A). The fluorescence was produced by the F01D4.5::mKate2 fusion protein because treatment with F01D4.5 RNAi reduced the fluorescence by about 50% (Fig. 6A). Because the translational reporter was only weakly expressed, perhaps due to being only present at two copies per cell, we also constructed a F01D4.5p::GFP transcriptional reporter transgene to determine whether the gene is expressed in other tissues. Stable lines obtained through bombardment of the transgene showed expression of this reporter within the intestine and early germline (Fig. 6B), and additional expression was observed in the vulva, hypodermis, nervous system, and male gonad in at least two but not all lines (Fig. S10). As the C. elegans germline is known to repress the expression of many transgenes, the absence of F01D4.5p::GFP signal within the adult germline is not unexpected despite this being the only tissue in which the translational reporter F01D4.5::mKate2 was observed (103, 104). Due to the differing transcriptional start sites for the two isoforms of F01D4.5, the transcriptional reporter included the first full exon of F01D4.5a, so the nuclear localization of the GFP likely indicates a nuclear localization signal within this exon.
Figure 6.
Expression and subcellular localization of F01D4.5. A, diagram of edit made by CRISPR to tag the C terminus of F01D4.5 with mKate and a representative image of these worms showing expression in the germline and localization to the germline nuclei. Fluorescence was produced by an F01D4.5::mKate2 fusion protein because the average fluorescence intensity was diminished with treatment of these animals with F01D4.5 RNAi compared with control RNAi. Shown are the mean fluorescence intensity and S.D. (error bars) for 10–13 worms/RNAi treatment normalized to control-treated worms. ***, p < 0.001 by a t test. B, diagram of the F01D4.5 transcriptional reporter construct used to assess gene expression and localization. Shown is a representative image of an L3 animal that exhibits expression of this fluorescent reporter in the intestine and early germline with localization in the nucleus as indicated by the arrows.
Because TCF20 has been shown to enhance Nrf2 transcriptional activity (75), we asked whether F01D4.5 could also function as a coactivator for the SKN-1 transcription factor. However, knockdown of F01D4.5 via RNAi had no effect on SKN-1 transcriptional activity as measured by the expression of a gst-4p::GFP reporter (Fig. S11A) (105). Given that retinoic acid can inhibit tyrosine aminotransferase gene expression and the homology of F01D4.5 with RAI1 (106), we sought to test the possibility that F01D4.5 may somehow regulate tatn-1 expression and produce the protective effects of F01D4.5 knockdown though increased TATN-1 protein levels. We treated worms expressing the tatn-1p::tatn-1::GFP reporter with F01D4.5 RNAi, but we failed to observe any differences in tatn-1 expression in these worms compared with controls (Fig. S11B).
To gain insights into the function of F01D4.5, we outcrossed the F01D4.5(baf20) allele into N2 to explore the function of F01D4.5 independently of tatn-1. RNA was isolated from N2 and F01D4.5(baf20) mutants and used for whole-transcriptome profiling by RNA-Seq, which identified 65 genes that were differentially expressed between the strains, including F01D4.5 (Table S1). Via the use of NanoString, we independently confirmed that the T27E7.1 gene was repressed in the F01D4.5 mutant relative to WT N2 worms (Fig. S12).
Functional annotation clustering utilizing the Database for Annotation, Visualization, and Integrated Discovery (DAVID) website revealed a statistically significant enrichment of genes related to the ribosome with both Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and gene ontology (GO) term analysis (Hochberg–Benjamini adjusted p values 0.0012 and 0.037, respectively). No other annotation group was significant when considering the false-discovery rate. Among the genes that were differentially expressed, five genes encoded isoforms of the large subunit of the ribosome: rpl-29, rpl-31, rpl-33, rpl-35, and Y37E3.8. All of these genes were down-regulated in the F01D4.5 mutant compared with the WT animals (Table S1). Reductions in ribosome protein levels have been shown to be a cellular response to the misfolding of proteins and proteotoxic stress (107, 108), and because m-tyrosine is erroneously substituted for phenylalanine during translation (46, 58, 59), the down-regulation of these genes suggests a possible mechanism that may explain the resistance of tatn-1 mutants to m-tyrosine and oxidative stress treatment when F01D4.5 is mutated or targeted via RNAi.
Other differentially expressed genes of interest that we identified via RNA-Seq include eif-2a, which encodes a translation initiation factor (109); elc-1, which encodes an E3 ligase and transcription elongation factor; aco-2, which encodes an aconitase; F37C12.3, which encodes an ortholog of NADH:ubiquinone oxidoreductase; fmo-4, which encodes a flavin-containing monooxygenase; and tbb-6, which encodes a class of tubulin proteins that for unclear reasons is highly induced with disruption to the electron transport chain (110).
Last, due to the proposed role for phenylalanyl-tRNA synthetase in mischarging phenylalanyl-tRNA with m-tyrosine (46, 58, 59), we were interested to see whether the F01D4.5 mutation altered the expression of this gene or others encoding tRNA synthetases. We probed the RNA-Seq data for 25 known C. elegans tRNA synthetases, and utilizing the unadjusted p value, only two genes were down-regulated: fars-1, encoding a phenylalanyl-tRNA-synthetase, and yars-2, encoding a tyrosyl-tRNA-synthetase (unadjusted p values 0.0084 and 0.027, respectively) (Table S2). Further studies are warranted to validate the significance of these results.
Discussion
Our work supports a novel role of tyrosine aminotransferase as part of the oxidative stress response, specifically through the metabolism of the abnormal tyrosine isomer m-tyrosine that can be formed after exposure to oxidative stress. In our working model, following exposure to oxidative stress, tatn-1 expression is induced by SKN-1, and TATN-1 is then able to metabolize m-tyrosine (Fig. 7A). When TATN-1 function is reduced, m-tyrosine accumulates and causes adverse effects within C. elegans, including developmental delay, reduced fertility, and shortened lifespan (Fig. 7B).
Figure 7.
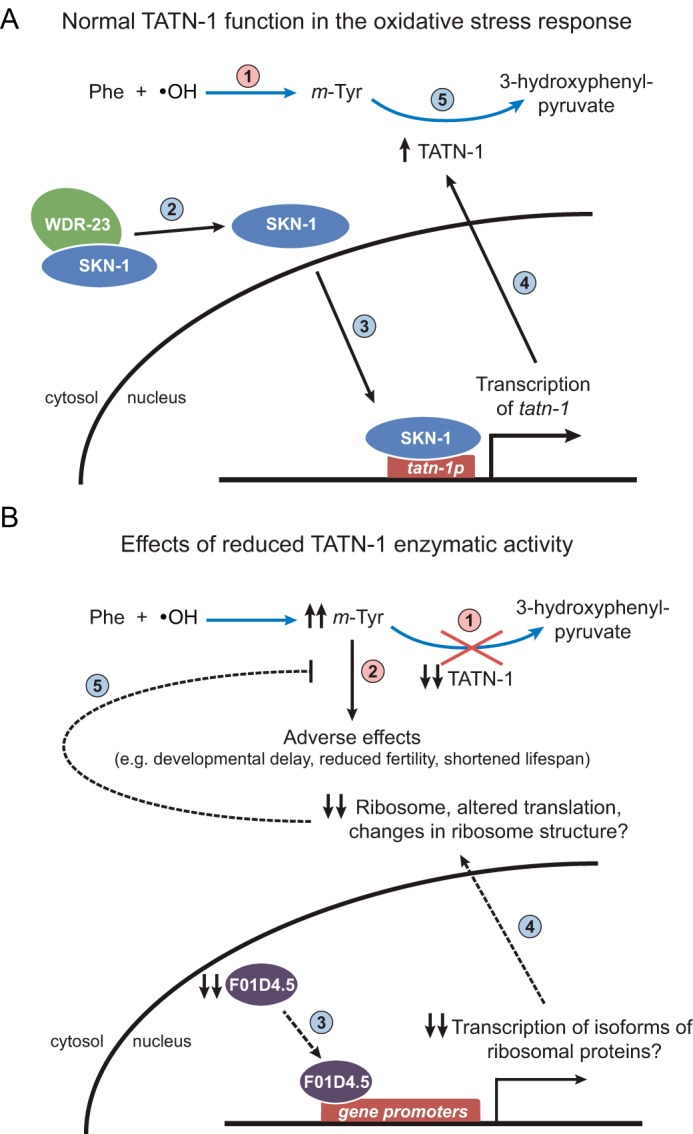
Proposed model of the role of TATN-1 in the oxidative stress response. A, in WT C. elegans, tyrosine aminotransferase prevents the accumulation of m-tyrosine following oxidative stress. In these diagrams, blue arrows represent chemical reactions, whereas black arrows represent biologic processes (1). Following exposure to oxidative stress, m-tyrosine is formed when hydroxyl radicals oxidize the phenyl ring of phenylalanine (blue arrow) (2). SKN-1 is activated as part of the cellular response to oxidative stress via post-translational modifications such as phosphorylation (3), and SKN-1 translocates to the nucleus, where it induces the expression of tatn-1, along with other key detoxification genes (4). This results in an increase in the levels of TATN-1 protein and enzymatic activity, and TATN-1 metabolizes m-tyrosine into the less toxic 3-hydroxyphenylpyruvate (blue arrow) (5). B, a decrease in tyrosine aminotransferase activity renders C. elegans susceptible to the adverse effects of m-tyrosine, but a mutation in the F01D4.5 gene can prevent this sensitivity (1). If tyrosine aminotransferase activity is inadequate, this can lead to the accumulation of m-tyrosine (2). This results in adverse effects, including developmental delay, reduced fertility, and a shortened lifespan, which could be due to the misincorporation of m-tyrosine into newly formed proteins (3). F01D4.5 is a putative transcription factor or coregulator for transcription factors, and RNA-Seq data suggest that an important consequence of the F01D4.5 mutation could be the down-regulation of specific ribosomal protein isoforms (4). This may result in a reduction in protein synthesis, altered mRNA translation, and/or altered ribosome structure, and perhaps a decreased rate of m-tyrosine substitution for phenylalanine, which could lead to a reduction in the production of unstable or misfolded proteins when m-tyrosine is present (5). It is possible that F01D4.5 could be regulated at the post-translational level in the setting of oxidative stress, although there are currently no data supporting this. Solid lines, processes with direct supporting evidence; dashed lines and/or question marks, hypothetical processes. Numbered red circles, detrimental pathways; numbered blue circles, protective pathways.
Not only does this work expand our view of pathways functioning within the oxidative stress response to include the tyrosine degradation pathway, but it may also help to explain the discrepancies related to the effects of oxidative stress on aging and disease. The adverse effects of m-tyrosine were only seen when tyrosine aminotransferase enzymatic activity was reduced due to a genetic mutation. A reduction in tyrosine aminotransferase activity also rendered the worms sensitive to the effects of oxidative stress. Considering that the transcript and activity levels of rat tyrosine aminotransferase have been reported to decrease with age (111, 112), it will be compelling to see whether tyrosine aminotransferase function correlates with any of the age-related diseases in which oxidative stress is thought to play a pathological role, including frailty, neurodegenerative diseases, and diabetes (2, 3, 5–9). In addition, because other amino acids may also be oxidized with oxidative stress, it would be of interest to investigate their potential roles in mediating oxidative stress and elimination by other metabolic pathways (12, 113, 114).
Beyond highlighting the important role of tyrosine aminotransferase within the oxidative stress response, we have identified a mutation in the previously uncharacterized gene F01D4.5 that renders tatn-1 mutants resistant to the effects of m-tyrosine. Localization studies utilizing a CRISPR-generated knock-in of the fluorescent protein mKate2 revealed expression of F01D4.5 within the germline, which is precisely where the adverse effects of m-tyrosine are most clearly observed. Furthermore, its nuclear localization supports a presumptive role as a transcription factor or coregulator for transcription factors based on homology with the human proteins TCF20 and RAI1. It is not clear why a mutation affecting F01D4.5 is beneficial in worms under stressed conditions, whereas mutations in the human homologs result in rare genetic syndromes, but further studies assessing the differences in their protein sequences may provide some answers. Additionally, the particular gene functions of TCF20 and RAI1 that account for their related disease syndrome manifestations are unknown.
The RNA-Seq data comparing N2 and F01D4.5 mutant worms may provide insights into how a reduction in F01D4.5 activity may potentially promote resistance to oxidative stress and m-tyrosine treatment in the tatn-1 mutant background. Most notably, DAVID functional annotation highlighted the ribosome as being affected by the F01D4.5 mutation, and specific isoforms of multiple genes encoding ribosomal large subunit proteins were down-regulated. This is surprising, considering the previous usage of some of them as reference genes in certain species (63, 115, 116); however, the down-regulation of the expression of these isoforms of ribosomal proteins may suggest a potential diminution of ribosome production, mRNA translation, and ultimately protein synthesis. Indeed, there is evidence from previously published work reporting that the reduced expression of ribosomal subunits results in a reduction of protein synthesis. A deletion in the gene encoding RPL29 in yeast reduced proper assembly of the 60S subunit, joining with the 40S subunit, and protein synthesis (117). Similarly, embryonic fibroblasts from RPL29 null mice demonstrated decreased rates of proliferation and protein synthesis (118). Knockdown of RPL35a expression also inhibited 60S subunit biogenesis and proliferation, and a deletion of this gene has been linked to the development of Diamond–Blackfan anemia (119). Furthermore, Steffen et al. (108) have shown that deletions in genes encoding ribosomal proteins correlate with resistance to ER stress in yeast, and reducing translation with cycloheximide treatment also provides resistance to tunicamycin treatment via a mechanism independent of the canonical ER stress response pathway. Likewise, work in yeast has shown that a reduction in the level of ribosomal proteins is a cellular response to proteotoxic stress induced by trivalent arsenic and that mutations in the genes encoding ribosomal proteins can protect against arsenic toxicity (107). Taken together with the proposed mechanism for m-tyrosine toxicity via its misincorporation into cellular proteins, it is reasonable to hypothesize that F01D4.5 may function to regulate the transcription of ribosomal proteins, and the inhibition of its function results in a reduction in the expression of these ribosomal proteins that then limits protein synthesis and potentially lowers the number of misfolded proteins produced by m-tyrosine misincorporation (Fig. 7B). This reduction in mRNA translation rate may also provide more time for phenylalanyl-tRNA synthetase to appropriately recognize and correct mischarged m-tyrosine-tRNAPhe molecules (46). Interestingly, the transcripts of tRNA synthetases for tyrosine and phenylalanine both were decreased in the F01D4.5 mutant, but these reductions failed to reach a genome-wide threshold after adjusting the p value to account for the desired false discovery rate. However, the selective reduction of tRNAPhe charging, due to lower phenylalanyl-tRNA synthetase expression, could be an alternate means of preventing the charging of m-tyrosine to tRNAPhe and thereby protect the cellular proteome from m-tyrosine misincorporation events. Further studies, such as measuring the rate of protein synthesis utilizing the radioisotope [35S]methionine, are necessary to examine this hypothesis in more detail.
Alternatively, the changes in the expression of ribosomal protein isoforms might have other effects on ribosome structure or function. These changes could result in selective changes in mRNA translation or the recruitment of a distinct set of ribosome-associated proteins. Further work will be needed to fully understand how the F01D4.5 mutations might alter the ribosomes.
Additionally, the affected ribosomal proteins may have functions beyond simply their involvement in the ribosome (120). For example, repression of either RPL27A or RPL29 has been reported to induce p53 expression (121–123), which has multiple functions in mediating the response to stress conditions (124, 125).
The RNA-Seq data also highlighted other genes of potential interest, including eif-2A, an initiation factor that also functions in regulating translation (109); elc-1, which encodes an E3 ligase and transcription elongation factor that prolongs lifespan and delays paralysis caused by Q35 aggregation in C. elegans when knocked down (126); and aco-2, a mitochondrial aconitase that significantly extends C. elegans lifespan when targeted by RNAi and has been implicated in aging (127–129). The F01D4.5 mutation may also stabilize mitochondria, as the expression of tbb-6 was also significantly decreased in the mutant worms. This gene, which is under control of the mitogen-activated protein kinase pmk-3, shows robust expression with electron transport chain disruption and mitochondria dysfunction (110). Studies are ongoing to confirm the role of F01D4.5 in inducing the expression of these genes and to assess how the F01D4.5 mutation might disrupt downstream pathways.
In conclusion, this work has defined novel functions for two genes, tatn-1 and F01D4.5, in response to oxidative stress. This adds to the growing body of literature supporting the potential role of m-tyrosine in mediating the adverse effects of oxidative stress but is among the first to show a direct biochemical defense mechanism to prevent its accumulation. It also identifies F01D4.5 as a potential regulator of the oxidative stress response in C. elegans and provides a hypothesis as to its function.
Experimental procedures
C. elegans strains and maintenance
All C. elegans strains were propagated on NGA plates spotted with OP50-1 E. coli.
The WT N2 C. elegans strain was obtained from the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources, as were the following strains: OD95 (unc-119(ed3); ltIs37[pie-1p::mCherry::his-58 + unc-119(+)]; ltIs38[pie-1p::GFP::PH(PLC1Δ) + unc-119(+)]); CL2166 ((pAF15)gst-4p::GFP::NLS); and HT1593 (unc-119(ed3)). LG335 (skn-1(zu135)/nT1[qIs51]) was a gift from Nicholas Bishop and Leonard Guarente. tatn-1(qd182) was described previously and is a gift from Daniel Pagano and Dennis Kim (62). The ALF131 (bafIs131[tatn-1p::tatn-1 cDNA::GFP]) strain, which expresses a TATN-1::GFP fusion protein, has been described previously (62). Standard genetic crosses were performed to introduce transgenes into mutant backgrounds with PCR being used to confirm the presence of the desired mutations.
Bioinformatic analysis
To determine the level of SKN-1 genomic DNA binding within the tatn-1 promoter, the ChIP-sequencing data of Niu et al. (60) was analyzed using GBrowse within WormBase. Tracks specific to SKN-1 were selected, and the scale was adjusted to display peaks with the y axis representing the S.D. value of the measured binding to each region from the mean SKN-1 binding measured across the genome. In these graphs, greater peak height represents more SKN-1 binding to this region compared with the levels seen in other parts of the genome.
To determine whether tyrosine aminotransferase is overexpressed when SKN-1 is active, we utilized the data of Peddibhotla et al. (61) in which they performed RNA-Seq to compare the gain-of-function mutant skn-1(k1023) with WT worms. We reported the normalized reads per kilobase per million mapped reads for tatn-1 for each of the three replicates of each strain. Statistical significance was measured using an unpaired t test via the software GraphPad Prism.
RNAi experiments
NGA plates were spotted with E. coli strains HT115 or OP50(xu363) expressing the desired RNAi clone (130). Bacteria were grown in Luria broth (LB) for 12–18 h, and then 300 μl was spotted onto NGA plates containing 50 μg/ml carbenicillin and 1 mm IPTG to promote RNAi expression. Due to a slower growth rate, OP50(xu363) was concentrated 2× prior to spotting on agar plates.
Quantification of fluorescent reporters
Approximately 20–30 worms from each treatment were mounted and imaged using fluorescent microscopy as described previously (131). Average fluorescence intensity was measured by outlining the tissue in which the fluorescent protein is expressed (e.g. intestinal tract for TATN-1::GFP, germline for F01D4.5::mKATE2, etc.) in the computer software ImageJ. Statistical significance was determined via GraphPad Prism using either a one-way analysis of variance (ANOVA) with Tukey post hoc test to correct for multiple comparisons when three or more conditions were tested per experiment or an unpaired t test when only two conditions were compared.
TATN-1 protein lysate assay
Synchronous populations of worms (∼1000 worms/treatment), treated with the indicated RNAi or empty vector control, were grown at 20 °C until day 1 of adulthood. Worms were then washed from the plates and rinsed twice with S-basal, and the pelleted worms were placed on ice. The worms were then resuspended in 250 μl of lysate buffer (50 mm Tris-HCl, pH 7.6, 1 mm EDTA, 1 mm DTT, 10% glycerol, and Roche cOmplete Mini EDTA-free Protease Inhibitor Mixture (1 tablet/10 ml)). The worms were transferred to a 2-ml Qiagen sample tube RB, and a single 7-mm steel bead was added to each tube. The samples were then subjected to two freeze-thaw cycles prior to homogenization. Homogenization was performed using a bead mill (Qiagen TissueLyser LT) run at 50 Hz for 5 min. Lysates were then transferred to a microcentrifuge tube and centrifuged at 4 °C at 10,000 × g for 10 min. The protein concentration of the soluble lysate fraction was measured using the Bradford assay. 20 μg of total protein lysate was used for the tyrosine aminotransferase assay.
TATN-1 activity was determined by following the protocol used by Ru et al. with minor modifications (132). 100 μl of reaction mixture (125 mm KH2PO4/K2HPO4, 5 mm l-tyrosine disodium salt hydrate, 0.75 mm EDTA, 0.1 mm pyridoxal phosphate, 10 mm α-ketoglutarate, pH 7.5) was put into each well of a 96-well plate and preincubated at 30 °C for 30 min. The reaction was initiated by adding 50 μl of lysate buffer containing 20 μg of total protein to each well and incubating at 30 °C for 30 min, at which time the reaction was stopped by adding 50 μl of 1 m potassium hydroxide. After an additional 30 min, the absorbance at 331 nm was measured using an UV-visible plate reader (SpectraMax M5) to quantify the presence of the product hydroxybenzaldehyde (133). Each condition was performed in either duplicate or triplicate as indicated. Statistical analysis was performed in GraphPad Prism using an unpaired t test.
Paraquat oxidative stress developmental assay
Approximately 150 eggs were spotted on NGA plates containing paraquat (0, 0.1, 0.2, 0.3, and 0.4 mm). Each plate was scored daily for the number of worms in each of the developmental stages. Statistical significance was determined in GraphPad Prism using a χ2 test comparing the counts of worms within each developmental stage per strain on the indicated day.
asb-1 RNAi oxidative stress assay
RNAi NGA plates were spotted with bacteria expressing control RNAi or asb-1 RNAi or a mixture of the two bacterial cultures at the indicated volume ratios. Eggs were isolated by hypochlorite treatment and placed on these plates. When worms on the control RNAi plates reached day 1 of adulthood, each plate was scored for sterility and normal germline morphology. Worms were scored as abnormal if it was clear on the basis of visual observation that there were fewer than 6 embryos present within the uterus. A χ2 test was used to measure statistical significance compared with worms treated with control RNAi.
Partial purification of TATN-1
Competent Rosetta DE3 bacteria were transformed with a pDEST 15 plasmid coding the full-length C. elegans tatn-1 cDNA fused to a GSH S-transferase (GST) tag. In parallel, additional bacteria were transformed with the pGEX-5X-2 plasmid encoding only GST to be used as a negative control for our enzymatic assay. Cells were grown at 37 °C in LB medium to an optical density of 0.5–1.0 at 600 nm. Cultures were cooled to room temperature, and protein expression was then induced by adding IPTG to a final concentration of 1 mm followed by overnight incubation at 20 °C with shaking. Cells were then harvested via centrifugation, and the pellet was washed with ice-cold PBS (pH 7.4). The pellet was subjected to a single freeze/thaw, and then the following reagents were added to lyse the cells: Novagen BugBuster Protein Extraction Reagent (5 ml/g of wet cell paste), Roche cOmplete Mini EDTA-free Protease Inhibitor Mixture tablets (1 tablet/10 ml), Benzonase (1 unit/1 ml), and lysozyme (1000 units/1 ml). Cell suspensions were incubated for 20 min at room temperature. Insoluble material was removed by centrifugation at 16,000 × g for 20 min at 4 °C, and the TATN-1 protein was semi-purified from the supernatant via GST-tagged affinity chromatography using the Millipore GST-Bind Purification Kit and the manufacturer's protocol. The eluted protein was then dialyzed at 4 °C in dialysis buffer (10 mm HEPES, pH 7.35, 100 mm NaCl, 1 mm EDTA, and 10% glycerol) for 15 h. Protein concentration measured by Bradford assay, and the expression of TATN-1 protein was verified via SDS-PAGE on a NuPage 10% BisTris gel followed by Coomassie Blue staining. As multiple bands were present, which likely represent degradation products and co-purified contaminants, the gel was analyzed using the software program ImageJ, and the concentration of full-length, GST-tagged TATN-1 protein was determined based on the density of the band present at the expected molecular mass (81.6 kDa).
TATN-1 biochemical assay and determination of Michaelis–Menten kinetics
The tyrosine aminotransferase reaction was coupled with glutamate dehydrogenase, and absorbance at 340 nm was recorded continuously in a SpectraMax M5 plate reader at room temperature to measure the formation of NADH product, using a modification of a previously published protocol (134–136). Reactions were performed at 27 °C in HEPES (200 mm), pH 7.4, KCl (100 mm) containing 28 units/ml lyophilized glutamate dehydrogenase (Sigma), 20 μm pyridoxal phosphate, 500 μm NAD+, 6 mm α-ketoglutarate, and varying concentrations (0.25–10 mm) of p-tyrosine, m-tyrosine, or phenylalanine. Background rates were measured in the absence of the TATN-1 enzyme. A negative control using purified GST alone was also conducted in parallel. Each reaction was initiated with the addition of semi-purified TATN-1 to a final concentration of 14 nm. The reactions were run in triplicate for each concentration of substrate. The measured absorbance data were converted to the concentration of NADH formed using Beer's law (absorbance = ϵLc, where ϵ is the molar extinction coefficient, L is the path length, and c is the concentration) with the coefficient of extinction for NADH at 340 nm being 6220 m−1 cm−1. The initial velocities of the reaction were determined for the various concentrations of the substrates, and the statistical software GraphPad Prism was used to calculate the kinetic parameters for each substrate using nonlinear regression with the built-in kcat equation with least squares (ordinary) fit: Y = Et × kcat × X/(Km + X), where X is the substrate concentration, Y is the enzyme velocity, kcat is the turnover number, Km is the Km in the same units as X, and Et is the concentration of the enzyme catalytic sites.
meta-Tyrosine treatment
A filter-sterilized solution of either l-m-tyrosine (Toronto Research Chemicals, Inc., catalog no. T910010, lot 1-TTK-126-1) or l-p-tyrosine (Sigma, catalog no. T3754, lot BCBT5226), which was used as a control, was added to NGA medium containing streptomycin (0.2 mg/ml) prior to pouring the plates. The plates were spotted with OP50(xu363) control RNAi bacteria that lacks resistance to streptomycin and that had been 2× concentrated. Immediately after the bacteria had dried on the plates, the bacteria were killed by UV irradiation as described by Burton et al. (137). Worms were grown on these plates at 20 °C from the egg stage. Statistical measurements between strains and treatments was performed using a χ2 test in GraphPad Prism.
LC/MS
Biological replicates of ∼1500 N2 WT and tatn-1(qd182) worms were grown on NGA plates supplemented with either p- or m-tyrosine as described above until day 1 of adulthood. Worms were then washed from plates, allowed to incubate in S-basal medium for 30 min to remove bacteria from the gut, and then washed three times. Worm pellets were then stored frozen. Samples were homogenized in 300 μl of hydrochloric acid (HCl) (0.1 n) using the FastPrep-24 homogenizer at a speed setting of 6.5 m/s for four 30-s cycles with a 1-min rest on ice between cycles. Samples were then centrifuged at 18,000 × g for 15 min, and the supernatant was dried in a vacuum centrifuge. The dried samples were redissolved in 50 μl of HCl (0.01 n), and the concentrations of meta-tyrosine, para-tyrosine, and phenylalanine were determined by LC/MS using multiple-reaction monitoring (MRM) as performed by Bullwinkle et al. (46), with minor differences—only 1 μl of solution was injected onto the reverse-phase HPLC column, and the typical retention times for p-tyrosine, m-tyrosine, and phenylalanine were 15.1, 17.5, and 20.9 min, respectively. Peak areas were measured using instrument manufacturer–supplied software (Agilent MassHunter). The amount of each amino acid in the samples was determined by interpolation from the curves constructed from standard samples and normalized by dividing by the amount of soluble protein in each sample as measured by the Bradford assay. Statistical significance was measured using an unpaired t test using GraphPad Prism.
Pacidamycin X treatment
The PacX gene was previously cloned into E. coli utilizing a pET30 expression vector and was received as a gift from Dr. Wenjun Zhang (78). The plasmid was transformed into OP50(xu363) E. coli. To supplement C. elegans with m-tyrosine via PacX expression, bacteria were grown overnight in LB medium, double-concentrated, and spotted to NGA plates containing kanamycin (50 μg/ml) to select for the pET30 vector and 2 mm IPTG to induce PacX expression. Bacteria harboring empty vector pET24 were used as a negative control. Worms were grown on these plates at 20 °C from the egg hatching. A χ2 test was used to measure statistical significance between worm strains and treatments.
Fertility assays
Worm embryos were collected via hypochlorite treatment, placed on specified plates, and allowed to develop until the L4 larval stage. At this time, individual worms (6–12 per condition) were moved to individual plates and transferred daily to fresh plates until each individual worm had ceased reproduction. Progeny were allowed to develop for 2 days prior to counting the number of worms per plate. The total number of progeny per worm was determined by summing the number of worms from each day of egg laying. A one-way ANOVA with Tukey post hoc test to correct for multiple comparisons was performed using GraphPad Prism to compare the number of progeny between strains and treatments.
Developmental time assay
Embryos were collected by hypochlorite treatment and allowed to hatch in S-basal at room temperature. Individual L1 larvae were moved onto individual NGA plates spotted with bacteria expressing either the control pET24 or PacX and incubated at 20 °C. Beginning 44 h later, worms were scored hourly for developmental stage, and the time for each worm to become a gravid adult was recorded. The developmental time of 5–10 worms was measured per strain per condition. Two independent experiments were performed to ensure reproducibility of the data. Statistical significance was calculated using a one-way ANOVA with the Tukey post hoc test to correct for multiple comparisons.
Embryonic lethality assay
Worms from the developmental time assay were allowed to lay eggs for 24 h, at which time they were removed from the plates, and the number of embryos and L1 worms were counted. Two days later, the number of viable worms were counted. This number was subtracted from and then divided by the number of embryos/L1s to calculate the percentage of lethal embryos. A one-way ANOVA with the Tukey post hoc test to correct for multiple comparisons was performed using to compare the number of progeny between strains and treatments.
Lifespan assays
Lifespan assays were conducted at 25 °C with tyrosine treatments as specified. Worm embryos were collected via hypochlorite treatment, placed on the specified plates, and allowed to develop at 20 °C until the young adult stage. At this time, worms were moved to treatment plates that contained 50 μm 5-fluoro-2′-deoxyuridine to inhibit growth of progeny and were moved to 25 °C. For each lifespan, 3–4 plates of 40 worms each per condition were followed for survival. Worms were scored every 1–2 days by examining for touch-provoked movement, and worms that did not respond to repeated touch stimuli were scored as dead. Worms that crawled off the plate or experienced herniation of the intestine through the vulva were censored. At the end of the assay when all worms had died, the data from the multiple plates for each condition were combined and analyzed with the Mantel–Cox log-rank test via GraphPad Prism, which also produced Kaplan–Meier survival curves. Life tables and mean survival were calculated using Stata 14. Each lifespan was replicated at least twice to verify the results.
Assessment of germline phenotypes with fluorescent reporters
The ltIs37[pie-1p::mCherry::his-58] and ltIs38[pie-1p::GFP::PH(PLC1Δ)] transgenes from the OD95 transgenic strain were crossed into the tatn-1 mutant background and treated with PacX-expressing bacteria or control bacteria or with 4 mm p-tyrosine or m-tyrosine. On day 1 of adulthood, worms were mounted and imaged. Phenotypes were determined by comparing with representative images from the publication of Green et al. (83).
Chemical mutagenesis screen
tatn-1(qd182) mutant worms were used for a forward genetic screen using the mutagen EMS as described previously (138). Briefly, the tatn-1 mutants were synchronized by hypochlorite treatment, grown until the L4 developmental stage, and then treated with 50 mm EMS for 4 h. After a short period of recovery, 100 worms were divided among 10 NGA plates. Once the F1 progeny were gravid, eggs representing the F2 generation were collected by hypochlorite treatment and put on plates spotted with bacteria expressing PacX. These F2 progeny were screened for normal fertility indicating resistance to m-tyrosine. Multiple worm lines that suppressed the reduced fertility phenotype were carried forward, and these lines were again tested for resistance to treatment with PacX. The most robust line was backcrossed twice to the tatn-1(qd182) strain, and then its genomic DNA was extracted utilizing Qiagen's DNeasy Blood and Tissue Kit according to the manufacturer's protocol. Whole-genome sequencing was performed for both the EMS mutant strain and the unmutagenized tatn-1(qd182) strain with an Illumina HiSeq2000 system. The sequencing data were then analyzed with the Variant Discovery Mapping workflow in the online CloudMap suite hosted on the Galaxy server as described previously by Minevich et al. (139).
CRISPR-edited F01D4.5 fluorescent tag
The C terminus of F01D4.5 was tagged with mKate using CRISPR (140). As a result of the differing translational start and stop sites of the different F01D4.5 isoforms, there was no obvious insertion site for a fluorescent tag, and we opted to insert it at the C-terminal end of the penultimate exon, which is identical for both isoforms. Single-guide RNA (sgRNA) targeting the C terminus of F01D4.5 was selected by utilizing an online design tool and published selection criteria (140, 141). This sequence was cloned into the pJW1285 plasmid vector, creating the Cas9-sgRNA construct, using the New England Biolabs Q5 Site-directed Mutagenesis kit. Homology arms ∼500 bp in length were generated by PCR and inserted into pDD287 containing the mKate2/self-excision cassette using AvrII and NgoMIV digestion and the NEB HiFi DNA Assembly Cloning kit. Correct assembly was verified via sequencing. N2 adults were then injected with a mixture of the following: 50 ng/μl Cas9-sgRNA plasmid, 10 ng/μl pDD287 with homology arms, 10 ng/μl pGH8 plasmid, 5 ng/μl pCFJ104 plasmid, and 2.5 ng/μl PCFJ90. pGH8, pCFJ104, and pCFJ90 were included as co-injection markers. F2 generation was screened for candidate knock-in worms by adding 500 μl of 5 mg/ml hygromycin to each plate. Resistant worms were then screened for knock-in by examining for 100% transmission of the roller marker followed by PCR for confirmation. The self-excision cassette was removed by subjecting L1 worms to heat shock at 34 °C for 4 h and picking nonrollers in the F1 generation. Primers utilized for this are shown in Table S3.
F01D4.5 transcriptional reporter
A 2-kb fragment containing the F01D4.5 promoter was generated via PCR using N2 genomic DNA using the primers displayed in Table S3. This PCR fragment included the promoter region through the first five codons of the first exon of F01D4.5b so as to contain the promoter regions for both isoforms of the gene. This fragment was cloned into the pPD95.75 vector. The resulting F01D4.5p::GFP reporter was modified by homologous recombination to express the unc-119 marker gene as described previously (142). This plasmid was used to bombard HT1593 (unc-119(ed3)) worms using a previously published protocol (143). Transgenic strains were identified by rescue of the unc-119 mutant phenotype, and stable lines that produced no uncoordinated progeny were imaged.
RNA-Seq analysis
Five biological replicates of N2 and F01D4.5(baf20) mutants were collected for each condition for a total of 1000–2000 worms/sample. Eggs were hatched in S-basal to arrest the animals in the L1 stage. The arrested worms were transferred to plates and incubated at 20 °C for 48 h, at which point late L4 worms were collected for subsequent RNA extraction. RNA extraction was performed by adding 1 ml of QIAzol Lysis Reagent to the worm pellet and freezing at −80 °C. After two freeze/thaw cycles, total RNA was then extracted and purified utilizing the Qiagen miRNeasy Mini Kit according to the manufacturer's protocol. The quality of total RNA was checked with an Advanced Analytical Fragment Analyzer. RNA quality numbers greater than 7 were accepted for library preparation and sequencing. Approximately 500 ng of total RNA was used for library preparation by following the Illumina TruSeq Stranded mRNA Sample Preparation Guide. The libraries were then subjected to quantification and pooled for cBot amplification and a subsequent 50-bp single-read sequencing run with the Illumina HiSeq 3000 platform. After the sequencing run, demultiplexing with CASAVA was employed to generate the fastq file for each sample. The fastq files were then uploaded to the Galaxy server for analysis.
Data analysis of the RNA-Seq reads was performed on the Galaxy server as follows: 1) sequencing reads were aligned to the C. elegans reference genome (ce11) using HISAT2; 2) the aligned reads were then counted via htseq-count in intersection (nonempty) mode without strandedness and default parameters; 3) differential expression of gene/transcript counts was then determined using DESeq2 with the local fit option and the outlier filtering and replacement options enabled. Genes showing an adjusted p value <0.05 were manually annotated using Wormbase, and these differentially expressed genes were input into the DAVID database for functional annotation and identify biological themes or pathways. Grouping terms and pathway analysis were considering meaningful if the false-discovery rate was 0.05 or less. The RNA-Seq data have been deposited at Gene Expression Omnibus under accession number GSE 115165.
Expression of the T27E7.1 gene was also measured via the use of NanoString technology using the RNA prepared above for RNA-Seq with the cdc-42, pmp-3, and Y45F10D.4 genes being used as reference genes for normalization as described previously (131).
Author contributions
B. R. I. and A. L. F. conceptualization; B. R. I. and K. F. F. data curation; B. R. I., R. A. G., K. F. F., and A. L. F. formal analysis; B. R. I. and A. L. F. funding acquisition; B. R. I., R. A. G., J. T. W., J. N. W., and K. F. F. investigation; B. R. I., R. A. G., J. T. W., J. N. W., and K. F. F. methodology; B. R. I., R. A. G., and A. L. F. writing-original draft; B. R. I., R. A. G., and A. L. F. writing-review and editing; A. L. F. supervision; A. L. F. project administration.
Supplementary Material
Acknowledgments
Whole-genome sequencing and RNA-Seq data were generated in the Genome Sequencing Facility at the University of Texas Health Science Center at San Antonio, which is supported by NCI, National Institutes of Health (NIH), Grant P30 CA054174, NIH Shared Instrument Grant 1S10OD021805-01, and Cancer Prevention and Research Institute of Texas (CPRIT) Core Facility Award RP160732.
This work was supported by funds from the South Texas Veterans Affairs Healthcare System; National Institutes of Health Grant AG053034 (to B. R. I.); NIA, National Institutes of Health, Grant AG044768 (to A. L. F.); NIEHS, National Institutes of Health, Grant ES017761 (to A. L. F.); and NINDS, National Institutes of Health, Grant NS102782 (to A. L. F.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Data set S1, Tables S1–S3, and Figs. S1–S12.
The RNA-Seq data have been deposited at the Gene Expression Omnibus under accession number GSE 115165.
- ARE
- antioxidant response element
- PacX
- pacidamycin X
- EMS
- ethyl methanesulfonate
- PHD
- plant homeodomain
- SPBP
- stromelysin-1 platelet-derived growth factor–responsive element–binding protein
- DAVID
- Database for Annotation, Visualization, and Integrated Discovery
- LB
- Luria broth
- NGA
- nematode growth agar
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- ANOVA
- analysis of variance
- GST
- GSH S-transferase
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- sgRNA
- single-guide RNA.
References
- 1. Stadtman E. R. (1992) Protein oxidation and aging. Science 257, 1220–1224 10.1126/science.1355616 [DOI] [PubMed] [Google Scholar]
- 2. Brownlee M. (2001) Biochemistry and molecular cell biology of diabetic complications. Nature 414, 813–820 10.1038/414813a [DOI] [PubMed] [Google Scholar]
- 3. Tamagno E., Bardini P., Obbili A., Vitali A., Borghi R., Zaccheo D., Pronzato M. A., Danni O., Smith M. A., Perry G., and Tabaton M. (2002) Oxidative stress increases expression and activity of BACE in NT2 neurons. Neurobiol. Dis. 10, 279–288 10.1006/nbdi.2002.0515 [DOI] [PubMed] [Google Scholar]
- 4. Pennathur S., Ido Y., Heller J. I., Byun J., Danda R., Pergola P., Williamson J. R., and Heinecke J. W. (2005) Reactive carbonyls and polyunsaturated fatty acids produce a hydroxyl radical-like species: a potential pathway for oxidative damage of retinal proteins in diabetes. J. Biol. Chem. 280, 22706–22714 10.1074/jbc.M500839200 [DOI] [PubMed] [Google Scholar]
- 5. Shen C., Chen Y., Liu H., Zhang K., Zhang T., Lin A., and Jing N. (2008) Hydrogen peroxide promotes Aβ production through JNK-dependent activation of γ-secretase. J. Biol. Chem. 283, 17721–17730 10.1074/jbc.M800013200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tamagno E., Guglielmotto M., Aragno M., Borghi R., Autelli R., Giliberto L., Muraca G., Danni O., Zhu X., Smith M. A., Perry G., Jo D. G., Mattson M. P., and Tabaton M. (2008) Oxidative stress activates a positive feedback between the γ- and β-secretase cleavages of the β-amyloid precursor protein. J. Neurochem. 104, 683–695 10.1111/j.1471-4159.2007.05072.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Giacco F., and Brownlee M. (2010) Oxidative stress and diabetic complications. Circ. Res. 107, 1058–1070 10.1161/CIRCRESAHA.110.223545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang X., Wang W., Li L., Perry G., Lee H. G., and Zhu X. (2014) Oxidative stress and mitochondrial dysfunction in Alzheimer's disease. Biochim. Biophys. Acta 1842, 1240–1247 10.1016/j.bbadis.2013.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Soysal P., Isik A. T., Carvalho A. F., Fernandes B. S., Solmi M., Schofield P., Veronese N., and Stubbs B. (2017) Oxidative stress and frailty: a systematic review and synthesis of the best evidence. Maturitas 99, 66–72 10.1016/j.maturitas.2017.01.006 [DOI] [PubMed] [Google Scholar]
- 10. Del Rio D., Stewart A. J., and Pellegrini N. (2005) A review of recent studies on malondialdehyde as toxic molecule and biological marker of oxidative stress. Nutr. Metab. Cardiovasc. Dis. 15, 316–328 10.1016/j.numecd.2005.05.003 [DOI] [PubMed] [Google Scholar]
- 11. Yakes F. M., and Van Houten B. (1997) Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 94, 514–519 10.1073/pnas.94.2.514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Berlett B. S., and Stadtman E. R. (1997) Protein oxidation in aging, disease, and oxidative stress. J. Biol. Chem. 272, 20313–20316 10.1074/jbc.272.33.20313 [DOI] [PubMed] [Google Scholar]
- 13. Davies K. J. (2000) Oxidative stress, antioxidant defenses, and damage removal, repair, and replacement systems. IUBMB Life 50, 279–289 10.1080/15216540051081010 [DOI] [PubMed] [Google Scholar]
- 14. Pérez V. I., Bokov A., Van Remmen H., Mele J., Ran Q., Ikeno Y., and Richardson A. (2009) Is the oxidative stress theory of aging dead? Biochim. Biophys. Acta 1790, 1005–1014 10.1016/j.bbagen.2009.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ristow M., and Schmeisser S. (2011) Extending life span by increasing oxidative stress. Free Radic. Biol. Med. 51, 327–336 10.1016/j.freeradbiomed.2011.05.010 [DOI] [PubMed] [Google Scholar]
- 16. Van Raamsdonk J. M., and Hekimi S. (2012) Superoxide dismutase is dispensable for normal animal lifespan. Proc. Natl. Acad. Sci. U.S.A. 109, 5785–5790 10.1073/pnas.1116158109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martindale J. L., and Holbrook N. J. (2002) Cellular response to oxidative stress: signaling for suicide and survival. J. Cell Physiol. 192, 1–15 10.1002/jcp.10119 [DOI] [PubMed] [Google Scholar]
- 18. Espinosa-Diez C., Miguel V., Mennerich D., Kietzmann T., Sánchez-Pérez P., Cadenas S., and Lamas S. (2015) Antioxidant responses and cellular adjustments to oxidative stress. Redox. Biol. 6, 183–197 10.1016/j.redox.2015.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Motohashi H., and Yamamoto M. (2004) Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol. Med. 10, 549–557 10.1016/j.molmed.2004.09.003 [DOI] [PubMed] [Google Scholar]
- 20. Rushmore T. H., Morton M. R., and Pickett C. B. (1991) The antioxidant responsive element: activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J. Biol. Chem. 266, 11632–11639 [PubMed] [Google Scholar]
- 21. Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., Hatayama I., Yamamoto M., and Nabeshima Y. (1997) An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 236, 313–322 10.1006/bbrc.1997.6943 [DOI] [PubMed] [Google Scholar]
- 22. Motohashi H., O'Connor T., Katsuoka F., Engel J. D., and Yamamoto M. (2002) Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene 294, 1–12 10.1016/S0378-1119(02)00788-6 [DOI] [PubMed] [Google Scholar]
- 23. Dinkova-Kostova A. T., Holtzclaw W. D., Cole R. N., Itoh K., Wakabayashi N., Katoh Y., Yamamoto M., and Talalay P. (2002) Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. U.S.A. 99, 11908–11913 10.1073/pnas.172398899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kobayashi A., Kang M. I., Watai Y., Tong K. I., Shibata T., Uchida K., and Yamamoto M. (2006) Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol. Cell Biol. 26, 221–229 10.1128/MCB.26.1.221-229.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. He X., and Ma Q. (2009) NRF2 cysteine residues are critical for oxidant/electrophile-sensing, Kelch-like ECH-associated protein-1-dependent ubiquitination-proteasomal degradation, and transcription activation. Mol. Pharmacol. 76, 1265–1278 10.1124/mol.109.058453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Walker A. K., See R., Batchelder C., Kophengnavong T., Gronniger J. T., Shi Y., and Blackwell T. K. (2000) A conserved transcription motif suggesting functional parallels between Caenorhabditis elegans SKN-1 and Cap'n'Collar-related basic leucine zipper proteins. J. Biol. Chem. 275, 22166–22171 10.1074/jbc.M001746200 [DOI] [PubMed] [Google Scholar]
- 27. An J. H., and Blackwell T. K. (2003) SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 17, 1882–1893 10.1101/gad.1107803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Choe K. P., Przybysz A. J., and Strange K. (2009) The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans. Mol. Cell Biol. 29, 2704–2715 10.1128/MCB.01811-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Powolny A. A., Singh S. V., Melov S., Hubbard A., and Fisher A. L. (2011) The garlic constituent diallyl trisulfide increases the lifespan of C. elegans via skn-1 activation. Exp. Gerontol. 46, 441–452 10.1016/j.exger.2011.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dickson A. J., Marston F. A., and Pogson C. I. (1981) Tyrosine aminotransferase as the rate-limiting step for tyrosine catabolism in isolated rat liver cells. FEBS Lett. 127, 28–32 10.1016/0014-5793(81)80333-X [DOI] [PubMed] [Google Scholar]
- 31. Natt E., Kao F. T., Rettenmeier R., and Scherer G. (1986) Assignment of the human tyrosine aminotransferase gene to chromosome 16. Hum. Genet. 72, 225–228 10.1007/BF00291882 [DOI] [PubMed] [Google Scholar]
- 32. Sgaravatti A. M., Magnusson A. S., de Oliveira A. S., Rosa A. P., Mescka C. P., Zanin F. R., Pederzolli C. D., Wyse A. T., Wannmacher C. M., Wajner M., and Dutra-Filho C. S. (2009) Tyrosine administration decreases glutathione and stimulates lipid and protein oxidation in rat cerebral cortex. Metab. Brain Dis. 24, 415–425 10.1007/s11011-009-9153-6 [DOI] [PubMed] [Google Scholar]
- 33. Macêdo L. G., Carvalho-Silva M., Ferreira G. K., Vieira J. S., Olegário N., Gonçalves R. C., Vuolo F. S., Ferreira G. C., Schuck P. F., Dal-Pizzol F., and Streck E. L. (2013) Effect of acute administration of l-tyrosine on oxidative stress parameters in brain of young rats. Neurochem. Res. 38, 2625–2630 10.1007/s11064-013-1180-3 [DOI] [PubMed] [Google Scholar]
- 34. De Prá S. D., Ferreira G. K., Carvalho-Silva M., Vieira J. S., Scaini G., Leffa D. D., Fagundes G. E., Bristot B. N., Borges G. D., Ferreira G. C., Schuck P. F., Andrade V. M., and Streck E. L. (2014) l-Tyrosine induces DNA damage in brain and blood of rats. Neurochem. Res. 39, 202–207 10.1007/s11064-013-1207-9 [DOI] [PubMed] [Google Scholar]
- 35. Teodorak B. P., Scaini G., Carvalho-Silva M., Gomes L. M., Teixeira L. J., Rebelo J., De Prá S. D., Zeni N., Schuck P. F., Ferreira G. C., and Streck E. L. (2017) Antioxidants reverse the changes in energy metabolism of rat brain after chronic administration of l-tyrosine. Metab. Brain Dis. 32, 557–564 10.1007/s11011-016-9936-5 [DOI] [PubMed] [Google Scholar]
- 36. Carvalho-Silva M., Gomes L. M., Scaini G., Rebelo J., Damiani A. P., Pereira M., Andrade V. M., Gava F. F., Valvassori S. S., Schuck P. F., Ferreira G. C., and Streck E. L. (2017) ω-3 fatty acid supplementation decreases DNA damage in brain of rats subjected to a chemically induced chronic model of tyrosinemia type II. Metab. Brain Dis. 32, 1043–1050 10.1007/s11011-017-9994-3 [DOI] [PubMed] [Google Scholar]
- 37. Streck E. L., De Prá S. D. T., Ferro P. R., Carvalho-Silva M., Gomes L. M., Agostini J. F., Damiani A., Andrade V. M., Schuck P. F., Ferreira G. C., and Scaini G. (2017) Role of antioxidant treatment on DNA and lipid damage in the brain of rats subjected to a chemically induced chronic model of tyrosinemia type II. Mol. Cell Biochem. 435, 207–214 10.1007/s11010-017-3070-5 [DOI] [PubMed] [Google Scholar]
- 38. Dundjerski J., Brkljacić J., Elaković I., Manitasević S., and Matić G. (2006) Mercury influences rat liver tyrosine aminotransferase activity and induction by dexamethasone. J. Appl. Toxicol. 26, 187–190 10.1002/jat.1123 [DOI] [PubMed] [Google Scholar]
- 39. Tabiri H. Y., Sato K., Takahashi K., Toyomizu M., and Akiba Y. (2002) Hepatic tyrosine aminotransferase activity is affected by chronic heat stress and dietary tyrosine in broiler chickens. Br. Poult. Sci. 43, 629–634 10.1080/0007166022000004561 [DOI] [PubMed] [Google Scholar]
- 40. Mager H. I. X., and Berends W. (1974) Activation and transfer of oxygen–IX: nonenzymatic hydroxylation of phenylalanine by model systems of dihydroalloxazine/O2 dihydroalloxazine/H2O2 and alloxazinium cation/H2O. Tetrahedron 30, 917–927 10.1016/S0040-4020(01)97154-6 [DOI] [Google Scholar]
- 41. Maskos Z., Rush J. D., and Koppenol W. H. (1992) The hydroxylation of phenylalanine and tyrosine: a comparison with salicylate and tryptophan. Arch. Biochem. Biophys. 296, 521–529 10.1016/0003-9861(92)90606-W [DOI] [PubMed] [Google Scholar]
- 42. Davies M. J., Fu S., Wang H., and Dean R. T. (1999) Stable markers of oxidant damage to proteins and their application in the study of human disease. Free Radic. Biol. Med. 27, 1151–1163 10.1016/S0891-5849(99)00206-3 [DOI] [PubMed] [Google Scholar]
- 43. Kaur H., and Halliwell B. (1994) Detection of hydroxyl radicals by aromatic hydroxylation. Methods Enzymol. 233, 67–82 10.1016/S0076-6879(94)33009-3 [DOI] [PubMed] [Google Scholar]
- 44. Smith L. C., Ravel J. M., Lax S. R., and Shive W. (1964) The effects of phenylalanine and tyrosine analogs on the synthesis and activity of 3-deoxy-d-arabino-heptulosonic acid 7-phosphate synthetases. Arch. Biochem. Biophys. 105, 424–430 10.1016/0003-9861(64)90026-8 [DOI] [PubMed] [Google Scholar]
- 45. Aronson J. N., and Wermus G. R. (1965) Effects of m-tyrosine on growth and sporulation of Bacillus species. J. Bacteriol. 90, 38–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bullwinkle T. J., Reynolds N. M., Raina M., Moghal A., Matsa E., Rajkovic A., Kayadibi H., Fazlollahi F., Ryan C., Howitz N., Faull K. F., Lazazzera B. A., and Ibba M. (2014) Oxidation of cellular amino acid pools leads to cytotoxic mistranslation of the genetic code. Elife 3, 10.7554/eLife.02501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bertin C., Weston L. A., Huang T., Jander G., Owens T., Meinwald J., and Schroeder F. C. (2007) Grass roots chemistry: meta-tyrosine, an herbicidal nonprotein amino acid. Proc. Natl. Acad. Sci. U.S.A. 104, 16964–16969 10.1073/pnas.0707198104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Movellan J., Rocher F., Chikh Z., Marivingt-Mounir C., Bonnemain J. L., and Chollet J. F. (2014) Synthesis and evaluation as biodegradable herbicides of halogenated analogs of l-meta-tyrosine. Environ. Sci. Pollut. Res. Int. 21, 4861–4870 10.1007/s11356-012-1302-5 [DOI] [PubMed] [Google Scholar]
- 49. Gurer-Orhan H., Ercal N., Mare S., Pennathur S., Orhan H., and Heinecke J. W. (2006) Misincorporation of free m-tyrosine into cellular proteins: a potential cytotoxic mechanism for oxidized amino acids. Biochem. J. 395, 277–284 10.1042/BJ20051964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ruggiero R. A., Bruzzo J., Chiarella P., di Gianni P., Isturiz M. A., Linskens S., Speziale N., Meiss R. P., Bustuoabad O. D., and Pasqualini C. D. (2011) Tyrosine isomers mediate the classical phenomenon of concomitant tumor resistance. Cancer Res. 71, 7113–7124 10.1158/0008-5472.CAN-11-0581 [DOI] [PubMed] [Google Scholar]
- 51. Ruggiero R. A., Bruzzo J., Chiarella P., Bustuoabad O. D., Meiss R. P., and Pasqualini C. D. (2012) Concomitant tumor resistance: the role of tyrosine isomers in the mechanisms of metastases control. Cancer Res. 72, 1043–1050 10.1158/0008-5472.CAN-11-2964 [DOI] [PubMed] [Google Scholar]
- 52. Mikolás E., Kun S., Laczy B., Molnár G. A., Sélley E., Kszegi T., and Wittmann I. (2013) Incorporation of ortho- and meta-tyrosine into cellular proteins leads to erythropoietin-resistance in an erythroid cell line. Kidney Blood Press Res. 38, 217–225 10.1159/000355770 [DOI] [PubMed] [Google Scholar]
- 53. Machuca D., Chiarella P., Montagna D., Dran G., Meiss R. P., and Ruggiero R. A. (2015) Meta-tyrosine: a powerful anti-metastatic factor with undetectable toxic-side effects. Medicina (B Aires) 75, 1–5 [PubMed] [Google Scholar]
- 54. Molnár G. A., Mikolás E. Z., Szijártó I. A., Kun S., Sélley E., and Wittmann I. (2015) Tyrosine isomers and hormonal signaling: a possible role for the hydroxyl free radical in insulin resistance. World J. Diabetes 6, 500–507 10.4239/wjd.v6.i3.500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Molnár G. A., Kun S., Sélley E., Kertész M., Szélig L., Csontos C., Böddi K., Bogár L., Miseta A., and Wittmann I. (2016) Role of tyrosine isomers in acute and chronic diseases leading to oxidative stress: a review. Curr. Med. Chem. 23, 667–685 10.2174/0929867323666160119094516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ipson B. R., and Fisher A. L. (2016) Roles of the tyrosine isomers meta-tyrosine and ortho-tyrosine in oxidative stress. Ageing Res. Rev. 27, 93–107 10.1016/j.arr.2016.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Smith I. K., and Fowden L. (1968) Studies on the specificities of the phenylalanyl- and tyrosyl-sRNA synthetases from plants. Phytochemistry 7, 1064–1075 10.1016/S0031-9422(00)88252-7 [DOI] [Google Scholar]
- 58. Klipcan L., Moor N., Kessler N., and Safro M. G. (2009) Eukaryotic cytosolic and mitochondrial phenylalanyl-tRNA synthetases catalyze the charging of tRNA with the meta-tyrosine. Proc. Natl. Acad. Sci. U.S.A. 106, 11045–11048 10.1073/pnas.0905212106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Popp O., Larraillet V., Kettenberger H., Gorr I. H., Hilger M., Lipsmeier F., Zeck A., and Beaucamp N. (2015) Molecular polygamy: the promiscuity of l-phenylalanyl-tRNA-synthetase triggers misincorporation of meta- and ortho-tyrosine in monoclonal antibodies expressed by Chinese hamster ovary cells. Biotechnol. Bioeng. 112, 1187–1199 10.1002/bit.25528 [DOI] [PubMed] [Google Scholar]
- 60. Niu W., Lu Z. J., Zhong M., Sarov M., Murray J. I., Brdlik C. M., Janette J., Chen C., Alves P., Preston E., Slightham C., Jiang L., Hyman A. A., Kim S. K., Waterston R. H., et al. (2011) Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res. 21, 245–254 10.1101/gr.114587.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Peddibhotla S., Fontaine P., Leung C. K., Maloney P., Hershberger P. M., Wang Y., Bousquet M. S., Luesch H., Mangravita-Novo A., Pinkerton A. B., Smith L. H., Malany S., and Choe K. (2015) Discovery of ML358, a selective small molecule inhibitor of the SKN-1 pathway involved in drug detoxification and resistance in nematodes. ACS Chem. Biol. 10, 1871–1879 10.1021/acschembio.5b00304 [DOI] [PubMed] [Google Scholar]
- 62. Ferguson A. A., Roy S., Kormanik K. N., Kim Y., Dumas K. J., Ritov V. B., Matern D., Hu P. J., and Fisher A. L. (2013) TATN-1 mutations reveal a novel role for tyrosine as a metabolic signal that influences developmental decisions and longevity in Caenorhabditis elegans. PLoS Genet. 9, e1004020 10.1371/journal.pgen.1004020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ahn K., Huh J. W., Park S. J., Kim D. S., Ha H. S., Kim Y. J., Lee J. R., Chang K. T., and Kim H. S. (2008) Selection of internal reference genes for SYBR green qRT-PCR studies of rhesus monkey (Macaca mulatta) tissues. BMC Mol. Biol. 9, 78 10.1186/1471-2199-9-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Blackwell T. K., Steinbaugh M. J., Hourihan J. M., Ewald C. Y., and Isik M. (2015) SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic. Biol. Med. 88, 290–301 10.1016/j.freeradbiomed.2015.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hassan H. M., and Fridovich I. (1978) Superoxide radical and the oxygen enhancement of the toxicity of paraquat in Escherichia coli. J. Biol. Chem. 253, 8143–8148 [PubMed] [Google Scholar]
- 66. Senchuk M. M., Dues D. J., and Van Raamsdonk J. M. (2017) Measuring oxidative stress in Caenorhabditis elegans: paraquat and juglone sensitivity assays. Bio. Protoc. 7, e2086 10.21769/BioProtoc.2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kawasaki I., Hanazawa M., Gengyo-Ando K., Mitani S., Maruyama I., and Iino Y. (2007) ASB-1, a germline-specific isoform of mitochondrial ATP synthase b subunit, is required to maintain the rate of germline development in Caenorhabditis elegans. Mech. Dev. 124, 237–251 10.1016/j.mod.2006.11.004 [DOI] [PubMed] [Google Scholar]
- 68. Braeckman B. P., Smolders A., Back P., and De Henau S. (2016) In vivo detection of reactive oxygen species and redox status in Caenorhabditis elegans. Antioxid. Redox Signal. 25, 577–592 10.1089/ars.2016.6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bai S. C., Rogers Q. R., Wong D. L., Sampson D. A., and Morris J. G. (1998) Vitamin B-6 deficiency and level of dietary protein affect hepatic tyrosine aminotransferase activity in cats. J. Nutr. 128, 1995–2000 10.1093/jn/128.11.1995 [DOI] [PubMed] [Google Scholar]
- 70. Sobrado V. R., Montemartini-Kalisz M., Kalisz H. M., De La Fuente M. C., Hecht H. J., and Nowicki C. (2003) Involvement of conserved asparagine and arginine residues from the N-terminal region in the catalytic mechanism of rat liver and Trypanosoma cruzi tyrosine aminotransferases. Protein Sci. 12, 1039–1050 10.1110/ps.0229403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mehere P., Han Q., Lemkul J. A., Vavricka C. J., Robinson H., Bevan D. R., and Li J. (2010) Tyrosine aminotransferase: biochemical and structural properties and molecular dynamics simulations. Protein Cell 1, 1023–1032 10.1007/s13238-010-0128-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Flibotte S., Edgley M. L., Chaudhry I., Taylor J., Neil S. E., Rogula A., Zapf R., Hirst M., Butterfield Y., Jones S. J., Marra M. A., Barstead R. J., and Moerman D. G. (2010) Whole-genome profiling of mutagenesis in Caenorhabditis elegans. Genetics 185, 431–441 10.1534/genetics.110.116616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rekdal C., Sjøttem E., and Johansen T. (2000) The nuclear factor SPBP contains different functional domains and stimulates the activity of various transcriptional activators. J. Biol. Chem. 275, 40288–40300 10.1074/jbc.M006978200 [DOI] [PubMed] [Google Scholar]
- 74. Sjøttem E., Rekdal C., Svineng G., Johnsen S. S., Klenow H., Uglehus R. D., and Johansen T. (2007) The ePHD protein SPBP interacts with TopBP1 and together they co-operate to stimulate Ets1-mediated transcription. Nucleic Acids Res. 35, 6648–6662 10.1093/nar/gkm739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Darvekar S. R., Elvenes J., Brenne H. B., Johansen T., and Sjøttem E. (2014) SPBP is a sulforaphane induced transcriptional coactivator of NRF2 regulating expression of the autophagy receptor p62/SQSTM1. PLoS One 9, e85262 10.1371/journal.pone.0085262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Fitzpatrick P. F. (1999) Tetrahydropterin-dependent amino acid hydroxylases. Annu. Rev. Biochem. 68, 355–381 10.1146/annurev.biochem.68.1.355 [DOI] [PubMed] [Google Scholar]
- 77. Fitzpatrick P. F. (2003) Mechanism of aromatic amino acid hydroxylation. Biochemistry 42, 14083–14091 10.1021/bi035656u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhang W., Ames B. D., and Walsh C. T. (2011) Identification of phenylalanine 3-hydroxylase for meta-tyrosine biosynthesis. Biochemistry 50, 5401–5403 10.1021/bi200733c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Audhya A., Hyndman F., McLeod I. X., Maddox A. S., Yates J. R. 3rd, Desai A., and Oegema K. (2005) A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J. Cell Biol. 171, 267–279 10.1083/jcb.200506124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. McNally K., Audhya A., Oegema K., and McNally F. J. (2006) Katanin controls mitotic and meiotic spindle length. J. Cell Biol. 175, 881–891 10.1083/jcb.200608117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kachur T. M., Audhya A., and Pilgrim D. B. (2008) UNC-45 is required for NMY-2 contractile function in early embryonic polarity establishment and germline cellularization in C. elegans. Dev. Biol. 314, 287–299 10.1016/j.ydbio.2007.11.028 [DOI] [PubMed] [Google Scholar]
- 82. Essex A., Dammermann A., Lewellyn L., Oegema K., and Desai A. (2009) Systematic analysis in Caenorhabditis elegans reveals that the spindle checkpoint is composed of two largely independent branches. Mol. Biol. Cell 20, 1252–1267 10.1091/mbc.e08-10-1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Green R. A., Kao H. L., Audhya A., Arur S., Mayers J. R., Fridolfsson H. N., Schulman M., Schloissnig S., Niessen S., Laband K., Wang S., Starr D. A., Hyman A. A., Schedl T., Desai A., et al. (2011) A high-resolution C. elegans essential gene network based on phenotypic profiling of a complex tissue. Cell 145, 470–482 10.1016/j.cell.2011.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Coulondre C., and Miller J. H. (1977) Genetic studies of the lac repressor. III. Additional correlation of mutational sites with specific amino acid residues. J. Mol. Biol. 117, 525–567 10.1016/0022-2836(77)90056-0 [DOI] [PubMed] [Google Scholar]
- 86. Darvekar S., Johnsen S. S., Eriksen A. B., Johansen T., and Sjøttem E. (2012) Identification of two independent nucleosome-binding domains in the transcriptional co-activator SPBP. Biochem. J. 442, 65–75 10.1042/BJ20111230 [DOI] [PubMed] [Google Scholar]
- 87. Darvekar S., Rekdal C., Johansen T., and Sjøttem E. (2013) A phylogenetic study of SPBP and RAI1: evolutionary conservation of chromatin binding modules. PLoS One 8, e78907 10.1371/journal.pone.0078907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Toulouse A., Rochefort D., Roussel J., Joober R., and Rouleau G. A. (2003) Molecular cloning and characterization of human RAI1, a gene associated with schizophrenia. Genomics 82, 162–171 10.1016/S0888-7543(03)00101-0 [DOI] [PubMed] [Google Scholar]
- 89. Carmona-Mora P., Canales C. P., Cao L., Perez I. C., Srivastava A. K., Young J. I., and Walz K. (2012) RAI1 transcription factor activity is impaired in mutants associated with Smith-Magenis syndrome. PLoS One 7, e45155 10.1371/journal.pone.0045155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Fragoso Y. D., Stoney P. N., Shearer K. D., Sementilli A., Nanescu S. E., Sementilli P., and McCaffery P. (2015) Expression in the human brain of retinoic acid induced 1, a protein associated with neurobehavioural disorders. Brain Struct. Funct. 220, 1195–1203 10.1007/s00429-014-0712-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Huang W. H., Guenthner C. J., Xu J., Nguyen T., Schwarz L. A., Wilkinson A. W., Gozani O., Chang H. Y., Shamloo M., and Luo L. (2016) Molecular and neural functions of Rai1, the causal gene for Smith-Magenis syndrome. Neuron 92, 392–406 10.1016/j.neuron.2016.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Smith A. C. M., Boyd K. E., Elsea S. H., Finucane B. M., Haas-Givler B., Gropman A., Laje G., Magenis E., and Potocki L. (1993) Smith-Magenis syndrome. in GeneReviews® (Adam M. P., Ardinger H. H., Pagon R. A., Wallace S. E., Bean L. J. H., Stephens K., and Amemiya A., eds) University of Washington, Seattle: [PubMed] [Google Scholar]
- 93. Elvenes J., Thomassen E. I., Johnsen S. S., Kaino K., Sjøttem E., and Johansen T. (2011) Pax6 represses androgen receptor-mediated transactivation by inhibiting recruitment of the coactivator SPBP. PLoS One 6, e24659 10.1371/journal.pone.0024659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Falco M., Amabile S., and Acquaviva F. (2017) RAI1 gene mutations: mechanisms of Smith-Magenis syndrome. Appl. Clin. Genet. 10, 85–94 10.2147/TACG.S128455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Cao L., Molina J., Abad C., Carmona-Mora P., Cárdenas Oyarzo A., Young J. I., and Walz K. (2014) Correct developmental expression level of Rai1 in forebrain neurons is required for control of body weight, activity levels and learning and memory. Hum. Mol. Genet. 23, 1771–1782 10.1093/hmg/ddt568 [DOI] [PubMed] [Google Scholar]
- 96. Carmona-Mora P., and Walz K. (2010) Retinoic acid induced 1, RAI1: a dosage sensitive gene related to neurobehavioral alterations including autistic behavior. Curr. Genomics 11, 607–617 10.2174/138920210793360952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. van der Zwaag B., Franke L., Poot M., Hochstenbach R., Spierenburg H. A., Vorstman J. A., van Daalen E., de Jonge M. V., Verbeek N. E., Brilstra E. H., van 't Slot R., Ophoff R. A., van Es M. A., Blauw H. M., Veldink J. H., et al. (2009) Gene-network analysis identifies susceptibility genes related to glycobiology in autism. PLoS One 4, e5324 10.1371/journal.pone.0005324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Gburcik V., Bot N., Maggiolini M., and Picard D. (2005) SPBP is a phosphoserine-specific repressor of estrogen receptor alpha. Mol. Cell Biol. 25, 3421–3430 10.1128/MCB.25.9.3421-3430.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Babbs C., Lloyd D., Pagnamenta A. T., Twigg S. R., Green J., McGowan S. J., Mirza G., Naples R., Sharma V. P., Volpi E. V., Buckle V. J., Wall S. A., Knight S. J., International Molecular Genetic Study of Autism Consortium (IMGSAC), Parr J. R., and Wilkie A. O. (2014) De novo and rare inherited mutations implicate the transcriptional coregulator TCF20/SPBP in autism spectrum disorder. J. Med. Genet. 51, 737–747 10.1136/jmedgenet-2014-102582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Schäfgen J., Cremer K., Becker J., Wieland T., Zink A. M., Kim S., Windheuser I. C., Kreiss M., Aretz S., Strom T. M., Wieczorek D., and Engels H. (2016) De novo nonsense and frameshift variants of TCF20 in individuals with intellectual disability and postnatal overgrowth. Eur. J. Hum. Genet. 24, 1739–1745 10.1038/ejhg.2016.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Smeland O. B., Frei O., Kauppi K., Hill W. D., Li W., Wang Y., Krull F., Bettella F., Eriksen J. A., Witoelar A., Davies G., Fan C. C., Thompson W. K., Lam M., Lencz T., et al. (2017) Identification of genetic loci jointly influencing schizophrenia risk and the cognitive traits of verbal-numerical reasoning, reaction time, and general cognitive function. JAMA Psychiatry 74, 1065–1075 10.1001/jamapsychiatry.2017.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Lee S. J., Hwang A. B., and Kenyon C. (2010) Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr. Biol. 20, 2131–2136 10.1016/j.cub.2010.10.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Seydoux G., and Strome S. (1999) Launching the germline in Caenorhabditis elegans: regulation of gene expression in early germ cells. Development 126, 3275–3283 [DOI] [PubMed] [Google Scholar]
- 104. Green R. A., Audhya A., Pozniakovsky A., Dammermann A., Pemble H., Monen J., Portier N., Hyman A., Desai A., and Oegema K. (2008) Expression and imaging of fluorescent proteins in the C. elegans gonad and early embryo. Methods Cell Biol. 85, 179–218 10.1016/S0091-679X(08)85009-1 [DOI] [PubMed] [Google Scholar]
- 105. Kahn N. W., Rea S. L., Moyle S., Kell A., and Johnson T. E. (2008) Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. Biochem. J. 409, 205–213 10.1042/BJ20070521 [DOI] [PubMed] [Google Scholar]
- 106. Pan C. J., Shelly L. L., Rabin D. S., and Chou J. Y. (1992) Inhibition of tyrosine aminotransferase gene expression by retinoic acid. Mol. Endocrinol. 6, 572–580 10.1210/mend.6.4.1350056 [DOI] [PubMed] [Google Scholar]
- 107. Guerra-Moreno A., Isasa M., Bhanu M. K., Waterman D. P., Eapen V. V., Gygi S. P., and Hanna J. (2015) Proteomic analysis identifies ribosome reduction as an effective proteotoxic stress response. J. Biol. Chem. 290, 29695–29706 10.1074/jbc.M115.684969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Steffen K. K., McCormick M. A., Pham K. M., MacKay V. L., Delaney J. R., Murakami C. J., Kaeberlein M., and Kennedy B. K. (2012) Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics 191, 107–118 10.1534/genetics.111.136549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zoll W. L., Horton L. E., Komar A. A., Hensold J. O., and Merrick W. C. (2002) Characterization of mammalian eIF2A and identification of the yeast homolog. J. Biol. Chem. 277, 37079–37087 10.1074/jbc.M207109200 [DOI] [PubMed] [Google Scholar]
- 110. Munkácsy E., Khan M. H., Lane R. K., Borror M. B., Park J. H., Bokov A. F., Fisher A. L., Link C. D., and Rea S. L. (2016) DLK-1, SEK-3 and PMK-3 are required for the life extension induced by mitochondrial bioenergetic disruption in C. elegans. PLoS Genet. 12, e1006133 10.1371/journal.pgen.1006133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Slagboom P. E., de Leeuw W. J., and Vijg J. (1990) mRNA levels and methylation patterns of the tyrosine aminotransferase gene in aging inbred rats. FEBS Lett. 269, 128–130 10.1016/0014-5793(90)81136-C [DOI] [PubMed] [Google Scholar]
- 112. Dellwo M., and Beauchene R. E. (1990) The effect of exercise, diet restriction, and aging on the pituitary–adrenal axis in the rat. Exp. Gerontol. 25, 553–562 10.1016/0531-5565(90)90021-S [DOI] [PubMed] [Google Scholar]
- 113. Ahmad S., Khan H., Shahab U., Rehman S., Rafi Z., Khan M. Y., Ansari A., Siddiqui Z., Ashraf J. M., Abdullah S. M., Habib S., and Uddin M. (2017) Protein oxidation: an overview of metabolism of sulphur containing amino acid, cysteine. Front. Biosci. (Schol Ed) 9, 71–87 10.2741/s474 [DOI] [PubMed] [Google Scholar]
- 114. Friguet B. (2006) Oxidized protein degradation and repair in ageing and oxidative stress. FEBS Lett. 580, 2910–2916 10.1016/j.febslet.2006.03.028 [DOI] [PubMed] [Google Scholar]
- 115. Wan G., Yang K., Lim Q., Zhou L., He B. P., Wong H. K., and Too H. P. (2010) Identification and validation of reference genes for expression studies in a rat model of neuropathic pain. Biochem. Biophys. Res. Commun. 400, 575–580 10.1016/j.bbrc.2010.08.106 [DOI] [PubMed] [Google Scholar]
- 116. Goodwin A. T., Khan M. A., Chester A. H., Amrani M., and Yacoub M. H. (2002) Up-regulation of endothelin-converting-enzyme mRNA expression following cardioplegic arrest. Clin. Sci. (Lond) 103, Suppl. 48, 206S–209S 10.1042/CS103S206S [DOI] [PubMed] [Google Scholar]
- 117. DeLabre M. L., Kessl J., Karamanou S., and Trumpower B. L. (2002) RPL29 codes for a non-essential protein of the 60S ribosomal subunit in Saccharomyces cerevisiae and exhibits synthetic lethality with mutations in genes for proteins required for subunit coupling. Biochim. Biophys. Acta 1574, 255–261 10.1016/S0167-4781(01)00372-4 [DOI] [PubMed] [Google Scholar]
- 118. Kirn-Safran C. B., Oristian D. S., Focht R. J., Parker S. G., Vivian J. L., and Carson D. D. (2007) Global growth deficiencies in mice lacking the ribosomal protein HIP/RPL29. Dev. Dyn. 236, 447–460 10.1002/dvdy.21046 [DOI] [PubMed] [Google Scholar]
- 119. Farrar J. E., Nater M., Caywood E., McDevitt M. A., Kowalski J., Takemoto C. M., Talbot C. C. Jr., Meltzer P., Esposito D., Beggs A. H., Schneider H. E., Grabowska A., Ball S. E., Niewiadomska E., Sieff C. A., et al. (2008) Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood 112, 1582–1592 10.1182/blood-2008-02-140012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Zhou X., Liao W. J., Liao J. M., Liao P., and Lu H. (2015) Ribosomal proteins: functions beyond the ribosome. J. Mol. Cell Biol. 7, 92–104 10.1093/jmcb/mjv014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Alkhatabi H. A., McLornan D. P., Kulasekararaj A. G., Malik F., Seidl T., Darling D., Gaken J., and Mufti G. J. (2016) RPL27A is a target of miR-595 and may contribute to the myelodysplastic phenotype through ribosomal dysgenesis. Oncotarget 7, 47875–47890 10.18632/oncotarget.10293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Liu J. J., Huang B. H., Zhang J., Carson D. D., and Hooi S. C. (2006) Repression of HIP/RPL29 expression induces differentiation in colon cancer cells. J. Cell Physiol. 207, 287–292 10.1002/jcp.20589 [DOI] [PubMed] [Google Scholar]
- 123. Terzian T., Dumble M., Arbab F., Thaller C., Donehower L. A., Lozano G., Justice M. J., Roop D. R., and Box N. F. (2011) Rpl27a mutation in the sooty foot ataxia mouse phenocopies high p53 mouse models. J. Pathol. 224, 540–552 10.1002/path.2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Budanov A. V. (2014) The role of tumor suppressor p53 in the antioxidant defense and metabolism. Subcell. Biochem. 85, 337–358 10.1007/978-94-017-9211-0_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Liu D., and Xu Y. (2011) p53, oxidative stress, and aging. Antioxid. Redox. Signal. 15, 1669–1678 10.1089/ars.2010.3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Hwang W., Artan M., Seo M., Lee D., Nam H. G., and Lee S. J. (2015) Inhibition of elongin C promotes longevity and protein homeostasis via HIF-1 in C. elegans. Aging Cell 14, 995–1002 10.1111/acel.12390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Hamilton B., Dong Y., Shindo M., Liu W., Odell I., Ruvkun G., and Lee S. S. (2005) A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 19, 1544–1555 10.1101/gad.1308205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Reis-Rodrigues P., Czerwieniec G., Peters T. W., Evani U. S., Alavez S., Gaman E. A., Vantipalli M., Mooney S. D., Gibson B. W., Lithgow G. J., and Hughes R. E. (2012) Proteomic analysis of age-dependent changes in protein solubility identifies genes that modulate lifespan. Aging Cell 11, 120–127 10.1111/j.1474-9726.2011.00765.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Yan L. J., Levine R. L., and Sohal R. S. (1997) Oxidative damage during aging targets mitochondrial aconitase. Proc. Natl. Acad. Sci. U.S.A. 94, 11168–11172 10.1073/pnas.94.21.11168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Xiao R., Chun L., Ronan E. A., Friedman D. I., Liu J., and Xu X. Z. (2015) RNAi interrogation of dietary modulation of development, metabolism, behavior, and aging in C. elegans. Cell Rep. 11, 1123–1133 10.1016/j.celrep.2015.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Keith S. A., Maddux S. K., Zhong Y., Chinchankar M. N., Ferguson A. A., Ghazi A., and Fisher A. L. (2016) Graded proteasome dysfunction in Caenorhabditis elegans activates an adaptive response involving the conserved SKN-1 and ELT-2 transcription factors and the autophagy-lysosome pathway. PLoS Genet. 12, e1005823 10.1371/journal.pgen.1005823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Ru M., Wang K., Bai Z., Peng L., He S., Wang Y., and Liang Z. (2017) A tyrosine aminotransferase involved in rosmarinic acid biosynthesis in Prunella vulgaris L. Sci. Rep. 7, 4892 10.1038/s41598-017-05290-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Diamondstone T. I. (1966) Assay of tyrosine transaminase activity by conversion of p-hydroxyphenylpyruvate to p-hydroxybenzaldehyde. Anal. Biochem. 16, 395–401 10.1016/0003-2697(66)90220-X [DOI] [Google Scholar]
- 134. Luong T. N., and Kirsch J. F. (1997) A continuous coupled spectrophotometric assay for tyrosine aminotransferase activity with aromatic and other nonpolar amino acids. Anal. Biochem. 253, 46–49 10.1006/abio.1997.2344 [DOI] [PubMed] [Google Scholar]
- 135. Rain-Guion M. C., and Chambon H. (1982) Tyrosine amino transferase as a teaching enzyme in a biochemistry class experiment. Biochem. Educ. 10, 88–92 10.1016/0307-4412(82)90082-6 [DOI] [Google Scholar]
- 136. Sivaraman S., and Kirsch J. F. (2006) The narrow substrate specificity of human tyrosine aminotransferase—the enzyme deficient in tyrosinemia type II. FEBS J. 273, 1920–1929 10.1111/j.1742-4658.2006.05202.x [DOI] [PubMed] [Google Scholar]
- 137. Burton E. A., Pendergast A. M., and Aballay A. (2006) The Caenorhabditis elegans ABL-1 tyrosine kinase is required for Shigella flexneri pathogenesis. Appl. Environ. Microbiol. 72, 5043–5051 10.1128/AEM.00558-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Jorgensen E. M., and Mango S. E. (2002) The art and design of genetic screens: Caenorhabditis elegans. Nat. Rev. Genet. 3, 356–369 10.1038/nrg794 [DOI] [PubMed] [Google Scholar]
- 139. Minevich G., Park D. S., Blankenberg D., Poole R. J., and Hobert O. (2012) CloudMap: a cloud-based pipeline for analysis of mutant genome sequences. Genetics 192, 1249–1269 10.1534/genetics.112.144204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Dickinson D. J., and Goldstein B. (2016) CRISPR-based methods for Caenorhabditis elegans genome engineering. Genetics 202, 885–901 10.1534/genetics.115.182162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Xu H., Xiao T., Chen C. H., Li W., Meyer C. A., Wu Q., Wu D., Cong L., Zhang F., Liu J. S., Brown M., and Liu X. S. (2015) Sequence determinants of improved CRISPR sgRNA design. Genome Res. 25, 1147–1157 10.1101/gr.191452.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Ferguson A. A., and Fisher A. L. (2009) Retrofitting ampicillin resistant vectors by recombination for use in generating C. elegans transgenic animals by bombardment. Plasmid 62, 140–145 10.1016/j.plasmid.2009.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Hochbaum D., Ferguson A. A., and Fisher A. L. (2010) Generation of transgenic C. elegans by biolistic transformation. J. Vis. Exp. 10.3791/2090 10.3791/2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.