Abstract
β-Catenin signaling is triggered by WNT proteins and is an important pathway that negatively regulates adipogenesis. However, the mechanisms controlling the expression of WNT proteins during adipogenesis remain incompletely understood. Lysine demethylase 5A (KDM5A) is a histone demethylase that removes trimethyl (me3) marks from lysine 4 of histone 3 (H3K4) and serves as a general transcriptional corepressor. Here, using the murine 3T3-L1 preadipocyte differentiation model and an array of biochemical approaches, including ChIP, immunoprecipitation, RT-qPCR, and immunoblotting assays, we show that Kdm5a is a target gene of CCAAT/enhancer-binding protein β (C/EBPβ), an important early transcription factor required for adipogenesis. We found that C/EBPβ binds to the Kdm5a gene promoter and transactivates its expression. We also found that siRNA-mediated KDM5A down-regulation inhibits 3T3-L1 preadipocyte differentiation. The KDM5A knockdown significantly up-regulates the negative regulator of adipogenesis Wnt6, having increased levels of the H3K4me3 mark on its promoter. We further observed that WNT6 knockdown significantly rescues adipogenesis inhibited by the KDM5A knockdown. Moreover, we noted that C/EBPβ negatively regulates Wnt6 expression by binding to the Wnt6 gene promoter and repressing Wnt6 transcription. Further experiments indicated that KDM5A interacts with C/EBPβ and that their interaction cooperatively inhibits Wnt6 transcription. Of note, C/EBPβ knockdown impaired the recruitment of KDM5A to the Wnt6 promoter, which had higher H3K4me3 levels. Our results suggest a mechanism involving C/EBPβ and KDM5A activities that down-regulates the Wnt/β-catenin pathway during 3T3-L1 preadipocyte differentiation.
Keywords: adipogenesis, CCAAT/enhancer-binding protein (C/EBP), histone demethylase, Wnt signaling, beta-catenin (B-catenin), epigenetics, post-translational modification (PTM), obesity, chromatin immunoprecipitation (ChiP), 3T3-L1 preadipocyte, adipogenesis, C/EBPbeta, lysine demethylase 5A (KDM5A), Wnt/beta-catenin pathway, fat cell, ChIP-on-chip
Introduction
Obesity has become an escalating global epidemic because people's diet and lifestyle changed during the 20th century (1, 2). The overexpansion of adipose tissue mass plays a central role in obesity-related complications, such as type 2 diabetes, hypertension, hyperlipidemia, and arteriosclerosis (3–5). Therefore, a comprehensive investigation into the molecular mechanisms underlying adipogenesis is critical for understanding obesity occurrence and progression and for the development of novel therapeutics against obesity and associated metabolic syndromes (6).
The 3T3-L1 preadipocyte cell line is an invaluable cellular model and has been widely used to investigate the adipocyte differentiation program (7, 8). In the 3T3-L1 adipogenesis model, cells differentiate in a rather synchronous manner after the hormonal induction (9–11). Peroxisome proliferator–activated receptor γ (PPARγ)3 agonists, the powerful adipogenic inducers, are not added during the whole process, which can avoid the possibility of PPARγ agonists overriding modest negative regulators of adipogenesis. Besides, most of the key adipogenic factors identified in the 3T3-L1 differentiation system have been proved to be true in vivo. Upon treatment with differentiation inducers, growth-arrested 3T3-L1 preadipocytes express CCAAT enhancer-binding protein β (C/EBPβ), which then activates the expression of CCAAT enhancer-binding protein α (C/EBPα) and PPARγ, the two principal adipogenic transcription factors that cooperate to switch on the adipocyte gene program (12, 13). Besides the role above, C/EBPβ is required for mitotic clonal expansion (MCE) and induction of autophagy by regulating the expression of a variety of genes during the early stage of 3T3-L1 preadipocyte differentiation (10, 11, 14–17). Thus, C/EBPβ functions as an important early adipogenic factor.
The Wnt family secreted proteins are important developmental regulators, which mediate autocrine and paracrine effects by binding to frizzled (Fzd) receptors and LDL-related protein 5/6 (LRP5/6) coreceptors (18, 19). In the Wnt/β-catenin signaling pathway, Wnt proteins mediate downstream effects by stabilizing β-catenin, leading to the accumulation of free β-catenin in the cytoplasm. The accumulated β-catenin translocates into the nucleus and coactivates the T-cell factor/lymphoid-enhancing factor family of transcription factors to regulate Wnt/β-catenin target genes for cell fate regulation. It is well-known that Wnt/β-catenin signaling, by preventing the induction of C/EBPα and PPARγ, is one of the major regulators of adipogenesis (20). Endogenous expression of Wnt6, Wn10a, and Wnt10b decreases during adipogenesis. Activation of Wnt/β-catenin signaling by overexpressing Wnt proteins, such as Wnt6, Wn10a, or Wnt10b, inhibits the induction of C/EBPα and PPARγ and blocks adipogenesis in a β-catenin–dependent manner (21, 22). On the contrary, inhibition of Wnt/β-catenin signaling by expressing Axin1 or dominant-negative TCF4 (dnTCF4) promotes adipogenesis (23). Although Wnt proteins are critical factors triggering the Wnt/β-catenin pathway and thereby inhibiting adipogenesis, the mechanism controlling the expression of the Wnt proteins during this process remains incompletely understood.
The epigenetic mechanism, especially histone modification, has been found to be an important modulator in regulating the expression of genes and cell differentiation. Histone acetylation and trimethylated histone 3 lysine 4, 9, and 27 (H3K4me3, H3K9me3, and H3K27me3) are among the major epigenetic histone signatures (24–26). Histone acetylation on lysine residues is generally correlated well with gene activation. However, histone methylation on lysine can be associated with either gene activation or inhibition, depending on which lysine is methylated. H3K9me3 and H3K27me3 are enriched on repressed genes, whereas H3K4me3 is implicated in transcriptional activation. Histone methylation has been demonstrated to be eliminated by histone lysine demethylases (KDMs) (27). Lysine-specific demethylase 5A (KDM5A) is a member of the family of Jumonji C (JmjC) domain–containing histone demethylases and demethylates H3K4me3 (28). Mounting evidence has shown an important role of KDM5A in cancer biology as an oncogene (29, 30). It is also reported that KDM5A is involved in regulating the repair of DNA breaks, cell cycle progression, cellular senescence, NK cell activation, mitochondrial function, circadian rhythm, etc. (31–37). Therefore, KDM5A plays an important role in a variety of cellular biological processes.
Previously, our laboratory has identified a series of potential target genes of C/EBPβ by using a promoter-wide ChIP coupled with microarrays (ChIP-on-chip) analysis at the early stage of 3T3-L1 preadipocyte differentiation (11). Among these candidate genes is KDM5A. In the present study, we confirm that KDM5A is transactivated by C/EBPβ. Further, we show that KDM5A interacts with C/EBPβ and is recruited to the Wnt6 promoter to decrease the H3K4me3 mark on its promoter and to repress Wnt6 transcription, thereby inhibiting the Wnt/β-catenin pathway and facilitating 3T3-L1 preadipocyte differentiation. These results provide new insights into the role of KDM5A in adipogenesis and its underlying mechanism.
Results
KDM5A is induced during 3T3-L1 preadipocyte differentiation, and C/EBPβ is required for KDM5A induction
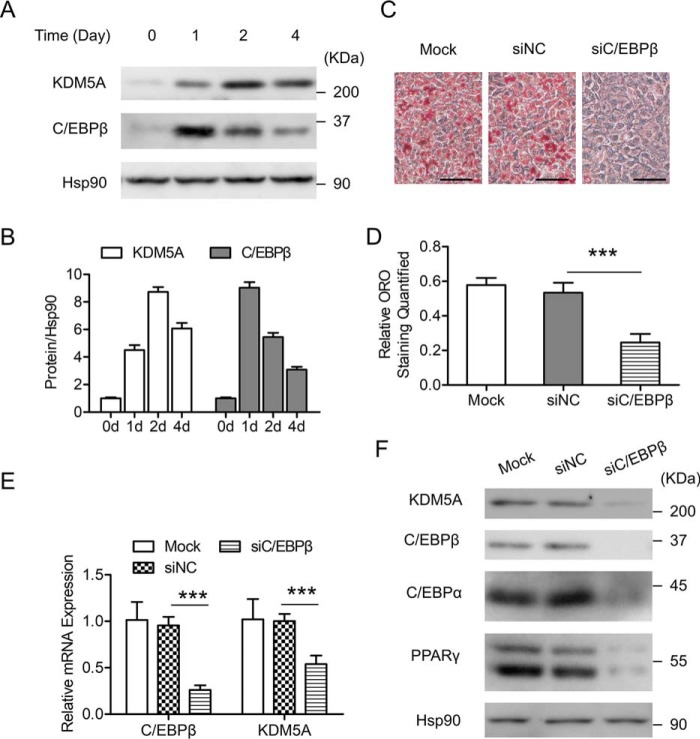
C/EBPβ has been shown to play a pivotal role at the early stage of 3T3-L1 preadipocyte differentiation (9, 13). In our previous work, a ChIP-on-chip at an early stage of 3T3-L1 preadipocyte differentiation was performed (11), which identified a putative C/EBPβ binding site on the proximal promoter of KDM5A (Table 1), a JmjC domain–containing histone demethylase. This suggests that KDM5A could be a transcriptional target of C/EBPβ. The expression of KDM5A during 3T3-L1 preadipocyte differentiation was investigated first. After adipogenic induction, KDM5A expression was induced, which positively correlates with the expression pattern of C/EBPβ (Fig. 1, A and B). To study whether C/EBPβ plays a role in the induction of KDM5A, siRNA-mediated knockdown of C/EBPβ was applied. C/EBPβ was knocked down in 3T3-L1 preadipocytes, and then the cells were induced to differentiation. As shown in Fig. 1, C and D, knockdown of C/EBPβ blocked adipogenesis because its knockdown led to a significant decrease in lipid accumulation in 3T3-L1 cells on day 6 post-induction, as evidenced by Oil Red O (ORO) staining. Meanwhile, down-regulation of C/EBPβ significantly impaired the expression of KDM5A at both the mRNA and protein levels (Fig. 1, E and F). Therefore, these data above demonstrate an important role of C/EBPβ in the induction of KDM5A during 3T3-L1 preadipocyte differentiation.
Table 1.
Summary of the Kdm5a gene that was identified to be potentially targeted by C/EBPβ in our previously reported ChIP-on-chip data
A promoter-wide ChIP-on-chip analysis was performed on 3T3-L1 cells harvested at 20 h after hormonal induction. Anti-C/EBPβ antibody and control IgG were used for the ChIP. For the region of C/EBPβ enrichment, the mm8 chromosomal coordinate is given, including the chromosome number (Chr), the start site (St), and the end of the region (End). “Fold” refers to the -fold change of anti-C/EBPβ signal over control IgG signal. FDR, false discovery rate. “Length” refers to the size of the continuous region across which the C/EBPβ signal was significantly enriched. The gene near the C/EBPβ binding sites is shown by “RefSeq” and “Gene.”
| Chr | St | End | -Fold | FDR (%) | Length | RefSeq | Gene | Strand | TSS |
|---|---|---|---|---|---|---|---|---|---|
| 6 | 120329051 | 120329755 | 2.71 | 1.71 | 704 | NM_145997 | Kdm5a | + | 120329717 |
Figure 1.
KDM5A is induced during 3T3-L1 preadipocyte differentiation, and C/EBPβ is required for KDM5A induction. A, protein expression of KDM5A and C/EBPβ at the indicated time points (days after MDI adipogenic induction) is shown. The targeted proteins were detected by Western blotting. Hsp90 is the loading control. B, Western blotting results in A were quantified against Hsp90 by using ImageJ. Data were normalized to data of day 0. C–F, 3T3-L1 preadipocytes were transfected with control siRNA (siNC) or C/EBPβ siRNA (siC/EBPβ) and induced to differentiation. C, on day 6 after adipogenic induction, cells were stained with Oil Red O, and representative images are shown. Scale bar, 100 μm. D, Oil Red O staining in C was quantified. E, on day 4 after adipogenic induction, the mRNA level of the indicated genes was determined by RT-qPCR. Data were normalized to the mock group. F, on day 4 after adipogenic induction, the indicated proteins were detected by Western blotting. Hsp90 was the loading control. All values are represented as means with error bars representing S.D. ***, p < 0.001. Mock, cells were not infected with viruses or transfected with siRNAs. For statistical analysis, one-way analysis of variance and Bonferroni's post hoc tests were carried out in D, and two-way analysis of variance and Bonferroni's post hoc tests were performed in E.
C/EBPβ transactivates KDM5A during 3T3-L1 preadipocyte differentiation
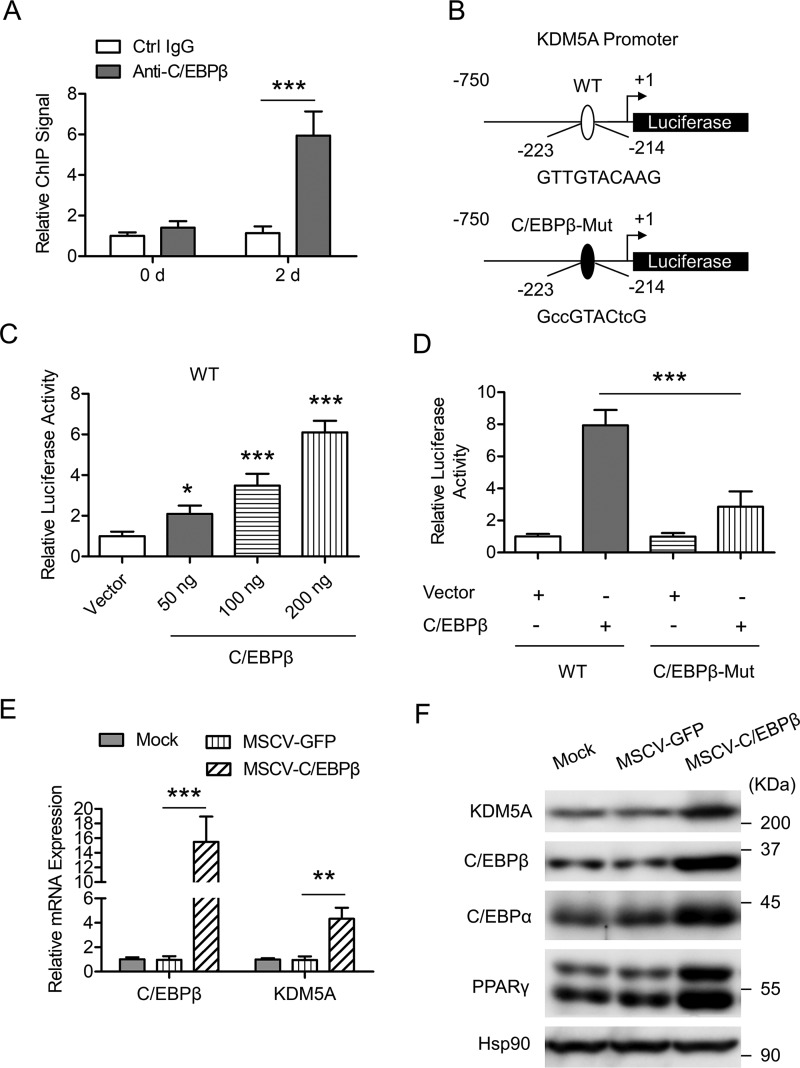
Our previous ChIP-on-chip data and the results in Fig. 1 strongly suggest that C/EBPβ could regulate KDM5A expression at the transcriptional level. ChIP plus real-time quantitative PCR (ChIP-qPCR) confirmed the significant binding of C/EBPβ to the proximal promoter of KDM5A in differentiating cells after induction (day 2) but not in quiescent cells before induction (day 0), as shown in Fig. 2A. On the basis of the results from the ChIP-on-chip analysis (Table 1), we analyzed the KDM5A proximal promoter and found a C/EBP binding site about −223 to −214 bp from the transcription start site (TSS) of KDM5A (Fig. 2B). Using a luciferase assay, we found that C/EBPβ could transactivate KDM5A in a dose-dependent manner in 3T3-L1 cells (Fig. 2C). When the predicted C/EBP-binding site was mutated, this transactivation was significantly blunted (Fig. 2D), suggesting a critical role of this C/EBP binding site in the function of C/EBPβ. To further validate the role of C/EBPβ in KDM5A induction, C/EBPβ was overexpressed in 3T3-L1 preadipocytes, and then the cells were induced to differentiation. Overexpression of C/EBPβ up-regulated both the mRNA level and the protein level of KDM5A on day 2 post-induction (Fig. 2, E and F). Together, these results demonstrate that KDM5A is a bona fide target of C/EBPβ during the adipogenesis of 3T3-L1 cells.
Figure 2.
KDM5A is transactivated by C/EBPβ during 3T3-L1 preadipocyte differentiation. A, enrichment of C/EBPβ on KDM5A proximal promoter was analyzed by ChIP-qPCR. 3T3-L1 cells were induced to differentiation. At the times indicated, ChIP-qPCR was performed by using control IgG and anti-C/EBPβ antibody. Data were normalized to the IgG controls at each time point. B, schematic representation of KDM5A proximal promoter constructs used for luciferase assays. The predicted consensus of C/EBP binding site is shown in the WT luciferase construct. The lowercase letters indicate mutations of the C/EBP-binding site in the C/EBPβ-Mut construct. C, 3T3-L1 preadipocyte was transiently transfected with WT reporter construct as shown in B, along with different amounts of C/EBPβ expression vector, and pRL-TK plasmid was used as an internal control; cell extracts were prepared, and luciferase activities were measured and normalized to Renilla activity. Data were then normalized to the vector group. D, 3T3-L1 preadipocyte was transiently transfected with WT or C/EBPβ-Mut reporter construct as shown in B, along with control vector or C/EBPβ expression vector, and pRL-TK plasmid was used as an internal control. Luciferase activities were measured as in C. E, 3T3-L1 preadipocyte was infected with retrovirus expressing GFP or C/EBPβ and induced to differentiation. The mRNA level of the indicated genes was determined by RT-qPCR on day 4 post-induction. F, cells were treated as in E, and Western blotting was performed by using the indicated antibodies. Hsp90 was the loading control. All values are represented as means with error bars representing S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Mock, cells were not infected with viruses or transfected with siRNAs. For statistical analysis, one-way analysis of variance and Bonferroni's post hoc tests were carried out in C (compared with vector) and D, and two-way analysis of variance and Bonferroni's post hoc tests were performed in A and E.
Knockdown of KDM5A inhibits 3T3-L1 preadipocyte differentiation
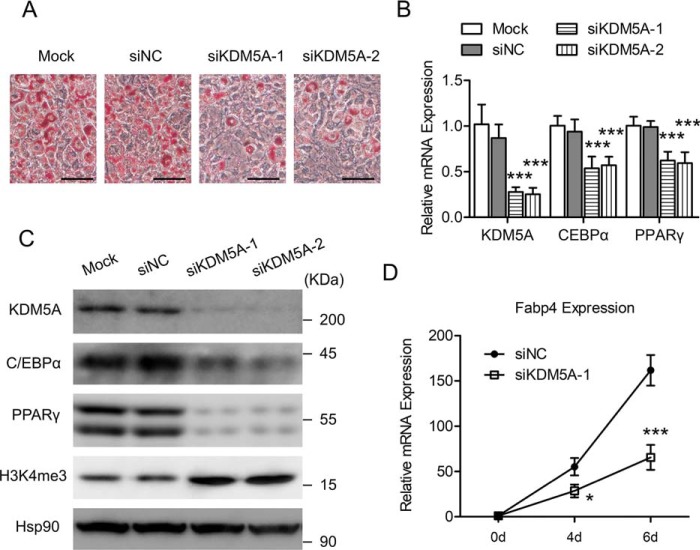
Because C/EBPβ is an important transcription factor for adipocyte differentiation, the transactivation of KDM5A by C/EBPβ suggests that KDM5A may also play a part in the adipogenic differentiation of 3T3-L1 preadipocyte. The siRNA-mediated knockdown of KDM5A was then performed to examine the role of KDM5A in adipogenesis. KDM5A was knocked down in 3T3-L1 preadipocytes, and then the cells were induced to differentiation. Knockdown of KDM5A led to a significant decrease in lipid accumulation in 3T3-L1 cells on day 6 post-induction, as evidenced by ORO staining (Fig. 3A). Moreover, both the mRNA level and the protein level of C/EBPα and PPARγ, the two key pro-adipogenic transcription factors, were decreased by the knockdown of KDM5A (Fig. 3, B and C). Consistent with its H3K4me3 demethylase activity, knockdown of KDM5A led to a global increase of H3K4me3 (Fig. 3C). The fatty acid–binding protein 4 (Fabp4) gene encodes a fatty acid–binding protein found in adipocytes. It is a transcriptional target gene of C/EBPα and PPARγ and is known to be an important adipocyte marker gene (8). As shown in Fig. 3D, the Fabp4 gene was dramatically induced during 3T3-L1 preadipocyte differentiation, but this induction was significantly suppressed by the knockdown of KDM5A. Collectively, these data above demonstrate that KDM5A could play an important role in facilitating the differentiation of 3T3-L1 preadipocytes.
Figure 3.
The siRNA-mediated down-regulation of KDM5A inhibits 3T3-L1 preadipocyte differentiation. 3T3-L1 preadipocyte was transfected with control siRNA (siNC) or KDM5A siRNAs (siKDM5A-1 and siKDM5A-2) and induced to differentiation. A, on day 6 after adipogenic induction, cells were stained with Oil Red O, and representative images are shown. Scale bar, 100 μm. B, on Day 4 after adipogenic induction, the mRNA level of the indicated genes was determined by RT-qPCR. Data were normalized to the mock group. C, on day 4 after adipogenic induction, the indicated proteins were detected by Western blotting. Hsp90 is the loading control. D, at the indicated time point, the mRNA level of Fabp4 was determined by RT-qPCR. Data were normalized to data of day 0. All values are represented as means with error bars representing S.D. *, p < 0.05; ***, p < 0.001. Mock, cells were not infected with viruses or transfected with siRNAs. For statistical analysis, two-way analysis of variance and Bonferroni's post hoc tests were performed in B (compared with siNC) and D (compared with siNC).
PPARγ is a master transcriptional factor that determines adipogenesis, and our results showed that knockdown of KDM5A inhibited adipogenesis with decreased expression of PPARγ (Fig. 3, B and C). We ask whether overexpression of PPARγ could overcome the KDM5A knockdown–mediated suppression of adipogenesis. KDM5A was knocked down in 3T3-L1 preadipocytes, with or without the overexpression of PPARγ, and then the cells were induced to differentiation. As expected, overexpression of PPARγ rescued adipogenesis, which was inhibited by KDM5A knockdown (Fig. S1).
Knockdown of KDM5A increases Wnt6 expression, and the down-regulation of Wnt6 significantly rescues adipogenesis, which is inhibited by the knockdown of KDM5A
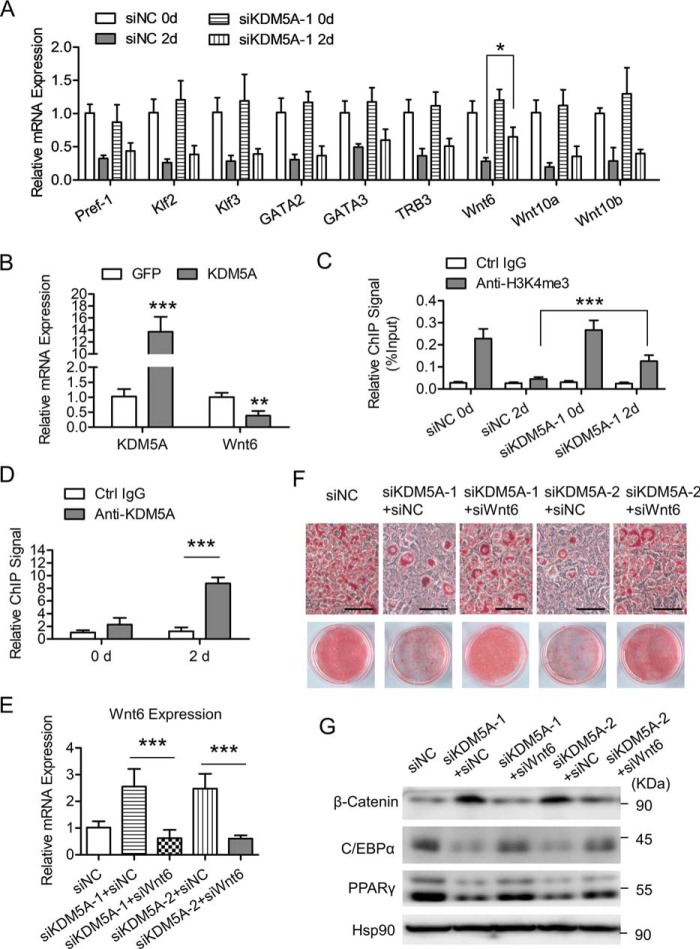
Adipocyte differentiation is controlled by the interplay of a series of positive and negative effectors. Pref-1, Klf2/3, GATA2/3, TRB3, and Wnt proteins are among the well-characterized negative regulators of adipogenesis (21, 22, 38–43). The timely decline of these negative regulators is required for the successful progression of adipocyte differentiation. However, the mechanisms governing the down-regulation of these negative effectors are not completely understood. Because KDM5A is a member of the family of JmjC domain–containing histone demethylases and is able to demethylate H3K4me3, which is a transcriptional activation mark on the gene promoters, we ask whether KDM5A could repress the transcription of the negative regulators during adipogenesis. After adipogenic induction in 3T3-L1 cells (day 2 versus day 0), the mRNA levels of those major negative regulators were dramatically declined (Fig. 4A). When KDM5A was knocked down, the down-regulation of Wnt6 was significantly impeded, whereas the decline of the other negative regulators was only slightly affected (Fig. 4A). Consistently, overexpression of KDM5A significantly inhibited the expression of Wnt6 (Fig. 4B). In addition, the H3K4me3 level on the Wnt6 proximal promoter was dramatically decreased after adipogenic induction, whereas the knockdown of KDM5A significantly restored the H3K4me3 level on the Wnt6 proximal promoter (Fig. 4C). Furthermore, ChIP-qPCR confirmed the significant binding of KDM5A to the proximal promoter of Wnt6 in differentiating cells (day 2 post-induction) but not in cells before induction (day 0), as shown in Fig. 4D. These data suggest that KDM5A could repress Wnt6 transcription through decreasing the H3K4me3 level on its proximal promoter.
Figure 4.
Knockdown of KDM5A increases Wnt6 expression, and the down-regulation of Wnt6 significantly rescues adipogenesis, which is inhibited by the knockdown of KDM5A. A, 3T3-L1 preadipocytes were transfected with control siRNA (siNC) or KDM5A siRNA (siKDM5A-1) and induced to differentiation. Cells were harvested at the indicated time point and subjected to mRNA extraction. The mRNA level of the indicated genes was determined by RT-qPCR. Data were normalized to the siNC day 0 group. B, 3T3-L1 preadipocyte was transfected with plasmids encoding GFP or KDM5A and induced to differentiation. On day 2 after adipogenic induction, the mRNA levels of the indicated genes were determined by RT-qPCR. Data were normalized to GFP group. C, H3K4me3 level on Wnt6 proximal promoter was analyzed by ChIP-qPCR. 3T3-L1 cells were treated as in A. At the times indicated, ChIP-qPCR was performed by using control IgG and anti-H3K4me3 antibody. Data were then normalized to input DNA. D, enrichment of KDM5A on Wnt6 proximal promoter was analyzed by ChIP-qPCR. 3T3-L1 cells were treated as in A. At the times indicated, ChIP-qPCR was performed by using control IgG and anti-KDM5A antibody. Data were normalized to the IgG controls at each time point. E, 3T3-L1 preadipocyte was transfected with the indicated siRNAs and induced to differentiation. On day 2 of adipogenic induction, the mRNA level of Wnt6 was determined by RT-qPCR. Data were normalized to the siNC group. F and G, 3T3-L1 cells were treated as in D, and the following experiments were performed. F, on day 6 after adipogenic induction, cells were stained with Oil Red O. Representative microscopic images and the culture dish images of Oil Red O staining are shown. Scale bar, 100 μm. G, on day 4 after adipogenic induction, Western blotting was performed by using the indicated antibodies. Hsp90 is the loading control. All values are represented as means, with error bars representing S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001. For statistical analysis, one-way analysis of variance and Bonferroni's post hoc tests were carried out in C and E, and two-way analysis of variance and Bonferroni's post hoc tests were performed in A, B, and D.
Wnt6 has been shown to be an important negative regulator during adipogenesis. Overexpression of Wnt6 inhibits adipocyte differentiation in a β-catenin–dependent mechanism (22). Here, we also confirmed that overexpressing Wnt6 in 3T3-L1 preadipocyte inhibited adipogenesis, with decreased C/EBPα and PPARγ expression (Fig. S2). But the expression of both KDM5A and the early adipogenic factor C/EBPβ was not affected by Wnt6 overexpression (Fig. S2). To investigate the role of Wnt6 in KDM5A-facilitated 3T3-L1 preadipocyte differentiation, siRNA-mediated knockdown of Wnt6 was performed. Kdm5A was knocked down in 3T3-L1 preadipocyte, with or without the knockdown of Wnt6, and then the cells were induced to differentiation (Fig. 4E). Knockdown of Wnt6 significantly rescued adipogenesis, which was inhibited by the knockdown of KDM5A, as assessed by ORO staining (Fig. 4F) and key adipogenic protein expression (Fig. 4G). Furthermore, knockdown of KDM5A alone led to increased protein level of β-catenin after adipogenic induction, whereas simultaneous knockdown of Wnt6 attenuated the increase of β-catenin (Fig. 4G). Collectively, these results above suggest that KDM5A could repress the transcription of Wnt6 via decreasing the H3K4me3 level on its proximal promoter, thereby inhibiting the β-catenin signaling to facilitate 3T3-L1 preadipocyte differentiation.
C/EBPβ inhibits Wnt6 transcription through binding to its promoter
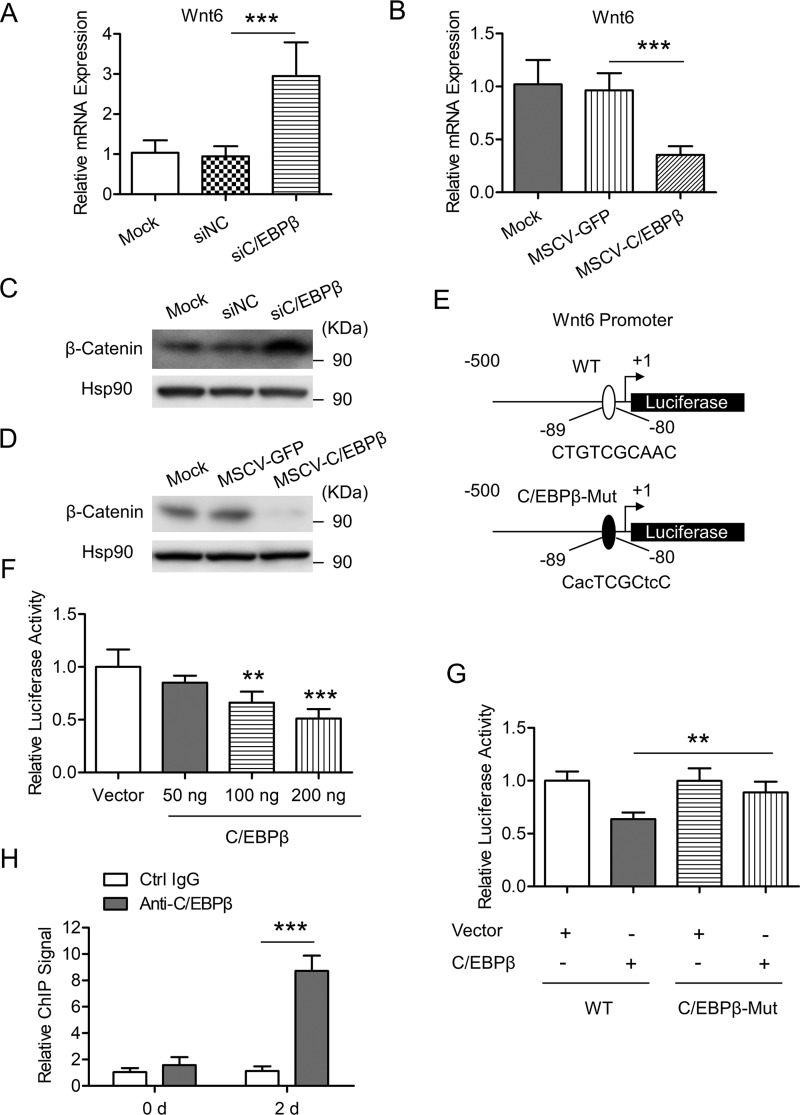
Besides transactivating gene expression, C/EBPβ can also have repressive activity on many target genes (44–46). Interestingly, we found that knockdown of C/EBPβ led to increased Wnt6 expression (Fig. 5A), and overexpression of C/EBPβ attenuated Wnt6 expression during 3T3-L1 preadipocyte differentiation (Fig. 5B). Consistently, knockdown of C/EBPβ increased, whereas overexpression of C/EBPβ decreased, the protein level of β-catenin (Fig. 5, C and D, respectively). Then we asked whether C/EBPβ could directly regulate Wnt6 transcription. 3T3-L1 preadipocyte was co-transfected with a plasmid expressing C/EBPβ and a firefly luciferase reporter plasmid containing Wnt6 proximal promoters (−500 to +1 bp from the TSS, as shown in Fig. 5E). The results showed that C/EBPβ inhibited Wnt6 promoter activity in a dose-dependent manner (Fig. 5F). Bioinformatics analysis predicted a potential binding site for C/EBPβ on the Wnt6 proximal promoter (−89 to −80 bp from the TSS), as shown in Fig. 5E. After this binding site was mutated (C/EBPβ-Mut), the inhibitory role of C/EBPβ in Wnt6 promoter activity was blunted (Fig. 5G), suggesting a critical role of this binding site in the function of C/EBPβ. ChIP-qPCR confirmed the binding of C/EBPβ to the Wnt6 proximal promoter after adipogenic induction in 3T3-L1 cells (Fig. 5H). Taken together, these data indicate that C/EBPβ could inhibit the transcription of Wnt6 through binding to a C/EBPβ consensus sequence on its promoter, which could contribute to the down-regulation of β-catenin signaling during 3T3-L1 preadipocyte differentiation.
Figure 5.
C/EBPβ inhibits Wnt6 transcription through binding to its promoter. A, 3T3-L1 preadipocyte was transfected with the indicated siRNAs and induced to differentiation. Cells were harvested on day 2 after adipogenic induction. The mRNA level of Wnt6 was determined by RT-qPCR. Data were normalized to the mock group. B, 3T3-L1 preadipocyte was infected with retrovirus expressing GFP or C/EBPβ and induced to differentiation. Cells were harvested on day 2 after adipogenic induction. The mRNA level of Wnt6 was determined by RT-qPCR. Data were normalized to the mock group. C, cells were treated as in A, and then the indicated proteins were detected by Western blotting. Hsp90 is the loading control. D, cells were treated as in B, and then the indicated proteins were detected by Western blotting. Hsp90 is the loading control. E, schematic representation of Wnt6 proximal promoter constructs used for luciferase assays. The predicted consensus of C/EBP binding site is shown in the WT luciferase construct. The lowercase letters indicate mutations of the C/EBP binding site in the C/EBPβ-Mut construct. F, 3T3-L1 preadipocyte was transiently transfected with a WT reporter construct as shown in E, along with different amounts of C/EBPβ expression vector, and pRL-TK plasmid was used as an internal control; cell extracts were prepared, and luciferase activities were measured and normalized to Renilla activity. Data were then normalized to the vector group. G, 3T3-L1 preadipocyte was transiently transfected with WT or C/EBPβ-Mut reporter construct as shown in E, along with control vector or C/EBPβ expression vector, and pRL-TK plasmid was used as an internal control. Luciferase activities were measured as in F. H, enrichment of C/EBPβ on Wnt6 proximal promoter was analyzed by ChIP-qPCR. 3T3-L1 cells were induced to differentiation. At the times indicated, ChIP-qPCR was performed by using control IgG and anti-C/EBPβ antibody. Data were normalized to the IgG controls at each time point. All values are represented as means with error bars representing S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001. For statistical analysis, one-way analysis of variance and Bonferroni's post hoc tests were carried out in A, B, and F (compared with vector) and G, and two-way analysis of variance and Bonferroni's post hoc tests were performed in H.
KDM5A interacts with C/EBPβ to cooperatively inhibit Wnt6 transcription
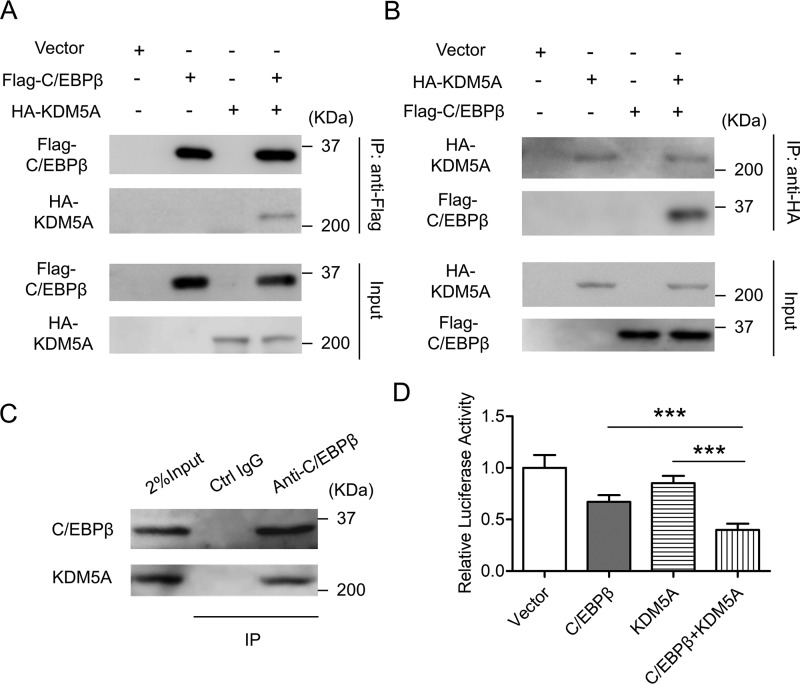
Because our data suggest that both KDM5A and C/EBPβ could be involved in the inhibition of Wnt6 transcription, the relationship between KDM5A and C/EBPβ in this process was then studied. We investigated the interaction between KDM5A and C/EBPβ in cells at both the overexpression level and the endogenous level. As shown in Fig. 6, A and B, HEK293T cells were transiently transfected with the plasmids encoding HA-KDM5A and/or FLAG-C/EBPβ. At 48 h post-transfection, cells were harvested, and co-immunoprecipitation experiments were performed. Immunoprecipitation with anti-FLAG antibody pulled down not only FLAG-C/EBPβ itself, but also HA-KDM5A (Fig. 6A). Similarly, immunoprecipitation with anti-HA antibody pulled down not only HA-KDM5A itself, but also FLAG-C/EBPβ (Fig. 6B). To further confirm the interaction between KDM5A and C/EBPβ, co-immunoprecipitation at the endogenous level was also performed. On day 2 post-induction, 3T3-L1 cells were harvested and subjected to immunoprecipitation with anti-C/EBPβ antibody, which resulted in the pulldown of both endogenous C/EBPβ and endogenous KDM5A (Fig. 6C). Moreover, luciferase assays demonstrated that KDM5A enhanced the ability of C/EBPβ to inhibit the promoter of Wnt6 in 3T3-L1 cells (Fig. 6D). These data suggest that KDM5A may function as a co-factor of C/EBPβ to repress the transcription of Wnt6.
Figure 6.
KDM5A interacts with C/EBPβ to cooperatively inhibit Wnt6 transcription. A and B, HEK293T cells were co-transfected with the indicated expression vectors. After 48 h, cells were harvested, lysed, and subjected to immunoprecipitation by using the indicated antibodies. The immunoprecipitates (IP) and the whole-cell lysates (Input) were subjected to Western blotting by using the anti-FLAG antibody or anti-HA antibody. C, on day 2 post-induction, 3T3-L1 cells were harvested, lysed, and subjected to immunoprecipitation by using control IgG and anti-C/EBPβ antibody. The immunoprecipitates and the whole-cell lysates were subjected to Western blotting by using the indicated antibodies. D, 3T3-L1 preadipocyte was transiently transfected with the reporter construct containing WT Wnt6 promoter, along with the indicated expression vectors. Luciferase activities were measured and plotted. Data were normalized to vector group. All values are represented as means with error bars representing S.D. ***, p < 0.001. For statistical analysis, one-way analysis of variance and Bonferroni's post hoc tests were carried out in D.
Knockdown of C/EBPβ impairs the recruitment of KDM5A to Wnt6 promoter, which has higher H3K4me3 levels
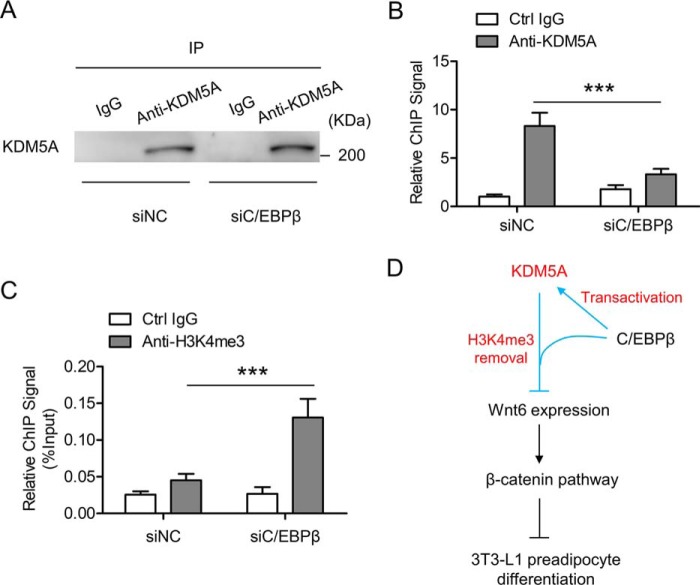
The interaction between KDM5A and C/EBPβ in 3T3-L1 cells prompted us to investigate the role of C/EBPβ in the recruitment of KDM5A to the Wnt6 promoter. C/EBPβ was knocked down in 3T3-L1 preadipocyte, and then the cells were induced to differentiation. Cells were collected on day 2 post-induction for ChIP-qPCR. As shown in Fig. 1F, knockdown of C/EBPβ decreased the expression of KDM5A. Therefore, based on the Western blotting results in Fig. 1F, cell lysates containing an approximately identical amount of KDM5A were taken from the siNC group and siC/EBPβ group for immunoprecipitation by using control IgG and anti-KDM5A antibody. This would guarantee that a similar amount of KDM5A (siNC versus siC/EBPβ) was pulled down in the immunoprecipitates (Fig. 7A), which helps us analyze the ChIP-qPCR results more appropriately. When C/EBPβ was knocked down, the binding of KDM5A to the Wnt6 promoter was significantly impaired (Fig. 7B). Consistently, enrichment of H3K4me3 level on the Wnt6 promoter was elevated upon knockdown of C/EBPβ (Fig. 7C). Taken together, these data suggest that KDM5A is recruited to the Wnt6 promoter in a C/EBPβ-dependent manner, which leads to the decrease of H3K4me3 levels on the Wnt6 promoter and the inactivation of the Wnt6 transcription during 3T3-L1 preadipocyte differentiation.
Figure 7.
Knockdown of C/EBPβ impairs the recruitment of KDM5A to Wnt6 promoter, which has higher H3K4me3 levels. A, 3T3-L1 preadipocyte was transfected with control siRNA (siNC) or C/EBPβ siRNA (siC/EBPβ) and induced to differentiation. On day 2 after adipogenic induction, cells were harvested. Based on the Western blotting results in Fig. 1F, cell lysates containing approximately identical amounts of KDM5A were taken from the siNC group and siC/EBPβ group for immunoprecipitation by using control IgG and anti-KDM5A antibody. The immunoprecipitates (IP) were subjected to Western blotting, showing that similar amounts of KDM5A were pulled down in both the siNC group and siC/EBPβ group. B, 3T3-L1 cells were treated as in A, and on day 2 post-induction, cell lysates containing similar amounts of KDM5A were taken from the siNC group and siC/EBPβ group for ChIP-qPCR to analyze the enrichment of KDM5A on the Wnt6 proximal promoter. Data were normalized to the IgG controls in the siNC group. C, 3T3-L1 cells were treated as in A, and on day 2 post-induction, cells were harvested and subjected to ChIP-qPCR to analyze to H3K4me3 level on the Wnt6 proximal promoter. Data were then normalized to input DNA. D, a proposed working model for the transcriptional network involving a DNA-binding transcription factor (C/EBPβ) and a chromatin regulator (KDM5A) in the regulation of the Wnt/β-catenin pathway during 3T3-L1 preadipocyte differentiation. All values are represented as means with error bars representing S.D. ***, p < 0.001. For statistical analysis, one-way analysis of variance and Bonferroni's post hoc tests were carried out in B and C.
Discussion
The transcription factor C/EBPβ is considered as an important regulator in the initiation of adipogenesis. It is expressed shortly after adipogenic induction and then activates the expression of a cascade of genes, including C/EBPα and PPARγ (13). Although some C/EBPβ-regulated genes have been identified, there are still a lot that are unknown. Our laboratory used ChIP-on-chip experiments to identify a series of C/EBPβ target genes during the process of 3T3-L1 preadipocyte differentiation, among which is KDM5A.
In the current study, we demonstrated that KDM5A is induced in 3T3-L1 cells after hormonal induction, and the expression profile of KDM5A is positively correlated with that of C/EBPβ during 3T3-L1 preadipocyte differentiation (Fig. 1, A and B). C/EBPβ is required for the induction of KDM5A, because knockdown of C/EBPβ decreased KDM5A expression (Fig. 1, E and F). We used both ChIP-qPCR and luciferase assays to confirm that C/EBPβ can bind to the promoter of KDM5A and transactivate it (Fig. 2, A–D). Consistently, overexpression of C/EBPβ increased KDM5A expression (Fig. 2, E and F). Then we used the loss-of-function experiments to examine the role of KDM5A in adipocyte differentiation (Fig. 3). Knockdown of KDM5A in 3T3-L1 preadipocyte inhibited lipid accumulation and was associated with down-regulated expression of C/EBPα and PPARγ, the two key adipogenic factors. Further studies indicate that KDM5A is required for 3T3-L1 preadipocyte differentiation at least partially because of its role in inhibiting Wnt6 gene expression and blocking the subsequent Wnt/β-catenin pathway (Fig. 4). Therefore, these data indicate that KDM5A funcitons as an important downstream target of C/EBPβ to facilitate 3T3-L1 preadipocyte differentiation, providing new insights into the role of C/EBPβ in adipogenesis.
The precise regulation of gene expression is critical for normal progression of cell differentiation. Gene expression can be regulated at the epigenetic level, which is achieved via various mechanisms, including different modifications on the lysine residues of histones. Growing evidence has indicated that adipogenesis is sensitive to epigenetic changes. During adipogenesis, the level of repressive epigenetic mark H3K27me3 is increased on the promoters of multiple negative adipogenic regulators, which is mediated by histone H3K27 methyltransferase Ezh2, thereby promoting adipogenesis (47). H3K9me2, another repressive epigenetic mark, is enriched on the entire PPARγ locus in preadipocytes, which is mediated by the histone methyltransferase G9a (48–50). After adipogenesis of 3T3-L1 cells, H3K9me2 levels on the entire PPARγ locus decreased markedly due to the down-regulation of G9a. The histone demethylase Kdm4b is induced at the early stage of 3T3-L1 preadipocyte differentiation and demethylates H3K9me3 on the regulatory regions of some cell cycle genes to promote their expression and the subsequent MCE, an important cell proliferation process during the early phase of 3T3-L1 cells adipogenesis (11). In our present study, it is shown that knockdown of KDM5A inhibits adipogenesis of 3T3-L1 cells, which is associated with increased Wnt6 expression, higher level of H3K4me3 on the Wnt6 promoter, and activated Wnt/β-catenin signaling (Figs. 3 and 4). ChIP-qPCR and luciferase assays indicate that KDM5A could be recruited to the Wnt6 promoter and inhibits its transcription in cooperation with C/EBPβ (Figs. 6 and 7). Moreover, siRNA-mediated down-regulation of Wnt6 in 3T3-L1 cells significantly rescues adipogenesis, which is inhibited by the knockdown of KDM5A (Fig. 4, E–G). These results suggest that KDM5A could inhibit the expression of Wnt6 through decreasing the active epigenetic mark H3K4me3 on its promoter, which leads to the inhibition of the Wnt/β-catenin pathway and activation of the adipogenic program. Thus, KDM5A could also be an important epigenetic regulator for adipogenesis.
Mounting evidence has shown that KDM5A is involved in cell fate determination. KDM5A is a critical epigenetic factor that maintains neural progenitor cell proliferation and multipotency by repressing astroglial differentiation (51). KDM5A is up-regulated in osteoporosis and impairs the ability of bone morphogenetic protein 2 (BMP2) to promote osteogenic differentiation (52). The role of KDM5A in promoting the proliferation and survival of cancer cells suggests that it may be critical for the proliferation of stem cells and progenitor cells and stemness maintenance (53). Conversely, KDM5A is required for cell differentiation in some other cases. KDM5A-mediated H3K4 demethylation represses cell cycle genes, leading to cell cycle exit and facilitating U937 differentiation induced by 12-O-tetradecanoylphorbol-13-acetate (33). Here in this paper, our results suggest that KDM5A could facilitate 3T3-L1 preadipocyte differentiation through inhibiting the Wnt/β-catenin pathway. According to the data in previous reports and in the present study, KDM5A appears to have the ability to either prevent or to facilitate cell differentiation, which depends on the cellular context. Besides, an inverse correlation between adipogenesis and osteogenesis has been convincingly demonstrated (54, 55). A previous report showed an inhibitory role of KDM5A in BMP2-induced osteogenesis (52), whereas our data indicated that KDM5A could facilitate adipogenesis of 3T3-L1 cells. Together, these results suggest that KDM5A could be an important regulator influencing the differentiation fate of mesenchymal precursors.
In this study, it is shown that C/EBPβ promotes KDM5A transcription through binding to its promoter but inhibits Wnt6 transcription through binding to its promoter. Similar cases have also been reported for some other transcriptional regulators. For example, glucocorticoid receptor facilitates 3T3-L1 preadipocyte differentiation by activating the transcription of E4 promoter–binding protein 4 (E4BP4) and repressing the transcription of runt-related transcription factor 2 (Runx2) (56, 57). The different roles of a transcriptional factor in the promoter activities of its target genes may be the result of different transcriptional complexes formed on the promoters. Further studies are needed to delineate the underlying mechanism.
Our ChIP-qPCR results show that KDM5A bound to the proximal promoter of Wnt6 and H3K4me3 modification was also located around this region. Other researchers have reported similar results indicating that KDM5A preferentially binds to proximal promoter regions and that the H3K4me3 is generally enriched around the TSSs (50, 51). KDM5A is known to interact with many partner proteins, such as the pRB family, TATA-binding protein, and a repressor of erythroid development (51). It usually functions as a co-factor of other transcription factors to regulate the epigenetic status of the promoters. Here, we demonstrate that C/EBPβ not only transactivates the expression of KDM5A, but also associates with KDM5A to facilitate KDM5A recruitment to the Wnt6 gene promoter, leading to the reduction of H3K4me3 levels on the Wnt6 promoter and the suppression of Wnt6 transcription. As illustrated in Fig. 7D, our results suggest a new function of C/EBPβ in the epigenetic regulation of adipogenesis and unveil a transcriptional network involving a DNA-binding transcription factor (C/EBPβ) and a chromatin regulator (KDM5A) in the regulation of the Wnt/β-catenin pathway during 3T3-L1 preadipocyte differentiation. These new insights into the epigenetic mechanism of adipogenesis may provide a functional pathway with therapeutic potential against obesity and its related metabolic disorders. As the epigenetic processes continue to receive more attention, more mechanisms underlying the epigenetic regulation during adipogenesis will be elucidated.
Experimental procedures
Cell culture and induction of differentiation
3T3-L1 preadipocyte was propagated and maintained in DMEM (Invitrogen) containing 10% calf serum (Gibco). The 2-day post-confluent 3T3-L1 preadipocytes (designated day 0) were induced to differentiation with DMEM containing 10% FBS (Gibco) and a mixture of inducers (MDI): 0.5 mm 3-isobutyl-1-methylxanthine (M; Sigma), 1 mm dexamethasone (D; Sigma), and 1 μg/ml insulin (I; Sigma), until day 2. Cells were then cultured in DMEM supplemented with 10% FBS and 1 μg/ml insulin for 2 days, after which they were fed every other day with DMEM containing 10% FBS. The cells expressed adipocyte-specific proteins beginning on day 3 and obtained the biochemical and morphological characteristics of mature adipocytes by day 6. Overexpression of the genes was performed in 3T3L-1 preadipocytes when the cells were about 50% confluent, and knockdown assays were performed on day −2, when the cells were confluent, and the cells were harvested at the indicated time points.
Oil Red O staining
At the indicated time points, to determine lipid accumulation, the medium was discarded, and cells were washed three times with PBS. Then the cells were fixed with 3.7% formaldehyde for 10 min and incubated with Oil Red O for 2 h at room temperature. Oil Red O stock solution (0.5% in isopropyl alcohol) was diluted with water (3:2) before using. Cells were washed with water, and the stained fat droplets in the cells were visualized by light microscopy and photographed. Finally, all of the water was removed, 1 ml of isopropyl alcohol was added into the cells for 10 min, and optical density was measured at 510 nm.
Plasmid constructs and siRNAs
C/EBPβ was cloned into MSCV vector, which was managed to generate stably transfected cell lines. MSCV retroviruses were prepared as described before (17). FLAG-C/EBPβ, HA-KDM5A, Wnt6, and PPARγ were cloned into PCDNA3.1 vector for transient transfection in 3T3-L1 cells by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The siRNAs were designed and synthesized by GenePharma. The target sequences for successful siRNAs were as follows: siC/EBPβ, GCCCTGAGTAATCACTTAAAG; siKDM5A-1, GCACAATCCTATGACACTTGG; siKDM5A-2, GCTCGTGAATGGACAGCTAAA; siWnt6, GCATTGGTGCAACTGCACAAC; siNC, TTCTCCGAACGTGTCACGT. 3T3-L1 cells were transfected at ∼100% confluence with siRNAs using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. 48 h after cells reached post-confluence, they were subjected to the standard differentiation protocol as described earlier, and at various times thereafter, cells were harvested for the tests. The siNC was used as a negative control.
ChIP
ChIP was performed as described before (11, 15). 3T3-L1 cells were fixed in 1% formaldehyde in a fume hood for 10 min, incubated with volume of 2.5 m glycine, gently mixed at room temperature for 5 min, and washed three times with ice-cold 1× PBS containing 1 mm phenylmethylsulfonyl fluoride (PMSF). Cells were scraped off and washed three times with 1 ml of lysis buffer with fresh PMSF. They were lysed in lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris-HCl (pH 8.0), 1 mm PMSF). Lysates were vortexed and sonicated with a Bioruptor (Diagenode, Denville, NJ). The average length of DNA fragments ranged between 300 and 800 bp. The lysates were clarified by centrifugation and diluted 10-fold in ChIP buffer (20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 2 mm EDTA, 1% Triton X-100, and complete protease inhibitor tablets from Roche Applied Science). The samples were precleared using protein A–Sepharose beads for 1 h at 4 °C, and 10% of each sample was used for input control. The samples were immunoprecipitated with the indicated antibodies: anti-C/EBPβ (Santa Cruz Biotechnology, Inc., sc-150), anti-KDM5A (Abcam, ab70892), anti-H3K4me3 (Abcam, ab8580), and control IgG (Abcam, ab46540). The immune complexes were washed with low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl (pH 8.0), 150 mm NaCl), followed by high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl (pH 8.0), 500 mm NaCl), washed with LiCl buffer (0.25 m LiCl, 1% Nonidet P-40, 1% deoxycholate, 1 mm EDTA, 10 mm Tris-HCl (pH 8.0)) followed by TE buffer (10 mm Tris-HCl (pH 8.0), 1 mm EDTA), and finally eluted with elution buffer (50 mm Tris-HCl (pH 8.0), 10 mm EDTA, 1% SDS). Immunoprecipitated DNA was reverse-cross-linked at 65 °C for 12 h and purified using a DNA purification kit (Qiagen). The same amount of purified input control DNA or ChIP DNA from IgG and specific antibodies was used to perform the RT-qPCR using primers specific to the promoters of the indicated genes. Primers used for ChIP-qPCR were as follows: 5′-AAGGCGACCGAGCGAAAC-3′ (KDM5A-forward), 5′-CGTTCTCCGAGACCTGTT-3′ (KDM5A-reverse), 5′-AACCCCGCAGAGGCTAGGAGA-3′ (Wnt6-forward), and 5′-GGGGTGGCAGTTGCGACAGT-3′ (Wnt6-reverse).
Immunoprecipitation assay
The immunoprecipitation assay was performed as described before (58). Cells were washed with PBS, scraped off, and collected by centrifugation. Then cells were suspended in radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS) in the presence of protease inhibitors (Roche Applied Science) for 3 h at 4 °C. After centrifugation, the supernates were incubated with the indicated antibodies at 4 °C overnight. The next day, protein A–agarose beads (Invitrogen) was added. After a 3-h incubation, the beads were washed with TBS-T (TBS + 0.05% Tween 20) in the presence of protease inhibitors (Roche Applied Science). The immunoprecipitates were separated by SDS-PAGE and subjected to Western blotting. Antibodies including anti-FLAG (Abclonal, AE005), anti-HA (Abclonal, AE036), anti-C/EBPβ (Santa Cruz Biotechnology, sc-150), anti-KDM5A (Abcam, ab70892), and control IgG (Abcam, ab46540) were used for the immunoprecipitation assay.
RNA isolation and qPCR
Total RNAs were extracted with TRIzol (Invitrogen) and transcribed to cDNA using the Superscript III kit (Invitrogen) according to the manufacturer's instructions. The cDNA was analyzed using the Power SYBR Green PCR kit on the ABI PRISM 7500 Q-PCR machine (Applied Biosystems). All qPCR data were normalized to 18S rRNA. Primers used for RT-qPCR were as follows: 5′-CGCCGCTAGAGGTGAAATTCT-3′ (18S rRNA-forward), 5′-CATTCTTGGCAAATGCTTTCG-3′ (18S rRNA-reverse), 5′-ACGACTTCCTCTCCGACCTCT-3′ (C/EBPβ-forward), 5′-CGAGGCTCACGTAACCGTAGT-3′ (C/EBPβ-reverse), 5′-CAAGAACAGCAACGAGTACCG-3′ (C/EBPα-forward), 5′-GTCACTCGTCAACTCCAGCAC-3′ (C/EBPα-reverse), 5′-TGCTGTTATGGGTGAAACTCT-3′ (PPARγ-forward), 5′-CGCTTGATGTCAAAGGAATGC-3′ (PPARγ-reverse), 5′-TGCCTTTGTGGGAACCTG-3′ (Fabp4-forward), 5′-GCTTGTCACCATCTCGTTTTC-3′ (Fabp4-reverse), 5′-CACAGACCCGCTGAGTTTTAT-3′ (KDM5A-forward), 5′-CTTCACAGGCAAATGGAGGTT-3′ (KDM5A-reverse), 5′-GCGGAGACGATGTGGACTTC-3′ (Wnt6-forward), and 5′-ATGCACGGATATCTCCACGG-3′ (Wnt6-reverse). Information on the other primers used for RT-qPCR is available upon request.
Western blotting analyses
To detect protein levels (except β-catenin), cells were lysed with lysis buffer containing 50 mm Tris (pH 7.5), 150 mm NaCl, 5 mm NaF, 25 mm β-glycerol phosphate, 1 mm sodium orthovanadate, 10% glycerol, 1% Triton X-100, 1 mm DTT, and freshly added protease inhibitors (Roche Applied Science). To detect β-catenin, cytosolic fractions of the cells were extracted as described (20) and were used for Western blotting. Equal amounts of protein were separated by SDS-PAGE and transferred to polyvinylidene fluoride membranes (Millipore), immunoblotted with the indicated antibodies: anti-KDM5A (Abcam, ab70892), anti-C/EBPβ (Santa Cruz Biotechnology, sc-150), anti-C/EBPα (Cell Signaling, catalog no. 2295), anti-PPARγ (Cell Signaling, catalog no. 2430), anti-β-catenin (Proteintech, 51067-2-AP), anti-Hsp90 (Santa Cruz Biotechnology, sc-7947), anti-FLAG (Abclonal, AE005), and anti-HA (Abclonal, AE036). Then they were visualized with horseradish peroxidase-coupled secondary antibodies.
Luciferase reporter assays
The proximal promoter regions of mouse KDM5A, Wnt6, and their artificial mutants were amplified via PCR and subcloned into the firefly luciferase reporter construct PGL3-basic (Promega). 3T3-L1 cells (2 × 105 cells/well) were transfected with 300 ng/well firefly luciferase reporter constructs, 2 ng/well Renilla luciferase reporter plasmids, in combination with pcDNA3.1(−) vector, C/EBPβ plasmid, or KDM5A plasmid, by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. After 48 h, luciferase activity was measured using a Dual-Luciferase Reporter Assay (Promega), normalizing firefly luciferase to Renilla activity.
Statistical analysis
Results are expressed as means with error bars representing S.D. Comparisons between groups (n = 5 in each group) were made using unpaired two-tailed Student's t tests. For comparisons of three or more independent groups (n = 5 in each group) with only one variable, one-way analyses of variance plus Bonferroni's post hoc tests were performed. For comparisons of two or more independent groups (n = 5 in each group) with two variables, two-way analyses of variance plus Bonferroni's post hoc tests were carried out. The statistical analyses are also indicated in the legends to each figure, with p < 0.05 being considered statistically significant. All experiments were repeated a minimum of three times, and representative data are shown.
Author contributions
L. G. was involved in study design, conducted the experiments, analyzed the data, and drafted the paper; Y.-Y. G., B.-Y. L., and W.-Q. P. performed the experiments; L. G. and Q.-Q. T. designed and supervised the study and wrote the paper.
Supplementary Material
This study was supported by National Natural Science Foundation (NSFC) Grants 31871435 and 31370027 (to L. G.), 81730021 and 31571471 (to Q.-Q. T.), and Ministry of Science and Technology of the People's Republic of China (MOST) Grant 2013CB530601 (to Q.-Q. T). The department is supported by 985 Project 985 III-YFX0302. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2.
- PPARγ
- peroxisome proliferator–activated receptor γ
- C/EBPα
- CCAAT/enhancer-binding protein α
- C/EBPβ
- CCAAT/enhancer-binding protein β
- KDM
- lysine-specific demethylase
- qPCR
- real-time quantitative PCR
- MSCV
- murine stem cell virus
- MCE
- mitotic clonal expansion
- H3K4
- H3K9, and H3K27, histone H3 Lys-4, Lys-9, and Lys-27, respectively
- me3
- trimethyl
- me2
- dimethyl
- ORO
- Oil Red O
- TSS
- transcription start site
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- PMSF
- phenylmethylsulfonyl fluoride.
References
- 1. Shukla A. P., Buniak W. I., and Aronne L. J. (2015) Treatment of obesity in 2015. J. Cardiopulm. Rehabil. Prev. 35, 81–92 10.1097/HCR.0000000000000112 [DOI] [PubMed] [Google Scholar]
- 2. Grundy S. M. (2016) Overnutrition, ectopic lipid and the metabolic syndrome. J. Investig. Med. 64, 1082–1086 10.1136/jim-2016-000155 [DOI] [PubMed] [Google Scholar]
- 3. Spiegelman B. M., and Flier J. S. (2001) Obesity and the regulation of energy balance. Cell 104, 531–543 10.1016/S0092-8674(01)00240-9 [DOI] [PubMed] [Google Scholar]
- 4. Rosen E. D., and MacDougald O. A. (2006) Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 7, 885–896 10.1038/nrm2066 [DOI] [PubMed] [Google Scholar]
- 5. Ferrante A. W., Jr. (2007) Obesity-induced inflammation: a metabolic dialogue in the language of inflammation. J. Intern. Med. 262, 408–414 10.1111/j.1365-2796.2007.01852.x [DOI] [PubMed] [Google Scholar]
- 6. Gesta S., Tseng Y. H., and Kahn C. R. (2007) Developmental origin of fat: tracking obesity to its source. Cell 131, 242–256 10.1016/j.cell.2007.10.004 [DOI] [PubMed] [Google Scholar]
- 7. Green H., and Kehinde O. (1975) An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell 5, 19–27 10.1016/0092-8674(75)90087-2 [DOI] [PubMed] [Google Scholar]
- 8. Tang Q. Q., and Lane M. D. (2012) Adipogenesis: from stem cell to adipocyte. Annu. Rev. Biochem. 81, 715–736 10.1146/annurev-biochem-052110-115718 [DOI] [PubMed] [Google Scholar]
- 9. Tang Q. Q., and Lane M. D. (1999) Activation and centromeric localization of CCAAT/enhancer-binding proteins during the mitotic clonal expansion of adipocyte differentiation. Genes Dev. 13, 2231–2241 10.1101/gad.13.17.2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang Y. Y., Li X., Qian S. W., Guo L., Huang H. Y., He Q., Liu Y., Ma C. G., and Tang Q. Q. (2011) Transcriptional activation of histone H4 by C/EBPβ during the mitotic clonal expansion of 3T3-L1 adipocyte differentiation. Mol. Biol. Cell 22, 2165–2174 10.1091/mbc.e10-11-0912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guo L., Li X., Huang J. X., Huang H. Y., Zhang Y. Y., Qian S. W., Zhu H., Zhang Y. D., Liu Y., Liu Y., Wang K. K., and Tang Q. Q. (2012) Histone demethylase Kdm4b functions as a co-factor of C/EBPβ to promote mitotic clonal expansion during differentiation of 3T3-L1 preadipocytes. Cell Death Differ. 19, 1917–1927 10.1038/cdd.2012.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu Z., Rosen E. D., Brun R., Hauser S., Adelmant G., Troy A. E., McKeon C., Darlington G. J., and Spiegelman B. M. (1999) Cross-regulation of C/EBPα and PPARγ controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol. Cell 3, 151–158 10.1016/S1097-2765(00)80306-8 [DOI] [PubMed] [Google Scholar]
- 13. Guo L., Li X., and Tang Q. Q. (2015) Transcriptional regulation of adipocyte differentiation: a central role for CCAAT/enhancer-binding protein (C/EBP) β. J. Biol. Chem. 290, 755–761 10.1074/jbc.R114.619957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tang Q. Q., Otto T. C., and Lane M. D. (2003) CCAAT/enhancer-binding protein beta is required for mitotic clonal expansion during adipogenesis. Proc. Natl. Acad. Sci. U.S.A. 100, 850–855 10.1073/pnas.0337434100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guo L., Huang J. X., Liu Y., Li X., Zhou S. R., Qian S. W., Liu Y., Zhu H., Huang H. Y., Dang Y. J., and Tang Q. Q. (2013) Transactivation of Atg4b by C/EBPβ promotes autophagy to facilitate adipogenesis. Mol. Cell. Biol. 33, 3180–3190 10.1128/MCB.00193-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jiang Y., Guo L., Xie L. Q., Zhang Y. Y., Liu X. H., Zhang Y., Zhu H., Yang P. Y., Lu H. J., and Tang Q. Q. (2014) Proteome profiling of mitotic clonal expansion during 3T3-L1 adipocyte differentiation using iTRAQ-2DLC-MS/MS. J. Proteome Res. 13, 1307–1314 10.1021/pr401292p [DOI] [PubMed] [Google Scholar]
- 17. Liu Y., Peng W. Q., Guo Y. Y., Liu Y., Tang Q. Q., and Guo L. (2018) Kruppel-like factor 10 (KLF10) is transactivated by the transcription factor C/EBPβ and involved in early 3T3-L1 preadipocyte differentiation. J. Biol. Chem. 293, 14012–14021 10.1074/jbc.RA118.004401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. MacDonald B. T., Tamai K., and He X. (2009) Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell 17, 9–26 10.1016/j.devcel.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clevers H., and Nusse R. (2012) Wnt/β-catenin signaling and disease. Cell 149, 1192–1205 10.1016/j.cell.2012.05.012 [DOI] [PubMed] [Google Scholar]
- 20. Bennett C. N., Ross S. E., Longo K. A., Bajnok L., Hemati N., Johnson K. W., Harrison S. D., and MacDougald O. A. (2002) Regulation of Wnt signaling during adipogenesis. J. Biol. Chem. 277, 30998–31004 10.1074/jbc.M204527200 [DOI] [PubMed] [Google Scholar]
- 21. Longo K. A., Wright W. S., Kang S., Gerin I., Chiang S. H., Lucas P. C., Opp M. R., and MacDougald O. A. (2004) Wnt10b inhibits development of white and brown adipose tissues. J. Biol. Chem. 279, 35503–35509 10.1074/jbc.M402937200 [DOI] [PubMed] [Google Scholar]
- 22. Cawthorn W. P., Bree A. J., Yao Y., Du B., Hemati N., Martinez-Santibañez G., and MacDougald O. A. (2012) Wnt6, Wnt10a and Wnt10b inhibit adipogenesis and stimulate osteoblastogenesis through a beta-catenin-dependent mechanism. Bone 50, 477–489 10.1016/j.bone.2011.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ross S. E., Hemati N., Longo K. A., Bennett C. N., Lucas P. C., Erickson R. L., and MacDougald O. A. (2000) Inhibition of adipogenesis by Wnt signaling. Science 289, 950–953 10.1126/science.289.5481.950 [DOI] [PubMed] [Google Scholar]
- 24. Shi Y. (2007) Histone lysine demethylases: emerging roles in development, physiology and disease. Nat. Rev. Genet. 8, 829–833 10.1038/nrg2218 [DOI] [PubMed] [Google Scholar]
- 25. ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. D'Urso A., and Brickner J. H. (2014) Mechanisms of epigenetic memory. Trends Genet. 30, 230–236 10.1016/j.tig.2014.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dimitrova E., Turberfield A. H., and Klose R. J. (2015) Histone demethylases in chromatin biology and beyond. EMBO Rep. 16, 1620–1639 10.15252/embr.201541113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Klose R. J., Yan Q., Tothova Z., Yamane K., Erdjument-Bromage H., Tempst P., Gilliland D. G., Zhang Y., and Kaelin W. G. Jr. (2007) The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell 128, 889–900 10.1016/j.cell.2007.02.013 [DOI] [PubMed] [Google Scholar]
- 29. Plch J., Hrabeta J., and Eckschlager T. (2019) KDM5 demethylases and their role in cancer cell chemoresistance. Int. J. Cancer 144, 221–231 10.1002/ijc.31881 [DOI] [PubMed] [Google Scholar]
- 30. Zeng J., Ge Z., Wang L., Li Q., Wang N., Björkholm M., Jia J., and Xu D. (2010) The histone demethylase RBP2 is overexpressed in gastric cancer and its inhibition triggers senescence of cancer cells. Gastroenterology 138, 981–992 10.1053/j.gastro.2009.10.004 [DOI] [PubMed] [Google Scholar]
- 31. Lopez-Bigas N., Kisiel T. A., Dewaal D. C., Holmes K. B., Volkert T. L., Gupta S., Love J., Murray H. L., Young R. A., and Benevolenskaya E. V. (2008) Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol. Cell 31, 520–530 10.1016/j.molcel.2008.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. DiTacchio L., Le H. D., Vollmers C., Hatori M., Witcher M., Secombe J., and Panda S. (2011) Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science 333, 1881–1885 10.1126/science.1206022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Beshiri M. L., Holmes K. B., Richter W. F., Hess S., Islam A. B., Yan Q., Plante L., Litovchick L., Gévry N., Lopez-Bigas N., Kaelin W. G. Jr., and Benevolenskaya E. V. (2012) Coordinated repression of cell cycle genes by KDM5A and E2F4 during differentiation. Proc. Natl. Acad. Sci. U.S.A. 109, 18499–18504 10.1073/pnas.1216724109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chicas A., Kapoor A., Wang X., Aksoy O., Evertts A. G., Zhang M. Q., Garcia B. A., Bernstein E., and Lowe S. W. (2012) H3K4 demethylation by Jarid1a and Jarid1b contributes to retinoblastoma-mediated gene silencing during cellular senescence. Proc. Natl. Acad. Sci. U.S.A. 109, 8971–8976 10.1073/pnas.1119836109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Váraljai R., Islam A. B., Beshiri M. L., Rehman J., Lopez-Bigas N., and Benevolenskaya E. V. (2015) Increased mitochondrial function downstream from KDM5A histone demethylase rescues differentiation in pRB-deficient cells. Genes Dev. 29, 1817–1834 10.1101/gad.264036.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhao D., Zhang Q., Liu Y., Li X., Zhao K., Ding Y., Li Z., Shen Q., Wang C., Li N., and Cao X. (2016) H3K4me3 demethylase Kdm5a is required for NK cell activation by associating with p50 to suppress SOCS1. Cell Rep. 15, 288–299 10.1016/j.celrep.2016.03.035 [DOI] [PubMed] [Google Scholar]
- 37. Price B. D. (2017) KDM5A demethylase: erasing histone modifications to promote repair of DNA breaks. J. Cell Biol. 216, 1871–1873 10.1083/jcb.201705005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tong Q., Dalgin G., Xu H., Ting C. N., Leiden J. M., and Hotamisligil G. S. (2000) Function of GATA transcription factors in preadipocyte-adipocyte transition. Science 290, 134–138 10.1126/science.290.5489.134 [DOI] [PubMed] [Google Scholar]
- 39. Banerjee S. S., Feinberg M. W., Watanabe M., Gray S., Haspel R. L., Denkinger D. J., Kawahara R., Hauner H., and Jain M. K. (2003) The Kruppel-like factor KLF2 inhibits peroxisome proliferator-activated receptor-γ expression and adipogenesis. J. Biol. Chem. 278, 2581–2584 10.1074/jbc.M210859200 [DOI] [PubMed] [Google Scholar]
- 40. Bezy O., Vernochet C., Gesta S., Farmer S. R., and Kahn C. R. (2007) TRB3 blocks adipocyte differentiation through the inhibition of C/EBPβ transcriptional activity. Mol. Cell. Biol. 27, 6818–6831 10.1128/MCB.00375-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sue N., Jack B. H., Eaton S. A., Pearson R. C., Funnell A. P., Turner J., Czolij R., Denyer G., Bao S., Molero-Navajas J. C., Perkins A., Fujiwara Y., Orkin S. H., Bell-Anderson K., and Crossley M. (2008) Targeted disruption of the basic Kruppel-like factor gene (Klf3) reveals a role in adipogenesis. Mol. Cell. Biol. 28, 3967–3978 10.1128/MCB.01942-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sul H. S. (2009) Minireview: Pref-1: role in adipogenesis and mesenchymal cell fate. Mol. Endocrinol. 23, 1717–1725 10.1210/me.2009-0160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jack B. H., and Crossley M. (2010) GATA proteins work together with friend of GATA (FOG) and C-terminal binding protein (CTBP) co-regulators to control adipogenesis. J. Biol. Chem. 285, 32405–32414 10.1074/jbc.M110.141317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dentesano G., Straccia M., Ejarque-Ortiz A., Tusell J. M., Serratosa J., Saura J., and Solà C. (2012) Inhibition of CD200R1 expression by C/EBPβ in reactive microglial cells. J. Neuroinflammation 9, 165 10.1186/1742-2094-9-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Verhoog N., Allie-Reid F., Vanden Berghe W., Smith C., Haegeman G., Hapgood J., and Louw A. (2014) Inhibition of corticosteroid-binding globulin gene expression by glucocorticoids involves C/EBPβ. PLoS One 9, e110702 10.1371/journal.pone.0110702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Arcidiacono M. V., Yang J., Fernandez E., and Dusso A. (2015) The induction of C/EBPβ contributes to vitamin D inhibition of ADAM17 expression and parathyroid hyperplasia in kidney disease. Nephrol. Dial. Transplant. 30, 423–433 10.1093/ndt/gfu311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang L., Jin Q., Lee J. E., Su I. H., and Ge K. (2010) Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 7317–7322 10.1073/pnas.1000031107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cristancho A. G., and Lazar M. A. (2013) Double SET point: G9a makes its mark in adipogenesis. EMBO J. 32, 4–6 10.1038/emboj.2012.320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li S. F., Guo L., Qian S. W., Liu Y., Zhang Y. Y., Zhang Z. C., Zhao Y., Shou J. Y., Tang Q. Q., and Li X. (2013) G9a is transactivated by C/EBPβ to facilitate mitotic clonal expansion during 3T3-L1 preadipocyte differentiation. Am. J. Physiol. Endocrinol Metab. 304, E990–E998 10.1152/ajpendo.00608.2012 [DOI] [PubMed] [Google Scholar]
- 50. Wang L., Xu S., Lee J. E., Baldridge A., Grullon S., Peng W., and Ge K. (2013) Histone H3K9 methyltransferase G9a represses PPARγ expression and adipogenesis. EMBO J. 32, 45–59 10.1038/emboj.2012.306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kong S. Y., Kim W., Lee H. R., and Kim H. J. (2018) The histone demethylase KDM5A is required for the repression of astrocytogenesis and regulated by the translational machinery in neural progenitor cells. FASEB J. 32, 1108–1119 10.1096/fj.201700780R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang C., Wang J., Li J., Hu G., Shan S., Li Q., and Zhang X. (2016) KDM5A controls bone morphogenic protein 2-induced osteogenic differentiation of bone mesenchymal stem cells during osteoporosis. Cell Death Dis. 7, e2335 10.1038/cddis.2016.238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lin W., Cao J., Liu J., Beshiri M. L., Fujiwara Y., Francis J., Cherniack A. D., Geisen C., Blair L. P., Zou M. R., Shen X., Kawamori D., Liu Z., Grisanzio C., Watanabe H., et al. (2011) Loss of the retinoblastoma binding protein 2 (RBP2) histone demethylase suppresses tumorigenesis in mice lacking Rb1 or Men1. Proc. Natl. Acad. Sci. U.S.A. 108, 13379–13386 10.1073/pnas.1110104108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schilling T., Nöth U., Klein-Hitpass L., Jakob F., and Schütze N. (2007) Plasticity in adipogenesis and osteogenesis of human mesenchymal stem cells. Mol. Cell. Endocrinol. 271, 1–17 10.1016/j.mce.2007.03.004 [DOI] [PubMed] [Google Scholar]
- 55. Sugimura R., and Li L. (2010) Shifting in balance between osteogenesis and adipogenesis substantially influences hematopoiesis. J. Mol. Cell. Biol. 2, 61–62 10.1093/jmcb/mjp030 [DOI] [PubMed] [Google Scholar]
- 56. Zhang Y. Y., Li X., Qian S. W., Guo L., Huang H. Y., He Q., Liu Y., Ma C. G., and Tang Q. Q. (2012) Down-regulation of type I Runx2 mediated by dexamethasone is required for 3T3-L1 adipogenesis. Mol. Endocrinol. 26, 798–808 10.1210/me.2011-1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yang Y., Wei H., Song T., Cai A., Zhou Y., Peng J., Jiang S., and Peng J. (2017) E4BP4 mediates glucocorticoid-regulated adipogenesis through COX2. Mol. Cell. Endocrinol. 450, 43–53 10.1016/j.mce.2017.04.015 [DOI] [PubMed] [Google Scholar]
- 58. Guo L., Zhang P., Chen Z., Xia H., Li S., Zhang Y., Kobberup S., Zou W., and Lin J. D. (2017) Hepatic neuregulin 4 signaling defines an endocrine checkpoint for steatosis-to-NASH progression. J. Clin. Invest. 127, 4449–4461 10.1172/JCI96324 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.