Summary
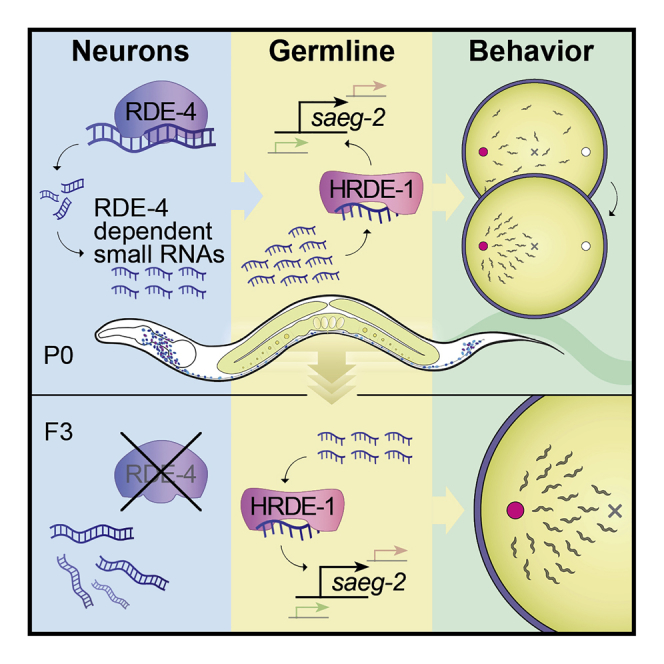
It is unknown whether the activity of the nervous system can be inherited. In Caenorhabditis elegans nematodes, parental responses can transmit heritable small RNAs that regulate gene expression transgenerationally. In this study, we show that a neuronal process can impact the next generations. Neurons-specific synthesis of RDE-4-dependent small RNAs regulates germline amplified endogenous small interfering RNAs (siRNAs) and germline gene expression for multiple generations. Further, the production of small RNAs in neurons controls the chemotaxis behavior of the progeny for at least three generations via the germline Argonaute HRDE-1. Among the targets of these small RNAs, we identified the conserved gene saeg-2, which is transgenerationally downregulated in the germline. Silencing of saeg-2 following neuronal small RNA biogenesis is required for chemotaxis under stress. Thus, we propose a small-RNA-based mechanism for communication of neuronal processes transgenerationally.
Keywords: transgenerational inheritance, epigenetic inheritance, small RNA inheritance, non-Mendelian inheritance, neuronal small RNAs, C. elegans
Graphical Abstract
Highlights
-
•
C. elegans neuronal small RNAs are characterized by RNA sequencing
-
•
RDE-4-dependent neuronal endogenous small RNAs communicate with the germline
-
•
Germline HRDE-1 mediates transgenerational regulation by neuronal small RNAs
-
•
Neuronal small RNAs regulate germline genes to control behavior transgenerationally
The idea that brain activity can impact the fate of the progeny goes against a central tenet of biology. Posner et al. describe an RNA-based mechanism for how neuronal responses to environmental cues can be translated into heritable information that affects the behavior of progeny.
Introduction
Among the different tissues of the body, the nervous system’s ability to collect and organize information about the environment is unmatched. Neuronal mechanisms of perception evolved to sense and interpret external and internal states and to orchestrate adaptive physiological responses fittingly. Ever since antiquity (Stubbe, 1972), many have speculated that brain activity could somehow generate heritable changes that would impact the fate of the next generations. The possibility that the nervous system can control the progeny could have far-reaching consequences.
This idea, however, despite its appeal, challenges one of the basic dogmas of biology, also known as “the second law of biology” (Mattick, 2012). The “Weismann Barrier” (Weismann, 1891) asserts that the heritable information in the germline is segregated from somatic influences. Accordingly, animals’ responses to environmental challenges should not become inherited. More specifically, if the “Weismann Barrier” is indeed impermeable, then the consequences of neuronal activity should never affect the progeny. Nevertheless, a number of studies suggested that neuronal responses in parents can affect the offspring’s behavior. While these examples are fascinating, the exact underling mechanisms remain unknown (Weaver et al., 2004, Remy, 2010, Vassoler et al., 2013, Dias and Ressler, 2014, Gapp et al., 2014a).
In the nematode Caenorhabditis elegans, small interfering RNAs that derive from artificial, exogenously supplied double-stranded RNA (exo-small interfering RNAs [siRNAs]) move from somatic cells, including neurons, to the germline (Fire et al., 1998, Devanapally et al., 2015). Further, an elaborate dedicated regulatory pathway has evolved to control transgenerational transmission of small RNA-initiated RNAi (Alcazar et al., 2008, Houri-Ze’evi et al., 2016, Houri-Zeevi and Rechavi, 2017, Lev et al., 2017, Spracklin et al., 2017). Transgenerational gene regulation depends on the amplification, by RNA-dependent RNA polymerases (RdRPs), of heritable small RNAs that bind specialized Argonautes in the germline, such as HRDE-1 (heritable RNAi deficient 1) (Aoki et al., 2007, Buckley et al., 2012). Environmental challenges (e.g., starvation and high temperatures) modulate the pool of heritable small RNAs and produce responses that last for multiple generations (Rechavi et al., 2014, Anava et al., 2015, Ni et al., 2016).
Like many other organisms, nematodes naturally produce, in the soma and in the germline, endogenous siRNAs (endo-siRNAs) that align to multiple loci across the genome. Endo-siRNAs target both protein-coding and non-protein coding loci (Gu et al., 2009, Vasale et al., 2010). Endo-siRNAs align in the antisense orientation to exons, can tile the entire length of the mature mRNA transcript, and complement the target perfectly (Blumenfeld and Jose, 2016). For simplicity, units of small RNAs targeting a specific gene, will be referred to here as STGs (see Rechavi et al. 2014) (see STAR Methods).
We hypothesized that biogenesis of neuronal endo-siRNAs could produce a heritable response. Endo-siRNAs were shown to control several neuronal functions affecting behavior and learning (Juang et al., 2013, Sims et al., 2016, Tonkin and Bass, 2003) and to mediate transgenerational gene regulation in the germline (Ashe et al., 2012, Shirayama et al., 2012, Rechavi and Lev, 2017). We focused on neuronal endo-siRNAs that depend on the double-stranded RNA (dsRNA)-binding protein RDE-4 (RNAi deficient 4). RDE-4 acts upstream in a biogenesis pathway that generates endo-siRNAs (Vasale et al., 2010, Welker et al., 2010, Lee et al., 2006, Duchaine et al., 2006, Gu et al., 2009) and is important for several neuronal functions, including migration of the HSN neuron, learning, and memory (Tonkin and Bass, 2003, Kennedy and Grishok, 2014, Juang et al., 2013).
To study the heritable effects of neuronal small RNAs, we engineered multiple transgenic strains in which we rescued RDE-4’s expression specifically in neurons of rde-4(−/−) worms. We found that RDE-4 controls the levels of various endo-siRNAs in neurons, but also, more intriguingly, in the germline. The function of the neuronal RDE-4-dependent germline endo-siRNAs depends on the germline-specific Argonaute HRDE-1 and regulates the expression levels of complementary mRNAs transgenerationally. Furthermore, we discovered that biogenesis of neuronal endo-siRNAs controls transgenerationally the capacity of worms to perform chemotaxis. We found that the conserved gene saeg-2 is regulated in the germline by neuronal RDE-4 in an HRDE-1-dependent manner across multiple generations, and saeg-2 silencing is key for proper chemotaxis. We propose that small RNA regulation is a mechanism that allows the nervous system to communicate with the germline affecting the behavior of the next generations.
Results
Identifying Neuronal Small RNAs
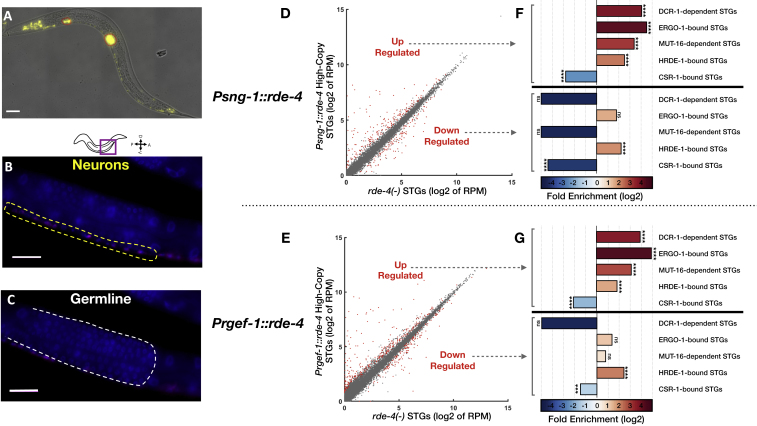
To create worms that produce RDE-4-dependent endo-siRNAs only in neurons, we rescued the expression of rde-4 specifically in neurons of rde-4(ne299) mutants (Tabara et al., 2002). In most experiments, we expressed rde-4 under the control of the pan-neuronal and neuron-specific promoter Psng-1 (Ruvinsky et al., 2007, Stefanakis et al., 2015). To complement these studies, we also rescued rde-4 in neurons using another pan-neuronal promoter, Prgef-1 (Figure S1).
Figure S1.
High-Copy Expression of RDE-4 in Neurons Regulates a Subset of STGs, Related to Figure 1
(A) A typical image demonstrating the neuronal expression pattern of the rescued RDE-4 (Prgef-1::rde-4::SL2::yfp), as monitored by examination of a trans-spliced YFP fluorescent reporter. Prgef-1::rde-4::SL2::yfp was co-injected with Punc122::GFP expressed in coelomocytes (marked in red). Bar = 20 μm.
(B and C) smFISH staining of yfp transcripts (magenta) and DAPI nuclei staining (blue) in one typical worm expressing Prgef-1::rde-4::SL2::yfp. Shown are focal plains focusing on the neuronal ventral chord (B), yellow dashed lines), and the germline (C), white dashed lines). Bar = 20 μm.
(D and E) Expression of STGs in rescued Psng-1::rde-4 (D) and Prgef-1::rde-4 (E) worms (y axis) compared to rde-4(ne299) mutants (x axis). Shown are the averaged expression values (log2 of RPM) of STGs (See also Table S2). Each dot represents an STG. Red dots: STGs that display differential expression between groups (analyzed with Deseq2, adjusted p value < 0.1).
(F and G) x-fold enrichment and depletion values of upregulated STGs (upper panel) and downregulated STGs (lower panel) following RDE-4 High-Copy rescue in neurons. P values for enrichment were calculated using 10,000 random gene sets identical in size to the tested group (See “STAR Methods” for details). For the clarity of display, complete depletion (linear enrichment = 0) appears with the smallest value in the scale. Enrichments were considered significant if p < 0.05. ns- p > 0.05. ∗∗∗- p < 0.001. ∗∗∗∗- p < 10−4.
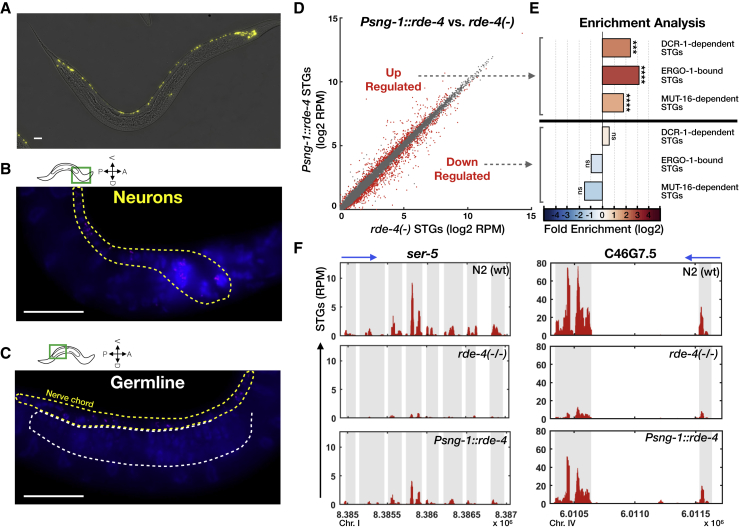
To monitor the expression of the rescued rde-4, we co-transcribed it with a trans-spliced yfp gene (Figure 1A; STAR Methods). When rde-4 was expressed in the nervous system, YFP fluorescence and mRNA molecules were detectable in neurons, as determined by both fluorescent microscopy and single molecule fluorescent in situ hybridization (smFISH) (Figures 1A and 1B). While we detected specific and robust expression in the nervous system, we could not detect yfp mRNA molecules in the germline (Figure 1C).
Figure 1.
Characterization of Small RNA Changes following Rescue of RDE-4 in Neurons
(A) Nervous system-specific rescue of RDE-4. A typical image demonstrating the neuronal expression pattern of the rescued RDE-4 (Psng-1::rde-4::SL2::yfp), as monitored by examination of a trans-spliced YFP fluorescent reporter. Bar, 20 μm.
(B and C) smFISH staining of yfp transcripts (magenta) and DAPI nuclei staining (blue) in one typical worm expressing the integrated single-copy Psng-1::rde-4::SL2::yfp pan-neuronal rescue transgene. Shown are focal plains focusing on the neuronal ventral chord (B, yellow dashed lines), and the germline (C, white dashed lines). Bar, 20 μm.
(D) Expression of STGs in rescued rde-4(n299);Psng-1::rde-4 worms (y axis) compared to rde-4(ne299) mutants (x axis). Shown are the averaged expression values (log2 of RPM) of STGs (see also Table S2). Each dot represents an STG. Red dots, STGs that display differential expression between groups (analyzed with Deseq2, adjusted p value < 0.1).
(E) x-fold enrichment and depletion values of upregulated STGs and downregulated STGs following RDE-4 rescue in neurons. We tested the enrichment of the RDE-4-dependent STGs against lists of STGs that are known to require DCR-1 for their biogenesis, to bind the Argonaute ERGO-1, and to depend on somatic mut-16 activity (Welker et al., 2010, Zhang et al., 2011, Vasale et al., 2010). p values for enrichment were calculated using 10,000 random gene sets identical in size to the tested group (see STAR Methods for details). Enrichments were considered significant if p < 0.05. Not significant [ns], p > 0.05; ∗∗∗p < 0.001; ∗∗∗∗p < 10−4.
(F) STGs distributions. Shown are STGs normalized read counts (y axis) as function of genomic location (x axis) of small RNAs targeting the genes ser-5 and C46G7.5 STGs (in red) in N2 wild-type worms, rde-4 mutants, and Psng-1::rde-4 rescue worms. Exons appear on a gray background. Blue arrow points to the direction of transcription.
See also Figure S1.
To characterize neuronal small RNAs, we extracted RNA and built libraries compatible with Illumina sequencing. Multiple biological replicates were created for each of the examined conditions, and altogether we sequenced 64 small RNA libraries and 27 mRNA libraries (see STAR Methods, GEO: GSE124049). We sequenced RNA extracted from N2 wild types, rde-4 mutants, and from transgenic worms that express rde-4 just in neurons. In additional experiments (described below), we sequenced RNA from dissected gonads and from fluorescence-activated cell sorted (FACS) neurons. Detailed descriptions of all datasets generated are available in Table S1. We hypothesized that the identification of neuronal endo-siRNAs could guide us toward the specific genes that they target, perhaps also non-cell autonomously, and by studying the regulation of these genes in the germline and across generations, we could examine if behavior can be controlled transgenerationally.
First, to identify bona fide RDE-4-dependent neuronal small RNAs, we applied stringent criteria to determine which STGs are upregulated when RDE-4 is rescued only in neurons using an integrated single copy Psng-1::rde-4 transgene (Figure 1D; Table S2; STAR Methods). We identified 476 such STGs and found that these endo-siRNAs display typical features of RDE-4-dependent small RNAs (Figures 1D–1F) (Welker et al., 2010, Vasale et al., 2010, Gu et al., 2009, Lee et al., 2006, Duchaine et al., 2006). During small RNA synthesis, RDE-4 binds DCR-1 that processes dsRNA molecules into primary endo-siRNAs, that in turn are further processed by the Argonaute ERGO-1 (Blanchard et al., 2011, Vasale et al., 2010, Welker et al., 2010). Accordingly, as is characteristic to rde-4-dependent small RNAs, we found that the 476 upregulated STGs are enriched for DCR-1-dependent siRNAs (5.3x enrichment, p < 0.001), as well as for ERGO-1-bound endo-siRNAs (8.9x enrichment, p < 10−4, Figure 1E) (Vasale et al., 2010). These STGs were also enriched (3.5x enrichment, p < 10−4) for endo-siRNAs that depend on somatic MUT-16, a factor required downstream of ERGO-1 to produce this class of small RNAs (Zhang et al., 2011). The same enrichments were obtained when we sequenced small RNAs from two additional lines of transgenic worms, this time overexpressing RDE-4 in neurons off high-copy transgenes, using two different pan-neuronal promoters, Psng-1 or Prgef-1 (Figure S1; Table S2).
Synthesis of RDE-4-dependent small RNAs in neurons led also to downregulation of 744 STGs (Figure 1D; Table S2). We reason that RDE-4 indirectly downregulated these small RNAs, as synthesis of RDE-4-dependent siRNAs in neurons likely comes at the expense of other types of small RNAs. It is well documented that different small RNA pathways compete over shared biosynthesis factors (Gu et al., 2009, Zhang et al., 2011, Zhuang and Hunter, 2011, Sarkies et al., 2013, Houri-Ze’evi et al., 2016). Indeed, in contrast to the upregulated STGs, the downregulated STGs did not display typical characteristics of RDE-4-dependent small RNAs (Figure 1E), again suggesting that RDE-4’s role in the downregulation of these STGs is indirect.
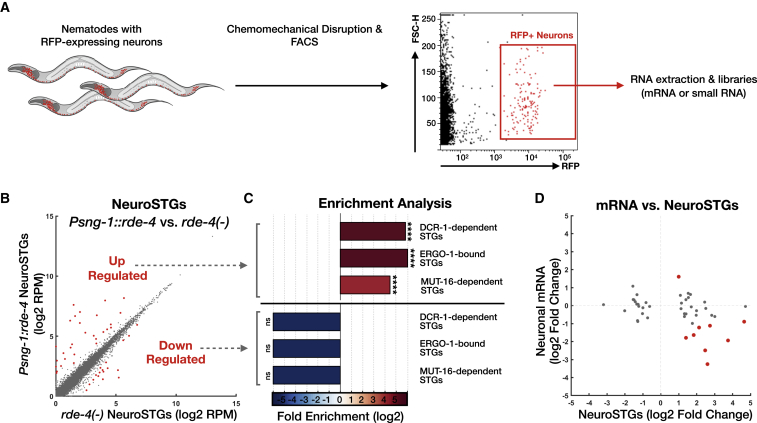
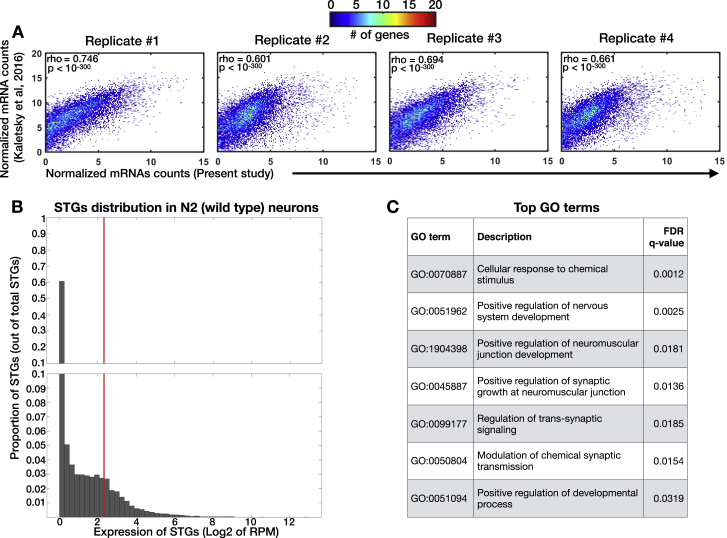
We hypothesized that rescuing RDE-4 in neurons could lead also to non-cell autonomous changes in small RNA levels. The experiments described above did not distinguish between neuronal and non-neuronal small RNAs, because we sequenced RNA from whole animals. Because it is challenging to lyse the worm’s cuticle without disrupting the integrity of its cells, small RNAs have not been sequenced from any isolated somatic tissue of C. elegans before. However, recently, mRNAs were successfully sequenced from isolated C. elegans neurons that were obtained using a specialized chemomechanical disruption protocol, followed by flow-cytometry cell sorting of single-cell suspensions (Kaletsky et al., 2016). We adapted this protocol for small RNA sequencing and isolated RFP-marked neurons from adult worms (Figure 2A, scheme; STAR Methods). We isolated neurons from N2 wild types, rde-4 mutants, and Psng-1::rde-4 worms and sequenced both small RNAs and mRNAs from the isolated cells. Our N2 mRNA data (Table S3) overlapped and correlated strongly with previously published neuronal mRNA data obtained by Kaletsky et al. (2016) (Figure S2, rho = 0.6–0.74 across replicates, p < 10−300). To create a list of confidently expressed neuronal small RNAs (dubbed “NeuroSTGs”), we applied a cut-off of >5 rpm per STG in all replicates of N2 worms (Figure S2; Table S3). The set of genes targeted by NeuroSTGs (in comparison to the set of genes targeted by small RNAs extracted from the entire animal) was enriched for genes that function in a variety of neuronal processes (6 out of the 7 top GO terms, Figure S2) (Eden et al., 2009). By comparing neurons extracted from rde-4 mutants and Psng-1::rde-4 worms, we identified 46 RDE-4-dependent NeuroSTGs (Figure 2B; Table S3) that exhibited typical characteristics of RDE-4-dependent small RNAs based on the analyses of their enrichments for STGs associated with DCR-1, ERGO-1, and MUT-16 (Figure 2C). Out of the 46 putative gene targets of these STGs, 9 were affected also at the mRNA level when RDE-4 was rescued in neurons (Figure 2D; Table S3).
Figure 2.
Sorting of Neurons Followed by RNA Sequencing Allows the Characterization of Neuronal RDE-4-Dependent Small RNAs
(A) Scheme depicting the production of RNA libraries specifically from neurons of C. elegans. Single-cell suspensions were produced out of wild type, rde-4(−), and Psng-1::rde-4 strains that express Prab-3::rfp in neurons (Kaletsky et al., 2016, Stefanakis et al., 2015), followed by immediate fluorescence-activated cell sorting (FACS). Sorted RFP+ neurons were collected for total RNA isolation.
(B) Expression levels of NeuroSTGs in rescued Psng-1::rde-4 worms (y axis) compared to rde-4(ne299) mutants (x axis). Shown are the averaged expression values (log2 of rpm) of NeuroSTGs (see also Table S5). Each dot represents a NeuroSTG. 46 NeuroSTGs (red) displayed differential expression between groups (analyzed with Deseq2, adjusted p value < 0.1).
(C) x-fold enrichment or depletion values of upregulated NeuroSTGs (left) and downregulated NeuroSTGs (right) following neuronal RDE-4 rescue. See also Figure 1E. For the clarity of display, complete depletion (linear enrichment = 0) appears with the smallest value in the scale. ns, p > 0.05; ∗∗∗∗p < 10−4.
(D) Changes in neuronal mRNA levels (y axis) in Psng-1::rde-4 compared to rde-4(−) neurons, plotted against changes in their associated NeuroSTGs (x axis). Each dot represents the values for one gene, and the 46 genes with significant changes in their corresponding NeuroSTGs are shown (analyzed with Deseq2, adjusted p value < 0.1). Nine genes (red) exhibited also differential mRNA expression (analyzed with Deseq2, adjusted p value < 0.1).
Figure S2.
Isolation and Sequencing of Neuronal Small RNAs and Neuronal mRNAs from N2 Wild-type Worms, Related to Figure 2
(A) Normalized mRNA levels reported by Kaletsky et al. (2016) (y axis), versus the mRNA levels (log2 of RPM) measured in four samples of N2 worms collected in independent experiments (x axis). Each dot represents a gene. As many points may overlap each other, and to better visualize the distribution of the data, we added a color code reflecting the number of genes in each bin.
(B) Shown is a histogram indicating the proportion of STGs (y axis) with different expression levels, displayed by log2 of RPM average (x axis). The vertical red line corresponds to value of 5 RPM (linear scale). We used a cut-off of > 5RPM to create a list of robustly expressed STGs in N2 neurons (See Table S3). Please note the scale in the y axis is changing, with steps of 0.01 in the range of 0 to 0.1, and steps of 0.1 for proportions higher than 0.1.
(C) Enriched GO terms for the sub-set of 412 genes targeted by NeuroSTGs with RPM > 5, which are upregulated in isolated neurons in comparison to STGs extracted from the entire animal. As can be seen, 6 of the 7 most enriched GO terms depict a variety of neuronal processes. Analysis done using the GOrilla tool (Eden et al., 2009); all the enriched GO terms with FDR < 0.05 are displayed.
In summary, neuronal RDE-4 rescuing experiments, coupled with isolation of neurons, enabled sequencing and identification of neuronal RDE-4-dependent endo-siRNAs. Further, these methods enabled us to continue and investigate whether neuronal rescue of RDE-4 changes germline small RNAs as well.
Neuronal RDE-4 Expression Leads to Transgenerational Inheritance of Endogenous Small RNAs and Germline Regulation of Cognate mRNA Targets
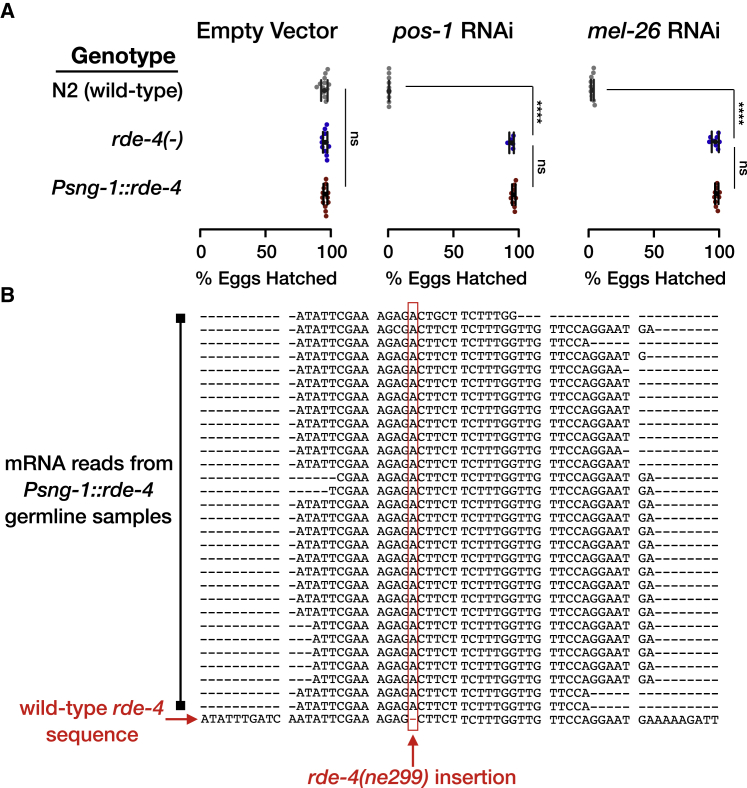
Next, we examined whether biogenesis of neuronal small RNAs could affect the germline and thus perhaps also the next generations. To ensure that any heritable effect that would be detected have originated in the soma, we validated that the rescued rde-4 is not mis-expressed in the germline, using four different complementary methods. First, we tested for RDE-4-dependent germline RNAi (silencing the genes pos-1 and mel-26, which induces embryonic lethality), and witnessed 100% efficiency in rde-4(+) control worms (none of the eggs hatched), while no germline RNAi activity could be detected in Psng-1::rde-4 worms (Figures 3A and S3). Second, the fluorescence of YFP (co-transcribed with rde-4 and used as its proxy) could not be detected in the germline of Psng-1::rde-4::yfp worms (Figures 1A and S1). Third, similarly, we could not detect any yfp transcripts in the germline using the very sensitive smFISH method (Figure 1C). smFISH cannot be used to distinguish between the wild-type rde-4 (rescue) allele and the mutated (ne299) allele, because these alleles differ only by a single nucleotide insertion (Tabara et al., 2002). Therefore, as a final measure, we used deep-sequencing to sequence mRNAs from isolated gonads of Psng-1::rde-4 animals. We found that all the reads (100%, 30 out of 30) that align to the relevant position (spanning the insertion site) in the rde-4(ne299) allele, contain the disabling insertion sequence (Figure 3B). Namely, no transcripts of the functional allele of rde-4 could be detected in the gonad.
Figure 3.
The Germline of Psng-1::rde-4 Worms Is Devoid of Functional RDE-4
(A) Worms with the indicated genotype (y axis) were allowed to lay eggs on plates containing dsRNA-producing bacteria targeting the germline-expressed genes pos-1 and mel-26 or an empty-vector control. Shown are the percentage of hatched eggs per plate (x axis) following exposure to RNAi. Each dot represents one tested plate (biological replicate). Bars represent mean ± SD. Each group was tested in at least three independent experiments. p values were determined by two-way ANOVA with Tukey’s post hoc correction for multiple comparison, ∗∗∗∗p < 10−4; ns, p > 0.05.
(B) Multiple sequence alignment of all the sequencing reads aligned to the genomic locations in the vicinity of the insertion defining the rde-4(ne299) allele. We combined all the reads (30) obtained from three independent replicate gonads samples from Psng-1::rde-4 worms. The wild-type rde-4 sequence is shown at the bottom row. We display only reads in which the insertion site is neither in the edge of the read nor included in soft clipping region of the CIGAR string. Shown is the complementary strand of the rde-4 gene, with the insertion position (chr-III: 10,218,186) marked in a red rectangle.
See also Figure S3.
Figure S3.
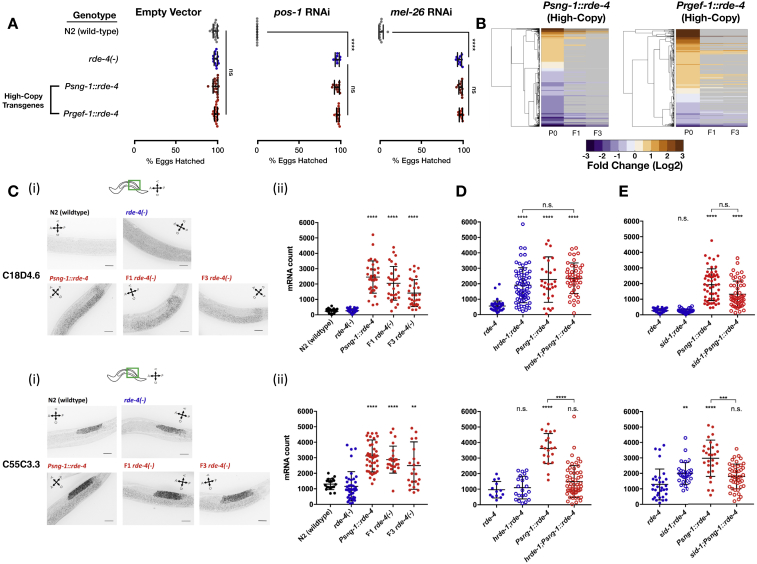
Worms Expressing Pan-Neuronal RDE-4 off High-Copy Transgenes Regulate mRNA Targets Transgenerationally, Related to Figures 3, 4, and 5
(A) Worms with the indicated genotype (y axis) were allowed to lay eggs on plates containing dsRNA-producing bacteria targeting the germline-expressed genes pos-1 & mel-26 or an empty-vector control. Shown are the percentage of hatched eggs per plate (x axis) following exposure to RNAi. Each dot represents one tested plate (biological replicate). Each group was tested in at least three independent experiments including n > = 2 biological replicates. P values were determined by Two-way ANOVA with Tukey’s post hoc correction for multiple comparison. ns- p > 0.05. ∗∗∗∗- p < 10−4. Related to Figure 3.
(B) Clustering of STGs based on changes in whole-animal samples from worms rescued with the indicated transgene, compared to rde-4(-) mutants. “P0” depicts the data for the rescued lines, and “F1” and “F3” depict the data for their progeny that have lost the High-Copy transgene (See “STAR Methods” section and Table S4). Shown are all STGs displaying significant differential expression in P0 (analyzed with Deseq2, adjusted p value < 0.1). Genes which did not show significant differential expression in F1 or F3 (adjusted p value > = 0.1) are colored in gray.
(C) (i) Representative images of smFISH staining against C18D4.6 (upper panel) and C55C3.3 (lower panel) in the posterior gonad of indicated genotypes. The stained worms were synchronized as late L4s. For representation, all images were filtered according to the FISH-quant software (Mueller et al., 2013), projected in the Z axis by maximum intensity and threshold adjusted, identically between conditions. Scale bars = 20μm (ii) Quantification of C18D4.6 and C55C3.3 germline mRNA expression (FISH-quant, see methods) in the indicated genotypes. Each dot represents one quantified worm, and worms were tested on three independent experiments. P values were determined by Kruskal-Wallis test with Dunn’s post hoc correction for multiple comparison and asterisks represent P values in comparison to rde-4(-). ∗∗∗∗- p < 10-4,∗∗- p < 0.01. Bars represent mean ± SD.
(D and E) smFISH quantification of worms with the indicated genotype (x axis) to determine the impact of hrde-1 (D) and sid-1 (E) on the germline regulation of C18D4.6 (upper panel) and C55C3.3 (lower panel) by High-Copy Psng-1::rde-4. n.s- p > 0.05. ∗∗∗- p < 0.001. ∗∗∗∗- p < 10−4.
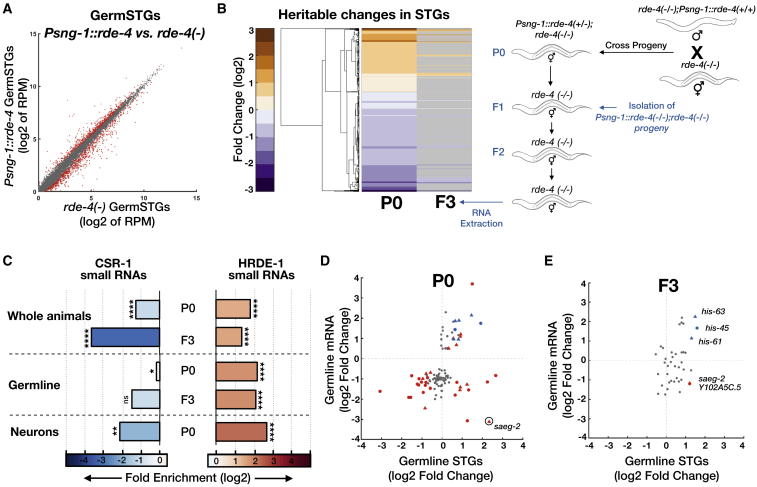
To examine if changes in NeuroSTGs could be communicated to the germline, we sequenced directly from dissected gonads both small RNAs (dubbed “GermSTGs”) and mRNAs (Table S4). We identified 1,287 GermSTGs that were affected by neuronal expression of RDE-4 (Figure 4A). To examine whether these changes are inherited, we sequenced small RNAs from F3 rde-4(−/−) progeny (from both whole worms and isolated gonads) derived from Psng-1::rde-4(+/−) great-grandparents. Whole-worm sequencing showed that 189 STGs were inherited to the F3 generation (Figure 4B; Table S4). Heritable, RdRP-amplified endo-siRNAs associate in the germline with two Argonautes performing opposite regulatory functions, HRDE-1 and CSR-1 (Ashe et al., 2012, Buckley et al., 2012, Luteijn et al., 2012, Claycomb et al., 2009, Seth et al., 2013). HRDE-1 is required for transgenerational inheritance of dsRNA-induced RNAi and for transgenerational inheritance of changes in endo-siRNAs levels in response to environmental challenges such as starvation and high temperatures (Ashe et al., 2012, Ni et al., 2016, Rechavi et al., 2014, Shirayama et al., 2012). CSR-1 was shown to inhibit silencing of germline genes and license expression (Conine et al., 2013). We found that inherited STGs are strongly enriched for HRDE-1-bound STGs (3.8x, p < 10−4 in whole worms samples and 4.7x, p < 10−4 in isolated germline samples) and are depleted from CSR-1-bound STGs (0.08x in whole worms samples, p < 10−4, and 0.4x in isolated germline samples, p = 0.07, ns) (Figure 4C) (Buckley et al., 2012, Claycomb et al., 2009).
Figure 4.
Neuronal RDE-4 Expression Leads to Transgenerational Inheritance of Endogenous Small RNAs and Germline Regulation of Cognate mRNA Targets
(A) Expression of GermSTGs in rescued rde-4(n299);Psng-1::rde-4 worms (y axis) compared to rde-4(ne299) mutants (x axis). Shown are the averaged expression values (log2 of RPM) of GermSTGs (see also Table S4). Each dot represents a GermSTG. Red dots, GermSTGs that display differential expression between groups (analyzed with Deseq2, adjusted p value < 0.1).
(B) Clustering of STGs based on changes in whole-animal samples from rescued rde-4(−);Psng-1::rde-4 worms compared to rde-4(−) mutants (left), and F3 rde-4(−) progeny of rde-4(−);Psng-1::rde-4(+/−) heterozygote rescue worms compared to rde-4(−) (right). Shown are all STGs displaying significant differential expression in P0 (analyzed with Deseq2, adjusted p value < 0.1) (see also Table S4). Genes that did not show significant differential expression in F3 (adjusted p value ≥0.1) are colored in gray.
(C) x-fold enrichment or depletion values of differentially expressed STGs, for small RNAs bound to the germline Argonautes CSR-1 (Claycomb et al., 2009) and HRDE-1 (Buckley et al., 2012). Tissue and generation of the analyzed STG samples are indicated. “P0” denotes samples extracted from rescued rde-4(−);Psng-1::rde-4 worms compared to rde-4(−) mutants. “F3” denotes samples extracted from F3 rde-4(−) progeny of rde-4(−);Psng-1::rde-4(+/−) compared to rde-4(−). Enrichments were considered significant if p < 0.05. ns, p > 0.05; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 10−4.
(D) Changes in germline mRNA levels (y axis) in Psng-1::rde-4 compared to rde-4(−), plotted against changes in their associated GermSTGs (x axis) (see also Table S4). Each dot represents the values for one gene, and the 124 genes with significant changes in germline mRNAs are shown (analyzed with Deseq2, adjusted p value < 0.1). 40 genes (colored) exhibited also differential STGs expression (analyzed with Deseq2, adjusted p value < 0.1). 18/40 genes (triangles) are regulated by the ZFP-1/DOT-1.1 complex. 7/40 genes (in blue) encode for histone proteins. saeg-2 is marked by a black circle.
(E) Changes in germline mRNA levels (y axis) in F3 rde-4(−) progeny of Psng-1::rde-4(+/−) compared to rde-4(−), plotted against changes in their associated GermSTGs (x axis) (see also Table S4). Shown are the 40 genes with differentially expressed mRNA and STGs from (D). Five genes (colored as in D, with their full name indicated) displayed differentially expressed mRNA and STGs also in the F3 generation (analyzed with Deseq2, adjusted p value < 0.1).
Further, examination of sequencing data of mRNA extracted from the isolated gonads showed that out of the 124 germline genes that were differentially expressed when RDE-4 was rescued in neurons, 40 (32%) were targeted by GermSTGs (Figure 4D; Table S4). These targets are enriched for genes regulated by the ZFP-1/DOT-1.1 complex (18/40, false discovery rate [FDR] <0.001), known to modulate the transcription rate of essential widely expressed genes (Cecere et al., 2013, Yang et al., 2016). Moreover, in this list of 40 genes, we found an enrichment for histone genes (7/40, FDR <0.001). We note that all seven histone genes had elevated levels of both STG and mRNA following neuronal expression of RDE-4 (Figure 4D, blue icons). It was previously shown that small RNA regulation of histone genes is important for proper transcription and maturation of histone mRNAs (Avgousti et al., 2012). Five genes were transgenerationally regulated by GermSTGs until the F3 generation (Figure 4E, adjusted p value <0.1): his-46, his-61, his-63, Y102A5C.5, and saeg-2. We investigated in depth the regulation of saeg-2, because its RDE-4-dependent STGs were highly abundant also in isolated neurons (Figures 2B and 5A; Table S3). Further, saeg-2 has been shown to affect foraging (exploration in search of food), and therefore we hypothesized that its regulation by neuronally induced parental small RNAs could allow transgenerational control over the progeny’s behavior (Hao et al., 2011).
Figure 5.
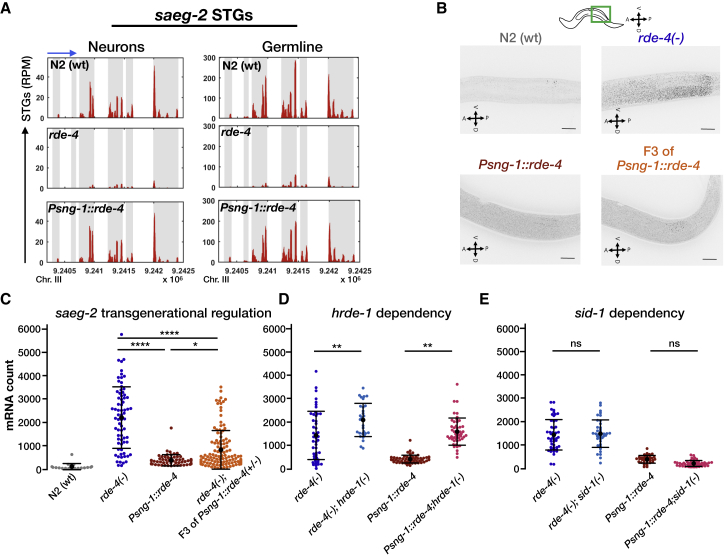
Neuronal RDE-4-Dependent Small RNAs Regulate Germline Expression of saeg-2 Transgenerationally in a HRDE-1-Dependent Manner
(A) STGs read distribution along the saeg-2 gene. Shown are STGs normalized read counts (y axis) against genomic location (x axis) of the small RNAs that target saeg-2 (in red) aligned to the gene locus in N2 wild-type worms, rde-4 mutants, and rde-4(−);Psng-1::rde-4 rescue worms. Exons appear on a gray background. Blue arrow points to the direction of transcription.
(B) Representative images of smFISH staining against saeg-2 in worms of the indicated genotype. The stained worms were synchronized as late L4s. For representation, all images were filtered according to the FISH-quant software (Mueller et al., 2013), projected in the z axis by maximum intensity and threshold adjusted, identically between conditions. Scale bar, 20 μm.
(C–E) Quantification of saeg-2 germline mRNA expression by smFISH in the indicated genotypes. Levels of saeg-2 mRNA in the germline are transgenerationally downregulated by neuronal RDE-4 (C), in a hrde-1-dependent (D) and sid-1-independent (E) manner. The groups were tested on three separate trials (except for N2s data in C obtained from two trials). Each dot represents one quantified worm. Black bars represent mean ± SD. p values were determined by Kruskal-Wallis test with Dunn’s post hoc correction for multiple comparison. ns, p > 0.05, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 10−4.
See also Figures S3 and S4 and Table S5.
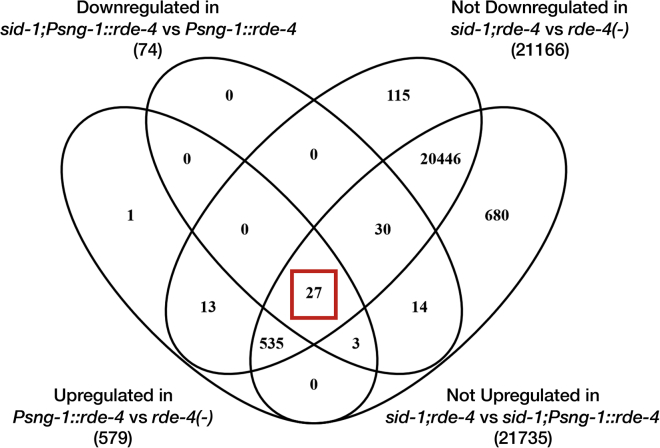
We confirmed the mRNA sequencing results using smFISH and found that saeg-2 is downregulated in the germline (−83%, p < 10−4), also transgenerationally (−63%, p < 10−4), when rde-4 is rescued in the nervous system (Figures 5B and 5C). Further, neuronal RDE-4 regulation of germline saeg-2 was found to be hrde-1-dependent, again strengthening the conclusion that this gene is transgenerationally regulated by germline-inherited anti-saeg-2 small RNAs (Figure 5D) (Buckley et al., 2012, Shirayama et al., 2012). Similar results were obtained when we validated using smFISH the changes in expression levels of C18D4.6 and C55C3.3, two additional target genes identified by global small RNA and mRNA analyses. C18D4.6 and C55C3.3 germline mRNAs were regulated transgenerationally upon neuronal rescue of RDE-4 using high-copy Psng-1::rde-4 transgenes (Figure S3; Table S4). The regulation of C18D4.6 and C55C3.3 was also affected by hrde-1 (Figure S3). Perhaps surprisingly, out of these genes, only the germline regulation of C55C3.3 was found to be significantly dependent on SID-1, a transmembrane RNA transporter shown to mediate spreading of exogenous siRNAs across tissues (Winston et al., 2002, Jose et al., 2011) (Figures 5E and S3). It was never examined whether SID-1 shuttles any endogenous small RNA species between cells (microRNAs, piwi-interacting RNAs [piRNAs], or endo-siRNAs). We sequenced small RNAs and mRNAs from the gonads of sid-1(+) and sid-1(−) animals and found only 27/579 upregulated GermSTGs that depend on both SID-1 and neuronal RDE-4 expression (Figure S4; Table S5). It is possible that other endo-siRNAs are shuttled by different transporters, as many genes came up in different screens for systemic RNAi, and sid-1 mutants are not entirely resistant to systemic silencing via RNAi (Winston et al., 2007, Rocheleau, 2012, Jose, 2015, Hinas et al., 2012). Alternatively, the dramatic effects of neuronal RDE-4 on the germline pool of endo-siRNAs could be triggered by secretion from neurons of other signaling molecules (e.g., hormones).
Figure S4.
SID-1 Regulates a Subset of GermSTGs that Depend on Neuronal RDE-4, Related to Figure 5
Shown is a Venn diagram depicting the criteria applied in order to detect sid-1-dependent GermSTGs in high confidence. The number of STGs passing each criteria appears in parenthesis. We kept only STGs that were: (1) upregulated in the germline in Psng-1::rde-4 versus rde-4(-), (2) downregulated in sid-1;Psng-1::rde-4 versus Psng-1::rde-4, (3) not downregulated in sid-1;rde-4(-) versus rde-4(-), (4) not upregulated in sid-1;rde-4(-) versus sid-1;Psng-1::rde-4.
Overall, our results reveal that expression of RDE-4 in neurons affects heritable HRDE-1 endo-siRNAs and leads to transgenerational regulation of germline genes.
Neuronal RDE-4 Controls Behavior Transgenerationally via Regulation of Germline RNA
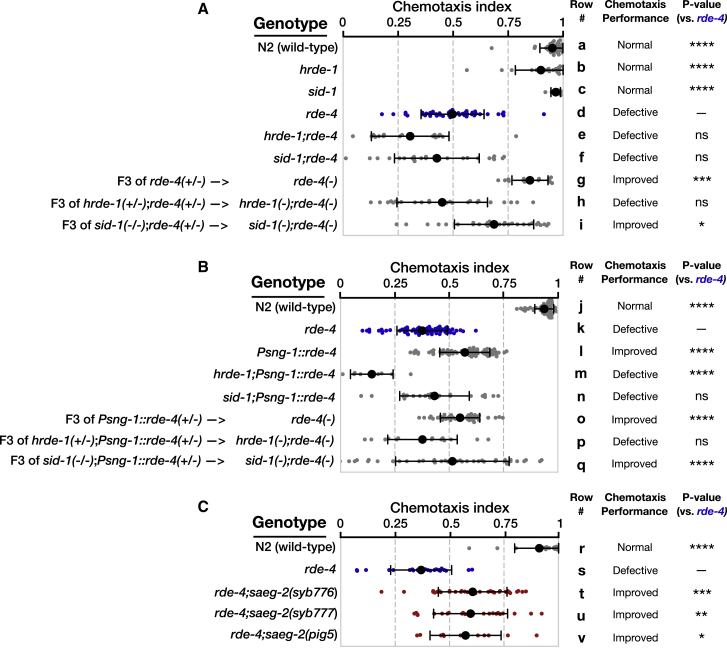
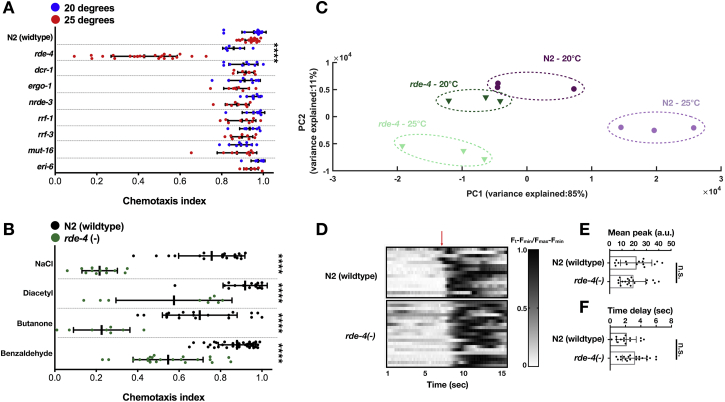
It was previously shown that RDE-4 plays a role in learning (adaptation) and migration of the HSN neuron, and endo-siRNAs are known to function in various neuronal processes (Bharadwaj and Hall, 2017, Kennedy and Grishok, 2014, Juang et al., 2013, Tonkin and Bass, 2003, Sims et al., 2016). We examined the chemotaxis capacity of 8 small RNA mutants and discovered that rde-4, but not the other mutants, display a strong defect in chemotaxis when cultivated in 25°C, but not in 20°C (Figures 6A, rows a and d, and S5). At 25°C, rde-4 mutants displayed defective chemotaxis toward multiple different stimuli, both volatile and soluble (Figure S5). A PCA projection showed that mis-expression of small RNAs in rde-4 mutants (compared to wild type) is more pronounced in 25°C than in 20°C (Figure S5), suggesting that RDE-4 activity is more important at higher temperatures.
Figure 6.
Neuronal RDE-4 Controls Behavior Transgenerationally via the Germline Small RNA Machinery
Results for experiments testing chemotaxis to benzaldehyde (1:100) at day 1 of adulthood of worms (ethanol was used as control odor). Chemotaxis index = ((# worms at benzaldehyde) − (# worms at ethanol))/((# total worms on plate) − (# worms at origin)). Each dot represents one plate with >200 worms. All groups were tested on at least three independent trials, each including several biological replicates. Black bars represent mean ± SD. For convenience, each biological group was assigned a letter label. p values were determined by Kruskal-Wallis test with Dunn’s post hoc correction for multiple comparison to the rde-4(−) group. ns, p > 0.05; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 10−4.
(A) Chemotaxis experiments on the F3 rde-4(−) progeny of rde-4(+/−) P0 ancestors and their hrde-1 and sid-1 double mutants, together with control strains.
(B) Chemotaxis experiments on the F3 rde-4(−) progeny of Psng-1::rde-4(+/−);rde-4(−/−) P0 ancestors and their hrde-1 and sid-1 double mutants, together with control strains.
(C) Chemotaxis experiments on the rde-4;saeg-2 double mutants alleles, together with control strains. Three double mutant strains were generated via CRISPR/Cas9.
See also Figure S5.
Figure S5.
rde-4 Mutants Are Defective in Chemotaxis under a Stressful Temperature but Display Normal Activity in the AWC Sensory Neuron, Related to Figure 6
(A and B) Results for experiments testing chemotaxis at day 1 of adulthood of worms. Chemotaxis index = ((# worms at stimulus)-(# worms at control)) / ((# total worms on plate)-(# worms at origin)). Each dot represents one plate with > 200 worms. All groups were tested on at least three independent trials (n = > 9).
(A) Chemotaxis screens were performed on RNAi factor mutants at both 20 (blue dots) and 25 degrees (red dots). Chemotaxis indices (x axis) were tested for the strains (y axis) N2(wild-type), rde-4(ne299), dcr-1(mg375), ergo-1 (tm1860), nrde-3 (gg66), rrf-1(ok589), rrf-3(pk1426), mut-16(pk710) and eri-6(mg379). The odor stimulus used was benzeldahyde (1:100). P values were determined by two-way ANOVA, ∗∗∗∗-p < 10−4.
(B) rde-4 mutants are defective in chemotaxis to multiple stimuli at high temperature. N2(wild-type) (black dots) and rde-4 mutants (green dots) raised at 25 degrees for chemotaxis to Benzaldehyde (1:102), Butanone (1:104), Diacetyl (1:103) and NaCl (50mM). P values were determined by Two-way ANOVA with Sidak’s post hoc correction for multiple comparisons. ∗∗∗∗- p < 10−4.
(C) A Principal Component Analysis (PCA) projection of 12 samples based on normalized STGs read counts. Each symbol represents one independent replicate. The corresponding genotype and temperature are indicated. The % variances, out of the total original variance in the high-dimensional space, spanned by the first and second Principal Components are indicated on the x- and y- axis, respectively. Related to Figure 6.
(D) Activity of sensory neuron AWC was quantified by GCaMP2 fluorescence intensity in a microfluidic device controlling stimulus exposure. N2 (wild-type) (n = 15) and rde-4 (n = 20) mutants were loaded into chips and exposed to the stimulus Isoamyl alcohol (1:104) for 1 minute followed by a switch to buffer (indicated by the red arrow). Each row represents an individual worm. Shown are the fluorescence intensity values normalized from 0 to 1 (Ft-Fmin)/(Fmax-Fmin) across time (seconds).
(E) Maximum fluorescence intensity increase (x axis) of the N2 and rde-4 worms (y axis) of AWCneurons in response to odor removal. Mean peak was defined as ΔF (2 s pre-stimuli) – max ΔF (post-stimuli).
(F) Time (x axis) it took from the moment of odor removal for AWC neurons in N2 and rde-4 worms (y axis) to reach maximum fluorescence intensity. P values were determined by Mann-Whitney tests. n.s.- p > 0.05.
To characterize the chemotaxis defect of the rde-4 mutants and understand which other factors could be involved, we examined a variety of genetic backgrounds. All the experiments were conducted by placing in each assay >200 worms in the center of a plate containing the volatile attractant benzaldehyde (sensed by the AWC neuron) on one side and ethanol (control) on the other side. We found that the capacity of rde-4 mutants to perform chemotaxis at 25°C was partially rescued by neuronal rde-4 expression (Figure 6B, rows k and l). To examine whether RDE-4 is required for sensation of external odor stimuli, we measured calcium influx in the AWC sensory neurons using a microfluidics device (Chronis et al., 2007) and could not detect any significant differences between rde-4 and wild-type (WT) worms (Figure S5). Because rde-4 mutants can sense external stimuli, their inability to perform chemotaxis at high temperatures could arise from defects in downstream neuronal circuits (e.g., interneurons) or even from physiological defects that arise in non-neuronal tissues (e.g., germline). Strikingly, while hrde-1 single mutants did not show defects in chemotaxis (Figure 6A, rows a and b), hrde-1 was necessary for the rescue of chemotaxis by neuronal RDE-4 (Figure 6B, rows k–m, and S6). As HRDE-1 is expressed exclusively in the germline (Buckley et al., 2012, Shirayama et al., 2012), these results suggest that neuronal-RDE-4 regulates chemotaxis by controlling the activity of germline small RNAs and regulation of germline-expressed genes.
Importantly, we found that RDE-4 affects chemotaxis in a transgenerational manner, because F3 rde-4(−/−) mutant progeny displayed improved chemotaxis when they derived from rde-4(+/−) great-grandparents (Figure 6A, rows d and g). Further, the activity of RDE-4 in the parents’ neurons is sufficient for rescuing the chemotaxis defects transgenerationally, since F3 rde-4(−/−) mutant progeny displayed improved chemotaxis also when they derived from Psng-1::rde-4(+/−) great-grandparents (Figure 6B, rows k and o). The ability of parental RDE-4 to rescue chemotaxis transgenerationally was also found to be hrde-1-dependent (Figures 6A rows d, g, and h, and 6B, rows k, o, and p).
As noted above, saeg-2 was previously identified in a screen as a gene that affects behavior (Hao et al., 2011). SAEG-2 is an ortholog of the mammalian Dnttip1, a terminal deoxynucleotidyl transferase interacting protein (Itoh et al., 2015). In a forward genetic screen, disruption of saeg-2 rescued the behavioral defects (foraging) of constitutively active EGL-4 kinase (Hao et al., 2011, L’Etoile et al., 2002). We hypothesized that knocking out saeg-2 in rde-4 mutants could rescue the mutant’s chemotaxis behavior, because in the absence of neuronal RDE-4, saeg-2 is not targeted by endo-siRNAs, and saeg-2 mRNA is strongly upregulated in the germline (Figure 5; Table S4). Therefore, we generated three rde-4;saeg-2 double mutant lines using CRISPR/Cas9 (STAR Methods). We found that all three rde-4;saeg-2 double mutants exhibited improved chemotaxis capacities in comparison to rde-4 mutants (Figure 6C, rows s–v).
Although saeg-2 was unaffected by SID-1 (Figure 5E), we examined if SID-1 influences chemotaxis, because saeg-2 mis-regulation does not explain the entire chemotaxis defect of rde-4 mutants (Figure 6C). Interestingly, we found that the regulation over behavior of neuronal RDE-4 is affected by SID-1 (Figure 6B, rows k, l, and n), and in sid-1 mutants the inherited ability of neuronal RDE-4 to rescue the progeny’s chemotaxis behavior is significantly more variable (Figure 6B, rows k, o, and q). SID-1 increased the consistency of the inherited behavior (reduces variability) both in F3 rde-4(−/−) mutants that derived from rde-4(+/−) great-grandparents (F = 4.67, p = 0.0038, F-test of equality of variances) (Figure 6A, rows g and i), and in F3 rde-4(−/−) mutants that derived from pan-neuronal Psng-1::rde-4(+/−) great-grandparents (F = 8.32, p < 10−4) (Figure 6B, rows o and q).
To conclude, neuronal RDE-4 controls chemotaxis for at least three generations via the germline-restricted Argonaute HRDE-1. Neuronal RDE-4 partially rescues the chemotaxis defect of rde-4 mutants by silencing saeg-2 in the germline. SID-1 also contributes to the ability of neuronal RDE-4 to rescue chemotaxis, probably by affecting genes other than saeg-2. In summary, biogenesis of neuronal small RNAs controls behavior by regulating the expression of germline-expressed genes.
Discussion
The ability to translate neuronal activity into heritable information could be adaptive. We discovered that RDE-4’s activity in the nervous system exerts systemic and transgenerational changes in endo-siRNAs, gene expression, and behavior. Previous studies have demonstrated that small RNAs are involved, in many animals, in neuronal differentiation, sensation, and behavior (Johnston and Hobert, 2003, Li and Jin, 2010, Chang et al., 2009). We propose here that changes in neuronal endo-siRNAs can be communicated to the offspring via regulation of germline RNA and the activity of the germline endo-siRNA inheritance machinery. Through this route, neuronal responses to external stimuli or internal physiological states could be translated into inheritable information and affect the progeny’s behavior and possibly fitness.
In recent years, several studies have suggested that the progeny’s behavior could be affected by the parent’s neuronal responses to specific stimuli. In mice, the behavior of F2 progenies was affected by odor fear conditioning or postnatal unpredictable care endured by the P0 parents (Dias and Ressler, 2014, Gapp et al., 2014b). In C. elegans, olfactory imprinting and, very recently, a learned pathogenic avoidance behavior, were shown to be maintained transgenerationally (Remy, 2010, Moore et al., 2018, Pereira et al., 2019). Changes in neuronal small RNAs could be related to these observations, and therefore in the future it would be interesting to identify specific external stimuli that modulate the activity of RDE-4 or endo-siRNAs in neurons and accordingly generate heritable changes in the germline.
In C. elegans, the nervous system coordinates a wide range of non-cell autonomous physiological processes (Styer et al., 2008, Cornils et al., 2011, Prahlad and Morimoto, 2011, Fletcher and Kim, 2017, Boulias and Horvitz, 2012, Srinivasan, 2015). How do endo-siRNA changes in neurons impact the germline’s RNA? We found that a small subset of the non-cell autonomous changes that we documented depend on the conserved and well-studied dsRNA transporter SID-1 (Winston et al., 2002, Devanapally et al., 2015, Jose, 2015, Hinas et al., 2012) (Figure S4; Table S5), and SID-1 affects the ability of neuronal RDE-4 to improve chemotaxis (Figures 6A and 6B). These results might suggest that certain endogenous small RNAs could move from neurons to other tissues. Likewise, we found that the non-cell autonomous regulation of the germline gene C55C3.3 is affected by SID-1 (Figure S3). However, the germline expression of C55C3.3 was regulated only when we overexpressed RDE-4 in neurons using high-copy arrays. It is hard to rule out the possibility that these results stem from indirect changes (Gu et al., 2009, Zhang et al., 2011, Zhuang and Hunter, 2011, Sarkies et al., 2013, Houri-Ze’evi et al., 2016). It is possible that movement of endo-siRNAs is achieved via the action of other transporters. Many genes that affect systemic silencing have been identified in screens (Winston et al., 2002, Jose, 2015), and sid-1 mutants are not entirely defective in systemic RNAi (e.g., gut cells can transport dsRNA-induced silencing in sid-1 mutants) (Rocheleau, 2012, Hinas et al., 2012). Alternatively, other signaling molecules may be involved: NeuroSTGs could trigger in neurons a variety of physiological responses that would lead to secretion of many different molecules that can in theory reach the germline. For example, aside from well-known secreted agents, such as neurotransmitters and neuropeptides, it has been shown very recently that neurons of both flies and mice repurpose a retrovirus-like Gag protein (Arc) to traffic RNA between cells (Ashley et al., 2018, Pastuzyn et al., 2018).
How do changes in heritable small RNAs affect the behavior of the next generations? Do heritable small RNAs that change in the germline move from the germline to the nervous system? We favor an alternative model. Previous studies have shown that heritable small RNAs regulate mostly germline-expressed genes (Ashe et al., 2012, Sapetschnig et al., 2015, Devanapally et al., 2015). Heritable small RNAs could change behavior by affecting the molecules that the progeny’s germline secretes. Numerous studies, across many species including humans, have shown that changes in germline homeostasis can affect nervous system-produced behavior through various signaling routes (Fujiwara et al., 2016, McCarthy et al., 2009, Bale, 2015). In C. elegans, for instance, the male germline affects temperature-dependent signaling in ASJ neurons (Sonoda et al., 2016).
It is likewise possible that the small RNAs transmitted to the zygote affect the nervous system’s development, thus affecting behavior. Inherited germline small RNAs have been shown to affect various stages of embryonic development (Gerson-Gurwitz et al., 2016, de Albuquerque et al., 2015). Even in mice, recent studies showed that microRNAs and tRNA fragments produced in the somatic testis tissue (epididymis) are delivered to maturing sperm and are crucial during post-implantation for proper embryonic development of the F1, probably through the regulation of specific targets (Sharma et al., 2018, Conine et al., 2018).
Our pipeline generated lists of small RNAs that could be investigated in depth. The strategy for characterizing RDE-4-dependent and neuronally controlled heritable small RNAs enabled, after applying stringent filters, to identify with high confidence five germline genes that are regulated for at least three generations. One of these, Y102A5C.5, is a predicted pseudogene, and 3 others are histone genes. It is possible that the regulation of these targets has functional consequences; however, each of these specific histones is encoded redundantly by numerous loci (14–16 different copies). We decided to further focus our inquiry on saeg-2. Intriguingly, saeg-2 is unique in that out of 80 endo-siRNAs that were found to be mis-regulated in DCR-1 helicase domain mutants, only saeg-2 was upregulated (the other 79 were downregulated) (Welker et al., 2010). We found endo-siRNAs that target saeg-2 in isolated gonads and in isolated neurons (Tables S3 and S4), and silencing of saeg-2 by neuronal RDE-4 is required for chemotaxis under stress. While saeg-2 levels are dramatically regulated in the germline following expression of RDE-4 in neurons, we did not observe changes in saeg-2 mRNA in neurons. We note, however, that overall saeg-2 levels in neurons are anyway very low across conditions. The regulation of saeg-2 by neuronal RDE-4 depends on HRDE-1, and HRDE-1 is also required for the control of neuronal RDE-4 over behavior. Because HRDE-1 expression is restricted to the germline, we suggest that neuronal RDE-4 influences behavior by regulating germline small RNA and genes.
In the future, it would be fascinating to investigate whether neuronal processes, and perhaps even decisions computed by neuronal circuits, can change inherited materials in meaningful ways. Namely, whether, by influencing heritable small RNAs, the nervous system can generate adaptive value.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| BsmAI | New England Bioloabs | R0529 |
| Mouth aspirator and Microcapillary tubes | Sigma | P0674 |
| Levamisole hydrochloride | Sigma | L0380000 |
| Trizol Reagent | Life Technologies | 15596026 |
| Pronase | Sigma | 10165921001 |
| 30 μm filters | Sysmex | SYS-04-004-2326 |
| Trizol LS Reagent | Life technologies | 10296028 |
| DTT | Sigma | 10197777001 |
| MiRneasy column RNA kit | QIAGEN | 217004 |
| Phenol Chloroform Isoamyl | Sigma | P2069 |
| Heavy Phase Lock tube | QuantaBio | 23028330 |
| RNA 5′ Polyphosphatase | Epicenter | RP8092H |
| NEBNext® Multiplex Small RNA Library Prep Set for Illumina | New England Biolabs | E7300 |
| TruSeq Small RNA Library Prep Kit | Illumina | RS-200-0012 |
| TapeStation screen tapes | Agilent | 5067-5582 5067-5588 |
| TapeStation reagents | Agilent | 5067-5583 5067-5589 |
| E-Gel 4% agarose | Life Technologies | G401004 |
| MinElute DNA purification kit | QIAGEN | 28006 |
| SMART-Seq v4 Ultra Low Input RNA Kit | Nextera | 634890 |
| Nextera XT DNA Library Preparation Kit | Illumina | FC-131-1024 |
| 10X Phosphate Buffered Saline (PBS) | Ambion | AM9624 |
| Formaldehyde | Sigma | F8775 |
| RNase free Nuclease-free water | Ambion | AM9932 |
| Dextran sulfate | Sigma | D8906-50G |
| Escherichia coli tRNA | ROCHE | 10109541001 |
| Vanadyl ribonucleoside complex | New England Biolabs | S1402S |
| RNase free BSA | Ambion | AM2618 |
| Formamide | Ambion | AM9342 |
| 20X SSC | Ambion | AM9763 |
| 1 M Tris-HCl, pH 8.0 | Ambion | AM9855G |
| Glucose oxidase stock | Sigma | G2133 |
| Catalase suspension | Sigma | C3515 |
| DAPI | Sigma | D9542 |
| Sodium Azide | Sigma | S2002 |
| Isoamyl alcohol | Sigma | W205702 |
| Benzaldehyde | Sigma | 418099 |
| Diacetyl | Sigma | 8.03528 |
| Butanone | Sigma | 360473 |
| polydimethylsiloxane (PDMS) | Dow Corning Sylgard | 4019862 |
| Milltex 1 mm biopsy punchers | Fisher | 12-460-401 |
| PTFE microbore tubing | Cole-Parmer | EW-06417-21 |
| Critical Commercial Assays | ||
| Affymetrix GeneChip® C. elegans Genome Array oligonucleotide arrays | Thermo Fisher | 900383 |
| Deposited Data | ||
| Small RNA-seq data | This study | GEO: GSE124049 |
| Experimental Models: Organisms/Strains | ||
| wild type Bristol isolate | CGC | N2 |
| rde-4(ne299) | Hobert lab | N/A |
| otIs356[rab-3p(prom1)::2xNLS::TagRFP] | Oren-Suissa lab | OH10690 |
| sid-1(qt9) | CGC | HC196 |
| ttTi5605 II; unc-119(ed3) III; oxEx1578 | CGC | EG6699 |
| rde-4(ne299) (6X outcrossed) | This study | BFF12 |
| sid-1(qt9); rde-4(ne299) | This study | BFF13 |
| rde-4(ne299); pigEx9[Psng-1::rde-4:SL2:yfp+Pmyo-3::Mcherry] | This study | BFF14 |
| rde-4(ne299); pigEx10[Prgef-1::rde-4:SL2:yfp+Punc-122::gfp] | This study | BFF15 |
| rde-4(ne299);pigSi3[cb-unc-119(+)Psng-1::rde-4:SL2:yfp] | This study | BFF16 |
| rde-4(ne299); hrde-1(pig4) | This study | BFF17 |
| rde-4(ne299); hrde-1(pig4); pigEx9[Psng-1::rde-4:SL2:yfp+Pmyo-3::Mcherry] | This study | BFF18 |
| rde-4(ne299); sid-1(qt9); pigEx9[Psng-1::rde-4:SL2:yfp+Pmyo-3::Mcherry] | This study | BFF19 |
| unc-119(ed3) III, oxTi38[cb-unc-119(+)Ppie-1::GFP] | Gift from Christian Frøkjær-Jensen | EG6089 |
| hrde-1(tm1200); EG6089 | This study | BFF20 |
| rde-4(ne299); EG6089 | This study | BFF21 |
| rde-4(ne299); hrde-1(pig4); EG6089 | This study | BFF22 |
| otIs356[rab-3p(prom1)::2xNLS::TagRFP] (3x outcrossed) | This study | BFF30 |
| rde-4(ne299); otIs356[rab-3p(prom1)::2xNLS::TagRFP] | This study | BFF31 |
| rde-4(ne299); pigSi3[cb-unc-119(+)Psng-1::rde-4:SL2:yfp]; otIs356[rab-3p(prom1)::2xNLS::TagRFP]. | This study | BFF32 |
| sid-1(qt9); rde-4(ne299); pigSi3[cb-unc-119(+)Psng-1::rde-4:SL2:yfp]; | This study | BFF33 |
| hrde-1(pig4); rde-4(ne299); pigSi3[cb-unc-119(+)Psng-1::rde-4:SL2:yfp)] | This study | BFF34 |
| kyEx2373[Pstr-2::Gcamp; Punc-122:GFP] | Hendricks lab | CX10281 |
| rde-4(ne299); kyEx2373 [Pstr-2::Gcamp; Punc-122:GFP] | This study | BFF35 |
| rde-4(ne299); saeg-2(syb776) | SunyBiotech | PHX776 |
| rde-4(ne299); saeg-2(syb777) | SunyBiotech | PHX777 |
| rde-4(ne299); saeg-2(syb776) (x2 outcrossed to rde-4) | This study | BFF36 |
| rde-4(ne299); saeg-2(syb777) (x2 outcrossed to rde-4) | This study | BFF37 |
| rde-4(ne299); saeg-2(pig5) | This study | BFF38 |
| rrf-1(ok589) | CGC | RB798 |
| dcr-1(mg375) | CGC | YY470 |
| ergo-1 (tm1860) | CGC | WM158 |
| rrf-3(pk1426) | CGC | NL2099 |
| mut-16(pk710) | CGC | NL1810 |
| eri-6(mg379) | CGC | GR181 |
| nrde-3 (gg66) | CGC | YY158 |
| Recombinant DNA | ||
| pos-1 RNAi plasmid | Vidal RNAi library | N/A |
| mel-26 RNAi plasmid | Vidal RNAi library | N/A |
| GFP RNAi plasmid | Hekimi lab- McGill | N/A |
| Oligonucleotides | ||
| PCR 1-FWD: genomic DNA rde-4 ATGGATTTAACCAAACTAACGTTTG | IDT | N/A |
| PCR 1-RV: genomic DNA rde-4 TCAATCCGTGAAATCATAGGTGTTG | IDT | N/A |
| PCR 2-FWD: Psng-1 AAACTGCAGTTAATTGTTAATTATCTAAGCTTG | IDT | N/A |
| PCR 2-RV: Psng-1 CGGGATCCGCTAAAATAAAAGAAATATAGAGG | IDT | N/A |
| PCR 3-FWD: Prgef-1 AAACTGCAGGCAGAATCGAGTCAACTGAAATCCG | IDT | N/A |
| PCR 3-RV: Prgef-1 AAAGGATCCCGTCGTCGTCGTCGATGC | IDT | N/A |
| PCR 4-FWD: hrde-1 (pig4) TCCACGTCCAATCCTTTGAGTG | IDT | N/A |
| PCR 4-RV: hrde-1 (pig4) AAGTTGCCCAGGGGGGT | IDT | N/A |
| PCR 5-FWD: saeg-2 (pig5) / saeg-2 (syb776) / saeg-2 (syb777)) TCGGCACGAAATATGTTG | IDT | N/A |
| PCR 5-RV: saeg-2 (pig5) / saeg-2 (syb776) / saeg-2 (syb777) AGATAGCATCTACCGGGTGCC | IDT | N/A |
| PCR 6-FWD: rde-4(ne299) CTAAGGCTGTCTATCAAAAGACGCCA | IDT | N/A |
| PCR 6-RV: rde-4(ne299) AAATACCAGGTGGAAATTCAGCACTTG | IDT | N/A |
| PCR -FWD: hrde-1 (tm1200) TCCACGTCCAATCCTTTGAG | N/A | |
| PCR -RV: hrde-1 (tm1200) AGTGGAAAGAGTCACCACTTC | IDT | N/A |
| PCR 7-FWD: pigSi3[cb-unc-119(+,Psng-1::rde-4:SL2:yfp] insert TCTCACTCGTTTAGGCTATTCC | IDT | N/A |
| PCR 7-RV-1: pigSi3[cb-unc-119(+,Psng-1::rde-4:SL2:yfp] insert ACCCGATGAAATACGGATGC | IDT | N/A |
| PCR 7-RV-2: pigSi3[cb-unc-119(+,Psng-1::rde-4:SL2:yfp] insert TGAGCACAATGGGAATACATCAG | IDT | N/A |
| PCR 75-RV-2: pigSi3[cb-unc-119(+,Psng-1::rde-4:SL2:yfp] insertPsng-1 TGAGCACAATGGGAATACATCAG | IDT | N/A |
| hrde-1 sgRNA 1: 5′ GGTGTCTATGGAAACCGAGG 3′ | IDT | N/A |
| hrde-1 sgRNA 2: 5′ CATACGATTAGCCTCCTCCT 3′, | IDT | N/A |
| rol-6sgRNA: 5′ GTGAGACGTCAACAATATGG 3′ | IDT | N/A |
|
rol-6(su1006) conversion template: 5′ TGTGGGTTGATATGGTTAAAC TTGGAGCAGGAACCGCTTCCAACCGT GTGCGCTGCCAACAATAT GGAGGATATGGAGCCACTGGTGTTCAGCCACCAGCACCAAC 3′. |
IDT | N/A |
| saeg-2 sgRNA 1: CCAGCTAGCATGACTGCCACTTT saeg-2 sgRNA 2: CCACGAATGTTTAGAGTAAGTAA | SunyBiotech | N/A |
|
saeg-2 sgRNA 3: AGACGATGCACCATCTTCGTGT TTTAG AGCTAGA AATAGCAAGT saeg-2 sgRNA 4: TGGATCTAATGAGACGGGTAGTTT TAGAGCTAGAAATAGCAAGT saeg-2 sgRNA 5: CAATGAACGAAGT CATTCGGGTTT TAGAGCTAGAAATAGCAAGT saeg-2 sgRNA 6: GAA TGTTTAGAGTAAGTAACGTTTT AGAGCTAGAAATAGCAAGT |
IDT | N/A |
| Software and Algorithms | ||
| RMA algorithm with Partek Genomic suite v6.6 | Partek | N/A |
| FastQC | Andrews, 2010 | N/A |
| Cutadapt | Martin, 2011 | N/A |
| Shortstack | Axtell, 2013 | N/A |
| Butter | Axtell, 2014 | N/A |
| HTSeq count | Anders et al., 2015 | N/A |
| R Deseq2 | Love et al., 2014 | N/A |
| FISH-quant | Mueller et al., 2013 | N/A |
| Fiji | ImageJ | N/A |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Oded Rechavi (odedrechavi@gmail.com).
Experimental Model and Subject Details
Cultivation of the Worms
All strains were maintained by standard methods (Stiernagle, 2006). Unless noted otherwise, we performed all experiments at 20 degrees. The chemotaxis experiments were all performed at 25 degrees (as mentioned in the results section), except for the experiment where we compared the chemotaxis performances in 20 versus 25 degrees (Figure S5). All strains used in the study are listed in the Key Resources Table.
Method Details
DNA constructs and transgenic animals
To express RDE-4 pan-neuronally we used two pan-neuronal promoters, sng-1 and rgef-1, known to drive expression in a large percent of neurons when 2kb promoters are utilized (Ruvinsky et al., 2007, Stefanakis et al., 2015). A trans-spliced Yellow Fluorescent Protein (yfp) gene was co-transcribed together with rde-4.
To create expression vectors that express rde-4 in neurons under the control of the sng-1 promoter: The GFP sequence in vector pPD95.67 was replaced with YFP, and the coding region of rde-4, amplified from genomic DNA (PCR 1) was fused to SL2 via fusion PCR, and cloned into the above-mentioned plasmid. An 2000bp upstream flank of sng-1 was amplified from genomic DNA (PCR 2) and cloned in to the plasmid with the restriction sites PstI and BamHI. The Psng-1::rde-4:SL2:yfp:unc-54 UTR construct and co-injection marker Pmyo-3::mcherry (pCFJ104) were linearized and injected in to rde-4(ne299) mutants (BFF12) generating BFF14: rde-4(ne299);pigEx9[Psng-1::rde-4:SL2:yfp+Pmyo-3::Mcherry]. The concentrations injected were Psng-1::rde-4:SL2:yfp:unc-54 UTR (5 ng/μl), Pmyo-3::mcherry (20 ng/μl) and sheared genomic DNA (60 ng/μl).
To express rde-4 in neurons under the rgef-1 promoter: The 2670 bp upstream flanking region of rgef-1 was amplified from genomic DNA (PCR 3) and cloned in to the plasmid with restriction sites PstI and BamHI. The Prgef-1::rde-4:SL2:yfp:unc-54 UTR construct and co-injection marker Punc122::GFP (pCFJ68) were linearized and injected in to rde-4 (ne299) mutants (BFF12) generating BBF15: rde-4(ne299);pigEx10[Prgef-1::rde-4::SL2”:yfp+Punc-122::gfp]. The concentrations injected were Prgef-1::rde-4::SL2::yfp::unc-54 UTR (5 ng/μl), Punc122::GFP (10 ng/μl) and sheared genomic DNA (60 ng/ μl).
Expression of Psng-1::rde-4 using mos1-mediated Single Copy Insertion (mosSCI):
Psng-1:rde-4:SL2:yfp:unc-54 was digested and cloned in to plasmid pCFJ350 with restriction sites BglII and SpeI to create a repair template. A mix of pGH8 (Prab-3::mCherry) (10ng/μl), pCFJ90 (Pmyo-2::mCherry) (2.5 ng/μl), pCFJ104 (Pmyo-3:mCherry) (5ng/μl), pMA122 (Phsp16.41::peel-1) (10 ng/μl), pCFJ601 (Peft-3:Mos1 transposase) (50 ng/μl), repair template (15 ng/μl) and Bluescript (60 ng/μl) was injected in to strain EG6699. Selection of transgenic strain was performed as previously described (Frøkjaer-Jensen et al., 2008). Construct insertion was verified by PCR amplification and sequencing. Worms were outcrossed four times to wild-type N2 before crossing to rde-4(ne299) mutant worms (BFF12) generating BFF16: rde-4(ne299); pigSi3[cb-unc-119(+)Psng-1::rde-4:SL2:yfp]. Worms were then nurtured for a minimum 100 generations until experiments were conducted.
CRISPR/Cas9
CRISPR/Cas9 directed mutagenesis was utilized to generate a mutant allele of hrde-1(pig4) in the rde-4(ne299) mutant background and mutant allele of saeg-2 (pig5) in the rde-4(ne299) mutant background. Single guide RNAs (sgRNAs) were inserted into pDD162 (Dickinson et al., 2013). A co-CRISPR approach, in which the wild-type rol-6 allele was replaced with the rol-6(su1006) allele, was used to detect successful genome editing events (Kim et al., 2014). Young adult worms were injected with an injection mix containing: 50ng/μl of each targeting sgRNA, 50ng/μl of rol-6 sgRNA, 20ng/μl rol-6(su1006) conversion template. Progeny of injected worms were screened for the roller phenotype and were examined by PCR (PCR 4 and 5) and sequencing to detect indels at edited locus. The sequences used are available in the Key Resources Table.
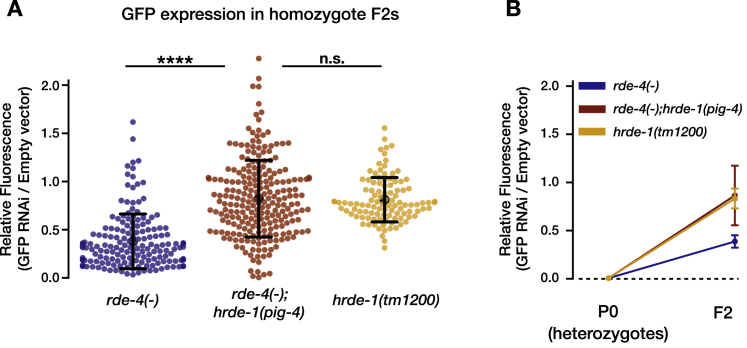
The generated allele hrde-1(pig4) contains a 399 nt deletion in exon 3 of hrde-1, and we called BFF17 the obtained strain (with the full genotype rde-4(ne299);hrde-1(pig4)). To examine whether the BFF17 double mutant lost HRDE-1 function we performed RNAi inheritance experiments and found that rde-4 worms homozygous to the hrde-1 mutant allele (pig4 allele) are defective in RNAi inheritance (silencing of germline expressed GFP driven by a pie-1 promoter), in comparison to rde-4 worms (the canonical HRDE-1 mutant phenotype). The BFF17 strain was outcrossed to the rde-4 mutant (BFF12) and then crossed to strain BFF14 to introduce pigEx9[Psng-1::rde-4:SL2:yfp+Pmyo-3::Mcherry], or with strain BFF16 to introduce pigSi3[cb-unc-119(+)Psng-1::rde-4:SL2:yfp]. All smFISH and chemotaxis experiments with the strains bearing the hrde-1(pig4) allele were performed at least 12 generations after the establishment of the relevant strain.
Two strains including mutant alleles of saeg-2, PHX776: rde-4(ne299); saeg-2(syb776) and PHX777: rde-4(ne299);saeg-2(syb777) were generated by SunyBiotech (China) and were verified with sequencing (PCR 5). The syb776 and syb777 alleles contain a 1255 nt deletion, eliminating 72% of the SAEG-2 coding sequence. PHX776 and PHX777 were outcrossed to rde-4 worms (BFF12) before conducting experiments and named BFF36 and BFF37 respectively. A third generated saeg-2 allele, pig5 was created in rde-4(ne299) mutants generating BFF38: rde-4(ne299); saeg-2(pig5). The allele has not been successfully genotyped yet. We suspect that the deletion created is larger than planned.
RNAi experiments testing hrde-1 functionality
To test the degree to which the CRISPR-induced hrde-1(pig4) allele disrupts RNAi inheritance (Figure S6) we crossed the mutants with an integrated germline GFP reporter (BFF22) and conducted RNAi inheritance experiments (heritable silencing of GFP). RNAi producing HT115 bacteria were inoculated into LB broth containing Ampicillin (100 μg/ml) at 37°C overnight with shaking. Bacterial cultures were seeded onto NGM plate containing IPTG (1 mM) and Ampicillin (100 μg/ml) and grown in the dark. hrde-1(pig4);rde-4(−/−) (BFF22) worms were first crossed to hrde-1(tm1200) (BFF20) worms and the cross progeny (considered as the ‘P0’ generation in this experiment) laid on RNAi bacteria in order to initiate the RNAi response, when the worms have one functional copy of rde-4 (in rde-4(+/−)hrde-1(pig-1/tm1200) heterozygotes). P0 worms were transferred to E.coli OP50 bacteria at the L4 stage and allowed to lay eggs. The F1 progeny, raised on E.coli OP50, were individually isolated, allowed to lay eggs and then genotyped in order to identify rde-4(ne299);hrde-1(pig4) homozygotes. Their F2 progeny were tested on day one of adulthood for germline GFP expression. In parallel, rde-4 mutants (BFF21) were crossed to N2 wild-type worms (EG6089) and their rde-4 homozygous progeny tested in an identical manner as a positive control. All genotypes were additionally raised on empty-vector bearing HT115 bacteria plates and tested.
Figure S6.
Worms that Are Homozygous for the hrde-1 Mutant Allele pig4 Are Defective in RNAi Inheritance, Related to STAR Methods
Worms heterozygote to the indicated genotype and expressing a germline GFP reporter were exposed to bacteria expressing anti-GFP dsRNA (P0 generation). We used heterozygotes so that the worms could initiate an RNAi response (rde-4 mutants do not initiate RNAi responses). GFP silencing levels were tested in the P0 heterozygotes and their homozygote mutated F2 progeny. These experiments were conducted in three independent replicates (n = > 30).
(A) Relative GFP (GFP/empty vector) fluorescence in F2 homozygotes. Each dot represents one worm. P values were determined by one-way ANOVA and Tukey’s post hoc correction for multiple comparison. ∗∗∗∗- p < 10−4; n.s.- p > 0.05. Data shown are means ± SD.
(B) Relative GFP fluorescence levels (GFP/empty vector) between worm populations exposed to GFP RNAi at the P0 generation were averaged across trials.
rde-4(ne299) DNA and cDNA analysis
The rde-4 allele was identified by PCR amplification of a 389 base pair amplicon (PCR 6) of rde-4 gene. The rde-4(ne299) mutant allele has a single-base insertion creating a restriction site for the enzyme BsmAI. PCR amplicons were digested with the BsmAI enzyme and DNA fragments run in a 2% electrophoresis gel to determine the genotype. rde-4(+) exhibits bands at 116 and 273 base pairs, while rde-4(ne299) exhibit bands at 102, 116 and 171 base pairs.
Experimental scheme for RNA extraction
All the experiments were conducted three times (independent biological triplicates) unless specified otherwise. Adults worms were allowed to lay eggs for 12 hours, and young adult worms (70 hours later) were collected and washed 4 times before total RNA extraction. To obtain RNA from gonads, we washed worms 4 times in M9 buffer and then transferred ∼30 worms to a cavity microscope slide in to 10ul of egg buffer (1M HEPES, 5M NaCl, 1M MgCl2, 1M CaCl2, 1M KCl and 20% tween-20) containing 2mM levamisole. Gonads were dissected with gauge needles by cutting right below the pharynx or the tip of the tail, and after the spermatheca. Gonads were collected from the slide using a mouth pipette (Sigma) into an Eppendorf on ice prior to addition of Trizol. To obtain samples from F3 rde-4(ne299) progeny of Psng-1::rde-4;rde-4(ne299) worms, we crossed males of the latter with rde-4(ne299) hermaphrodites, isolated the Psng-1::rde-4(+/−) worms (at the L4 stage) in the next generation that we considered as the P0 generation (PCR 7). The P0 heterozygotes were allowed to lay eggs, and we isolated F1 individuals into separate plates. After two days of adulthood, we collected the F1 mothers for genotyping (PCR 6) and continued only with rde-4(ne299) lines. We used the F2s for synchronization of the F3 generation, as depicted above. For the High-Copy rescue strain: As extrachromosomal arrays in transgenic C. elegans strains are not inherited to 100% of the worms’ progeny, we were able to collect non-array expressing worms whose parents expressed neuronal RDE-4. For the collection of Psng-1::rde-4 and F1-rde-4(−/−) worms, Psng-1::rde-4 adult worms were allowed to lay eggs, and their progeny were separately picked in to (1) array expressing worms (Psng-1::rde-4) and (2) non-array expressing worms (F1-rde-4(−/−)) under an MVX10 Olympus microscope, enabling detection of the array by both the co-transcribed YFP reporter and a co-injection marker (Pmyo-3:mcherry). Worms were allowed to recover for an hour at 20°C after picking before collection for RNA extraction. All genotypes were identically picked under the MVX10 Olympus microscope prior to collection. F1-rde-4(−/−) worms were allowed to lay eggs to give the F2 progeny, that in turn produced the worms of the F3 generation that were collected for RNA extraction. The very same scheme was applied with worms expressing the other high-copy array Prgef-1::rde-4 generated with co-injection marker Punc-122::GFP. To verify that the presence of the fluorescent co-injection markers, assembled into the same extrachromosomal array following DNA injection, faithfully reports the presence of the pan-neuronal RDE-4 construct, we tested for rde-4(ne299) allele expression in Psng-1:rde-4 and Prgef-1:rde-4 worms expressing Pmyo-3::mcherry and Punc-122::GFP respectively, and their (not fluorescent) F1-rde-4(−/−) progeny (PCR 6).
Collection of isolated neurons for RNA extraction
Isolation of neurons was performed using Fluorescent-Activated Cell Sorting (FACS) based on a previously described protocol (Kaletsky et al., 2016) with minor modifications. We introduced by genetic crossing a rab-3p::2xNLS::TagRFP transgene into the rde-4(ne299) (BFF12) and Psng-1::rde-4 (BFF 16) strains, in order to mark their neurons with RFP expression (we used the OH10690 strain kindly provided by Dr. Meital Oren-Suissa, and backcrossed it x3 before further use). Synchronized Day 1 adult worms were washed 4 times in M9 buffer and transferred into a 1.7ml eppendorf tube. The pellet of worms was then quickly washed (resuspension, short spin-down centrifugation and removal of supernatant) with 500 μL of lysis buffer (200 mM DTT, 0.25% SDS, 20 mM HEPES pH 8.0, 3% sucrose), and resuspended in 750 μL lysis buffer. The worms were lysed in room temperature for 6.5 minutes, then washed rapidly 5 times in M9 (mixing by finger-tapping and quick spin down in each round of wash). Next, we removed the supernatant and applied 500 μL of 20mg/ml Pronase (Sigma) on the worms. We incubated the worms with Pronase for 15-20 minutes, during which we vigorously pipetted them every 2 minutes. Following each round of pipetting, 1 μL of worms was transferred to a slide for inspection using a table microscope. We ended enzymatic incubation when most worm bodies were dissociated leaving only small debris and eggs. The digestion was stopped by adding 250 μL of ice-cold PBS with 2% Fetal Bovine Serum and transferring the tubes to ice. The samples were then passed through a 30 μm filter (Sysmex) into a FACS tube (still on ice), followed by additional 250 μL of PBS(2%FBS) passing through the filter to collect remnants. Sorting was performed immediately in 4 degrees using a BDFacs Aria IIu Sorter (Beckton Dickinson), and RFP+ cells were collected directly into a tube containing 850 μL of TRIzol LS (Life technologies). RNA isolation has been performed using the miRNeasy Mini Kit (QIAGEN, Cat No./ID: 217004) according to the manufacturer’s protocol.
Small RNA Library Preparation and Sequencing
For RNA isolation worms were lysed using the TRIzol reagent. 300 μL of TRIzol was added to 50ul of adult worms and were subjected to three cycles of freezing in −80C and vortexing at RT for 15mins. 60 μL of chloroform was added and samples were transferred to a pre-spun Heavy Phase Lock tube and centrifuged at 12,000 g for 15mins at 4C. The aqueous phase was transferred to a pre-spun Heavy Phase Lock tube and 1:1 Phenol:Chloroform:Isoamyl Alcohol was added and centrifuged at 16,000 g for 5mins at RT. The aqueous layer was transferred to a 1.5ml Eppendorf tube and 20 μg of Glycogen and 1:1 Isopropanol was added. The samples were incubated at −20C for 30 mins and then centrifuged for 30mins at 16,000 g at 4C. The pellet was washed 2 times with 70% ethanol and then air-dried for 10 minutes. The pellet was re-suspended in 10ul of RNase free water. The total RNA samples (at least 150ng) were treated with RNA 5′ Polyphosphatase (Epicenter), to ensure 5′ monophosphate-independent capturing of small RNAs. Libraries were prepared using NEBNext® Multiplex Small RNA Library Prep Set for Illumina (New England Biolabs) or TruSeq Small RNA Library Prep Kit (Illumina) according to the manufacturer’s protocol. Samples were tested for their quality and concertation on an Agilent 2200 TapeStation and then pooled together. Pools were separated on a 4% agarose E-Gel (Life Technologies) and the 140–160 nt length bands were excised and purified using MiniElute Gel Extraction kit (QIAGEN). Libraries were sequenced using an Illumina HighSeq2500 instrument or Nextseq 500. (Details about the sequencer used for each sample is available in Table S1 and GEO: GSE124049).
Gene arrays
The microarray experiments were performed using Affymetrix GeneChip® C. elegans Genome Array oligonucleotide arrays (Thermofisher). 500 ng of mRNA from each sample was used to generate cDNA followed by in vitro transcription with biotinylated UTP and CTP resulting in approximately 300-fold amplification of RNA. Spike controls were added to 15 μg fragmented cRNA before overnight hybridization. Arrays were then washed and stained with streptavidin-phycoerythrin, before being scanned on an Affymetrix GeneChip scanner. 3′/5′ ratios for GAPDH and beta-actin were confirmed to be within acceptable limits (pos_vs_neg_auc > 0.9), and BioB spike controls were found to be present on all chips, with BioC, BioD and CreX also present in increasing intensity.
mRNA Library Preparation and Sequencing
cDNA libraries were prepared with the SMART-Seq v4 Ultra Low Input RNA Kit (Clontech). A total of 1 ng RNA was used as input and processed according to the manufacturer’s protocol. Double strand cDNA was amplified by a 15 cycle PCR. cDNA quantity and quality was verified using a TapeStation 2200 (Agilent). One ng of cDNA was used as input for preparation of sequencing libraries using the Nextera XT DNA Sequencing kit (Illumina). Libraries were prepared according to manufacturer’s instructions. Quality and concentration were verified with TapeStation 2200 and libraries were pooled according to molarity for sequencing on the NextSeq 500.
Single Molecule Fluorescent In Situ Hybridization
Probes of 20nt in length targeting mature mRNAs were designed with Stellaris RNA FISH probe designer, and probe sets of 32-40 probes selected based on gene target specificity, defined by a maximum complementary sequence of 16 nt to genes other than the defined target transcript (ensembl BLAST tool). An exemption to this was C18D4.6, as it has a paralog, F31E9.6 (Woods et al., 2013) with 86% identity in sequence, thus it was not possible to discriminate between these two transcripts. Probes were produced, conjugated to Quasar 670 and purified by Biosearch Technologies (details of probe sets are available in Table S6). Populations of worms were treated with a sodium hypochlorite protocol and left to incubate at 20°C overnight in M9 buffer to reach a synced L1 population. Psng-1::rde-4 array expressing worms were picked under a MVX10 Olympus microscope prior to bleaching - ensuring the examination of their F1 rde-4(−/−) progeny. F3 rde-4(ne299) progeny of worms expressing an integrated single copy of Psng-1::rde-4 were obtained as previously described above, only F2′s were bleached to generate F3 progeny. L1 larvae were transferred to plates with E.coli OP50 and allowed to develop at 20°C for 48 hours reaching mid-late L4 stage. SNR (signal to noise ratio) of probes detecting tissue specific expression was optimal at the L4 stage. Synced worms were collected in M9 buffer and washed three times in 1.5 mL Eppendorf tubes. Supernatant was removed and 1 mL of fixation buffer (100ul 37% formaldehyde solution, 100ul 10X PBS and 800ul nuclease free water) added, followed by an incubation of 45 minutes on a rotator at room temperature. Worms were then washed twice with 1XPBS and suspended in 70% Ethanol and incubated on a rotator for 30 hours at 4°C. Ethanol was removed and 1 mL wash buffer (100ul 20XSSC, 100ul deionized formamide and 800ul nuclease free water) added and let to stand for 5 minutes. Wash buffer was aspirated away and 100ul hybridization buffer (for 10mL: 1g dextran sulfate, 10 mg Escherichia coli tRNA, 100 μL 200 mM vanadyl ribonucleoside complex, 40 μL 50 mg/mL RNase free BSA, 1ml Formamide and nuclease free water to 10 mL final volume) with a concertation of 0.05uM single-molecule fluorescent in situ hybridization (smFISH) probes added, and left for overnight incubation at 37C in the dark. Prior to imaging, hybridization buffer was removed and 1 mL of wash buffer added and left to incubate at 37C in the dark for 30 minutes. Samples were then suspended in 1 mL of fresh wash buffer with 5ug/mL DAPI, and left to incubate at 37C in the dark for 30 minutes. Wash buffer was removed and worms suspended in 1 mL 2XSSC. Buffer was aspirated and samples were incubated in 100ul GLOX buffer (for 1 mL: 40 μL 10% glucose, 10 μL 1 M Tris-HCl, pH 8.0, 100 μL 20X SSC and 850 μL nuclease-free water) for 2 minutes. Buffer was aspirated and 100ul GLOX buffer with the addition of 1ul glucose oxidase (3.7mg/mL) and 1ul of mildly vortexed Catalase suspension, samples were kept on ice until imaging. Images were acquired with an Olympus IX83 motorized inverted microscope coupled with an ORCA flash4.0 V2 Hamamatsu camera. Exposure times and acquisition settings were identical between replicates. Individual worms were chosen for acquisition based on normal morphology and developmental stage was verified based on vulva morphology (Mok et al., 2015). Regions of interest for acquisition were defined by nuclei DAPI staining. Z stacks were acquired with a 0.3 um step size ranging between 70-85 slices per stack.
pos-1 and mel-26 RNAi
HT115 Escherichia coli strains expressing dsRNAs were obtained from the Vidal RNAi library (Rual et al., 2004). L4 stage worms were transferred to plates for 24 hours and allowed to lay eggs. After 72 hours their surviving progeny were transferred to fresh plates (IPTG 1M, Ampicillin 100ug/ml) seeded with RNAi bacteria at day one adult and allowed to lay eggs overnight for 12 hours. After 36 hours, L3 worms were counted (2 generations on RNAi). High-Copy Psng-1::rde-4 and Prgef-1::rde-4 strains were counted under a MVX10 Olympus microscope to differentiate between array expressing and non-array expressing worms.
Chemotaxis assays
Behavioral chemotaxis assays were conducted according to the standard procedures (Hart 2006). Gravid worms were allowed to lay eggs for 12 hours in conditioning temperature (20 or 25 degrees) and then removed. When synced populations of worms reached day 1 adult they were collected in wash buffer (same content as chemotaxis assay plates: 5 mM KP04, 1mM CaCl2 and 1mM MgS04), allowing worms to sediment by gravity, and washed three times to remove bacteria. > 200 worms were placed in 90mm chemotaxis plates (2% agar, 5 mM KP04, 1mM CaCl2 and 1mM MgS04) in a drop of wash buffer. For volatile odors 1 μL of benzaldehyde (Sigma), diacetyl (Sigma) or butanone (Sigma) (diluted in 95% ethanol) was applied to one point on the assay plate while 95% ethanol was placed on the opposite side. For NaCl tests, agar plugs consisting 50mM of NaCl were left to diffuse overnight in order to create a gradient. 1 μL of 1M sodium azide (Sigma) was previously placed on odor points and allowed to dry prior to assay. Excess fluid was collected with a kimpwipe, plates were wrapped with parafilm, and worms were allowed to move for 1 hour in the incubator at the correlating conditioning temperature. Plates were transferred to 4°C and worms scored after 2 days.
Microfluidic device fabrication
Standard soft lithography methods were used to fabricate photoresist (SU8) masters for microfluidic devices (San-Miguel and Lu, 2013-wormbook) as described in Ouellette et al., (2018). Two-component polydimethylsiloxane (PDMS) (Dow Corning Sylgard) was mixed at 10:1 w/w, degassed under vacuum, poured over masters, degassed again, and cured at 50°C overnight. Inlet holes were made with a Milltex 1 mm biopsy punches (Fisher). Chips were cleaned with Isopropyl alcohol and Scotch tape (3M) and then bonded to glass coverslips using air plasma generated by a handheld Electro-Technic Products). Chips were then left at 50°C overnight. Coupling to fluid reservoirs was done by inserting PTFE microbore tubing (Cole-Parmer) directly into chip inlet holes.
Calcium Imaging
Fluorescence time lapse imaging was performed as described (Hendricks et al., 2012) on restrained animals in microfluidic chips (Chronis et al., 2007). Images were acquired at 5 fps with a 100ms exposure using a 40X silicone-immersion objective on an Olympus IX83 inverted microscope. Animals were washed in buffer (5mM KP04, 1mM CaCl2 and 1mM MgS04) for 15 minutes prior to acquisition. In the chip, worms were exposed to 1 minute of isoamyl alcohol (1:104), followed by a switch to wash buffer. Approximately 30 s of image acquisition documented the event of odor removal, which induces responses in sensory neuron AWC in wild-type animals. Fluid flow was driven by negative pressure from a vacuum pump and switches were controlled with a Valvebank solenoid pinch valve array (AutoMate Scientific).
Quantification and Statistical Analysis
Small RNA-seq analysis
Illumina fastq output files were assessed for quality with FastQC (Andrews, 2010). Cutadapt (Martin, 2011) was then used for adaptor removal:
cutadapt -m 15–discard-untrimmed -a [3′ adaptor sequence] input.fastq > output.fastq
Clipped reads were aligned to the ce10 assembly of C. elegans genome using Butter (Axtell, 2014) or Shortstack (Axtell, 2013):
Butter ‘or’ Shortstack–mismatches 1 input.fq genome _reference.fa
We next filtered for small RNA reads of 20-23 nucleotide in length (Blumenfeld and Jose, 2016). Then we counted reads in antisense orientation to genes as defined by the corresponding Ensembl .gff file, using a python based script HTSeq count (Anders et al., 2015):
HTSeq.scripts.count–stranded = reverse–minaqual = 0–mode = intersection-nonempty input.sam GENES.gff > output.txt
Next, we used the HTSeq output as an input file for R Deseq2 (Love et al., 2014). We defined differentially expressed genes using cutoff of adjusted p value < 0.1.
Bioinformatic gene enrichment analysis
The enrichment values denote the ratio between the observed representation of a specific gene set within a defined differentially expressed genes group, to the expected one, i.e., the representation of the examined gene set among all protein-coding genes in C. elegans. The analysis was done for 5 gene sets: (1) targets regulated by DCR-1 helicase domain (Welker et al., 2010), (2) 177 STGs downregulated in mut-16(mg461) worms (Zhang et al., 2011) (3) 87 targets of ERGO-1 dependent 22Gs, (4) 4191 targets of CSR-1 endo-siRNAs (Claycomb et al., 2009), (5) 1587 targets of HRDE-1 endo-siRNAs (Buckley et al., 2012).
Specifically, the enrichment value of a given gene set i in differentially expressed STGs was calculated using the following formula:
Obtaining the observed-to-expected ratios, we then calculated the corresponding p values using 10,000 random gene groups identical in size to that of the examined group of differentially expressed genes. For instance, if we found enrichment of 2 (linear scale) for a given gene set among the 476 genes which had statistically significant differentially-expressed targeting STGs in Psng-1::rde-4 (compared to the rde-4 mutants, Figure 1D), we randomly sampled from all the genes 10,000 groups of 476 genes, and calculated their enrichment for the dataset of interest. We then ranked the govern enrichment values, and determined the p value of the expressed genes with upregulated STGs in Psng-1::rde-4 by the fraction of the enrichment values higher than 2 among the 10,000 random sets.
smFISH Image Analysis
Images were analyzed using FISH-quant software (Mueller et al., 2013). For each experimental trial, images of individual worms were first filtered with FISH-quant and then processed on Fiji (ImageJ) to produce Z stack by maximum intensity. Images with high background signal due to non-optimal fixation were disregarded. Next, according to the FISH-quant software protocol (Mueller et al., 2013), a pre-detection process was conducted on random images for each probe set and an optimal intensity threshold was chosen for mRNA dot detection. According to the FISH-quant pipeline, the detected spots were then fit with a 3D Gaussian and detection settings optimized and saved. The saved settings were then used for a batch process to analyze all images for the relevant probe set. Following image analysis of all experiments per probe set, for each genotype tested, images with average, minimum and maximum values were further examined for accuracy of dot detection to confirm that thresholds could be applied across trials. Additionally, for images with values that were statistical outliers dot detection was conducted and examined to rule out possible technical factors due to over fixation (low SNR) or inefficient probe staining.
Calcium Imaging analysis
GCaMP2 fluorescence intensity was measured with FIJI (ImageJ). Time lapse images were corrected for movements in the xy axis using stack registration. For each worm the ROI was selected manually, defined by the cell body of AWC projected in z by maximum intensity. GCaMP2 (Tian et al., 2009) intensity of the ROI was measured and normalized with the formula:
(Ft - Fmin) / (Fmax - Fmin), transforming fluorescent intensity to values within a range of 0 to 1. Mean Peak was calculated as the average delta of intensity 2 s prior to stimuli removal, minus the maximum delta intensity following odor removal: F0: average of the first 10 frames. ΔF: ((Ft-F0)/F0)∗100.
Mean peak: average ΔF(2 s pre-stimuli) - max ΔF(post-stimuli). Time delay was calculated as the number of seconds from odor removal it took for the florescence to reach maximum intensity.
Germline GFP expression analysis
For each condition, at least 50 animals were mounted on a 2% agarose slide and paralyzed in a drop of M9 with 25mM sodium azide. The worms were imaged with a 10x objective on a IX83 Olympus microscope (Identical exposure times and led intensities between conditions and replicates). The images were analyzed with ImageJ software. For at least 40 worms per condition, the CTCF values of three oocyte nuclei were averaged per worm. CTCF values for worms raised on dsRNA-GFP expressing bacteria were normalized to the average CTCF value of the genotype population raised on empty-vector.
Pan-neuronal YFP expression analysis
Day one adult worms of Psng-1::rde-4 and Prgef-1::rde-4 strains were mounted a 2% agarose slides and paralyzed in a drop of M9 with 25mM sodium azide. Images were acquired with a 10x objective on a IX83 Olympus microscope, acquiring a 10-12 frame Z stack. Images were then processed with ImageJ software, producing Z-projections of the maximum intensity.
Statistics and graphing
Statistics and graphing were conducted on MATLAB, GraphPad Prism 6 or using Python with the Seaborn data visualization library. Corrections for multiple comparisons were performed as indicated in legends, using Tukey’s or Dunn’s multiple comparisons corrections, and adjusted P values are denoted. Comparisons of variance were performed with the F-test of equality of variances.
Data and Software Availability
The accession number for the sequencingd data reported in this paper is GEO: GSE124049.
Acknowledgments
We thank all the Rechavi lab members for helpful discussions. This project was initiated when O.R. was a postdoctoral fellow in Oliver Hobert’s lab, and we thank Oliver for his wise advice and for contributing useful ideas. We are also thankful to Sze Yen (John) Kerk from the O.H. lab for his advice. We thank members of Michael Hendricks’s lab for their support and discussion. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We thank Meital Oren-Suissa for kindly providing us with strains. We thank Siegfried Hekimi and his lab for providing us with strains and plasmids. Thanks to Hugo Aguilaniu and his lab for assisting with preparation of transgenic strains. We are grateful to Jasmine Jacob, Karen Cesarkas, Sarit Farage Barhom, and Efrat Glick Saar from the Sheba Cancer Research Institute for sequencing services. We thank Irena Shur for scientific assistance with flow cytometry and cell sorting. Special thanks to Dror Cohen for the illustrations that he contributed. We thank the Eldee Foundation and the Bloomfield family of Montreal for their support. O.R. gratefully acknowledges the support of the Allen Discovery Center (part of the Paul G. Allen Frontiers Group) and the support of the Adelis Foundation (0604916191). This work was funded by ERC (335624).
Author Contributions
R.P., I.A.T., and O.R. conceived and designed the experiments. R.P., I.A.T., O.A., E.S., and S.A. performed the experiments. H.G., R.P., I.A.T., and S.B. performed genomic data analysis. E.A., R.P., O.A., and E.S. performed microscopy analysis. I.A.T., R.P., H.G., and O.R. wrote the manuscript with input from S.A., O.A., and M.H.
Declaration of Interests
The authors declare no competing interests.
Published: June 6, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2019.04.029.
Supplemental Information
RNA samples used in the present study.
DESeq2 fold-change results for the detection of differentially expressed STGs in Psng-1::rde-4 worms compared to rde-4(-) (Sheet #1), and in the Psng-1::rde-4 and Prgef-1::rde-4 High-Copy rescue strains versus rde-4(-) (Sheets #2 & #3).
NeuroSTGs (Sheet #1) and mRNAs (Sheet #2) counts detected in sorted neurons of N2 (wildtype) worms. DESeq2 fold-change results for the detection of differentially expressed NeuroSTGs (Sheet #3) and mRNAs (Sheet #4) in neurons of Psng-1::rde-4 worms compared to rde-4(-).
Transgenerational regulation of STGs in the germline of Psng-1::rde-4 neuronal rescue worms (Sheets #1-#6). Transgenerational regulation of STGs in High-Copy RDE-4 rescue worms (Sheets #7 & #8).
GermSTGs whose regulation by neuronal RDE-4 requires sid-1. See also Figure S4.
List of probes used for smFISH experiments for staining of the genes rde-4, yfp, saeg-2, C18D4.6 and C55C3.3.
References
- Alcazar R.M., Lin R., Fire A.Z. Transmission dynamics of heritable silencing induced by double-stranded RNA in Caenorhabditis elegans. Genetics. 2008;180:1275–1288. doi: 10.1534/genetics.108.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]; Alcazar, R.M., Lin, R., and Fire, A.Z. (2008). Transmission dynamics of heritable silencing induced by double-stranded RNA in Caenorhabditis elegans. Genetics 180, 1275-1288. [DOI] [PMC free article] [PubMed]
- Anava S., Posner R., Rechavi O. The soft genome. Worm. 2015;3:e989798. doi: 10.4161/21624054.2014.989798. [DOI] [PMC free article] [PubMed] [Google Scholar]; Anava, S., Posner, R., and Rechavi, O. (2015). The soft genome. Worm 3, e989798. [DOI] [PMC free article] [PubMed]
- Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]; Anders, S., Pyl, P.T., and Huber, W. (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166-169. [DOI] [PMC free article] [PubMed]
- Aoki K., Moriguchi H., Yoshioka T., Okawa K., Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007;26:5007–5019. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]; Aoki, K., Moriguchi, H., Yoshioka, T., Okawa, K., and Tabara, H. (2007). In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 26, 5007-5019. [DOI] [PMC free article] [PubMed]
- Ashe A., Sapetschnig A., Weick E.M., Mitchell J., Bagijn M.P., Cording A.C., Doebley A.L., Goldstein L.D., Lehrbach N.J., Le Pen J. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ashe, A., Sapetschnig, A., Weick, E.M., Mitchell, J., Bagijn, M.P., Cording, A.C., Doebley, A.L., Goldstein, L.D., Lehrbach, N.J., Le Pen, J., et al. (2012). piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150, 88-99. [DOI] [PMC free article] [PubMed]
- Ashley J., Cordy B., Lucia D., Fradkin L.G., Budnik V., Thomson T. Retrovirus-like Gag Protein Arc1 Binds RNA and Traffics across Synaptic Boutons. Cell. 2018;172:262–274. doi: 10.1016/j.cell.2017.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ashley, J., Cordy, B., Lucia, D., Fradkin, L.G., Budnik, V., and Thomson, T. (2018). Retrovirus-like Gag Protein Arc1 Binds RNA and Traffics across Synaptic Boutons. Cell 172, 262-274. [DOI] [PMC free article] [PubMed]
- Avgousti D.C., Palani S., Sherman Y., Grishok A. CSR-1 RNAi pathway positively regulates histone expression in C. elegans. EMBO J. 2012;31:3821–3832. doi: 10.1038/emboj.2012.216. [DOI] [PMC free article] [PubMed] [Google Scholar]; Avgousti, D.C., Palani, S., Sherman, Y., and Grishok, A. (2012). CSR-1 RNAi pathway positively regulates histone expression in C. elegans. EMBO J. 31, 3821-3832. [DOI] [PMC free article] [PubMed]
- Axtell M.J. ShortStack: comprehensive annotation and quantification of small RNA genes. RNA. 2013;19:740–751. doi: 10.1261/rna.035279.112. [DOI] [PMC free article] [PubMed] [Google Scholar]; Axtell, M.J. (2013). ShortStack: comprehensive annotation and quantification of small RNA genes. RNA 19, 740-751. [DOI] [PMC free article] [PubMed]
- Axtell M.J. Butter: High-precision genomic alignment of small RNA-seq data. bioRxiv. 2014 [Google Scholar]; Axtell, M.J. (2014). Butter: High-precision genomic alignment of small RNA-seq data. bioRxiv, doi: 10.1101/007427.
- Bale T.L. Epigenetic and transgenerational reprogramming of brain development. Nat. Rev. Neurosci. 2015;16:332–344. doi: 10.1038/nrn3818. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bale, T.L. (2015). Epigenetic and transgenerational reprogramming of brain development. Nat. Rev. Neurosci. 16, 332-344. [DOI] [PMC free article] [PubMed]
- Bharadwaj P.S., Hall S.E. Endogenous RNAi Pathways Are Required in Neurons for Dauer Formation in Caenorhabditis elegans. Genetics. 2017;205:1503–1516. doi: 10.1534/genetics.116.195438. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bharadwaj, P.S., and Hall, S.E. (2017). Endogenous RNAi Pathways Are Required in Neurons for Dauer Formation in Caenorhabditis elegans. Genetics 205, 1503-1516. [DOI] [PMC free article] [PubMed]
- Blanchard D., Parameswaran P., Lopez-Molina J., Gent J., Saynuk J.F., Fire A. On the nature of in vivo requirements for rde-4 in RNAi and developmental pathways in C. elegans. RNA Biol. 2011;8:458–467. doi: 10.4161/rna.8.3.14657. [DOI] [PMC free article] [PubMed] [Google Scholar]; Blanchard, D., Parameswaran, P., Lopez-Molina, J., Gent, J., Saynuk, J.F., and Fire, A. (2011). On the nature of in vivo requirements for rde-4 in RNAi and developmental pathways in C. elegans. RNA Biol. 8, 458-467. [DOI] [PMC free article] [PubMed]
- Blumenfeld A.L., Jose A.M. Reproducible features of small RNAs in C. elegans reveal NU RNAs and provide insights into 22G RNAs and 26G RNAs. RNA. 2016;22:184–192. doi: 10.1261/rna.054551.115. [DOI] [PMC free article] [PubMed] [Google Scholar]; Blumenfeld, A.L., and Jose, A.M. (2016). Reproducible features of small RNAs in C. elegans reveal NU RNAs and provide insights into 22G RNAs and 26G RNAs. RNA 22, 184-192. [DOI] [PMC free article] [PubMed]
- Boulias K., Horvitz H.R. The C. elegans microRNA mir-71 acts in neurons to promote germline-mediated longevity through regulation of DAF-16/FOXO. Cell Metab. 2012;15:439–450. doi: 10.1016/j.cmet.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; Boulias, K., and Horvitz, H.R. (2012). The C. elegans microRNA mir-71 acts in neurons to promote germline-mediated longevity through regulation of DAF-16/FOXO. Cell Metab. 15, 439-450. [DOI] [PMC free article] [PubMed]
- Buckley B.A., Burkhart K.B., Gu S.G., Spracklin G., Kershner A., Fritz H., Kimble J., Fire A., Kennedy S. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489:447–451. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]; Buckley, B.A., Burkhart, K.B., Gu, S.G., Spracklin, G., Kershner, A., Fritz, H., Kimble, J., Fire, A., and Kennedy, S. (2012). A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489, 447-451. [DOI] [PMC free article] [PubMed]
- Cecere G., Hoersch S., Jensen M.B., Dixit S., Grishok A. The ZFP-1(AF10)/DOT-1 complex opposes H2B ubiquitination to reduce Pol II transcription. Mol. Cell. 2013;50:894–907. doi: 10.1016/j.molcel.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cecere, G., Hoersch, S., Jensen, M.B., Dixit, S., and Grishok, A. (2013). The ZFP-1(AF10)/DOT-1 complex opposes H2B ubiquitination to reduce Pol II transcription. Mol. Cell 50, 894-907. [DOI] [PMC free article] [PubMed]
- Chang S., Wen S., Chen D., Jin P. Small regulatory RNAs in neurodevelopmental disorders. Hum. Mol. Genet. 2009;18(R1):R18–R26. doi: 10.1093/hmg/ddp072. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chang, S., Wen, S., Chen, D., and Jin, P. (2009). Small regulatory RNAs in neurodevelopmental disorders. Hum. Mol. Genet. 18 (R1), R18-R26. [DOI] [PMC free article] [PubMed]
- Chronis N., Zimmer M., Bargmann C.I. Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nat. Methods. 2007;4:727–731. doi: 10.1038/nmeth1075. [DOI] [PubMed] [Google Scholar]; Chronis, N., Zimmer, M., and Bargmann, C.I. (2007). Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nat. Methods 4, 727-731. [DOI] [PubMed]
- Claycomb J.M., Batista P.J., Pang K.M., Gu W., Vasale J.J., van Wolfswinkel J.C., Chaves D.A., Shirayama M., Mitani S., Ketting R.F. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; Claycomb, J.M., Batista, P.J., Pang, K.M., Gu, W., Vasale, J.J., van Wolfswinkel, J.C., Chaves, D.A., Shirayama, M., Mitani, S., Ketting, R.F., et al. (2009). The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139, 123-134. [DOI] [PMC free article] [PubMed]
- Conine C.C., Moresco J.J., Gu W., Shirayama M., Conte D., Jr., Yates J.R., III, Mello C.C. Argonautes Promote Male Fertility and Provide a Paternal Memory of Germline Gene Expression in C. elegans. Cell. 2013;155:1532–1544. doi: 10.1016/j.cell.2013.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]; Conine, C.C., Moresco, J.J., Gu, W., Shirayama, M., Conte Jr., D., Yates III, J.R., and Mello, C.C. (2013). Argonautes Promote Male Fertility and Provide a Paternal Memory of Germline Gene Expression in C. elegans. Cell 155, 1532-1544. [DOI] [PMC free article] [PubMed]
- Conine C.C., Sun F., Song L., Rivera-Pérez J.A., Rando O.J. Small RNAs Gained during Epididymal Transit of Sperm Are Essential for Embryonic Development in Mice. Dev. Cell. 2018;46:470–480.e3. doi: 10.1016/j.devcel.2018.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]; Conine, C.C., Sun, F., Song, L., Rivera-Perez, J.A., and Rando, O.J. (2018). Small RNAs Gained during Epididymal Transit of Sperm Are Essential for Embryonic Development in Mice. Dev. Cell 46, 470-480.e3. [DOI] [PMC free article] [PubMed]
- Cornils A., Gloeck M., Chen Z., Zhang Y., Alcedo J. Specific insulin-like peptides encode sensory information to regulate distinct developmental processes. Development. 2011;138:1183–1193. doi: 10.1242/dev.060905. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cornils, A., Gloeck, M., Chen, Z., Zhang, Y., and Alcedo, J. (2011). Specific insulin-like peptides encode sensory information to regulate distinct developmental processes. Development 138, 1183-1193. [DOI] [PMC free article] [PubMed]
- de Albuquerque B.F.M., Placentino M., Ketting R.F. Maternal piRNAs Are Essential for Germline Development following De Novo Establishment of Endo-siRNAs in Caenorhabditis elegans. Dev. Cell. 2015;34:448–456. doi: 10.1016/j.devcel.2015.07.010. [DOI] [PubMed] [Google Scholar]; de Albuquerque, B.F.M., Placentino, M., and Ketting, R.F. (2015). Maternal piRNAs Are Essential for Germline Development following De Novo Establishment of Endo-siRNAs in Caenorhabditis elegans. Dev. Cell 34, 448-456. [DOI] [PubMed]
- Devanapally S., Ravikumar S., Jose A.M. Double-stranded RNA made in C. elegans neurons can enter the germline and cause transgenerational gene silencing. Proc. Natl. Acad. Sci. USA. 2015;112:2133–2138. doi: 10.1073/pnas.1423333112. [DOI] [PMC free article] [PubMed] [Google Scholar]; Devanapally, S., Ravikumar, S., and Jose, A.M. (2015). Double-stranded RNA made in C. elegans neurons can enter the germline and cause transgenerational gene silencing. Proc. Natl. Acad. Sci. USA 112, 2133-2138. [DOI] [PMC free article] [PubMed]
- Dias B.G., Ressler K.J. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat. Neurosci. 2014;17:89–96. doi: 10.1038/nn.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dias, B.G., and Ressler, K.J. (2014). Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat. Neurosci. 17, 89-96. [DOI] [PMC free article] [PubMed]
- Dickinson D.J., Ward J.D., Reiner D.J., Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods. 2013;10:1028–1034. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dickinson, D.J., Ward, J.D., Reiner, D.J., and Goldstein, B. (2013). Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10, 1028-1034. [DOI] [PMC free article] [PubMed]
- Duchaine T.F., Wohlschlegel J.A., Kennedy S., Bei Y., Conte D., Jr., Pang K., Brownell D.R., Harding S., Mitani S., Ruvkun G. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]; Duchaine, T.F., Wohlschlegel, J.A., Kennedy, S., Bei, Y., Conte, D., Jr., Pang, K., Brownell, D.R., Harding, S., Mitani, S., Ruvkun, G., et al. (2006). Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124, 343-354. [DOI] [PubMed]
- Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]; Eden, E., Navon, R., Steinfeld, I., Lipson, D., and Yakhini, Z. (2009). GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48. [DOI] [PMC free article] [PubMed]
- Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]; Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806-811. [DOI] [PubMed]
- Fletcher M., Kim D.H. Age-dependent neuroendocrine signaling from sensory neurons modulates the effect of dietary restriction on longevity of Caenorhabditis elegans K. PLoS Genet. 2017;13:e1006544. doi: 10.1371/journal.pgen.1006544. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fletcher, M., and Kim, D.H. (2017). Age-dependent neuroendocrine signaling from sensory neurons modulates the effect of dietary restriction on longevity of Caenorhabditis elegans K. PLoS Genet. 13, e1006544. [DOI] [PMC free article] [PubMed]
- Frøkjaer-Jensen C., Davis M.W., Hopkins C.E., Newman B.J., Thummel J.M., Olesen S.P., Grunnet M., Jorgensen E.M. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]; Frokjaer-Jensen, C., Davis, M.W., Hopkins, C.E., Newman, B.J., Thummel, J.M., Olesen, S.P., Grunnet, M., and Jorgensen, E.M. (2008). Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40, 1375-1383. [DOI] [PMC free article] [PubMed]
- Fujiwara M., Aoyama I., Hino T., Teramoto T., Ishihara T. Gonadal Maturation Changes Chemotaxis Behavior and Neural Processing in the Olfactory Circuit of Caenorhabditis elegans. Curr. Biol. 2016;26:1522–1531. doi: 10.1016/j.cub.2016.04.058. [DOI] [PubMed] [Google Scholar]; Fujiwara, M., Aoyama, I., Hino, T., Teramoto, T., and Ishihara, T. (2016). Gonadal Maturation Changes Chemotaxis Behavior and Neural Processing in the Olfactory Circuit of Caenorhabditis elegans. Curr. Biol. 26, 1522-1531. [DOI] [PubMed]
- Gapp K., Jawaid A., Sarkies P., Bohacek J., Pelczar P., Prados J., Farinelli L., Miska E., Mansuy I.M. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat. Neurosci. 2014;17:667–669. doi: 10.1038/nn.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gapp, K., Jawaid, A., Sarkies, P., Bohacek, J., Pelczar, P., Prados, J., Farinelli, L., Miska, E., and Mansuy, I.M. (2014a). Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat. Neurosci. 17, 667-669. [DOI] [PMC free article] [PubMed]
- Gapp K., Soldado-Magraner S., Alvarez-Sánchez M., Bohacek J., Vernaz G., Shu H., Franklin T.B., Wolfer D., Mansuy I.M. Early life stress in fathers improves behavioural flexibility in their offspring. Nat. Commun. 2014;5:5466. doi: 10.1038/ncomms6466. [DOI] [PubMed] [Google Scholar]; Gapp, K., Soldado-Magraner, S., Alvarez-Sanchez, M., Bohacek, J., Vernaz, G., Shu, H., Franklin, T.B., Wolfer, D., and Mansuy, I.M. (2014b). Early life stress in fathers improves behavioural flexibility in their offspring. Nat. Commun. 5, 5466. [DOI] [PubMed]
- Gerson-Gurwitz A., Wang S., Sathe S., Green R., Yeo G.W., Oegema K., Desai A. A Small RNA-Catalytic Argonaute Pathway Tunes Germline Transcript Levels to Ensure Embryonic Divisions. Cell. 2016;165:396–409. doi: 10.1016/j.cell.2016.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gerson-Gurwitz, A., Wang, S., Sathe, S., Green, R., Yeo, G.W., Oegema, K., and Desai, A. (2016). A Small RNA-Catalytic Argonaute Pathway Tunes Germline Transcript Levels to Ensure Embryonic Divisions. Cell 165, 396-409. [DOI] [PMC free article] [PubMed]
- Gu W., Shirayama M., Conte D., Jr., Vasale J., Batista P.J., Claycomb J.M., Moresco J.J., Youngman E.M., Keys J., Stoltz M.J. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gu, W., Shirayama, M., Conte, D., Jr., Vasale, J., Batista, P.J., Claycomb, J.M., Moresco, J.J., Youngman, E.M., Keys, J., Stoltz, M.J., et al. (2009). Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell 36, 231-244. [DOI] [PMC free article] [PubMed]
- Hao Y., Xu N., Box A.C., Schaefer L., Kannan K., Zhang Y., Florens L., Seidel C., Washburn M.P., Wiegraebe W., Mak H.Y. Nuclear cGMP-Dependent Kinase Regulates Gene Expression via Activity-Dependent Recruitment of a Conserved Histone Deacetylase Complex. PLoS Genet. 2011;7:e1002065. doi: 10.1371/journal.pgen.1002065. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hao, Y., Xu, N., Box, A.C., Schaefer, L., Kannan, K., Zhang, Y., Florens, L., Seidel, C., Washburn, M.P., Wiegraebe, W., and Mak, H.Y. (2011). Nuclear cGMP-Dependent Kinase Regulates Gene Expression via Activity-Dependent Recruitment of a Conserved Histone Deacetylase Complex. PLoS Genet. 7, e1002065. [DOI] [PMC free article] [PubMed]
- Hart A. Behavior. WormBook. 2006:1–67. [Google Scholar]; Hart, A. (2006). Behavior. WormBook 1-67.
- Hendricks M., Ha H., Maffey N., Zhang Y. Compartmentalized calcium dynamics in C. elegans interneuron encode head movement. Nature. 2012;487:99–103. doi: 10.1038/nature11081. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hendricks, M., Ha, H., Maffey, N., and Zhang, Y. (2012). Compartmentalized calcium dynamics in C. elegans interneuron encode head movement. Nature 487, 99-103. [DOI] [PMC free article] [PubMed]
- Hinas A., Wright A.J., Hunter C.P. SID-5 is an endosome-associated protein required for efficient systemic RNAi in C. elegans. Curr. Biol. 2012;22:1938–1943. doi: 10.1016/j.cub.2012.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hinas, A., Wright, A.J., and Hunter, C.P. (2012). SID-5 is an endosome-associated protein required for efficient systemic RNAi in C. elegans. Curr. Biol. 22, 1938-1943. [DOI] [PMC free article] [PubMed]
- Houri-Ze’evi L., Korem Y., Sheftel H., Faigenbloom L., Toker I.A., Dagan Y., Awad L., Degani L., Alon U., Rechavi O. A Tunable Mechanism Determines the Duration of the Transgenerational Small RNA Inheritance in C. elegans. Cell. 2016;165:88–99. doi: 10.1016/j.cell.2016.02.057. [DOI] [PubMed] [Google Scholar]; Houri-Ze’evi, L., Korem, Y., Sheftel, H., Faigenbloom, L., Toker, I.A., Dagan, Y., Awad, L., Degani, L., Alon, U., and Rechavi, O. (2016). A Tunable Mechanism Determines the Duration of the Transgenerational Small RNA Inheritance in C. elegans. Cell 165, 88-99. [DOI] [PubMed]
- Houri-Zeevi L., Rechavi O. A Matter of Time: Small RNAs Regulate the Duration of Epigenetic Inheritance. Trends Genet. 2017;33:46–57. doi: 10.1016/j.tig.2016.11.001. [DOI] [PubMed] [Google Scholar]; Houri-Zeevi, L., and Rechavi, O. (2017). A Matter of Time: Small RNAs Regulate the Duration of Epigenetic Inheritance. Trends Genet. 33, 46-57. [DOI] [PubMed]
- Itoh T., Fairall L., Muskett F.W., Milano C.P., Watson P.J., Arnaudo N., Saleh A., Millard C.J., El-Mezgueldi M., Martino F., Schwabe J.W. Structural and functional characterization of a cell cycle associated HDAC1/2 complex reveals the structural basis for complex assembly and nucleosome targeting. Nucleic Acids Res. 2015;43:2033–2044. doi: 10.1093/nar/gkv068. [DOI] [PMC free article] [PubMed] [Google Scholar]; Itoh, T., Fairall, L., Muskett, F.W., Milano, C.P., Watson, P.J., Arnaudo, N., Saleh, A., Millard, C.J., El-Mezgueldi, M., Martino, F., and Schwabe, J.W. (2015). Structural and functional characterization of a cell cycle associated HDAC1/2 complex reveals the structural basis for complex assembly and nucleosome targeting. Nucleic Acids Res. 43, 2033-2044. [DOI] [PMC free article] [PubMed]
- Johnston R.J., Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]; Johnston, R.J., and Hobert, O. (2003). A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426, 845-849. [DOI] [PubMed]
- Jose A.M. Movement of regulatory RNA between animal cells. Genesis. 2015;53:395–416. doi: 10.1002/dvg.22871. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jose, A.M. (2015). Movement of regulatory RNA between animal cells. Genesis 53, 395-416. [DOI] [PMC free article] [PubMed]
- Jose A.M., Garcia G.A., Hunter C.P. Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Nat. Struct. Mol. Biol. 2011;18:1184–1188. doi: 10.1038/nsmb.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jose, A.M., Garcia, G.A., and Hunter, C.P. (2011). Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Nat. Struct. Mol. Biol. 18, 1184-1188. [DOI] [PMC free article] [PubMed]
- Juang B.-T., Gu C., Starnes L., Palladino F., Goga A., Kennedy S., L’Etoile N.D. Endogenous nuclear RNAi mediates behavioral adaptation to odor. Cell. 2013;154:1010–1022. doi: 10.1016/j.cell.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; Juang, B.-T., Gu, C., Starnes, L., Palladino, F., Goga, A., Kennedy, S., and L’Etoile, N.D. (2013). Endogenous nuclear RNAi mediates behavioral adaptation to odor. Cell 154, 1010-1022. [DOI] [PMC free article] [PubMed]
- Kaletsky R., Lakhina V., Arey R., Williams A., Landis J., Ashraf J., Murphy C.T. The C. elegans adult neuronal IIS/FOXO transcriptome reveals adult phenotype regulators. Nature. 2016;529:92–96. doi: 10.1038/nature16483. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kaletsky, R., Lakhina, V., Arey, R., Williams, A., Landis, J., Ashraf, J., and Murphy, C.T. (2016). The C. elegans adult neuronal IIS/FOXO transcriptome reveals adult phenotype regulators. Nature 529, 92-96. [DOI] [PMC free article] [PubMed]
- Kennedy L.M., Grishok A. Neuronal migration is regulated by endogenous RNAi and chromatin-binding factor ZFP-1/AF10 in Caenorhabditis elegans. Genetics. 2014;197:207–220. doi: 10.1534/genetics.114.162917. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kennedy, L.M., and Grishok, A. (2014). Neuronal migration is regulated by endogenous RNAi and chromatin-binding factor ZFP-1/AF10 in Caenorhabditis elegans. Genetics 197, 207-220. [DOI] [PMC free article] [PubMed]
- Kim H., Ishidate T., Ghanta K.S., Seth M., Conte D., Jr., Shirayama M., Mello C.C. A co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics. 2014;197:1069–1080. doi: 10.1534/genetics.114.166389. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kim, H., Ishidate, T., Ghanta, K.S., Seth, M., Conte, D., Jr., Shirayama, M., and Mello, C.C. (2014). A co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics 197, 1069-1080. [DOI] [PMC free article] [PubMed]
- L’Etoile N.D., Coburn C.M., Eastham J., Kistler A., Gallegos G., Bargmann C.I. The cyclic GMP-dependent protein kinase EGL-4 regulates olfactory adaptation in C. elegans. Neuron. 2002;36:1079–1089. doi: 10.1016/s0896-6273(02)01066-8. [DOI] [PubMed] [Google Scholar]; L’Etoile, N.D., Coburn, C.M., Eastham, J., Kistler, A., Gallegos, G., and Bargmann, C.I. (2002). The cyclic GMP-dependent protein kinase EGL-4 regulates olfactory adaptation in C. elegans. Neuron 36, 1079-1089. [DOI] [PubMed]
- Lee R.C., Hammell C.M., Ambros V. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA. 2006;12:589–597. doi: 10.1261/rna.2231506. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lee, R.C., Hammell, C.M., and Ambros, V. (2006). Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA 12, 589-597. [DOI] [PMC free article] [PubMed]
- Lev I., Seroussi U., Gingold H., Bril R., Anava S., Rechavi O. MET-2-Dependent H3K9 Methylation Suppresses Transgenerational Small RNA Inheritance. Curr. Biol. 2017;27:1138–1147. doi: 10.1016/j.cub.2017.03.008. [DOI] [PubMed] [Google Scholar]; Lev, I., Seroussi, U., Gingold, H., Bril, R., Anava, S., and Rechavi, O. (2017). MET-2-Dependent H3K9 Methylation Suppresses Transgenerational Small RNA Inheritance. Curr. Biol. 27, 1138-1147. [DOI] [PubMed]
- Li X., Jin P. Roles of small regulatory RNAs in determining neuronal identity. Nat. Rev. Neurosci. 2010;11:329–338. doi: 10.1038/nrn2739. [DOI] [PubMed] [Google Scholar]; Li, X., and Jin, P. (2010). Roles of small regulatory RNAs in determining neuronal identity. Nat. Rev. Neurosci. 11, 329-338. [DOI] [PubMed]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Love, M.I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed]
- Luteijn M.J., van Bergeijk P., Kaaij L.J., Almeida M.V., Roovers E.F., Berezikov E., Ketting R.F. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J. 2012;31:3422–3430. doi: 10.1038/emboj.2012.213. [DOI] [PMC free article] [PubMed] [Google Scholar]; Luteijn, M.J., van Bergeijk, P., Kaaij, L.J., Almeida, M.V., Roovers, E.F., Berezikov, E., and Ketting, R.F. (2012). Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J. 31, 3422-3430. [DOI] [PMC free article] [PubMed]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. [Google Scholar]; Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10-12.
- Mattick J.S. Rocking the foundations of molecular genetics. Proc. Natl. Acad. Sci. USA. 2012;109:16400–16401. doi: 10.1073/pnas.1214129109. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mattick, J.S. (2012). Rocking the foundations of molecular genetics. Proc. Natl. Acad. Sci. USA 109, 16400-16401. [DOI] [PMC free article] [PubMed]
- McCarthy M.M., Auger A.P., Bale T.L., De Vries G.J., Dunn G.A., Forger N.G., Murray E.K., Nugent B.M., Schwarz J.M., Wilson M.E. The epigenetics of sex differences in the brain. J. Neurosci. 2009;29:12815–12823. doi: 10.1523/JNEUROSCI.3331-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]; McCarthy, M.M., Auger, A.P., Bale, T.L., De Vries, G.J., Dunn, G.A., Forger, N.G., Murray, E.K., Nugent, B.M., Schwarz, J.M., and Wilson, M.E. (2009). The epigenetics of sex differences in the brain. J. Neurosci. 29, 12815-12823. [DOI] [PMC free article] [PubMed]
- Mok D.Z.L., Sternberg P.W., Inoue T. Morphologically defined sub-stages of C. elegans vulval development in the fourth larval stage. BMC Dev. Biol. 2015;15:26. doi: 10.1186/s12861-015-0076-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mok, D.Z.L., Sternberg, P.W., and Inoue, T. (2015). Morphologically defined sub-stages of C. elegans vulval development in the fourth larval stage. BMC Dev. Biol. 15, 26. [DOI] [PMC free article] [PubMed]
- Moore R.S., Kaletsky R., Murphy C.T. C. elegans pathogenic learning confers multigenerational pathogen avoidance. bioRxiv. 2018 [Google Scholar]; Moore, R.S., Kaletsky, R., and Murphy, C.T. (2018). C. elegans pathogenic learning confers multigenerational pathogen avoidance. bioRxiv, doi: 10.1101/476416.
- Mueller F., Senecal A., Tantale K., Marie-Nelly H., Ly N., Collin O., Basyuk E., Bertrand E., Darzacq X., Zimmer C. FISH-quant: automatic counting of transcripts in 3D FISH images. Nat. Methods. 2013;10:277–278. doi: 10.1038/nmeth.2406. [DOI] [PubMed] [Google Scholar]; Mueller, F., Senecal, A., Tantale, K., Marie-Nelly, H., Ly, N., Collin, O., Basyuk, E., Bertrand, E., Darzacq, X., and Zimmer, C. (2013). FISH-quant: automatic counting of transcripts in 3D FISH images. Nat. Methods 10, 277-278. [DOI] [PubMed]
- Ni J.Z., Kalinava N., Chen E., Huang A., Trinh T., Gu S.G. A transgenerational role of the germline nuclear RNAi pathway in repressing heat stress-induced transcriptional activation in C. elegans. Epigenetics Chromatin. 2016;9:3. doi: 10.1186/s13072-016-0052-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ni, J.Z., Kalinava, N., Chen, E., Huang, A., Trinh, T., and Gu, S.G. (2016). A transgenerational role of the germline nuclear RNAi pathway in repressing heat stress-induced transcriptional activation in C. elegans. Epigenetics Chromatin 9, 3. [DOI] [PMC free article] [PubMed]
- Ouellette M.H., Desrochers M.J., Gheta I., Ramos R., Hendricks M. A Gate-and-Switch Model for Head Orientation Behaviors in Caenorhabditis elegans. eNeuro. 2018;5 doi: 10.1523/ENEURO.0121-18.2018. ENEURO.0121–18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ouellette, M.H., Desrochers, M.J., Gheta, I., Ramos, R., and Hendricks, M. (2018). A Gate-and-Switch Model for Head Orientation Behaviors in Caenorhabditis elegans. eNeuro 5, ENEURO.0121-18.2018. [DOI] [PMC free article] [PubMed]
- Pastuzyn E.D., Day C.E., Kearns R.B., Kyrke-Smith M., Taibi A.V., McCormick J., Yoder N., Belnap D.M., Erlendsson S., Morado D.R. The Neuronal Gene Arc Encodes a Repurposed Retrotransposon Gag Protein that Mediates Intercellular RNA Transfer. Cell. 2018;172:275–288. doi: 10.1016/j.cell.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pastuzyn, E.D., Day, C.E., Kearns, R.B., Kyrke-Smith, M., Taibi, A.V., McCormick, J., Yoder, N., Belnap, D.M., Erlendsson, S., Morado, D.R., et al. (2018). The Neuronal Gene Arc Encodes a Repurposed Retrotransposon Gag Protein that Mediates Intercellular RNA Transfer. Cell 172, 275-288. [DOI] [PMC free article] [PubMed]
- Pereira A., Gracida X., Kagias K., Zhang Y. Learning of pathogenic bacteria in adult C. elegans bidirectionally regulates pathogen response in the progeny. bioRxiv. 2019 [Google Scholar]; Pereira, A., Gracida, X., Kagias, K., and Zhang, Y. (2019). Learning of pathogenic bacteria in adult C. elegans bidirectionally regulates pathogen response in the progeny. bioRxiv, doi: 10.1101/500264v2.
- Prahlad V., Morimoto R.I. Neuronal circuitry regulates the response of Caenorhabditis elegans to misfolded proteins. Proc. Natl. Acad. Sci. USA. 2011;108:14204–14209. doi: 10.1073/pnas.1106557108. [DOI] [PMC free article] [PubMed] [Google Scholar]; Prahlad, V., and Morimoto, R.I. (2011). Neuronal circuitry regulates the response of Caenorhabditis elegans to misfolded proteins. Proc. Natl. Acad. Sci. USA 108, 14204-14209. [DOI] [PMC free article] [PubMed]
- Rechavi O., Lev I. Principles of Transgenerational Small RNA Inheritance in Caenorhabditis elegans. Curr. Biol. 2017;27:R720–R730. doi: 10.1016/j.cub.2017.05.043. [DOI] [PubMed] [Google Scholar]; Rechavi, O., and Lev, I. (2017). Principles of Transgenerational Small RNA Inheritance in Caenorhabditis elegans. Curr. Biol. 27, R720-R730. [DOI] [PubMed]
- Rechavi O., Houri-Ze’evi L., Anava S., Goh W.S.S., Kerk S.Y., Hannon G.J., Hobert O. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell. 2014;158:277–287. doi: 10.1016/j.cell.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rechavi, O., Houri-Ze’evi, L., Anava, S., Goh, W.S.S., Kerk, S.Y., Hannon, G.J., and Hobert, O. (2014). Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell 158, 277-287. [DOI] [PMC free article] [PubMed]
- Remy J.-J. Stable inheritance of an acquired behavior in Caenorhabditis elegans. Curr. Biol. 2010;20:R877–R878. doi: 10.1016/j.cub.2010.08.013. [DOI] [PubMed] [Google Scholar]; Remy, J.-J. (2010). Stable inheritance of an acquired behavior in Caenorhabditis elegans. Curr. Biol. 20, R877-R878. [DOI] [PubMed]
- Rocheleau C.E. RNA interference: Systemic RNAi SIDes with endosomes. Curr. Biol. 2012;22:R873–R875. doi: 10.1016/j.cub.2012.08.039. [DOI] [PubMed] [Google Scholar]; Rocheleau, C.E. (2012). RNA interference: Systemic RNAi SIDes with endosomes. Curr. Biol. 22, R873-R875. [DOI] [PubMed]
- Rual J.-F., Ceron J., Koreth J., Hao T., Nicot A.S., Hirozane-Kishikawa T., Vandenhaute J., Orkin S.H., Hill D.E., van den Heuvel S., Vidal M. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rual, J.-F., Ceron, J., Koreth, J., Hao, T., Nicot, A.S., Hirozane-Kishikawa, T., Vandenhaute, J., Orkin, S.H., Hill, D.E., van den Heuvel, S., and Vidal, M. (2004). Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14, 2162-2168. [DOI] [PMC free article] [PubMed]
- Ruvinsky I., Ohler U., Burge C.B., Ruvkun G. Detection of broadly expressed neuronal genes in C. elegans. Dev. Biol. 2007;302:617–626. doi: 10.1016/j.ydbio.2006.09.014. [DOI] [PubMed] [Google Scholar]; Ruvinsky, I., Ohler, U., Burge, C.B., and Ruvkun, G. (2007). Detection of broadly expressed neuronal genes in C. elegans. Dev. Biol. 302, 617-626. [DOI] [PubMed]
- San-Miguel A., Lu H. Microfluidics as a tool for C. elegans research. WormBook. 2013:1–19. doi: 10.1895/wormbook.1.162.1. [DOI] [PMC free article] [PubMed] [Google Scholar]; San-Miguel, A., and Lu, H. (2013). Microfluidics as a tool for C. elegans research. WormBook 1-19.journal [DOI] [PMC free article] [PubMed]
- Sapetschnig A., Sarkies P., Lehrbach N.J., Miska E.A. Tertiary siRNAs mediate paramutation in C. elegans. PLoS Genet. 2015;11:e1005078. doi: 10.1371/journal.pgen.1005078. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sapetschnig, A., Sarkies, P., Lehrbach, N.J., and Miska, E.A. (2015). Tertiary siRNAs mediate paramutation in C. elegans. PLoS Genet. 11, e1005078. [DOI] [PMC free article] [PubMed]
- Sarkies P., Ashe A., Le Pen J., McKie M.A., Miska E.A. Competition between virus-derived and endogenous small RNAs regulates gene expression in Caenorhabditis elegans. Genome Res. 2013;23:1258–1270. doi: 10.1101/gr.153296.112. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sarkies, P., Ashe, A., Le Pen, J., McKie, M.A., and Miska, E.A. (2013). Competition between virus-derived and endogenous small RNAs regulates gene expression in Caenorhabditis elegans. Genome Res. 23, 1258-1270. [DOI] [PMC free article] [PubMed]
- Seth M., Shirayama M., Gu W., Ishidate T., Conte D., Jr., Mello C.C. The C. elegans CSR-1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Dev. Cell. 2013;27:656–663. doi: 10.1016/j.devcel.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; Seth, M., Shirayama, M., Gu, W., Ishidate, T., Conte, D., Jr., and Mello, C.C. (2013). The C. elegans CSR-1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Dev. Cell 27, 656-663. [DOI] [PMC free article] [PubMed]
- Sharma U., Sun F., Conine C.C., Reichholf B., Kukreja S., Herzog V.A., Ameres S.L., Rando O.J. Small RNAs Are Trafficked from the Epididymis to Developing Mammalian Sperm. Dev. Cell. 2018;46:481–494. doi: 10.1016/j.devcel.2018.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sharma, U., Sun, F., Conine, C.C., Reichholf, B., Kukreja, S., Herzog, V.A., Ameres, S.L., and Rando, O.J. (2018). Small RNAs Are Trafficked from the Epididymis to Developing Mammalian Sperm. Dev. Cell 46, 481-494. [DOI] [PMC free article] [PubMed]
- Shirayama M., Seth M., Lee H.C., Gu W., Ishidate T., Conte D., Jr., Mello C.C. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shirayama, M., Seth, M., Lee, H.C., Gu, W., Ishidate, T., Conte, D., Jr., and Mello, C.C. (2012). piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150, 65-77. [DOI] [PMC free article] [PubMed]
- Andrews S. Babraham Bioinformatics; 2010. FastQC-A Quality Control tool for High Throughput Sequence Data. [Google Scholar]; Andrews, S. (2010). FastQC-A Quality Control tool for High Throughput Sequence Data (Babraham Bioinformatics).
- Sims J.R., Ow M.C., Nishiguchi M.A., Kim K., Sengupta P., Hall S.E. Developmental programming modulates olfactory behavior in C. elegans via endogenous RNAi pathways. Elife. 2016;5:e11642. doi: 10.7554/eLife.11642. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sims, J.R., Ow, M.C., Nishiguchi, M.A., Kim, K., Sengupta, P., and Hall, S.E. (2016). Developmental programming modulates olfactory behavior in C. elegans via endogenous RNAi pathways. Elife 5, e11642. [DOI] [PMC free article] [PubMed]
- Sonoda S., Ohta A., Maruo A., Ujisawa T., Kuhara A. Sperm Affects Head Sensory Neuron in Temperature Tolerance of Caenorhabditis elegans. Cell Rep. 2016;16:56–65. doi: 10.1016/j.celrep.2016.05.078. [DOI] [PubMed] [Google Scholar]; Sonoda, S., Ohta, A., Maruo, A., Ujisawa, T., and Kuhara, A. (2016). Sperm Affects Head Sensory Neuron in Temperature Tolerance of Caenorhabditis elegans. Cell Rep. 16, 56-65. [DOI] [PubMed]
- Spracklin G., Fields B., Wan G., Vijayendran D., Wallig A., Shukla A., Kennedy S. Identification and Characterization of C. elegans RNAi Inheritance Machinery. Genetics. 2017;206:1403–1416. doi: 10.1534/genetics.116.198812. [DOI] [PMC free article] [PubMed] [Google Scholar]; Spracklin, G., Fields, B., Wan, G., Vijayendran, D., Wallig, A., Shukla, A., and Kennedy, S. (2017). Identification and Characterization of C. elegans RNAi Inheritance Machinery. Genetics 206, 1403-1416. [DOI] [PMC free article] [PubMed]
- Srinivasan S. Regulation of body fat in Caenorhabditis elegans. Annu. Rev. Physiol. 2015;77:161–178. doi: 10.1146/annurev-physiol-021014-071704. [DOI] [PMC free article] [PubMed] [Google Scholar]; Srinivasan, S. (2015). Regulation of body fat in Caenorhabditis elegans. Annu. Rev. Physiol. 77, 161-178. [DOI] [PMC free article] [PubMed]
- Stefanakis N., Carrera I., Hobert O. Regulatory Logic of Pan-Neuronal Gene Expression in C. elegans. Neuron. 2015;87:733–750. doi: 10.1016/j.neuron.2015.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stefanakis, N., Carrera, I., and Hobert, O. (2015). Regulatory Logic of Pan-Neuronal Gene Expression in C. elegans. Neuron 87, 733-750. [DOI] [PMC free article] [PubMed]
- Stiernagle T. Maintenance of C. elegans. WormBook. 2006:1–11. doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stiernagle, T. (2006). Maintenance of C. elegans. WormBook 1-11. [DOI] [PMC free article] [PubMed]
- Stubbe H. MIT Press; 1972. History of Genetics: From Prehistoric Times to the Rediscovery of Mendel’s Laws. [Google Scholar]; Stubbe, H. (1972). History of Genetics: From Prehistoric Times to the Rediscovery of Mendel’s Laws (MIT Press).
- Styer K.L., Singh V., Macosko E., Steele S.E., Bargmann C.I., Aballay A. Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science. 2008;322:460–464. doi: 10.1126/science.1163673. [DOI] [PMC free article] [PubMed] [Google Scholar]; Styer, K.L., Singh, V., Macosko, E., Steele, S.E., Bargmann, C.I., and Aballay, A. (2008). Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science 322, 460-464. [DOI] [PMC free article] [PubMed]
- Tabara H., Yigit E., Siomi H., Mello C.C. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]; Tabara, H., Yigit, E., Siomi, H., and Mello, C.C. (2002). The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell 109, 861-871. [DOI] [PubMed]
- Tian L., Hires S.A., Mao T., Huber D., Chiappe M.E., Chalasani S.H., Petreanu L., Akerboom J., McKinney S.A., Schreiter E.R. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tian, L., Hires, S.A., Mao, T., Huber, D., Chiappe, M.E., Chalasani, S.H., Petreanu, L., Akerboom, J., McKinney, S.A., Schreiter, E.R., et al. (2009). Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods 6, 875-881. [DOI] [PMC free article] [PubMed]
- Tonkin L.A., Bass B.L. Mutations in RNAi rescue aberrant chemotaxis of ADAR mutants. Science. 2003;302:1725. doi: 10.1126/science.1091340. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tonkin, L.A., and Bass, B.L. (2003). Mutations in RNAi rescue aberrant chemotaxis of ADAR mutants. Science 302, 1725. [DOI] [PMC free article] [PubMed]
- Vasale J.J., Gu W., Thivierge C., Batista P.J., Claycomb J.M., Youngman E.M., Duchaine T.F., Mello C.C., Conte D., Jr. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc. Natl. Acad. Sci. USA. 2010;107:3582–3587. doi: 10.1073/pnas.0911908107. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vasale, J.J., Gu, W., Thivierge, C., Batista, P.J., Claycomb, J.M., Youngman, E.M., Duchaine, T.F., Mello, C.C., and Conte, D., Jr. (2010). Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc. Natl. Acad. Sci. USA 107, 3582-3587. [DOI] [PMC free article] [PubMed]
- Vassoler F.M., White S.L., Schmidt H.D., Sadri-Vakili G., Pierce R.C. Epigenetic inheritance of a cocaine-resistance phenotype. Nat. Neurosci. 2013;16:42–47. doi: 10.1038/nn.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vassoler, F.M., White, S.L., Schmidt, H.D., Sadri-Vakili, G., and Pierce, R.C. (2013). Epigenetic inheritance of a cocaine-resistance phenotype. Nat. Neurosci. 16, 42-47. [DOI] [PMC free article] [PubMed]
- Weaver I.C.G., Cervoni N., Champagne F.A., D’Alessio A.C., Sharma S., Seckl J.R., Dymov S., Szyf M., Meaney M.J. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]; Weaver, I.C.G., Cervoni, N., Champagne, F.A., D’Alessio, A.C., Sharma, S., Seckl, J.R., Dymov, S., Szyf, M., and Meaney, M.J. (2004). Epigenetic programming by maternal behavior. Nat. Neurosci. 7, 847-854. [DOI] [PubMed]
- Weismann A. Oxford Clarendon Press; 1891. Essays Upon Heredity and Kindred Biological Problems. [Google Scholar]; Weismann, A. (1891). Essays Upon Heredity and Kindred Biological Problems (Oxford Clarendon Press).
- Welker N.C., Pavelec D.M., Nix D.A., Duchaine T.F., Kennedy S., Bass B.L. Dicer’s helicase domain is required for accumulation of some, but not all, C. elegans endogenous siRNAs. RNA. 2010;16:893–903. doi: 10.1261/rna.2122010. [DOI] [PMC free article] [PubMed] [Google Scholar]; Welker, N.C., Pavelec, D.M., Nix, D.A., Duchaine, T.F., Kennedy, S., and Bass, B.L. (2010). Dicer’s helicase domain is required for accumulation of some, but not all, C. elegans endogenous siRNAs. RNA 16, 893-903. [DOI] [PMC free article] [PubMed]
- Winston W.M., Molodowitch C., Hunter C.P. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456–2459. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]; Winston, W.M., Molodowitch, C., and Hunter, C.P. (2002). Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295, 2456-2459. [DOI] [PubMed]
- Winston W.M., Sutherlin M., Wright A.J., Feinberg E.H., Hunter C.P. Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc. Natl. Acad. Sci. USA. 2007;104:10565–10570. doi: 10.1073/pnas.0611282104. [DOI] [PMC free article] [PubMed] [Google Scholar]; Winston, W.M., Sutherlin, M., Wright, A.J., Feinberg, E.H., and Hunter, C.P. (2007). Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc. Natl. Acad. Sci. USA 104, 10565-10570. [DOI] [PMC free article] [PubMed]
- Woods S., Coghlan A., Rivers D., Warnecke T., Jeffries S.J., Kwon T., Rogers A., Hurst L.D., Ahringer J. Duplication and retention biases of essential and non-essential genes revealed by systematic knockdown analyses. PLoS Genet. 2013;9:e1003330. doi: 10.1371/journal.pgen.1003330. [DOI] [PMC free article] [PubMed] [Google Scholar]; Woods, S., Coghlan, A., Rivers, D., Warnecke, T., Jeffries, S.J., Kwon, T., Rogers, A., Hurst, L.D., and Ahringer, J. (2013). Duplication and retention biases of essential and non-essential genes revealed by systematic knockdown analyses. PLoS Genet. 9, e1003330. [DOI] [PMC free article] [PubMed]
- Yang W., Dierking K., Schulenburg H. WormExp: a web-based application for a Caenorhabditis elegans-specific gene expression enrichment analysis. Bioinformatics. 2016;32:943–945. doi: 10.1093/bioinformatics/btv667. [DOI] [PubMed] [Google Scholar]; Yang, W., Dierking, K., and Schulenburg, H. (2016). WormExp: a web-based application for a Caenorhabditis elegans-specific gene expression enrichment analysis. Bioinformatics 32, 943-945. [DOI] [PubMed]
- Zhang C., Montgomery T.A., Gabel H.W., Fischer S.E., Phillips C.M., Fahlgren N., Sullivan C.M., Carrington J.C., Ruvkun G. mut-16 and other mutator class genes modulate 22G and 26G siRNA pathways in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2011;108:1201–1208. doi: 10.1073/pnas.1018695108. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhang, C., Montgomery, T.A., Gabel, H.W., Fischer, S.E., Phillips, C.M., Fahlgren, N., Sullivan, C.M., Carrington, J.C., and Ruvkun, G. (2011). mut-16 and other mutator class genes modulate 22G and 26G siRNA pathways in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 108, 1201-1208. [DOI] [PMC free article] [PubMed]
- Zhuang J.J., Hunter C.P. Tissue specificity of Caenorhabditis elegans enhanced RNA interference mutants. Genetics. 2011;188:235–237. doi: 10.1534/genetics.111.127209. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhuang, J.J., and Hunter, C.P. (2011). Tissue specificity of Caenorhabditis elegans enhanced RNA interference mutants. Genetics 188, 235-237. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RNA samples used in the present study.
DESeq2 fold-change results for the detection of differentially expressed STGs in Psng-1::rde-4 worms compared to rde-4(-) (Sheet #1), and in the Psng-1::rde-4 and Prgef-1::rde-4 High-Copy rescue strains versus rde-4(-) (Sheets #2 & #3).
NeuroSTGs (Sheet #1) and mRNAs (Sheet #2) counts detected in sorted neurons of N2 (wildtype) worms. DESeq2 fold-change results for the detection of differentially expressed NeuroSTGs (Sheet #3) and mRNAs (Sheet #4) in neurons of Psng-1::rde-4 worms compared to rde-4(-).
Transgenerational regulation of STGs in the germline of Psng-1::rde-4 neuronal rescue worms (Sheets #1-#6). Transgenerational regulation of STGs in High-Copy RDE-4 rescue worms (Sheets #7 & #8).
GermSTGs whose regulation by neuronal RDE-4 requires sid-1. See also Figure S4.
List of probes used for smFISH experiments for staining of the genes rde-4, yfp, saeg-2, C18D4.6 and C55C3.3.
Data Availability Statement
The accession number for the sequencingd data reported in this paper is GEO: GSE124049.