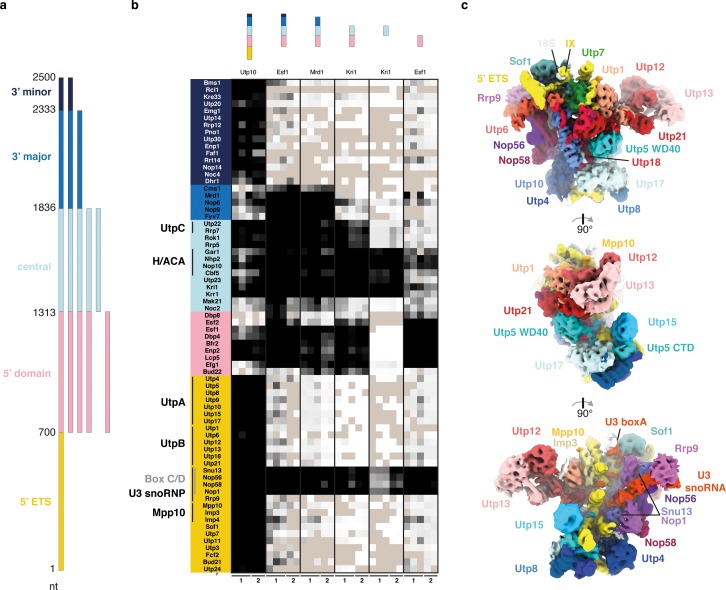
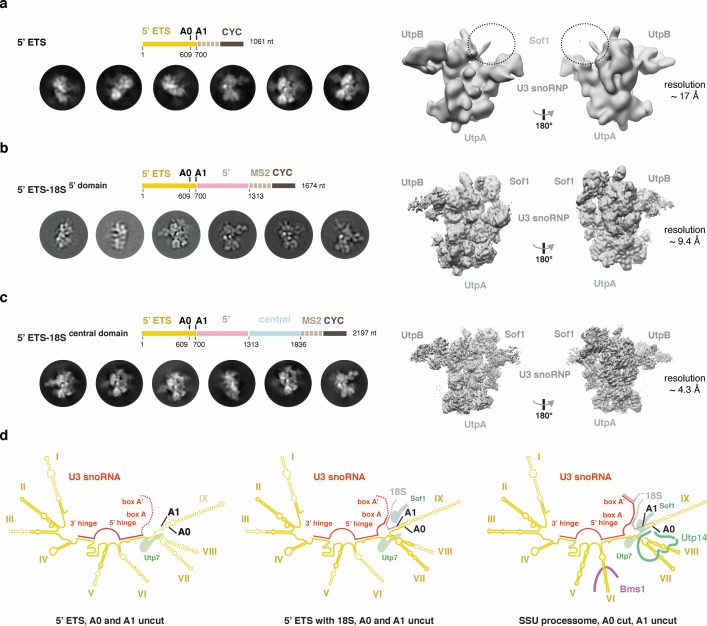
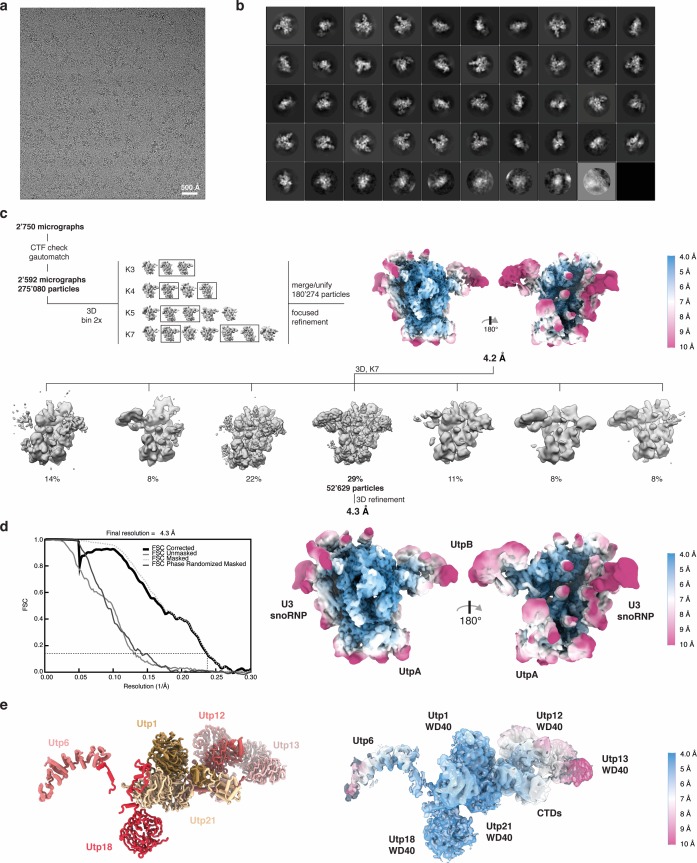
Figure 1. The 18S rRNA domains recruit assembly factors independent of the 5’ ETS.
(a) Schematics of rRNA mimics with color-coded rRNA domains. (b) Bait-normalized (MS2-protein) iBAQ based heat-map (proteins not detected in light brown, low abundance to high abundance in gradient from white to black) of ribosome biogenesis factors co-purified with pre-rRNA constructs shown in (a). Each biological replicate (n = 2) is labeled at the bottom and all technical replicates (n = 3, n = 2) are shown. (c) Three 90° related views of the cryo-EM structure of the 5’ ETS RNP lowpass-filtered to 5 Å with density colored according to subunits. Subunits of UtpA (shades of blue), UtpB (shades of red) and U3 snoRNP (shades of purple with U3 snoRNA in red) are shown.