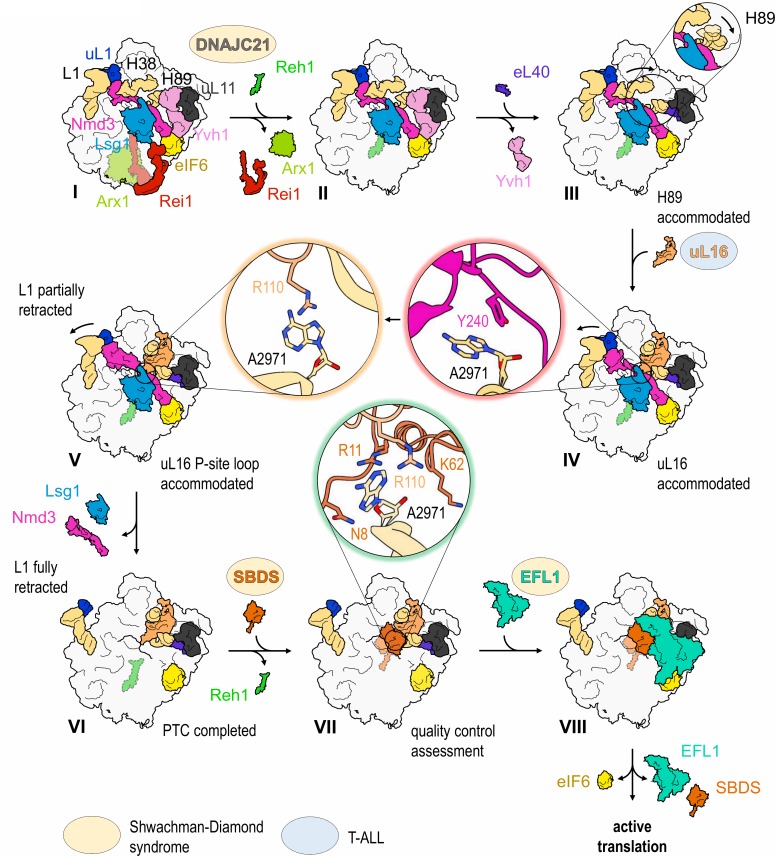
Figure 7. Mechanism of completion of PTC assembly and its quality control.
Insets show the sequential interactions of base A2971 with Nmd3, uL16 and SBDS during PTC completion. Models for 60S-bound EFL1 and SBDS are based on EMD-3145, EMD-3146 and EMD-3147 (Weis et al., 2015). Proteins targeted by mutations in Shwachman-Diamond syndrome (yellow) and paediatric T-ALL (blue) are highlighted.