Abstract
Objectives
Circular RNAs (circRNA) expression aberration has been identified in various human diseases. In this study, we investigated whether circRNAs could act as competing endogenous RNAs to regulate the pathological process of osteoarthritis (OA).
Methods
CircRNA deep sequencing was performed to the expression of circRNAs between OA and control cartilage tissues. The regulatory and functional role of CircSERPINE2 upregulation was examined in OA and was validated in vitro and in vivo, downstream target of CircSERPINE2 was explored. RNA pull down, a luciferase reporter assay, biotin-coupled microRNA capture and fluorescence in situ hybridisation were used to evaluate the interaction between CircSERPINE2 and miR-1271-5 p, as well as the target mRNA, E26 transformation-specific-related gene (ERG). The role and mechanism of CircSERPINE2 in OA was also explored in rabbit models.
Results
The decreased expression of CircSERPINE2 in the OA cartilage tissues was directly associated with excessive apoptosis and imbalance between anabolic and catabolic factors of extracellular matrix (ECM). Mechanistically, CircSERPINE2 acted as a sponge of miR-1271-5 p and functioned in human chondrocytes (HCs) through targeting miR-1271-5 p and ERG. Intra-articular injection of adeno-associated virus-CircSERPINE2-wt alleviated OA in the rabbit model.
Conclusions
Our results reveal an important role for a novel circRNA-CircSERPINE2 in OA progression. CircSERPINE2 overexpression could alleviate HCs apoptosis and promote anabolism of ECM through miR-1271-ERG pathway. It provides a potentially effective therapeutic strategy for OA progression.
Keywords: osteoarthritis, chondrocytes, knee osteoarthritis
Key messages.
What is already known about this subject?
Circular RNAs (CircRNAs) have important roles in many diseases, including osteoarthritis (OA).
Targeting dysfunctional circRNA-micro-RNA-mRNA interactions fulfil a critical therapeutic promise.
What does this study add?
A powerful protective circRNA-CircSERPINE2 and its ‘CircSERPINE2-miR-1271-5p-E26 transformation-specific-related gene (ERG)’ axis involved in OA pathogenesis.
How might this impact on clinical practice?
‘CircSERPINE2-miR-1271-5p-ERG’ axis could be a promising therapeutic target in the treatment of OA.
Introduction
Osteoarthritis (OA) is an age-related or post-traumatic degenerative joint disease that affects 12% of people over the age of 25 years and 50% of population over the age of 65 years1. OA is characterised with the progressive loss of hyaline articular cartilage, concomitant sclerotic changes in the subchondral bone and advancement in osteophytes.2 The identified risk factors include ageing, previous joint injury, obesity, genetics, sex and anatomical factors related to joint shape and alignment; however, the exact pathogenesis of OA remains undefined.3–6 Current therapy is limited and there is no effective treatment for OA. Therefore, elucidation of the mechanisms that regulate the expression of extracellular matrix (ECM) degradation-related genes is critical for the development of effective therapies for OA.
There is now overwhelming evidence that alterations in microRNA (miRNA) expression levels are linked to a variety of disease processes, including OA progression.7 8 For example, expression of miR-140 is reduced in OA tissue and miR-140 regulates ADAMTS-5 expression9 and MMP13,10 which are involved in OA pathology in cartilage. Importantly, in addition to miRNA, RNA sequencing (RNAseq) has identified multiple families of noncoding RNAs, including antisense RNAs, long intergenic noncoding RNAs and circular RNAs (circRNAs).11–13 Of note, circRNAs are characterised by high tissue-specific expression and stable structure, and exist ubiquitously in eukaryotic cells.14 Compared with linear RNAs, circRNAs can undergo noncanonical splicing without a free 3′ or 5′ end.15 Recent reports show that some circRNAs are enriched in miRNA-binding sites and can function as miRNA sponges or competing endogenous RNAs (ceRNAs) that naturally sequester and competitively inhibit miRNA activity.16 Of interest, some researches have indicated that circRNAs may also be central regulators of biological processes.17 Currently, little is known about the functional role of circRNAs in OA joint tissue. Their potential importance is indicated in a recent report by Liu et al,18 who identified 104 circRNAs that were differentially expressed in damage cartilage samples using a microarray-based approach. Although this preliminary study did not examine the mechanism and function of these circRNAs in depth, we hypothesise that circRNAs may be central regulators in cartilage tissue. Therefore, the aim of this study was to investigate the functional circRNAs in OA progress.
In the current study, we identified a circRNA (hsa_circ_0008365, termed CircSERPINE2) in OA, and systemically investigated its role in in vitro and animal models of OA.
Methods
Detailed experimental procedures are described in the online supplementary materials and methods and table S4.
annrheumdis-2018-214786supp001.docx (42.4KB, docx)
annrheumdis-2018-214786supp002.docx (23.8KB, docx)
Results
CircRNA expression patterns in human OA and control tissues
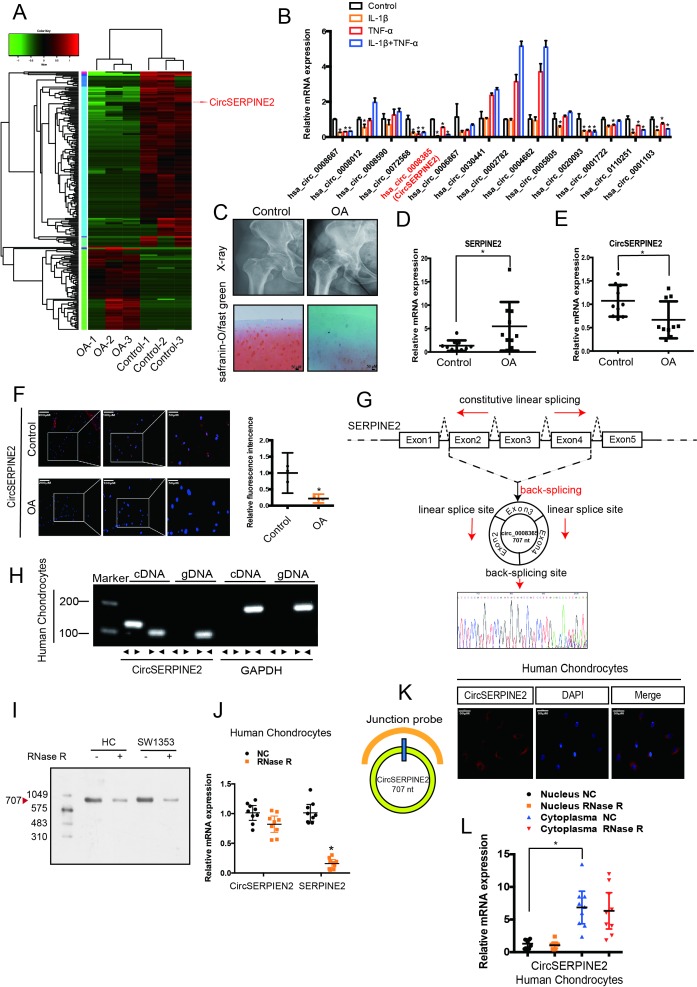
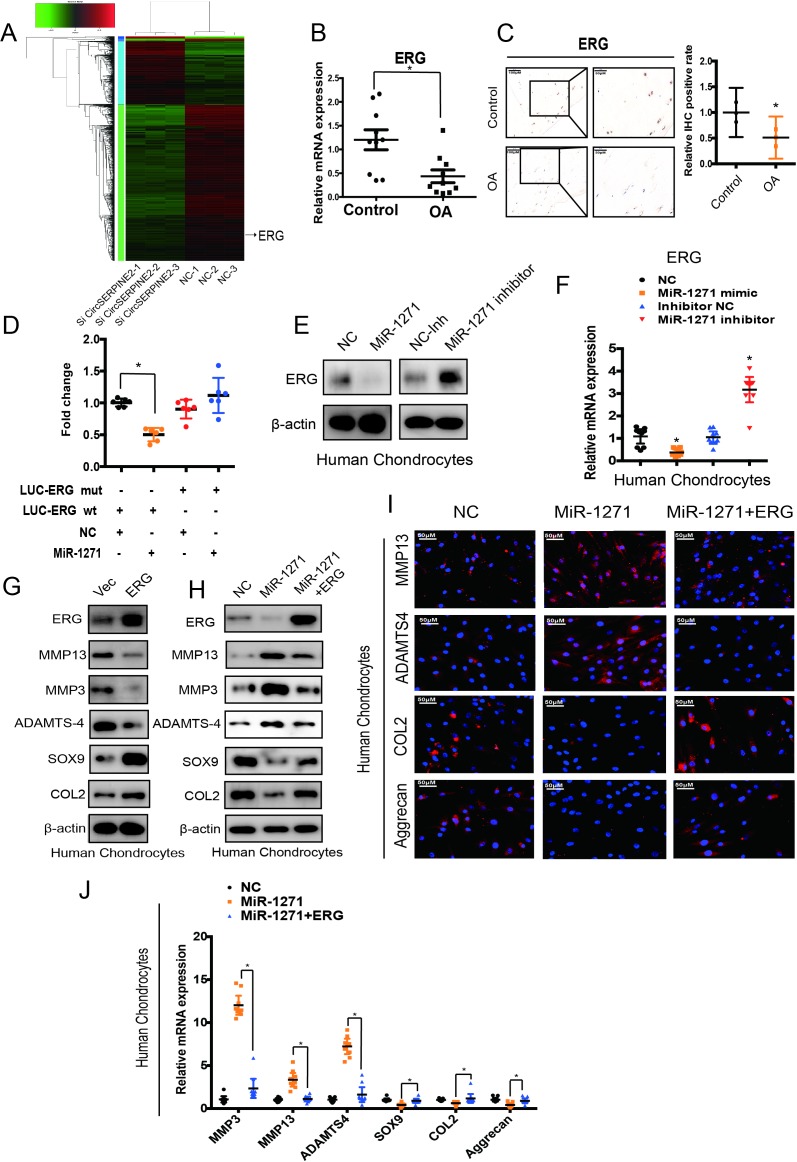
To generate a circRNA profiling database, we performed RNAseq analyses of ribosomal RNA-depleted total RNA from three clinical OA and three control tissues. Each sample was sequenced on an Illumina HiSeq yielding at least 30 million reads, which mapped to reference genome (hg38, human genome)by BWA19 and STAR20 separately. A total of 12 738 circRNAs were identified by both CIRI2 and CIRCexplorer, the majority of identified circRNAs in the study were supported by more than 10 reads (online supplementary figure S1A). We annotated these identified candidates using the RefSeq database,21 22 most of the circRNAs originated from protein-coding exons, and others aligned with 5′-UTR and 3′-UTR introns (online supplementary figure S1B). The majority of the identified exonic circRNAs were <1000 nucleotides (nt) in length (online supplementary figure S1C). The chromosome distribution of the identified circRNAs showed no obvious difference between OA and control groups (online supplementary figure S1D and E). Consistent with a previous report,23 analysis of the number of circRNAs in their host genes indicated that one gene could produce multiple circRNAs (online supplementary figure S1F). We normalised the back-spliced reads (support for circRNA) by read length and number of mapped reads (spliced reads per billion mapping,24 which permits quantitative comparisons between back splicing from different RNAseq data. There is often one predominantly expressed circRNA isoform from one gene locus (online supplementary figure S1G). OA and control groups showed differential expression pattern of circRNAs (figure 1A). To validate the RNA deep sequencing results, we performed real-time (RT) quantitative (qPCR) analysis to identify the 25 most differentially expressed circRNAs (online supplementary table S1) and confirmed 14 downregulated circRNAs (online supplementary figure S2A). In the subsequent step, we used interleukin (IL)-1β-treated and tumour necrosis factor (TNF)-α-treated primary human chondrocytes (HCs) and confirmed the downregulated expression of hsa_circ_00008667, hsa_circ_0072568, hsa_circ_0008365, hsa_circ_0020093, hsa_circ_0110251 and hsa_circ_0001103 in both IL-1β- and TNF-α-treated HCs (figure 1B & supplementary figure1B & S2B, *p<0.05). Moreover, western blotting (WB) for COL2A1, MMP3, MMP13 and SOX9 expression revealed the protective role of hsa_circ_0008365, which is formed by the circularisation of exons 2–4 of Serpine2 gene (hereafter referred to as CircSERPINE2) (online supplementary figure S2C).
Figure 1.
CircSERPINE2 validation and expression in OA cartilage tissue and cells. (A) Heat map of all differentially expressed circRNAs between OA and control cartilage samples. (B) The expression of circRNAs in the chondrocytes stimulated with TNF-α (10 ng/mL) and IL-1β (10 ng/mL) for 24 hours. n=3 (three different experiments). *p<0.05. (C) X-ray (upper) and safranin-O/fast green (lower) staining of the cartilage from OA or control patients. Scale bar: 50 µm. (D) SERPINE2 expression was higher in human OA cartilage than in control cartilage. n = 10. *p<0.05. (E) CircSERPINE2 expression was lower in human OA cartilage than in control cartilage tissue. n = 10. *p<0.05. (F) CircSERPINE2 expression was lower in human OA cartilage than in control cartilage tissue. Representative images are shown (scale bar, 50–200 µm). n=3 (three different donors). *p<0.05, mean with 95% CI. (G) Schematic illustration showing SERPINE2 exons 2–4 circularisation to form CircSERPINE2 (black arrow). The presence of CircSERPINE2 was validated by RT-PCR, followed by Sanger sequencing. Red arrow represents head-to-tail CircSERPINE2 splicing sites. (H) The presence of CircSERPINE2 was validated in HCs by RT-PCR. Divergent primers amplified CircSERPINE2 from cDNA, but not from genomic DNA. GAPDH was used as a negative control. (I) Northern blot analysis for the detection of CircSERPINE2 expression in HC and SW1353 cells treated with or without RNase R digestion. The panels show the blots probed with CircSERPINE2, and the red triangle indicates CircSERPINE2 band size (707 nt). (J) The expression of CircSERPINE2 and SERPINE2 mRNA in HC cells treated with or without RNase R was detected by RT-qPCR. The relative levels of CircSERPINE2 and SERPINE2 mRNA were normalised to the values measured for mock treatment. n=9 (three different donors for three different experiments). *p<0.05. (K) RNA FISH revealed the predominant localisation of CircSERPINE2 in the cytoplasm. CircSERPINE2 probes were labelled with Cy-3. Nuclei were stained with DAPI. Scale bar, 50 µm. Upper panel: FISH with junction-specific probes indicative of the cellular localisation of CircSERPINE2. n = 3 (three different donors). Scale bars=50 µM. Lower panel: CircSERPINE2 expression was detected in different cell fractions. Nuclear and cytoplasmic RNAs were extracted, and junction primers were used for CircSERPINE2 detection. U6 was used as an internal control for nuclear RNA, whereas GAPDH served as the control for cytoplasmic RNA. n=9 (three different donors for three different experiments). *p<0.05. CircRNAs, circular RNAs; DAPI, 4′,6-diamidino-2-phenylindole; FISH, fluorescence in situ hybridisation; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HCs, human chondrocytes; IL, interleukin; OA, osteoarthritis; RT-qPCR, real-time quantitative PCR; TNF, tumour necrosis factor.
annrheumdis-2018-214786supp003.jpg (2.7MB, jpg)
annrheumdis-2018-214786supp004.docx (27.6KB, docx)
annrheumdis-2018-214786supp005.jpg (4.1MB, jpg)
CircSERPINE2 expression is relatively low in OA tissues and cell lines and is predominantly localised in the cytoplasm
We detected the expression of CircSERPINE2 in 10 human OA cartilage and 10 control cartilage (figure 1C & online supplementary figure1C & S2D-E). RT-qPCR and RNA fluorescence in situ hybridisation (FISH) showed that unlike the upregulated SERPINE2 gene expression (figure 1D, differentials of means=4.135; 95% CI 0.412 to 7.857; *p<0.05), CircSERPINE2 expression was downregulated in OA samples (figure 1E–F, differentials of means=0.407, 95% CI 0.043 to 0.772, *p<0.05). We used Sanger sequencing to confirm the head-to-tail splicing (a special splice reaction formed by a 5′-end splice site and the corresponding site at 3′-end of an exon) in the RT-qPCR product of CircSERPINE2 identified by its expected size (figure 1G). However, head-to-tail splicing may be produced by trans-splicing (a phenomenon naturally existing in different organisms, wherein the 5′-UTR of pre-mRNA is trimmed by a spliced leader RNA, followed by the attachment of a short sequence to the 5′-terminus) or genomic rearrangement. Therefore, we took several steps to rule out these possibilities as per the previously described methodology.25 26 We first designed convergent primers to amplify SERPINE2 mRNA and divergent primers to amplify CircSERPINE2 using cDNA and genomic DNA (gDNA). CircSERPINE2 was amplified by divergent primers in cDNA but not in gDNA (figure 1H). In the second step, we performed northern blot analysis, as shown in figure 1I, a single band at expected size (707 nt) was detected by CircSERPINE2-specific probe targeting the junction region in cells with or without RNase R digestion. We confirmed that CircSERPINE2 was resistant to RNase R, whereas SERPINE2 mRNA level was significantly decreased after RNase R treatment (figure 1J). FISH and RT-qPCR analysis indicated the abundant expression of cytoplasmic CircSERPINE2 in HCs (figure 1K,L).
CircSERPINE2 regulates apoptosis and ECM metabolism in chondrocytes
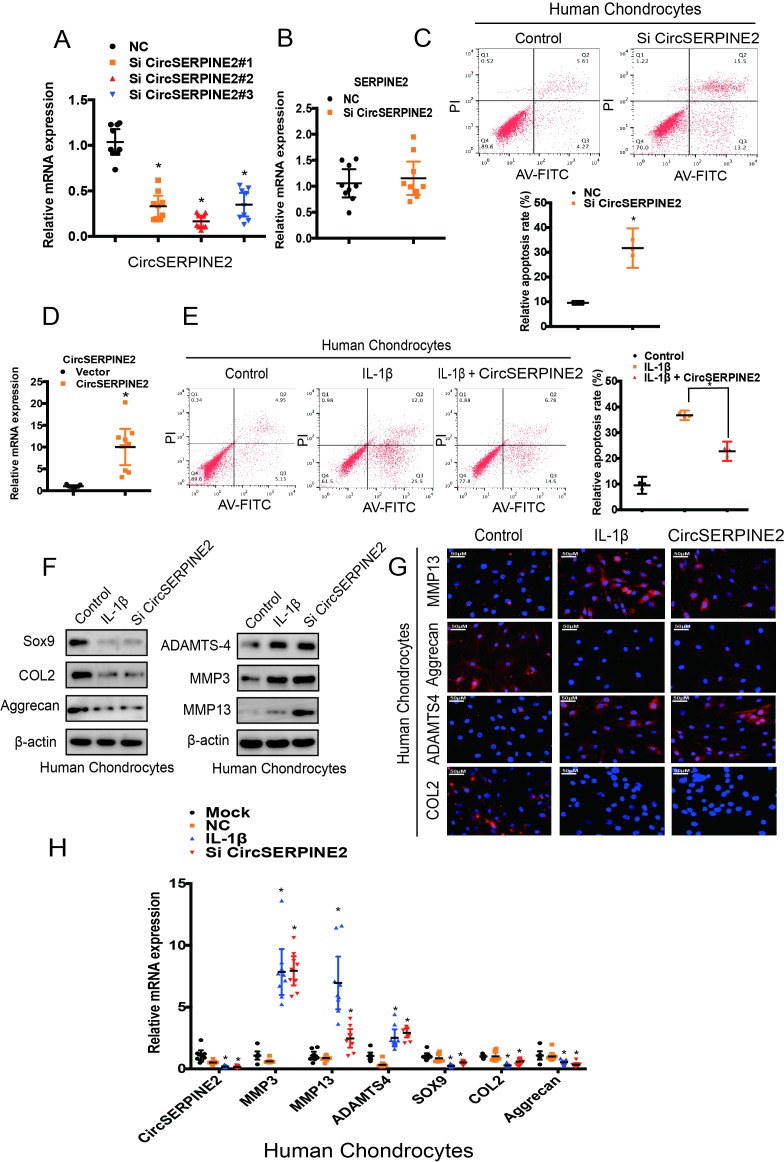
To assess the involvement of CircSERPINE2 in the regulation of matrix-degrading enzymes, we transfected HCs with CircSERPINE2 small-interfering RNA (figure 2A). Knockdown of CircSERPINE2 expression had no effect on SERPINE2 mRNA level (figure 2B). We examined the effects of CircSERPINE2 inhibition or overexpression on IL-1β-induced HC apoptosis. As shown in figure 2C, the knockdown of CircSERPINE2 expression increased the rate of chondrocyte apoptosis while the increase in the apoptotic rate after IL-1β treatment markedly suppressed in response to CircSERPINE2 overexpression (figure 2D,E). In addition, the inhibition of CircSERPINE2 expression promoted the expression of MMP3, MMP13 and ADAMTS4, and decreased the levels of SOX9, COL2A1 and aggrecan, as observed in WB (figure 2F). The knockdown of CircSERPINE2 expression obviously affected MMP13, ADAMTS4, COL2A1 and aggrecan levels in HCs and SW1353 cells, as evident from immunofluorescence (IF) assay (figure 2G & supplementary figure 3A) and RT-qPCR results (figure 2H & supplementary figure 3B). These data clearly indicate the proapoptotic and procatabolic effects of CircSERPINE2 knockdown in HCs and SW1353 cells.
Figure 2.
CircSERPINE2 expression knockdown induced the expression of matrix-degrading enzymes in chondrocytes. (A) HCs were transfected with CircSERPINE2 siRNA or negative control siRNA at a final concentration of 20 nM. After 48 hours of transfection, the expression level of CircSERPINE2 was measured by RT-qPCR and normalised to β-actin level. n=9 (three different donors for three different experiments). *p<0.05. (B) HCs were transfected with CircSERPINE2#2 siRNA or negative control siRNA at a final concentration of 20 nM. After 48 hours of transfection, the expression level of SERPINE2 was measured by RT-qPCR and normalised to β-actin level. n=9 (three different donors for three different experiments). *p<0.05. (C) HCs were transfected with CircSERPINE2#2 siRNA or negative control siRNA at a final concentration of 20 nM. After 48 hours, cell apoptosis was detected with annexin V-FITC/PI double staining using quantitative FACS analysis. n=3 (three different donors). *p<0.05. (D) Cells were infected with CircSERPINE2 lentivirus or control virus. Overexpression was detected by RT-qPCR. n=9 (three different donors for three different experiments). *p<0.05. (E) HCs were infected with CircSERPINE2 lentivirus or control virus and stimulated with IL-1β for 48 hours. Cell apoptosis was detected with annexin V-FITC/PI double staining using quantitative FACS analysis. n=3 (three different donors). *p<0.05. (F–H) HCs were transfected with CircSERPINE2#2 siRNA or negative control siRNA at a final concentration of 20 nM, followed by the treatment with or without IL-1β. After 48 hours, WB for (F, n=3) MMP13, MMP3, aggrecan, SOX9, COL2A1 and ADAMTS4 detection; IF (G, scale bar: 50 µm) for MMP13, aggrecan, COL2A1 and ADAMTS4 expression detection; and RT-qPCR (H, n=9; three different donors for three different experiments; *p<0.05) for MMP3, MMP13, ADAMTS4, COL2A1, SOX9 and aggrecan expression evaluation were performed. FACS, fluorescence-activated cell sorting; FITC, fluorescein isothiocyanate; HCs, human chondrocytes; IF, immunofluorescence; IL; interleukin; PI, propidium iodide; RT-qPCR, real-time quantitative PCR; siRNA, small interfering RNA; WB, western blotting.
CircSERPINE2 serves as sponge for miRNAs
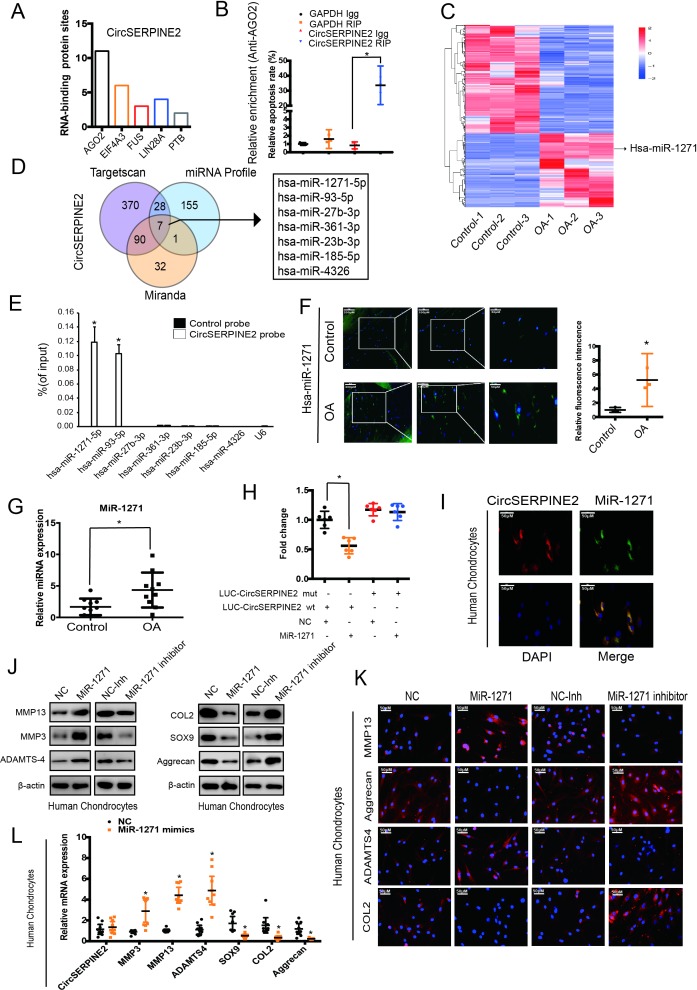
As CircSERPINE2 level is abundant and stable in the cytoplasm, we investigated the ability of CircSERPINE2 to bind to miRNAs. A search in the Circular RNA Interactome database revealed that CircSERPINE2 has 11 conserved Argonaute 2 (AGO2)-binding sites (figure 3A). We demonstrated the specific enrichment of endogenous CircSERPINE2 pulled down from Flag-AGO2 cells by RNA immunoprecipitation (RIP) analysis (figure 3B). The differential expression of miRNAs in control and OA tissues was investigated using deep sequencing. In all, 191 miRNAs showed upregulated expression in OA tissues as compared with control tissues (figure 3C, supplementary figure 3S 3C, table S2, and online supplementary figure S4A-C). We identified seven candidate miRNAs by overlapping the predicted miRNA recognition elements (MREs) in CircSERPINE2 sequence using miRanda, TargetScan and miRNA sequence (figure 3D). The levels of the seven candidate miRNAs were determined using a pull-down assay. As shown in figure 3E and supplementary figure S4D and E, miR-1271 and miR-93 were pulled down by CircSERPINE2. Furthermore, WB results demonstrated the association of miR-1271 with the protein expression of matrix-degrading and synthesising components (supplementary figure S4F). Figure 3F,G shows the upregulated expression of miR-1271 in the 10 human OA cartilage samples (differentials of means=2.728; 95% CI 0.623 to 4.832, *p<0.05), consistent with the results of deep sequencing (figure 3C). Therefore, miR-1271 was selected for further analysis. Luciferase assay confirmed the binding of CircSERPINE2 to miR-1271 (figure 3H & supplementary figure3H & S4G). FISH experiment further confirmed the co-localisation of CircSERPINE2 and miR-1271 (figure 3I). The effects of miR-1271 were investigated. miR-1271-overexpressing cells demonstrated the increase in the percentage of apoptotic cells (supplementary figure S4H-I), enhanced expression of MMP3, MMP13 and ADAMTS4, and decreased levels of SOX9, COL2A1 and aggrecan, as determined by WB (figure 3J), IF (figure 3K & supplementary figure3K & S5A-B) and RT-qPCR (figure 3L & supplementary figure3L & S5C). On the contrary, miR-1271 expression inhibition exerted opposite effects on matrix-degrading and synthesising components (figure 3J–L & supplementary figure S5A-C).
Figure 3.
CircSERPINE2 serves as a sponge for miR-1271 in HC cells. (A) Number of RBP sites between RBPs and CircSERPINE2 is shown. (B) AGO2 RIP assay was performed to detect CircSERPINE2 levels in SW1353 cells stably expressing AGO2. n=3. *p<0.05. (C) miRNA-mediated differential dysregulation of miRNAs was detected by miRNA high-throughput sequencing in OA cartilage samples and control cartilage samples (n=3). (D) Schematic illustration to show the overlapping of the target miRNAs of CircSERPINE2, as predicted by Miranda, TargetScan and miRNAs upregulated in OA identified by sequencing analysis. (E) Lysates prepared from SW1353 cells were subjected to an RNA pull-down assay and tested with RT-qPCR. Relative levels of CircSERPINE2 were normalised to the levels of input. n = 3. *p<0.05 versus control (lac Z) probe. (F–G) miR-1271 expression was higher in human OA cartilage than in control cartilage tissue, as determined by FISH (F, n=3; three different donors; *p<0.05; mean with 95% CI) and RT-qPCR (G, n=10, *p<0.05). Representative images are shown (scale bar: 50–200 µm). *p<0.05. (H) HEK-293T cells were co-transfected with miR-1271 mimic or NC and a luciferase reporter construct containing WT or MUT CircSERPINE2. n = 6. *p<0.05. Mean with 95% CI. (I) Fish images showing the co-localisation of CircSERPINE2 and miR-1271 in HCs. miR-1271 probes were labelled with Alexa fluor 488, whereas CircSERPINE2 probes were tagged with Cy3. Nuclei were stained with DAPI. n = 3 (three different donors). Scale bar, 50 µm. (J–L) HCs were transfected with miR-1271 mimic, inhibitor or NC at a final concentration of 20 nM. After 48 hours of transfection (J, n = 3; three different donors), protein levels of MMP3, MMP13, ADAMTS4, SOX9, COL2A1 and aggrecan in chondrocytes were detected; IF assay for MMP13, aggrecan, COL2A1 and ADAMTS4 expression detection in chondrocytes was performed (K, n = 3; three different donors; scale bar: 50 µm). mRNA levels of MMP3, MMP13, ADAMTS4, SOX9, COL2A1 and aggrecan in chondrocytes were evaluated (l, n=9; three different donors for three different experiments, *p<0.05). AGO2, Argonaute 2; DAPI, 4′,6-diamidino-2-phenylindole; FISH, fluorescence in situ hybridisation; HC, human chondrocyte; IF, immunofluorescence; miRNAs, microRNAs; MUT, mutant; NC, negative control; OA, osteoarthritis; RBP, RNA-binding protein; RIP, RNA immunoprecipitation; RT-qPCR; real-time quantitative PCR; WT, wild type.
annrheumdis-2018-214786supp008.jpg (2.9MB, jpg)
annrheumdis-2018-214786supp009.jpg (3.9MB, jpg)
CircSERPINE2 functions in OA by targeting miR-1271 expression
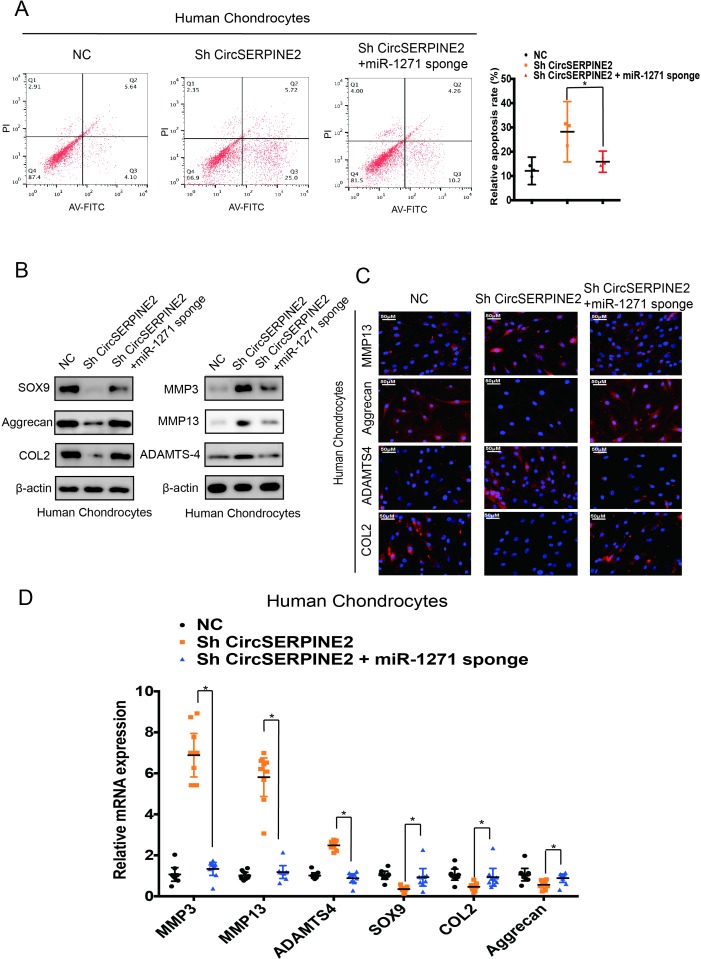
To examine whether CircSERPINE2 functions in OA via interaction with miR-1271, we co-infected cells with sh-CircSERPINE2 and miR-1271 sponge adenovirus. The miR-1271 sponge adenovirus contains repeated complementary-binding sites to miR-1271 and has continuous miR-1271 loss of function in cells. As shown in figure 4A, the knockdown of CircSERPINE2 expression increased the rate of HC apoptosis while this rate was markedly rescued by the inhibition of the expression of both miR-1271 and CircSERPINE2. The results showed that protein and mRNA levels of MMP13, MMP3 and ADAMTS4 significantly decreased while those of SOX9, COL2A1 and aggrecan increased in the cells co-infected with sh-CircSERPINE2 and miR-1271 sponge adenovirus as compared with the cells infected with sh-CircSERPINE2 alone (figure 4B,D & online supplementary figure S6A-C). IF analysis confirmed that the expression of COL2A1 and aggrecan increased and MMP13 and ADAMTS4 levels decreased in the cells with CircSERPINE2 and miR-1271 expression inhibitors as compared with the cells treated with CircSERPINE2 inhibitor (figure 4C & supplementary figure4C & S6D). Taken together, these results suggest that CircSERPINE2 functions by targeting miR-1271 in vitro.
Figure 4.
Knockdown of miR-1271 expression reverses the CircSERPINE2-induced cartilage degradation in HC cells. (A) Downregulation of both CircSERPINE2 and miR-1271 resulted in fewer apoptotic HCs than those observed with the inhibition of CircSERPINE2 alone. Apoptosis rates were determined with annexin V-FITC/PI staining and FACS. n = 3 (three different donors). *p<0.05. (B) The expression of MMP3, MMP13, ADAMTS4, SOX9, aggrecan and COL2A1 in HC cells was detected by WB. Cells were co-infected with sh-CircSERPINE2 and miR-1271 sponge adenovirus or control virus. n = 3 (three different donors). (C) The expression of MMP13, ADAMTS4, aggrecan and COL2A1 in HC cells was detected by IF analysis. Cells were infected with control virus or sh-CircSERPINE2 with or without miR-1271 sponge adenovirus. n = 3 (three different donors). Scale bars=50 µm. (D) mRNA expression of CircSERPINE2, MMP3, MMP13, ADAMTS4, SOX9, aggrecan and COL2A1 in HC cells was detected by RT-qPCR analysis. Cells were infected with control virus or sh-CircSERPINE2 with or without miR-1271 sponge adenovirus. n=9 (three different donors for three different experiments). *p<0.05, cells co-infected with miR-1271 sponge and sh-CircSERPINE2 adenovirus versus cells infected with sh-CircSERPINE2 adenovirus only. FACS, fluorescence-activated cell sorting; FITC, fluorescein isothiocyanate; HCs, human chondrocytes; IF, immunofluorescence; PI, propidium iodide; RT-qPCR, real-time quantitative PCR; WB, western blotting.
annrheumdis-2018-214786supp010.jpg (3.8MB, jpg)
miR-1271 directly targets ETS-related gene
We investigated the possible targets of miR-1271 and CircSERPINE2 by overlapping the predicted results of TargetScan, miRDB and RNA-seq (figure 5A, online supplementary figure S7A and B, and supplementary table S3). We first selected 10 significantly differentially expressed genes; RT-qPCR results showed that eight genes were regulated by CircSERPINE2 (supplementary figure S7C). Moreover, E26 transformation-specific (ETS)-related gene (ERG) had the most obvious effect on COL2A1, aggrecan, MMP3 and MMP13 expression (online supplementary figure S7D), as indicated in recent reports.27 28 ERG was therefore selected for further investigation and was found to be markedly downregulated in OA-affected, damaged regions of human cartilage (figure 5B,C, differentials of means=0.766, 95% CI 0.213 to 1.319, *p<0.05), suggestive of the potential role of ERG in OA pathogenesis. Furthermore, bioinformatic analysis using miRNA target prediction software revealed that ERG is a putative target of miR-1271 (online supplementary figure S7E). Luciferase assay results showed that miR-1271 mimic significantly inhibited the luciferase activity in the cells transfected with WT 3′-UTR of ERG, but not in those transfected with mutant (MUT) 3′-UTR of ERG (figure 5D). Furthermore, figure 5E,F demonstrated that miR-1271 mimic reduced the expression of ERG protein and mRNA while miR-1271 inhibitor exerted opposite effects. In addition, ERG overexpression decreased the expression of MMP3, MMP13 and ADAMTS4 and increased the levels of SOX9 and COL2A1 (figure 5G). These results suggest that miR-1271 targets ERG, and endogenous ERG expression is closely related to OA pathogenesis.
Figure 5.
ERG serves as a direct target of miR-1271 and mediates the function of miR-1271 in HC cells. (A) The differential dysregulation of mRNAs by CircSERPINE2 knockdown was detected by RNA-seq in HCs (n=3). (B) ERG expression levels were lower in human OA cartilage than in control tissue. n = 10. *p<0.05. (C) ERG expression levels were lower in human OA cartilage than in control tissue. Representative images are shown. n=3; *p<0.05; mean with 95% CI. (D) HEK-293T cells were co-transfected with miR-1271 mimics or NC and luciferase reporter constructs containing WT or MUT 3′-UTR of ERG. n = 6; *p<0.05; mean with 95% CI. (E–F) miR-1271 overexpression reduced ERG (E) protein and (F) mRNA levels while miR-1271 inhibition increased ERG (E) protein and (F) mRNA levels. Cells were transfected with NC or miR-1271 mimic/inhibitor, and mRNA or protein levels were evaluated. Protein expression was evaluated by WB; mRNA levels were evaluated by RT-qPCR. n=9 (three different donors for three different experiments). *p<0.05. (G–H) The expression of ERG, MMP3, MMP13, ADAMTS4, SOX9 and COL2A1 in HC cells was detected by WB. Cells were infected with ERG adenovirus (G) co-infected with pre-miR-1271 (H) or control virus. (I) Expression of MMP13, ADAMTS4, aggrecan and COL2A1 in HC cells was detected by IF. Cells were co-infected with ERG and pre-miR-1271 adenovirus or control virus. n = 3 (three different donors). Scale bars=50 µm. (J) mRNA expression of MMP3, MMP13, ADAMTS4, SOX9, aggrecan and COL2A1 in HC cells was detected by RT-qPCR analysis. Cells were co-infected with ERG and pre-miR-1271 adenovirus or control virus. n=9 (three different donors for three different experiments). *p<0.05, cells co-infected with ERG and pre-miR-1271 adenovirus versus cells infected with pre-miR-1271 adenovirus. ERG, E26 transformation-specific (ETS)-related gene; HC, human chondrocyte; MUT, mutant; NC, negative control; OA, osteoarthritis; RT-qPCR; real-time quantitative PCR; WB, western blotting; WT, wild type.
annrheumdis-2018-214786supp011.jpg (2.4MB, jpg)
annrheumdis-2018-214786supp012.docx (18.1KB, docx)
We designed a rescue experiment to verify whether the effects of miR-1271 expression on chondrogenic phenotypes were achieved through ERG. Figure 5H and online supplementary figure S7F and G showed that the infection of pre-miR-1271 adenovirus resulted in the inhibition of ERG expression. Furthermore, as shown in figure 5H–J, the infection of pre-miR-1271 adenovirus remarkably decreased the expression levels of COL2A1, SOX9 and aggrecan and increased MMP3, MMP13 and ADAMTS4 levels in HCs as compared with the negative control cells. However, these effects were abolished after ERG overexpression. Taken together, our results indicate that miR-1271 overexpression may impair chondrogenic phenotypes via ERG.
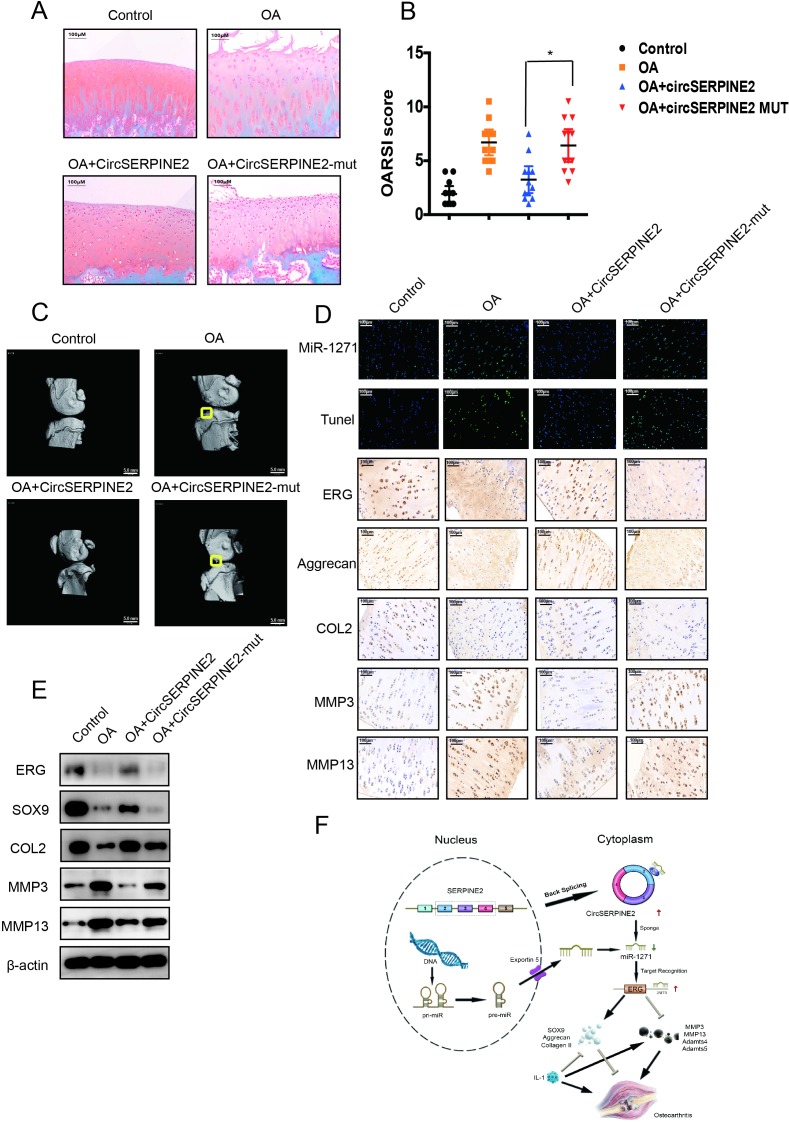
Injection of CircSERPINE2 alleviates OA in a rabbit model
To determine whether CircSERPINE2 plays a role in OA progression in vivo, WT or MUT adeno-associated virus (AAV) CircSERPINE2 was intra-articularly administered to anterior cruciate ligament transection (ACLT)-induced OA rabbits. Overexpression efficiency was confirmed by FISH and RT-qPCR (online supplementary figure S8A and B). Safranin O and fast green staining (figure 6A) showed cartilage surfaces in ACLT-induced OA rabbits improved after the injection of WT AAV CircSERPINE2 but not MUT AAV CircSERPINE2. Quantitative analysis with Osteoarthritis Research Society International (OARSI) scoring showed that WT AAV CircSERPINE2 significantly lowered OARSI scores, whereas MUT AAV CircSERPINE2 treatment increased OARSI scores (figure 6B). Three-dimensional reconstruction of micro-CT images demonstrated the increase in osteophytes in OA rabbits, whereas CircSERPINE2 intra-articular injection decreased osteophytes (figure 6C). The injection of WT AAV CircSERPINE2 alleviated the degenerative changes in the cartilage matrix, such as enhanced apoptotic (TUNEL) and catabolic response and increased ECM composition in the rabbit model of OA, as indicated by immunohistochemistry and WB results (figure 6D–E). Taken together, these results revealed the positive effects of elevated CircSERPINE2 on the inhibition of ECM catabolism to promote anabolism and alleviate OA in vivo (figure 6F).
Figure 6.
CircSERPINE2 alleviates OA in vivo. (A) Safranin-O/fast green staining of the cartilage in the indicated groups at 8 weeks after surgery. Scale bar=200 µm. ACLT-induced OA rabbits were injected with AAV negative control, WT AAV CircSERPINE2, or MUT AAV CircSERPINE2, and the degree of knee OA was evaluated by safranin-O/fast green staining (n=12). (B) OARSI scoring was performed according to staining results; *p<0.05. (C) Micro-CT images of OA rabbits. (D) FISH and IHC staining of ACLT-induced OA rabbits injected with AAV negative control, WT AAV CircSERPINE2 or MUT AAV CircSERPINE2. (E) WB for the expression of ERG, SOX9, catabolic enzymes (MMP-3 and MMP-13), and ECM composition (COL2A1) in rabbit cartilage tissues. the injection of WT AAV CircSERPINE2 alleviated the progression of OA and induced the expression of Col2a1 in the rabbit model of OA. Scale bar=100 µm. (F) Schematic of the working hypothesis. AAV, adeno-associated virus; ACLT, anterior cruciate ligament transection; ECM, extracellular matrix; ERG, E26 transformation-specific-related gene; FISH, fluorescence in situ hybridisation; IHC, immunohistochemistry; MUT mutant; OA, osteoarthritis; OARSI, Osteoarthritis Research Society International; TUNEL, terminal dexynucleotidyl transferase(TdT)-mediated dUTP nick end labeling; WB, western blotting; WT, wild type.
annrheumdis-2018-214786supp013.jpg (1.5MB, jpg)
Discussion
At present, no effective therapeutic drugs have been approved for the clinical treatment of OA. To develop effective therapeutics, it is important to investigate the underlying mechanisms of OA.29 30 CircRNAs are endogenous noncoding RNAs that are widespread, tissue specific and conserved in mammalian cells and have a great potential that remains to be explored. Evidences have shown that certain circRNAs are related to the occurrence and progression of various diseases. However, little is known about their functions in the development and progression of OA and their roles in the regulation of the vitality and functions of HCs. Therefore, evaluation of the cellular mechanisms of circRNAs in OA may provide a new perspective for the treatment of OA. In the present study, we first identified CircSERPINE2 as a key downregulated circRNA involved in OA. In addition, we used gain-of-function and loss-of-function approaches in vitro to demonstrate the participation of CircSERPINE2 in the proapoptotic response and protection of ECM components from degradation. We revealed a new function of CircSERPINE2 in OA progression and identified it as a promising therapeutic target for OA treatment.
CircSERPINE2 is generated by the back splicing of the exons 2 to 4 of SERPINE2 gene. It mainly comprises the protein-coding sequence of SERPINE2 mRNA; hence, its sequence is relatively conserved between human and rabbit. Moreover, the ERG-binding site of miR-1271 is highly conserved in rabbit, justifying the use of a rabbit animal model. SERPINE2 is a subtype of the serpin superfamily and plays a role in the modification of matrix metalloproteinase activity in ECM homeostasis.31–33 In our study, SERPINE2 expression was upregulated and CircSERPINE2 levels were downregulated in patients with OA. However, both molecules played a protective role in OA progression. In addition, CircSERPINE2 knockdown or overexpression had no effect on SERPINE2 expression, indicative of the independent role of CircSERPINE2 in OA progression.
Accumulating evidences have indicated the key mechanistic role of ceRNA networks in many diseases, including OA.34 Various types of RNAs, including circRNAs, pseudogenes, long noncoding RNAs and mRNAs, may function as ceRNAs in distinct physiological and pathophysiological conditions. Computational models and analyses of clinical microarray data to explore ceRNA regulation network showed that ceRNA essentially operates in an miRNA-target titration mechanism,35 36 and the abundance of competing RNAs and miRNAs, the binding energy of ceRNAs and miRNAs, and the number of MREs are the key determinants of the range and strength of ceRNA regulation.37 38 Many validated ceRNA networks participate in the initiation and progression of various diseases.39 However, the overall pathophysiological contributions of circRNAs to OA remain largely unknown. In the present study, CircSERPINE2 contains a conserved miR-1271 target site that was validated by pull-down, RIP, luciferase and FISH analyses. Furthermore, the expression of ERG (miR-1271 target) was positively regulated by CircSERPINE2. Therefore, we propose a mechanism wherein CircSERPINE2 acts as an miR-1271 sponge to inhibit HC apoptosis, promote ECM anabolism and suppress ECM catabolism, thereby delaying the progression of OA.
Several miRNAs are known to exhibit aberrant expression levels in OA.40 41 With increasing evidences highlighting the link between miRNAs and OA pathogenesis, miRNAs have been recognised as the potential therapeutic targets for OA treatment. In the present study, we found that miR-1271 expression was significantly upregulated in OA and inversely related to the expression of SOX9, aggrecan and COL2A1. Subsequent analyses showed that miR-1271 suppressed ERG expression, a mechanism reported herein for the first time. ERG was recently shown to play an important role in inducing the permanent differentiation of chondrocytes into joint-forming cells.42 It has been proposed that the lack of ERG expression may render joints more susceptible to surgical challenge. In addition, the beneficial roles of ERG in OA have been verified in mice.43 The current study also shows a distinctive decrease in ERG expression levels in the articular cartilage of patients with OA. Moreover, ERG overexpression induced by miR-1271 knockdown was found to play a protective role in OA. Therefore, we propose the targeting of the CircSERPINE2/miR-1271/ERG axis as a preventative strategy for OA treatment. However, we do not exclude the possibility of the involvement of other critical gene targets of CircSERPINE2 besides ERG that may play important roles in the progression of OA.
In conclusion, we have identified the CircSERPINE2-miR-1271-ERG axis as a novel target for the prevention and treatment of OA. CircSERPINE2 and ERG overexpression may alleviate the catabolism of ECM. Besides, miR-1271 inhibition may serve as an effective therapeutic strategy for OA treatment.
annrheumdis-2018-214786supp006.jpg (3.2MB, jpg)
annrheumdis-2018-214786supp007.docx (30.9KB, docx)
annrheumdis-2018-214786supp014.pdf (14.1MB, pdf)
annrheumdis-2018-214786supp015.docx (19.4KB, docx)
Footnotes
SS, YW and JC contributed equally.
Handling editor: Josef S Smolen
Contributors: SF, YM and SS designed the experiments. SS, YM, YW and JC performed the experiments and acquired the data. SS, ZX, KH, GW, YY, WN, ZC and PS analysed the data. SF, YM and SS supervised the project and wrote the manuscript.
Funding: The study was sponsored by National Natural Science Foundation of China (81802680, 81871797, 81472064, 81873985, 81874015 and 81601925), Key Project of Zhejiang Medical Science and Technology Plan (2015145597 and 2016145597), Key Project of Zhejiang Provincial Natural Science Foundation (LZ15H06002), Medical Science and Technology Project of Zhejiang Province (2017179447). No benefits in any form have been or will be received from a commercial party related directly or indirectly to the subject of this study.
Competing interests: None declared.
Patient consent for publication: Obtained.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Hayami T, Pickarski M, Zhuo Y, et al. Characterization of articular cartilage and subchondral bone changes in the rat anterior cruciate ligament transection and meniscectomized models of osteoarthritis. Bone 2006;38:234–43. 10.1016/j.bone.2005.08.007 [DOI] [PubMed] [Google Scholar]
- 2. Dieppe P, Kirwan J. The localization of osteoarthritis. Br J Rheumatol 1994;33:201–3. 10.1093/rheumatology/33.3.201 [DOI] [PubMed] [Google Scholar]
- 3. Goldring MB. The role of the chondrocyte in osteoarthritis. Arthritis Rheum 2000;43:1916–26. [DOI] [PubMed] [Google Scholar]
- 4. Goldring MB. Update on the biology of the chondrocyte and new approaches to treating cartilage diseases. Best Pract Res Clin Rheumatol 2006;20:1003–25. 10.1016/j.berh.2006.06.003 [DOI] [PubMed] [Google Scholar]
- 5. Conaghan PG, Kloppenburg M, Schett G, et al. Osteoarthritis research priorities: a report from a EULAR ad hoc expert Committee. Ann Rheum Dis 2014;73:1442–5. 10.1136/annrheumdis-2013-204660 [DOI] [PubMed] [Google Scholar]
- 6. Lotz M. Cytokines in cartilage injury and repair. Clin Orthop Relat Res 2001;391(391 Suppl):S108–15. 10.1097/00003086-200110001-00011 [DOI] [PubMed] [Google Scholar]
- 7. Nugent M. MicroRNAs: exploring new horizons in osteoarthritis. Osteoarthritis Cartilage 2016;24:573–80. 10.1016/j.joca.2015.10.018 [DOI] [PubMed] [Google Scholar]
- 8. Vicente R, Noël D, Pers Y-M, et al. Deregulation and therapeutic potential of microRNAs in arthritic diseases. Nat Rev Rheumatol 2016;12 10.1038/nrrheum.2016.119 [DOI] [PubMed] [Google Scholar]
- 9. Tardif G, Hum D, Pelletier J-P, et al. Regulation of the IGFBP-5 and MMP-13 genes by the microRNAs miR-140 and miR-27a in human osteoarthritic chondrocytes. BMC Musculoskelet Disord 2009;10 10.1186/1471-2474-10-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Araldi E, Schipani E. MicroRNA-140 and the silencing of osteoarthritis. Genes Dev 2010;24:1075–80. 10.1101/gad.1939310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell 2013;152:1298–307. 10.1016/j.cell.2013.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014;157:77–94. 10.1016/j.cell.2014.03.008 [DOI] [PubMed] [Google Scholar]
- 13. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014;505:344–52. 10.1038/nature12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012;7:e30733 10.1371/journal.pone.0030733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hentze MW, Preiss T. Circular RNAs: splicing's enigma variations. Embo J 2013;32:923–5. 10.1038/emboj.2013.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384–8. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 17. Chen YT, Rettig WJ, Yenamandra AK, et al. Cerebellar degeneration-related antigen: a highly conserved neuroectodermal marker mapped to chromosomes X in human and mouse. Proc Natl Acad Sci U S A 1990;87:3077–81. 10.1073/pnas.87.8.3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu Q, Zhang X, Hu X, et al. Emerging roles of circRNA related to the mechanical stress in human cartilage degradation of osteoarthritis. Mol Ther Nucleic Acids 2017;7:223–30. 10.1016/j.omtn.2017.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dobin A, Davis CA, Schlesinger F, et al. StAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haft DH, DiCuccio M, Badretdin A, et al. RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res 2018;46:D851–D60. 10.1093/nar/gkx1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. O'Leary NA, Wright MW, Brister JR, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 2016;44:D733–45. 10.1093/nar/gkv1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang X-O, Wang H-B, Zhang Y, et al. Complementary sequence-mediated exon circularization. Cell 2014;159:134–47. 10.1016/j.cell.2014.09.001 [DOI] [PubMed] [Google Scholar]
- 24. Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 2016;7 10.1038/ncomms11215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333–8. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 26. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453–61. 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li L, Yang C, Liu X, et al. Elevated expression of microRNA-30b in osteoarthritis and its role in ERG regulation of chondrocyte. Biomed Pharmacother 2015;76:94–9. 10.1016/j.biopha.2015.10.014 [DOI] [PubMed] [Google Scholar]
- 28. Ohta Y, Okabe T, Larmour C, et al. Articular cartilage endurance and resistance to osteoarthritic changes require transcription factor ERG. Arthritis Rheumatol 2015;67:2679–90. 10.1002/art.39243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Berenbaum F. Osteoarthritis year 2010 in review: pharmacological therapies. Osteoarthritis Cartilage 2011;19:361–5. 10.1016/j.joca.2011.01.019 [DOI] [PubMed] [Google Scholar]
- 30. Wieland HA, Michaelis M, Kirschbaum BJ, et al. Osteoarthritis - an untreatable disease? Nat Rev Drug Discov 2005;4:331–44. 10.1038/nrd1693 [DOI] [PubMed] [Google Scholar]
- 31. Boulaftali Y, François D, Venisse L, et al. Endothelial protease nexin-1 is a novel regulator of a disintegrin and metalloproteinase 17 maturation and endothelial protein C receptor shedding via furin inhibition. Arterioscler Thromb Vasc Biol 2013;33:1647–54. 10.1161/ATVBAHA.113.301494 [DOI] [PubMed] [Google Scholar]
- 32. Pagliara V, Adornetto A, Mammì M, et al. Protease nexin-1 affects the migration and invasion of C6 glioma cells through the regulation of urokinase plasminogen activator and Matrix Metalloproteinase-9/2. Biochim Biophys Acta 2014;1843:2631–44. 10.1016/j.bbamcr.2014.07.008 [DOI] [PubMed] [Google Scholar]
- 33. Rao JS, Kahler CB, Baker JB, et al. Protease nexin I, a serpin, inhibits plasminogen-dependent degradation of muscle extracellular matrix. Muscle Nerve 1989;12:640–6. 10.1002/mus.880120805 [DOI] [PubMed] [Google Scholar]
- 34. Xiao K, Yang Y, Bian Y, et al. Identification of differentially expressed long noncoding RNAs in human knee osteoarthritis. J Cell Biochem 2019;120 10.1002/jcb.27750 [DOI] [PubMed] [Google Scholar]
- 35. He J, Xie Q, Xu H, et al. Circular RNAs and cancer. Cancer Lett 2017;396:138–44. 10.1016/j.canlet.2017.03.027 [DOI] [PubMed] [Google Scholar]
- 36. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 2016;17:272–83. 10.1038/nrg.2016.20 [DOI] [PubMed] [Google Scholar]
- 37. Cheng X, Zhang L, Zhang K, et al. Circular RNA VMA21 protects against intervertebral disc degeneration through targeting miR-200c and X linked inhibitor-of-apoptosis protein. Ann Rheum Dis 2018;77:770–9. 10.1136/annrheumdis-2017-212056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer 2018;17 10.1186/s12943-018-0827-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer 2017;16 10.1186/s12943-017-0663-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nakamura A, Rampersaud YR, Nakamura S, et al. microRNA-181a-5p antisense oligonucleotides attenuate osteoarthritis in facet and knee joints. Ann Rheum Dis 2019;78:111–21. 10.1136/annrheumdis-2018-213629 [DOI] [PubMed] [Google Scholar]
- 41. Wu C, Tian B, Qu X, et al. MicroRNAs play a role in chondrogenesis and osteoarthritis (review). Int J Mol Med 2014;34:13–23. 10.3892/ijmm.2014.1743 [DOI] [PubMed] [Google Scholar]
- 42. Jones SW, Watkins G, Le Good N, et al. The identification of differentially expressed microRNA in osteoarthritic tissue that modulate the production of TNF-alpha and MMP13. Osteoarthritis Cartilage 2009;17:464–72. 10.1016/j.joca.2008.09.012 [DOI] [PubMed] [Google Scholar]
- 43. Iwamoto M, Tamamura Y, Koyama E, et al. Transcription factor ERG and joint and articular cartilage formation during mouse limb and spine skeletogenesis. Dev Biol 2007;305:40–51. 10.1016/j.ydbio.2007.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
annrheumdis-2018-214786supp001.docx (42.4KB, docx)
annrheumdis-2018-214786supp002.docx (23.8KB, docx)
annrheumdis-2018-214786supp003.jpg (2.7MB, jpg)
annrheumdis-2018-214786supp004.docx (27.6KB, docx)
annrheumdis-2018-214786supp005.jpg (4.1MB, jpg)
annrheumdis-2018-214786supp008.jpg (2.9MB, jpg)
annrheumdis-2018-214786supp009.jpg (3.9MB, jpg)
annrheumdis-2018-214786supp010.jpg (3.8MB, jpg)
annrheumdis-2018-214786supp011.jpg (2.4MB, jpg)
annrheumdis-2018-214786supp012.docx (18.1KB, docx)
annrheumdis-2018-214786supp013.jpg (1.5MB, jpg)
annrheumdis-2018-214786supp006.jpg (3.2MB, jpg)
annrheumdis-2018-214786supp007.docx (30.9KB, docx)
annrheumdis-2018-214786supp014.pdf (14.1MB, pdf)
annrheumdis-2018-214786supp015.docx (19.4KB, docx)