Abstract
EGFL6, a member of the EGF like superfamily, plays an important role during embryonic development and has been implicated in promotion of tumor angiogenesis without affecting wound healing. There is very little known about the function of EGFL6 in cancer cells. Here, we investigated whether EGFL6 plays a direct role in cancer cells in addition to the promotion of tumor angiogenesis. Our study showed that EGFL6 promoted epithelial mesenchymal transition (EMT) and stemness of breast cancer cells and increased cell migration and invasion in cell culture studies. We also found that EGFL6 reduced apoptotic signaling in cancer cells and promoted tumor growth in vivo. Importantly, expression of EGFL6 in cancer cells and tumor endothelial cells not only increased tumor angiogenesis but also promoted migration of cancer cells. Such dual engagement of cancer and stromal cells suggests crosstalk mediated by EGFL6 in the tumor microenvironment. Blockade of EGFL6 using our novel anti-EGFL6 monoclonal antibody significantly reduced cancer cell migration, tumor angiogenesis and tumor growth in mouse xenograft tumor models. Silencing EGFL6 mRNA by shRNA transfection of cancer cells also significantly reduced cancer cell migration, tumor angiogenesis, and tumor growth in mouse xenograft tumor models. Taken together, the results of this study indicate that targeting EGFL6 is a unique strategy for inhibiting both cancer cell metastasis and tumor angiogenesis.
Keywords: EGFL6, antibody, breast cancer, metastasis, tumor angiogenesis
INTRODUCTION
EGF-like domain 6 (EGFL6), one of the EGF-like repeat superfamily members, plays important roles in cell cycle regulation and angiogenesis (1). The tumor angiogenic function of EGFL6 was first implicated in hepatitis C virus associated hepatocellular carcinoma (2). Recently, this function has also been implicated to play a role in ovarian cancers (3–5). A study also showed that EGFL6 functions in a paracrine manner to promote migration of endothelial cells and mediate crosstalk between osteoblastic-like cells and adjacent endothelial cells (6). We recently reported that blocking EGFL6 inhibited ovarian cancer tumor growth and tumor angiogenesis without affecting wound healing (3). Another member of the EGF-like repeat superfamily, EGFL7, is also involved in vascular development as an angiogenic factor (7, 8). Targeting EGFL7 with the monoclonal antibody parsatuzumab has been tested in a phase II clinical trial. Results showed no improvement with the addition of parsatuzumab to bevacizumab or chemotherapy for metastatic colon cancer (9, 10). Complete understanding of the biological functions of EGF like proteins is critical for development of biomarker-guided targeting of EGF like proteins in cancer treatment.
After EGFL6 was found to be correlated with tumorigenesis, angiogenesis, and vasculogenesis, most studies of EGFL6 were focused on ovarian cancer (3, 4, 11, 12). Although EGFL6 was revealed as a tumor vasculature ligand for breast cancer (5), the role of EGFL6 in promoting breast cancer tumorigenesis is still unclear. Systematic investigation of the functions of EGFL6 in breast cancer is important for further targeting EGFL6 in cancer therapy. In the present study, we investigated the roles of EGFL6 in breast cancer cell migration and crosstalk with tumor endothelial cells. Results showed that EGFL6 significantly enhanced migration and invasion of breast cancer cells and increased the population of stem-like cancer cells. We also demonstrated that neutralization of EGFL6 using our novel anti-EGFL6 monoclonal antibody reduced migration and invasion of breast cancer cells and significantly inhibited tumor growth and angiogenesis in mouse xenograft tumor models. Taken together, the results of this study demonstrate that targeting EGFL6 in breast cancer can provide a valuable therapeutic strategy for blocking both tumor angiogenesis and potential cancer metastasis.
RESULTS
EGFL6 expression stimulates mobility of cancer cells and is associated with poor prognosis in breast cancer patients with regional metastasis
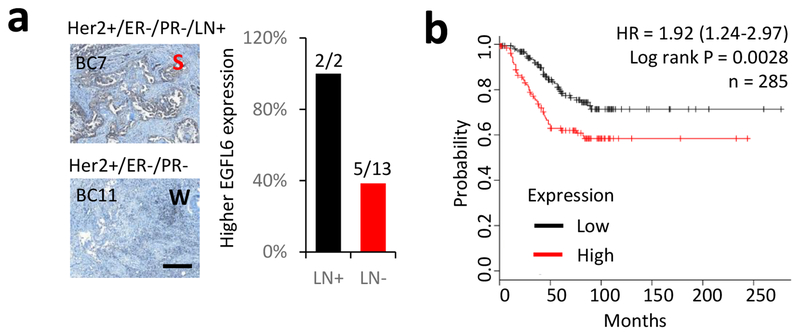
We and others have shown that EGFL6 is expressed in ovarian cancer cells and tumor endothelial cells (3, 6, 12). To determine whether EGFL6 is also produced in breast cancer, we examined expression of EGFL6 in tumor tissues from breast cancer patients (Fig. 1a) and in multiple cancer cell lines. We detected strong (S) expression of EGFL6 in 47% (7 out 15) of breast tumor tissues (Fig. S1a, b) by immunohistochemistry (IHC) staining. Expression of EGFL6 in multiple breast cancer cell lines was detected using qRT-PCR (Fig. S1c). It is worth noting that tumor tissues from the lymph node positive cancer patients (2 out 15) in this small cohort had strong EGFL6 expression (2 out 2) (Fig. 1a, Fig. S1a). More significantly, analysis using a database from Kaplan Meier Plotter (13) shows high EGFL6 expression is associated with poor survival in a cohort of breast cancer patients with lymph node metastasis (Fig. 1b) with hazard ratio (HR) = 1.92, p=0.0028. For a cohort including all breast cancer patients in the analysis, the correlation of EGFL6 expression and poor survival is also significant (Fig. S1d) with HR=1.24 and p=0.031.
Figure 1.
EGFL6 expression correlates with poor survival prognosis for patients with lymph node positive breast cancer. a. FFPE-fixed breast tumor tissues were stained with anti-EGFL6 antibody. Representative images of EGFL6 staining in lymph node positive and negative patients are shown. The percentage of strong staining of EGFL6 in lymph node positive (LN+) and negative patients (LN−) is plotted. Scale bar, 100 μm. b. Kaplan-Meier survival analysis of EGFL6 in lymph node positive breast cancer patients, n = 285.
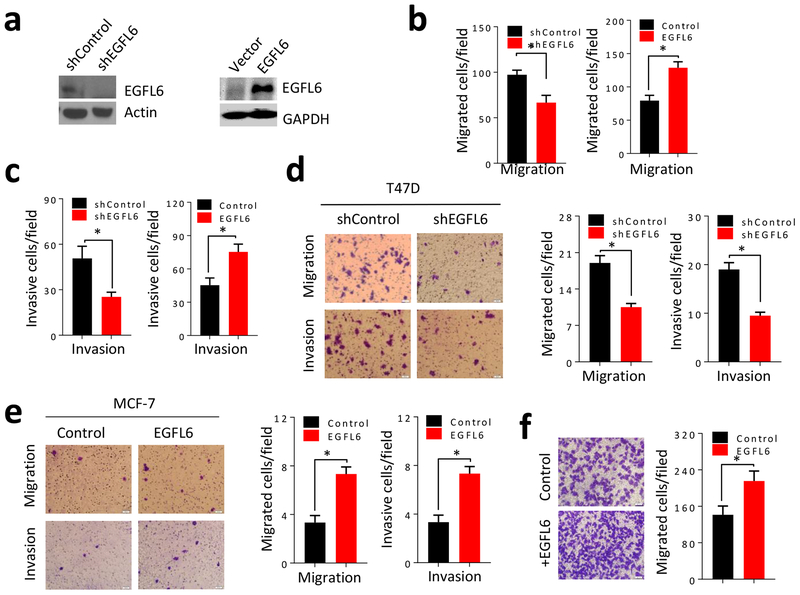
To investigate a possible link between EGFL6 expression and breast cancer metastasis, we overexpressed or knocked down EGFL6 in MDA-MB-231 breast cancer cells using a lenti-vector transfection system (Fig. 2a). The cancer cells with EGFL6 knockdown (shEGFL6) had significantly reduced cell migration and invasion, while breast cancer cells with ectopic expression of EGFL6 (+EGFL6) exhibited increased migration and invasion (Fig. 2b and 2c; Fig. S2a and S2b). Similar results were observed in EGFL6 transfected MCF-7 and T47D breast cancer cell lines (Fig. 2d and 2e; Fig. S2c and d). Addition of recombinantly produced EGFL6 protein to the culture also enhanced cancer cell (MDA-MB-231) migration (Fig. 2f). In addition, endothelial cell (RF24) conditioned media containing EGFL6 secreted protein (3), EGFL6 expression, and EGFL6 knockdown affected cell migration in wound healing assays (Fig. S2e, f, g and h). Taken together, our results demonstrate that EGFL6 expression in cancer cells stimulates the mobility of breast cancer cells and may play an important role in breast cancer metastasis.
Figure 2.
EGLF6 stimulates mobility of breast cancer cells. a, Western blot of MDA-MB-231/shEGFL6 and MDA-MB-231/EGFL6 cells. b, Migration assays of MDA-MB-231 cells and EGFL6 knockdown (MDA-MB-231/shEGFL6) or expression (MDA-MB-231/EGFL6) were performed in 24-well transwell plates. 5 × 104 cells were seeded in the up-chamber. After 8 hours of incubation, the migrated cells were stained and quantified. c, Invasion assays of MDA-MB-231/shEGFL6 and MDA-MB-231/EGFL6 cells were performed in 24-well transwell plates. 5 × 104 cells were seeded in the upper-chamber with Matrigel. After 24 hours of incubation, the invaded cells were stained and quantified. d, Migration and invasion assays of T47D cells with knockdown of EGFL6 (T47D/shEGFL6) were performed in 24-well transwell plates. 1 × 105 cells were seeding in the up-chamber with or without Matrigel. After 8 hours incubation (migration) or 24 hours incubation (invasion), the migrated or invasive cells were stained and quantified. Representative images and quantitative results are shown at the bottom. e, Migration and invasion assays of MCF-7 cells with expression of EGFL6 (MCF-7/EGFL6) were performed in 24-well transwell plates. 5 × 104 cells were seeding in the upper-chamber with or without Matrigel. After 8 hours incubation (migration) or 24 hours incubation (invasion), the migrated or invasive cells were stained and quantified. Representative images and quantitative results were shown at the bottom. f, MDA-MB-231 cells were incubated with EGFL6 protein and migration assay was performed in 24-well transwell plates. After 8 hours of incubation, the migrated cells were stained and quantified. Representative images and quantitative results were shown. *, p < 0.05. Error bar, standard deviation (SD).
EGFL6 induces EMT in breast cancer cells
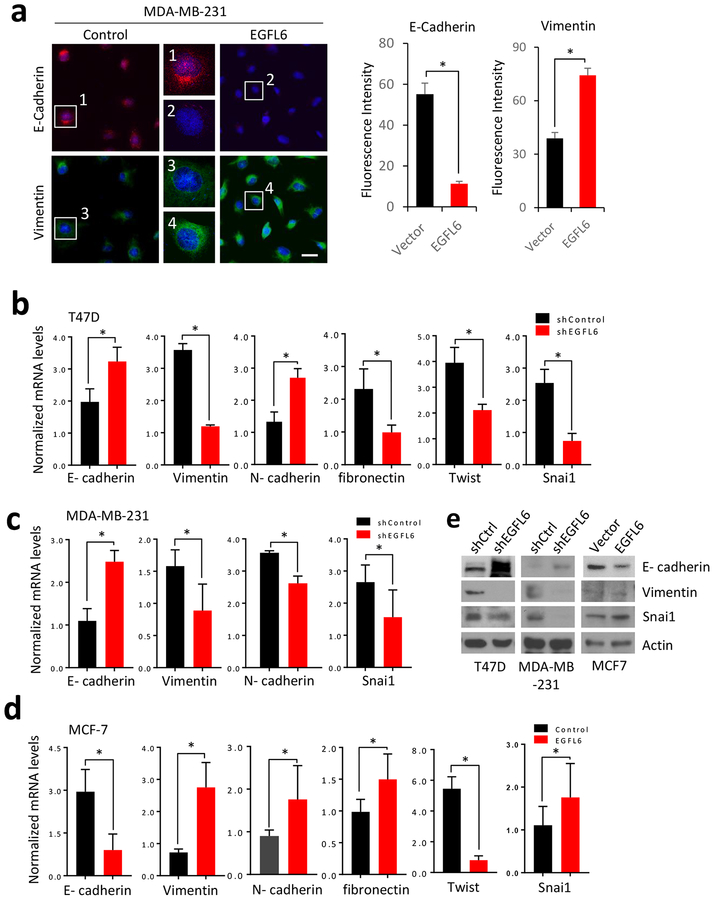
Epithelial–mesenchymal transition (EMT) is often associated with metastasis of cancer cells (14). We observed cancer cell morphology changes with knockdown or ectopic expression of EGFL6 (Fig. S3a). Cells with higher EGFL6 showed mesenchymal phenotype (Fig. S3b). We further used IF assay to evaluate expression of molecular markers associated with EMT. We found that ectopic expression of EGFL6 decreased protein levels of epithelial marker E-cadherin and increased levels of vimentin (Fig. 3a; Fig. S3a, S3d). Consistently, mRNA levels of several EMT markers including Snai1, another important molecular marker associated with EMT, were impacted by EGFL6 expression in breast cancer cells (Fig 3b, 3c, and 3d). Collectively, these results suggest that EGFL6 expression is associated with EMT of breast cancer cells.
Figure 3.
EGFL6 induces EMT in breast cancer cells. a, Immunofluorescence staining of E-cadherin and vimentin in paired MDA-MB-231 and MDA-MB-231/shEGFL6 cells. Nuclei were visualized with DRAQ5 staining (blue). Representative images and quantitative results are shown. Scale bar, 20 μm. b, qRT-PCR analysis showed the gene expression of E-cadherin, vimentin, fibronectin, Twist and snai1 in T47D and T47D/shEGFL6 cells. c, qRT-PCR analysis showed the gene expression of E-cadherin, vimentin, N-cadherin and snai1 inMDA-MB-231 and MDA-MB-231/shEGFL6 cells. d, qRT-PCR analysis showed the gene expression of E-cadherin, vimentin, fibronectin, Twist and snai1 in MCF-7 and MCF-7/EGFL6 cells. e, Western blot detection of E-cadherin, Vimentin and snail in the paired T47D and T47D/shEGFL6, MDA-MB-231 and MDA-MB-231/shEGFL6, and MCF-7 and MCF-7/EGFL6 cells. *, p < 0.05. Error bar, standard deviation (SD).
EGFL6 maintains breast cancer stem-like cell population
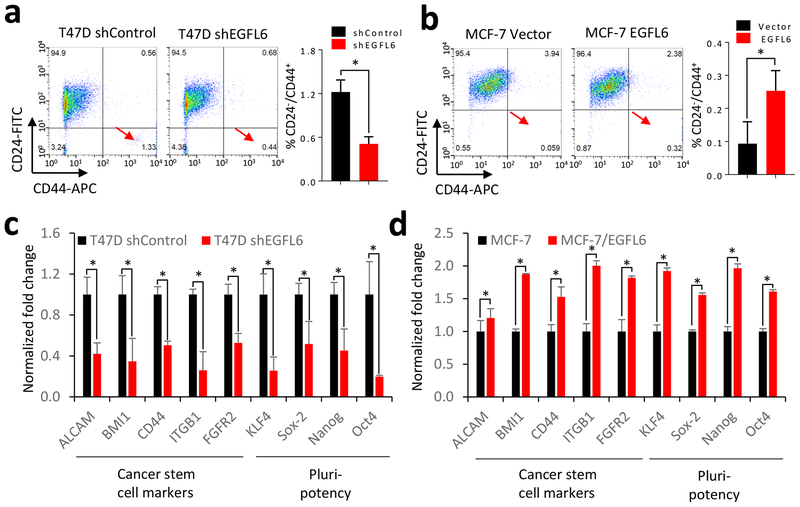
Cancer stem cells are associated with EMT and cancer metastasis (15). We hypothesized that EGFL6 expression promotes the cancer stem-like cell population. To test this hypothesis, we analyzed the breast cancer stem-like cell population (CD24− /CD44+) using a flow cytometry assay. EGFL6 knockdown significantly decreased the stem-like cancer cell (CD24− /CD44+) population (Fig. 4a). Ectopic expression of EGFL6 increased this population (Fig. 4b). In addition, levels of several gene transcripts for cancer stem cell or pluripotency and self-renewal markers were lower in cancer cells with EGFL6 knocked down than in control cells (Fig. 4c). On the other hand, upregulation of those markers was detected by qRT-PCR analysis of cancer cells with ectopic expression of EGFL6 (Fig. 4d). These data indicate that EGFL6 expression promotes the population of breast cancer stem-like cells.
Figure 4.
EGFL6 mediates cancer stem cell population of breast cancer. a and b, T47D, T47D/shEGFL6 and MCF-7, MCF-7/EGFL6 cells were double stained with CD24 and CD44 antibody. Flow cytometry assay was used to analyze the population of CD24 negative and CD44 positive cells. Representative images and quantitative results were shown. c and d, qRT-PCR analysis indicated gene expression of cancer stem cell markers in T47D, T47D/shEGFL6 and MCF-7, MCF-7/EGFL6 cells. Quantitative results are shown. *, p < 0.05. Error bar, SD.
EGFL6 expression promoted colony formation and reduced apoptosis of breast cancer cells
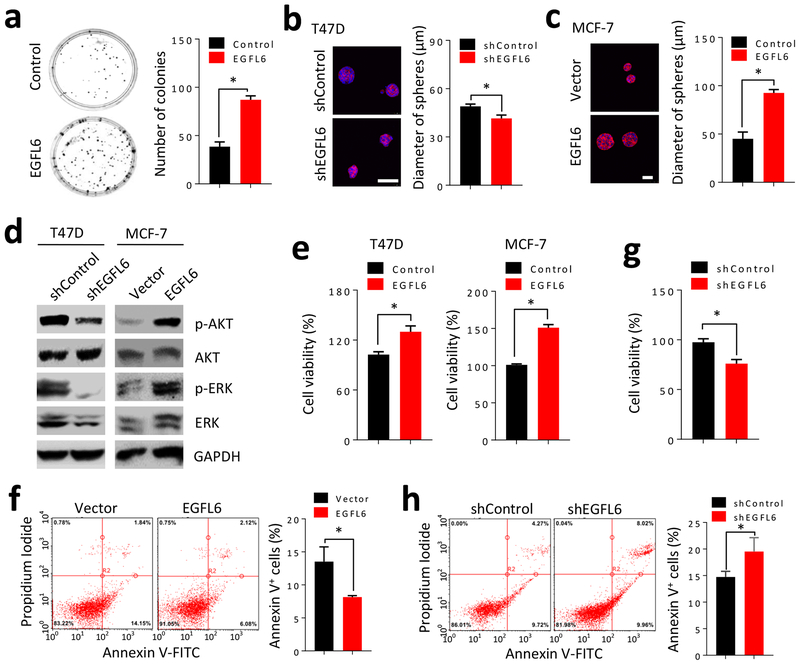
In order to determine if cancer cells with EGFL6 expression can directly affect cell growth in 3-dimensional (3D) cultures, we compared colony formation in the presence and absence of EGFL6 in 3D cell cultures. Cancer cells were cultured in a low density condition and colonies were counted after 10 days of culture. EGFL6 expression increased colony formation (Fig. 5a). We performed further experiments with 3D cultures and found that knockdown of EGFL6 reduced the sphere size of T47D cancer cells (Fig. 5b), while expression of EGFL6 increased the sphere size of MCF-7 cells (Fig. 5c). It has been reported that EGFL6 can increase phosphorylation of ERK (p-ERK) and Akt (p-Akt) in fibroblastic meningioma (16). Knockdown of EGFL6 in breast cancer cells reduced p-AKT and p-ERK, while high expression of EGFL6 enhanced p-Akt and p-ERK (Fig. 5d).
Figure 5.
EGFL6 enhances cell growth and reduces apoptosis of breast cancer cells. a, Colony formation assay in T47D/shEGFL6 cultured with recombinant EGFL6 protein. Representative images and quantitative results in a bar graph are shown. b and c, 3D cell culture for sphere formation in a soft agar with matrigel was performed with T47D and T47D/shEGFL6 or MCF-7/EGFL6 cells. Representative images and quantitative results are shown. Scale bar, 50 μm. d, Detection of pAKT and pERK by Western blotting using cell lysates prepared with T47D vs. T47D/shEGFL6 and MCF-7 vs. MCF-7/EGFL6. e. EGFL6 expression in MDA-MB-231 (+EGFL6) and MCF-7 (+EGFL6) cells increased cell growth in comparison with the counterpart parental cells. Cancer cells were seeded in 96-well plates, 2000 cells/well and cell growth was measured by alamarBlue assay (Invitrogen) after 72 hours culture. f, Flow scattering plots show percentage of apoptosis cells using annexin-V assay kit in MCF-7 and MCF-7/EGFL6 cancer cells. g, Reduced cell growth in EGFL6 knockdown cells in comparison with parental T47D cells using alamarBlue assay. h, Apoptosis assay of T47D and T47D/EGFL6 knockdown cells using annexin-V by flow cytometry assay. All assays were repeated three times (n=3), * p < 0.05. Error bar, SD.
Using annexin-V flow cytometry assay, we investigated effects of EGFL6 on cell proliferation and apoptosis. Expression of EGFL6 increased cell proliferation (Fig. 5e) and reduced apoptosis (Fig. 5f). On the other hand, knockdown of EGFL6 reduced cell proliferation (Fig. 5g) and enhanced apoptosis (Fig. 5h). Several apoptosis markers showed changes at mRNA levels, as assessed by RT-qPCR. Higher levels of Fas and Caspase3 and lower Bcl-2 were detected in EGFL6 knockdown cancer cells (Fig. S4a), while lower Fas and higher Bcl-2 were detected in cancer cells with EGFL6 expression (Fig. S4b).
EGFL6 expression in tumor endothelial cells promotes migration of cancer cells and tumor angiogenesis in vivo
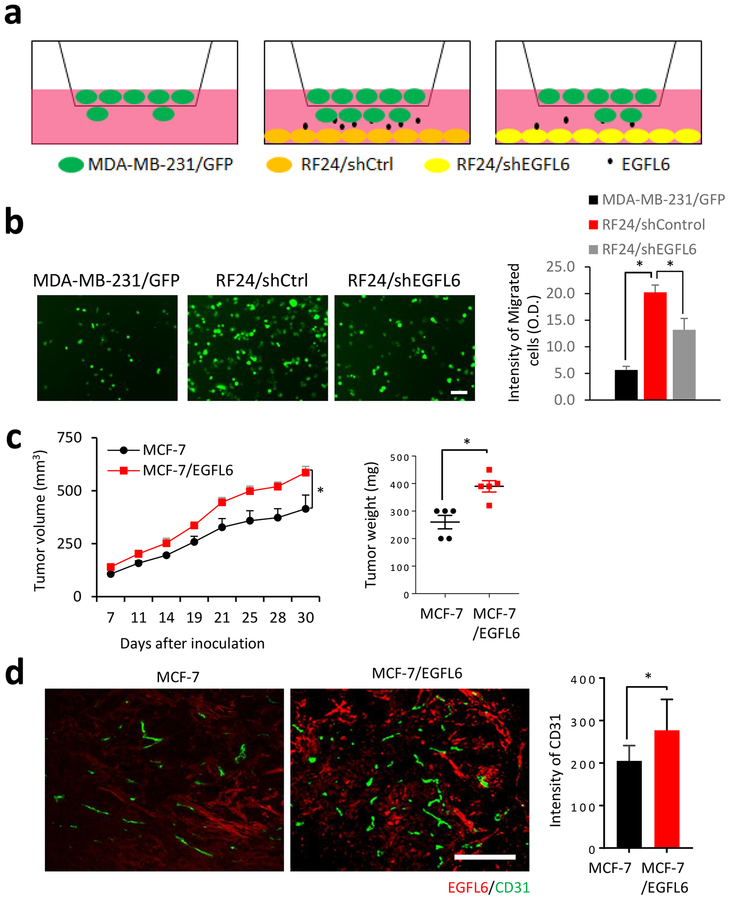
We reported previously (7) that ovarian tumor endothelial cells showed EGFL6 expression. We sought to determine breast cancer cell response to EGFL6 produced by endothelial cells in co-culture. Endothelial cells (RF24) were seeded in the bottom of transwells. EGFL6 secreted by the endothelial cells promoted migration of cancer cells (with GFP-expression) placed in the upper chamber. When EGFL6 was knocked down in RF24 cells, the cell migration was reduced (Fig. 6a and 6b). We compared tumor growth of MCF7 cells with EGFL6 expression with the parental control cells in a mouse xenograft model. MCF7 with EGFL6 expression were correlated with increased tumor growth in vivo (Fig. 6c). To determine the effect of EGFL6 expression on tumor angiogenesis, we used CD31 as a marker for detecting tumor endothelial cells (CD31, green) by immunofluorescence staining (Fig. 6d). Tumor angiogenesis (CD31 staining) was significantly higher in tumor tissues with EGFL6 expression (stained in red) than in parental control tumor tissues.
Figure 6.
EGFL6 expression in endothelial cells promotes breast cancer cell migration and increases tumor angiogenesis. a, Diagram shows the layout of co-culture assays. b, Endothelial cells (RF24) promoted migration of cancer cells. MDA-MB-231 cancer cells were seeded on transwell upper inserts and RF24/shControl and RF24/shEGFL6 cells were seeded in the lower chamber. After 8 hours incubation, migrated cancer cells were imaged and quantified in the bar graph. A representative image for each conditions is shown from 3 replication, n=3. Scale bar, 50 μm c, MCF-7/EGFL6 cells showed increased tumor growth than the parental control. Tumor sizes were measured in the indicated days. Tumor weights were measured at the end point of the experiment. d, EGFL6 expression in cancer cells promoted tumor angiogenesis in vivo. IF staining of EGFL6 (in red) and CD31 (green) in tumor tissues, representative images with overlay of nuclear staining (blue) in MCF7/EGFL6 and MCF7 xenograft tumor tissues are shown and quantified. Scale bar, 100 μm. All experiments were repeated 3 to 5 times, *, p < 0.05. Error bar, SD.
Blocking EGFL6 tumor promoting function by neutralizing antibodies
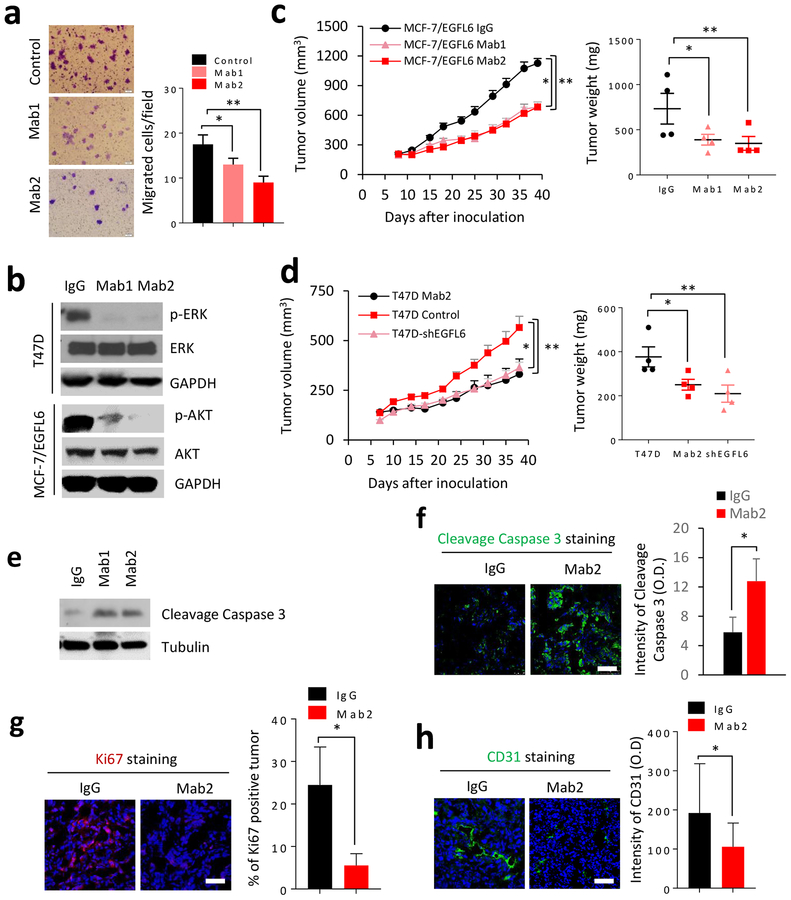
We identified a panel of novel EGFL6 targeting monoclonal antibodies that reduced tumor angiogenesis in ovarian tumor models (7). We tested the anti-tumor efficacy of the EGFL6 neutralizing antibodies in vitro and in vivo using breast cancer models (Fig. 7). Treatment with anti-EGFL6 antibody reduced cancer cell migration (Fig. 7a; Fig. S4c and S4d) and neutralized EGFL6 effects on wound healing (Fig. S4e). Treatment of breast cancer cells with EGFL6 neutralizing antibody (mAb1 and mAb2) decreased p-ERK and p-AKT in comparison with antibody (IgG) isotype control (Fig. 7a). Weekly treatment (10μg/kg) of mice with xenograft tumor by the anti-EGFL6 antibody in vivo showed significant inhibition of tumor growth and reduced tumor weight (Fig. 7c). The anti-EGFL6 antibody treatment had a similar tumor inhibition as the knockdown of EGFL6 in T47D breast cancer cells (Fig. 7d). As expected, tumor samples showed increased level of cleaved caspase 3 with antibody treatment by western blot or immunofluorescence staining (Fig. 7e, f) Treatment also decreased tumor cell proliferation (Ki67 staining, Fig. 7g) and micro-vasculature formation (CD31 staining) in tumor tissues (Fig. 7h).
Figure 7.
Anti-EGFL6 monoclonal antibodies inhibited mobility and tumor growth of breast cancer cells. a, Anti-EGFL6 antibody (Mab1 and Mab2) inhibited migration of T47D cancer cells. T47D cells were treated with EGFL6 antibody at 10 μg/ml concentration for 24 hours and cell migration was determined by imaging quantitation using image J. Representative images are shown. b, Treatment of cancer cells by anti-EGFL6 antibodies Mab1 and Mab2 reduced pERK and pAKT in T47D/EGFL6 and MCF-7/EGFL6 cells, respectively by Western blot analysis. c, MCF-7/EGFL6 cells were inoculated into the mammary fat pad of nude mice, antibodies were administered weekly at 10mg/kg for 4 weeks by interperitoneal (ip.) injection and tumor sizes were measured in the indicated days. Tumor weights were measured at the end point of the experiment. Five mice (n=5) per group and error bars indicate SEM. d, Treatment of anti-EGFL6 antibody Mab2 showed similar inhibition to T47D with knockdown EGFL6 (T47D/shEGFL6). Antibodies were administered weekly at 10 mg/kg for 4 weeks by ip injection and isotype IgG was used as control. Tumor sizes were measured in the indicated days and tumor weight were measured at the end point of the experiment, n=5. e. Xenograft tumor tissues from Fig. 7c were lysed and performed SDS-PAGE and Western blot with indicated antibodies f. IF staining of cleaved caspase 3 in xenograft tumor samples from Fig. 7c. Representative images were shown for each group and n=3. Scale bar, 50 μm. g. IF staining of Ki67 in T47D xenograft tumor samples, Representative images are shown for each group and n=3. Scale bar, 50 μm. h. Anti-EGFL6 antibody (mAb2) inhibited tumor angiogenesis. IF staining of CD31 in T47D tumor samples and a representative image is shown for each group and n=3. Scale bar, 50 μm. All experiments were repeated 3 to 5 times and error bars show SD, **, p<0.01 and *, p<0.05.
DISCUSSION
In this study, we revealed roles of EGFL6 in tumor growth and potential metastasis of breast cancer. We demonstrated, for the first time, that EGFL6 promotes breast cancer cell migration and invasion. Our data also suggest that EGFL6 plays an important role in the maintenance of cancer stem cells and the promotion of EMT of breast cancer cells. This finding is in agreement with a recent study that showed high expression of EGFL6 correlates positively with metastasis of ovarian carcinoma through regulation of asymmetric cell division and maintenance of cancer stem cells (4). EGFL6 expression increased expression of Snail, a key transcriptional factor related to EMT. This result suggests that EGFL6 promotes EMT through Snail transcriptional regulation (17).
We demonstrated that EGFL6 promotes breast cancer tumor growth. We showed that blocking EGFL6 by knockdown of the gene or using an anti-EGFL6 neutralizing antibody increased cancer cell apoptosis. Our recent study of EGFL6 in ovarian cancer cells identified Tie2 as a receptor that interacts with EGFL6 and integrin for downstream EGFL6 mediated signaling in tumor endothelial cells (3). Other studies have indicated that EGFL6 may bind to integrin proteins through RGD domain interaction with integrin (18, 19). We found that blocking EGFL6 in breast cancer cells inhibited phosphorylation of Akt and ERK, which plays an important role in tumor growth and invasion. In addition to the reported receptors such as Tie2 and integrin alpha 8, EGFL6 may interact with other surface receptors in breast cancer cells. The identification of such prospective receptors requires further investigation.
Tumor endothelial cells produce EGFL6, particularly under hypoxic conditions (3). This study showed that EGFL6 promoted cancer cell migration towards tumor endothelial cells in a co-culture study. We also showed that breast cancer cells produced EGFL6. Different levels of EGFL6 expression in cancer cells and tumor associated endothelial cells may regulate cancer cell migration and metastasis in the tumor microenvironment. Based on the signal peptide of EGFL6 protein, we hypothesized that it functions primarily as a secreted protein. The important question of whether intercellular EGFL6 is also involved tumor progression should be addressed in continuing studies. Another member of the EGF like superfamily, EGFL7, has also been reported to be involved in vascular development as an angiogenic factor (7, 20). Other reports assert that EGFL7 is involved in multiple cancer types including glioma, myeloid leukemia, and hepatocellular carcinoma (21–24). Those reports suggest that EGFL7 may play a role similar to that of EGFL6. This raises a critical scientific question about distinguishing EGFL6 and EGFL7 during tumor development. Nevertheless, EGFL6 is a good target for inhibition of both cancer cells and tumor-associated angiogenesis.
In summary, we demonstrated that EGFL6 promotes tumor growth and cell mobility, which suggests that EGFL6 is involved in breast cancer metastasis. Mechanistically, EGFL6 can activate both Akt and ERK signaling pathways and cross-talk with Snail, a zinc figure transcriptional regulator for EMT and cancer cell metastasis. Our novel anti-EGFL6 antibodies can block EGFL6 function and inhibited breast tumor growth and cancer cell mobility. This study provides a strong foundation for rational development of antibody cancer therapeutics targeting EGFL6.
MATERIALS AND METHODS
Cell lines and reagents
HEK293T was purchased from Life Technologies (Carlsbad, CA). MCF-7, MDA-MB-231, and T47D human breast carcinoma cells were obtained from the American Type Culture Collection (Manassas, VA) and maintained in a humidified atmosphere of 5% CO2 at 37°C in medium and supplemented with penicillin, streptomycin (Life Technologies) and 10% fetal bovine serum (FBS) (GE Healthcare Life Sciences, Chicago, Illinois). Antibodies to detect AKT, ERK, GAPDH (Cell Signaling Technology, Danvers, MA) and anti-FLAG antibody (Sigma-Aldrich, St. Louis, Missouri) were used at 1:1000 in 5% milk. Phospho-Akt (ser473) and phospho-Erk (Thr202/Tyr204) were purchased from Cell Signaling and used at 1:1000 in 1% bovine serum albumin (BSA). The EGFL6 neutralizing antibodies Mab1 and Mab2 were generated in house as we reported previously (3).
Construction of stable cell lines with ectopic EGFL6 expression and shRNA knockdown
In construction of stable shRNA knockdown cell lines, lentiviral particles encoding shRNA targeting EGFL6 (Sigma-Aldrich, St. Louis, Missouri) were used for transfection. Cells were cultured in RPMI media containing puromycin (3μg/ml) (Thermo Fisher Scientific, Waltham, Massachusetts) for 3 weeks to select for successful transformants. For expression of EGFL6 in cancer cells, EGFL6 cDNA purchased from Origene (Rockville, MD) was first cloned into the PLVX lenti-viral based expression vector (PLVX-EGFL6). Then, EGFL6 stable cell lines were selected under puromycin after lentiviral particle infection.
Preparation of conditional media
RF24 endothelial cells were cultured in T175 tissue culture flasks at normal cell culture conditions in a 37°C humidified incubator, 5% CO2. When they reached confluence, cells were cultured in serum-free media (SFM) for 24 h and the media was centrifuged and filtered through 0.2 μm syringe filters (Corning Inc., Corning, NY) to be used as conditioned media.
Colony formation assay
200 cells were seeded in the 35mm×10mm dish. After culturing for 10 days, colonies were fixed with 100% methanol for 15 min and then stained with crystal violet (0.5%) for 20 min. The numbers of colonies were counted and analyzed.
Migration and invasion assay
Migration and invasion assays were conducted as described previously (25). Briefly, 1×105 cells were seeded on the top chambers of a transwell plate (8 μm pores, Corning Inc., Corning, NY) in medium with 1% FBS. After 12 hours, cells in the top were removed, and the cells on the bottom of the filter were fixed and stained with crystal violet. For the invasion assay, the top chamber was pre-coated with Matrigel (BD Biosciences, San Jose, CA). Cells were seeded on the coated top chamber in serum-free media. The bottom chamber was filled with normal medium. After 24 hours cells on the top chamber were removed and invaded cells were fixed and stained with 0.5% crystal violet.
3D sphere cultures
Cells were seeded on top of a 1:1 diluted matrigel in culture media at a cell density of 5000 cells/cm2. After 10 days culture, cells were stained with rhodamine-labeled phalloidin (Invitrogen, Carlsbad, CA) for 20 mins and nuclei were stained with DRAQ5 (Molecular Probes, Carlsbad, CA) for 20 min then imaged using a confocal microscope (Leica, Wetzlar, Germany).
Flow cytometry analysis
Cells were trypsinized, suspended into single cells, washed with PBS and incubated on ice for 30 min with EGFL6 protein with his tag. After incubation with the anti-his antibody (R&D, Minneapolis, MN) or anti-CD24-FITC and anti-CD44-APC (BD Biosciences, San Jose, CA) and washed twice with PBS, cells were analyzed using a Guava flow cytometer (Millipore-Sigma, Burlington, Massachusetts).
Immunofluorescence (IF) staining
IF staining of cells was conducted as described previously (26). Briefly, 1×104 cells were plated into Lab-Tek 8 chamber slides (Thermo Fisher Scientific, Waltham, MA) for 24 hours, then washed with PBS, fixed in 4% paraformaldehyde and permeabilized in 0.1% Triton X-100. The cells were washed with PBS and blocked with 5% BSA. Then, the cells were incubated with primary antibodies overnight at 4 °C. Then cells were washed with PBS and incubated with the secondary antibody for 1 hour at room temperature in the dark. Samples were imaged using confocal microscopy.
IF staining of tissue samples was conducted as described previously (27). Briefly, tumor samples (0.2−0.3 grams) were embedded in Optimal Cutting Temperature (OCT) solution (SAKURA), snap-frozen in liquid nitrogen, and stored at −80°C until making tissue sections (0.4 μm). The sectioned tissues were fixed with 4% Formaldehyde washed in PBS and blocked with 1% BSA containing 10% normal goat serum. Ki67 antibody (Abcam) and CD31 (BD Bioscience) were used. EGFL6 (made in house) was labeled with DyLight 550. All paired samples were imaged using confocal microscopy.
Immunohistochemistry
For immunohistochemistry, 4 μm sections obtained from the paraffin-embedded samples were used. Slides were dewaxed with xylenes, hydrated with graded alcohols, and treated with 0.6% H2O2 to eliminate endogenous peroxidase activity. Then the slides were immersed in boiling 10 mmol/L sodium citrate (pH 6.5) in a pressure cooker heated for the antigen retrieval step. Slides were blocked with 5% normal goat serum in 0.01 M PBS for 30 min at 37°C, then incubated with primary antibodies, anti-EGFL6 antibodies produced in rabbit (HPA001838, Sigma), diluted at 1:100 dilutions in blocking solution, overnight at 4°C in a humidified chamber. After washing, samples were successively incubated with biotinylated secondary antibody and streptavidin–horseradish peroxidase (HRP) avidin working solution and mounted on slides using Permount mounting media (Fisher Scientific, USA).
RNA isolation, reverse transcription and qPCR
Total RNA was isolated from target cells using RNeasy Mini kit (QIAGEN). cDNA was synthesized using iScript Select cDNA Synthesis Kit (Bio-Rad). The oligonucleotide forward and reverse primers used for qRT-PCR analysis were listed in the supplement data. All primers were designed and synthesized by Sigma listed in Additional file 1: Table S1.
Western blotting
Western blot was conducted as described previously (28). Briefly, Cells were lysed in RIPA buffer. Equal amounts of protein were used for sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidenedifluoride (PVDF) membranes blotting with indicated antibodies.
Co-cultures of cancer cells and endothelial cells in a transwell plate
Transwell cell culture chambers (Corning Inc., Corning, NY) consisting of polycarbonate filters (8 μm pore size; 0.33 cm2 area) were used for the co-culture assay. Cancer cells (in upper chamber insert) and RF24 cells in lower culture wells were seeded at a ratio of 1:10: cancer cells: RF24 cells. After 12 hours co-culture, tumor cells were removed from top chamber and migrated cells were fixed with methanol and stained for imaging detection.
Wound healing assay
Cells were plated in a 6-well plate and incubated for 24 hours. The monolayer was disrupted by a single scratch using a yellow pipette tip (200 μl). Images were obtained at 0 hour and 16 hours post-scratch using a phase-contrast microscope (Olympus USA, Waltham, MA).
Mouse tumor models
Animal experimental protocols were approved by the Animal Welfare Committee (AWC) at the University of Texas Medical School at Houston. Six-week-old female nude mice (BALB/c) were randomly divided into indicated groups (5 mice/group) before inoculation. For MCF-7 ectopic expression and control groups, T47D knockdown and control groups, cells were subcutaneously injected with 1×107 cells suspended at 1:1 ratio of PBS and Matrigel (BD Biosciences, San Jose, CA, USA). For antibody treatment, mice were randomly divided into groups (n = 5) before antibody treatments on day 10 post-implanting cancer cells. Anti-EGFL6 antibodies, IgG or PBS were administered weekly using intraperitoneal injections. Tumors were collected at the end of in vivo experiment for ex vivo analysis. Evaluation was performed measuring tumor weight by an electronic balance and volume by a digital caliper as we reported previously (29).
Statistical analysis
The data are expressed as means ± standard deviation (S.D.) or standard error of the mean (SEM) as indicated in the figure legends. The Student’s t-test (two-tail) was used for comparison between groups. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgements
We thank Dr. Wei Xiong and Ms. Hui Deng for their technical assistance in cell culture and antibody preparation. We also want to thank Dr. Yunfei Wen and Dr. Prahlad Ram for their suggestions and discussion on data analysis during the manuscript preparation. We thank Dr. Georgina Salazar for her critical editing of the manuscript. This study was supported in part by Welch Foundation grant (AU-0042–20030616) to Dr. Zhiqiang An, and Cancer Prevention and Research Institute of Texas (CPRIT) Grants (RP150230 and RP150551) to Dr. Zhiqiang An and Dr. Ningyan Zhang. This research was also supported by NIH support (P50 CA217685) to Dr. Anil K Sood, the Frank McGraw Memorial Chair in Cancer Research, and the ACS Research Professor Award.
Footnotes
Conflict of interest
The authors declare no potential conflicts of interest.
References
- 1.Yeung G, Mulero JJ, Berntsen RP, Loeb DB, Drmanac R, Ford JE. Cloning of a novel epidermal growth factor repeat containing gene EGFL6: expressed in tumor and fetal tissues. Genomics. 1999. December 1;62(2):304–7. [DOI] [PubMed] [Google Scholar]
- 2.Mas VR, Maluf DG, Archer KJ, Yanek KC, Fisher RA. Angiogenesis soluble factors as hepatocellular carcinoma noninvasive markers for monitoring hepatitis C virus cirrhotic patients awaiting liver transplantation. Transplantation. 2007. November 27;84(10):1262–71. [DOI] [PubMed] [Google Scholar]
- 3.Noh K, Mangala LS, Han HD, Zhang N, Pradeep S, Wu SY, et al. Differential Effects of EGFL6 on Tumor versus Wound Angiogenesis. Cell reports. 2017. December 5;21(10):2785–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai S, Ingram P, Chen YC, Deng N, Pearson A, Niknafs Y, et al. EGFL6 Regulates the Asymmetric Division, Maintenance, and Metastasis of ALDH+ Ovarian Cancer Cells. Cancer research. 2016. November 1;76(21):6396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larimer BM, Deutscher SL. Identification of a Peptide from In vivo Bacteriophage Display with Homology to EGFL6: A Candidate Tumor Vasculature Ligand in Breast Cancer. Journal of molecular biomarkers & diagnosis. 2014. May;5(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chim SM, Qin A, Tickner J, Pavlos N, Davey T, Wang H, et al. EGFL6 promotes endothelial cell migration and angiogenesis through the activation of extracellular signal-regulated kinase. The Journal of biological chemistry. 2011. June 24;286(25):22035–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nichol D, Stuhlmann H. EGFL7: a unique angiogenic signaling factor in vascular development and disease. Blood. 2012. February 9;119(6):1345–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nichol D, Shawber C, Fitch MJ, Bambino K, Sharma A, Kitajewski J, et al. Impaired angiogenesis and altered Notch signaling in mice overexpressing endothelial Egfl7. Blood. 2010. December 23;116(26):6133–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Carbonero R, van Cutsem E, Rivera F, Jassem J, Gore I Jr., Tebbutt N, et al. Randomized Phase II Trial of Parsatuzumab (Anti-EGFL7) or Placebo in Combination with FOLFOX and Bevacizumab for First-Line Metastatic Colorectal Cancer. The oncologist. 2017. October;22(10):1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Pawel J, Spigel DR, Ervin T, Losonczy G, Barlesi F, Juhasz E, et al. Randomized Phase II Trial of Parsatuzumab (Anti-EGFL7) or Placebo in Combination with Carboplatin, Paclitaxel, and Bevacizumab for First-Line Nonsquamous Non-Small Cell Lung Cancer. The oncologist. 2018. June;23(6):654–e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu C, Bonome T, Li Y, Kamat AA, Han LY, Schmandt R, et al. Gene alterations identified by expression profiling in tumor-associated endothelial cells from invasive ovarian carcinoma. Cancer research. 2007. February 15;67(4):1757–68. [DOI] [PubMed] [Google Scholar]
- 12.Buckanovich RJ, Sasaroli D, O’Brien-Jenkins A, Botbyl J, Hammond R, Katsaros D, et al. Tumor vascular proteins as biomarkers in ovarian cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2007. March 1;25(7):852–61. [DOI] [PubMed] [Google Scholar]
- 13.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast cancer research and treatment. 2010. October;123(3):725–31. [DOI] [PubMed] [Google Scholar]
- 14.Brabletz T EMT and MET in metastasis: where are the cancer stem cells? Cancer cell. 2012. December 11;22(6):699–701. [DOI] [PubMed] [Google Scholar]
- 15.Biddle A, Mackenzie IC. Cancer stem cells and EMT in carcinoma. Cancer metastasis reviews. 2012. February 3. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Gong Y, Wang D, Xie Q, Zheng M, Zhou Y, et al. Analysis of gene expression profiling in meningioma: deregulated signaling pathways associated with meningioma and EGFL6 overexpression in benign meningioma tissue and serum. PloS one. 2012;7(12):e52707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Shi J, Chai K, Ying X, Zhou BP. The Role of Snail in EMT and Tumorigenesis. Current cancer drug targets. 2013. November;13(9):963–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong LJ, Hsieh JC, Chung AE. Two distinct cell attachment sites in entactin are revealed by amino acid substitutions and deletion of the RGD sequence in the cysteine-rich epidermal growth factor repeat 2. The Journal of biological chemistry. 1995. June 30;270(26):15838–43. [DOI] [PubMed] [Google Scholar]
- 19.Fujiwara H, Ferreira M, Donati G, Marciano DK, Linton JM, Sato Y, et al. The basement membrane of hair follicle stem cells is a muscle cell niche. Cell. 2011. February 18;144(4):577–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parker LH, Schmidt M, Jin SW, Gray AM, Beis D, Pham T, et al. The endothelial-cell-derived secreted factor Egfl7 regulates vascular tube formation. Nature. 2004. April 15;428(6984):754–8. [DOI] [PubMed] [Google Scholar]
- 21.Gong C, Fang J, Li G, Liu HH, Liu ZS. Effects of microRNA-126 on cell proliferation, apoptosis and tumor angiogenesis via the down-regulating ERK signaling pathway by targeting EGFL7 in hepatocellular carcinoma. Oncotarget. 2017. August 8;8(32):52527–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang FY, Kang CS, Wang-Gou SY, Huang CH, Feng CY, Li XJ. EGFL7 is an intercellular EGFR signal messenger that plays an oncogenic role in glioma. Cancer letters. 2017. January 1;384:9–18. [DOI] [PubMed] [Google Scholar]
- 23.Dudvarski Stankovic N, Bicker F, Keller S, Jones DT, Harter PN, Kienzle A, et al. EGFL7 enhances surface expression of integrin alpha5beta1 to promote angiogenesis in malignant brain tumors. EMBO molecular medicine. 2018. September;10(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papaioannou D, Shen C, Nicolet D, McNeil B, Bill M, Karunasiri M, et al. Prognostic and biological significance of the proangiogenic factor EGFL7 in acute myeloid leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2017. June 6;114(23):E4641–E7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dhupkar P, Zhao H, Mujoo K, An Z, Zhang N. Crk II silencing down-regulates IGF-IR and inhibits migration and invasion of prostate cancer cells. Biochemistry and biophysics reports. 2016. December;8:382–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salameh A, Fan X, Choi BK, Zhang S, Zhang N, An Z. HER3 and LINC00052 interplay promotes tumor growth in breast cancer. Oncotarget. 2017. January 24;8(4):6526–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan X, Brezski RJ, Deng H, Dhupkar PM, Shi Y, Gonzalez A, et al. A novel therapeutic strategy to rescue the immune effector function of proteolytically inactivated cancer therapeutic antibodies. Molecular cancer therapeutics. 2015. March;14(3):681–91. [DOI] [PubMed] [Google Scholar]
- 28.Shi Y, Fan X, Meng W, Deng H, Zhang N, An Z. Engagement of immune effector cells by trastuzumab induces HER2/ERBB2 downregulation in cancer cells through STAT1 activation. Breast cancer research : BCR. 2014. April 2;16(2):R33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang S, Mukherjee S, Fan X, Salameh A, Mujoo K, Huang Z, et al. Novel association of DJ-1 with HER3 potentiates HER3 activation and signaling in cancer. Oncotarget. 2016. October 4;7(40):65758–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.