Abstract
Directed differentiation of pluripotent stem cells provides an accessible system to model development. However, the distinct cell types that emerge, their dynamics, and their relationship to progenitors in the early embryo has been difficult to decipher because of the cellular heterogeneity inherent to differentiation. Here, we used a combination of bulk RNA-Seq, single cell RNA-Seq, and bioinformatics analyses to dissect the cell types that emerge during directed differentiation of mouse embryonic stem cells as embryoid bodies and we compared them to spatially and temporally resolved transcriptional profiles of early embryos. Our single cell analyses of the day 4 embryoid bodies revealed three populations which had retained related yet distinct pluripotent signatures that resemble the pre- or post-implantation epiblast, one population of presumptive neuroectoderm, one population of mesendoderm, and four populations of neural progenitors. By day 6, the neural progenitors predominated the embryoid bodies, but both a small population of pluripotent-like cells and an anterior mesoderm-like Brachyury-expressing population were present. By comparing the day 4 and day 6 populations, we identified candidate differentiation paths, transcription factors, and signaling pathways that mark the in vitro correlate of the transition from the mid-to-late primitive streak stage.
Keywords: Directed differentiation, Embryoid body, Mesendoderm, Single cell rna-seq, Noggin, Transcriptional profiling, Neuroectoderm
1. Introduction
Gastrulation, the process of extensive cell reorganization that specifies the three major germ layers, is often considered the most formative event during mammalian embryonic development (Tam and Behringer, 1997; Tam and Loebel, 2007). In mice, gastrulation begins during the blastocyst stage at approximately embryonic day 6.5 (E6.5) and is initiated by the formation of a transient structure called the primitive streak (Tam and Loebel, 2007; Wang and Chen, 2016). The primitive streak begins to form on the posterior side of the epiblast and is marked by the expression of the T-box transcription factor Brachyury (Kispert and Herrmann, 1994). During primitive streak formation, uncommitted epiblast cells migrate through the primitive streak and then exit to form either mesoderm or endoderm, while the remaining cells in the epiblast form the ectoderm. It is believed that extraembryonic tissue provides signals to initiate the formation of the primitive streak as well as to direct other forms of embryonic patterning (Rivera-Pérez and Hadjantonakis, 2014; Tam and Behringer, 1997).
As cells differentiate from pluripotency and commit to distinct lineages, their gene expression profiles diverge. Tracking this divergence uncovers the sequential molecular events that ultimately lead to the establishment of different cell types. In the past, gene expression microarrays successfully identified differentially expressed genes during developmental processes (Hemberger et al., 2001). The advent of transcriptomic sequencing has allowed us to obtain greater resolution of genetic programs active during gastrulation. For example, RNA-Seq of approximately 20 cells from 20 distinct regions of E7.0 embryos resulted in a spatially resolved transcriptional atlas of the mid-gastrula stage embryo (Peng et al., 2016). Moreover, correlating the expression signaling pathway targets to regions with previously determined fates revealed an absence of signaling activity enrichment in presumptive neuroectoderm and an enrichment of Wnt and Nodal signaling activity in presumptive mesendoderm regions.
More recently, molecular profiling of single cells has proven useful for understanding development and cell fate acquisition by enabling the identification of rare and transitory cell types, the mapping of cells spatially or temporally, and the reconstruction of gene regulatory networks (Kumar et al., 2017). For example, single cell qPCR of early murine development from zygote to blastocyst stage uncovered that the expression of mutually antagonistic transcription factors in single cells at early stages is allocated into distinct cells at later stages (Guo et al., 2010). Single cell RNA-Seq (scRNA-Seq) of pre-implantation to early gastrulation embryos revealed that elevated transcriptional noise in the epiblast coincides with exit from pluripotency (Mohammed et al., 2017), and scRNA-Seq of > 1000 cells from E6.5 to E7.75 identified regulators of distinct mesoderm sub-types (Scialdone et al., 2016). None of these findings would have been possible with traditional, bulk population derived measurements, and thus they demonstrate the power of single cell analysis in delineating crucial features and mechanisms of development in vivo.
Just as single cell profiling is beginning to provide new insights into in vivo development, it has also been used to characterize regulatory dynamics during in vitro directed differentiation of pluripotent stem cell populations into target cell types. scRNA-Seq of ~2500 mouse embryonic stem cells (mESC) undergoing retinoic acid-induced neural differentiation identified two distinct populations (neuroectoderm-like and extra-embryonic endoderm-like), pin-pointing a temporal window of increased transcriptional noise and increased signaling responsiveness preceding fate bifurcation (Semrau et al., 2016). scRNA-Seq of 4950 mESCs differentiated by manipulation of growth factors or directly converted by the ectopic expression of neural-promoting transcription factors documented different lineage paths that both converged on the same motor neuron fate (Briggs et al., 2017). Both of these studies explored directed differentiation towards neural fates using adherent culture systems. In contrast, many differentiation protocols begin with the formation of non-adherent aggregates of cells called embryoid bodies (EBs). EBs, in many ways, recapitulate representative events of early embryogenesis, including gastrulation (Doetschman et al., 1985). For example, EBs form a primitive streaklike structure with migrating cells that express Brachyury, and the expression of Fgf8, Wnt3, and Nodal, genes integral to gastrulation signaling pathways, are upregulated (Murry and Keller, 2008). Thus, while the directed differentiation of mouse ESCs as EBs can approximate several aspects of early development, it has yet to be characterized using high-throughput scRNA-Seq.
Here, we use scRNA-Seq to address several remaining questions of the EB-based differentiation system. For example, to what extent do pluripotent cells remain in differentiating EBs? To what extent is the detection of signatures of multiple, distinct lineages (from bulk profiling) attributable to population heterogeneity versus transient hybrid intermediates? What becomes of primitive streak stage cell populations at later stages of in vitro differentiation, and what are the candidate regulators of these processes? Here, we address these questions via scRNA-Seq of EBs at four days post-induction of differentiation in the primitive streak-promoting conditions of exogenous Wnt, Activin, and Noggin, and two days later upon further differentiation in bFGF. Our computational analysis of the scRNA-Seq data and comparison to data derived from the early embryo has revealed the distinct populations that emerge in this experimental context, the regulators of their further differentiation, and how they compare to in vivo populations of the gastrulating embryo.
2. Materials and methods
2.1. Cell maintenance and differentiation
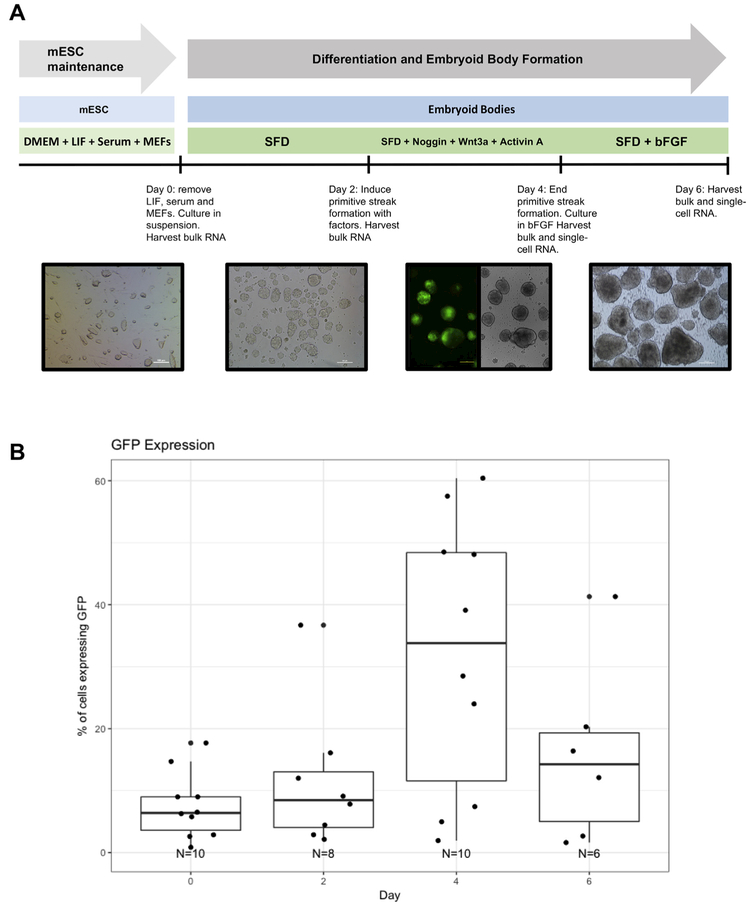
GFP-Brachyury (Bry) reporter mESCs (Gadue et al., 2006) were maintained on mouse embryonic fibroblast (mEF) feeder cells in Dul-becco's Modified Eagles' Medium (DMEM; Gibco) containing 15% FBS (Sigma-Aldrich), 1% PSG (Gibco), LIF (MTI Global Stem; 1000 U/mL), and 2-mercaptoethanol (Sigma; 0.1 mM). mESCs were passaged every other day via trypsinization (TrypLE; Gibco) and complete dissociation of colonies by pipetting. To begin differentiation (day 0), cells were trypsinized, removed from feeder cells, and cultured in suspension in serum-free differentiation media (SFD; Craft et al., 2013) at a density of 75,000 cells/mL for 48 h to form embryoid bodies (EBs). At this point (day 2), we induced primitive streak formation by culturing EBs in SFD containing the following growth factors and inhibitors: Noggin (150 ng/mL), Wnt3a (25 ng/mL), and Activin A (9 ng/mL). After the 48-hour primitive streak induction (day 4), EBs were removed from primitive streak-inducing factors and cultured in SFD containing bFGF (10 ng/mL) for an additional 48 h. This differentiation protocol was adapted from Craft et al., with the following modifications: mESCs were maintained on mEFs in the presence of serum (as opposed to serum-free and feeder-free culture) prior to the start of differentiation, primitive streak induction was maintained for 48 h instead of 24 h, and EBs were not dissociated and re-aggregated after primitive streak induction. All growth factors were purchased from R&D Systems.
2.2. GFP quantification and bulk RNA collection and sequencing
We collected bulk RNA samples from small sub-populations of the ESCs or EBs at days 0, 2, 4, and 6 throughout the differentiation process. To collect RNA, we first dissociated the EBs to single cells by incubating them in TrypLE for 2 min at 37°C and then vortexing them briefly. Total RNA was extracted from the counted cells using the Sigma GenElute Mammalian Total RNA kit, and then prepared for sequencing according the TruSeq Stranded mRNA Sample Preparation Guide (Part # 15031047 Rev. E). Samples were sequenced on the Illumina MiSeq platform.
2.3. 10 × single cell sequencing
We performed single-cell RNA sequencing on day 4 and day 6 EBs. Single-cell samples were prepared by dissociating EBs with TrypLE as described previously and straining them with a 30 μm cell strainer (Miltenyi Biotec). The strained cells were washed twice with dPBS containing 0.04% bovine serum albumin (BSA, Sigma) and then counted. The concentration of the cells was adjusted to 450–500 cells/μL and then 10,000 cells were loaded onto the 10× Chromium for isolation and pairing with oligo-coated beads, as described previously (Zheng et al., 2017). Briefly, single cells were isolated in droplets and lysed, and mRNA was reverse transcribed, primed off of the bead-conjugated oligo which includes both a bead-specific sequence and a unique molecular identifier. Then droplets were pooled, and cDNA was amplified and subjected to high-throughput sequencing. Libraries from our two samples were sequenced at a depth of 476 million reads. The 10× Genomics CellRanger pipeline (version 2.0) was used to align reads to the reference genome (mm10). Reads were assigned to individual cells based on barcode sequences, and gene expression levels were estimated based on UMI counts (Zheng et al., 2017). The MAGIC algorithm was used to correct for drop out using the following parameter settings number of principle components = 15, k = 15, ka = 5, t = 10 (van Dijk et al., 2017).
2.4. Bioinformatics
2.4.1. Bulk RNA-Seq
Reads from bulk RNA-Seq data were analyzed as previously described (Radley et al., 2017). In short, reads were trimmed and pseudo aligned to the mouse transcriptome with Salmon (Patro et al., 2017). Gene expression estimates were obtained by summing counts of transcripts that map to common canonical genes, and estimates were then normalized by down-sampling to 100,000 mapped reads per sample, and then transformed by taking the natural logarithm of the normalized read counts plus 1. We used the bulk RNA-Seq CellNet resource cnProc_MM_RS_Oct_24_2016.rda from https://github.com/pcahan1/CellNet for classification and gene regulatory network status analysis. Genes that were preferentially expressed at each time-point were identified using the Template Matching method, which tests for an association between each profile and an artificial profile that represents an ideal, cluster- or condition-specific, profile using the Pearson's product moment correlation coefficient (Pavlidis and Noble, 2001). Nominal P values were corrected for multiple testing using Holm's method (Holm, 1979). Gene sets with higher or lower expression in each cluster than expected by chance were identified by using the gene set enrichment analysis (GSEA) (Subramanian et al., 2005) as implemented in the fgsea package (Sergushichev, 2016) with Benjamini-Hochberg-corrected p-values < .05.
2.4.2. Clustering and analysis of scRNA-Seq data
We used our singleCellNet tool to cluster the day 4 and day 6 EB cells separately (manuscript in preparation). singleCellNet performs dimension reduction on the most variable genes by Principle Component Analysis (PCA), then uses hierarchical clustering and cutree (Becker et al., 1988), partitioning among medoids (PAM) (Kaufman and Rousseeuw, 1990), density peak finding (Rodriguez and Laio, 2014), and optionally, mclust (Scrucca et al., 2016) to assign cells to distinct clusters. singleCellNet selects the clustering that maximizes a metric of cluster quality, the average silhouette (Rousseeuw, 1987). Using this pipeline on all cells finds the large-scale structure of the data, however, we have observed that large, single cell clusters often contain sub-types or sub-states that are obscured in contrast to the stark differences between widely divergent cell types. To enable the simultaneous detection of diverse cell types, and sub-states therein, we designed singleCellNet to iteratively apply the dimension reduction/clustering procedure on each cluster for either a pre-specified number of iterations, or until the quality of the clustering results degrades significantly, as measured by the average silhouette. Application of this method to the day 4 EB data identified eight clusters, and application to the day 6 EB data identified ten clusters. Day 4 and Day 6 clusters were compared by first averaging the expression profile of all cells within each cluster to obtain mean cluster profiles. Then, we computed the Euclidean distance between each Day 4 and Day 6 cluster. To derive Fig. 7, we drew an edge from the day 6 cluster to the day 4 cluster with the minimal distance. GSEA of gene signatures from Gene Ontology (GO), mouse embryo (Mohammed et al., 2017; Peng et al., 2016) and human embryoid body data (Han et al., 2018) sets was performed as described above in the bulk RNA-Seq section. To re-analyze the early embryo scRNA-Seq data, we downloaded the QC-filtered gene count file count_table_QC_filter-ed.txt from GEO accession GSE100597. We then applied the single-CellNet clustering function to the 588 single cells in this data set, followed by template matching to identify genes preferentially expressed in each of the seven resulting clusters. To identify mouse orthologs for the human EB gene sets, we used the DRSC integrative ortholog prediction tool (DIOPT; http://www.flyrnai.org/cgi-bin/DRSC_orthologs.pl). The number of orthologs identified by this process are listed in Supp Table 2.
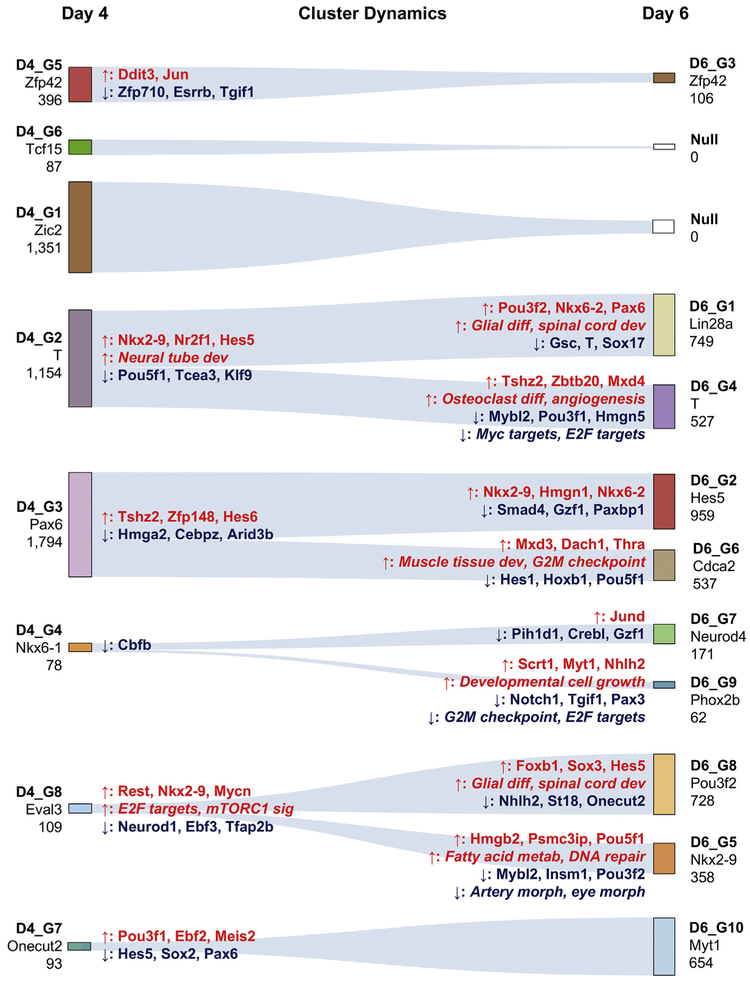
Fig. 7.
Cluster dynamics. Euclidean distances between D4 and D6 clusters were used to identify the most likely origin of each D6 cluster. A line was drawn from each D6 cluster to the closest D4 cluster. The number under the cluster name represents the number of cells in that cluster. The gene named under the cluster name is a gene that is representative of the population. Genes and pathways listed on the left (closest to day 4 clusters) are differentially expressed or enriched versus the derived D6 clusters. Arrows indicate direction of change. For example, Nkx2–9 is higher in D4_G2 versus both D6_G1 and D6_G4. Similarly, genes and pathways listed on the right (closest to day 6 clusters) are differentially expressed or enriched versus the originating D4 cluster. For example, Pou3f2 is up-regulated in D6_G1 (and not in D6_G4) versus D4_G2.
3. Results
3.1. In vitro differentiation captures major transcriptional states of in vivo E3.5-E6.5 embryogenesis
Monitoring T dynamics in EBs is possible with the GFP-Bry reporter mESC line (Gadue et al., 2006). We used this cell line to investigate the sequence of transcriptional states as pluripotent cells undergo early stages of differentiation as EBs. We subjected GFP-Bry mESCs to a differentiation protocol that promotes paraxial mesoderm formation (Craft et al., 2013). First, the pluripotent state is released by removing LIF, serum, and feeder cells for 2 days. Then, a stage analogous to the primitive streak is induced via exposure to Wnt3a and Activin A for 48 h. In addition, emergence of cardiac and hematopoietic fates is restrained by inhibiting BMP signaling with Noggin during induction. The differentiating cells are then removed from primitive streak-inducing factors and exposed to bFGF for an additional 48 h (Fig. 1A). During the course of differentiation, we monitored the dynamics of GFP-Bry by flow cytometry to ensure that a primitive streak-like population was induced. On day 4, approximately 32% (n = 10) of cells expressing GFP-Bry were detected by flow cytometry (Fig. 1B). Though the percentage of cells expressing GFP-Bry in these differentiation cultures was lower than previous reports (Craft et al., 2013; Gadue et al., 2006) the peak of GFP-Bry at day four and subsequent loss of expression is consistent with published findings and with the expected pattern during gastrulation.
Fig. 1.
Overview of the differentiation protocol. (A) Time course and methodology of differentiation protocol. (B) GFP-Bry quantification by flow cytometry (n = 1) and automated fluorescent cell counter.
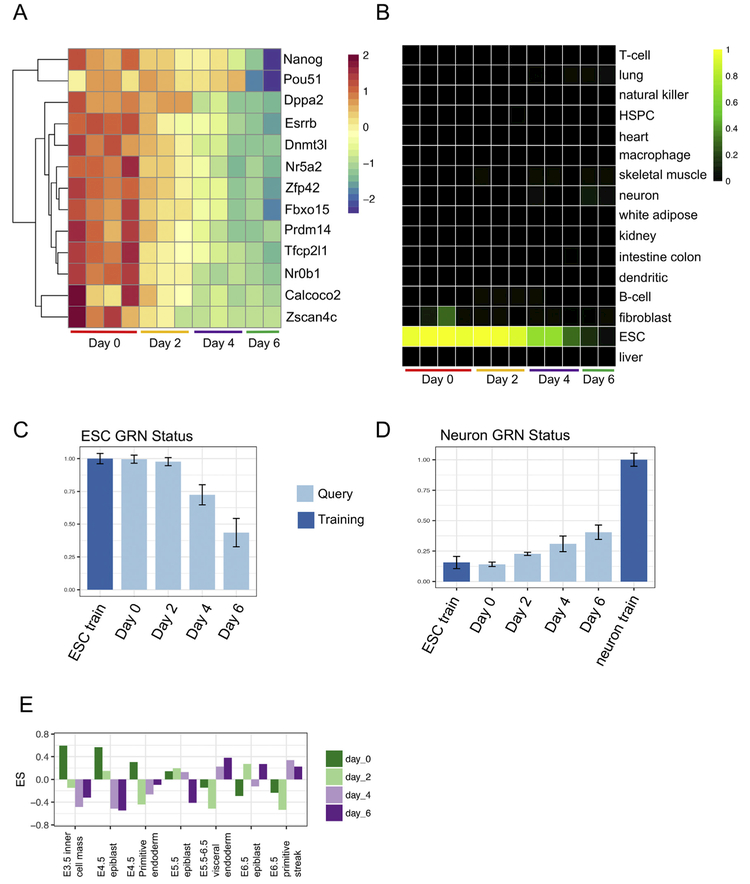
Next, we determined the transcriptional states of the cells from day 0 through day 6 of the differentiation. We first examined the expression of pluripotency marker genes, including Nanog, Pou5f1, and Esrrb, and confirmed that there was a gradual decrease, but not a complete loss of expression of genes from day 0 to day 6 (Fig. 2A). To gain a more global perspective on the identity of the cells, we subjected the RNA-Seq data to CellNet analysis (Cahan et al., 2014; Radley et al., 2017). Consistent with the analysis of individual pluripotency genes, CellNet classification analysis showed a gradual reduction in ESC classification through day 4 and total loss of this classification only at day 6 (Fig. 2B). Notably, cells at day 2 were still classified as ‘ESC’, consistent with their transition through a primed or post-implantation epiblast state (Tesar et al., 2007).
Fig. 2.
Bulk RNA analysis. (A) Heat map showing decreasing expression levels of genes associated with pluripotency for day 0–6 EBs. (B) CellNet classification analysis of bulk RNA samples at days 0,2,4 and 6 showing decreasing ESC classification.(C) Barplot showing reduction of ESC GRN status to 50% by day 6. (D) Barplot showing increase in Neuron GRN status through day 6 of the protocol. Shows to what extent the GRNs of the samples look like the GRN of the neuron training. (E) Barplot showing enrichment of gene sets related to different days of embryonic development in our bulk RNA samples of day 0–6 EBs.
In addition to cell and tissue type classification, CellNet also quantifies the extent to which Cell/Tissue-specific (C/T-specific) gene regulatory networks (GRNs) are established, a metric termed ‘GRN status’. The GRN status is more sensitive than the classification analysis, and thus it is able to detect intermediate or hybrid profiles that may reflect progenitor states (Kumar et al., 2017; Morris et al., 2014). Indeed, the ESC GRN status tracked the ESC classification with reduced (i.e. 50%) yet detectable ESC GRN status in the day 6 populations (Fig. 2C). By surveying the status of all fifteen other C/T GRNs, we found that only the neuron GRN status increased over the time course by > 2-fold at day 6 as compared to day 0 (Fig. 2D). Since the average proportion of GFP+ (and thus T+ mesendoderm) cells we achieved was 32% (Fig. 1B), we suspect the partial establishment of the neural GRN was likely due to the addition of the BMP inhibitor, Noggin, causing neural induction of the non-primitive streak-like cells (Chambers et al., 2009). Based on the observations that epiblast cells that do not migrate through the primitive streak are fated for the ectoderm (Lawson et al., 1991; Tam et al., 1993) and that differentiating ESCs have a tendency towards neuroectoderm-like derivatives (Muñoz-Sanjuán and Brivanlou, 2002), we speculate that the neural signature we observed arose from neural precursor-like cells that either did not transit through a T+ phase, or that transited through a neuromesodermal progenitor-like phase (Gouti et al., 2014).
Next, we asked which developmental stage differentiating GFP-Bry cells most resembled. To achieve this, we performed GSEA using gene signatures defined by analyzing previously reported scRNA-Seq of cells from four stages of early embryogenesis ranging from pre-implantation inner cell mass (ICM) of E3.5 through E6.5 (Mohammed et al., 2017). We re-analyzed this data and identified seven distinct clusters (Supp Fig. 1A). These clusters corresponded to the originally-described groups annotated as E3.5 ICM (high Calcoco2 expression), E4.5 epiblast (high Notum expression), E5.5 primitive endoderm (high Sox7 expression), E5.5 epiblast (high Aire expression), E5.5-E6.5 visceral endoderm (high Ang expression), E6.5 epiblast (high Zic5 expression), and E6.5 primitive streak (high Foxc2 expression) (Supp Fig. 1B). We call these gene sets the ‘chronological early embryo signatures’. We then performed GSEA (Mootha et al., 2003; Subramanian et al., 2005) to determine the extent to which each stage of directed differentiation up-regulated the early embryo signatures. We found that the day 0 population was significantly enriched in the E3.5 ICM and the E4.5 epiblast signatures, whereas the day 6 population was significantly enriched in the E5.5–6.5 visceral endoderm signatures and E6.5 epiblast signatures (Fig. 2E). Lastly and as expected due to the high expression of GFP-Bry, the day 4 population had the greatest enrichment in the E6.5 primitive streak signatures.
3.2. Single cell RNA-seq of day 4 embryoid bodies
Collectively, the results of the bulk RNA-Seq indicate that the broad transcriptional signatures of early embryogenesis are detectable and upregulated in EB-mediated directed differentiation. However, the bulk nature of RNA-seq limits the ability to assess heterogeneity within a sample. For example, it is difficult to parse the extent to which pluripotent cells remain in EBs and to what extent the partial establishment of the neuron GRN is due to population heterogeneity. Moreover, the extensive plasticity of the epiblast and gastrulating embryo between E4.5-E6.5 (Tam and Behringer, 1997) suggests that partially specified individual cells may still harbor latent expression signatures of diverse fates. To what extent can we detect this phenomenon in EBs? To address these questions, we subjected day 4 EB-derived cells to 10× Genomics droplet-based single cell capture and RNA sequencing (see Methods). Of the approximately 10,000 cells that were loaded into the Chromium, sequencing libraries from 5062 barcodes were determined by Cell-Ranger to reflect single cells. This includes post-Cell Ranger filtering to exclude the 5% of cells with the highest number of UMIs to exclude profiles that are more likely to include RNA from more than one cell, and to exclude cells with fewer than 1000 UMIs.
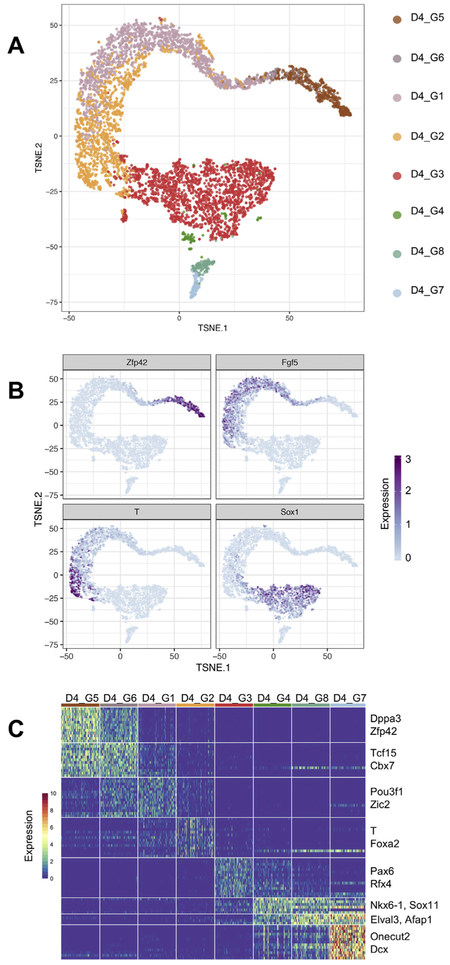
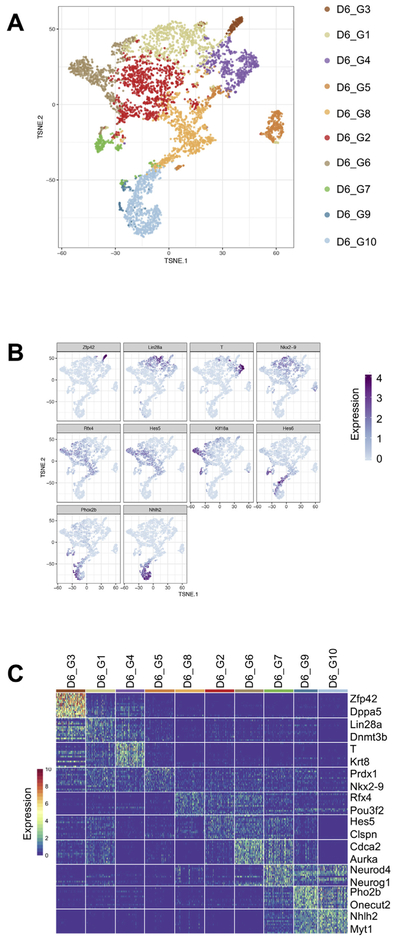
We used our singleCellNet computational platform to cluster the day 4 cells identifying eight distinct clusters (manuscript in preparation). We visualized these clusters using t-distributed stochastic neighbor embedding (t-SNE) (Van Der Maaten and Hinton, 2008), which shows the global structure of the day 4 cells (Fig. 3A). As an initial attempt to determine the lineages present in the day 4 clusters, we examined the expression of genes characteristic of pluripotent cells (Zfp42), epiblast stem cells (Fgf5), neuroectoderm (Sox1), and primitive streak (T) (Fig. 3B). Consistent with the bulk CellNet analysis, which showed both ESC classification and neural GRN status increase, we observed both ground state and Fgf5-expressing EpiSC-like cells in the day 4 EBs, and Sox1 neuroectoderm progenitors. We detected T in 303 (6%) day 4 cells. There are several possible contributors to the apparent discrepancy between flow cytometry and scRNA-Seq estimates of %GFP and T expression. One contributor is likely the longer half-life of protein as compared to mRNA. GFP has a half-life around 26 h (Corish and Tyler-Smith, 1999), whereas an mRNA transcript has an average half-life around 7.1 h (Sharova et al., 2009). Another contributor is that the sensitivity of droplet scRNA-seq is estimated to be 10–20% (Macosko et al., 2015; Zheng et al., 2017). If T is indeed expressed in ~30% of the cells that we sequenced at day 4 but, due to ‘drop out’ (Grün et al., 2014) is detected in only 10–20% of these cells, then we would expect to detect T in approximately 300–600 of the cells (3 to 6%).
Fig. 3.
Clustering and marker gene identification of day 4 EBs. (A) tSNE visualization of 8 distinct clusters in day 4 EBs. (B) Visualization of the expression of a marker genes for pluripotent cells (Zfp42), epiblast stem cells(Fgf5), neuroectoderm (Sox1), and primitive streak (T). (C) Heatmap of lineage marker gene expression for each of the eight day 4 clusters.
Next, we identified genes that were more highly expressed in one cluster as compared to all other clusters. This analysis confirmed the marker-driven inspection of gene patterns described above, and the identification of biologically relevant genes specific to each cluster (Fig. 3C). Clusters D4_G5 and D4_G6 were characterized by high levels of pluripotency genes such as Zfp42, Dppa3, Tcf15, and Cbx7, the ESC-specific Polycomb repressive complex 1 (PRC1) member (O'Loghlen et al., 2012). Cluster D4_G1 contains genes characteristic of post-implantation epiblast cells and EpiSCs such as Fgf5 (Khoa et al., 2016), Dnmt3b (Watanabe et al., 2002), and Pou3f1 (Song et al., 2016). Cluster D4_G2 contains genes indicative of the primitive streak stage gastrula including T and Fgf8 (Mikawa et al., 2004). Cluster D4_G3 contains the neuroectoderm marker Sox1 (Bylund et al., 2003; Kan et al., 2004). The remaining three clusters, D4_G4, D4_G8, and D4_G7 were marked by upregulation of more specified neural lineages such as Nkx6-1 (Qiu et al., 1998), Lhx1 (Avraham et al., 2009), and Lhx5 (Zhao et al., 1999).
To more comprehensively characterize the day 4 EB populations, we performed differential gene expression analysis and GSEA of biological processes relevant to development (Supplemental Table 1), of gene signatures indicative of distinct cell populations of the early embryo (as described in the section in vitro differentiation captures major transcriptional states of in vivo E3.5-E6.5 embryogenesis), and of gene signatures indicative of distinct regions of the E7.0 embryo (Peng et al., 2016). Below we describe the results of these analyses for the major clusters.
3.3. Day 4 EBs contain three distinct populations of pluripotent stem cells
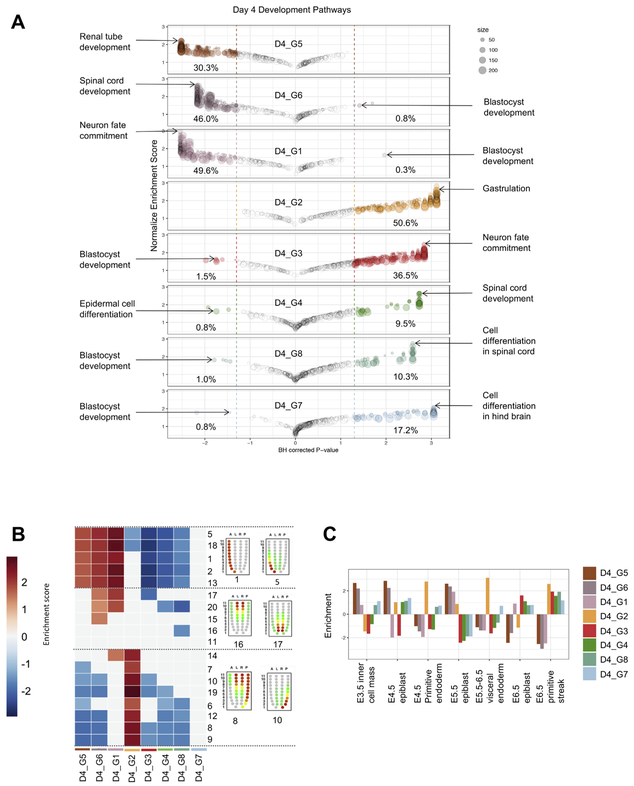
Based on the ESC GRN status of the bulk RNA-Seq data, we expected that either all cells at day 4 had reduced, but not extinguished, expression of pluripotency genes or that only a subset of the population of cells retained this signature. To discriminate between these two possibilities, we examined the expression of Zfp42 (a.k.a Rex1), a regulator of the naive pluripotent state in mouse PSCs (Son et al., 2013). We found that Zfp42 was expressed in D4_G5 (n = 396 cells), and to a lesser extent in D4_G6 (n = 87 cells) (Fig. 3B). Moreover, other pluripotency-related factors such as Pou5f1, Nanog, Tbx3, Dppa3, and Essrb were expressed in a similar pattern, suggesting that these clusters of cells maintained the pluripotency transcriptional network. GSEA of GO categories associated with development and differentiation showed that in D4_G5, D4_G6, and D4_G1 30–49% of all development categories were lower than expected by chance, suggesting an active repression of differentiation programs in these clusters (Fig. 4A). Surprisingly, when we performed GSEA for cell cycle-related gene sets, we found meiosis-related sets to be enriched in both D4_G5 and D4_G6, including genes such as Sycp1 and Sycp3 (Supp Fig. 4). There is a transcriptional overlap between primordial germ cells and ESCs (Mise et al., 2008), and ectopic depletion of Max, Smarca4, Mga, or Atf7ip in mESCs de-represses PGC-related genes such as Sycp3 (Maeda et al., 2013). We speculated that cells in clusters D4_G5 and D4_G6 may have lost expression of one of these genes, thus allowing for de-repression of the PGC/meiosis related transcriptional program. Indeed, we found that this was the case for Smarca4 expression. Our analysis of signaling pathways revealed a consistent downregulation of the BMP pathway and a substantial lack of enrichment of signaling pathways (Supp Fig. 2A), consistent with the observation that BMP is downregulated in the ground state (Boroviak et al., 2014) and that overall this state represents a state of relative signaling unresponsiveness (Kumar et al., 2014).
Fig. 4.
GSEA of day 4 EBs. (A) Bubble chart representing enrichment score and adjusted p-values for 389 developmental gene sets in each of the 8 clusters. Bubble size represents the number of genes in that gene set. Percentages represent the percent of gene sets significantly up-regulated or down-regulated in each cluster. Two gene sets of interest were labeled for each cluster. (B) Heatmap indicating which clusters are enriched in gene sets that represent different spatial regions of a developing embryo. Red/Blue squares indicate significant enrichment scores. (C) Bar plot indicating which clusters are enriched in gene sets related to different stages of embryo development. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Recently, two non-proliferating, dormant states in mouse pluripotent stem cells have been described. One of the dormant states emerges during serum-free neural differentiation and is dependent on Foxo3 expression (Ikeda and Toyoshima, 2017). The other is a dormancy induced by Myc depletion, with a resulting molecular profile that resembles E4.5 diapaused epiblasts (Takahashi and Yamanaka, 2006). We examined the expression of Foxo transcription factors and Myc to explore whether the D4_G5 and D4_G6 clusters resembled either of these previously described pluripotent states. We found that, consistent with the Myc-mediated diapause state, D4_G5 and D4_G6 had no detectable levels of Myc expression (Supp Fig. 3). While Foxo1 and Foxo3 were detected only sporadically in the pluripotent clusters (Supp Fig. 3), it is possible that both the Foxo and Myc pathways contributed independently to the acquisition of this state. Moreover, Foxo3 regulation of a dormant state was detected previously at later stages of directed differentiation, so it is possible that D4_G5 and D4_G6 precede this state. We also found that both D4_G5 and D4_G6 were enriched in genes related to glutathione metabolism (Supp Fig. 5A), consistent with the observation that in the ground state PSCs increase glutamine catabolism as a means to maintain lower levels of global epigenomic methylation (Carey et al., 2015), supporting the notion that these clusters were either residual PSCs or cells that had transiently moved through a primed but not fully committed stage prior to regressing to a pluripotent state.
Next, we asked what developmental stage each cluster most reflected by comparison to the single cell profiles described above. We found that D4_G5 and D4_G6 were significantly enriched in E3.5 ICM, E4.5 epiblast, and E5.5 epiblast signatures, whereas D4_G1 was only enriched in the E5.5 signature (Fig. 4C).
Finally, we sought to determine the region in the embryo that each cluster most resembled (Fig. 4B). We performed enrichment analysis using all 20 patterns of expression defined in the iTranscriptome database, which was derived by RNA-Seq of discrete sectors of the E7.0 epiblast (Peng et al., 2016). All three of these pluripotency-related clusters where enriched in signatures derived from the anterior region of the embryo and included genes such as Utf1. This observation is consistent with the observation that at the mid-gastrulation stage the un-patterned anterior region of the embryo would be expected to most resemble temporally earlier and less specified time-points of embryogenesis.
Taken together, these observations suggest that D4_G5 and D4_G6 cells represent ground state PSCs, whereas D4_G1 represents a post-implantation primed PSC state.
3.4. T-expressing cells in Day 4 EBs are similar to mesendoderm or axial mesoderm cells
In the embryo, T is expressed in nascent embryonic mesoderm, the node and notochord, and posterior neuroectoderm and is also a classical marker of the primitive streak (Inman and Downs, 2006). We found that T expression was confined almost exclusively to cluster D4_G2 (272/1154 or 24%). T was also sporadically expressed in the primed pluripotent cluster D4_G1 (25/1351 or 2%), consistent with previous reports of T expression in EpiSC (Song et al., 2016), and it was also rarely detected in cluster D4_G3 (6/1794 or < 1%). To better understand the biological pathways active in the T-expressing cluster D4_G2, we examined the gene set enrichment results. In contrast to the pluripotent clusters, D4_G2 was enriched in 50% of the development associated categories, with ‘Gastrulation’, ‘Endoderm’, and ‘Mesoderm’ as some of the most highly enriched gene sets (Fig. 4A), consistent with the notion that the cells in this cluster resemble mesoderm or mesendoderm cells of the primitive streak-stage embryo. In addition to T, several other markers of mesoderm (e.g. Mixl1 and Eomes) and endoderm (e.g. Foxa2, and Cer1) were co-expressed in many of the D4_G2 cells, suggesting a similarity to the mixed mesoderm/endoderm progenitor that can give rise to definitive endoderm and anterior mesoderm derivatives, and is referred to as the mesendoderm (Tada et al., 2005). D4_G2 cells also expressed genes associated with the mesendoderm, including Chrd, Gsc, and Lhx1 (Tada et al., 2005). To further understand these cells, we examined the results of the other enrichment analysis of the other categories. In contrast to the pluripotent clusters, D4_G2 was not enriched in any cell cycle category (Supp Fig. 4), and the sole metabolic category that was enriched (Multicellular organism metabolic process) precludes a meaningful interpretation (Supp Fig. 5). The most significantly enriched signaling pathway was “Positive regulation by Smoothened” (Supp Fig. 3). This result is biologically relevant as the Hedgehog pathway, of which Smoothened or Smo is the effector, is active in late streak-stage anterior mesendoderm (Echelard et al., 1993). Finally, we assessed whether the chronological early embryo signatures and spatial pattern signatures were enriched in the D4_G2 cluster. We found that this cluster was significantly enriched in E4.5 primitive endoderm, E5.5–6.5 visceral endoderm, and E6.5 primitive streak, and in spatial patterns from the posterior of the E7.0 embryo (Fig. 4B-C). Even though the timing of primitive streak formation in EB differentiation differs from that of in vivo development (day 4 for EBs vs. day 6.5 in vivo), D4_G2 still showed similarities to E6.5 primitive streak where compared to the chronological embryo signatures. Taken together, these results strongly support the notion that D4_G2 cells bear the transcriptional hallmarks of mesendoderm cells that are found in the posterior primitive streak in mid-to-late primitive streak-stage embryos.
3.5. Day 4 EBs contain a neuroectoderm population and three neural populations
The final four populations had high levels of expression of well-established neuroectoderm and more specialized neural progenitors. For example, the cluster D4_G3 expressed Sox1, a neuroectoderm specific gene, while D4_G4 expressed Foxn4, a regulator of both retinal cell diversity and spinal cord differentiation (Xiang and Li, 2013), D4_G8 expressed Neurog1, and D4_G7 expressed the pan-neuronal gene Myt1 (Matsushita et al., 2002, 2014). GSEA corroborated this gene-by-gene attribution of broad lineage to these clusters. The top developmental categories enriched in these clusters were “Neuron fate commitment” in D4_G3 and “Spinal Cord development” and “Neural retina development” in each of the D4_G4, D4_G8, and D4_G7 clusters (Fig. 4A). From a spatial and temporal perspective, these three clusters most resemble pre-patterned epiblast rather than posterior embryo (Fig. 4B-C). Although we used a directed differentiation protocol designed to promote mesoderm, we observed a substantial number of neural progenitors. This is not, however, completely unexpected for the following reasons. D2 EBs were treated with Noggin, a BMP pathway antagonist, in order to reduce lateral plate mesoderm progenitors such as those that give rise to hematopoietic and cardiac progenitors. However, BMP inhibition likely promoted uncommitted cells that did not undergo primitive streak induction in these cultures towards the neuroectoderm lineage. It is likely that Noggin also served to induce presumptive neuroectoderm and thus resulted in the neural populations that we observed.
3.6. A subset of Day 6 EBs have a pluripotent stem cell signature
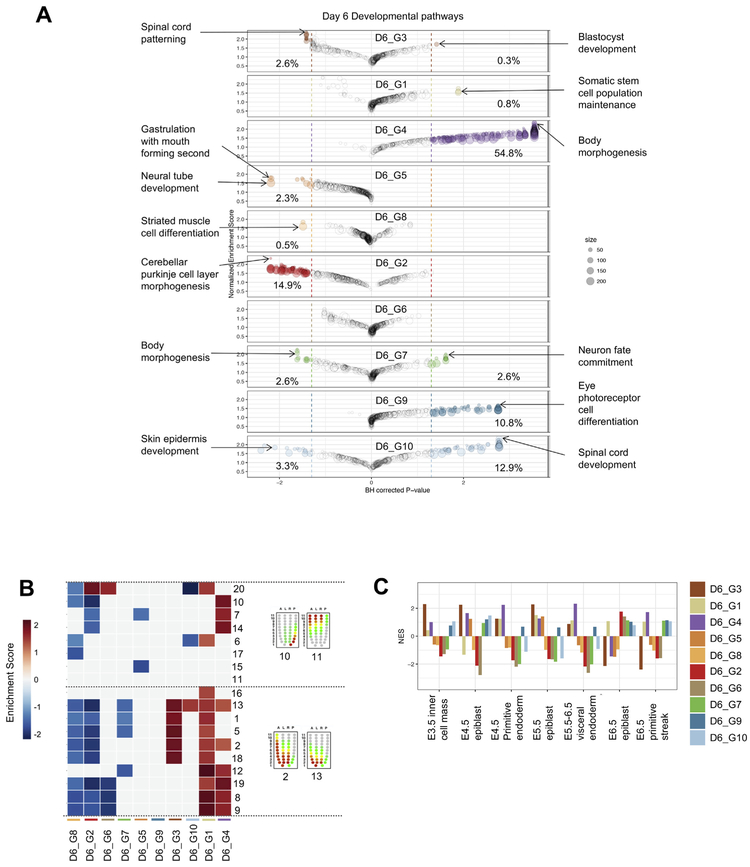
Next, we sought to determine how the distinct D4 populations changed and further diversified over time in the differentiation cultures. To achieve this, we performed scRNA-Seq on D6 cells (n = 4482). We used the same analysis procedure as described above to define distinct clusters of cells, to identify differentially expressed genes, and to identify enriched gene categories and embryonic signatures (Supplemental Table 2). We identified ten clusters (Fig. 5A) and sought to assign a putative and broad lineage identity to each by examining the expression of specific genes. D6_G3 cells expressed some ground state pluripotency genes including Zfp42 (Fig. 5B-C), however, in contrast to the D4 naive-like clusters, D6_G3 cluster did not have as substantial of an active repression of differentiation-related transcriptional programs (Fig. 6A). On the other hand, and similar to the D4 PSC clusters, D6_G3 was enriched in meiosis cell cycle gene sets (Supp Fig. 4B), enriched in E3.5 ICM, E4.5 epiblast, and E5.5 epiblast embryonic chronological signatures (Fig. 6C), and enriched in more anterior pre-patterned signatures of the E7.0 embryo (Fig. 6B). Collectively, these analyses suggested that the D6_G3 cluster was likely to have come from the D4_G5 and or D4_G6 pluripotent-like clusters. To explore this more thoroughly, we computed the Euclidean distance between the mean profiles of each cluster and determined the closest D4 cluster for each D6 cluster (Fig. 7). Based on this simple analysis, we did find that D4_G5 was closest to the D6_G3 cluster. We performed both differential gene expression and GSEA comparing these two clusters of cells to identify potential functional changes and the transcriptional mediators of these changes. In this comparison, we found that the transcriptional regulators Ddit3 and Jun were upregulated as the cells transitioned from D4_G5 to D6_G3 and down regulated both Zfp710, the ground state regulator Esrrb, and Tgif1, transcriptional repressor that inhibits the Tgf-ß and retinoic acid pathways (Yan et al., 2013). Taken together, this analysis revealed that the PSC-like cluster in day 4 is maintained (if somewhat diminished in its proportional representation) in day 6 rather than the alternative possibility that these cells enter a primed state. The persistence of PSC-like cells at day 6 is unlike in vivo development in that it is not possible to derive pluripotent stem cells from gastrulation- or later-stage embryos.
Fig. 5.
Clustering and marker gene identification of day 6 EBs (A) tSNE visualization of 10 distinct clusters in day 6 EBs. (B) Visualization of the expression of lineage marker genes. (C) Heatmap of lineage marker gene expression for each of the ten day 6 clusters.
Fig. 6.
GSEA of day 6 EBs. (A) Bubble chart representing enrichment score and adjusted p-value for 389 developmental gene sets in each of the 10 day 6 clusters. Bubble size represents the number of genes in that gene set. Percentages represent the percent of gene sets significantly up-regulated or down-regulated in each cluster. Two gene sets of interest were labeled for each cluster. (B) Heatmap indicating which clusters are enriched in gene sets representing different spatial regions of a developing embryo. Red/Blue squares indicate significant enrichment scores. (C) Barplot indicating which clusters are enriched in gene sets representing different stages of embryo development. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The comparison of D4 to D6 clusters also revealed that the two other PSC-related clusters (D4_G6 and D4_G1) had no matching derivative cluster at D6, implying either that these clusters gave rise to populations more similar to other D4 clusters, or that the cells in these clusters were not represented at D6 due to due to cell death or reduction of proliferation. Cell death may have contributed to D4_G6 cluster cells, which was enriched in the “Intrinsic apoptotic signaling pathway (Supp Table 1). Finally, based on our current data and analysis, the fate of the D4_G1 (or primed PSC) cluster is unclear and is a subject of further exploration.
3.7. Day 4 mesendoderm bifurcates to a Pax6+/T− population and a Krt9+/T+ population
In D6 EBs, only the D6_G4 cluster of cells maintained expression of T (Fig. 5B-C). Similar to the T+ D4_G2 cluster, D6_G4 was also enriched in differentiation pathways such as “Body morphogenesis” and moreover was enriched in “TGF-beta signaling” (Supp Fig. 2B), in E4.5 primitive endoderm, E5.5–6.5 visceral endoderm, and E6.5 primitive streak embryonic chronological signatures, and in posterior spatial signatures of the E7.0 embryo (Fig. 6B-C). Our comparison to the D4 profiles confirmed that D6_G4 was closest and thus most likely derived from the D4_G2 mesendoderm cluster (Fig. 7). A second D6 cluster, D6_G1, also had a similar global gene expression pattern to the D4_G2 mesoendoerm cluster. D6_G1 expressed high levels of Lin28a, enriched in “Somatic stem cell population maintenance” and “Folic acid metabolism” (Supp Fig. 5B), and was positive for the neural progenitor transcription factor Pax6 but negative for T. To better understand the genes driving this putative bifurcation, we first performed differential expression analysis to find the genes that were commonly changed between D4_G2 and the two D6 clusters together. Neural- and Notch- related genes were upregulated along with an enrichment of ‘Neural tube development’ genes, and with concomitant down-regulation of Pou5f1, Tcea3, and Klf9 expression. Next, we performed differential expression analysis to find genes and pathways that specifically changed between either D4_G2 and D6_G1 or between D4_G2 and D6_G4, but not both. We found that D6_G1 specifically down-regulated Gsc, T, and Sox17, while it up-regulated neural progenitor factors such as Pou3f2, Nkx6–2, and Pax6. Upregulation of these factors was not sporadic as both ‘glial differentiation’ and ‘spinal cord’ development categories were also enriched, indicating the execution of a coordinated transcriptional program. D6_G4 specifically up-regulated Tshz2 (homolog of the Drosphila homeotic patterning gene tsh), Zbtb20, and Mxd4, and was enriched in mesoderm-derivative lineage pathways ‘osteoclast differentiation’ and ‘angiogenesis’. The potential emergence of the neural-like D6_G1 from the mesendoderm-like D4_G2 suggests the day 4 EBs may harbor neuromesodermal-like progenitors, which appear during gastrulation under the influence of Wnt and FGF signaling, and which ultimately contribute to spinal cord and paraxial mesoderm (Gouti et al., 2014; Turner et al., 2014). Prospective isolation of the D4 populations and subsequent differentiation and analysis will help to resolve this intriguing possibility.
3.8. Most day 6 cells are neural-like
The majority of the day 6 clusters, comprising 4218 of the 4851 day 6 cells (87%), exhibited neural hallmarks, which is in stark contrast to cell compositions from normal embryos at analogous stages of development. D6_G5 expressed Nkx2–9, Rfx4 is broadly expressed in clusters D6_G8 through D6_G10, and indicators of more specialized neural cell types are expressed in D6_G7, D6_G9, and D6_G10 (Fig. 5B-C). Moreover, these clusters are enriched in neural categories such as “Spinal cord development” (D6_G10), “Eye photoreceptor cell differentiation” (D6_G9), and “Neuron fate commitment (D6_G7) (Fig. 6A). These neural-like clusters are not enriched in any of the embryonic chronological signatures, and are enriched in either a single pre-patterned spatial signature or none at all (Fig. 6B-C). The bottom of Fig. 7 illustrates the relationships between the D4 clusters and these clusters of neural populations, along with lists of the corresponding top regulators and pathways that are changed. We noticed a large increase in the number of neural-like cells in the D6 EBs and a corresponding decrease in mesendoderm-like cells and their derivatives. We speculate that the proliferative state of the neural progenitors at day 4 along with the apoptotic state of other cell populations at this time point allowed for the neural-like cells to become the most prominent population.
3.9. Comparison of mouse and human embryoid bodies
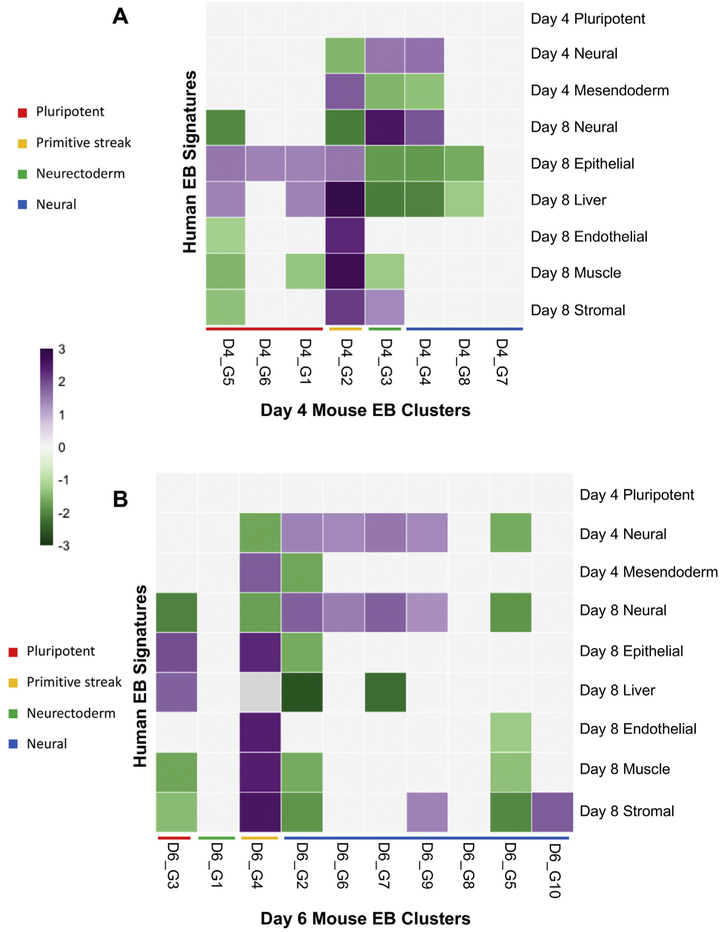
Many aspects of development are conserved across humans and mice (Gabdoulline et al., 2015); therefore, a comparison between species can be informative. Despite some phenotypic differences that exist when growing ESCs in vitro, there are many conserved molecular pathways. To better understand the similarities and differences between human and mouse ESC differentiation, we compared our data to that of differentially expressed genes obtained from day 4 and day 8 embryoid bodies differentiated from human pluripotent stem cells (hPSCs) (Han et al., 2018). After scRNA-seq, differentially expressed genes were defined for three progenitor cell types (neural, mesendoderm, and pluripotent) from day 4 EBs and six progenitor cell types (neural, epithelial, liver, muscle, endothelial and stromal) from day 8 EBs for a total of 9 gene sets. Using the mouse orthologs of these gene sets, we performed GSEA to compare our day 4 and day 6 EB clusters to the human EB gene signatures (Fig. 8A-B).
Fig. 8.
GSEA comparing mouse EBs to human EBs. (A) Heatmap showing gene set comparison of day 4 mouse EBs to day 4 and day 8 human EBs. (B) Heatmap showing gene set comparison of day 6 mouse EBs to day 4 and day 8 human EBs. Purple and green squares indicate significant enrichment scores. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
When comparing day 4 mouse EBs to day 4 human EBs we found that our D4_G2 cluster was highly enriched in genes representing mesendodermal cells types in human EBs (Fig. 8A). This finding parallel-sour previous assessment of the D4_G2 cluster where we propose that it resembles mesendoderm cells from the primitive streak. Neural signatures appeared in D4_G3 and D4_G4 which aligns with our previous assessment that these clusters represent neurectoderm and neural lineages respectively. In addition to neural genes, D4_G3 was also enriched in stromal (mesoderm-derived) genes as was D4_G2. In contrast to our previous findings of enrichment in spinal development and neural retina development genes, D4_G7 and D4_G8 did not show enrichment in neural genes when compared to day 4 or day 8 human EBs. When comparing day 4 mouse EBs to day 8 human EBs, D4_G2 showed enrichment for all progenitor cells types except neural. D4_G3 and D4_G4 were even more similar to day 8 neural progenitors than to the day 4 neural progenitors. Interestingly, D4_G1, D4_G2, D4_G5, and D4_G6 all showed enrichment in genes for epithelial progenitors, and all these clusters except D4_G6 showed enrichment for liver genes. In summary, D4_G2 primitive streak-like clusters were most enriched in genes highly expressed in day 8 human mesoderm- and endoderm-derived lineages. Lastly, D4_G3 most resembles day 8 human neural progenitors.
Next, we compared our day 6 EBs to the human EB data (Fig. 8B). D6_G4 was enriched in genes for mesendoderm and most of the day 8 progenitor types except neural and liver. This is consistent with the maintenance of T expression in D6_G4 and with our analysis that predicts this cluster is derived from the T-expressing cluster D4_G2, which showed similar patterns when compared to the human EB data. D6_G4 was also the only day 6 cluster to be enriched in mesendoderm genes. D6_G3, potentially derived from D4_G5, showed enrichment in epithelial and liver genes and repression of neural, muscle and stromal genes, similar to D4_G5. D6_G2, D6_G6, D6_G7, and D6_G9 showed enrichment in day 4 and day 8 neural genes. D6_G7 and D6_G9 had previously been identified as neural-like cells by GSEA. Lastly, D6_G10 showed enrichment in only stromal genes, which differs from our previous analysis where it showed enrichment for spinal cord genes. This finding may suggest that D6_G10 is similar to a neuromesodermal progenitor population due to the expression of neural and mesoderm genes. However, our Euclidean distance analysis suggested that D6_G10 was derived from D4_G7, which did not show enrichment for any of the human EB gene sets.
Overall, the comparison of mouse EBs to human EBs showed that our day 4 mesendoderm cluster resembles day 4 human EB mesendoderm. Additionally, our neural clusters resemble both day 4 and day 8 human EB neural progenitors. A few of our clusters (D4_G7, D6_G1, D6_G8) did not resemble any of the human EB cell types. There are many possible contributors to these differences, including the distinct differentiation conditions and the different times scales of mouse and human development where the first 10 days of mESC development correspond to first 21 days of hESC development (Gabdoulline et al., 2015).
4. Conclusions
Our data and analyses have characterized the transient cell populations that emerge during murine embryoid body differentiation using a protocol designed to favor mesoderm and endoderm formation. CellNet analysis of bulk RNA showed a decreasing ESC GRN status and an increasing neuron GRN status as differentiation progressed. At the same time, a comparison to transcriptomic data from early embryos showed that day 4 EBs are enriched in a gene set indicative of the primitive streak.
To better resolve the cell types that emerge in EB differentiation, we used scRNA-Seq to further investigate the heterogeneity of in vitro cell populations and we compared them to spatial and temporal profiles of embryos. In the day 4 EBs, we observed both ground state PSCs and post-implantation primed PSCs. T expression was confined almost completely to cluster D4_G2, a cluster that was enriched in 50% of developmental categories, suggesting that these cells represent mesoderm or mesendoderm cells from the primitive streak of an early embryo. Four D4 populations expressed markers of neuroectoderm and neural progenitors. These clusters showed upregulation of gene sets for the development of neural retina and spinal cord as well as neuron fate commitment.
Analysis of day 6 EBs showed that the D6_G3 cluster mostly likely came from the pluripotent-like cluster (D4_G5) because it expressed ground state pluripotency genes like Zfp42, it was enriched in meiosis cell cycle gene sets, and was enriched in anterior pre-patterned signatures of an E7.0 embryo. This was corroborated by our analysis comparing the average expression profiles of the day 4 and 6 clusters (Fig. 7). The remaining two PSC-like clusters at day 4 did not appear to have a matching derivative cluster at day 6. Only one day 6 cluster maintained T expression, while the majority of day 6 clusters were neural-like especially clusters D6_G7, D6_G9 and D6_G10 which were enriched in gene sets for neuron fate commitment, eye photoreceptor cell differentiation, and spinal cord development respectively.
There are several caveats to our findings. First, current scRNA-seq methods capture only 10–20% of the transcriptome. While there are computational methods to correct for dropout (van Dijk et al., 2017; Kwak et al., 2017; Li and Li, 2018), these methods are nascent and have yet to be rigorously and comprehensively validated. The sensitivity of scRNA-Seq is important in investigations of cell state and cell type heterogeneity where distinguishing transcriptional features either are of modest magnitude or are from lowly expressed genes (such as transcription factors). Therefore, it is possible that our analysis did not detect important genes in EB differentiation or did not completely resolve the distinct cell groups. Secondly, our clustering approach uses a heuristic to determine a stopping point and thus, independent of the sensitivity issue raised above, it may fail to fully resolve the cell groups. For example, one day 4 cluster (D4_G2), expressed both neurectoderm genes like Sox1 as well as T. When we re-clustered D4_G2 alone, it separated into three clusters with significantly different levels of T expression. Nonetheless, even at the broad level of clustering that we performed, we were able to draw meaningful conclusions about the cell types that emerge during embryoid body differentiation. In summary, we have presented data and analysis that will serve as a reference of the cell types that emerge during EB differentiation.
Supplementary Material
Acknowledgements
Many thanks to the members of the Cahan Lab for helping with this project. In particular, thank you, Remy Schwab and Stephanie Cai for assistance with data analysis, Emily Lo and Sydney Mason for assistance with the tissue culture procedures required for this study. PC was funded by R35GM124725 and K01DK096013. Bulk and single-cell sequence data are available at GEO under the super series accession number: GSE114220.
References
- Avraham O, Hadas Y, Vald L, Zisman S, Schejter A, Visel A, Klar A, 2009. Transcriptional control of axonal guidance and sorting in dorsal interneurons by the Lim-HD proteins Lhx9 and Lhx1. Neural Dev. 4, 21. 10.1186/1749-8104-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker RA, Chambers JM, Wilks AR, 1988. The New S Language: A Programming Environment for Data Analysis and Graphics. Wadsworth & Brooks/Cole. [Google Scholar]
- Boroviak T, Loos R, Bertone P, Smith A, Nichols J, 2014. The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat. Cell Biol. 16, 516–528. 10.1038/ncb2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs J, Li V, Lee S, Woolf C, Klein A, Kirschner MW, 2017. Mouse embryonic stem cells can differentiate via multiple paths to the same state. bioRxiv 1–31. 10.1101/124594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylund M, Andersson E, Novitch BG, Muhr J, 2003. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat. Neurosci. 6, 1162–1168. 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- Cahan P, Li H, Morris SA, Lummertz Da Rocha E, Daley GQ, Collins JJ, 2014. CellNet: network biology applied to stem cell engineering. Cell 158, 903–915. https://doi.Org/10.1016/j.cell.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey BW, Finley LWS, Cross JR, Allis CD, Thompson CB, 2015. Intracellular α- ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518, 413–416. 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L, 2009. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275–280. 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corish P, Tyler-Smith C, 1999. Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng. Des. Sel 12, 1035–1040. 10.1093/protein/12.12.1035. [DOI] [PubMed] [Google Scholar]
- Craft AM, Ahmed N, Rockel JS, Baht GS, Alman BA, Kandel RA, Grigoriadis AE, Keller GM, 2013. Specification of chondrocytes and cartilage tissues from embryonic stem cells. Development 140, 2597–2610. 10.1242/dev.087890. [DOI] [PubMed] [Google Scholar]
- Doetschman TC, Eistetter H, Katz M, Schmidt W, Kemler R, 1985. The In Vitro Development of Blastocyst-Derived Embryonic Stem Cell Lines: Formation of Visceral Yolk Sac, Blood Islands and Myocardium, vol. 87. pp. 27–45. [PubMed] [Google Scholar]
- Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, McMahon JA, McMahon AP, 1993. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell 75, 1417–1430. [DOI] [PubMed] [Google Scholar]
- Gabdoulline R, Kaisers W, Gaspar A, Meganathan K, Doss MX, Jagtap S, Hescheler J, Sachinidis A, Schwender H, 2015. Differences in the early development of human and mouse embryonic stem cells. PLoS One 10. 10.1371/journal.pone.0140803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadue P, Huber TL, Paddison PJ, Keller GM, 2006. Wnt and TGF-beta signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A 103, 16806–16811. 10.1073/pnas.0603916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouti M, Tsakiridis A, Wymeersch FJ, Huang Y, Kleinjung J, Wilson V, Briscoe J, 2014. In vitro generation of neuromesodermal progenitors reveals distinct roles for wnt signalling in the specification of spinal cord and paraxial mesoderm identity. PLoS Biol. 12. 10.1371/journal.pbio.1001937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grün D, Kester L, Van Oudenaarden A, 2014. Validation of noise models for single-cell transcriptomics. Nat. Methods 11, 637–640. 10.1038/nmeth.2930. [DOI] [PubMed] [Google Scholar]
- Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P, 2010. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell 18, 675–685. https://doi.Org/10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Han X, Chen H, Huang D, Chen H, Fei L, Cheng C, Huang H, Yuan G-C, Guo G, 2018. Mapping human pluripotent stem cell differentiation pathways using high throughput single-cell RNA-sequencing. Genome Biol. 19, 47. 10.1186/s13059-018-1426-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemberger M, Cross JC, Ropers HH, Lehrach H, Fundele R, Himmelbauer H, 2001. UniGene cDNA array-based monitoring of transcriptome changes during mouse placental development. Proc. Natl. Acad. Sci. U. S. A 98, 13126–13131. 10.1073/pnas.231396598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm S, 1979. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 6, 65–70. 10.2307/4615733. [DOI] [Google Scholar]
- Ikeda M, Toyoshima F, 2017. Dormant pluripotent cells emerge during neural differentiation of embryonic stem cells in a FoxO3-dependent manner. Mol. Cell. Biol. 37, e00417–16. 10.1128/MCB.00417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman KE, Downs KM, 2006. Localization of Brachyury (T) in embryonic and extra-embryonic tissues during mouse gastrulation. Gene Expr. Patterns 6, 783–793. https://doi.Org/10.1016/j.modgep.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Kan L, Israsena N, Zhang Z, Hu M, Zhao LR, Jalali A, Sahni V, Kessler JA, 2004. Sox1 acts through multiple independent pathways to promote neurogenesis. Dev. Biol. 269, 580–594. https://doi.Org/10.1016/j.ydbio.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Kaufman L, Rousseeuw PJ, 1990. Finding Groups in Data: An Introduction to Cluster Analysis. Wiley, New York. [Google Scholar]
- Khoa LTP, Azami T, Tsukiyama T, Matsushita J, Tsukiyama-Fujii S, Takahashi S, Ema M, 2016. Visualization of the epiblast and visceral endodermal cells using Fgf5-P2A-Venus BAC transgenic mice and epiblast stem cells. PLoS One 11, e0159246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispert A, Herrmann BG, 1994. Immunohistochemical analysis of the Brachyury protein in wild-type and mutant mouse embryos. Dev. Biol 161, 179–193. 10.1006/dbio.1994.1019. [DOI] [PubMed] [Google Scholar]
- Kumar RM, Cahan P, Shalek AK, Satija R, DaleyKeyser AJ, Li H, Zhang J, Pardee K, Gennert D, Trombetta JJ, Ferrante TC, Regev A, Daley GQ, Collins JJ, 2014. Deconstructing transcriptional heterogeneity in pluripotent stem cells. Nature 516, 56–61. 10.1038/nature13920.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Tan Y, Cahan P, 2017. Understanding development and stem cells using single cell-based analyses of gene expression. Development 144, 17–32. 10.1242/dev.133058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak I-Y, Gong W, Koyano-Nakagawa N, Garry D, 2017. DrImpute: imputing dropout events in single cell RNA sequencing data. bioRxiv 181479. 10.1101/181479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson KA, Meneses JJ, Pedersen RA, 1991. Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development 113, 891–911 (VL - 113). [DOI] [PubMed] [Google Scholar]
- Li WV, Li JJ, 2018. An accurate and robust imputation method scImpute for singlecell RNA-seq data. Nat. Commun. 9, 997. 10.1038/s41467-018-03405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA, 2015. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214. https://doi.Org/10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda I, Okamura D, Tokitake Y, Ikeda M, Kawaguchi H, Mise N, Abe K, Noce T, Okuda A, Matsui Y, 2013. Max is a repressor of germ cell-related gene expression in mouse embryonic stem cells. Nat. Commun. 4. 10.1038/ncomms2780. [DOI] [PubMed] [Google Scholar]
- Matsushita F, Kameyama T, Marunouchi T, 2002. NZF-2b is a novel predominant form of mouse NZF-2/MyT1, expressed in differentiated neurons especially at higher levels in newly generated ones. Mech. Dev. 118, 209–213. [DOI] [PubMed] [Google Scholar]
- Matsushita F, Kameyama T, Kadokawa Y, Marunouchi T, 2014. Spatiotemporal expression pattern of Myt/NZF family zinc finger transcription factors during mouse nervous system development. Dev. Dyn. 243, 588–600. 10.1002/dvdy.24091. [DOI] [PubMed] [Google Scholar]
- Mikawa T, Poh AM, Kelly KA, Ishii Y, Reese DE, 2004. Induction and patterning of the primitive streak, an organizing center of glastrulation in the amniote. Dev. Dyn 10.1002/dvdy.10458. [DOI] [PubMed] [Google Scholar]
- Mise N, Fuchikami T, Sugimoto M, Kobayakawa S, Ike F, Ogawa T, Tada T, Kanaya S, Noce T, Abe K, 2008. Differences and similarities in the developmental status of embryo-derived stem cells and primordial germ cells revealed by global expression profiling. Genes Cells 13, 863–877. 10.1111/j.1365-2443.2008.01211.x. [DOI] [PubMed] [Google Scholar]
- Mohammed H, Hernando-Herraez I, Savino A, Scialdone A, Macaulay I, Mulas C, Chandra T, Voet T, Dean W, Nichols J, Marioni JC, Reik W, 2017. Single-cell landscape of transcriptional heterogeneity and cell fate decisions during mouse early gastrulation. Cell Rep. 20, 1215–1228. https://doi.Org/10.1016/j.celrep.2017.07. 009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC, 2003. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267. [DOI] [PubMed] [Google Scholar]
- Morris SA, Cahan P, Li H, Zhao AM, San Roman AK, Shivdasani RA, Collins JJ, Daley GQ, 2014. Dissecting engineered cell types and enhancing cell fate conversion via Cellnet. Cell 158, 889–902. 10.1016/j.cell.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Sanjuán I, Brivanlou AH, 2002. Neural induction, the default model and embryonic stem cells. Nat. Rev. Neurosci. 3, 271–280. 10.1038/nrn786. [DOI] [PubMed] [Google Scholar]
- Murry CE, Keller G, 2008. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- O'Loghlen A, Muñoz-Cabello AM, Gaspar-Maia A, Wu HA, Banito A, Kunowska N, Racek T, Pemberton HN, Beolchi P, Lavial F, Masui O, Vermeulen M, Carroll T, Graumann J, Heard E, Dillon N, Azuara V, Snijders AP, Peters G, Bernstein E, Gil J, 2012. MicroRNA regulation of Cbx7 mediates a switch of polycomb orthologs during ESC differentiation. Cell Stem Cell 10, 33–46. 10.1016/j.stem.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C, 2017. Salmon provides fast and bias-aware quantification of transcript expression using dual-phase inference. Nat. Methods 14, 417–419. 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, Noble WS, 2001. Analysis of strain and regional variation in gene expression in mouse brain. Genome Biol. 2 10.1186/gb-2001-2-10-research0042. (RESEARCH0042). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G, Suo S, Chen J, Chen W, Liu C, Yu F, Wang R, Chen S, Sun N, Cui G, Song L, Tam PPL, Han JDJ, Jing N, 2016. Spatial transcriptome for the molecular annotation of lineage fates and cell identity in mid-gastrula mouse embryo. In: Developmental Cell. Elsevier Inc.. https://doi.Org/10.1016/j.devcel.2016.02.020. [DOI] [PubMed] [Google Scholar]
- Qiu M, Shimamura K, Sussel L, Chen S, Rubenstein JL, 1998. Control of anteroposterior and dorsoventral domains of Nkx-6.1 gene expression relative to other Nkx genes during vertebrate CNS development. Mech. Dev. 72, 77–88. [DOI] [PubMed] [Google Scholar]
- Radley AH, Schwab RM, Tan Y, Kim J, Lo EKW, Cahan P, 2017. Assessment of engineered cells using CellNet and RNA-seq. Nat. Protoc 12, 1089–1102. 10.1038/nprot.2017.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Pérez JA, Hadjantonakis AK, 2014. The dynamics of morphogenesis in the early mouse embryo. Cold Spring Harb. Perspect. Biol. 7, 1–18. 10.1101/cshperspect.a015867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Laio A, 2014. Clustering by fast search and find of density peaks. Science (80-.) 344, 1492 LP–1496. [DOI] [PubMed] [Google Scholar]
- Rousseeuw PJ, 1987. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 20, 53–65. 10.1016/0377-0427(87)90125-7. [DOI] [Google Scholar]
- Scialdone A, Tanaka Y, Jawaid W, Moignard V, Wilson NK, Macaulay IC, Marioni JC, Göttgens B, 2016. Resolving early mesoderm diversification through single-cell expression profiling. Nature 535, 4–6. 10.1038/nature18633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrucca L, Fop M, Murphy TB, Raftery AE, 2016. Mclust 5: clustering, classification and density estimation using Gaussian finite mixture models. R J. 8, 289–317. 10.1007/s00210-015-1172-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semrau S, Goldmann J, Soumillon M, Mikkelsen TS, Jaenisch R, van Oudenaarden A, 2016. Dynamics of lineage commitment revealed by single-cell transcriptomics of differentiating embryonic stem cells. bioRxiv, 068288. 10.1101/068288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergushichev A, 2016. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv. [Google Scholar]
- Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, Ko MSH, 2009. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 16, 45–58. 10.1093/dnares/dsn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son MY, Choi H, Han YM, Cho YS, 2013. Unveiling the critical role of REX1 in the regulation of human stem cell pluripotency. Stem Cells 31, 2374–2387. 10.1002/stem.1509. [DOI] [PubMed] [Google Scholar]
- Song L, Chen J, Peng G, Tang K, Jing N, 2016. Dynamic heterogeneity of Brachyury in mouse epiblast stem cells mediates distinct response to extrinsic bone morphogenetic protein (BMP) signaling. J. Biol. Chem 291, 15212–15225. 10.1074/jbc.M115.705418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. 102, 15545–15550. 10.1073/PNAS.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada S, Era T, Furusawa C, Sakurai H, Nishikawa S, Kinoshita M, Nakao K, Chiba T, Nishikawa S-I, 2005. Characterization of mesendoderm: a diverging point of the definitive endoderm and mesoderm in embryonic stem cell differentiation culture. Development 132, 4363–4374. 10.1242/dev.02005. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S, 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tam PPL, Behringer RR, 1997. Mouse gastrulation: the formation of a mammalian body plan. Mech. Dev 10.1016/S0925-4773(97)00123-8. [DOI] [PubMed] [Google Scholar]
- Tam PPL, Loebel DAF, 2007. Gene function in mouse embryogenesis: get set for gastrulation. Nat. Rev. Genet. 8, 368–381. 10.1038/nrg2084. [DOI] [PubMed] [Google Scholar]
- Tam PPL, Williams EA, Chan WY, 1993. Gastrulation in the mouse embryo: ul-trastructural and molecular aspects of germ layer morphogenesis. Microsc. Res. Tech. 26, 301–328. 10.1002/jemt.1070260405. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD, 2007. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199. 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Turner DA, Hayward PC, Baillie-Johnson P, Rué P, Broome R, Faunes F, Martinez Arias A, 2014. Wnt/β-catenin and FGF signalling direct the specification and maintenance of a neuromesodermal axial progenitor in ensembles of mouse embryonic stem cells. Development, 10.1242/dev.112979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Maaten L, Hinton G, 2008. Visualizing data using t-SNE. J. Mach. Learn. Res. 1 (620), 267–284. 10.1007/s10479-011-0841-3. [DOI] [Google Scholar]
- van Dijk D, Nainys J, Sharma R, Kathail P, Carr AJ, Moon KR, Mazutis L, Wolf G, Krishnaswamy S, Pe'er D, 2017. MAGIC: a diffusion-based imputation method reveals gene-gene interactions in single-cell RNA-sequencing data. bioRxiv 111591. 10.1101/111591. [DOI] [Google Scholar]
- Wang L, Chen Y-G, 2016. Signaling control of differentiation of embryonic stem cells toward Mesendoderm. J. Mol. Biol. 428, 1409–1422. https://doi.Org/10.1016/j.jmb. 2015.06.013. [DOI] [PubMed] [Google Scholar]
- Watanabe D, Suetake I, Tada T, Tajima S, 2002. Stage- and cell-specific expression of Dnmt3a and Dnmt3b during embryogenesis. Mech. Dev. 118, 187–190. 10.1016/S0925-4773(02)00242-3. [DOI] [PubMed] [Google Scholar]
- Xiang M, Li S, 2013. Foxn4: a multi-faceted transcriptional regulator of cell fates in vertebrate development. Sci. China Life Sci. 56, 985–993. 10.1007/s11427-013-4543-8. [DOI] [PubMed] [Google Scholar]
- Yan L, Womack B, Wotton D, Guo Y, Shyr Y, Dave U, Li C, Hiebert S, Brandt S, Hamid R, 2013. Tgif1 regulates quiescence and self-renewal of hematopoietic stem cells. Mol. Cell. Biol. 33, 4824–4833. 10.1128/MCB.01076-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Sheng HZ, Amini R, Grinberg A, Lee E, Huang SP, Taira M, Westphal H, 1999. Control of hippocampal morphogenesis and neuronal differentiation by the LIM homeobox gene Lhx5. Science (80-.) 284, 1155–1158. 10.1126/science.284.5417.1155. [DOI] [PubMed] [Google Scholar]
- Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ, Bielas JH, 2017. Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049. 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.