Abstract
Retinal gene therapy has had unprecedented success in generating treatments that can halt vision loss. However, immunogenic response and long-term toxicity with the use of viral vectors remain a concern. Non-viral vectors are relatively non-immunogenic, scalable platforms that have limited success with DNA delivery to the eye. Messenger RNA (mRNA) therapeutics has expanded the ability to achieve high gene expression while eliminating unintended genomic integration or the need to cross the restrictive nuclear barrier. Lipid-based nanoparticles (LNPs) remain at the forefront of potent delivery vectors for nucleic acids. Herein, we tested eleven different LNP variants for their ability to deliver mRNA to the back of the eye. LNPs that contained ionizable lipids with low pKa and unsaturated hydrocarbon chains showed the highest amount of a reporter gene transfection in the retina. The kinetics of gene expression showed a rapid onset (within 4 h) that persisted for 96 h. The gene delivery was cell-type specific with majority of the expression in the retinal pigmented epithelium (RPE) and limited expression in the Müller glia. LNP-delivered mRNA can be used to treat monogenic retinal degenerative disorders of the RPE. The transient nature of mRNA-based therapeutics makes it desirable for applications that are directed towards retinal reprogramming or genome editing. Overall, non-viral delivery of RNA therapeutics to diverse cell types within the retina can provide transformative new approaches to prevent blindness.
Keywords: lipid nanoparticle, retina, eye, retinal degeneration, mRNA, transfection efficiency
INTRODUCTION
The posterior eye contains the retina, a multi-layered tissue, comprised mostly of neurons and one layer of epithelial cells. Each retinal cell type plays an important role in visual processing and retinal maintenance. When proteins within these cells are dysfunctional, retinal function and health can be disturbed resulting in retinal degeneration [1]. Currently, retinal degeneration, arising from advanced glaucoma, atrophic macular degeneration, advanced diabetic retinopathy, and inherited retinal degenerations, affects millions of people worldwide. Gene-therapy based approaches have been widely investigated for the treatment of these disorders [2,3]. Recently, an adeno-associated virus (AAV)-based gene therapy, Vortigern Neparvovec (Luxturnatm) was approved by the FDA for the treatment of Leber Congenital Amaurosis (LCA), making subretinal delivery of AAV the new gold standard [4]. Despite these advances, AAVs have safety concerns associated with mutagenic integration and immune activation, limiting repeated administration [5]. In addition, their inability to deliver large transgenes (>5kb) restricts their broader application. There is a need to develop delivery systems that overcome these limitations and expand the utility of gene therapy for retinal degeneration.
Nanoparticle-based gene delivery to the eye has garnered attention as a potentially safer and highly modular alternative to viral gene delivery. Nanoparticles have evolved into various forms such as polyplexes, lipoplexes, lipid-nanoparticles (LNPs), dendrimers, inorganic nanoparticles, and hybrid nanoparticles [6,7]. They allow immense flexibility regarding engineering to achieve desirable physicochemical characteristics such as size, shape, surface chemistry, charge, interfacial properties, and hydrophobicity. As a consequence of the extensive varieties of chemical structures offered by nanoparticles, they can be specifically designed to overcome biological barriers such as cell entry and endosomal escape [8–12]. They make ideal carriers for the delivery of transgenes due to low immunogenicity and the ability to deliver large transgenes (>5kb). Their biodegradability can be optimized to reduce toxicity while maintaining efficacy [13–15]. The nanoparticle surface can also be decorated with targeting ligands for cell-specific delivery [16–18]. Moreover, nanoparticles are relatively cheaper to produce and easy to scale up for large-scale formulation.
The recent success of the LNPs in the clinic has made it the premier nanoparticle system for nucleic acid delivery. Many clinical trials are underway for RNA delivery using LNPs for the treatment of myriad disorders [19–21]. While many nanoparticles have been evaluated in the retina, to the best of our knowledge, few have shown potent nucleic acid delivery [22–26]. Current nanoparticle-based gene delivery in the eye has focused on using DNA as the therapeutic of choice. However, DNA delivery using nanoparticles is inherently inefficient due to the nuclear entry requirement in post-mitotic retinal cells. There is a pressing need to develop potent non-viral vectors which can generate therapeutic levels of transfection efficiency and protein production. The emergence of in vitro transcribed (IVT) mRNA has substantially expanded our ability to achieve high gene expression. These IVT mRNA are chemically modified to prevent activation of immune response and stabilize the mRNA for improved half-life. mRNA-based gene delivery eliminates the possibility of unintended genomic integration and associated safety concerns. The presence of the translational machinery in the cytoplasm erases the nuclear entry barrier faced by DNA, thus allowing higher efficiencies and rapid protein synthesis [27–29]. Its relatively short half-life makes mRNA uniquely suitable for applications which require transient protein expression, as in the case of endo/exonucleases for genetic manipulation or transcription factors for regenerative applications [28–30]. These characteristics have propelled mRNA to the forefront of pharmaceutical research with abundant preclinical and clinical investigations underway for applications including protein replacement therapy, immunotherapy, cancer, and infectious disease vaccines, gene editing, and tissue engineering [19,31– 42].
LNPs are nanostructures that are composed of a combination of different classes of lipids such as a cationic or ionizable lipid (CIL), structural lipids (phospholipid and sterol lipid) and PEG-conjugated lipid (PEG-lipid). These lipids self-assemble into LNPs under controlled microfluidic mixing with an aqueous phase containing the nucleic acids [43]. The most critical component of LNP, the CIL is responsible for electrostatically binding with the nucleic acids and encapsulating them. These CILs are also responsible for facilitating the endosomal escape of the nucleic acids [44–46]. PEG-lipids prevent aggregation, degradation, and opsonization of the LNPs, while the structural lipids promote the stability and integrity of the nanoparticle. Herein, we have screened several known CILs to determine the cell specificity of gene transfection in the retina and to gain a better understanding of their function and efficacy for the delivery of mRNA to the back of the eye.
METHODS
Materials
Dlin-MC3-DMA (MC3) and Dlin-KC2-DMA (KC2) were custom synthesized. 1,2-dioleyloxy-3-dimethylaminopropane (DODMA), 1,2-dioleoyl-3-trimethylammonium-propane (chloride salt) (DOTAP), N-(4-carboxybenzyl)-N,N-dimethyl-2,3-bis(oleoyloxy)propan-1-aminium (DOBAQ), 1,2-di-O-octadecenyl-3-trimethylammonium propane (chloride salt) (DOTMA), Dimethyldioctadecylammonium (Bromide Salt) (DDAB), 1,2-distearoyl-sn-glycero-3-ethylphosphocholine (chloride salt) (18:0 EPC), 1,2-dioleoyl-sn-glycero-3-ethylphosphocholine (chloride salt) (18:1 EPC), 1,2-distearoyl-3-dimethylammonium-propane (18:0 DAP), 1,2-stearoyl-3-trimethylammonium-propane (chloride salt) (18:0 TAP), and 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC)were acquired from Avanti Polar Lipids. Cholesterol and 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000 (DMG-PEG2k) was purchased from MP Biomedicals and NOF America Corporation, respectively. Pierce D-Luciferin was purchased from Thermo Fisher Scientific. Firefly luciferase (L-7202), EGFP (L-7201), and mCherry (L-7203) mRNA were obtained from Trilink Biotechnologies.
Animals
Albino BALB/c mice were either bred in-house or purchased from The Jackson Laboratory (Bar Harbor, ME, USA). Both male and female mice aged 1 to 4 months were used in experiments. All the experimental procedures followed the protocols approved by the Institutional Animal Care and Use Committee at Oregon Health & Science University and were in adherence to the Association for Research in Vision and Ophthalmology (ARVO) Statement for the Use of Animals in Ophthalmic and Vision Research.
Nanoparticle Formulation and Characterization
LNPs were formulated via microfluidic mixing of one-part ethanol phase (containing the lipids) and three parts aqueous phase (containing the mRNA). The ethanol phase contains the ionizable/cationic lipid, DSPC, DMG-PEG2k, and cholesterol at a molar ratio of 50:10:1.5:38.5, respectively. The aqueous phase consists of the mRNA in 50 mM Citrate buffer pH 4. Following microfluidic mixing, the nanoparticle solution was subjected to buffer exchange with PBS (pH 7.2) and concentrated using Amicon Ultra-4 100k MWCO (EMD Millipore) centrifugal filters. The nanoparticles were stored at 4°C and used within 24 h of formulation. The LNPs remain stable for more than a year [47,48]. The various LNPs were characterized for hydrodynamic radius and polydispersity index (PDI) using dynamic light scattering (DLS) (Zetasizer Nano ZSP (Malvern Instruments)) and mRNA encapsulation efficiency using a modified Quant-iT RiboGreen RNA reagent (Life Technologies)[9].
Injections
Prior to subretinal injections, mice were topically administered 0.5% proparacaine, 1% tropicamide, and 2.5% phenylephrine and anesthetized with ketamine (100 mg/kg)/xylazine (10 mg/kg). To initiate the injection, 2.5% hypromellose was placed over the eye and a 30-gauge needle was used to make an incision in the limbus. A glass coverslip was then placed over the eye to allow for visualization of the retina. Going through the scleral incision in the limbus, using a Hamilton syringe with a 33-gauge blunt needle, 1 μL of PBS or LNP-Luciferase (200 ng mRNA/injection for initial screen, 400 ng mRNA/injection for MC3-LNP expression kinetics), LNP-EGFP (200 ng mRNA/injection) or LNP-mCherry (400 ng mRNA/injection) was delivered to the subretinal space. Each mouse received LNP in one eye and PBS (control) in the contralateral eye. A 2% fluorescein solution was added to the PBS and LNPs so retinal detachment could be observed and documented. For most injections, scleral incisions in the limbus were created nasally and PBS or LNPs were delivered temporally.
In vivo Bioluminescent Imaging and Quantification
Mice were injected intraperitoneally with 150 mg Luciferin/kg body weight according to manufacturer’s protocol. Bioluminescent imaging was conducted on the IVIS Spectrum In vivo Imaging System (PerkinElmer). Image analysis for region of interest (ROI) measurement was performed on Living Image Software (PerkinElmer) and was reported as average radiance (the sum of the radiance from each pixel inside the ROI/number of pixels or super pixels (photons/sec/cm2/sr)). Statistical analysis comparing average radiance between each LNP at each time point was performed using an unpaired multiple t-test in GraphPad Prism. Statistical analysis for MC3 kinetic profile was performed using Ordinary one-ay ANOVA with Tukey’s multiple comparison test in GraphPad Prism. A p value < 0.05 was considered significant.
Immunohistochemistry
At specified time points, mouse eyes were enucleated and fixed in 4% paraformaldehyde (pH 7.4) overnight at 4 °C. To create retinal sections both cryopreservation and paraffin embedding were used. For cryopreservation, posterior eyecups were dissected and incubated in 30% sucrose solution for 2 h prior to embedding in OCT media. Retinal cryosections were sectioned 12 °m thick. For paraffin embedding, after fixation eyes were placed in cassettes and stored in 70% ethanol at room temperature. Orientated eyes were processed and embedded for sectioning (Tissue-Tek VIP®6, Tissue-Tek® TEC™ 5, Sakura Finetek USA, Inc., California). Sections were cut with a microtome to a thickness of 4 microns. Paraffin-embedded retinal sections were dipped in 100% Xylene, 100% ethanol, 95% ethanol, 80% ethanol, running water and deionized water to deparaffinize the tissue prior to staining. All retinal sections were then washed with PBS, permeabilized with 0.3% Triton X-100 for 10 min and blocked with 1% BSA for 1 h. Cryopreserved retinal sections were incubated with an anti-RPE65 or anti-glutamine synthetase antibody (1:1000, mouse, Cat# NB100-355 & Cat# 610517) and an anti-GFP antibody (1:1000, chicken, Cat# ab13970) overnight at 4°C. The next day, sections were washed with PBS and incubated in secondary antibody (goat anti-chicken Alexa 647, 1:800, Cat#A21449 and donkey anti-mouse Alexa 594, 1:800, Cat#A21203, Life Technologies, Eugene, OR). Paraffin-embedded sections were incubated with the same anti-RPE65 antibody (1:1000, mouse, Cat#NB 100-355) and an anti-mCherry antibody (1:500, rabbit, Cat#NBP2-25157) overnight at 4°C. The next day, sections were washed with PBS and incubated in secondary antibody (goat anti-mouse Alexa 647, 1:800, Cat#A21235 and donkey antirabbit Alexa 594, 1:800, Cat#A21207, Life Technologies, Eugene, OR). Images were obtained with either the TCS SP8 X (Leica Microsystems, Buffalo Grove, IL) or the ZEISS LSM 880 with Airyscan (Carl Zeiss Microscopy, Thornwood, NY) laser scanning microscope. Z-stacks (spanned 10 μm with 1 μm interval) were collected using a 40X objective, and maximum intensity projections were made for further analysis. For panretinal images, stitched Z-stacks (spanned 9 μm with 1.5 μm interval) were collected using a 20X objective, and maximum intensity projections were made for further analysis.
Image Analysis
IHC images that were captured with the same exposure settings were analyzed for fluorescence intensity using ImageJ (version 1.45; National Institutes of Health, Bethesda, MD). First, the retinal pigment epithelium (RPE) layer was outlined. Then, ImageJ calculated the pixel intensity (averaging 1,023 points) and area. The pixel intensity was divided by the area and plotted. Three images were analyzed for each condition. Using GraphPad Prism, an Ordinary one-way ANOVA Tukey’s multiple comparisons test was used to compare pixel intensity/area between each condition. A p value < 0.05 was considered significant.
Cryo-Transmission Electron Microscopy
400 mesh Cu grids with thin carbon film supported by lacey carbon substrate (Ted Pella) were glow discharged and administered with 5 μI of sample. The sample was allowed to settle on the grid for 10 sec before blotting for 3 sec and plunging into liquid ethane using a Vitribot Mark IV (FEI) set to 22 °C and 100% humidity. Grids were imaged on a Talos Arctica microscope (FEI) operated at 200 kV, with movie stacks recorded using a K2 Summit camera (Gatan) in counting mode. MotionCor2 was used to gain normalize, align, and sum movie frames, using 5×5 patches with dose weighting.
RESULTS
The CILs utilize their amphiphilicity and positive charge to bind and encapsulate mRNA into organized LNP structures. Besides the CIL, LNPs also contain structural lipids (DSPC and cholesterol) and PEG-lipid (DMG-PEG2k) (Fig. 1a). We grouped CILs based on the cationic or ionizable nature as determined by its polar head group or saturation of the hydrocarbon tails (Fig. 1b). These two regions of CILs have been implicated in the efficacy of LNPs by playing a role in encapsulation and endosomal escape. Group I (MC3, KC2, DODMA) represent ionizable lipids with a tertiary amine and unsaturation in the hydrocarbon chains. Group II (DOBAQ, DOTMA, 18:1 EPC, DOTAP) consists of cationic lipids with a quaternary amine and unsaturated hydrocarbon chains. Group III (DDAB, 18:0 EPC, 18:0 DAP, 18:0 TAP) includes cationic or ionizable lipids which possess a tertiary or quaternary amine but no unsaturation in the hydrocarbon tails. Eleven LNPs were formulated using microfluidic mixing of the lipids and mRNA. These LNPs were subjected to characterization based on hydrodynamic size, polydispersity index, and mRNA encapsulation efficiency (Fig. 1c). Group I and II CILs showed good LNP characteristics (diameter < 200 nm, encapsulation efficiency > 90%, PDI < 0.2), with few exceptions. DOBAQ showed low encapsulation (67.5%) and high PDI (0.42). DOTMA formed LNPs with a high PDI (0.39) and relatively large size (242 nm). Finally, 18:1 EPC also exhibited a high PDI (0.36). These were all tested nonetheless to investigate their characteristics in the retina. Group III lipids failed to formulate into LNPs and as such, were eliminated from further studies.
Figure 1. Lipid nanoparticle variants and their characterization.
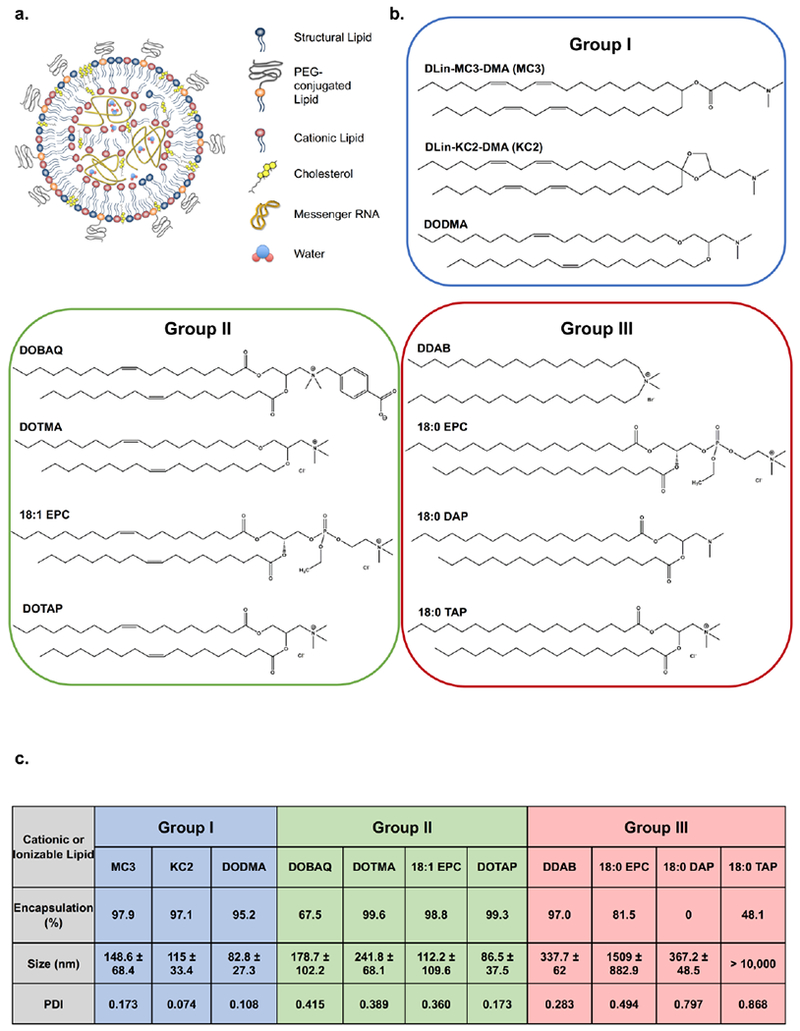
a) Schematic representation of an LNP made up of 4 different categories of lipids and mRNA encapsulated in its core, b) Chemical structures of the CILs utilized in the study categorized by head group protonation state and fatty acid chain saturation. Group I and Group II consists of ionizable and cationic lipids, respectively, with unsaturated hydrocarbon chains. Group III is made up of cationic or ionizable lipids without unsaturated hydrocarbon tails. c) Table representing the mRNA encapsulation efficiency, hydrodynamic size, and polydispersity index of the tested Luciferase-LNPs containing different cationic or ionizable lipids. Group I LNPs – blue, Group II LNPs – green, Group III – red.
Seven LNPs (Group I and II) were injected into mouse eyes via subretinal injections, and luciferase protein expression was imaged every day for up to 5 days (Fig. 2a; Supplementary Fig. 1). The images were analyzed to measure the average radiance per eye (Fig. 2b, Supplementary Fig. 2). We observed the highest expression from MC3 and KC2 followed closely by DODMA. At the 24 h time point, MC3 and KC2 expression was 2.8- and 3.2-fold higher than DODMA expression, respectively. Overall Group I had statistically higher luciferase expression compared to Group II at the 24 h time point (Supplementary Fig. 2b, p < 0.05). All LNPs in Group I followed a similar trend in expression kinetics with the maximum signal at 24 h which deteriorated daily over the 120 h. Moreover, each LNP in Group II had a different expression kinetics profile. DOBAQ’s peak expression persisted for 72 h before deteriorating swiftly. On the other hand, DOTMA reached peak expression at 72 h post-injection, which persisted up to 120 h. 18:1 EPC reached peak expression at 48 h, which remained stable up to 96 h and then diminished by 120 h post-injection. Lastly, DOTAP demonstrated the lowest signal out of the seven LNPs. At the 24 h time point, expression was approximately 200- and 10-fold lower than average Group I and II expression, respectively. Its expression persisted for 72 h before deteriorating.
Figure 2. LNP screen for mRNA delivery to the eye.
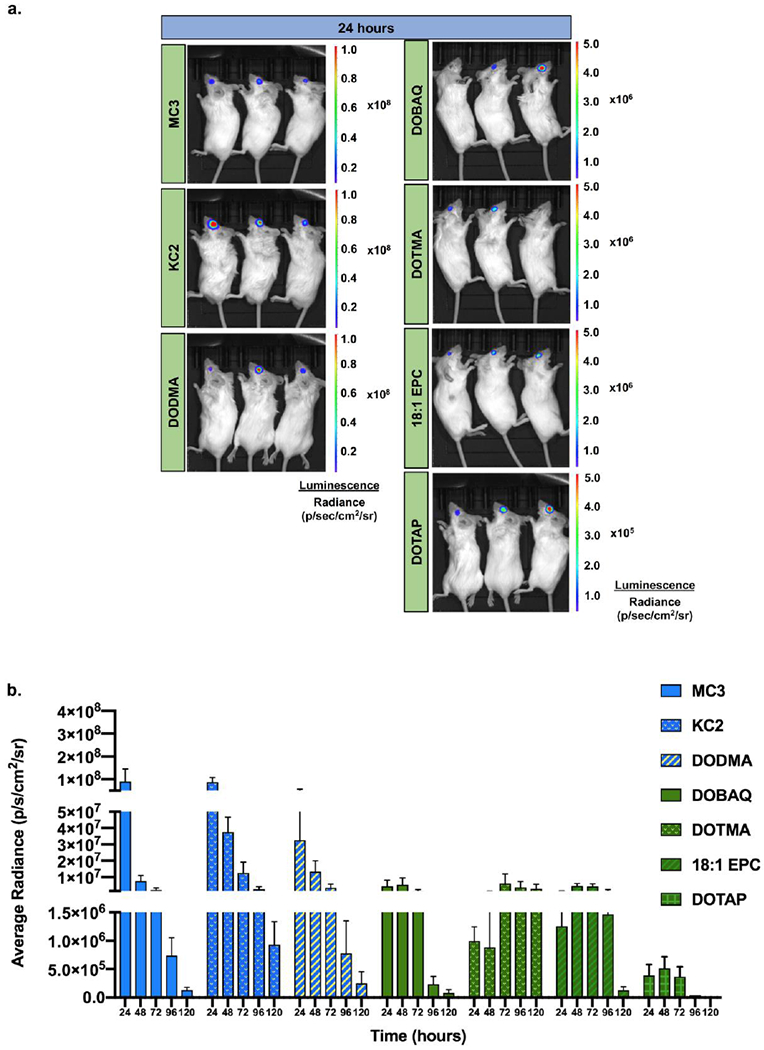
a) Representative images of bioluminescent imaging showing luciferase expression in mouse eyes at 24 h post-subretinal injections of 7 different cationic/ionizable lipids at a dose of 200 ng luciferase mRNA per injection, b) Bar graph showing luciferase expression (average radiance) as a function of time for each of the 7 CIL LNPs. n = 3-7; mean ± SEM. Group I LNPs – blue, Group II LNPs – green.
We further tested Group I and II LNPs to determine their localization in the retina. Reporter mRNA encoding EGFP was encapsulated in LNPs and delivered subretinally. Encapsulation efficiency, hydrodynamic size, and PDI of these nanoparticles were characterized and are reported in Supplementary Fig. 3. At 48 h post-injection, immunohistochemistry with antibodies specific to RPE (retinal pigment epithelium) 65, a retinoid isomerohydrolase, and glutamine synthetase (GS), an enzyme that facilitates nitrogen metabolism, were used to distinguish localization of expression in the RPE and Müller glia, respectively. Group I LNPs, namely MC3, KC2, and DODMA exhibited a strong EGFP signal co-localized with RPE65 demonstrating expression in the RPE layer (Fig. 3a, b). However, EGFP expression was not detected above background staining for the Group II LNPs (Fig. 3a, b). MC3-based LNPs showed significantly higher signal in the RPE compared to all other LNPs (Fig. 3b, p ≤ 0.05). Although most of the EGFP expression observed was localized to the RPE, amongst Group I lipids, both MC3 and KC2 also facilitated limited expression in Müller glia (Fig. 3c). This is demonstrated through EGFP co-localization with GS and the distinct Müller glia expression pattern, which spans across the entire retina with end feet at the base of the ganglion cell layer and projections extending up to the top of the photoreceptor cell bodies.
Figure 3. Localization of mRNA expression in the retina.
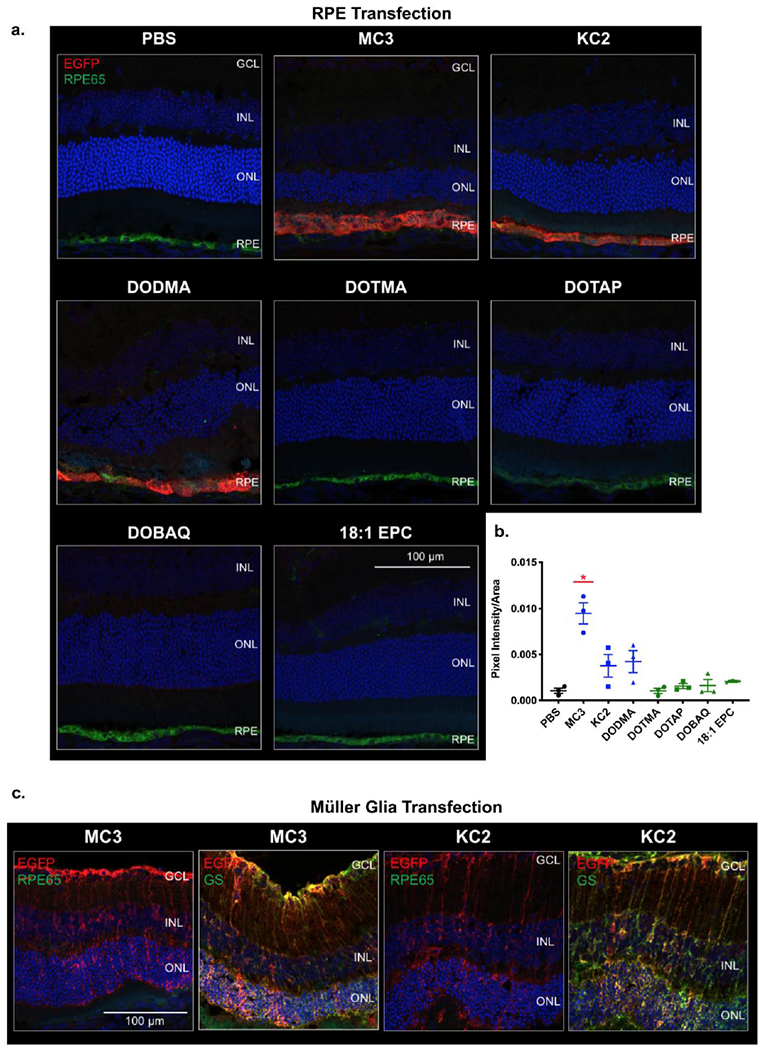
Nanoparticles were injected subretinally at 200 ng mRNA/injection. Eyes were harvested at the 48-h time point, sectioned and stained for EGFP, RPE65 and GS. a) Representative confocal images showing RPE transfection. RPE cells - green, and EGFP - red b) Plot showing the fluorescence intensities of RPE transfection quantified from confocal images of EGFP IHC. Group I LNPs – blue, Group II LNPs – green. MC3-based LNPs showed significantly higher signal in the RPE compared to all other LNPs. n = 3; mean ± SEM. *p ≤ 0.05. c) MC3 and KC2 demonstrated an ability to transfect Müller glia. Nuclei - blue, RPE cells - green, GS - green, and EGFP - red. GS-glutamine synthetase, GCL-ganglion cell layer, INL-inner nuclear layer, ONL-outer nuclear layer, RPE-retinal pigment epithelium.
We chose MC3 as our lead CIL for our LNPs as this formulation is an FDA-approved non-viral nucleic acid carrier for the treatment of amyloidosis and had exhibited the highest RPE transfection (Fig. 3a, b) [49]. Moreover, it demonstrates potent transfection and high formulation reproducibility. Cryo-transmission electron microscopy (cryo-TEM) images of MC3-based LNPs exhibited spherical nanoparticles with electron dense cores (Fig. 4a). We conducted a more thorough kinetic study on these LNPs and found that gene expression ensues within 4 h and lasts for at least 168 h (Fig. 4b i, ii; Supplementary Fig. 4a). The luciferase expression significantly increases by ~13-fold from 4 to 24 h and subsequently drops ~36-fold by 96 h (Fig. 4b ii, p ≤ 0.05). The expression continued to decline from 24 to 168 h. For retinal localization, reporter mRNA encoding mCherry was encapsulated and injected subretinally. mCherry expression was co-localized with RPE65 from 24 to 120 h post-injection (Fig. 4c, Supplementary Fig. 4b). Robust expression was maintained up to 72 h and diminished at the 120 h time point. Subretinal delivery leads to the formation of a subretinal bleb detaching approximately 60% of the retina [50]. This commonly results in localized gene expression to the site of the bleb. However, in the case of LNPs, a pan-retinal expression of mCherry was observed throughout the retina up to 72 h after a single subretinal injection of MC3 based LNPs (Fig. 4d i, ii, Supplementary Fig. 4c).
Figure 4. Expression kinetics of MC3 based LNPs.
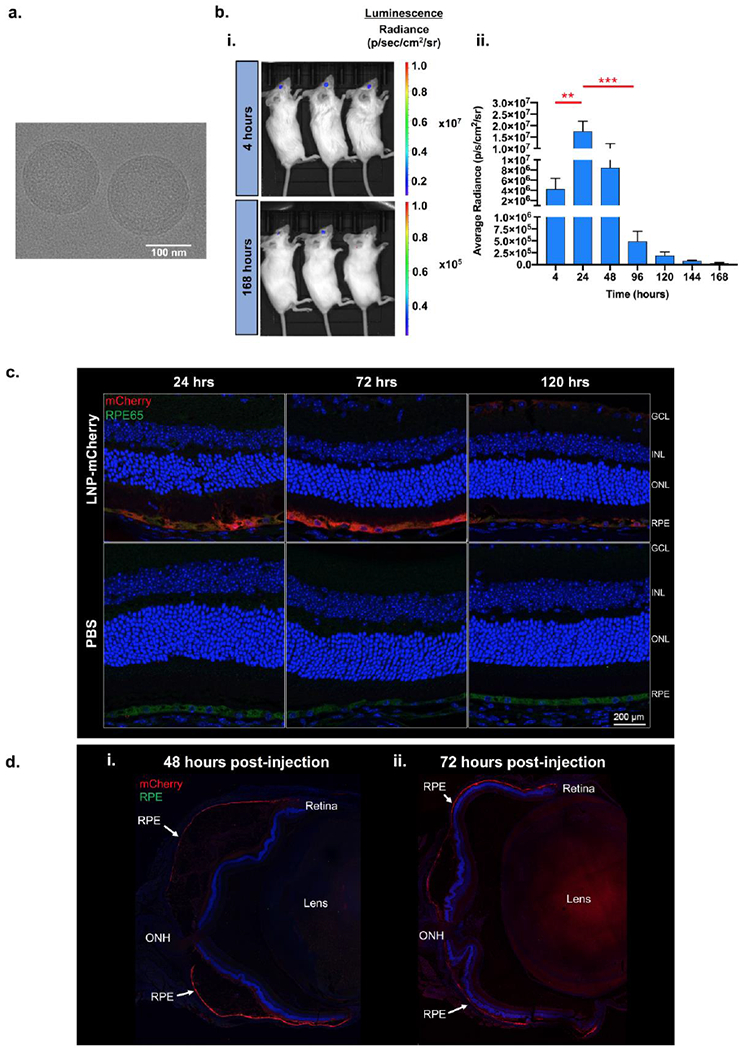
a) CryoTEM image of MC3 LNPs exhibiting spherical nanoparticles with electron dense cores, b) i) Representative bioluminescent images at 24 and 168 h showing localization of luciferase expression to the eye and ii) Bar graph showing the quantified expression (average radiance) at each time point following subretinal injections of MC3 LNPs at a dose of 400 ng mRNA per injection, n = 5; mean ± SEM. ** p ≤ 0.01, *** p ≤ 0.001. c) IHC sections of mouse retina showing RPE localization of mCherry expression which persists for up to at least 120 h. Nuclei - blue, RPE cells - green, and mCherry - red. d) Pan-retinal expression of mCherry at (i) 48 h and (ii) 72 h following subretinal injection of MC3-LNP. Nuclei – blue and mCherry - red. GCL - ganglion cell layer, INL - inner nuclear layer, ONL - outer nuclear layer, RPE - retinal pigment epithelium, ONH – optic nerve head.
DISCUSSION
The successful treatment of retinal degenerative disorders will likely require a combination of gene therapy, stem cell therapy, and neuroprotective approaches. Thus, optimizing gene delivery vehicles is one crucial part of therapy development. Nanoparticles, owing to their diversity and modularity, possess diverse characteristics and superior control over toxicity, carrier size, bioavailability, biodistribution, specificity, efficacy, and type of cargo, making them an attractive therapeutic. In this study, we evaluated LNP systems with differing characteristics for enhanced transfection of the retina. LNPs that contained ionizable lipids with low pKa and unsaturated hydrocarbon chains showed the highest amount of reporter gene transfection in the eye after a subretinal injection. The kinetics of gene expression showed a rapid onset (within 4 h) that persisted for 96 h. The gene delivery was cell-type specific with the majority of expression in the RPE and limited expression in the Müller glia. Pan-retinal, RPE expression was sustained for 72 h and decreased by 120 h post-delivery.
The efficiency of Group I CILs is attributed to the unsaturation of the hydrocarbon tail and the pKa of the ionizable lipid headgroup, both properties which are thought to facilitate endosomal escape [45,46,51]. Unsaturation in the hydrocarbon chain has been demonstrated to enhance membrane destabilization to mediate endosomal escape [51]. The pKa of the lipids should be low enough to make the lipid neutral at physiological pH (pH 7.4) to avoid the mononuclear phagocyte system and toxicity, but high enough to allow protonation in the endosome (pH 5.5-6.5) to enable electrostatic interactions with the anionic membrane of the endosome leading to escape to the cytosol [45,52]. MC3 and KC2 both have pKa values around 6.4 and two double bonds on each hydrocarbon chain, while pKa of DODMA is around 7.0 and has one double bond on each hydrocarbon tail [45,46,51]. We hypothesize that the slightly higher transfection observed with MC3 and KC2 compared to DODMA may be due to better endosomal escape as previously described [45,46]. Group II consists of cationic lipids with one double bond on the hydrocarbon chains. It has been observed that ionizable lipids form LNPs with an amorphous electron-dense core, whereas, the cationic lipids tend to lack electron dense solid cores and possess a more bilayer system [53]. These LNPs may lack the ability to effectively fuse and facilitate endosomal escape within the retina as well [51]. Moreover, their cationic charge may cause aggregation and dissociation in biological fluids. A high cationic charge can also lead to unintended toxicity [54]. The endosomal escape efficiency is further impacted by the presence of only a single double bond on each hydrocarbon tail [51], These factors are likely culpable in the low efficacy of these cationic systems. Finally, Group III failed to formulate, presumably due to the absence of unsaturation in the hydrocarbon chain preventing efficient packaging of mRNA into an LNP.
Group I and II LNPs mediated luciferase expression in the eye. However, only Group I LNPs showed protein expression that localized to the retina. To account for this discrepancy, whole eyes were sectioned and stained to determine if expression was localized to different portions of the eye. Group I LNPs have expression primarily limited to the RPE and we did not see expression in the anterior portion of the eye. Group II based LNP variants failed to show any localization in the retina even though we detected enzymatically driven luciferase expression due to relatively lower limit of immunofluorescence detection. Due to the local administration, we did not observe any systemic side effects associated with LNP delivery. MC3-mediated mCherry expression did not show signs of retinal toxicity up to 72 h post-injection illustrated in Fig. 4. However, we did observe outer nuclear layer thinning in some retinas. When observed, it was most prominently associated with EGFP signal and mCherry signal after five days of expression. It is unclear if the retinal thinning observed was due to high reporter gene expression or LNP toxicity. Future studies will be performed to asses any LNP-related retinal toxicity.
Ionizable LNPs (such as MC3, KC2, and DODMA) have been previously demonstrated to enter hepatocytes and neurons via the low-density lipoprotein (LDL) receptor in an ApoE-dependent manner [16,55]. In a biological medium, the LNPs rapidly lose the surface PEG-lipid allowing adsorption of ApoE, a ligand to LDL receptors. In the retina, ApoE is secreted from the Müller glia and can be internalized in both RPE and Müller glia via the LDL receptor [56–58]. Since the injection was performed subretinally, it is likely that the small pore size of the outer limiting membrane compartmentalized the LNPs in the subretinal space in close proximity to the epithelial layer, mediating robust RPE expression while preventing distribution [59]. The high concentration of LNPs sequestered near the RPE and presence of ApoE-based cellular targeting are perhaps responsible for the pan-retinal expression of the mRNA in the RPE. Achieving pan-retinal expression may be optimal for therapeutic gene delivery as it has the potential for maximal functional restoration.
Our ability to transfect RPE and Müller glia establishes a range of applications within our grasp. These nanoparticle systems could be used to treat RPE-specific inherited retinal degenerations, such as MERTK-RP, RPE65-LCA and LRAT-LCA [1]. Müller cells are resident glial cells responsible for supporting retinal function. Recent evidence points to the regenerative potential of Müller glia, thus presenting the possibility that these cells can be used to regenerate the neural retina in the event of damage or degeneration [60]. The differentiation, proliferation, and reprogramming of Müller glia could be favorably facilitated through the transient expression of transcription factors and signaling proteins using mRNA. LNPs could also mediate transient expression of transcription factors that up-regulate neuroprotective pathways in the RPE, protecting RPE integrity and photoreceptor degeneration in age-related macular degeneration (AMD) and inherited retinal degeneration (IRD) models [61]. Moreover, the CRISPR-cas9 system has already been exploited for treating Retinitis Pigmentosa [62– 64], glaucoma [65] and LCA [66] using AAVs and lentivirus. Utilizing LNPs to transiently express Cas9 mRNA offers a safer approach for gene editing. Our future aim is to develop materials that would enable transfection of the neural retina after intravitreal delivery, which is less invasive and allows for repeated administration, to broaden the scope of therapeutic applications.
Supplementary Material
Highlights:
Vision loss due to genetic abnormalities resulting in retinal degeneration is a global health problem.
Lipid nanoparticles and mRNA are at the forefront of non-viral gene therapy.
Ionizable lipids with unsaturated hydrocarbon tails demonstrate the best efficacy at mRNA delivery to the retinal pigment epithelium and Müller glia.
Gene expression following mRNA delivery using the best performing lipid nanoparticle persists for up to 5 days.
ACKNOWLEDGEMENTS
We would like to thank the Casey Eye Institute, Leonard Christenson Eye Pathology Laboratory for their support with tissue processing and sectioning. This work was supported by the Medical Research Foundation of Oregon (G.S), OSU Startup funds (G.S), National Institute of Biomedical Imaging and Bioengineering (N.I.B.I.B) 1R15EB021581 −01 (G.S), unrestricted departmental funding from Research to Prevent Blindness (M.P) and grant P30 EY010572 from the National Institutes of Health (M.P). Electron microscopy was performed at the Multiscale Microscopy Core (MMC) with technical support from the Oregon Health and Science University (OHSU) and the OHSU Center for Spatial Systems Biomedicine (OCSSB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST:
None
References:
- [1].Veleri S, Lazar CH, Chang B, Sieving PA, Banin E, Swaroop A, Biology and therapy of inherited retinal degenerative disease: insights from mouse models, Disease Models & Mechanisms. 8(2015) 109–129. doi: 10.1242/dmm.017913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Auricchio A, Smith AJ, AM RR, The Future Looks Brighter After 25 Years of Retinal Gene Therapy, Human Gene Therapy. 28 (2017) 982–987. doi: 10.1089/hum.2017.164. [DOI] [PubMed] [Google Scholar]
- [3].Han Z, Conley SM, Naash MI, AAV and Compacted DNA Nanoparticles for the Treatment of Retinal Disorders: Challenges and Future Prospects, Invest. Ophthalmol. Vis. Sci. 52 (2011) 3051–3059. doi: 10.1167/iovs.10-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Russell S, Bennett J, Wellman JA, Chung DC, Yu Z-F, Tillman A, Wittes J, Pappas J, Elci O, McCague S, Cross D, Marshall KA, Walshire J, Kehoe TL, Reichert H, Davis M, Raffini L, George LA, Hudson FP, Dingfield L, Zhu X, Haller JA, Sohn EH, Mahajan VB, Pfeifer W, Weckmann M, Johnson C, Gewaily D, Drack A, Stone E, Wachtel K, Simonelli F, Leroy BP, Wright JF, High KA, Maguire AM, Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial, The Lancet. 390 (2017) 849–860. doi: 10.1016/S0140-6736(17)31868-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Russell DW, Grompe M, Adeno-associated virus finds its disease, Nat Genet. 47 (2015) 1104–1105. doi: 10.1038/ng.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Oliveira AV, Rosa da Costa AM, Silva GA, Non-viral strategies for ocular gene delivery, Materials Science and Engineering: C. 77 (2017) 1275–1289. doi: 10.1016/j.msec.2017.04.068. [DOI] [PubMed] [Google Scholar]
- [7].Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG, Non-viral vectors for gene-based therapy, Nat Rev Genet. 15 (2014) 541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- [8].Stewart MP, Lorenz A, Dahlman J, Sahay G, Challenges in carrier-mediated intracellular delivery: moving beyond endosomal barriers, WIREs Nanomed Nanobiotechnol. 8(2016) 465–478. doi: 10.1002/wnan.1377. [DOI] [PubMed] [Google Scholar]
- [9].Patel S, Ashwanikumar N, Robinson E, DuRoss A, Sun C, Murphy-Benenato KE, Mihai C, Almarsson O, Sahay G, Boosting Intracellular Delivery of Lipid Nanoparticle-Encapsulated mRNA, Nano Lett. 17 (2017)5711–5718. doi: 10.1021/acs.nanolett.7b02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, Manygoats K, Seifert S, Andree C, Stoter M, Epstein-Barash H, Zhang L, Koteliansky V, Fitzgerald K, Fava E, Bickle M, Kalaidzidis Y, Akinc A, Maier M, Zerial M, Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape, Nat Biotech. 31 (2013) 638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- [11].Gilleron J, Paramasivam P, Zeigerer A, Querbes W, Marsico G, Andree C, Seifert S, Amaya P, Stoter M, Koteliansky V, Waldmann H, Fitzgerald K, Kalaidzidis Y, Akinc A, Maier MA, Manoharan M, Bickle M, Zerial M, Identification of siRNA delivery enhancers by a chemical library screen, Nucleic Acids Res. 43 (2015) 7984–8001. doi: 10.1093/nar/gkv762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sahay G, Alakhova DY, Kabanov AV, Endocytosis of nanomedicines, Journal of Controlled Release. 145 (2010) 182–195. doi: 10.1016/j.jconrel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kaczmarek JC, Kauffman KJ, Fenton OS, Sadtler K, Patel AK, Heartlein MW, DeRosa F, Anderson DG, Optimization of a Degradable Polymer-Lipid Nanoparticle for Potent Systemic Delivery of mRNA to the Lung Endothelium and Immune Cells, Nano Lett. (2018). doi: 10.1021/acs.nanolett.8b02917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Whitehead KA, Dorkin JR, Vegas AJ, Chang PH, Veiseh O, Matthews J, Fenton OS, Zhang Y, Olejnik KT, Yesilyurt V, Chen D, Barros S, Klebanov B, Novobrantseva T, Langer R, Anderson DG, Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity, Nature Communications. 5 (2014). doi: 10.1038/ncomms5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tzeng SY, Green JJ, Subtle Changes to Polymer Structure and Degradation Mechanism Enable Highly Effective Nanoparticles for siRNA and DNA Delivery to Human Brain Cancer, Advanced Healthcare Materials. 2(2013)468–480. doi: 10.1002/adhm.201200257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Akinc A, Querbes W, De S, Qin J, Frank-Kamenetsky M, Jayaprakash KN, Jayaraman M, Rajeev KG, Cantley WL, Dorkin JR, Butler JS, Qin L, Racie T, Sprague A, Fava E, Zeigerer A, Hope MJ, Zerial M, Sah DW, Fitzgerald K, Tracy MA, Manoharan M, Koteliansky V, de Fougerolles A, Maier MA, Targeted Delivery of RNAi Therapeutics With Endogenous and Exogenous Ligand-Based Mechanisms, Molecular Therapy. 18(2010) 1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Singh S, Grossniklaus H, Kang S, Edelhauser H, Ambati B, Kompella U, Intravenous transferrin, RGD peptide and dual-targeted nanoparticles enhance anti-VEGF intraceptor gene delivery to laser-induced CNV, Gene Ther. 16 (2009) 645–659. doi: 10.1038/gt.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pollinger K, Hennig R, Ohlmann A, Fuchshofer R, Wenzel R, Breunig M, Tessmar J, Tamm ER, Goepferich A, Ligand-functionalized nanoparticles target endothelial cells in retinal capillaries after systemic application, PNAS. 110 (2013) 6115–6120. doi: 10.1073/pnas.1220281110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hajj KA, Whitehead KA, Tools for translation: non-viral materials for therapeutic mRNA delivery, Nature Reviews Materials. 2(2017) natrevmats201756. doi: 10.1038/natrevmats.2017.56. [DOI] [Google Scholar]
- [20].Kaczmarek JC, Kowalski PS, Anderson DG, Advances in the delivery of RNA therapeutics: from concept to clinical reality, Genome Medicine. 9 (2017). doi: 10.1186/s13073-017-0450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Cullis PR, Hope MJ, Lipid Nanoparticle Systems for Enabling Gene Therapies, Molecular Therapy. 25 (2017) 1467–1475. doi: 10.1016/j.ymthe.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kelley RA, Conley SM, Makkia R, Watson JN, Han Z, Cooper MJ, Naash MI, DNA nanoparticles are safe and nontoxic in non-human primate eyes, Int J Nanomedicine. 13 (2018) 1361–1379. doi: 10.2147/IJN.S157000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gomes dos Santos AL, Bochot A, Tsapis N, Artzner F, Bejjani RA, Thillaye-Goldenberg B, de Kozak Y, Fattal E, Behar-Cohen F, Oligonucleotide-Polyethylenimine Complexes Targeting Retinal Cells: Structural Analysis and Application to Anti-TGFp-2 Therapy, Pharm Res. 23 (2006) 770–781. doi: 10.1007/s11095-006-9748-0. [DOI] [PubMed] [Google Scholar]
- [24].Sunshine JC, Sunshine SB, Bhutto I, Handa JT, Green JJ, Poly(p-Amino Ester)- Nanoparticle Mediated Transfection of Retinal Pigment Epithelial Cells In Vitro and In Vivo, PLOSONE. 7 (2012) e37543. doi: 10.1371/journal.pone.0037543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang Y, Rajala A, Rajala RVS, Nanoparticles as Delivery Vehicles for the Treatment of Retinal Degenerative Diseases, in: Ash JD, Anderson RE, LaVail MM, Rickman C. Bowes, Hollyfield JG, Grimm C (Eds.), Retinal Degenerative Diseases, Springer International Publishing, 2018: pp. 117–123. [Google Scholar]
- [26].Adijanto J, Naash M.l., Nanoparticle-based technologies for retinal gene therapy, European Journal of Pharmaceutics and Biopharmaceutics. 95 (2015) 353–367. doi: 10.1016/j.ejpb.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zou S, Scarfo K, Nantz MH, Hecker JG, Lipid-mediated delivery of RNA is more efficient than delivery of DNA in non-dividing cells, Int J Pharm. 389 (2010) 232–243. doi: 10.1016/j.ijpharm.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Warren L, Manos PD, Ahfeldt T, Loh Y-H, Li H, Lau F, Ebina W, Mandal P, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ, Highly efficient reprogramming to pluripotency and directed differentiation of human cells using synthetic modified mRNA, Cell Stem Cell. 7 (2010) 618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bugeon S, de Chevigny A, Boutin C, Core N, Wild S, Bosio A, Cremer H, Beclin C, Direct and efficient transfection of mouse neural stem cells and mature neurons by in vivo mRNA electroporation, Development. 144 (2017) 3968–3977. doi: 10.1242/dev.151381. [DOI] [PubMed] [Google Scholar]
- [30].Yin H, Song C-Q, Dorkin JR, Zhu LJ, Li Y, Wu Q, Park A, Yang J, Suresh S, Bizhanova A, Gupta A, Bolukbasi MF, Walsh S, Bogorad RL, Gao G, Weng Z, Dong Y, Koteliansky V, Wolfe SA, Langer R, Xue W, Anderson DG, Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo, Nat Biotech. 34 (2016) 328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bernal JA, RNA-Based Tools for Nuclear Reprogramming and Lineage-Conversion: Towards Clinical Applications, J Cardiovasc Transl Res. 6 (2013) 956–968. doi: 10.1007/s12265-013-9494-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Patel S, Athirasala A, Menezes PP, Ashwanikumar N, Zou T, Sahay G, Bertassoni LE, Messenger RNA Delivery for Tissue Engineering and Regenerative Medicine Applications, Tissue Engineering Part A. (2018). doi: 10.1089/ten.tea.2017.0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Li B, Zhao W, Luo X, Zhang X, Li C, Zeng C, Dong Y, Engineering CRISPR-Cpf1 crRNAs and mRNAs to maximize genome editing efficiency, Nature Biomedical Engineering. 1 (2017)0066. doi: 10.1038/s41551-017-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].DeRosa F, Guild B, Karve S, Smith L, Love K, Dorkin JR, Kauffman KJ, Zhang J, Yahalom B, Anderson DG, Heartlein MW, Therapeutic efficacy in a hemophilia B model using a biosynthetic mRNA liver depot system, Gene Therapy. 23 (2016) 699–707. doi: 10.1038/gt.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Alberer M, Gnad-Vogt U, Hong HS, Mehr KT, Backert L, Finak G, Gottardo R, A Bica M, Garofano A, Koch SD, Fotin-Mleczek M, Hoerr I, Clemens R, von Sonnenburg F, Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: an open-label, non-randomised, prospective, first-in-human phase 1 clinical trial, The Lancet. 390 (2017) 1511–1520. doi: 10.1016/S0140-6736(17)31665-3. [DOI] [PubMed] [Google Scholar]
- [36].Pardi N, Hogan MJ, Pelc RS, Muramatsu H, Andersen H, DeMaso CR, Dowd KA, Sutherland LL, Scearce RM, Parks R, Wagner W, Granados A, Greenhouse J, Walker M, Willis E, Yu J-S, McGee CE, Sempowski GD, Mui BL, Tam YK, Huang Y-J, Vanlandingham D, Holmes VM, Balachandran H, Sahu S, Litton M, Higgs S, Hensley SE, Madden TD, Hope MJ, Kariko K, Santra S, Graham BS, Lewis MG, Pierson TC, Haynes BF, Weissman D, Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination, Nature. 543 (2017) 248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Robinson E, MacDonald KD, Slaughter K, McKinney M, Patel S, Sun C, Sahay G, Lipid Nanoparticle-Delivered Chemically Modified mRNA Restores Chloride Secretion in Cystic Fibrosis, Molecular Therapy. 26 (2018) 2034–2046. doi: 10.1016/j.ymthe.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bahl K, Senn JJ, Yuzhakov O, Bulychev A, Brito LA, Hassett KJ, Laska ME, Smith M, Almarsson O, Thompson J, Ribeiro A. (Mick), Watson M, Zaks T, Ciaramella G, Preclinical and Clinical Demonstration of Immunogenicity by mRNA Vaccines against H10N8 and H7N9 Influenza Viruses, Molecular Therapy. 25 (2017) 1316–1327. doi: 10.1016/j.ymthe.2017.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ramaswamy S, Tonnu N, Tachikawa K, Limphong P, Vega JB, Karmali PP, Chivukula P, Verma IM, Systemic delivery of factor IX messenger RNA for protein replacement therapy, PNAS. 114 (2017) E1941–E1950. doi: 10.1073/pnas.1619653114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kormann MSD, Hasenpusch G, Aneja MK, Nica G, Flemmer AW, Herber-Jonat S, Huppmann M, Mays LE, Illenyi M, Schams A, Griese M, Bittmann I, Handgretinger R, Hartl D, Rosenecker J, Rudolph C, Expression of therapeutic proteins after delivery of chemically modified mRNAin mice, Nat Biotech. 29 (2011) 154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- [41].McNamara MA, Nair SK, Holl EK, RNA-Based Vaccines in Cancer Immunotherapy, Journal of Immunology Research. (2015). doi: 10.1155/2015/794528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Schlake T, Thess A, Thran M, Jordan I, mRNA as novel technology for passive immunotherapy, Cell. Mol. Life Sci. (2018). doi: 10.1007/s00018-018-2935-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Belliveau NM, Huft J, Lin PJ, Chen S, Leung AK, Leaver TJ, Wild AW, Lee JB, Taylor RJ, Tam YK, Hansen CL, Cullis PR, Microfluidic Synthesis of Highly Potent Limit-size Lipid Nanoparticles for In Vivo Delivery of siRNA, Molecular Therapy - Nucleic Acids. 1 (2012). doi: 10.1038/mtna.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nguyen J, Szoka FC, Nucleic acid delivery: the missing pieces of the puzzle?, Acc Chem Res. 45 (2012) 1153–1162. doi: 10.1021/ar3000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Jayaraman M, Ansell SM, Mui BL, Tam YK, Chen J, Du X, Butler D, Eltepu L, Matsuda S, Narayanannair JK, Rajeev KG, Hafez IM, Akinc A, Maier MA, Tracy MA, Cullis PR, Madden TD, Manoharan M, Hope MJ, Maximizing the Potency of siRNA Lipid Nanoparticles for Hepatic Gene Silencing In Vivo, Angewandte Chemie International Edition. 51 (2012) 8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, Sah DWY, Stebbing D, Crosley EJ, Yaworski E, Hafez IM, Dorkin JR, Qin J, Lam K, Rajeev KG, Wong KF, Jeffs LB, Nechev L, Eisenhardt ML, Jayaraman M, Kazem M, Maier MA, Srinivasulu M, Weinstein MJ, Chen Q, Alvarez R, Barros SA, De S, Klimuk SK, Borland T, Kosovrasti V, Cantley WL, Tam YK, Manoharan M, Ciufolini MA, Tracy MA, de Fougerolles A, MacLachlan I, Cullis PR, Madden TD, Hope MJ, Rational design of cationic lipids for siRNA delivery, Nat Biotech. 28 (2010) 172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- [47].Suzuki Y, Hyodo K, Tanaka Y, Ishihara H, siRNA-lipid nanoparticles with long-term storage stability facilitate potent gene-silencing in vivo, Journal of Controlled Release. 220 (2015) 44–50. doi: 10.1016/j.jconrel.2015.10.024. [DOI] [PubMed] [Google Scholar]
- [48].Ball RL, Bajaj P, Whitehead KA, Achieving long-term stability of lipid nanoparticles: examining the effect of pH, temperature, and lyophilization, International Journal of Nanomedicine. (2016). doi: 10.2147/IJN.S123062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hoy SM, Patisiran: FirstGlobal Approval, Drugs. (2018). doi: 10.1007/s40265-018-0983-6. [DOI] [PubMed] [Google Scholar]
- [50].Boye SL, Conlon T, Erger K, Ryals R, Neeley A, Cossette T, Pang J, Dyka FM, Hauswirth WW, Boye SE, Long-term Preservation of Cone Photoreceptors and Restoration of Cone Function by Gene Therapy in the Guanylate Cyclase-1 Knockout (GC1KO) Mouse, Invest. Ophthalmol. Vs. Sci. 52 (2011) 7098–7108. doi: 10.1167/iovs.11-7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Heyes J, Palmer L, Bremner K, MacLachlan I, Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids, Journal of Controlled Release. 107 (2005) 276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- [52].Semple SC, Klimuk SK, Harasym TO, Dos Santos N, Ansell SM, Wong KF, Maurer N, Stark H, Cullis PR, Hope MJ, Scherrer P, Efficient encapsulation of antisense oligonucleotides in lipid vesicles using ionizable aminolipids: formation of novel small multilamellar vesicle structures, Biochimicaet Biophysica Acta (BBA) - Biomembranes. 1510 (2001) 152–166. doi: 10.1016/S0005-2736(00)00343-6. [DOI] [PubMed] [Google Scholar]
- [53].Kulkarni JA, Darjuan MM, Mercer JE, Chen S, van der Meel R, Thewalt JL, Tam YYC, Cullis PR, On the Formation and Morphology of Lipid Nanoparticles Containing Ionizable Cationic Lipids and siRNA, ACS Nano. 12 (2018) 4787–4795. doi: 10.1021/acsnano.8b01516. [DOI] [PubMed] [Google Scholar]
- [54].Lv H, Zhang S, Wang B, Cui S, Yan J, Toxicity of cationic lipids and cationic polymers in gene delivery, Journal of Controlled Release. 114 (2006) 100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- [55].Rungta RL, Choi HB, Lin PJ, Ko RW, Ashby D, Nair J, Manoharan M, Cullis PR, MacVicar BA, Lipid Nanoparticle Delivery of siRNA to Silence Neuronal Gene Expression in the Brain, Molecular Therapy—Nucleic Acids. 2 (2013) e136. doi: 10.1038/mtna.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Amaratunga A, Abraham CR, Edwards RB, Sandell JH, Schreiber BM, Fine RE, Apolipoprotein E Is Synthesized in the Retina by Müller Glial Cells, Secreted into the Vitreous, and Rapidly Transported into the Optic Nerve by Retinal Ganglion Cells, Journal of Biological Chemistry. 271 (1996)5628–5632. doi: 10.1074/jbc.271.10.5628. [DOI] [PubMed] [Google Scholar]
- [57].Gordiyenko N, Campos M, Lee JW, Fariss RN, Sztein J, Rodriguez IR, RPE Cells Internalize Low-Density Lipoprotein (LDL) and Oxidized LDL (oxLDL) in Large Quantities In Vitro and In Vivo, Invest. Ophthalmol. Vis. Sci. 45 (2004) 2822–2829. doi: 10.1167/iovs.04-0074. [DOI] [PubMed] [Google Scholar]
- [58].Tserentsoodol N, Sztein J, Campos M, Gordiyenko NV, Fariss RN, Lee JW, Fliesler SJ, Rodriguez IR, Uptake of cholesterol by the retina occurs primarily via a low density lipoprotein receptor-mediated process, Mol. Vis. 12 (2006) 1306–1318. [PubMed] [Google Scholar]
- [59].Omri S, Omri B, Savoldelli M, Jonet L, Thillaye-Goldenberg B, Thuret G, Gain P, Jeanny JC, Crisanti P, Behar-Cohen F, The outer limiting membrane (OLM) revisited: clinical implications, Clin Ophthalmol. 4 (2010) 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Yao K, Qiu S, Wang YV, Park SJH, Mohns EJ, Mehta B, Liu X, Chang B, Zenisek D, Crair MC, Demb JB, Chen B, Restoration of vision after de novo genesis of rod photoreceptors in mammalian retinas, Nature. (2018) 1. doi: 10.1038/s41586-018-0425-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Pardue MT, Allen RS, Neuroprotective strategies for retinal disease, Progress in Retinal and Eye Research. 65 (2018) 50–76. doi: 10.1016/j.preteyeres.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Tsai Y-T, Wu W-H, Lee T-T, Wu W-P, Xu CL, Park KS, Cui X, Justus S, Lin C-S, Jauregui R, Su P-Y, Tsang SH, Clustered Regularly Interspaced Short Palindromic Repeats-Based Genome Surgery for the Treatment of Autosomal Dominant Retinitis Pigmentosa, Ophthalmology. 125 (2018) 1421–1430. doi: 10.1016/j.ophtha.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Giannelli SG, Luoni M, Castoldi V, Massimino L, Cabassi T, Angeloni D, Demontis GC, Leocani L, Andreazzoli M, Broccoli V, Cas9/sgRNA selective targeting of the P23H Rhodopsin mutant allele for treating retinitis pigmentosa by intravitreal AAV9.PHP.B-based delivery, Hum Mol Genet. 27 (2018) 761–779. doi: 10.1093/hmg/ddx438. [DOI] [PubMed] [Google Scholar]
- [64].Yu W, Mookherjee S, Chaitankar V, Hiriyanna S, Kim J-W, Brooks M, Ataeijannati Y, Sun X, Dong L, Li T, Swaroop A, Wu Z, Nrl knockdown by AAV-delivered CRISPR/Cas9 prevents retinal degeneration in mice, Nature Communications. 8(2017) 14716. doi: 10.1038/ncomms14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Jain A, Zode G, Kasetti RB, Ran FA, Yan W, Sharma TP, Bugge K, Searby CC, Fingert JH, Zhang F, Clark AF, Sheffield VC, CRISPR-Cas9-based treatment of myocilin-associated glaucoma, PNAS. 114 (2017) 11199–11204. doi: 10.1073/pnas.1706193114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Ruan G-X, Barry E, Yu D, Lukason M, Cheng SH, Scaria A, CRISPR/Cas9-Mediated Genome Editing as a Therapeutic Approach for Leber Congenital Amaurosis 10, Molecular Therapy. 25 (2017) 331–341. doi: 10.1016/j.ymthe.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


