Abstract
Inverted urothelial papilloma (IUP) and urothelial papilloma (UP) are rare urothelial neoplasms that typically follow a benign clinical course. Oncogenic mutations in FGFR3, HRAS and the TERT promoter have been reported in these entities but no comprehensive molecular analysis has been performed. We sought to characterize the genomic landscape of IUP and UP using whole exome and targeted next generation sequencing. In IUP, 10 of 11 tumors harbored oncogenic hotspot mutations in HRAS and the remaining tumor had an oncogenic KRAS mutation. None of the IUP tumors harbored TERT promoter or FGFR3 mutations. In UP, 8 of 11 tumors had oncogenic KRAS mutations and 2 had oncogenic HRAS mutations. One UP tumor had oncogenic mutations in FGFR3, PIK3CA and the TERT promoter and arose in a patient with recurrent non-invasive papillary urothelial carcinomas. In contrast to urothelial carcinoma, the APOBEC mutational signature was not present in any IUP and UP tumors and oncogenic alterations in chromatin remodeling genes were uncommon in both IUP and UP. The current study suggests that IUP and UP are driven primarily by RAS pathway activation and lack the more common genomic features of urothelial cancers.
Keywords: Genomics, Inverted Urothelial Papilloma, Urothelial Papilloma, Urothelial Carcinoma, RAS pathway
Introduction
Initially described by Paschkis in 1927 and named by Potts and Hirst in 1963, inverted urothelial papilloma (IUP) is an uncommon urothelial neoplasm that rarely recurs following complete surgical resection [1]. IUP is characterized by endophytic growth of urothelial nests and cords with normal/bland histology. In contrast, urothelial papilloma (UP), another rare urothelial neoplasm, is a benign proliferation of the urothelium characterized by discrete, exophytic papillary growth with central fibrovascular cores lined by urothelium of normal thickness and cytology [2]. IUP and UP are distinguishable from carcinomas as they lack architectural and cellular disorder or atypia. Prior studies of IUP and UP using single gene assays reported FGFR3 and TERT promoter mutations at various rates [3–7]. One prior study of 5 IUP tumors using a targeted next generation sequencing panel reported activating HRAS mutations in 3 tumors and an oncogenic FGFR3 mutation in one [8]. In this study, we performed whole exome and/or targeted next generation sequencing to characterize the genomic landscape of IUP and UP with the goal of identifying similarities and differences between these clinically benign entities and urothelial carcinomas.
Materials and Methods
We identified 11 IUP and 11 UP cases with sufficient tissue for molecular analysis and confirmed the diagnosis in all cases by consensus pathologic review among 7 genitourinary pathologists (YBC, AG, SWF, SKT, SJS, VER, and HAA) (Table 1). Patient ages ranged from 31 to 90 years and the male:female distribution was 16:6. For 3 IUP and 3 UP tumors in which matched normal tissue was available, we initially performed whole exome sequencing (WES) using paired tumor and germline DNA. To validate the WES results, we performed targeted deep sequencing of 468 cancer-associated genes using the MSK-IMPACT assay to a target mean coverage of 586x for the 6 cases that had undergone WES and an additional 8 IUP and 8 UP cases (11 total for each histology). For all cases, formalin-fixed, paraffin embedded (FFPE) tissue was the source of DNA used for exome as well as targeted sequencing. Genomic alterations were annotated using the OncoKB knowledge base (www.OncoKB.org) [9]. See supplementary material, Supplementary materials and methods for additional details regarding WES and the MSK-IMPACT assay.
Table 1.
shows clinicopathologic details on 11 IUP and 11 UP cases included in the study.
| Case | Histology | Sex | Age (Years) | Matched Normal | Estimated Tumor Purity (%) | Target Mean Coverage IMPACT Assay | VAF of Driver Alterations (%) |
|---|---|---|---|---|---|---|---|
| IUP 1 | Inverted Papilloma | Male | 69 | No | 60 | 481 | 19 |
| IUP 2 | Inverted Papilloma | Male | 77 | No | 75 | 501 | 45 |
| IUP 3 | Inverted Papilloma | Female | 66 | No | 80 | 613 | 45 |
| IUP 4 | Inverted Papilloma | Male | 57 | No | 80 | 677 | 46 |
| IUP 5 | Inverted Papilloma | Female | 60 | No | 85 | 539 | 47 |
| *IUP 6 | Inverted Papilloma | Male | 61 | Yes | 80 | 575 | 41 |
| IUP 7 | Inverted Papilloma | Male | 66 | No | 80 | 594 | 43 |
| IUP 8 | Inverted Papilloma | Male | 90 | Yes | 75 | 572 | 41 |
| *IUP 9 | Inverted Papilloma | Male | 53 | Yes | 80 | 579 | 41 |
| *IUP 10 | Inverted Papilloma | Male | 65 | Yes | 70 | 459 | 36 |
| IUP 11 | Inverted Papilloma | Male | 86 | No | 80 | 690 | 56 |
| UP 1 | Papilloma | Male | 70 | No | 85 | 600 | 51 |
| UP 2 | Papilloma | Male | 54 | No | 80 | 711 | 59 |
| UP 3 | Papilloma | Female | 81 | No | 80 | 405 | 59 |
| *UP 4 | Papilloma | Female | 51 | Yes | 50 | 570 | 16 |
| UP 5 | Papilloma | Male | 69 | Yes | 70 | 667 | 54 |
| UP 6 | Papilloma | Male | 65 | Yes | 20 | 409 | 3 |
| *UP 7 | Papilloma | Male | 71 | Yes | 60 | 579 | 31 |
| UP 8 | Papilloma | Female | 31 | No | 70 | 597 | 39 |
| UP 9 | Papilloma | Female | 62 | Yes | 70 | 671 | 25 |
| *UP 10 | Papilloma | Male | 51 | Yes | 50 | 875 | 22 |
| UP 11 | Papilloma | Male | 66 | Yes | 60 | 518 | 33 |
Cases that underwent WES also
Results and Discussion
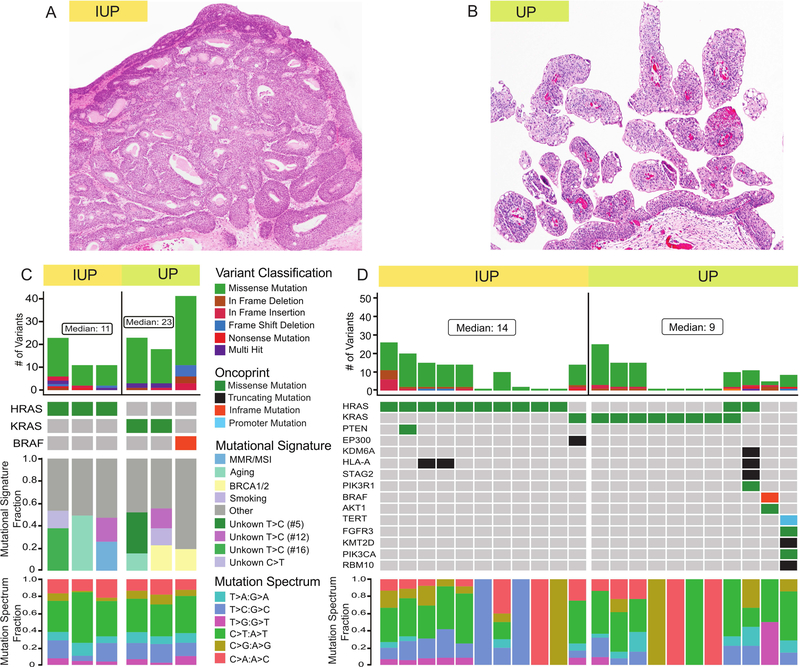
WES revealed a relatively low tumor mutational burden (TMB) with a median TMB for IUP and UP of 0.334 and 0.698 (mutations/Mb), respectively (Figure 1). This low TMB was significantly less than that reported for non-invasive bladder cancers by Hurst et al (1.64 mut/Mb based on WES of 24 tumors) [10], or muscle invasive urothelial carcinoma by the TCGA (5.8 mut/Mb based on WES of 412 tumors) [11]. Oncogenic hotspot mutations in HRAS were identified in all 3 IUPs and oncogenic hotspot KRAS mutations were present in 2 of the 3 UPs (Figure 1C). The third UP had an oncogenic BRAF mutation (T599dup). Based on the OncoKB annotation system [9], these were the only known or likely oncogenic driver mutations in these IUP and UP tumors (supplementary material, Table S1). Missense mutations were more prevalent than nonsense mutations, insertions or deletions and these single nucleotide variants (SNV) were predominately C>T transitions. Notably, the APOBEC (apolipoprotein B mRNA editing catalytic polypeptide-like) mutational signature prevalent in urothelial carcinomas [11] was absent in all IUP and UP cases (Figure 1C).
Figure 1.
Genomic landscape of inverted urothelial papilloma (IUP) and urothelial papilloma (UP). (A) H&E image of IUP at 40X magnification. (B) H&E image of UP at 40X magnification. (C) Variant classification, OncoPrint, mutational signature and mutation spectrum of 3 IUP and 3 UP cases that underwent whole exome sequencing. (D) Variant classification, OncoPrint and mutation spectrum of the 11 IUP and 11 UP cases analyzed using the MSK-IMPACT assay.
To validate and expand upon the WES results, we performed targeted NGS of the same 6 samples and 16 additional tumors (8 each of IUP and UP, Figure 1D). The results of WES and MSK-IMPACT were highly concordant in the cases that underwent both analyses (supplementary material, Figure S1). Similar to WES, missense mutations were more prevalent than nonsense mutations, insertions or deletions and SNVs were predominantly C>T transitions (supplementary material, Table S2). In this expanded cohort, all IUP tumors harbored oncogenic RAS mutations (10 HRAS and 1 KRAS mutations). TERT promoter and FGFR3 mutations, which are common in urothelial carcinomas, were not detected in any of the IUP cases. Ten of 11 UP tumors also had oncogenic mutations in the RAS/ERK signaling pathway (8 KRAS, 2 HRAS and 1 BRAF mutations). Notably, one of the UP tumors had both KRAS and HRAS mutations (with similar allele frequencies of 0.39 and 0.35). Further investigation as to whether these co-occurring RAS mutations were sub-clonal was hampered by the lack of matched normal DNA for this sample.
Only one of the cases, a UP, harbored oncogenic FGFR3 or TERT promoter mutations. This tumor, which also had oncogenic PIK3CA, KMT2D, and CDKN1A mutations, had a mutational profile common to that observed in urothelial carcinoma [10, 12]. While morphologic evaluation of this tumor, which consisted of a single small papillary lesion, was compatible with the diagnosis of papilloma, it arose in a patient who had several low-grade non-invasive papillary urothelial carcinomas prior and subsequent to the index UP tumor. This clinical history suggested that this tumor was likely a recurrence of the prior NMIBC despite the bland histopathologic appearance and that the correct histologic diagnosis was confounded by its small size. To explore this possibility, we performed MSK-IMPACT analysis on tumor tissue from two subsequent recurrent tumors from this patient, both diagnosed as non-invasive, low grade papillary urothelial carcinomas. The mutational profiles of the two recurrent tumors were nearly identical to the index UP and confirmed that all three lesions were clonally related (Figure 2 and supplementary material, Table S3). The results suggest that in the presence of a prior history of papillary urothelial carcinoma, the diagnosis of UP or IUP should be made with extreme caution and that tumor molecular profiling could help clarify whether the tumor represented a recurrence of the prior low grade urothelial cancer or a new clonally unrelated primary tumor.
Figure 2.
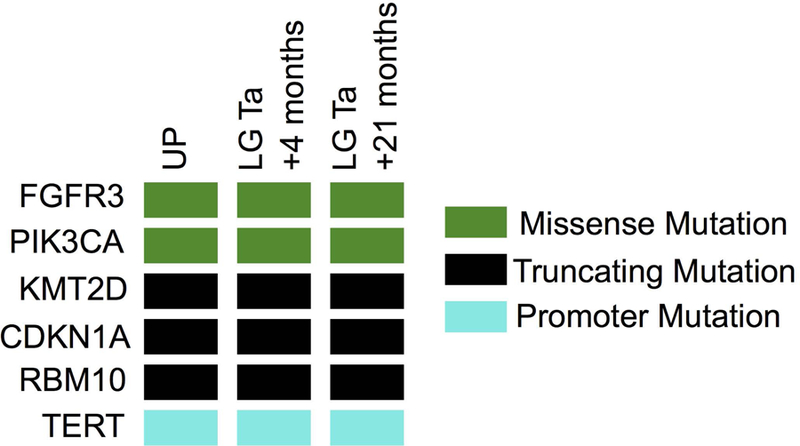
OncoPrint showing clonally related identical oncogenic mutations in index UP and two subsequent recurrent low grade non-invasive tumors (LG pTa) in a patient with history of several LG pTa urothelial cancers.
With the exception of the case outlined above and contrasting prior reports in the literature [3–7], none of the other IUP or UP tumors had TERT promoter or activating FGFR3 mutations. Similarly, in contrast to urothelial carcinomas, oncogenic alterations in chromatin remodeling genes were uncommon in both IUP and UP, being identified in only 1 IUP and 2 UP cases. In addition to the case described above, only two other patients in this series had a concurrent or subsequent bladder tumor. In one, a low grade urothelial carcinoma was diagnosed 16 months following the resection of IUP, whereas the second had a synchronous urothelial carcinoma elsewhere in the bladder. In the latter case (supplementary material, Table S3), the concurrent tumor was a plasmacytoid variant of urothelial carcinoma, which upon tumor sequencing was found to share no somatic alterations with the IUP but was rather a second genomically unrelated primary tumor [13].
In sum, our data suggest that IUP and UP are distinct molecular entities from urothelial carcinoma. Clinically, both IUP and UP are characterized by a benign clinical course with rare recurrence if completely excised. Pathologically, IUP and UP have morphologic mimics that may make the diagnosis challenging and likely explain the lack of inter-observer agreement in prior studies. For example, Eiber et al [3] reported that considerable inter-observer variability exists in histopathologic diagnoses of IUP with only 62 of 89 cases (70%) having an initial IUP diagnosis confirmed by subsequent pathology review whereas 23 cases (26%) were reclassified as inverted non-invasive urothelial carcinomas. The remaining four cases were diagnosed as cystitis cystica or glandularis with Brunn nests upon secondary review. These findings highlight the challenges in establishing the correct diagnosis of papilloma in difficult cases and underscore the importance of ancillary studies such as next generation sequencing that could be applied to properly classify such urothelial proliferations.
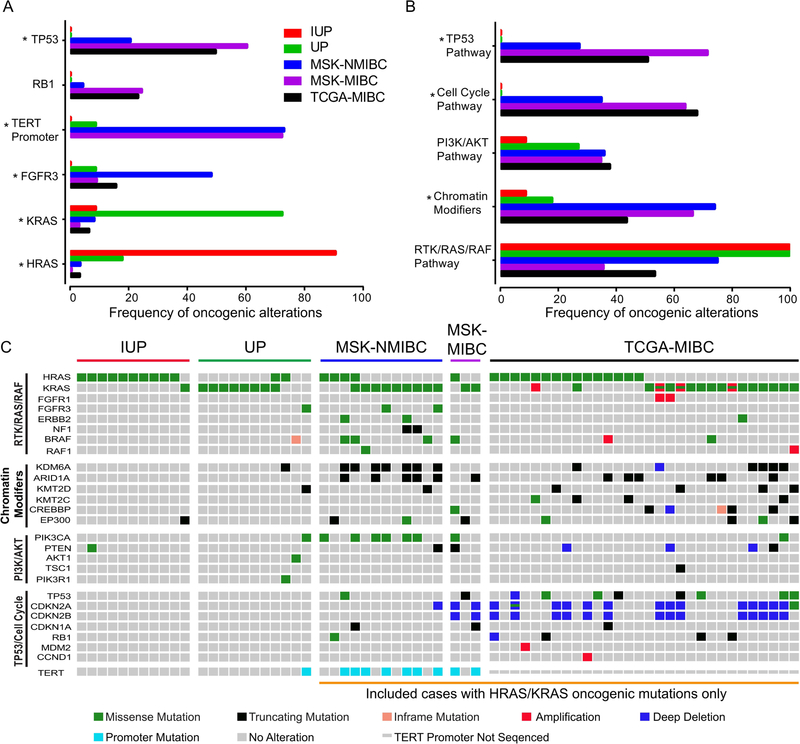
Identifying a unique molecular profile in IUP and UP, as we have shown in this study, suggests that tumor molecular profiling can help to correctly characterize these tumors and help to distinguish them from morphologic mimics. Specifically, we identified activating RAS pathway mutations as the defining feature of IUP and UP. HRAS mutations were much more common in IUP, whereas KRAS mutations were more prevalent in UP. Mutations in both KRAS and HRAS were present at the highest allele frequencies and, therefore, are likely to be early events in these tumors. The average VAF of KRAS and HRAS mutations was 41%. In addition, the median tumor cell fractions associated with KRAS and HRAS mutations in the 6 WES samples were mostly 100%, supporting that these are clonal and initiating alterations. At the nucleotide level, mutations in RAS genes in IUP or UP did not have a specific pattern. Beyond these specific alterations, these tumors were typically genetically silent with very low TMB. In contrast to non-invasive and invasive urothelial carcinomas, IUP and UP were not characterized by frequent mutations in the TERT promoter or in FGFR3, TP53, RB1 or chromatin remodeling genes or associated pathways (Figure 3A, Figure 3B and supplementary material, Table S4). Our findings contrast with prior reports of activating FGFR3 and TERT promoter mutations in IUP and UP [4–7]. Such differences could be attributed to selection bias or due to the methodologies employed in prior studies, typically single gene-based assays versus comprehensive next generation sequencing platforms. It is important to note that a small subset of urothelial carcinomas harbor hotspot mutations in HRAS and KRAS [11, 12]. These RAS mutant urothelial carcinomas, however, display considerable morphologic atypia, do not exhibit exclusive inverted growth pattern and have other genomic alterations, which distinguish them from IUP and UP (Figure 3C). While the oncogenic RAS mutations observed in IUP and UP are occasionally found in invasive urothelial carcinomas and are also common in several other highly aggressive solid tumors, the fact that none of the papillomas in this study progressed to frank urothelial carcinoma suggests that the co-mutational background or epigenetic differences in the cell of origin may dictate the likelihood that a RAS mutant cell will progress to carcinoma. Lacking a specific co-mutational background, RAS mutations in urothelial cells may induce oncogene-induced senescence and thus result in a benign lesion incapable of further progression to carcinoma [14].
Figure 3.
Genomic comparison of inverted urothelial papilloma (IUP), urothelial papilloma (UP), non-muscle invasive bladder cancer (NMIBC) and muscle invasive bladder cancer (MIBC). (A) Frequency of TP53, RB1, TERT promoter, FGFR3, KRAS and HRAS alterations in IUP (n=11), UP (n=11), MSK-NMIBC n=105 [12], MSK-MIBC (n=117; unpublished data) and TCGA-MIBC n=412 [11]. * indicates p value <0.05. (B) Frequency of oncogenic alterations in molecular pathways in IUP (n=11), UP (n=11), MSK-NMIBC n=105 [12], MSK-MIBC (n=117; unpublished data) and TCGA-MIBC n=412 [11] cohort. * indicates p value <0.05. (C) OncoPrint showing molecular pathways in IUP (n=11), UP (n=11) and cases with oncogenic HRAS/KRAS mutations in MSK-NMIBC (n=12), MSK-MIBC (n=3) and TCGA-MIBC (n=30).
One of the limitations of the current study was its power to detect infrequently mutated genes due to small sample size. This is, however, the first study to report on the genomic landscape of IUP and UP using whole exome sequencing approach as well as the largest study of IUP and UP using targeted next generation sequencing. As IUP and UP are rare entities, larger studies of IUP and UP would require multi-institutional collaboration. To facilitate such efforts, all clinical and genomic data reported here will be made available through cBioPortal for Cancer Genomics [15].
In summary, oncogenic HRAS and KRAS mutations are present in almost all IUP and UP tumors. The benign nature of IUP and UP tumors may be attributed to the lack of cooperative co-mutations in the TERT promoter, TP53 and in chromatin modifying genes, oncogenic drivers that are common in urothelial carcinomas.
Supplementary Material
Figure S1. Variant allele frequency (VAF) of genomic alterations in whole exome sequencing and the MSK-MPACT targeted panel assay
Table S1. All non-synonymous somatic alterations detected in 3 IUP and 3 UP cases by WES. Alterations listed in black and grey colors are oncogenic /likely oncogenic and putative passenger, respectively, per OncoKB
Table S2. All non-syn-onymous somatic alterations detected in 11 IUP and 11 UP cases by MSK-IMPACT assay. Alterations listed in black and grey colors are oncogenic/likely oncogenic and putative passenger respectively per OncoKB
Table S3. All non-synonymous somatic alterations detected in paired samples from two patients. One represents an index UP and two subsequent recurrent low grade non-invasive tumors (LG pTa) in a patient with history of several LG pTa. The other represents alterations in a IUP and concurrent urothelial carcinoma, plasmacytoid variant. Alterations listed in black and grey colors were classified as oncogenic/likely oncogenic versus putative passenger, respectively, per OncoKB
Table S4. List of genes that are part of five molecular pathways included in Figure 3
Acknowledgments
This study was supported by Cycle for Survival (HAA, DBS), a Ruth L. Kirschstein National Research Service Award T32CA082088 (SI), the Sloan Kettering Institute for Cancer Research Cancer Center Support Grant P30CA008748 and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology.
Footnotes
Conflict of Interest: None for the work presented in the current study.
Pietzak: Scientific Advisory Board: Merck.
Bochner: Chair of data safety monitoring committee: Genentech; Course Instructor: Olympus.
Berger: Consultant: Roche; Research Support: Illumina.
Iyer: Consultant: Bayer.
Solit: Consultant for Pfizer, Loxo Oncology, Intezyne, Vivideon Therapeutics and Illumina.
Al-Ahmadie: Consultant: Bristol-Myers-Squibb, AstraZeneca, EMD Serono.
References
*Cited only in supplementary material.
- 1.Picozzi S, Casellato S, Bozzini G, et al. Inverted papilloma of the bladder: a review and an analysis of the recent literature of 365 patients. Urol Oncol 2013; 31: 1584–1590. [DOI] [PubMed] [Google Scholar]
- 2.Miyamoto H, Miller JS, Fajardo DA, et al. Non-invasive papillary urothelial neoplasms: the 2004 WHO/ISUP classification system. Pathol Int 2010; 60: 1–8. [DOI] [PubMed] [Google Scholar]
- 3.Eiber M, van Oers JM, Zwarthoff EC, et al. Low frequency of molecular changes and tumor recurrence in inverted papillomas of the urinary tract. Am J Surg Pathol 2007; 31: 938–946. [DOI] [PubMed] [Google Scholar]
- 4.Lott S, Wang M, Zhang S, et al. FGFR3 and TP53 mutation analysis in inverted urothelial papilloma: incidence and etiological considerations. Mod Pathol 2009; 22: 627–632. [DOI] [PubMed] [Google Scholar]
- 5.Cheng L, Davidson DD, Wang M, et al. Telomerase reverse transcriptase (TERT) promoter mutation analysis of benign, malignant and reactive urothelial lesions reveals a subpopulation of inverted papilloma with immortalizing genetic change. Histopathology 2016; 69: 107–113. [DOI] [PubMed] [Google Scholar]
- 6.Cheng L, Montironi R, Lopez-Beltran A. TERT promoter mutations occur frequently in urothelial papilloma and papillary urothelial neoplasm of low malignant potential. Eur Urol 2017; 71: 497–498. [DOI] [PubMed] [Google Scholar]
- 7.van Rhijn BW, Montironi R, Zwarthoff EC, et al. Frequent FGFR3 mutations in urothelial papilloma. J Pathol 2002; 198: 245–251. [DOI] [PubMed] [Google Scholar]
- 8.McDaniel AS, Zhai Y, Cho KR, et al. HRAS mutations are frequent in inverted urothelial neoplasms. Hum Pathol 2014; 45: 1957–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakravarty D, Gao J, Phillips SM, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol 2017; 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed]
- 10.Hurst CD, Alder O, Platt FM, et al. Genomic subtypes of non-invasive bladder cancer with distinct metabolic profile and female gender bias in KDM6A mutation frequency. Cancer Cell 2017; 32: 701–715 e707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robertson AG, Kim J, Al-Ahmadie H, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell 2017; 171: 540–556 e525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pietzak EJ, Bagrodia A, Cha EK, et al. Next-generation sequencing of nonmuscle invasive bladder cancer reveals potential biomarkers and rational therapeutic targets. Eur Urol 2017; 72: 952–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Ahmadie HA, Iyer G, Lee BH, et al. Frequent somatic CDH1 loss-of-function mutations in plasmacytoid variant bladder cancer. Nat Genet 2016; 48: 356–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minoo P, Jass JR. Senescence and serration: a new twist to an old tale. J Pathol 2006; 210: 137–140. [DOI] [PubMed] [Google Scholar]
- 15.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *16.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017; 23: 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *17.Johnson BE, Mazor T, Hong C, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 2014; 343: 189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *18.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *19.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011; 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *20.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *21.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013; 31: 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *22.Ye K, Schulz MH, Long Q, et al. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 2009; 25: 2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *23.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013; 500: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *24.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Deciphering signatures of mutational processes operative in human cancer. Cell Rep 2013; 3: 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Variant allele frequency (VAF) of genomic alterations in whole exome sequencing and the MSK-MPACT targeted panel assay
Table S1. All non-synonymous somatic alterations detected in 3 IUP and 3 UP cases by WES. Alterations listed in black and grey colors are oncogenic /likely oncogenic and putative passenger, respectively, per OncoKB
Table S2. All non-syn-onymous somatic alterations detected in 11 IUP and 11 UP cases by MSK-IMPACT assay. Alterations listed in black and grey colors are oncogenic/likely oncogenic and putative passenger respectively per OncoKB
Table S3. All non-synonymous somatic alterations detected in paired samples from two patients. One represents an index UP and two subsequent recurrent low grade non-invasive tumors (LG pTa) in a patient with history of several LG pTa. The other represents alterations in a IUP and concurrent urothelial carcinoma, plasmacytoid variant. Alterations listed in black and grey colors were classified as oncogenic/likely oncogenic versus putative passenger, respectively, per OncoKB
Table S4. List of genes that are part of five molecular pathways included in Figure 3