Abstract
Various methods for understanding the structural and dynamic properties of proteins rely on the analysis of their NMR chemical shifts. These methods require the initial assignment of NMR signals to particular atoms in the sequence of the protein, a step that can be very time-consuming. The PINE (Probabilistic Interaction Network of Evidence) algorithm for automated assignment of backbone and side chain chemical shifts utilizes a Bayesian probabilistic network model that analyzes sequence data and peak lists from multiple NMR experiments. PINE, which is one of the most popular and reliable automated chemical shift assignment algorithms, has been available to the protein NMR community for longer than a decade. We announce here a new web server version of PINE, called I-PINE (Integrative PINE), which supports more types of NMR experiments than PINE (including three-dimensional nuclear Overhauser enhancement (NOE) and four-dimensional J-coupling experiments) along with more comprehensive visualization of chemical shift based analysis of protein structure and dynamics. The I-PINE server is freely accessible at http://i-pine.nmrfam.wisc.edu. Help pages and tutorial including browser capability are available at: http://i-pine.nmrfam.wisc.edu/instruction.html Sample data that can be used for testing the web server are available at: http://i-pine.nmrfam.wisc.edu/examples.html
Keywords: AUTOMATION, PROTEIN NMR CHEMICAL SHIFT ASSIGNMENT, ANALYSIS OF PROTEIN NMR CHEMICAL SHIFTS, INTEGRATIVE NMR
Introduction
One of the fundamental steps in NMR spectroscopy is to correlate NMR signals to atoms of certain residues. Once the `chemical shift assignment` is completed, a variety of applied studies can follow. Information on structure and function can be gained from changes in assigned NMR observables (chemical shifts, line widths, peak intensities, exchange effects, etc.) as a function of alterations in pH, temperature, protein concentration, or added ligand. Constraints required for calculation of three-dimensional structures (e.g., distance, dihedral angle, residual dipolar coupling) are obtained from additional NMR experiments analyzed in terms of the assigned chemical shifts.
However, the task of assigning NMR chemical shifts from proteins can be very time-consuming and laborious, not only for non-specialists but also for experienced spectroscopists. Hundreds to thousands of signals from different NMR spectra need to be detected and assigned to extract scientifically meaningful information. The PINE (Probabilistic Interaction Networking of Evidence) algorithm and associated computer program that automates chemical shift assignments1 has become one of the most popular protein assignment tools over the past decade. In PINE, chemical shift assignments are defined as a bipartite matching problem, and Bayesian networks are used to integrate multiple pieces of evidence with applied belief propagation utilized to achieve maximum accuracy and reliability. To make this program more user friendly, we developed an associated toolkit called PINE-SPARKY2, which supports graphical user interfaces (GUIs) that display the probabilistic assignments recommended by PINE against experimental spectra. These tools were developed as extension plugins for NMRFAM-SPARKY3, our upgraded version of Sparky (Goddard and Kneller, SPARKY 3). Because PINE-SPARKY and other useful automation and visual integration tools are pre-installed in NMRFAM-SPARKY, an extra installation step is not required and the software works seamlessly with other tools that are part of the Integrative NMR research platform4. We also used PINE along with reduced dimensionality and automated signal detection algorithms to build ADAPT-NMR, which combines protein data acquisition on a Varian or Bruker spectrometer and peak assignments5,6.
We report here a new version of PINE, Integrative PINE (I-PINE) along with its web server (Fig. 1). Like its predecessor, I-PINE utilizes a probabilistic interaction network under a Bayesian framework to assign protein NMR data. In comparison to PINE, I-PINE supports more types of NMR experiments, including three- and four-dimensional nuclear Overhauser enhancement (NOE) and J-coupling experiments, integrates real-time statistical analysis of the PACSY database7, and supports more comprehensive visualization of chemical shift based analysis of protein structure and dynamics. Analysis of data from a variety of proteins of varying size has shown that I-PINE produces higher assignment coverage and accuracy than PINE.
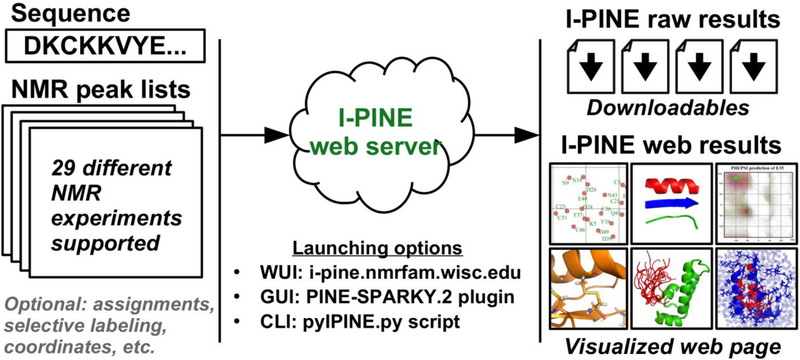
Fig. 1.
Cartoon representation of the I-PINE web server. The input is a sequence file and peak lists from up to 29 different NMR experiments. The three different job submission methods are available: WUI (web-based user interface), GUI (graphical user interface) and CLI (command-line user interface). After the I-PINE job complete, the users has access, through the job ID, to the raw results and visualizations of the results in web pages.
The I-PINE web server not only assigns chemical shifts but also conducts chemical shift-based structural analysis in an automated manner. The provision of I-PINE through a web server eliminates the complex steps required to install the different programs it uses to prepare input files and analyze output files.
I-PINE web server
We optimized the I-PINE program through repetitive, systematic analysis of experimental protein NMR data from 85 selected BMRB entries. The data sets for these proteins represent different combinations of NMR experiments. We have posted the evaluation results to the I-PINE user group web site (http://i-pine.nmrfam.wisc.edu/usergroup.html) and to the bottom of the I-PINE example page (http://i-pine.nmrfam.wisc.edu/examples.html) along with algorithmic details. The beta version of the I-PINE web server was launched in March, 2018, and the release candidate version was introduced in November, 2018.
Job submission.
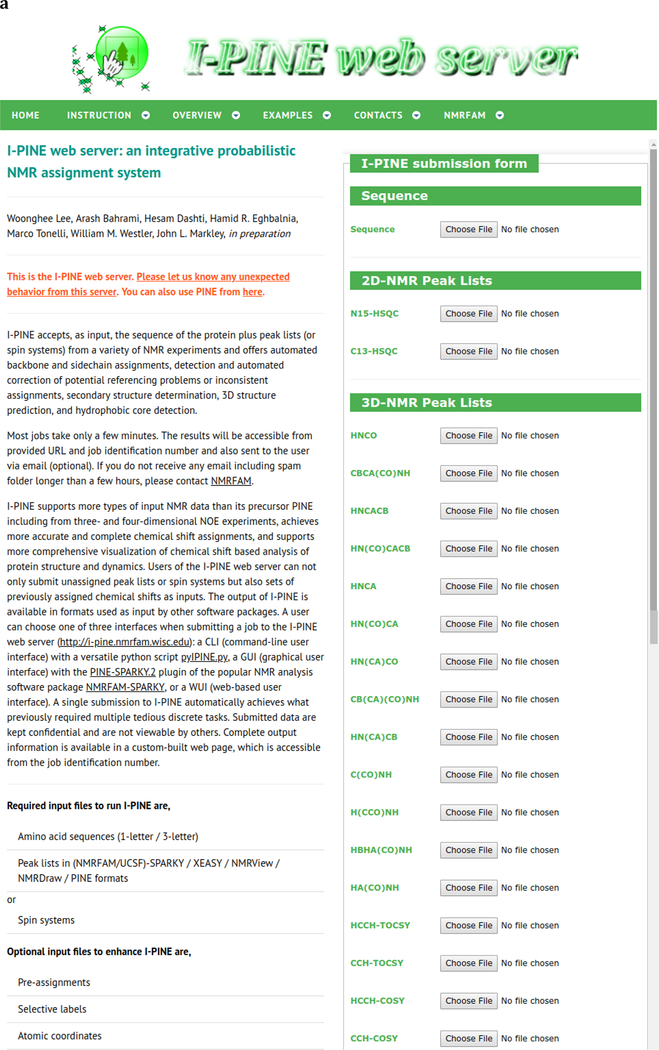
Jobs can be submitted to the I-PINE web server by three different ways: i) by a CLI (command line interface) with a versatile Python script pyIPINE.py; ii) by a WUI (web-based user interface); or iii) by the GUI (graphical user interface) provided by the PINE-SPARKY.2 plugin from NMRFAM-SPARKY. According to statistics from 2018, the GUI provided by PINE-SPARKY.2 plugin from NMRFAM-SPARKY is most popular choice (~55% of all submitted jobs). This approach avoids the need for input preparation, integrates signal detection with the assignment engine, and supports the verification of assignments against experimental data by a few mouse clicks.
Immediately after a submission, the I-PINE web server assigns a unique Job ID of the format (YYMMDD_HHMMSS_XXXXXXX: date, time and random numbers). Complete output information is available in a web page associated with that Job ID. This enables the I-PINE web server to be used anonymously without providing user information. However, if the user supplies an email address, the results will be delivered by via email. Submitted data are kept confidential and are not accessible by others without the Job ID.
The I-PINE web server provides in its “example page” data sets for four different proteins (http://i-pine.nmrfam.wisc.edu/examples.html). Input files and results for these examples are available for examination. The data sets for three of the proteins (ubiquitin, vitamin D receptor, sterol carrier protein-2) consist of simple backbone peak lists and sequence files. Data for the fourth protein (brazzein) is provided as a pyIPINE.py script. Furthermore, any BMRB entry containing protein NMR data can be used as input to the I-PINE web server by using the deposited information to prepare the required sequence and peak list files.
Input data.
As described in the I-PINE instruction page (http://i-pine.nmrfam.wisc.edu/instruction.html), the minimal input required for I-PINE are three files: a sequence file, a peak list containing pairs of 1HNi and 15Ni chemical shifts from individual residues as generated by an experiment such as 1H,15N HSQC, 3D HNCO, 3D HN(CO)CA, or 4D HNCOCA, and a 3D or 4D peak list from an experiment that generates sets of (1HNi, 15Ni, 13Cαi) and (1HNi-1, 15Ni-1, 13Cαi-1) chemical shifts such as 3D HNCA or 3D HNCACB. Data from additional experiments, including TOCSY and NOESY, will enhance the overall quality of the assignments.
Sequence file.
Both one- and three-letter sequence codes are accepted. A FASTA one-letter code file12 or an NMR-STAR 3.1 file with the “_Entity_comp_index.Comp_ID” tag are also supported. Although, I-PINE expects an ASCII text file, the software recognizes files with RTF (Rich Text Format; .rtf), ODT (OpenDocument Text; .odt), and DOCX (Office Open XML; .docx) file extensions and converts them automatically to ASCII. Currently, only standard amino acids are accepted; however, I-PINE will soon support common non-standard residues occurring in PDB and BMRB as defined in the Chemical Component Dictionary (https://www.wwpdb.org/data/ccd).
Peak lists.
Peak lists in Sparky (NMRFAM-SPARKY), XEASY13, NMRView14, NMRDraw15 or PINE simple formats are accepted as inputs. CARA and CCPNmr Analysis users can use Sparky format files by using the WriteSparkyPeakList.lua script or Format Converter program, respectively16,17. Peak lists from one NH root experiment and one experiment providing CA for previous and current residues of each NH peak are required unless the user provides a spin system file or a pre-assignment file. The most common peak list combinations are 1H,15N HSQC, CBCA(CO)NH, and HNCACB for backbone assignments and 1H,13C-HSQC, C(CO)NH, H(CCO)NH, and HCCH-TOCSY for side chain assignments. We recommend including peak intensity information in peak lists, especially for the constant time version of 1H,13C-HSQC, because the software makes use of this information. Future plans are for the I-PINE web server to import and export peak lists in the NMR-STAR 3.2 file format18 used by BMRB.
Spin systems.
The software uses peak lists to generate spin system matrices. Thus, if the user provides already established spin systems as input, peak lists are not required. The provision of spin systems by users can be useful in cases where overlapped resonances confuse the software, for example, when root nitrogen and amide hydrogen signals from different residues overlap.
Pre-assignment.
Pre-assignments can be uploaded when a user wants to improve assignment quality by reducing the number of assignment choices. If complete pre-assigned chemical shifts are uploaded, the I-PINE software can be used to carry out chemical shift-based analysis. In this case, the I-PINE web server will use the assigned chemical shifts to simulate 1H,15N HSQC, HNCO, HN(CA)CO, HN(CO)CA, HNCA, HN(CO)CACB, HNCACB, HA(CO)NH, HBHA(CO)NH, HBHANH, C(CO)NH, H(CCO)NH, HCCH-TOCSY, CCH-TOCSY, 1H,13C HSQC and TOCSY-1H,15N HSQC peak lists and run a regular job as if these experiments and a pre-assignment had been submitted. Accepted formats for pre-assigned chemical shifts are NMR-STAR 2.1 or newer versions.
Atomic coordinates.
If the user already has a 3D structure of a protein, atomic coordinates can be also submitted to improve assignment quality. The I-PINE web server accepts atomic coordinates in PDB or PDBx/mmCIF format and uses this information as input to STRIDE, which extracts secondary structural features for use by I-PINE in refining the initial probability density functions. In addition, the availability of 3D coordinates improves the efficiency of AUDANA19, the database assisted NOE assignment algorithm that interfaces with NMRFAM-SPARKY and I-PINE.
Outputs
Running time is determined by the size of protein and complexity of the problem. For example, the backbone assignments of human ubiquitin, from the 1H,15N-HSQC, CBCA(CO)NH and HNCACB data set provided in I-PINE web server, usually takes two to three minutes. If data from side chain experiments are submitted, the job will require several more minutes. Once a job finishes, results are posted in a customized web page assigned to the Job ID. Two URLs are provided immediately after submitting a job: I-PINE raw results and I-PINE web results. I-PINE raw results consists of a web page with a list of output files for the user to download or view in a web browser (this is similar to PINE results but with many more files). I-PINE web results are more visually intuitive and interactive. All the plots are SVG (Scalable Vector Graphics) files that are not pixelated. For reference, the I-PINE web server displays I-PINE web results for the four protein examples.
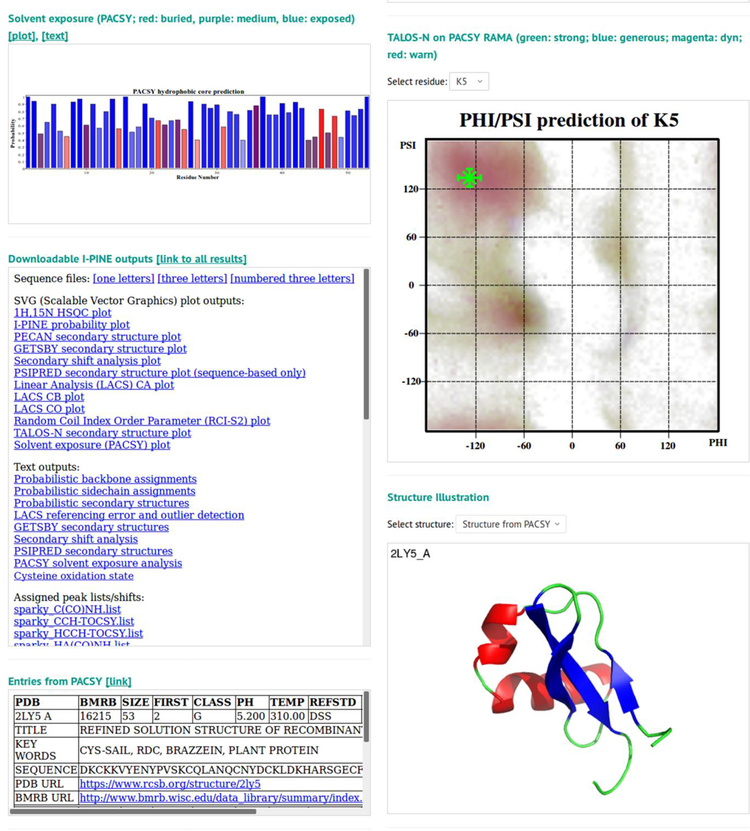
Identification of related information.
If the user-submitted protein sequence matches records in the SEQ_DB table or PDBSEQ_DB table of the PACSY database, a section called “Entries from PACSY” appears. If the match is to SEQ_DB, which carries entries deposited in PDB and BMRB, the software returns the relevant PDB / BMRB accession codes, as well as details including title, pH, temperature, keywords, and URL links to the PDB / BMRB archives. If the match is to PDBSEQ_DB, which contains only PDB-deposited sequences, the software will provide PDB accession codes and URL links to the PDB archive.
Reference correction and outliers.
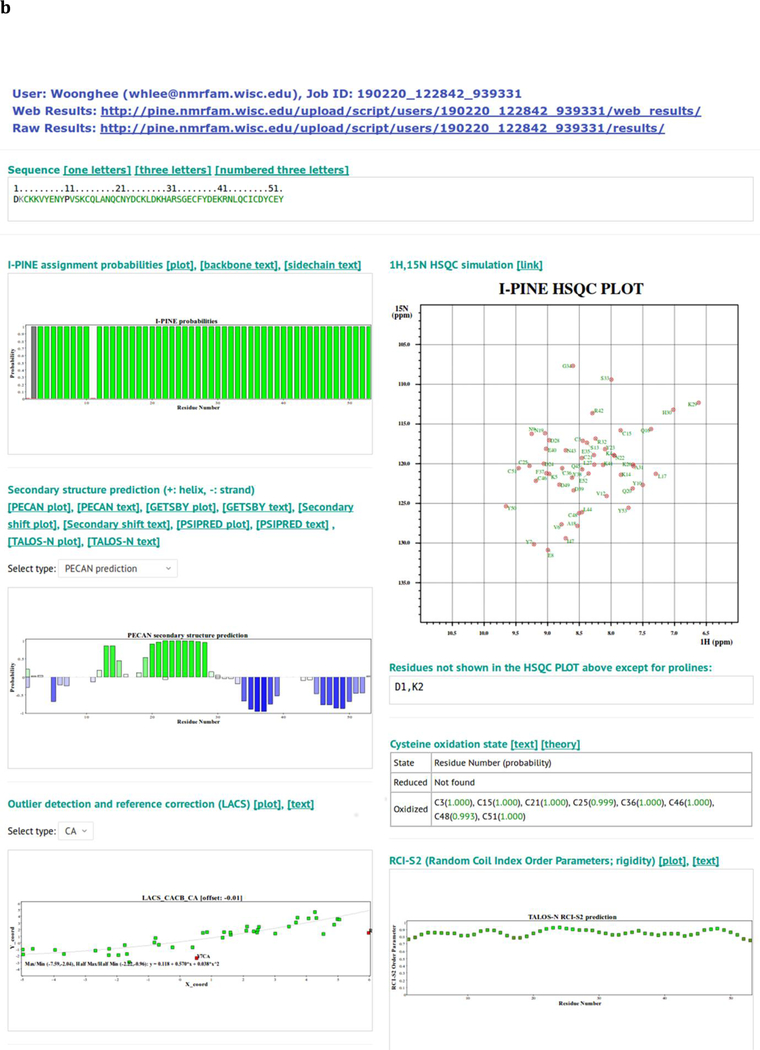
LACS (Linear Analysis of Chemical Shifts)21 results from assigned chemical shifts of CA, CB and CO are visualized in a 2D plot; these report on potential referencing errors and erroneous assignments. An option in NMRFAM-SPARKY (two-letter-code st) enables re-referencing on the basis of LACS results, which has been found to improve assignment accuracy and completeness.
Assignment probabilities.
A bar graph at the top of the page displays color-coded assignment probabilities for the backbone amides of each residue in the protein sequence. Green indicates assignment probabilities higher than 0.99, cyan between 0.85 and 0.99, yellow between 0.50 and 0.85, and red smaller than 0.50. A gray bar indicates no assignment available, and proline residues, which lack an amide hydrogen, are shown in black. Sequence files in ASCII format are also available for download with user-selected format: one-letter, three-letter, or three-letter codes with indices.
Simulated 1H,15N correlation spectrum.
A 2D plot displays a simulated 1H,15N correlation spectrum with assigned peaks color-coded to indicate assignment probability. The color-code is similar to that used in the assignment probability bar graph, but with darker colors on a white background for better visual recognition (dark green: higher than 0.99; blue: between 0.85 and 0.99; mustard: between 0.50 and 0.85; red: lower than 0.50). Because the plot is SVG (scalable vector graphics), it is not pixelated. A “find” and “zoom” tool is available in the web browser. Residues that have not been assigned (excluding prolines) are listed below the simulated spectrum.
Secondary structure analysis.
The web browser displays secondary structure analysis results from five different programs: PECAN20, GetSBY21, Secondary Shift Analysis, and TALOS-N22, which utilize NMR chemical shifts, and PSI-PRED20, which utilized peptide sequence alone. 2D SVG plots and ASCII text are provided as links.
Analysis of cysteine oxidation state.
The sulfur atom in the cysteine side chain is a highly reactive nucleophile. In its reduced form, it may be involved in catalysis, and in its oxidized, disulfide, form it plays roles in structural stabilization and dimerization. On the basis of statistics relating cysteine CB chemical shifts to oxidation state26,27, I-PINE provides probabilistic oxidation states for all assigned cysteine residues.
Flexibility.
Berjanskii and Wishart.22 developed a simple method, called random coil index (RCI) that predicts protein flexibility from secondary chemical shifts of backbone atoms. Predicted order parameters (RCI-S2) are similar to model-free order parameters (S2)23 calculated from T1, T2 and hetNOE data. I-PINE web results provide RCI-S2 values as a function of sequence in a 2D plot.
Solvent exposure.
The I-PINE web server provides an estimate of solvent exposure. The assigned backbone CA, CB, and CO chemical shifts are used in querying the PACSY database, which utilizes known assigned chemical shifts associated with structure to predict solvent accessible surface area (SASA)24. In terms of Glycine-X-Glycine SASA, PACSY defines a residue presenting 30% or higher as water exposed, between 10% and 30% as a medium, and below 10% as buried. Probabilities are derived from histograms generated from the correlation of PACSY DB information on each residue type X: X_DB, X_STRC_DB, and X_CS_DB with a bin size of 2.0 ppm. Solvent exposure results are available as an ASCII text file and as a color-coded 2D SVG plot (blue, exposed; purple, medium; red, buried; gray, no prediction).
Dihedral angles.
Protein backbone φ/ψ dihedral angles predicted by the TALOS-N program22 are provided as PACSY RAMA4 plots. The dark area in PACSY RAMA indicates frequently appearing regions for an amino acid type. Upon selecting a residue type, the I-PINE results are displayed interactively on the PACSY RAMA plot. Reliabilities are color-coded (green: strong; blue: generous; magenta: dynamic; red: warning). Horizontal and vertical crosses show ±σ (standard deviation) while diagonal crosses show predicted φ/ψ angles. Full TALOS-N results, including angle constraint files in XPLOR and DYANA formats25,26 for 3D structure calculation, are available for download.
3D structures.
If any 3D structure is available for the protein of interest, it can be shown as a ribbon diagram. A structure may be available from various sources: a) a structure of the same sequence found in PACSY; b) atomic coordinates submitted by the user; c) a CS-ROSETTA structure27 calculated from I-PINE chemical shifts; d) an AUDANA-calculated 3D structure from I-PINE chemical shifts and user-submitted 13C-NOESY and 15N-NOESY data using PONDEROSA-C/S19,28. Because CS-ROSETTA and AUDANA require a few hours in general, I-PINE web results will be returned before the structure calculations are completed. Once the structural results are in, the I-PINE web server automatically incorporates them into the I-PINE web results. If an email is provided, the user will be notified; otherwise, the user needs to check the I-PINE web results page periodically to determine whether the results are available.
Discussion and future prospects
In PINE, the initial probability density function for chemical shifts is generated by normalizing occurrences for curated entries in BMRB (BioMagResBank)8. For I-PINE, we developed a simple real-time preprocessor that customizes the initial probability density function by querying PACSY with information available from the submission: protein sequence and atomic coordinates, if available. An additional improvement of I-PINE over PINE results from the use of peak intensities. Some NMR experiments (e.g., constant time 1H,13C-HSQC, HNCACB, HN(CO)CACB) produce peaks that are positive or negative depending on atom types or the number of attached covalent bonds. I-PINE recognizes intensity columns in submitted peak list files and uses them when they are available. In parallel, we have improved the overall assignment quality in terms of both accuracy and completeness by optimizing I-PINE parameters thorough the systematic evaluation of entries in the BMRB. I-PINE now supports 29 different NMR experiments, including three-dimensional nuclear Overhauser enhancement (NOE) and four-dimensional J-coupling experiments (Table 1).
Table 1.
The twenty-nine NMR experiments currently supported by I-PINE. Expected peak profiles are available from the I-PINE web page (http://i-pine.nmrfam.wisc.edu/files/nmr_experiment_profile.txt).
| 2D experiments | 1H,15N HSQC, 1H,13C HSQC |
| 3D experiments | HNCO, HN(CA)CO, HN(CO)CA, HNCA, CBCA(CO)NH, CB(CA)(CO)NH, HN(CO)CACB, HNCACB, HN(CA)CB, HNHA,a HBHANH,a HA(CO)NH,a HBHA(CO)NH, HN(CA)NNH,a H(CCO)NH, C(CO)NH, (H)CCH-TOCSY,a H(C)CH-TOCSY, (H)CCH-COSY,a H(C)CH-COSY,a TOCSY−15N HSQC,a 13C-filtered NOESY,a 15N-filtered NOESYa |
| 4D experiments | HNCOCA,a HNCACO,a simultaneous HNCOCA,a HNCO−1CAa |
Additionally supported experiments in I-PINE program.
I-PINE serves as a component of an evolving NMR research system that integrates more than fifty auxiliary programs and databases. Discrete tasks requiring complex manipulation of inputs and outputs are no longer necessary. Results are visualized on a web page that can be readily accessed from a web browser. The PACSY database is updated monthly with new information from the PDB and BMRB.
Future plans are to develop I-PINE versions that support more NMR experiments and automatically analyze raw data so as to eliminate peak picking and spectral alignment steps. The goal is to be able to handle more difficult biological systems, such as those that exhibit multiple conformations or asymmetric dimers. In addition, we plan to support experiments for aromatic side chain atoms, such as HBCBCGCDCEHE and HBCBCGCDHD. These experiments, in our experience, do not always provide sharp and well-resolved peaks. Therefore, we are developing a separate flexible program as both an I-PINE plug-in and as a free-standing program. A solid-state version is also under development and will be available soon.
Web server availability.
I-PINE web server is freely available from http://i-pine.nmrfam.wisc.edu for academic users.
Fig. 2.
Screen shots of interfaces provided by I-PINE web server. a Web user interface (WUI) collecting input data prior to launching I-PINE. b I-PINE web results page. Included are the protein sequence color coded by assignment probability, a bar graph with assignment probabilities, a bar graph with PECAN analysis of secondary structure, results from LACS analysis, a simulated 2D 1H,15N HSQC spectrum color coded by assignment probability with list on non-assigned residues (excluding proline), predictions of oxidation states of cysteine residues, random coil index order parameter plot, predicted solvent exposure, table with link to I-PINE output, available PDB and BMRB entries from PACSY, interactive predicted (phi, psi) dihedral angles from TALOS-N on Ramachandran plot, structure predicted from PACSY.
ACKNOWLEDGEMENTS
This work was supported by a grant (P41GM103399) from the Biomedical Technology Research Resources (BTRR) Program of the National Institute of General Medical Sciences (NIGMS), National Institutes of Health (NIH).
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
REFERENCES
- 1.Bahrami A, Assadi AH, Markley JL & Eghbalnia HR Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLoS Comput. Biol 5, e1000307 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee W, Westler WM, Bahrami A, Eghbalnia HR & Markley JL PINE-SPARKY: graphical interface for evaluating automated probabilistic peak assignments in protein NMR spectroscopy. Bioinformatics 25, 2085–2087 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee W, Tonelli M & Markley JL NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee W et al. Integrative NMR for biomolecular research. J. Biomol. NMR 64, 307–332 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bahrami A et al. Robust, Integrated Computational Control of NMR Experiments to Achieve Optimal Assignment by ADAPT-NMR. PloS One 7, e33173 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee W et al. Fast automated protein NMR data collection and assignment by ADAPT-NMR on Bruker spectrometers. J. Magn. Reson 236, 83–88 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee W et al. PACSY, a relational database management system for protein structure and chemical shift analysis. J. Biomol. NMR 54, 169–179 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ulrich EL et al. BioMagResBank. Nucleic Acids Res. 36, D402–408 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLano WL & Lam JW PyMOL: A communications tool for computational models. Abstr Pap Am Chem S 230, U1371–U1372 (2005). [Google Scholar]
- 10.Frishman D & Argos P Knowledge-based protein secondary structure assignment. Proteins 23, 566–579 (1995). [DOI] [PubMed] [Google Scholar]
- 11.Berman H, Henrick K, Nakamura H & Markley JL The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 35, D301–303 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pearson WR [5] Rapid and sensitive sequence comparison with FASTP and FASTA in Methods in Enzymology 183, 63–98 (Academic Press, 1990). [DOI] [PubMed] [Google Scholar]
- 13.Bartels C, Xia T, Billeter M, Güntert P & Wüthrich K The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR 6, 1–10 (1995). [DOI] [PubMed] [Google Scholar]
- 14.Johnson BA Using NMRView to Visualize and Analyze the NMR Spectra of Macromolecules in Protein NMR Techniques 278, 313–352 (Humana Press, 2004). [DOI] [PubMed] [Google Scholar]
- 15.Delaglio F et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 16.Keller R Optimizing the process of nuclear magnetic resonance spectrum analysis and computer aided resonance assignment. (ETH Zurich, 2004). [Google Scholar]
- 17.Vranken WF et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59, 687–696 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Ulrich EL et al. NMR-STAR: comprehensive ontology for representing, archiving and exchanging data from nuclear magnetic resonance spectroscopic experiments. J. Biomol. NMR (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee W, Petit CM, Cornilescu G, Stark JL & Markley JL The AUDANA algorithm for automated protein 3D structure determination from NMR NOE data. J. Biomol. NMR 65, 51–57 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones DT Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol 292, 195–202 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Wang L, Eghbalnia HR, Bahrami A & Markley JL Linear analysis of carbon-13 chemical shift differences and its application to the detection and correction of errors in referencing and spin system identifications. J. Biomol. NMR 32, 13–22 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Berjanskii MV & Wishart DS A simple method to predict protein flexibility using secondary chemical shifts. J. Am. Chem. Soc 127, 14970–14971 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Lipari G & Szabo A Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc 104, 4546–4559 (1982). [Google Scholar]
- 24.Lee B & Richards FM The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol 55, 379–400 (1971). [DOI] [PubMed] [Google Scholar]
- 25.Brünger AT et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr 54, 905–921 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Güntert P, Mumenthaler C & Wüthrich K Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol 273, 283–298 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Shen Y et al. Consistent blind protein structure generation from NMR chemical shift data. Proc. Natl. Acad. Sci 105, 4685–4690 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee W, Stark JL & Markley JL PONDEROSA-C/S: client–server based software package for automated protein 3D structure determination. J. Biomol. NMR 60, 73–75 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]