Abstract
Gene therapy of malignant gliomas has shown a lack of clinical success to date due in part to inability of conventional gene vectors to achieve widespread gene transfer throughout highly disseminated tumor areas within the brain. Here, we demonstrate that newly engineered polymer-based DNA-loaded nanoparticles (DNA-NP) possessing small particle diameters (~50 nm) and non-adhesive surface polyethylene glycol (PEG) coatings efficiently penetrate brain tumor tissue as well as healthy brain parenchyma. Specifically, this brain-penetrating nanoparticle (BPN), following intracranial administration via convection enhanced delivery (CED), provides widespread transgene expression in heathy rodent striatum and an aggressive brain tumor tissue established orthotopically in rats. The ability of BPN to efficiently traverse both tissues is of great importance as the highly invasive glioma cells infiltrated into normal brain tissue are responsible for tumor recurrence. Of note, the transgene expression within the orthotopic tumor tissue occurred preferentially in glioma cells over microglial cells. We also show that three-dimensional (3D) multicellular spheroids established with malignant glioma cells, unlike conventional two-dimensional (2D) cell cultures, serve as an excellent in vitro model reliably predicting gene vector behaviors in vivo. Briefly, DNA-NP possessing greater surface PEG coverage exhibited more uniform and higher-level transgene expression both in the 3D model and in vivo, whereas the trend was opposite in 2D culture. The finding here alerts that gene transfer studies based primarily on 2D cultures should be interpreted with caution and underscores the relevance of 3D models for screening newly engineered gene vectors prior to their in vivo evaluation.
Keywords: malignant gliomas, synthetic gene vectors, tumor spheroids, convection enhanced delivery, volume of transgene expression
1. Introduction
Despite the most advanced multimodal therapeutic regimen, median survival of malignant gliomas remains ~ 14 months [1]. Gene therapy potentially provides an alternative means to achieve more specific and powerful as well as longer-lasting therapeutic benefits. However, its clinical relevance is yet to be addressed due in large part to suboptimal gene transfer efficacy. Given the highly aggressive, invasive and infiltrative nature of malignant gliomas, inability of conventional gene vectors to provide a widespread therapeutic transgene expression is an utmost challenge [2–4]. We have previously determined the brain extracellular matrix (ECM) to be a critical adhesive (i.e. hydrophobic and negatively charged) and steric diffusional barrier, and established design criteria for engineering nanoparticle-based therapeutic delivery systems that can efficiently spread through the ECM [5–7]. Specifically, we found that nanoparticles with diameters small enough (≤ ~110 nm) to fit through the ECM pores while possessing non-adhesive dense surface polyethylene glycol (PEG) coatings rapidly penetrated rodent and human brain tissues [5, 8–10].
While being well-characterized for outstanding efficacies as a polymeric gene delivery platform, widespread use of polyethylenimine (PEI) is largely discouraged by its safety concern [11, 12]. However, we note that PEI toxicity is often overstated based on in vitro studies where conventional cytotoxicity is measured with only one or two cell types at very high doses that would unlikely be translatable in vivo. In reality, PEI remains one of those few polymer-based platforms most widely explored in clinic [13, 14], including an ongoing phase II trial (NCT02806687), with a proven favorable safety profile (NCT01274455) [15]. Further, PEI has been commonly derivatized with PEG to improve its pharmacokinetic behaviors and/or safety profiles as a gene delivery system [9, 16–19]. To this end, we have recently demonstrated that DNA-loaded nanoparticles (DNA-NP) formulated with an inclusion of densely PEGylated PEI (PEG-PEI) provide widespread transgene expression in healthy rat brain while exhibiting good in vivo safety profiles, namely brain-penetrating nanoparticles (BPN) [9, 19]. However, the finding does not readily ensure their ability to penetrate brain tumor tissues, given that the barrier properties of tumor ECM are unlikely analogous to those of normal ECM due to significant pathological changes in cellularity and macromolecular compositions in tumors [20]. In this study, we thus sought to engineer and evaluate PEI-based DNA-NP possessing a broader range of surface PEG contents to identify a lead system providing widespread transgene expression in brain tumor in vivo.
Most of the studies developing gene vectors for cancer therapy employ routine sequential experimental steps of screening newly designed systems using two-dimensional (2D) cell cultures [21–23], followed by in vivo evaluation. However, conventional 2D cultures are limited in recapitulating the highly complex and heterogeneous nature of tumors established in living organisms, including humans [24, 25]. Indeed, virtually no correlation has been observed between performances of gene vectors in 2D cultures versus in animals in a recent study [26]. Alternatively, three-dimensional (3D) models of multicellular tumor spheroids mimic several features found in tumors in vivo, including but not limited to dense ECM formation [27, 28] and induction of hypoxia and necrosis [29, 30], thereby providing a reliable surrogate for screening gene vectors, particularly in terms of predicting their in vivo behaviors [31]. Thus, we here evaluate and compare performances of newly designed aforementioned PEI-based DNA-NP in a 3D spheroid model as well as in an orthotopically-established rat glioma.
2. Materials and methods
2.1. DNA-NP formulation and characterization
2.1.1. Polymer preparation.
Methoxy PEG N-hydroxysuccinimide (mPEG-NHS, 5 kDa, Sigma-Aldrich, St. Louis, MO) was conjugated to 25 kDa branched PEI (Sigma-Aldrich) to yield PEG-PEI copolymers, as previously described [9, 16]. Briefly, PEI was dissolved in ultrapure distilled water, pH was adjusted to 7.5–8.0 and then mPEG-NHS was added to the PEI solution at various molar ratios, followed by an overnight reaction at 4 °C under a constant mixing condition. The resultant polymer solution was dialyzed extensively against ultrapure distilled water for 3 days at a molecular weight cut-off (MWCO) of 50,000 kDa (Spectrum Laboratories, Inc., Rancho Dominguez, CA) and subsequently lyophilized. Nuclear magnetic resonance (NMR) analysis was conducted to confirm PEG to PEI molar ratios of 8, 30, 50, 60, and 85. 1H NMR (500 MHz, D2O): δ 2.48–3.20 (br, CH2CH2NH), 3.62–3.72 (br, CH2CH2O). The lyophilized polymers were dissolved in ultrapure distilled water with a pH adjustment to ~6.5 – 7 at 30 mg/ml and stored at 4 °C until use.
2.1.2. DNA-NP formulation.
The luciferase-expressing plasmids driven by the short-acting cytomegalovirus (CMV) promoter (i.e. pd1GL3-RL) was a kind gift from Professor Alexander M. Klibanov (M.I.T). The ZsGreen- and green-fluorescent protein (GFP)-expressing plasmids driven by the CMV promoter were purchased from Clontech Laboratories Inc. (Mountainview, CA). The luciferase- and mCherry-expressing plasmids driven by human β-actin promoter (i.e. pBAL and pBACH, respectively) were produced and provided by Copernicus Therapeutics (Cleveland, OH). Plasmids were propagated and purified, as previously described [16, 32]. Briefly, plasmids were transformed into E. coli DH5α competent bacterial cells using a heat shock method, and following the bacterial expansion in LB media, plasmids were purified using EndoFree Plasmid Giga Kit (QIAGEN, Valencia, CA), as per manufacturer’s protocol. Mirus Label IT Tracker Intracellular Nucleic Acid Localization Kit (MirusBio, Madison, WI) was used to fluorescently tag plasmids with Cy5 fluorophores. DNA-NP were formed by dropwise addition of 10 volumes of labeled or unlabeled plasmids (0.2 mg/mL) to 1 volume of a swirling polymer solution. All DNA-NP were prepared at an optimized nitrogen to phosphate (N/P) ratio of 6. For the formulation of conventional PEGylated nanoparticles (CPN), PEG-PEI copolymers prepared at a PEG to PEI molar ratio of 8 alone were used to condense plasmids. BPN formulations were engineered by condensation of plasmids by a mixture of non-PEGylated PEI (25%) and PEG-PEI (75%) synthesized at higher PEG to PEI molar ratios of 30 and 50. For microscopic observations, Cy5-labeled plasmids were used to assemble fluorescently-labeled DNA-NP. The plasmid/polymer solution was incubated for 30 minutes at room temperature to spontaneously form DNA-NP. DNA-NP were washed twice with 3 volumes of ultrapure distilled water, and re-concentrated to 1 mg/ml using Amicon Ultra Centrifugal Filters (100,000 MWCO; Millipore Corp., Billerica, MA). Plasmid concentration was determined via absorbance at 260 nm using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE).
2.1.3. Physicochemical characterization of DNA-NP.
The hydrodynamic diameters as well as polydispersity index (PDI) and ζ-potentials of DNA-NP were measured by dynamic light scattering (DLS) and laser Doppler anemometry, respectively, in 10 mM NaCl solution at pH 7.0 using a Nanosizer ZS90 (Malvern Instruments, Southborough, MA). The size and morphology of DNA-NP were also confirmed by transmission electron microscopy (TEM; Hitachi H7600, Japan). The DNA complexation was confirmed by conventional agarose gel-based electrophoretic analysis. The colloidal stability was assessed by monitoring the change in hydrodynamic diameters and PDI in artificial cerebrospinal fluid (aCSF; Harvard Apparatus, Holliston, MA) over time at 37 °C via DLS. Number of plasmid copies in each DNA-NP was CICS, as previously reported [33]. Briefly, DNA-NP carrying Cy5-labeled plasmids were passed through a custom-made polydimethylsiloxane-based microfluidic device to spatially constrict particles to ensure single particle detection using a HeNe laser. The fluorescence data was collected with DNA-NP immediately after the preparation or 4 hours after an incubation in aCSF. Then fluorescence readings were detected and analyzed using Labview interface (National Instruments, Austin, TX) and a MATLAB script, respectively, to determine the number of plasmid copies for each DNA-NP population. Of note, quenching artifact was carefully assessed using a particle series of increasing ratio of labeled DNA to unlabeled free-labeled plasmid DNA and including a correction factor in the final plasmid calculation.
2.2. In vitro experiments in 2D cell cultures
2.2.1. Cell culture.
Highly aggressive rat glioma F98 cells (ATCC, Manassas, VA) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen Corp., Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Invitrogen Corp.) and 1% penicillin/streptomycin (P/S; Invitrogen Corp.). All the following in vitro studies were conducted when cells reached 70% - 80% confluency in respective plates.
2.2.2. Cell viability.
Cells were seeded onto 96-well plates at an initial density of 5,000 cells/well and grown overnight at 37 °C. Cells were treated with various DNA-NP at a wide range of plasmid doses for 48 hours at 37 °C. Cell viability was then assessed using Dojindo cell counting kit-8 (Dojindo Molecular Technologies, Inc., Rockville, MD). Absorbance at 450 nm was measured spectrophotometrically using a Synergy Mx Multi-Mode Microplate Reader (Biotek, Instruments Inc. Winooski, VT), and cell viability was determined by normalizing the readouts to absorbance measured with untreated control cells.
2.2.3. Cellular uptake.
Cells were seeded onto 6-well plates at an initial density of 200,000 cells/well and grown overnight at 37 °C. Cells were treated with various DNA-NP carrying Cy5-labeled plasmids (5 μg DNA/well) and after 5 hours of incubation, the media was removed. Cells were then thoroughly washed with 1X phosphate buffered saline (PBS) and trypsinized. Flow cytometry was conducted to quantify the cellular uptake of DNA-NP using a Sony Cell Sorter SH800 (Sony, San Jose, CA), followed by data analyzed with a FlowJo software (FlowJo LLC, Ashland, OR).
2.2.4. Transgene expression.
Cells were seeded at an initial density of 50,000 cells/well onto 24-well plates and grown overnight at 37 °C. Cells were then treated with various DNA-NP carrying luciferase-expressing plasmids (i.e. pd1GL3-RL) at a plasmid concentration of 1 μg/well, and the culture media was replaced with fresh media 5 hours after the incubation. After 48 hours of additional incubation, media was removed and 0.5 ml of 1X Reporter Lysis Buffer (Promega, Madison, WI) was added. Subsequently, cells were subjected to three freeze-and-thaw cycles to achieve complete lysis followed by the collection of supernatants by centrifugation. Luciferase activity was measured in relative light units (RLU) using a standard Luciferase Assay Kit (Promega) and a 20/20n luminometer (Turner Biosystems, Sunnyvale, CA). The RLU values were normalized to the total protein content measured by a Bicinchoninic Acid (BCA) Protein Assay Kit (Thermo scientific, Rockford, IL).
In parallel, transgene expression was also evaluated by confocal microscopy. Cells were seeded in 35 mm glass-bottom dishes of which surfaces were coated with poly-D-lysine (MatTek Corporation, Ashland, MA) at an initial density of 30,000 cells/dish and grown overnight at 37 °C. Cells were then treated with various DNA-NP carrying ZsGreen- or GFP-expressing plasmids at a plasmid concentration of 1 μg/dish, and the culture media was replaced with fresh media 5 hours after the incubation. After 48 hours of additional incubation, one drop of NucBlue™ Live Cell Stain (4’,6-diamidino-2-phenylindole or DAPI; Invitrogen Corp.) was added to each dish to visualize cell nuclei. Then fluorescence was captured from DAPI and ZsGreen or GFP, respectively, using an LSM 710 confocal microscope (Carl Zeiss; Hertfordshire, UK) under 20X magnification. Subsequently, images were analyzed using an ImageJ software (NIH, Bethesda, MD).
2.3. In vitro experiments in 3D multicellular tumor spheroids
2.3.1. Preparation of spheroids.
3D tumor spheroids were established by the hanging-drop method in a house-made spheroid media. The spheroid media was prepared by dissolving autoclaved methylcellulose (Sigma-Aldrich) at 2.4% (w/v) in preheated (60 °C), serum-free DMEM followed by dilution with an equivolume of DMEM supplemented with 10% FBS and an overnight stirring at 4 °C. The solution was then centrifuged at 5,000 xg for 2 hours at 4 °C to precipitate undissolved methylcellulose and the supernatant stored at 4 °C until use. To form each spheroid, we cultured 5,000 cells, either F98 glioma cells or F98 cells engineered to constitutively express a red fluorescent protein, mKate (F98-mKate; provided by Dr. Surojit Sur; Johns Hopkins University), in 20 μL spheroid media on the lid of a petri-dish. Spheroids were then grown upside down by covering a PBS-filled/moisturized petri-dish with the cell-seeded lid for 2 days. Finally, spheroids were transferred onto a sterile U-bottom 96-well plate (CELLSTAR®; Sigma-Aldrich) along with DMEM and incubated at 37⁰ C for 24 hours prior to their use for subsequent experiments.
2.3.2. Distribution of DNA-NP in spheroids.
To evaluate the ability of DNA-NP to penetrate 3D tumor spheroids, we treated F98-mKate-based spheroids with various DNA-NP carrying Cy5-labeled plasmids (1 μg plasmids/spheroid). The media was removed 5 hours after the incubation and replaced with the background-minimizing FluoroBrite DMEM. Subsequently, spheroids were fixed with 4% formaldehyde, washed with PBS, embedded in Tissue-Tek® optimal cutting temperature compound (Sakura Finetek, Torrance, CA) and sectioned into 20 μm-thick slices using Leica CM 1905 cryostat (Leica Biosystems, Buffalo Grove, IL). Slices were imaged for the fluorescence originated from F98 glioma cells (i.e. mKate) and DNA-NP (i.e. Cy5) using an LSM 710 confocal microscope under 10X magnification. The radial distribution profile of DNA-NP was obtained by measuring the mean fluorescence intensity throughout the radial coordinates from the edge to the core of the spheroid using an ImageJ software, as previously described [34].
2.3.3. Transgene expression by DNA-NP in spheroids.
F98-based spheroids were grown and treated in a sterile U-bottom 96-well plate with various DNA-NP carrying ZsGreen-expressing plasmids (1 μg plasmids/spheroid) to evaluate the ability of DNA-NP to mediate transgene expression in spheroids After 48 hours of additional incubation without media refreshment, the spheroids were transferred on to custom-made gel-wells. To prepare a stock gel media, low gelling temperature agarose (Sigma-Aldrich) was dissolved at 5% (w/v) in sterile PBS and subsequently autoclaved. The working gel media was then made by diluting the stock with FluoroBrite DMEM to reach the final agarose concentration of 0.5% (w/v). Subsequently, a 4-well Millicell EZ slide (Millipore Corp) was filled with the working gel media and spheroids were carefully placed in each well. Fluorescence images were then captured using an LSM 710 confocal microscope under 10X magnification and analyzed for spheroid area-normalized mean fluorescence intensity using an ImageJ software.
2.4. Multiple particle tracking – ex vivo diffusion of DNA-NP in rat brain
Multiple particle tracking (MPT) experiments were conducted to quantify the diffusion rates of various DNA-NP carrying Cy5-labeled plasmids in healthy rat brain parenchyma ex vivo, as previously reported [9, 16]. Briefly, whole brain tissues were harvested from 6-to-8-week-old female Fischer 344 rats (Harlan Laboratories, Frederick, MD) and incubated in aCSF on ice for 10 minutes. The brain tissues were then sliced using a Zivic brain matrix slicer (Zivic Instruments, Pittsburgh, PA) and the resultant 1.5-mm coronal slices were placed on custom-made slides. Subsequently, we bolus-injected 0.5 μL of DNA-NP solution at a plasmid concentration of 2 μg/mL into the cerebral cortex at a depth of 1 mm using a Neuros syringe (50 μL; Hamilton, Reno, NV) mounted on an ultra-precise small-animal stereotactic frame (Stoelting Co., Wood Dale, IL). Following a 10-minute incubation, trajectories of DNA-NP were recorded over 20 seconds at an exposure time of 66.7 milliseconds (i.e. 15 fames/s) by an Evolve 512 EMCCD camera (Photometrics, Tucson, AZ) mounted on an inverted epifluorescence microscope (Axio Observer D1; Carl Zeiss, Hertfordshire, UK) equipped with a 100X/1.46 NA oil-immersion objective. Movies were then analyzed with a custom-made automated particle tracking MATLAB script to extract x, y-coordinates of DNA-NP centroids over time from which the mean square displacement (MSD) values of individual DNA-NP were calculated as a function of timescale [35]. Median MSD was determined based on the measured MSD values of individual DNA-NP at a timescale of 1 second, at which both static and dynamic errors are minimized in MPT experiments [35].
2.5. Animal studies
We used 6-to-8-week-old female Fischer 344 rats for the assessment of in vivo gene transfer efficacy of various DNA-NP. Animals were treated in accordance with the guidelines and policies of the Johns Hopkins University Animal Care and Use Committee. Surgical procedures were performed using standard sterile surgical techniques. Animals were anesthetized using a mixture of 75 mg/kg ketamine and 7.5 mg/kg xylazine, as previously described.[36] A midline scalp incision was made to expose the coronal and sagittal sutures and a burr whole was drilled 3 mm lateral to the sagittal suture and 0.5 mm posterior to the bregma. Following the tumor cell inoculation or DNA-NP administration, the skin was sealed using biodegradable sutures (Polysorb™ Braided Absorbable Sutures 5–0) and bacitracin was applied.
2.5.1. Orthotopic tumor cell inoculation.
Orthotopic tumor cell inoculation was performed, as previously described [36]. Briefly, 100,000 cells, either F98 or F98-mKate cells, were administered in 10 μL of DMEM over 5 minutes and at a depth of 3.5 mm using a Neuros Syringe mounted on an ultra-precise small-animal stereotactic frame. Of note, we inoculated rat brains with a high numbers of tumor cells, relevant to clinical translation [37, 38], to establish a model mimicking malignant gliomas characterized by rapid tumor growth rates.
2.5.2. Intracranial administration - convection enhanced delivery (CED).
To assess the distribution of transgene expression, various DNA-NP carrying ZsGreen- or luciferase-expressing plasmids were intracranially administered via CED, as previously described [9, 10]. Briefly, a Neuros syringe connected to a 33-gauge needle was filled with 20 μL of DNA-NP solution at a plasmid concentration of 1 mg/mL and lowered to a depth of 3.5 or 2.5 mm of a healthy or an orthotopic tumor-bearing rat brain, targeting the striatum or the tumor core, respectively. DNA-NP solution was then infused at a rate of 0.33 μL/min as controlled by a Chemyx Nanojet Injector Module (Chemyx, Stafford, TX).
2.5.3. Distribution and overall level of transgene expression.
For assessing the distribution of transgene expression, brain tissues of rats received DNA-NP carrying ZsGreen-expressing plasmids were harvested 48 hours after the administration and subsequently fixed with 4% formaldehyde. Tissues were then sectioned using a Leica CM 1905 cryostat into 100 or 50 μm coronal slices for healthy or tumor-bearing rats at ± 3 mm of the infusion plane in striatum or until the tumor tissue was no longer visible, respectively. Slices of F98-based orthotopic tumor tissues were stained with DAPI (Molecular Probes, Eugene, OR) to visualize tumor cell nuclei. All fluorescence images were taken using an LSM 710 confocal microscope under 5X, 20X and/or 40X magnification. We carefully optimized the settings to avoid background fluorescence based on the microscopy of untreated control rat brains. The volume of transgene expression was quantified using a custom-made MATLAB script that subtracted background fluorescence by Otsu’s method of thresholding [39, 40]. The area of transgene expression in each slice was integrated to calculate the total volume of transgene expression. To reconstruct 3D-rendered images of volumetric transgene expression, we stacked and aligned the acquired images using Metamorph ® Microscopy Automation & Image Analysis Software (Molecular Devices, CA). Finally, we used an Imaris Software (Bitplane, CT) to create 3D isosurfaces of the reconstructed images.
The kinetics of transgene expression was also determined based on the distribution of transgene expression over time. For the assessment of a short- and a long-term transgene expression, animals were treated with a BPN formulation carrying ZsGreen- and mCherry-expressing plasmids driven by CMV and human β-actin promoters, respectively. In parallel, we determine the volume of transgene expression mediated by a BPN formulation stored at 4 °C for one month, following a routine physicochemical characterization as described above.
To quantify the overall level of transgene expression, we treated rats using the identical dosing method applied to the distribution study but with various DNA-NP carrying luciferase-expressing plasmids (i.e. pBAL). Seven days after the administration, tissues were harvested and subjected to homogenate-based luciferase assay using a standard Luciferase Assay Kit and a 20/20n luminometer. The RLU values were normalized to the total protein content measured by BCA protein assay.
2.5.4. Immunofluorescence.
To assess the transgene expression in different cell types, animals bearing orthotopic F98-mKate-based tumor were treated with a BPN formulation carrying ZsGreen-expressing plasmids as described above. Two days after the administration, brain tissues were harvested, sectioned and subjected to immunochemistry. Specifically, astrocytes and microglial cells were immunostained by antibodies against GFAP (Abcam, Cambridge, UK) and Iba1 (Wako Chem USA Corp., Richmond, VA). Images were taken using an LSM 710 confocal microscope under 20X and/or 40X magnification. The transgene expression in F98-mKate glioma and microglial cells was quantified using an ImageJ software based on the co-localization of different fluorescence. At least three representative fields of view from at least three different coronal sections were used for the analysis.
2.6. Statistical analysis
Statistical analysis between two groups was conducted using a two-tailed Student’s t-test assuming unequal variances. If multiple comparisons were involved, one-way analysis of variance (ANOVA), followed by Sidak’s multiple comparisons test, was employed, using GraphPad Software (GraphPad Software Inc., La Jolla, CA). Differences were determined to be statistically significant at p < 0.05.
3. Results and discussion
3.1. Formulation and characterization of DNA-NP
We first engineered three PEI-based DNA-NP possessing different surface PEG contents. CPN were formulated by condensing plasmids solely with PEG-PEI synthesized at a moderate PEG to PEI ratio of 8, as previously reported [9]. In parallel, we engineered two additional DNA-NP with greater surface PEG contents using a method that we have previously developed to endow gene vectors with excellent compaction and colloidal stability in physiological conditions while providing capability to efficiently penetrate healthy brain parenchyma, BPN [9]. Specifically, a blend of non-PEGylated PEI and PEG-PEI synthesized at a high PEG to PEI ratio of 30 or 50 was used to condense plasmids to yield BPNL (i.e. lower PEG BPN) or BPNH (i.e. higher PEG BPN), respectively (Supplementary Figure 1). Of note, BPNL is a close relative to a PEI-based DNA-NP that we have confirmed for efficient penetration through healthy rat brain tissues ex vivo and in vivo in our previous study [9]. We here hypothesized that a greater PEGylation, unless interferes with a capacity of PEI to form stable particles, might further enhance the ability of the resultant DNA-NP to percolate not only in healthy brain parenchyma but also in brain tumor tissues.
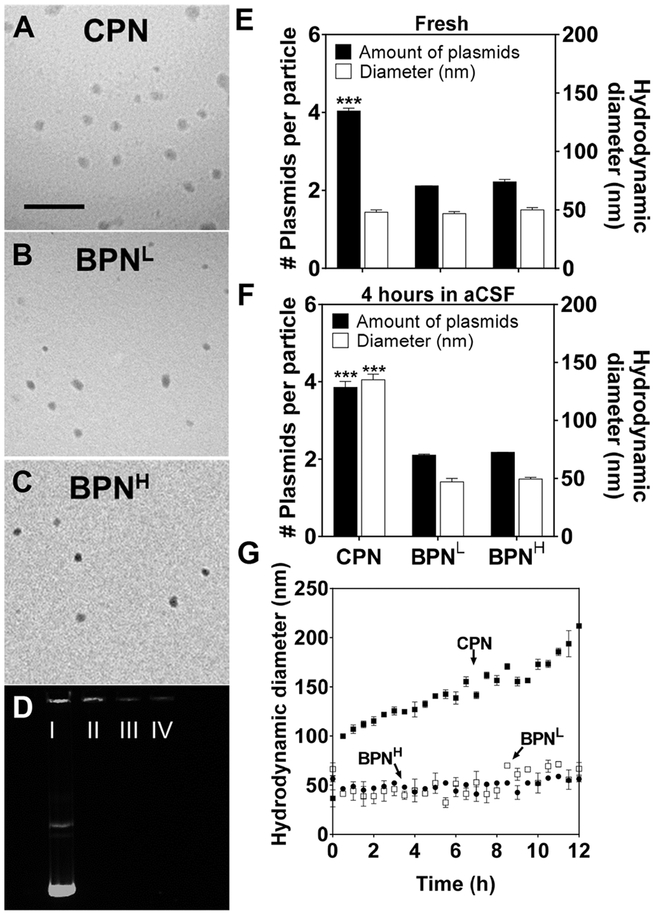
We found that all three freshly prepared DNA-NP exhibited similarly small sizes (i.e. ~50 nm in hydrodynamic diameters) and excellent PDI values (≤ 0.20) (Table 1). On the other hand, the ζ-potential, an indicative of particle surface charge, decreased from positive towards neutral values with increased PEG contents (i.e. 18.0 ± 0.2, 7.0 ± 0.3 and 2.0 ± 0.1 mV for CPN, BPNL and BPNH, respectively; Table 1), indicating that the otherwise cationic DNA-NP surfaces were effectively masked by PEG corona, particularly more so for BPNH that showed near-neutral ζ-potential values. The particle morphology was confirmed by TEM to be ~50 nm spheres regardless of PEG contents (Figure 1A–C), in consistent with the measured hydrodynamic diameters (Table 1). We found that BPN candidates formulated with an inclusion of PEG-PEI synthesized at even higher PEG to PEI ratios of 60 and 85 exhibited similar physiochemical properties as BPNH (Supplementary Table 1), and thus excluded in subsequent evaluations. We then verified robust DNA compaction by all three different DNA-NP via a conventional agarose-gel migration assay; however, CPN, regardless of its lower PEG content, showed a brighter signal in the well compared to BPNL and BPNH, suggesting that an inclusion of non-PEGylated PEI in the blended BPN formulations might have enhanced their abilities to compact the plasmid payloads (Figure 1D).
Table 1.
Physicochemical properties of DNA-NP.
| Hydrodynamic diameter ± SEM (nm)a | PDIa | ζ-Potential ± SEM (mV)b | Hydrodynamic diameter ± SEM at 12 hours in aCSF (nm)c | PDI at 12 hours in aCSFc | |
|---|---|---|---|---|---|
| CPN | 50 ± 1 | 0.16 | 18.0 ± 0.2 | 212 ± 3 | 0.17 |
| BPNL | 49 ± 1 | 0.19 | 7.0 ± 0.3 | 67 ± 6 | 0.16 |
| BPNH | 48 ± 5 | 0.20 | 2.0 ± 0.1 | 56 ± 3 | 0.15 |
a Hydrodynamic diameter and polydispersity index (PDI) were measured by dynamic light scattering (DLS) in 10 mM NaCl. Mean ± SEM (n ≥ 3).
b ζ-potential was measured by laser Doppler anemometry in 10mM NaCl at pH 7.0. Mean ± SEM (n ≥ 3).
c Hydrodynamic diameter and PDI were measured by DLS following a 12-hour incubation in aCSF at 37° C.
Figure 1. Physicochemical properties and stability of DNA-NP.
Transmission electron microscopy images of (A) CPN, (B) BPNL, (C) BPNH freshly made in ultrapure water. Scale bar = 400 nm. and (D) Compaction of plasmids in DNA-NP shown by electrophoresis: I) free DNA, II) CPN, III) BPNL, IV) BPNH. Number of plasmids in each DNA-NP and hydrodynamic diameter of DNA-NP when (E) freshly made in water and (F) when incubated in aCSF for 4 hours. (G) Change in hydrodynamic diameters over 12 hours in aCSF at 37°C measured by DLS. Differences are statistically significant as indicated (***p < 0.001), of CPN compared to BPNL and BPNH.
In addition to these conventional physicochemical characterization, we conducted an additional analysis to quantify the average number of plasmid copies per different DNA-NP using cylindrical illumination confocal spectroscopy (CICS) technique [33]. We found that each CPN contained an average of approximately four plasmid copies, whereas BPNL and BPNH packaged roughly two copies on average (Figure 1E). The finding suggests that double the amount (i.e. number of particles) of both BPN formulations will be administered compared to CPN at a fixed treatment dose of plasmids; this may be beneficial for enhancing gene transfer efficacy given a prior observation that the total amount of plasmid-containing particles rather than the overall plasmid quantity is the limiting factor for PEI-mediated transfection [41]. We next assessed the hydrodynamic diameters and plasmid copies per particle of different DNA-NP following a 4-hour incubation in aCSF at 37° C to predict potential alteration of particle properties in the physiological brain environment. We found that while both BPN formulations retained their hydrodynamic diameters in aCSF, the size of CPN incremented near to ~150 nm (Figure 1F) and continue to grow afterward (Figure 1G), suggesting that CPN would be unlikely to efficiently penetrate brain tissues due to large particle size as well as positively charged particle surfaces. In contrast, the average number of plasmid copies per particle did not change upon aCSF incubation regardless of the DNA-NP type. The finding reveals that the increase in CPN size in aCSF is most likely attributed to particle swelling rather than aggregation, providing an additional evidence that inclusion of non-PEGylated PEI in the blended BPN formulations endowed DNA-NP with an enhanced plasmid compaction stability.
3.2. In vitro transfection efficiency of DNA-NP in 2D culture versus 3D spheroid model
We initially sought to investigate the in vitro transfection efficiency of different DNA-NP formulated to carry the most widely used reporter, GFP. However, evidences suggest that immunogenicity and cytotoxicity of GFP potentially confound the interpretation of in vitro and/or vivo transfection data [42–44]. Therefore, we compared the cytotoxicity and transfection efficiency, as measured by mean fluorescence intensity, of GFP with those of ZsGreen in F98 rat glioma cells following the treatment of cells with lipofectamine carrying respective plasmids at an identical dose. We found that GFP exhibited greater (i.e. ~3-fold) toxicity and lower (i.e. ~5-fold) transfection efficiency than ZsGreen (Supplementary Figure 2B), in agreement with previous reports [45, 46]. We thus pursed the following studies with different PEI-based DNA-NP, including CPN, BPNL and BPNH, carrying ZsGreen-expressing plasmids. We first evaluated the cytotoxicity of DNA-NP at various concentrations of plasmid payloads following 48 hours of incubation. All DNA-NP showed comparably good cell viability (i.e. ~100%) at the plasmid concentrations of 1 and 5 μg/mL, but CPN, but not BPN formulations, resulted in over 40% cell death at 10 μg/mL (p < 0.001) (Supplementary Figure 3A). However, we note that the 10 μg/mL plasmid concentration is markedly great than conventional working concentrations for in vitro transfection experiments.
We then assessed in vitro cellular uptake and transfection efficiency of DNA-NP carrying Cy5-labeled and unlabeled plasmids, respectively, using an F98-based 2D cell culture. The percentage of cells that took up detectable amounts of Cy5-labeled plasmid payloads were all ~100% regardless of the type of DNA-NP (Supplementary Figure 3B), in agreement with our and others’ observations that PEGylation does not completely block individual cells to engulf small nanoparticles [9, 10, 47, 48]. However, the amounts of internalized DNA-NP per cell, as indicated by fluorescence intensity, were significantly lower for BPNL and BPNH in comparison to CPN (p <0.001; Supplementary Figure 3B). This is most likely due to the reduced interactions of charge-shielded (Table 1) BPN formulations with negatively charged cell surfaces, although potential impacts of dense PEG on other intracellular processes cannot be fully excluded.[49] Accordingly, in vitro transfection efficiencies of BPNL and BPNH, as confirmed by confocal microscopy and homogenate-based luminescence assay (p < 0.001), were lower than those of CPN (Supplementary Figure 3C–D).
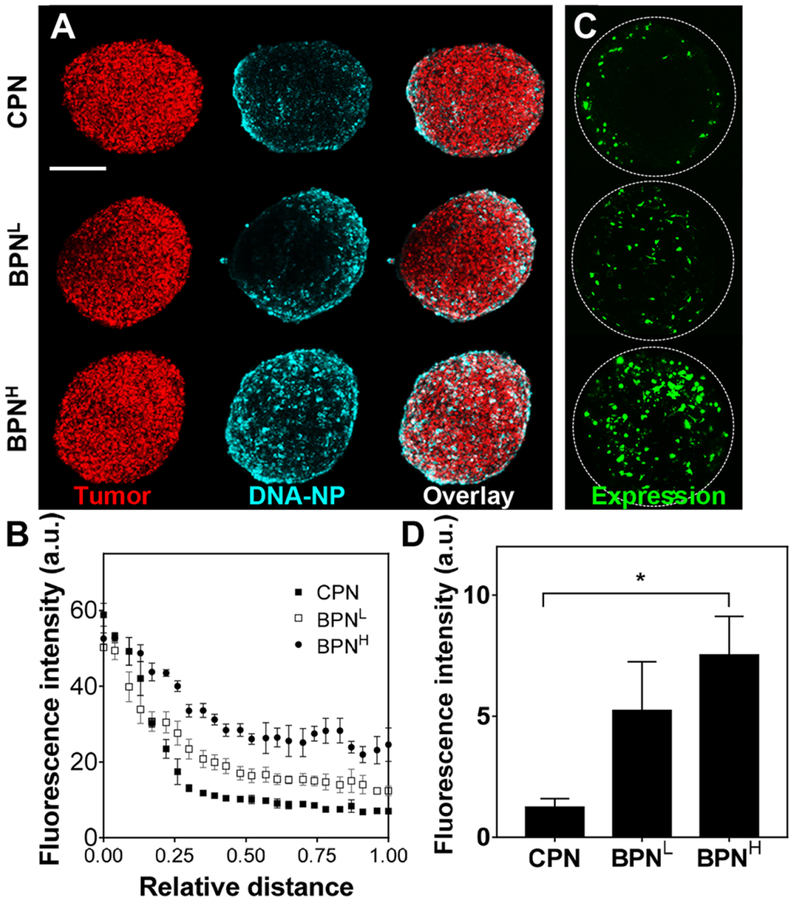
However, 2D cell cultures, due in part to the absence of extracellular components, do not provide a reliable model for predicting in vivo performances of gene vectors. We thus investigated in vitro behaviors of DNA-NP in an F98-based 3D model of multicellular tumor spheroid that better recapitulates architecture and biology of solid tumors observed in vivo [50]. In particular, 3D spheroid models simulate an ECM-filled intercellular permeability barrier [50–53], and thus serve as an excellent model to evaluate the penetration of gene vectors through solid tumor tissues [54]. To this end, we constructed a 3D spheroid model with F98-mKate cells and assessed distribution of different DNA-NP carrying Cy5-labeled plasmids. As shown by representative confocal images, DNA-NP with denser surface PEG coronas exhibited greater inward penetration compared to those with less PEG contents (BPNH > BPNL > CPN; Figure 2A). Quantitatively, BPNH showed the greatest spheroid penetration in the radial profile from the edge (coordinate 0) to the core (coordinate 1) of the spheroid, followed by the BPNL and CPN (Figure 2B). We then evaluated the distribution and level of transgene expression following the treatment of the spheroids with DNA-NP carrying ZsGreen-expressing plasmids. Similar to the vector distribution study (Figure 2A), representative confocal images revealed a greatest distribution of transgene expression by BPNH compared BPNL and CPN with CPN exhibiting a weakest distribution (Figure 2C and Supplementary Figure 4). In addition, BPNH, but not BPNL, showed significantly greater level of transgene expression, as measured by mean fluorescence intensity, compared to CPN (p <0.05; Figure 2D). While the average fluorescence intensity of BPNL was ~4-fold greater than that of CPN, statistical significance was not achieved presumably due to variability of the transgene expression mediated by BPNL in different individual spheroids (Figure 2D and Supplementary Figure 4). The findings here underscore that, in contrast to the observations with 2D cultures (Supplementary Figure 3), DNA-NP with greater surface PEG coverage provides superior transgene expression compared to those with lesser PEG, due to the ability to efficiently penetrate the extracellular barrier established in tumor spheroids.
Figure 2. In vitro DNA-NP distribution and transgene expression in F98-based 3D spheroids.
(A) Representative images showing F98 tumor spheroids (red; mKate) 5 hours after treatment with DNA-NP (cyan; Cy5). Scale bar = 250 μm. (B) Quantification of radial distribution of DNA-NP within the spheroids. The coordinates 0 and 1 indicate the spheroid edge and core, respectively. (C) Representative reporter transgene expression (green; ZsGreen) by DNA-NP carrying ZsGreenexpressing plasmids 48 hours after NP-treatment. (D) Quantification of ZsGreen transgene expression within the spheroids. Differences are statistically significant as indicated (*p < 0.05).
3.3. Ex vivo diffusion of DNA-NP in rodent brain tissues
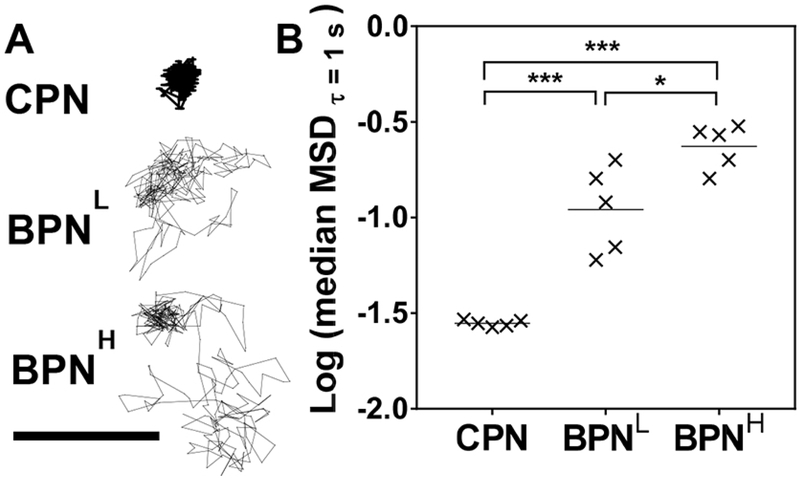
We have previously demonstrated that diffusion rates of nanoparticles, measured by MPT, in brain tissues ex vivo reliably predict their spread in brain parenchyma in vivo [5, 8–10]. We thus conducted MPT experiments to confirm that a greater surface PEG coverage resulted in enhanced brain penetration by DNA-NP. Specifically, DNA-NP carrying Cy5-labled plasmids were injected into a freshly harvested healthy rat brain tissue and their diffusion was monitored over a time period of 20 seconds. As evidenced by the highly constrained trajectory, CPN were immobilized in the brain parenchyma (Figure 3A). In contrast, BPNL and BPNH demonstrated relatively unhindered motions, leading to trajectories that span several microns in distance (Figure 3A). We then quantified their diffusion rates of DNA-NP by measuring the MSD; which represents an averaged square of distances traveled by individual particles within a given time interval and thus MSD values are directly proportional to respective particle diffusion rates [55]. Consistent with our prior observation [9], BPNL exhibited significantly greater MSD values compared to CPN (p < 0.001), but we found here that increasing the surface PEG coverage further enhanced the ability of PEI-based DNA-NP to penetrate brain tissue ex vivo (i.e. BPNH > BPNL > CPN; Figure 3B). We also note that BPNH exhibited more uniform high MSD values than BPNL, suggesting that BPNH may provide a more consistent ability to penetrate through the highly heterogeneous tumor microenvironment.
Figure 3. Ex vivo diffusion of different DNA-NP in rat brain tissues.
(A) Representative trajectories of DNA-NP in rat brain tissue over 20 seconds. Scale bar = 1 μm. (B) Logarithms of median mean square displacements (MSD) of DNA-NP at a time scale of 1 second. Data represent 5 independent experiments with n ≥ 200 particles tracked for each experiment. Differences are statistically significant as indicated (*p < 0.05; ***p < 0.001).
3.4. In vivo transgene expression mediated by DNA-NP in healthy brain parenchyma
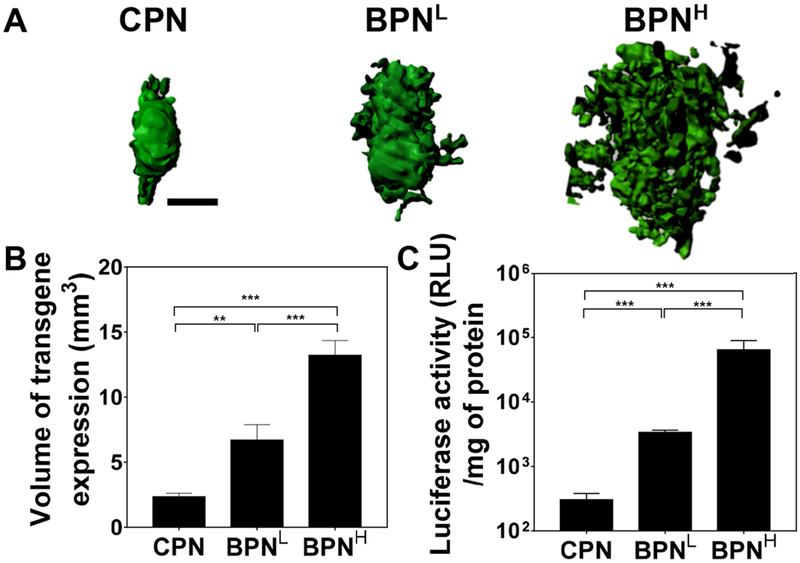
We next investigated whether ex vivo diffusion behaviors of DNA-NP translated to their in vivo distribution patterns in healthy rat brain. Different DNA-NP carrying ZsGreen-expressing plasmids were administered into the rat striatum via CED that has been clinically applied to enhance the therapeutic distribution within brain by creating a continuous pressure-driven bulk flow [39, 56]. Transgene expression was then assessed 48 hours after the treatment. The 3D reconstruction of confocal images of serial coronal sections of rat brains revealed enhanced volumes of transgene expression when rats were treated with DNA-NP possessing greater surface PEG coverage (Figure 4A). The transgene expression mediated by CPN was confined to the site of administration despite the pressure gradient provided by CED. Inability of conventional cationic gene vectors to provide widespread transgene expression regardless of CED may have attributed to a limited success in a phase I/II clinical trial in which patients with glioblastoma received non-PEGylated DNA-NP formulated with cationic lipids via CED [57, 58]. In agreement with our previous observation [9], BPNL exhibited significantly greater volumetric distribution of transgene expression than CPN (p < 0.01), but importantly, BPNH, mostly likely due to its superior ability to penetrate the brain parenchyma (Figure 4), roughly doubled the transfected volume on average in the rat striatum compared to BPNL (p < 0.001; Figure 4B). In parallel, we quantitatively determined the overall levels of transgene expression mediated by CED of BPNH, BPNL and CPN carrying luciferase-expressing plasmids, using a homogenate-based luciferase assay. Like the trend observed with the transfected volume, DNA-NP possessing greater surface PEG contents provided greater overall levels of transgene expression; BPNH exhibited statistically significant improvements (p < 0.001) with ~20- and ~210-fold greater luciferase activities in comparison to BPNL and CPN, respectively (Figure 4C). The findings here suggest that the unique ability of BPNH to further increase the volumetric distribution of transgene expression over BPNL led to quantum enhancement of the overall gene transfer efficacy. Efforts towards greater transfection efficiencies often focus on enhancing cellular gene delivery to a specific cell type of interest, but this study provides an example underscoring the importance of achieving a greater coverage of transgene expression in a target tissue.
Figure 4. In vivo transgene expression mediated by CED of different DNA-NP carrying ZsGreen- or luciferase-expressing plasmids in healthy rat brain tissues.
(A) Representative isosurface 3D images depicting volumetric distribution of ZsGreen reporter transgene expression (green) in healthy rat brains, obtained by stacking multiple sequential confocal images 48 hours after NP-treatment carrying ZsGreen-expressing plasmids. Scale bar = 1 mm. (B) Volume of ZsGreen transgene expression (n ≥ 6 rats). (C) Overall level of luciferase transgene expression (n ≥ 6 rats). Differences are statistically significant as indicated (**p < 0.01;***p < 0.001).
To evaluate the expression kinetics of BPNH in healthy rat brains, we treated rats via CED with BPNH carrying plasmids controlled either by the CMV or by the long-acting human β-actin promoter. We found that peak volume of transgene expression lasted up to 2 days or 3 weeks upon a single treatment when regulated by CMV or human β-actin promoter (Supplementary Figure 5), respectively, suggesting that the life span of transgene expression mediated by can be tuned by a careful selection of a promoter. We also confirmed that BPNH retained the brain-penetrating physiochemical properties (i.e. small particle diameters and near-neutral surface charges; Supplementary Table 2) and the ability to yield widespread transgene expression in rodent striatum at least for 1 month when stored at 4⁰ C (Supplementary Figure 6)
3.5. In vivo transgene expression mediated by DNA-NP in orthotopically established malignant glioma
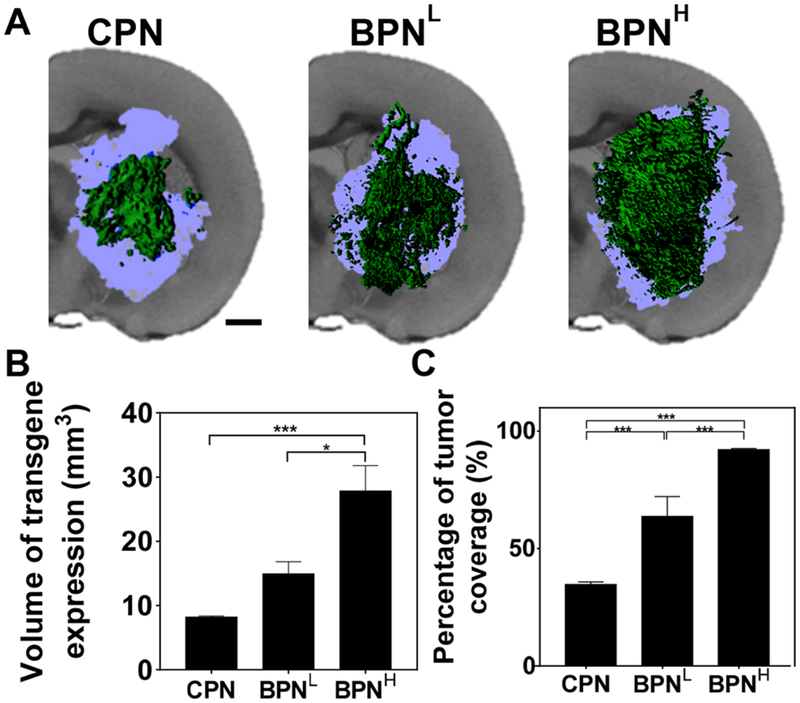
We next evaluated in vivo gene transfer efficacy of BPNH, BPNL and CPN carrying ZsGreen-expressing plasmids in an orthotopic model of aggressive brain tumor following CED. The model was established by intracranial stereotactic inoculation of 1 × 105 F98 cells and a CED experiment was conducted 10 days after the inoculation when a large volume (~30 mm3) of orthotopic tumor was established in the rat striatum. As shown by representative 3D-reconstructed images, BPNH exhibited the greatest volumetric distribution of transgene expression, followed by BPNL and CPN (Figure 5A). The volume of transgene expression achieved by BPNH was ~2-and ~3.5-fold greater than BPNL (p < 0.05) and CPN (p < 0.001), respectively, in the orthotopic brain tumor (Figure 5B). We then quantified the volumetric fractions of the orthotopic tumor with reporter transgene expression mediated by different DNA-NP. The percentages of total tumor volumes exhibited positive reporter transgene expression were ~90%, ~65% and ~35% for BPNH, BPNL and CPN, respectively, with statistically significant differences between all different DNA-NP groups (Figure 5C). Importantly, the finding here is in good accordance with our observation with a 3D tumor spheroid model (Figure 2) but opposite to the trend shown in a 2D culture (Supplementary Figure 3). To this end, 3D tumor spheroid models, providing good in vivo correlation unlike 2D culture, may serve as a reliable in vitro surrogate to screen newly engineered gene vectors prior to their evaluation for in vivo cancer gene delivery.
Figure 5. In vivo transgene expression mediated by CED of different DNA-NP carrying ZsGreen-expressing plasmids in F98-based orthotopic rat brain tumor tissues.
(A) Representative isosurface 3D images depicting volumetric distribution of ZsGreen reporter transgene expression (green) in orthotopic brain tumors (light blue), obtained by stacking multiple sequential confocal images. Scale bar = 1 mm. (B) Volume of transgene expression (n ≥ 4 rats). (C) Percentage of tumor volume covered by transgene expression (n ≥ 4 rats). Differences are statistically significant as indicated (*p < 0.05; ***p < 0.001).
In this study, we noticed that the volume of transgene expression mediated by identically administered DNA-NP was significantly greater in F98-based orthotopic tumor than in healthy rat brain (Figure 4 and 5). This is not readily expected a prior as the high cellularity commonly found in tumors would render the intercellular spaces narrower, thereby potentially reinforcing the tumor ECM as a steric barrier. We speculate that the greater volumetric transgene expression shown in the orthotopic model may be in part attributed to the heterogeneous nature of tumor microenvironment [32], specifically the necrotic regions through which BPN formulations can spread out relatively unhindered. In addition, the characteristic high intratumoral pressure [59], may have facilitated outward diffusion of DNA-NP from the core to the tumor edge.
Highly invasive tumor cells that migrate beyond the tumor edge and infiltrate into normal brain tissue are responsible for recurrence of malignant gliomas [60]. Thus, the unique ability of BPNH to efficiently penetrate both normal brain parenchyma and brain tumor tissue may serve favorably for this specific application. However, an additional method to confine therapeutic transgene expression to tumor cells would be desired, particularly when therapy can exert a toxic effect to normal tissues (e.g. cytotoxic genes).
3.6. In vivo cell tropism of BPNH in orthotopically established malignant glioma
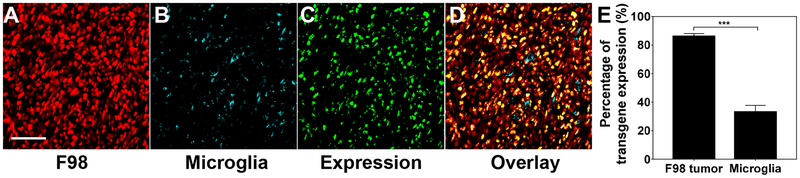
While we showed robust reporter transgene expression by BPNH in an orthotopically established malignant glioma, the tumor volume and edge were estimated based on the high tumor-cell cellularity, uncovered by a conventional DAPI staining (Figure 5). Therefore, we established an orthotopic model as described above but with F98 cells constitutively expressing fluorescent mKate (i.e. F98-mKate) to confirm whether the transgene expression indeed took place in tumor cells in vivo. We first determined F98 cells to be the most abundant cells within the orthotopically-established tumor tissue (Supplementary Figure 7). Microglial cells were also ubiquitously present within the tumor bulk, as expected from their roles in glioma maintenance and progression [61], whereas neuron-supporting astrocytes were rarely found (Supplementary Figure 7). We then treated rats harboring orthotopic mKate-expressing F98 tumor with BPNH carrying ZsGreen-expressing plasmids via CED and evaluated relative transgene expression in those two cells most highly prevalent in the tumor, including F98 and microglial cells. We found that ~90% and ~30% of F98 and microglial cells, respectively, were positive to BPNH-mediated ZsGreen transgene expression, as determined by image-based analysis of randomly selected confocal image fields (Figure 6). The BPNH is not specifically designed to preferentially target cancer cells and thus a ~3-folder greater number of transfected cells for F98 over microglial cells was not readily assumed. It is most likely that passive mechanisms have been involved. A relatively larger number of F98 cells in the tumor tissue (Supplementary Figure 7), as well as high endocytic activities often observed with tumor cells [62–64], may render them a better “particle sink” compared to other brain-resident cells, such as microglial cells. In addition, mitotic activities of brain-resident cells are very low, whereas glioma cells are highly mitotic [39], thereby enhancing the probability of plasmid payloads entering into the nucleus to be transcribed. We note that while the described are reasonable scenarios, further mechanistic underpinning should be followed to elucidate a full picture.
Figure 6. In vivo transgene expression mediated by BPNH carrying ZsGreen-expressing plasmids in F98-mKate orthotopic rat tumor tissues.
Representative confocal images at 40X magnification showing (A) F98 tumor cells (red; mKate), (B) microglia (cyan; Iba1), (C) transgene expression (green; ZsGreen) and (D) overlay. Scale bar = 50 μm. (E) Quantification of percentage of F98 tumor and microglia cells with the transgene expression. Differences are statistically significant as indicated (***p < 0.001).
4. Conclusions
Here, we introduce a novel polymer-based gene vector capable of efficiently penetrating both healthy brain parenchyma and brain tumor tissue. Specifically, a lead formulation, BPNH, exhibited the greatest ability to provide widespread and high-level transgene expression in 3D tumor spheroids and orthotopically-established brain tumors compared to formulations with inferior surface PEG coatings, with an excellent in vitro-to-in vivo correlation. We note that while the transgene expression occurred preferentially in gliomas cells within orthotopically-established tumor tissue, a significant expression was observed in microglial cells as well. Thus, an additional strategy is likely needed if highly selective cancer cell transfection is required, while retaining the ability to cover widely scattered tumor areas within the brain. To this end, our current effort focuses on achieving widespread but cancer-selective therapeutic transgene expression via a marriage of BPNH and promoters designed to drive transgene expression specifically in cancer cells.
Supplementary Material
Acknowledgements
The funding was provided by the National Institutes of Health (R01CA164789, R01EB020147, R01CA197111, R01CA220841, R01CA204968, and P30EY001765), Ruth L. Kirschstein National Research Service Award Individual Predoctoral Fellowship F31 (K.N.) and W.W. Smith Charitable Trust (J.S.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
There is no conflict of interest to declare.
5. References
- [1].Davis ME, Glioblastoma: Overview of Disease and Treatment, Clin J Oncol Nurs, 20 (2016) S2–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chira S, Jackson CS, Oprea I, Ozturk F, Pepper MS, Diaconu I, Braicu C, Raduly LZ, Calin GA, Berindan-Neagoe I, Progresses towards safe and efficient gene therapy vectors, Oncotarget, 6 (2015) 30675–30703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Pulkkanen KJ, Yla-Herttuala S, Gene therapy for malignant glioma: current clinical status, Mol Ther, 12 (2005) 585–598. [DOI] [PubMed] [Google Scholar]
- [4].Murphy AM, Rabkin SD, Current status of gene therapy for brain tumors, Transl Res, 161 (2013) 339–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nance EA, Woodworth GF, Sailor KA, Shih TY, Xu Q, Swaminathan G, Xiang D, Eberhart C, Hanes J, A dense poly(ethylene glycol) coating improves penetration of large polymeric nanoparticles within brain tissue, Sci Transl Med, 4 (2012) 149ra119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sykova E, Nicholson C, Diffusion in brain extracellular space, Physiological reviews, 88 (2008) 1277–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nance E, Brain-Penetrating Nanoparticles for Analysis of the Brain Microenvironment, in: Petrosko SH, Day ES (Eds.) Biomedical Nanotechnology: Methods and Protocols, Springer New York, New York, NY, 2017, pp. 91–104. [DOI] [PubMed] [Google Scholar]
- [8].Zhang C, Mastorakos P, Sobral M, Berry S, Song E, Nance E, Eberhart CG, Hanes J, Suk JS, Strategies to enhance the distribution of nanotherapeutics in the brain, J Control Release, 267 (2017) 232–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mastorakos P, Zhang C, Berry S, Oh Y, Lee S, Eberhart CG, Woodworth GF, Suk JS, Hanes J, Highly PEGylated DNA Nanoparticles Provide Uniform and Widespread Gene Transfer in the Brain, Adv Healthc Mater, 4 (2015) 1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mastorakos P, Song E, Zhang C, Berry S, Park HW, Kim YE, Park JS, Lee S, Suk JS, Hanes J, Biodegradable DNA Nanoparticles that Provide Widespread Gene Delivery in the Brain, Small, 12 (2016) 678–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chen J, Guo Z, Tian H, Chen X, Production and clinical development of nanoparticles for gene delivery, Mol Ther Methods Clin Dev, 3 (2016) 16023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Merkel OM, Urbanics R, Bedocs P, Rozsnyay Z, Rosivall L, Toth M, Kissel T, Szebeni J, In vitro and in vivo complement activation and related anaphylactic effects associated with polyethylenimine and polyethylenimine-graft-poly(ethylene glycol) block copolymers, Biomaterials, 32 (2011) 4936–4942. [DOI] [PubMed] [Google Scholar]
- [13].Shi B, Zheng M, Tao W, Chung R, Jin D, Ghaffari D, Farokhzad OC, Challenges in DNA Delivery and Recent Advances in Multifunctional Polymeric DNA Delivery Systems, Biomacromolecules, 18 (2017) 2231–2246. [DOI] [PubMed] [Google Scholar]
- [14].Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG, Non-viral vectors for gene-based therapy, Nat Rev Genet, 15 (2014) 541–555. [DOI] [PubMed] [Google Scholar]
- [15].Buscail L, Bournet B, Vernejoul F, Cambois G, Lulka H, Hanoun N, Dufresne M, Meulle A, Vignolle-Vidoni A, Ligat L, Saint-Laurent N, Pont F, Dejean S, Gayral M, Martins F, Torrisani J, Barbey O, Gross F, Guimbaud R, Otal P, Lopez F, Tiraby G, Cordelier P, First-in-man phase 1 clinical trial of gene therapy for advanced pancreatic cancer: safety, biodistribution, and preliminary clinical findings, Mol Ther, 23 (2015) 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Suk JS, Kim AJ, Trehan K, Schneider CS, Cebotaru L, Woodward OM, Boylan NJ, Boyle MP, Lai SK, Guggino WB, Hanes J, Lung gene therapy with highly compacted DNA nanoparticles that overcome the mucus barrier, J Control Release, 178 (2014) 8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Petersen H, Fechner PM, Martin AL, Kunath K, Stolnik S, Roberts CJ, Fischer D, Davies MC, Kissel T, Polyethylenimine-graft-Poly(ethylene glycol) Copolymers: Influence of Copolymer Block Structure on DNA Complexation and Biological Activities as Gene Delivery System, Bioconjugate Chemistry, 13 (2002) 845–854. [DOI] [PubMed] [Google Scholar]
- [18].Zintchenko A, Philipp A, Dehshahri A, Wagner E, Simple modifications of branched PEI lead to highly efficient siRNA carriers with low toxicity, Bioconjug Chem, 19 (2008) 1448–1455. [DOI] [PubMed] [Google Scholar]
- [19].Mead BP, Mastorakos P, Suk JS, Klibanov AL, Hanes J, Price RJ, Targeted gene transfer to the brain via the delivery of brain-penetrating DNA nanoparticles with focused ultrasound, J Control Release, 223 (2016) 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Seo YE, Bu T, Saltzman WM, Nanomaterials for convection-enhanced delivery of agents to treat brain tumors, Curr Opin Biomed Eng, 4 (2017) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].van Gaal EV, van Eijk R, Oosting RS, Kok RJ, Hennink WE, Crommelin DJ, Mastrobattista E, How to screen non-viral gene delivery systems in vitro?, J Control Release, 154 (2011) 218–232. [DOI] [PubMed] [Google Scholar]
- [22].Anderson DG, Peng W, Akinc A, Hossain N, Kohn A, Padera R, Langer R, Sawicki JA, A polymer library approach to suicide gene therapy for cancer, Proc Natl Acad Sci U S A, 101 (2004) 16028–16033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yamano S, Dai J, Moursi AM, Comparison of Transfection Efficiency of Nonviral Gene Transfer Reagents, Molecular Biotechnology, 46 (2010) 287–300. [DOI] [PubMed] [Google Scholar]
- [24].Zanoni M, Piccinini F, Arienti C, Zamagni A, Santi S, Polico R, Bevilacqua A, Tesei A, 3D tumor spheroid models for in vitro therapeutic screening: a systematic approach to enhance the biological relevance of data obtained, Scientific Reports, 6 (2016) 19103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Edmondson R, Broglie JJ, Adcock AF, Yang L, Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors, Assay Drug Dev Technol, 12 (2014) 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Paunovska K, Sago CD, Monaco CM, Hudson WH, Castro MG, Rudoltz TG, Kalathoor S, Vanover DA, Santangelo PJ, Ahmed R, Bryksin AV, Dahlman JE, A Direct Comparison of in Vitro and in Vivo Nucleic Acid Delivery Mediated by Hundreds of Nanoparticles Reveals a Weak Correlation, Nano Lett, 18 (2018) 2148–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Cukierman E, Pankov R, Stevens DR, Yamada KM, Taking cell-matrix adhesions to the third dimension, Science, 294 (2001) 1708–1712. [DOI] [PubMed] [Google Scholar]
- [28].Sutherland R, Cell and environment interactions in tumor microregions: the multicell spheroid model, Science, 240 (1988) 177–184. [DOI] [PubMed] [Google Scholar]
- [29].Hambardzumyan D, Bergers G, Glioblastoma: Defining Tumor Niches, Trends Cancer, 1 (2015) 252–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lu H, Stenzel MH, Multicellular Tumor Spheroids (MCTS) as a 3D In Vitro Evaluation Tool of Nanoparticles, Small, 14 (2018) e1702858. [DOI] [PubMed] [Google Scholar]
- [31].Thakuri PS, Liu C, Luker GD, Tavana H, Biomaterials-Based Approaches to Tumor Spheroid and Organoid Modeling, Adv Healthc Mater, 7 (2018) e1700980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mastorakos P, Zhang C, Song E, Kim YE, Park HW, Berry S, Choi WK, Hanes J, Suk JS, Biodegradable brain-penetrating DNA nanocomplexes and their use to treat malignant brain tumors, J Control Release, 262 (2017) 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Beh CW, Pan D, Lee J, Jiang X, Liu KJ, Mao HQ, Wang TH, Direct interrogation of DNA content distribution in nanoparticles by a novel microfluidics-based single-particle analysis, Nano Lett, 14 (2014) 4729–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kopanska KS, Alcheikh Y, Staneva R, Vignjevic D, Betz T, Tensile Forces Originating from Cancer Spheroids Facilitate Tumor Invasion, PLoS One, 11 (2016) e0156442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Duncan GA, Jung J, Joseph A, Thaxton AL, West NE, Boyle MP, Hanes J, Suk JS, Microstructural alterations of sputum in cystic fibrosis lung disease, JCI Insight, 1 (2016) e88198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Recinos VR, Tyler BM, Bekelis K, Sunshine SB, Vellimana A, Li KW, Brem H, Combination of intracranial temozolomide with intracranial carmustine improves survival when compared with either treatment alone in a rodent glioma model, Neurosurgery, 66 (2010) 530–537; discussion 537. [DOI] [PubMed] [Google Scholar]
- [37].Tyler B, Fowers KD, Li KW, Recinos VR, Caplan JM, Hdeib A, Grossman R, Basaldella L, Bekelis K, Pradilla G, Legnani F, Brem H, A thermal gel depot for local delivery of paclitaxel to treat experimental brain tumors in rats, J Neurosurg, 113 (2010) 210–217. [DOI] [PubMed] [Google Scholar]
- [38].Vellimana AK, Recinos VR, Hwang L, Fowers KD, Li KW, Zhang Y, Okonma S, Eberhart CG, Brem H, Tyler BM, Combination of paclitaxel thermal gel depot with temozolomide and radiotherapy significantly prolongs survival in an experimental rodent glioma model, Journal of Neuro-Oncology, 111 (2013) 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Song E, Gaudin A, King AR, Seo YE, Suh HW, Deng Y, Cui J, Tietjen GT, Huttner A, Saltzman WM, Surface chemistry governs cellular tropism of nanoparticles in the brain, Nat Commun, 8 (2017) 15322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Greensted A, Otsu Thresholding, in, The Lab Book Pages, 2010. [Google Scholar]
- [41].van Gaal EV, Oosting RS, Hennink WE, Crommelin DJ, Mastrobattista E, Junk DNA enhances pEI-based non-viral gene delivery, Int J Pharm, 390 (2010) 76–83. [DOI] [PubMed] [Google Scholar]
- [42].Ansari AM, Ahmed AK, Matsangos AE, Lay F, Born LJ, Marti G, Harmon JW, Sun Z, Cellular GFP Toxicity and Immunogenicity: Potential Confounders in in Vivo Cell Tracking Experiments, Stem Cell Rev, 12 (2016) 553–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gambotto A, Dworacki G, Cicinnati V, Kenniston T, Steitz J, Tuting T, Robbins PD, DeLeo AB, Immunogenicity of enhanced green fluorescent protein (EGFP) in BALB/c mice: identification of an H2-Kd-restricted CTL epitope, Gene Ther, 7 (2000) 2036–2040. [DOI] [PubMed] [Google Scholar]
- [44].Liu HS, Jan MS, Chou CK, Chen PH, Ke NJ, Is green fluorescent protein toxic to the living cells?, Biochem Biophys Res Commun, 260 (1999) 712–717. [DOI] [PubMed] [Google Scholar]
- [45].Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA, Fluorescent proteins from nonbioluminescent Anthozoa species, Nature Biotechnology, 17 (1999) 969. [DOI] [PubMed] [Google Scholar]
- [46].Alieva NO, Konzen KA, Field SF, Meleshkevitch EA, Hunt ME, Beltran-Ramirez V, Miller DJ, Wiedenmann J, Salih A, Matz MV, Diversity and evolution of coral fluorescent proteins, PLoS One, 3 (2008) e2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Pamujula S, Hazari S, Bolden G, Graves RA, Chinta DD, Dash S, Kishore V, Mandal TK, Cellular delivery of PEGylated PLGA nanoparticles, J Pharm Pharmacol, 64 (2012) 61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hu Y, Xie J, Tong YW, Wang CH, Effect of PEG conformation and particle size on the cellular uptake efficiency of nanoparticles with the HepG2 cells, J Control Release, 118 (2007) 7–17. [DOI] [PubMed] [Google Scholar]
- [49].Mishra S, Webster P, Davis ME, PEGylation significantly affects cellular uptake and intracellular trafficking of non-viral gene delivery particles, Eur J Cell Biol, 83 (2004) 97–111. [DOI] [PubMed] [Google Scholar]
- [50].Sant S, Johnston PA, The production of 3D tumor spheroids for cancer drug discovery, Drug Discov Today Technol, 23 (2017) 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Baker BM, Chen CS, Deconstructing the third dimension: how 3D culture microenvironments alter cellular cues, J Cell Sci, 125 (2012) 3015–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Shin CS, Kwak B, Han B, Park K, Development of an in vitro 3D tumor model to study therapeutic efficiency of an anticancer drug, Mol Pharm, 10 (2013) 2167–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Lovitt CJ, Shelper TB, Avery VM, Miniaturized three-dimensional cancer model for drug evaluation, Assay Drug Dev Technol, 11 (2013) 435–448. [DOI] [PubMed] [Google Scholar]
- [54].Oishi M, Nagasaki Y, Nishiyama N, Itaka K, Takagi M, Shimamoto A, Furuichi Y, Kataoka K, Enhanced growth inhibition of hepatic multicellular tumor spheroids by lactosylated poly(ethylene glycol)-siRNA conjugate formulated in PEGylated polyplexes, ChemMedChem, 2 (2007) 1290–1297. [DOI] [PubMed] [Google Scholar]
- [55].Schuster BS, Ensign LM, Allan DB, Suk JS, Hanes J, Particle tracking in drug and gene delivery research: State-of-the-art applications and methods, Adv Drug Deliv Rev, 91 (2015) 70–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Jahangiri A, Chin AT, Flanigan PM, Chen R, Bankiewicz K, Aghi MK, Convection-enhanced delivery in glioblastoma: a review of preclinical and clinical studies, J Neurosurg, 126 (2017) 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Voges J, Weber F, Reszka R, Sturm V, Jacobs A, Heiss WD, Wiestler O, Kapp JF, Clinical protocol. Liposomal gene therapy with the herpes simplex thymidine kinase gene/ganciclovir system for the treatment of glioblastoma multiforme, Hum Gene Ther, 13 (2002) 675–685. [DOI] [PubMed] [Google Scholar]
- [58].Voges J, Reszka R, Gossmann A, Dittmar C, Richter R, Garlip G, Kracht L, Coenen HH, Sturm V, Wienhard K, Heiss WD, Jacobs AH, Imaging-guided convection-enhanced delivery and gene therapy of glioblastoma, Ann Neurol, 54 (2003) 479–487. [DOI] [PubMed] [Google Scholar]
- [59].Kayama Takamasa, Yoshimoto Takashi, Fujimoto Shunichi, Sakurai Yoshiharu, Intratumoral oxygen pressure in malignant brain tumor, Journal of Neurosurgery, 74 (1991) 55–59. [DOI] [PubMed] [Google Scholar]
- [60].Giese A, Bjerkvig R, Berens ME, Westphal M, Cost of Migration: Invasion of Malignant Gliomas and Implications for Treatment, Journal of Clinical Oncology, 21 (2003) 1624–1636. [DOI] [PubMed] [Google Scholar]
- [61].Hambardzumyan D, Gutmann DH, Kettenmann H, The role of microglia and macrophages in glioma maintenance and progression, Nat Neurosci, 19 (2016) 20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Amoedo ND, Valencia JP, Rodrigues MF, Galina A, Rumjanek FD, How does the metabolism of tumour cells differ from that of normal cells, Biosci Rep, 33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Romero-Garcia S, Lopez-Gonzalez JS, Baez-Viveros JL, Aguilar-Cazares D, Prado-Garcia H, Tumor cell metabolism: an integral view, Cancer Biol Ther, 12 (2011) 939–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Mellman I, Yarden Y, Endocytosis and cancer, Cold Spring Harb Perspect Biol, 5 (2013) a016949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.