Abstract
This manuscript describes protocols for separation of complex protein samples using two-dimensional polyacrylamide gel electrophoresis (2D-PAGE). Electrophoresis in a single dimension, e.g., 1D SDS polyacrylamide gels, has the potential to rapidly separate hundreds of proteins. When two orthogonal high-resolution electrophoretic methods are efficiently combined in perpendicular dimensions, complex protein mixtures can be separated into thousands of discrete spots. The most common 2D gel separation for intact proteins involves a first dimensional separation using isoelectric focusing (IEF) followed by separation based on protein size (SDS-PAGE). Currently, most 2D gel studies rely on the use of commercially available immobilized pH gradient (IPG) gels, which provide improved ease of use and reproducibility compared with older methods. IPG gels are available in a range of sizes and different pH ranges. Resolution typically increases as the 2D gel size increases; however, difficulty of use increases sharply and throughput decreases as gel size increases.
Keywords: two-dimensional gel electrophoresis, isoelectric focusing, proteomics, protein profiling
INTRODUCTION
Two-dimensional gel electrophoresis combines two different electrophoretic separation modes in perpendicular directions to provide a much greater separation of complex protein mixtures than either of the individual separation methods. Typically, separation based on protein isoelectric point (pI) is followed by size separation, which is the most powerful single tool for protein separations currently available. After staining, proteins appear on the final 2D gel as round or elliptical spots instead of the rectangular bands observed on 1D gels. Although the total separating power of full-sized or standard 2D (∼16–18 cm × ∼16–18 cm) is estimated to exceed 5000 spots per gel, in practice a single 2D separation of a complex mixture such as a whole-cell or tissue extract will typically produce on the order of 1000 to 2000 well-resolved spots when a sensitive detection method is used. This less than predicted resolution is in part due to non-uniform distribution of proteins across the available separation range as well as actual ranges in protein abundances for most proteomes that exceed the dynamic range of any single detection method. The most common 2D technique uses isoelectrofocusing (IEF) under denaturing conditions in commercially prepared immobilized pH gradient (IPG) gels (see Basic Protocol 1). In this system, the buffering side chains are covalently incorporated into the acrylamide matrix to produce a stable pH range. An advantage of this approach is that any pH range and curve shape can be generated by pouring a gradient gel using two solutions that differ in covalent ampholyte composition rather than acrylamide concentration. The initial electrophoresis is followed by a second separation using SDS-PAGE in a perpendicular direction (see Basic Protocol 2). Initial screening of the sample pH range using Immobiline DryPlates is described in Support Protocol 1. The popularity of IPG gels has steadily increased since their introduction, for at least three major reasons: reproducible premade IPG gels are now commercially available in multiple pH ranges and sizes; most technical problems associated with their use have been solved or substantially minimized; and the narrow thickness of the IPG stripes make transfer to the second dimension relatively seamless.
In contrast to use of IPG gels, the traditional method for the first dimension IEF separation, which still has the advantage of higher protein load capacity, is based on the use of soluble ampholytes. These are relatively small organic molecules with various isoelectric points that act as buffers at their isoelectric point. The pH gradient for these IEF gels, which are cast in glass tubes of varying diameter, is produced when an electric field is applied, which causes the soluble ampholytes to migrate in the gel matrix until they reach their isoelectric point where their buffering capacity sets up the pH gradient. This soluble ampholyte IEF in a tube gel (see Basic Protocol 3) is followed by SDS PAGE in a perpendicular direction (Basic Protocol 4). Tube gels, particularly larger format gels have substantially more total protein capacity than IPG stripes; however, the efficient transfer from a tube gel to the second-dimension slab gel can present additional technical challenges, particularly for inexperienced users.
The most common separation conditions for either IPG or soluble ampholyte IEF gels are to separate the proteins until equilibrium is reached; that is, all proteins and soluble ampholytes have reached their isoelectric point and migration has (ideally) stopped. Because stable pH gradients outside the pH 4.0 to 8.0 range are difficult to create in soluble ampholyte IEF tube gels, alternative protocols using nonequilibrium conditions are required to resolve proteins with pI values below 3.0 to 4.0 (see Alternate Protocol 1 for acidic proteins) or above 8.0 (see Alternate Protocol 2 for basic proteins). One of the more important limitations of soluble ampholytes is the difficulty in obtaining highly reproducible pH profiles, especially when very narrow pH ranges are needed.
Another common two-dimensional electrophoresis format is a nonreducing/reducing electrophoretic separation (see Alternate Protocol 3). This method provides useful information about either intrinsic inter-subunit disulfides or protein-protein complexes that have been cross-linked using a bifunctional chemical cross-linker that contains a disulfide bond within the linker region.
Several support protocols are also provided that describe: use of IPG slab gels to compare multiple samples after IEF separation only (see Support Protocol 1), preparation of pI standards and pH profile measurements (see Support Protocol 2), preparation of molecular weight standards for two-dimensional gels (see Support Protocol 3), and 2D protein databases (see Support Protocol 4). Sample preparation for 2D gels is described in unit 22.4. Additional details for preparing samples for 2-D DIGE, which is a method that differentially fluorescently tags multiple samples prior to mixing and separation in a single 2D gel, are described in unit 22.2. Regardless of whether samples are unlabeled or fluorescently tagged for 2D DIGE, the gel separation methods are the same, e.g. Basic Protocols 1 and 2.
NOTE: High-purity water (e.g., Milli-Q water or equivalent) is essential for all solutions. For cautions relating to electricity and electrophoresis, refer to Safety Considerations in the introduction to UNIT 10.1.
BASIC PROTOCOL 1: ISOELECTROFOCUSING USING IMMOBILIZED pH GRADIENT GEL STRIPS
In IPG gels, the ampholytes are covalently linked to the acrylamide matrix, which facilitates production of highly reproducible gradients, as well as very narrow pH gradients for optimal resolution of minor charge differences. A variety of precast gels and all the necessary equipment are commercially available from BioRad or Thermo Fisher Scientific. Narrow strips of precast IEF gels (IPG Strips) may be used to achieve a first-dimension separation for two-dimensional gel electrophoresis, and broader precast slab gels (Immobiline DryPlates) can be used to compare multiple samples after IEF separation only (see Support Protocol 1 and Table 3). In this protocol, samples are loaded to precast IPG Strips using active rehydration (low voltage is applied during the rehydration step) in the i12 focusing tray using the Protean i12 IEF System (one to twelve sample strips of the same length may be handled at a time. The rehydration is programmed as the first step of the protocol. This procedure has been adapted from instruction booklets provided by Bio-Rad. See Basic Protocol 2 for details concerning preparing and running the second-dimension gel.
Table 3.
Size Options for Two-Dimensional Gel Electrophoresis
| First-dimension gel |
Second-dimension gela |
|||||
|---|---|---|---|---|---|---|
| Gel type | Diameter D (mm) | Length L (cm) | Thickness T (mm) | Height H (cm) | Purpose | Commentsb |
| Microgels/minigelsc | <1.5 | <10 | <D | <10 | Analytical | 1–4 |
| <1.5 | <10 | >D | <10 | Analytical | 3–6 | |
| >1.5 | <10 | <Dd | <10 | Analytical/preparative | 1,2,7,8 | |
| Full-size gelse | <1.5 | 12–18 | <D | 12–18 | Analytical | 1–4 |
| <1.5 | 12–18 | >D | 12–18 | Analytical | 3–6 | |
| >1.5 | 12–18 | <Dd | 12–18 | Analytical/preparative | 1,2,7,8 | |
| Giant gelsf | <1.5 | >20 | >D | >20 | Analytical | 4–6,9 |
| >1.5 | >20 | <Dd | >20 | Analytical/preparative | 1–3,7 | |
The second-dimension gel width has to be at least equal to the IEF tube gel height.
Key to comments: (1) tube gel cannot be placed directly on top of second-dimension gel, and use of agarose is recommended; (2) use of stacking gel is recommended; (3) extrusion and handling are relatively difficult; (4) total protein load is limited to usually ≤50 μg for whole-cell or tissue extracts; (5) tube gel can be placed directly on top of second-dimension gel, and use of agarose is not necessary; (6) use of a stacking gel is not necessary; (7) total protein load capacity is relatively large; (8) extrusion and handling are relatively simple; (9) extrusion and handling are very difficult.
Minigel systems provide rapid separations with moderate resolution. Microgels (Phastgels) are precast gels that are slightly smaller than most minigels.
Use of second-dimension gels thicker than 1.5 mm is generally not recommended owing to difficulty with either efficient staining or efficient electroblotting.
Full-size gels provide resolution satisfactory for most applications.
Giant gels provide very good resolution. Specialized equipment is required, such as Ettan-Dalt (Thermo Fisher Scientific), or homemade giant-size gel systems.
Wear gloves throughout the procedure and handle the IPG Strips with clean forceps where feasible to prevent extraneous protein contamination of the gels and gel solutions. Thoroughly clean all equipment with a mild laboratory detergent solution, rinse well with Milli-Q water, and allow to dry before using. When air drying and storing equipment and accessories, they should be protected from airborne contamination which contains skin keratins and other environmental proteins that can interfere with high sensitivity staining patterns or protein identification by MS. Solutions containing 10 M urea may be heated briefly to 30° to 40°C to aid in solubilization, but excessive heating should be avoided so that decomposition to form cyanate is minimized.
Materials
Urea (ultrapure)
CHAPS or Triton X-100
IPG Buffer corresponding to IPG Strip pH range (see Table 1)
Table 1.
Rehydration Solutions for IPG Stripsa
| IPG Strip type |
||||
|---|---|---|---|---|
| Component | Final conc. | 3–10L | 3–10NL | 4–7L |
| Ultrapure urea | 9 Mb | 2.7 g | 2.7 g | 2.7 g |
| CHAPSc | 2% | 0.1 g | 0.1 g | 0.1 g |
| IPG Buffer pH 3–10d | 1:50 | 100 μl | ||
| IPG Buffer pH 3–10NLd | 100 μl | |||
| IPG Buffer pH 4–7d | 100 μl | |||
| DTT | 0.3% | 75 mg | 75 mg | 75 mg |
| Bromphenol blue | Trace | A few grains | A few grains | A few grains |
| Milli-Q water | To 5 ml | To 5 ml | To 5 ml | |
Rehydration solutions should be prepared fresh immediately before use and should be filtered using a 0.2-μm filter. Minimize total time the solution is at room temperature prior to use to minimize decomposition of urea. If the reswelling tray is used, ~250 or 400 μl rehydration solution is required per 11- or 18-cm IPG Strip, respectively.
Urea concentrations between 8 M and 10 M are typically used or a combination of 2M thiourea with 7M or 8M urea. Higher concentrations promote better protein solubility during isofocusing while increasing the risk of urea crystallization. If 10 M urea is used, extra care should be taken to minimize evaporation and the temperature should be maintained at 20°C or slightly higher at all times to avoid crystallization of the urea. Improved protein solubilization can be achieved by using a combination of 7M urea and 2 M thiourea.
The optimal detergent and detergent concentration should be empirically determined. Other common alternatives are Triton X-100 and octyl-glucoside. The detergent used must be nonionic or zwitterionic to avoid high current and consequent overheating during isoelectrofocusing.
The IPG buffer is an ampholyte containing buffer and should be matched to the pH range of the IPG strip.
DTT (dithiothreitol)
Bromophenol blue
Protein sample to be analyzed
Lysis buffer (see recipe)
Protean i12 IEF System and instruction manual (Bio-Rad)
Precast IPG Strips (Bio-Rad or Thermo Fisher Scientific) (See Table 2)
Table 2.
Commercially Available Precast IPG Gelsa
| Name | Use | Available pH range |
|---|---|---|
| Immobiline DryPlate | Running one-dimensional IPG gels | pH 4.0–7.0 |
| Immobiline IPG Strip | Running first dimension in 2D gels | |
| 7 cm | pH 5.3–6.5, pH 6.2–7.5, pH 3–5.6NL, pH4–7, pH 6–11, pH 7–11NL, pH3–10, pH 3–11NL,pH 3–10NL | |
| 11 cm | pH 5.3–6.5, pH 6.2–7.5, pH 3–5.6NL, pH 4–7, pH 6–11, pH 7–11NL pH 3–10, pH 3–11NL, | |
| 13 cm | pH 5.3–6.5, pH 6.2–7.5, pH 3–5.6NL, pH 4–7, pH 6–11, pH 7–11NL pH 3–10, pH 3–11NL, pH 3–10NL | |
| 18 cm | pH 5.3–6.5, pH 6.2–7.5, pH 3–5.6NL, pH 4–7, pH 6–9, pH 6–11, pH 7–11NL, pH 3–10b, pH 3–10NLc, pH 3–10NL | |
| 24 cm | pH 3.5–4.5, pH 5.3–6.5, pH 6.2–7.5, pH 3–5.6NL, pH 3–7NL, pH 4–7, pH 6–9, pH 7–11NL, pH 3–10, pH |
From Thermo Fisher Scientific. Additional narrow range pH 3.9–5.1, pH 4.7–5.9, pH 5.5–6.7 and pH 6.3–8.3 are available from Bio-Rad in all strip lengths.
A linear gradient with maximum resolution above pH 7.0.
A nonlinear gradient with best resolution at pH 5.0–7.0.
Forceps
Mineral oil
Filter paper
Glass plate
Petri dishes
Additional reagents and equipment for protein detection by staining (UNIT 10.5) and/or for electroblotting (UNIT 10.7, optional)
Rehydrate the IPG Strip(s)
-
Prepare an appropriate rehydration solution for the type of IPG Strip to be used as described in Table 1 (~315 μl rehydration solution per 18-cm IPG Strip).
The urea concentration in the rehydration solution should be 8 – 10 M, and 2% CHAPS or another appropriate detergent (zwitterionic or nonionic) such as Triton X-100, NP-40, or n-octylglucoside should be included in the rehydration solution to aid in sample solubility. The optimal detergent and detergent concentration may vary with type of sample and may need to be determined empirically. The ampholyte containing IPG Buffer should match the pH range of the IPG strips.
One possible method of loading large sample volumes onto IPG gels is to add the sample directly to the rehydration solution. This sample loading method is recommended since there is no discrete application point and it eliminates the formation of protein precipitates (most proteins are least soluble at or near their isoelectric points). Cup loading or paper-bridge loading allow a maximum sample volume of 100 μl to be loaded and are recommended for use with basic or acidic strips (see the manufacturer’s literature).
Samples should either be lyophilized and then solubilized in lysis buffer, or diluted 9 parts lysis buffer to 1 part sample. The complexity of the sample, the sample solubility at the loading concentration and pH used, the thickness of the second-dimension gel, and the detection method to be employed should be considered when deciding how much protein to load. As a starting reference, typical loading ranges for 1.0- to 1.5-mm-thick 18-cm × 18-cm gels would be ~5 to 20 ng per major spot for silver staining and ~1 to 5 μg per major spot for Coomassie blue staining. When very complex samples are used such as whole cell extracts, total protein loads are likely to be ~20 to 100 μg for silver staining and ~200 to 1000 μg for Coomassie blue staining. The salt concentration in samples should be kept <50 mM and, if the sample contains SDS, the final SDS concentration should be <0.25%.
Solutions containing urea should be filtered using a 0.2-μm filter before use.
-
Position the electrode assemblies in the focusing tray. Push down on the tabs until they lock into place.
The focusing tray should be the same size as the IPG strip.
For an 18-cm gel, pipet 315 μl of rehydration solution containing the sample into a channel of the focusing tray. Move the pipet along the length of the well while adding the solution to spread it evenly throughout the length of the slot. Avoid excessive air bubble formation while pipetting this solution.
-
Using forceps, remove the protective cover from the IPG Strips and gently place them, gel side down, into the prepared slot.
The strips should be oriented with the “+” and “pH 4–7” against the positioning stop in each channel. Be careful not to trap any air bubbles under the gel strips. Gently move the IPG Strips back and forth to ensure even distribution of the solution along the length of the IPG strips.
Place the focusing tray on the Peltier cooling platform and connect the electrodes to the instrument.
-
Overlay each strip with 7 ml of mineral oil to prevent evaporation and urea crystallization.
Apply the mineral oil to each end of the strip and allow it to flow toward the middle of each channel.
Position the IPG Strip retainers on top of the IPG strips at both the anode and cathode.
-
Rehydration with in-gel sample application can be programmed as part of the IEF run. Select the protocol for each lane containing an IPG Strip. Program the desired rehydration conditions (~12h) in the Run Settings and start the run. Focus the gels with constant voltage for 30 min at 250 V followed by gradual ramp to 10,000 V (2h) then focus at 10,000 V for a total of 43 kVhr. Followed by a hold at 1,000 V. Refer to the user manual for exact recommended voltage conditions for each type of IPG Strip.
Shorter rehydration times may not completely and reproducibly rehydrate the gels, but do not substantially exceed 16 hr as extensive incubation increases potential problems due to evaporation and urea decomposition, which increases the risk of amino group modification by cyanate produced from the decomposition of the urea.
Sample loading using active rehydration will facilitate better entry of high-molecular weight proteins.
The optimal number of Vhr will depend upon the pH range of the IPG Strip used, the type of sample, and the sample load and volume; therefore, the optimal Vhr may need to be empirically determined for different applications, although recommendations provided by the IPG strip provided are a good place to start.
-
When isoelectrofocusing is complete, disconnect the power supply and remove IPG Strip retainers.
If gels are to be run in the second dimension immediately after isoelectrofocusing, steps 1 to 3 of Basic Protocol 2 should be completed prior to terminating isoelectrofocusing.
-
Using forceps, remove the IPG Strips from the tray. If the gels are to be run immediately in the second dimension, place in a petri dish with the support film along the wall of the dish and proceed directly to equilibration of the gel (see Basic Protocol 2, step 4). Alternatively, gels may be stored sealed in a plastic bag at −80°C until ready to run the second-dimension gel.
Gels may be stored at least 2 to 3 months at −80°C. Do not place in the equilibration buffers required for the second dimension prior to storage.
SUPPORT PROTOCOL 1: ELECTROPHORESIS ON IMMOBILIZED pH GRADIENT GELS
In this protocol, after precast IEF gels (Immobiline DryPlates) from Thermo Fisher Scientific are rehydrated, samples are loaded and subjected to isoelectrofocusing. Gels are typically run at 2500 to 3500 V and require focusing times of 2 to 7 hr. Protein samples may be detected by conventional methods such as Coomassie blue or silver staining (UNIT 10.5). Isoelectric points can be determined with the use of pI calibration proteins; alternatively, because the gradient is linear and immobilized, one can measure the migration distance across the gel and estimate the pI at each location. Some applications of the Immobiline plates include initial screening of samples to determine the optimal pH gradient prior to running more time-consuming two-dimensional gels, prescreening fractions from a chromatographic purification step prior to running two-dimensional gels, and evaluation of charge heterogeneity of purified proteins.
Additional Materials (also see Basic Protocol 1)
Precast pH 4–7 DryPlate gel (Thermo Fisher Scientific)
Repel-Silane (Thermo Fisher Scientific)
Paraffin oil
Protein samples to be analyzed
Reswelling Cassette kit (Thermo Fisher Scientific) including:
125 × 260 × 3-mm glass plate with 0.5-mm U frame
125 × 260 × 3-mm glass plate
Silicone tubing
Pinchcock
Clamps
20-ml syringe
Roller (Thermo Fisher Scientific)
Whatman no. 1 filter paper
Flatbed electrophoresis unit
10° or 15°C cooling water bath
Electrode strips
Sample applicator strip or sample application pieces
Power supply (minimum capacity 3000 to 3500 V)
Additional reagents and equipment for protein detection by staining (UNIT 10.5) and for electroblotting (UNIT 10.7; optional)
Rehydrate the gel
-
1
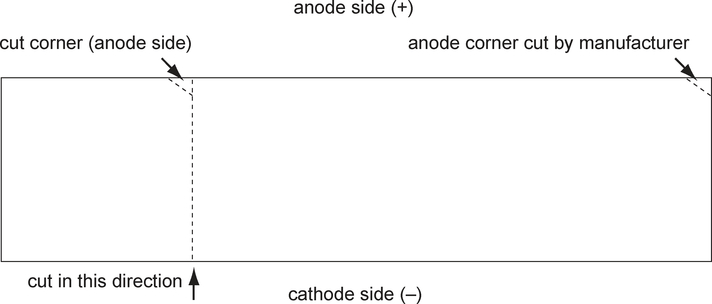
Remove precast gel from packaging. If the entire gel is not needed, cut off the required number of lanes and reseal the unused gel. Mark the polarity of the gel section to be used by cutting a small triangle off the anode corner. Handle the gel by the support film only.
It is critical that the lanes are cut from the gel in the proper orientation to preserve the pH gradient (see Fig. 1), and polarity must be indicated for proper orientation of electrodes later in the procedure.
-
2
Use the Reswelling Cassette to rehydrate the gel. Connect silicone tubing through the hole in the bottom corner of the U-frame plate, seal with silicone glue, and connect the pinchcock to the other end of the tubing. Place a glass plate on a clean flat surface and wet with a few drops of water. Place the gel on the plate, gel side up. Gently roll with a clean rubber roller to remove any air bubbles.
-
3
Cover the plate and gel with the plate fitted with the U frame.
The U-frame plate should be coated with a thin layer of Repel-Silane to prevent the gel from sticking to the plate.
-
4
Place clamps around the edges of the plates, making sure the seal is tight.
-
5
Slowly fill the cassette with the desired rehydration solution using a 20-ml syringe connected to the silicone tubing and let stand for the recommended amount of time. Precool electrophoresis unit 1 to 2 hr prior to electrophoresis (see step 9).
Reswelling with water for 2 to 3 hr is normally sufficient. If using additives such as urea, Triton, glycerol, or reducing agents, allow the gel to rehydrate overnight. Additives can be used to improve solubility of proteins near their isoelectric point. Reducing reagents such as 2-mercaptoethanol or DTT are used to reduce disulfide bonds.
-
6
When gel has been allowed to rehydrate completely, remove the clamps and gently pry the plates apart.
-
7
Moisten a piece of filter paper with water and place on top of the gel, then layer with a piece of dry filter paper.
-
8
Gently blot the gel by rolling over the dry filter paper with the rubber roller to remove excess water. The gel is now ready to be placed on the cooling plate.
Do not let the gel dehydrate prior to placing it on the cooling plate in step 11.
Figure 1.
Marking orientation of a precast IPG gel when only a portion of the gel is used.
Run the gel
-
9
Connect the flatbed electrophoresis unit to a recirculating cooling water bath. Allow to cool to 10°C for 1 to 2 hr to ensure even cooling. If the gel has been rehydrated in the presence of urea, do not cool below 15°C so that urea does not precipitate.
-
10
Pipet 2 to 3 ml paraffin oil onto the surface of the cooling plate.
-
11
Position the gel on the cooling plate, being careful not to trap air bubbles between the gel and the plate. Orient the gel so that the polarity of the gel matches the polarity of the cooling plate.
-
12
Soak two electrode strips with ~3 ml water, then blot to remove excess water.
-
13
Lay a blotted electrode strip along each long edge of the gel. Cut off the ends of the electrode strip so that it does not extend beyond the edge of the gel.
-
14
Load protein samples to be analyzed onto the gel. Use an applicator strip for sample volumes between 5 to 20 μl (make sure contact between the strip and the gel is uniform). Use sample application pieces for sample volumes >20 μl. Remove the application pieces halfway through focusing. For sample volumes of 2 to 10 μl, samples may be spotted directly on the gel without using applicator strips. See manufacturer’s instructions for further details.
An important experimental consideration is the position in the pH gradient where the sample is applied. The acidic end of the gel can usually be used for sample application; however, the optimal loading position may need to be determined empirically for different types of samples. At high protein concentrations and/or at nonoptimal pH’s, samples may precipitate in the gel at the loading position.
Samples should contain <50 mM salt or buffer components; greater concentrations will cause local overheating of the gel. If possible, salt-free samples should be solubilized or dialyzed in the rehydration buffer.
-
15
Align the electrodes with the electrode strips, put the safety lid in place, and connect the apparatus to the power supply. Conduct electrophoresis at 3000 V for 2 to 4 hr.
-
16
After removing gels from the electrophoresis apparatus, detect proteins using any conventional staining technique such as Coomassie blue or silver staining.
-
17
Preserve the gels by sealing in a plastic bag or by drying for a permanent record. Alternatively, electrotransfer the proteins on the gel to a PVDF membrane.
To dry a gel, presoak it first in a preservation solution. For silver-stained gels, use a solution of 5% to 10% (w/v) glycerol/30% (v/v) ethanol; for Coomassie blue-stained gels, use a solution of 5% to 10% (w/v) glycerol/16% (v/v) ethanol/8% (w/v) acetic acid. After soaking the gel, place it on a glass plate gel side up, cover with a cellophane sheet soaked in preservation solution, and allow to dry at room temperature.
For electrotransfer (UNIT 10.7), use film remover to remove the plastic support film from the gel. Electrotransfer of proteins to a polyvinylidene difluoride (PVDF) membrane using a Multiphor II NovaBlot transfer kit (Thermo Fisher Scientific) or equivalent is recommended. Transferring IPG gels requires special procedures; see the transfer kit manual for instructions.
BASIC PROTOCOL 2: SECOND-DIMENSION ELECTROPHORESIS OF IPG GELS
In this protocol, vertical slab gels are used as the second dimension for IPG gels. Depending upon the IPG strip length and the slab gel apparatus, special second-dimension gel spacers may be needed to fit the IPG strip. For example, Bio-Rad offers a conversion kit to increase the gel width from 16 cm to 18 cm in order to accommodate 18 cm IPG strips, and Thermo Fisher Scientific offers the Ettan-Dalt gel system. A variety of precast gels that can accommodate IPG strips are also available from both Bio-Rad and Thermo Fisher Scientific. A two-step equilibration of the strips prior to electrophoresis is required.
IPG Strip equilibration solutions 1 and 2 (see recipes; prepare fresh in step 4)
IPG Strip with focused protein (see Basic Protocol 1)
Platform shaker
Additional reagents and equipment for linear or gradient Laemmli gels or other slab gels (UNIT 10.1)
2% agarose (see recipe)
Cast the second-dimension gel
-
1
Assemble the glass-plate sandwich of an electrophoresis apparatus, using gel plates wide enough to accommodate an 18-cm-long IPG Strip gel.
If the spacers are not wide enough to accommodate an 18 cm gel, the ends of the gel strip may be trimmed away from the IPG gel so that it will fit on top of the second dimension; however, some very basic or acidic proteins may be lost.
-
2
Pour a separating gel of the desired acrylamide concentration and immediately overlay with water to produce a smooth surface.
The separating gel should be a minimum of ~2.5 cm below the top of the inner plate to accommodate a 2-cm stacking gel.
-
3
After the separating gel has polymerized, remove the water overlay, rinse the gel surface with water to remove any unpolymerized acrylamide, and pour the stacking gel to a height of 0.5 cm from the top of the plate. Overlay with water to produce a smooth surface.
A water overlay provides a smooth surface for better contact between the IPG Strip and the second dimension gel. The stacking gel height should be ~2 cm.
Load the IPG Strip gel onto the second-dimension gel
-
4
Prepare IPG Strip equilibration solutions 1 and 2 (see recipe).
-
5
Assemble the second-dimension gels in an electrophoresis chamber. Do not pour electrophoresis buffer into the upper chamber.
-
6
Melt 2% (w/v) agarose in a boiling water bath. Mix a solution of 1 part 2% agarose to 2 parts equilibration solution 2.
Keep agarose/equilibration buffer mixture in boiling water bath until step 11 is completed.
The agarose prevents the IPG Strip from shifting position and ensures good contact between the IEF and second-dimension gels.
-
7
Using forceps, remove the IPG gels from the electrophoresis tray after isoelectrofocusing is complete or from the −80°C freezer (see Basic Protocol 1, step 19) and place each strip in a separate petri dish with the support film side of the strip facing the petri dish wall. Add 15 ml of IPG Strip equilibration buffer 1. Cover and place on a platform shaker for 10 min.
Strips may be run in the second dimension immediately after isoelectrofocusing or after storage at −80°C. If the strips have been stored at −80°C, remove them from the freezer, then place in petri dish as stated and continue with the equilibration procedure.
-
8
Discard equilibration buffer 1 and add 15 ml of equilibration buffer 2. Cover and place on a platform shaker for 10 min.
-
9
Dampen a piece of filter paper and place on a glass plate. Remove the IPG Strips from equilibration buffer 2. Place each strip on its edge on the filter paper to remove any excess buffer.
Strips should not be left in this position for >10 min, or spot sharpness may be affected.
-
10
Add a small amount of SDS electrode buffer along the glass plate above the second-dimension gel. Place the IPG Strip gel in the well with the gel facing out and the basic side to the left. Push the IPG Strip down so that it is firmly in contact with the stacking gel of the second-dimension gel. Remove excess running buffer.
-
11
Overlay the IPG gel strip with the agarose/equilibration buffer (from step 6) and allow agarose to solidify.
-
12
Carefully pour electrophoresis buffer into the upper reservoir, taking care to avoid disturbing the agarose-embedded IPG Strip.
-
13
Connect electrodes and run the gels.
See UNIT 10.1 for electrophoresis conditions and UNIT 10.5 for gel staining conditions.
BASIC PROTOCOL 3: HIGH-RESOLUTION EQUILIBRIUM ISOELECTROFOCUSING IN TUBE GELS
This protocol describes the preparation of broad-range first-dimension gels using soluble ampholytes that resolve proteins with pI values between approximately 3.0 and 8.0, and is based on the original procedure described by O’Farrell (1975). The procedure presented here refers specifically to 3-mm IEF tube gels (first-dimension) combined with 1.5-mm-thick 16 × 16-cm (size of separating gel) second-dimension gels (see Basic Protocol 4) and may be easily adapted to a variety of different gel sizes (see Table 3). A 3-mm IEF gel has a total protein capacity of ~500 μg for complex protein mixtures such as whole-cell extracts. The maximum capacity of any single protein spot is ~0.5 to 5 μg, depending on the solubility of the protein near its isoelectric point and the separation distance from any near neighbors.
In this protocol, gels are cast and prefocused before the sample is loaded. The proteins are then separated according to isoelectric point, and the gels are extruded from the tubes and stored. Measuring pH profiles in IEF gels is a convenient and accurate method for determining pI (see Support Protocol 2). To provide optimal reproducibility, multiple gels should be cast and run simultaneously. This is especially important for comparative studies involving complex mixtures of proteins.
The IEF gels may be cast either by pouring the gel solution into the gel tubes (steps 3a to 7a) or by using hydrostatic pressure (steps 3b to 7b). Pouring the gel solution into the gel tubes is convenient for 3-mm-diameter IEF gels and requires only a minimal excess of reagents. Because the gels are cast using a long needle and syringe, for narrower gels, where the needle does not fit inside the gel tube, casting using hydrostatic pressure is more appropriate. This method requires a larger excess of reagents and special casting cylinders. Many types of ampholytes are readily available from different suppliers to form the desired pH profiles. As ampholytes may vary significantly in their performance, careful selection of the appropriate ampholytes is usually necessary (see Commentary).
Materials
Chromic acid, in acid-resistant container (use extreme caution; highly corrosive)
Urea (ultrapure)
30% acrylamide/0.8% bisacrylamide (see recipe)
20% (w/v) Triton X-100 (see recipe)
Ampholytes (e.g., pH 3–10; BioRad)
TEMED (N,N,N′,N′-tetramethylethylenediamine)
2.5% (w/v) ammonium persulfate (see recipe; prepare immediately before use)
8 M urea (see recipe; prepare immediately before use)
0.1 M orthophosphoric acid (H3PO4; see recipe)
0.1 M NaOH (APPENDIX 2E; make fresh daily)
Lysis buffer (see recipe)
Protein samples to be analyzed
Equilibration buffer (see recipe)
2-Mercaptoethanol
Isoelectrofocusing apparatus (e.g., Protean II xi 2D from Bio-Rad or equivalent) with glass tubes, casting stand, buffer chambers, rubber grommets, and plugs
37°C water bath
110°C oven
10-ml syringe equipped with filter capsule (0.22 or 0.45 um)
10-ml syringe equipped with blunt needle [e.g., 20-G × 6 in. (15 cm) or 18-G × 6 in. (15 cm)]
Large glass cylinder sealed at bottom with Parafilm (optional, for hydrostatic pressure casting method only)
2000-V power supply
60-ml syringe
Metal or plastic scoop
Dry ice pellets
Wash tubes and prepare the gel mixture
-
Remove the glass tubes from a chromic acid-filled container (use extreme caution, highly corrosive). Extensively wash the tubes with water, using high-purity water for the last wash. Dry the tubes at least 1 hr in an oven at 110°C and store them at room temperature, covered with aluminum foil.
To prevent gels from sticking to the glass tubes, gel tubes have to be very clean. Satisfactory results are obtained by storing the tubes in chromic acid between uses and washing them shortly before use. Because drying the tubes requires at least 1 hr, cleaning steps should be performed the day before gels will be cast.
CAUTION: Chromic acid is highly corrosive; follow supplier’s precautions carefully.
-
Prepare the gel solution by mixing:
16.9 g urea
4.0 ml of 30% acrylamide/0.8% bisacrylamide
3.0 ml of 20% (w/v) Triton X-100
7.5 ml water
3.0 ml ampholytes.
Briefly warm the mixture in a 37°C water bath to solubilize urea if needed.
To minimize decomposition of urea, never warm any solutions containing urea above 37°C, use ultrapure urea, and prepare solutions immediately before use.
Choice of ampholyte composition is one of the key factors determining the quality of isoelectrofocusing separations. Substantial differences in performance, resolution, and shape of the pH gradient formed may be observed with different combinations of ampholytes and with ampholytes from different suppliers.
Although purity of all reagents is important, the purity of urea and choice of ampholytes are among the most critical factors for the quality and performance of isoelectrofocusing. Most commercially available reagents marketed specifically for two-dimensional gel electrophoresis should be suitable, although individual lots of reagents from any supplier may provide variability and/or unacceptable results.
Cast gels by pouring
3a. Wrap one end of each glass tube with Parafilm and mount the tube in a casting stand. Mark all the tubes to indicate the desired gel height.
For reproducible results, all gels should be the same height.
4a. Filter the gel solution using a 10-ml syringe equipped with a syringe-tip filter capsule. Briefly degas the gel solution (~5 min) either by sonication or under vacuum. Then add 42.5 μl TEMED and 187.5 μl of 2.5% (w/v) ammonium persulfate solution to the filtered gel mixture and swirl gently to mix.
5a. Using a 10-ml syringe with a blunt needle, fill each glass tube with gel solution to the desired height. Make sure there are no air bubbles trapped in the gel.
A needle is the best choice for casting gels if tubes of 3-mm inner diameter are used. For narrower tubes, the use of hydrostatic pressure is more appropriate (see steps 3b to 7b, below). For long gels the needle can be extended by inserting a piece of capillary polyethylene tubing over the needle tip. The amount of gel solution described in step 2 is sufficient for sixteen 3-mm tube gels that are 16 cm long.
6a. Immediately overlay each gel with ~50 μl of 8 M urea.
A pipettor with a capillary pipet tip is a convenient tool for overlaying with urea. Avoid mixing the overlay and gel solutions. Polymerization starts to occur ~15 min after the addition of TEMED and ammonium persulfate. It is essential that the gels be poured and overlaid before significant polymerization has occurred.
7a. Let the gels polymerize at least 3 hr prior to use.
Urea decomposes at a substantial rate at room temperature; therefore, the gels should be used the same day they are cast.
Alternative casting of gels using hydrostatic pressure
3b. Place a rubber band around the gel tubes so they form a tight bundle. Place the bundle inside a larger glass cylinder that is sealed at the bottom with several layers of Parafilm. All tubes must be precisely vertical.
The dimensions of the larger cylinder depend on the dimensions and number of gel tubes. Excessive space will require more gel solution to cast the gels.
4b. Filter the gel solution using a 10-ml syringe and filter capsule. Degas the gel solution briefly (~5 min) either with sonication or under vacuum. Add 42.5 μl TEMED and 187.5 μl of 2.5% ammonium persulfate solution and swirl.
5b. Pipet the gel solution into the bottom of the glass cylinder. Gently run water down the outside of the tube bundle using a wash bottle. Keep adding water until the gel mix reaches the desired height.
Hydrostatic pressure will force the gel solution into the tubes. Sufficient gel solution must be used to obtain the desired gel height while avoiding forcing any water into the tubes. The volume of gel solution required can be estimated as follows: number of gels × 3.14 × (tube internal radius in cm)2 × height in cm + ~10 ml to keep a safe level of gel mix at the bottom of the casting cylinder. As water is less dense than the gel solution, the water level will be slightly higher than the level of gel solution inside the tubes.
6b. Overlay the gels with 8 M urea.
Urea decomposes at a substantial rate at room temperature; therefore, the gels should be used the same day they are cast.
7b. Let the gels polymerize at least 3 hr prior to use.
Mount the gels in the electrophoresis unit
8. Prepare the lower electrode solution by degassing the proper amount of 0.1 M H3PO4 under vacuum with stirring for at least 5 min. Fill the bottom electrophoresis chamber.
The amount of phosphoric acid depends on the length of the gel tubes and the type of electrophoresis unit. The solution should cover the entire gel for good heat dissipation. Approximately 3 liters are required for Protean II xi 2D electrophoresis units.
9. Remove the gel tubes from the casting stand, remove the Parafilm from the tube bottoms, and inspect gels for irregularities or trapped air bubbles. Discard imperfect gels. If using gels cast with hydrostatic pressure, remove the bundle of tubes en bloc, cut off excess acrylamide with a razor blade, and then rinse away remaining acrylamide particles from the outside of each tube.
10. Place a rubber grommet on the top of the tube. Approximately 5 mm of the tube should be visible above the upper edge of the grommet.
11. Mount the tube with the grommet in the upper reservoir and plug any unused holes.
After the tube is seated, its lower end must be submerged in the lower electrode solution. Be sure to remove any air bubbles trapped at the bottom of the tube by shaking or tapping the tube gently. Alternatively, with some units bubbles can be dislodged by raising and lowering the tubes or by using a long curved needle and syringe.
Prefocus the gels
12. Prepare the 0.1 M NaOH upper electrode solution by degassing under vacuum with stirring for at least 5 min.
The amount of upper electrode solution necessary depends on the type of electrophoresis chamber. If a Bio-Rad Protean II xi 2D apparatus is used, 1 liter of 0.1 M NaOH is sufficient for both prefocusing and the separation.
13. Remove the 8 M urea overlay from the top of the gels using a Pasteur pipet and place ~50 μl lysis buffer on the top of each gel.
14. Overlay lysis buffer with the degassed 0.1 M NaOH to fill the gel tubes. Avoid mixing of NaOH with the lysis buffer.
15. Pour the degassed 0.1 M NaOH into the upper chamber, making sure that all the gel tubes are covered with the electrode solution. Check carefully for leaks and air bubbles, then place lid on apparatus.
16. Connect the electrodes to a power supply by the red (+) lead to the lower chamber and the black (−) lead to the upper chamber.
The voltages and currents used during electrophoresis are dangerous and potentially lethal. Safety considerations are given in the Electricity and Electrophoresis section of UNIT 10.1.
17. Prefocus for 30 min using 500 V constant voltage.
Load the samples
18. Turn off power supply (refer to Safety Considerations in UNIT 10.1), disconnect leads, and remove lid. Using a 60-ml syringe, remove the electrode solution (0.1 M NaOH) from the upper chamber.
19. Remove the electrode solution and the overlay solution from each tube. Be careful not to damage the gel surface.
20. Place ~50 μl lysis buffer on the top of each gel to rinse the surface. Wait at least 2 min.
21. Remove the lysis buffer from the tubes.
Rinsing the gels with lysis buffer removes any residual NaOH and protects the samples against exposure to high pH.
22. Load protein samples to be analyzed and carefully overlay each sample with ~50 μl lysis buffer diluted with water 8:2 (v/v). Avoid mixing the buffer with the sample.
The overlay solution protects samples from direct contact with the strong base used as an upper electrode solution. Dilution of the lysis buffer with water is necessary to decrease the density so the overlay does not mix with the sample.
A 3-mm-i.d. × 16-cm-long IEF gel has a total protein capacity of ~500 μg for whole-cell extracts and other complex protein mixtures. The maximum capacity for any single protein spot is ~0.5 to 5 μg, depending on its solubility near its isoelectric point and the separation distance from any near neighbors. Preparation of relatively pure protein samples for isoelectrofocusing is generally straightforward. The sample usually may be prepared in one of the following ways: dialyze into any compatible low-ionic-strength buffer, lyophilize in a volatile or compatible low-ionic-strength buffer and dissolve in lysis buffer, or precipitate the protein using trichloroacetic acid (TCA) and redissolve in lysis buffer. For preparing extracts from cultured cells and from tissue samples, see Unit 22.4.
The minimum sample concentration of protein or radioactivity has to be sufficient for the desired detection method. For complex protein mixtures such as tissue or cell extracts, a 500-μg total load is recommended for Coomassie blue staining (UNIT 10.5) or electroblotting (UNIT 10.7) for subsequent structural analysis, a 50-μg total protein load should be sufficient for silver staining (UNIT 10.5) or immunoblotting, and no less than 100,000 counts/gel is recommended for proteins labeled with 3H, 14C, or 35S for autoradiography purposes. Sample volumes should be <150 μl for 3-mm gels and <40 μl/ for 1.5-mm gels. This implies at least a 5 μg/μl protein concentration in the sample for gels to be stained with Coomassie blue.
23. Carefully fill all tubes with 0.1 M NaOH. Avoid mixing the NaOH solution with the overlay solution and the sample.
24. Fill the upper reservoir with 0.1 M NaOH. Be sure that all gel tubes are covered with the solution.
Run the gels
25. Connect the electrodes to a power supply with red (+) to the lower chamber and black (−) to the upper chamber.
26. Focus for a total of 12,000 Vhr.
Unlike other electrophoretic techniques, in IEF the volt-hour is the most common unit describing the “time” of isoelectrofocusing. The initial voltage is usually set according to the desired number of volt-hours in a way that is convenient for the operator (i.e., so that the separation will run overnight), but it should not be <400 V. The upper voltage limit is restricted by heat released in the gels during isoelectrofocusing. At constant voltage the current will be the highest during the first hour of separation. The initial current will be strongly influenced by the ionic strength of the samples loaded onto the gels. An initial voltage of <800 V is recommended for 3-mm gels loaded with samples containing less than 100 mM salts/buffers; the voltage could be increased to 1200 V after ~1 hr, if cooling is used. The current is a derivative of voltage and is never preset for isoelectrofocusing purposes. Some power supplies allow preprogramming the desired number of volt-hours and continuously adjust voltage and current during the isoelectrofocusing procedure (constant power). The total number of volt-hours is the major factor that affects separation in the first dimension. Optimal focusing time will vary for different ampholyte combinations, but 12,000 Vhr is a reasonable value for most systems. To achieve a total of 12,000 Vhr set the power supply to 667 V for 18 hr. These conditions are convenient for an overnight separation and do not require use of a cooling unit. Higher voltages can be used but may cause overheating of gels unless a highly efficient cooling system is employed. The maximum practical voltage decreases with increased gel tube inner diameter. Focusing for too long may cause cathodic drift and result in a shifted pH profile in the gel, whereas focusing for a short time will decrease resolution.
Extrude and store gels
27. Turn off power supply and carefully disconnect leads. Detach the lid and remove the NaOH solution from the upper reservoir of the electrophoresis chamber using a 60-ml disposable plastic syringe.
28. Remove one gel tube at a time from the chamber.
29. Using a 10-ml syringe equipped with a blunt needle, slowly and carefully inject water between the gel and glass tube. Start from the bottom of the tube, then repeat the procedure from the top. The gel should slide out of the glass tube.
It is convenient to let the gel slide from the glass tube onto a metal or plastic scoop, which facilitates transfer of the gel into a storage vial. It is relatively easy to break the gel during extrusion, and practicing on several unused gels is recommended. To extrude smaller-diameter gels, use water pressure generated by a syringe connected to the gel tube with Tygon tubing. If clean, unscratched glass tubes are used, extrusion should be easy.
30. Using the scoop, slide the gel into a 4.5-ml cryovial containing 3 ml equilibration buffer and 50 μl 2-mercaptoethanol. Close the vial, incubate exactly 5 min at room temperature, then freeze by placing the tube horizontally on top of dry ice pellets. Do not move or agitate the tube while the sample is freezing.
The IEF gels may be run on a second-dimension gel immediately (see Basic Protocol 4), or can be stored at −80°C for many weeks. Even when the second-dimension is to be run immediately, extruded gels should be frozen after a carefully controlled incubation time at room temperature, such as the 5 min equilibration cited above for 3-mm-i.d. gels, to minimize diffusion of proteins out of the IEF gel. This short incubation before freezing will allow glycerol to diffuse into the gel. Too short an incubation or agitation during freezing can result in gel breakage. The total incubation time in equilibration buffer (sum of the time prior to freezing and after thawing) is critical and should be carefully controlled. Insufficient incubation time in equilibration buffer will not allow sufficient time for SDS to diffuse into the gel and saturate sites on the proteins. Excessive incubation times can result in appreciable protein losses due to diffusion out of the highly porous IEF gel.
SUPPORT PROTOCOL 2: CONDUCTING pH PROFILE MEASUREMENTS
Standards with well-defined different isoelectric points can help in evaluating the performance of a specific system and determining the effective pH range in the isoelectrofocusing gel. Many pI standards are commercially available from different suppliers. It is most useful to separate a mixture of standard proteins that is prepared from several individual proteins or purchased as a preformulated kit. This mixture should be run in parallel with experimental samples on a separate reference gel. It is generally not recommended to run pI standards together in the same gel with samples because of possible interference with migration and identification of proteins of interest. Instead of analyzing standard proteins, a more precise evaluation of the pH profile for tube gels with soluble ampholytes can be made by directly measuring the pH throughout the gel using either a surface pH electrode or the following procedure.
Prepare and focus one or two gels (see Basic Protocol 3, steps 1 to 26) without any sample in parallel with experimental samples.
-
Prepare 20 to 40 glass test tubes each containing 1 ml high-purity, degassed water for each gel that will be used to measure the pH gradient (measurements on duplicate gels are recommended).
The number of test tubes required per gel equals twice the gel length (in cm).
-
After electrofocusing is completed, extrude the blank gels (see Basic Protocol 3, steps 27 to 29). Briefly rinse the gels with water.
After extrusion, gel surfaces may be contaminated with electrode solutions. Rinsing with water is essential for obtaining reliable pH profiles.
Place the gel on a glass plate with a plastic ruler below the plate. Cut the gel into 0.5-cm pieces using a sharp razor blade.
-
Place each gel piece in a test tube containing 1 ml water.
Be careful to not mix the order of samples because each gel piece represents a single pH profile data point.
Place all test tubes on a shaker and shake gently for 1 hr at room temperature.
Read the pH of each solution and plot the pH profile as a function of the distance from the top of the gel.
ALTERNATE PROTOCOL 1: NONEQUILIBRIUM ISOELECTROFOCUSING OF VERY ACIDIC PROTEINS
Basic Protocol 3 is sufficient for separating proteins with isoelectric points greater than ~3.0 to 3.5. For very acidic proteins, however, a nonequilibrium system is needed. The major features of this method are utilization of a shorter focusing time (without reaching equilibrium), a modified ampholyte mixture, and different electrode solutions.
Additional Materials (also see Basic Protocol 3)
10% (w/v) ammonium persulfate (prepare immediately before use)
Concentrated sulfuric acid (used in lower chamber electrode solution)
Ampholytes, pH 2–11 (used in upper chamber electrode solution)
To analyze very acidic proteins, follow Basic Protocol 3 with these exceptions in the indicated steps:
2. When preparing the gel solution, use the following mixture of ampholytes: 2.4 ml ampholytes pH 2.5–4 and 0.6 ml ampholytes pH 2–11.
4. Following the procedure for casting gels by pouring, add 100 μl of 10% ammonium persulfate solution, swirl, add 42.5 μl TEMED, and swirl again.
Gel mixtures containing entirely or predominantly very acidic or very basic ampholytes are generally difficult to polymerize. Use of an increased ammonium persulfate concentration and adherence to the proper order of adding the reagents should ensure polymerization.
8. Prepare the bottom chamber electrode solution by adding 4.5 ml concentrated sulfuric acid to 3 liters water. Degas at least 5 min.
Omit steps 12 to 19 (do not prefocus the gels).
20. Remove the 8 M urea (polymerization overlay solution) and place ~50 μl lysis buffer on top of each gel. Wait at least 2 min, then remove the lysis buffer.
23. Carefully fill all tubes with the upper chamber electrode (anode) solution prepared by mixing pH 2–11 ampholytes with water in a 1:40 ratio.
24. Fill the upper buffer chamber (anode) with the solution described in step 23.
Iminodiacetic acid (10 mM) may be a more economical alternative anode solution.
26. Focus for a total of 4000 Vhr.
ALTERNATE PROTOCOL 2: NONEQUILIBRIUM ISOELECTROFOCUSING OF BASIC PROTEINS
In general, most equilibrium IEF gel systems using soluble ampholytes produce pH gradients that do not exceed pH 8.0 on the basic end, yet many proteins have higher pI values. For this reason samples containing very basic proteins are usually focused on IPG strips or by using a nonequilibrium system with soluble ampholytes. In an equilibrium system, proteins are loaded on the basic end of the gel and migrate toward the acidic end until they reach a pH equal to their pI. In nonequilibrium systems, the sample is loaded on the acidic end of the gel, and focusing is terminated after a relatively short time (fewer volt-hours).
To run nonequilibrium IEF gels, follow the procedure previously described (see Basic Protocol 3) with these alterations in the indicated steps:
8. Use 0.1 M NaOH as the lower electrode solution.
Electrode solutions and electrodes are reversed in this procedure relative to equilibrium isoelectrofocusing.
Omit steps 12 to 19 (do not prefocus the gels)
20. Remove the 8 M urea (polymerization overlay solution) and place 50 μl lysis buffer on top of each gel. Wait at least 2 min, then remove the lysis buffer.
23. After loading the samples and overlaying with lysis buffer diluted with water 8:2 (v/v) as in Basic Protocol 1, use 0.1 M H3PO4 instead of NaOH to fill all gel tubes.
24. Use 0.1 M H3PO4 as the upper electrode solution.
25. Reverse the connection of electrodes—i.e., connect the red (+) lead to the upper chamber and the black (−) lead to the lower chamber.
26. Focus for a total of 3000 to 5000 Vhr.
The optimal number of volt-hours depends on the nature of the sample and the ampholytes used. The values recommended above may need to be adjusted empirically.
BASIC PROTOCOL 4: SECOND-DIMENSION ELECTROPHORESIS OF IEF TUBE GELS
Second-dimension gels are identical to those described in UNIT 10.1 except for sample loading, which requires a broad, flat well. A broad well can be cast using an appropriate two-dimensional comb if the second-dimension gel thickness is slightly larger than that of the first-dimension gel. Alternatively, when the second-dimension gel is being cast, water can be layered over the entire surface of the gel to produce a flat surface that will accommodate the first-dimension gel.
Narrow analytical isoelectrofocusing gels (≤1.5 mm) that fit between the glass plates of the second-dimension gel do not generally require a stacking gel, although a stacking gel may improve resolution under some circumstances. Stacking gels are essential when first-dimension gels >1.5 mm are loaded on reduced-thickness second-dimension gels, for example, when 3-mm first-dimension gels are loaded on 1.5-mm second-dimension gels. To ensure the best reproducibility, casting multiple second-dimension gels in a multigel casting stand is strongly recommended. This is especially important when gradient gels are used for the second-dimension and/or critical comparisons of multiple samples are planned.
This protocol describes all the specific steps required for successfully casting and running the second-dimension gel. The use of beveled plates and an agarose overlay is especially important when 3-mm IEF gels are loaded onto 1.5-mm second-dimension gels.
Materials
2% (w/v) agarose (see recipe)
Equilibration buffer (see recipe)
Isoelectrofocusing gels containing protein samples to be analyzed (see Basic Protocol 3)
Piece of agarose containing molecular weight standards (see Support Protocol 3)
Beveled glass plates
Boiling water bath
Metal or plastic scoop
Additional reagents and equipment for linear and gradient Laemmli gels (UNIT 10.1)
Cast the second-dimension gels
-
1
Assemble the glass-plate sandwich of an electrophoresis apparatus, using a beveled plate for the shorter side of the gel sandwich.
A beveled plate provides more space for a thicker IEF gel and will accommodate a first-dimension gel that is at least 1 to 2 mm larger than the thickness of the second-dimension gel.
-
2
If the thickness of the first-dimension gel exceeds that of the second-dimension gel, pour a separating gel of the desired acrylamide concentration and immediately overlay with water to produce a smooth surface. The separating gel height should be a minimum of 2 cm below the top of the beveled plate to accommodate the stacking gel.
-
3
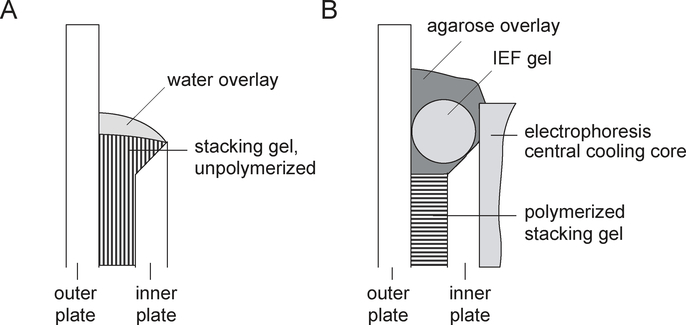
After the separating gel has polymerized (a sharp interface between the polymerized gel and the water overlay will reappear), remove the overlay, rinse the gel surface with water, and pour the stacking gel. The stacking gel solution should reach to the top of the bevel. Immediately overlay the stacking gel solution with a minimum amount of water, which will adhere owing to the surface tension (see Fig. 2A).
A water overlay of the stacking gel provides a smooth surface and better contact between the IEF gel and second-dimension gel. A small volume of water has to be used to avoid lowering the upper edge of the stacking gel below the edge of the beveled plate. The stacking gel height must be between 1.5 and 2 cm. The solution is filled to the top of the bevel so that after the slight shrinkage that occurs during polymerization the top of the polymerized gel will be near the bottom of the bevel (see Fig. 2B).
Figure 2.
Casting the second-dimension gel and loading the IEF gel. (A) The stacking gel solution should reach to the upper edge of the beveled plate, and then the gel solution has to be overlaid with a minimum volume of water. The water will stay on the surface because of surface tension. (B) After polymerization, the gel is mounted on the central cooling core of the electrophoresis unit, and the equilibrated IEF gel is placed on top of the polymerized stacking gel. Excess buffer is removed, and the IEF gel is overlaid with hot agarose/equilibration buffer mixture. After the agarose solidifies, the upper electrophoresis chamber is filled with buffer.
Load the isoelectrofocusing gels onto the second-dimension gels
-
4
Assemble second-dimension gels in an electrophoresis chamber. Do not pour electrophoresis buffer into the upper chamber.
-
5
Melt 2% (w/v) agarose in a boiling water bath and add an equal volume of equilibration buffer for use in step 11. Keep agarose/equilibration buffer in the boiling water bath until step 11 is completed.
-
6
Retrieve isoelectrofocusing gels containing protein samples to be analyzed from storage. Incubate cryotubes containing frozen IEF gels in a 37°C water bath for 15 min for a 3-mm tube gel. A 5- to 7-min incubation is sufficient for 1.5-mm or thinner IEF gels. Do not agitate during thawing, as vigorous agitation of a partially thawed gel can break the gel.
During this thawing/equilibration step, SDS in the equilibration solution in which the gels were frozen diffuses into the gel matrix and binds to proteins in the IEF gel. The length of incubation in the equilibration buffer is critical because insufficient saturation of proteins with SDS will contribute to vertical streaks after staining the gel. On the other hand, extended incubation in equilibration buffer will result in excessive loss of proteins due to excessive diffusion of protein out of the gel, which is especially critical for thin IEF gels. For this reason, it is recommended that after extrusion from the IEF tube IEF gels be initially incubated for 5 min to allow adequate diffusion of glycerol into the gel to minimize gel breakage, followed by freezing on dry ice (see Basic Protocol 3, step 30). This is desirable even if the second-dimension gel will be run directly after isoelectrofocusing, as it is the most feasible way of precisely controlling the equilibration time while the remaining gels in the IEF run are extruded.
-
7
Pour the gel and equilibration solution out of the cryovial onto a metal or plastic scoop. Carefully remove excess equilibration buffer with a pipet.
-
8
Place a few milliliters of electrophoresis buffer on the top of the second-dimension gel.
-
9
Slowly slide the IEF gel off the scoop and onto the top of the second-dimension gel. Remove all air bubbles trapped between the gels. Remove excess electrophoresis buffer from the top of the second-dimension gel.
The basic end of the gel may be placed on either the left or right side of the second-dimension gel depending upon operator preference. However, once a convention is established, all gels should be oriented the same way. The acidic end of the IEF gel can be recognized in two ways: the bromphenol blue will usually be yellow, and a bulge (increased gel diameter) will be present.
-
10
Place a piece of agarose containing molecular weight standards (see Support Protocol 6) beside, but not contacting the basic side of the IEF gel (optional).
Note that when molecular weight standards are used, the isoelectrofocusing gel has to be shorter than the width of the second-dimension gel.
-
11
Carefully overlay the IEF gel (and the optional gel piece with standard proteins) with the hot agarose/equilibration buffer mixture (~2 ml/gel) prepared in step 5. Let the agarose solidify.
The agarose prevents the IEF gel from shifting position and ensures good contact between the IEF and second-dimension gels.
-
12
Carefully pour electrophoresis buffer into the upper reservoir, taking care to avoid disturbing the agarose-covered IEF gel.
-
13
Connect electrodes and run the gels.
See UNIT 10.1 for electrophoresis conditions.
SUPPORT PROTOCOL 3: PREPARING MOLECULAR WEIGHT STANDARDS FOR TWO-DIMENSIONAL GELS
Molecular weight markers are usually necessary for the identification of proteins or as references to describe experimental proteins on 2D gels. In many cases, molecular weight markers are required only at the beginning of a project. Once the system is established, commonly observed major proteins in the sample (e.g., actin, tubulin, etc.) provide sufficient references for molecular weight identification on subsequent gels. To minimize any differences in migration of the molecular weight standards and isoelectrofocused proteins, the standard proteins should be loaded on the second-dimension gel in the same manner as the IEF gel. This protocol describes the preparation of standards in solidified agarose. The agarose pieces may be stored at −80°C for at least a year and provide a convenient source of standards for the second-dimension gel. The procedure described is recommended for 3-mm IEF gels. Narrow standards in solidified agarose (made in tubes ≤1.5 mm in diameter) can be prepared by the same method, but extrusion of the thinner agarose gel without breaking is more difficult. The protocol supplies molecular weight markers containing ~2.5 μg of each standard suitable for Coomassie blue staining or 0.25 μg of each standard for silver staining.
Materials
Molecular weight standards (Table 2)
1× SDS sample buffer (UNIT 10.1)
2% (w/v) agarose (see recipe)
Boiling water bath
Glass tubes (3-mm inner diameter)
Plastic or metal tray
-
Prepare 3 ml molecular weight standards in 1× SDS sample buffer using 250 μg of each standard.
The stated amount is appropriate for Coomassie blue staining of gels. If silver staining is planned, use 25 μg of each standard.
Mix the standards with 2 ml of 2% (w/v) agarose melted in a boiling water bath.
Prepare clean glass tubes by wrapping one end with Parafilm. Pour the hot mixture into the tubes and let the agarose solidify.
Carefully extrude the agarose from the tubes.
Cut agarose rods into 5-mm pieces using a razor blade.
Freeze all pieces separately on a plastic or metal tray using dry ice.
Collect frozen pieces in a plastic bottle and store at −80°C. The standards may be stored ≥1 year.
ALTERNATE PROTOCOL 3: DIAGONAL GEL ELECTROPHORESIS (NONREDUCING/ REDUCING GELS)
The composition of protein complexes where subunit are linked by disulfide bond or protein complexes crosslinked by reagents with a disulfide in the linker region can be analyzed by two-dimensional gel electrophoresis using separation under nonreducing conditions in the first dimension followed by reduction of disulfide bonds and separation under reducing conditions in the second dimension. Most proteins will migrate equal distances in both dimensions, forming a diagonal pattern. Proteins containing interchain disulfide bonds will be dissociated into individual subunits and will be resolved in the second-dimension gel.
The approach is similar to that described for two-dimensional (IEF/SDS) gel electrophoresis (see Basic Protocols 1–4) except, in this protocol, the first-dimension gels are nonreducing (i.e., 2-mercaptoethanol or dithiothreitol is omitted from sample buffer) SDS-denaturing gels instead of isoelectrofocusing gels. Use of 1.0 – 1.2-mm tube gels for the first-dimension separation and 1.5-mm slab gels for the second-dimension run is recommended.
Additional Materials (also see Basic Protocol 4)
Separating and stacking gel solutions (see Table 1)
1× SDS sample buffer without reducing agents (UNIT 10.1)
Reducing buffer (see recipe)
1.5% (w/v) agarose in reducing buffer (see recipe; optional, for securing first-dimension gel on second-dimension gel)
Two-dimensional comb (optional)
Additional reagents and equipment for casting tube gels (see Basic Protocol 3), SDS-PAGE (UNIT 10.1), and protein staining (UNIT 10.5)
Pour and run the first-dimension gel
-
1
Clean and dry 1.2-mm glass gel tubes for the first-dimension gel (see Basic Protocol 3, step 1).
-
2
Prepare a separating gel solution with the desired percentage acrylamide (Table 1); omit the stacking gel for the first dimension.
Stacking gels can usually be avoided in the first dimension by keeping sample volumes small (i.e., ≤10 μl).
Less than 200 μl of gel solution is required to cast a single 1.2-mm tube gel 12 cm in length. Adjust the amounts from Table 1 accordingly.
-
3
Cast the first-dimension polyacrylamide gels in 1.2-mm tubes using a syringe with a long needle (see Basic Protocol 3, step 5a). Overlay with water and allow the gels to polymerize.
-
4
Prepare samples in 1× SDS sample buffer without any reducing reagents (i.e., no 2-mercaptoethanol or DTT). Load the samples and electrophorese until the tracking dye is ~1 cm from the bottom of the tube.
Reduce sample and run the second-dimension gel
-
5
Extrude the gel from the tube (see Basic Protocol 3, steps 27 to 29).
-
6
Place the extruded gel in a test tube containing 5 ml reducing buffer. Equilibrate 15 min at 37°C with gentle agitation.
-
7
Cast the second-dimension separating and stacking gels (see Basic Protocol 4, steps 1 to 3), making sure that the top of the stacking gel is at least 5 mm below the top of the short glass plate. Layer water across the entire stacking gel or use a two-dimensional comb.
Most two-dimensional gel combs have a separate small well for a standard or reference sample.
The use of beveled plates (see Basic Protocol 4, steps 1 to 3) is not essential but is still preferred because it will facilitate loading of the first-dimension gel. In this procedure, the first-dimension gel will fit between the glass plates if 1.2-mm tubes are used for the first dimension and 1.5-mm gels are used for the second dimension.
-
8
Load the first-dimension gel onto the second-dimension gel. Remove any air bubbles trapped between the gels.
If the first-dimension gel does not remain securely in place, it can be embedded using 1.5% (w/v) agarose in reducing buffer.
-
9
Carefully pour electrophoresis buffer into the upper electrophoresis chamber and electrophorese using voltages and times appropriate for the gel type selected.
Parameters for electrophoresis are given in UNIT 10.1.
SUPPORT PROTOCOL 4: USING TWO-DIMENSIONAL PROTEIN DATABASES
Computerized image acquisition and manipulation constitute the only practical method for systematic qualitative and quantitative evaluation of complex protein patterns from different samples that are to be compared by high-resolution two-dimensional gel analysis. Examples of experimental applications include comparisons of tumor cells or tissues with appropriate normal controls and comparisons of a single cell line under different experimental conditions.
There are currently a number of commercially available image acquisition/computer systems specifically designed for comparing two-dimensional gels and storing associated information in a database (UNIT 10.12). The systems include both hardware and the necessary software for comparing different gels and producing databases containing the two-dimensional protein patterns, with options for annotating specific spots and producing quantitative comparisons among large numbers of different samples. With most systems, images can be acquired from either stained gels or autoradiographs. The equipment used to obtain two-dimensional gel images includes laser scanners, video cameras, and phosphoimagers. After image acquisition, software running on a microcomputer or workstation is used to refine the image, detect spots, and match spots between different gels.
It is essential that very high-quality, reproducible gels be used for computerized comparisons. The greatest dynamic range in protein abundance for a single two-dimensional gel can be obtained using autoradiography (UNIT 10.12) or phosphoimaging. With these methods, up to several thousand spots can be compared and tracked. A representative reference gel or a composite image can be stored and used as a reference for future experiments.
Information related to each spot on the two-dimensional pattern, including the quantity of protein in the indicated spot on different gels used in the comparison, can be archived and updated. Other known information related to a specific spot can also be added to the investigator-built database, including the pI, molecular weight, amino acid composition, sequence, and/or identity of the protein and any other important attributes correlated with the indicated spot. A number of research groups, including those of Garrels and Celis (Garrels, 1989; Garrels and Franza, 1989; Celis et al., 1991), have extensively characterized hundreds of spots from specific cell lines and have used multiple methods to characterize proteins of interest. An annotated 2D gel electrophoresis database can be found at http://world-2dpage.expasy.org/swiss-2dpage/. The most definitive methods for establishing the identities for proteins of interest detected by computer-assisted comparisons are excision of the spot, trypsin digestion, and LC-MS/MS analysis followed by a database search containing the protein sequences translated from the complete genome of the species being analyzed. If an organism is being analyzed where the genome has not been completely sequenced, identifications can often still be made by searching the sequence database from an evolutionarily closely related species but in this case, only peptides with exact matches will be identified as most amino acid substitutions will not be isobaric. An alternative approach for species where good representation of protein sequences are not available is to use N-terminal protein sequence analysis.
REAGENTS AND SOLUTIONS
Note: Use Milli-Q-purified water or equivalent for the preparation of all buffers. For common stock solutions, see APPENDIX 2E; for suppliers, see SUPPLIERS APPENDIX.
30% acrylamide/0.8% bisacrylamide
30 g acrylamide
0.8 g bisacrylamide
H2O to 100 ml
Filter solution through 0.2- to 0.45-μm filter (e.g., Thermo Fisher Scientific, cellulose nitrate, 0.2 μm). Store at 4°C (stable at least 3 months).
Acrylamide is a neurotoxin. Wear gloves and a dust mask when handling solid acrylamide. Wear gloves when working with acrylamide solution. Never pipet acrylamide solutions (or any reagent) by mouth.
Agarose in reducing buffer, 1.5% (w/v)
Mix 0.15 g agarose and 10 ml reducing buffer (see recipe). Heat in boiling water bath until dissolved. Prepare immediately before use.
Agarose, 2% (w/v)
Mix 2 g agarose and 100 ml water. Stir on a hot plate until dissolved. Keeping the solution near 100°C, divide by placing 5-ml aliquots in 25-ml glass screw-cap tubes. Let the aliquots solidify. Store at 4°C (stable at least 3 months).
Ammonium persulfate, 2.5% (w/v)
0.25 g ammonium persulfate
H2O to 10 ml
Prepare immediately before use
IPG Strip equilibration solutions
20 ml 1 M Tris·Cl, pH 6.8 (APPENDIX 2E)
72 g ultrapure urea
60 ml glycerol
2 g sodium dodecyl sulfate (SDS)
67 ml Milli-Q water
For solution 1: Add 50 mg DTT per 10 ml of equilibration buffer
For solution 2: Add 0.45 g iodoacetamide and a few grains bromphenol blue per 10 ml of equilibration buffer
Make fresh immediately before use
Final concentrations are 50 mM Tris·Cl, pH 6.8; 6 M urea; 30% glycerol; and 1% SDS, in a final volume of 200 ml.
Pre-mixed solutions are also available from commercial suppliers (e.g. Bio-Rad)
EDTA, 2% (w/v)
2 g Na2EDTA
H2O to 100 ml
Adjust to pH 7.0 with NaOH
Store at room temperature (stable several months)
Titrate while dissolving. EDTA is difficult to dissolve without addition of NaOH even when the disodium salt is used.
Equilibration buffer
3 g SDS
7.4 ml 2% (w/v) EDTA, pH 7.0 (see recipe)
10 ml glycerol
2 ml 1.0 M Tris·Cl, pH 8.65 (APPENDIX 2E)
0.3 ml bromphenol blue (saturated solution in H2O)
H2O to 100 ml
Store at room temperature (stable for several weeks)
Final concentrations are 3% (w/v) SDS, 0.4 mM EDTA, 10% (v/v) glycerol, and 20 mM Tris·Cl, pH 8.65.
Leupeptin, 2 mg/ml
20 mg leupeptin
10 ml water
Divide into convenient volumes
Store at −20°C (stable at least 1 year)
Lysis buffer
2.59 g urea (ultrapure)
1.6 ml H2O
0.25 ml 2-mercaptoethanol
0.3 ml ampholytes
1.0 ml 20% (w/v) Triton X-100 solution (see recipe)
Prepare immediately before use
Use same ampholytes as for the IEF gel formulation. To dissolve urea, warm the mixture in a 30°C water bath if necessary.
Orthophosphoric acid (H3PO4), 0.1 M
13.7 ml 85% phosphoric acid
Water to 2 liters
Make fresh daily
Must be degassed prior to use.
Reducing buffer
0.5 g dithiothreitol (DTT)
0.1 g SDS
1.51 g Tris base
Adjust to pH 6.8 with HCl
Add H2O to 100 ml
Prepare fresh every time
Final concentrations are 0.5% (w/v) DTT, 0.1% (w/v) SDS, and 125 mM Tris·Cl, pH 6.8.
Triton X-100 solution, 20% (w/v)
3 g Triton X-100
12 ml H2O
Warm in 37°C water bath to dissolve Triton X-100
Store at 4°C (stable ~2 weeks)
Urea, 8 M
0.75 g ultrapure urea
1.0 ml H2O
Prepare immediately before use
Avoid heating above room temperature.
COMMENTARY
Background Information
The use of 2D PAGE remains a powerful analytical tool despite the fact that continuing advances over the past two decades in bottom up proteomics using LC-MS/MS instruments have surpassed 2D PAGE for many proteomics studies (Rogowska-Wrzesinka, A.et al., 2013; Meleady 2018). From a separations perspective, 2D PAGE, using denaturing IEF followed by SDS-PAGE, is the single most powerful analytical separations method currently available for separating complex sets of intact proteins. It can also be used to distinguish small differences and biologically important shifts in charge of a single protein of interest, such as multiple phosphorylation sites on a single protein as well as resolve some protein isoforms such as those produced by alternative splicing. In addition to phosphorylation changes, other changes that can usually be effectively tracked using 2D gels include: glycosylation, acetylation, deamidation, and chemical modifications that result in charge shifts. Also, as noted above, this approach can be useful when studying proteomes of organisms with incomplete genome sequences. This is because standard LC-MS/MS based methods identify peptides and proteins by matching MS data to known protein sequences in databases translated from the gene sequences. An incomplete protein sequence database can substantially compromise reliability of LC-MS/MS based identifications. In such applications, an alternative approach is to identify biologically interesting protein changes using 2D gels followed by MS identification of pertinent spots using de novo sequencing. Several recent reviews further highlight the importance of 2D gels (Rabilloud et al., 2010; Rogowska-Wrzesinka, A.et al., 2013; Strohkamp, 2016; Meleady 2018).
Despite the exceptionally high theoretical resolving power of 2D PAGE, the total number of protein spots that can be resolved in a single 16–18 cm × 16–18 cm 2D gel is typically only ~1,000 to 2,000. For more complex organisms where extensive and variable PTMs occur, 2000 spots will typically represent less than 500 unique proteins due to the detection of different isoforms of the most abundant proteins. Unless an enrichment strategy is used, the proteins detected with general protein stains such as Colloidal Coomassie or silver stains will be primarily limited to high abundance structural and housekeeping proteins, whereas a single human cell type typically expresses 10,000 or more genes. Therefore, the 2D gel is providing information about only 5% of expressed genes for human cells or other mammals unless a more focused sample preparation or detection method is used. In this regard, because it is easy to transfer 2D gels to PVDF membranes, one can focus on either protein classes such as glycosylation or isoforms of specific proteins by probing the membranes with appropriate lectins or antibodies, respectively.
In addition to physiologically meaningful charge changes, 2D gels can detect artifactual modifications to charged amino acid side chains that may occur during sample preparation or separation. For this reason, care must be exercised when preparing and using reagents containing urea. While urea is a highly effective neutral chaotropic reagent that is essential for maintaining solubility of proteins near their isoelectric point, it decomposes to form cyanate, which readily reacts with amino groups, especially above pH 7. This reactivity increases 10-fold for each pH unit between 7 and 10, which represents a particular challenge when attempting to focus very basic proteins.
Commercially available equipment for running 2D gels can be roughly divided into four groups based on size: microgels (Thermo Fisher Scientific Phast system), minigels (e.g., Bio-Rad or Thermo Fisher Scientific), standard or full-sized gels (e.g., Bio-Rad or Thermo Fisher Scientific), and large or “giant” gels (Thermo Fisher Scientific Ettan-Dalt gel system and custom systems). In general, the larger the gel, the better the final resolution, but as gel size increases so do costs, difficulty of gel handling, and time requirements.
Standard-size gels (∼16–18 cm × ∼16–18 cm) provide adequate resolution for most applications and are relatively easy to handle, thereby representing the most commonly used compromise between speed/cost and resolution. A 0.5 mm × 3 mm × 18-cm IPG strip or a 1.5-mm soluble ampholyte tube IEF gel has a total protein capacity of ~500 μg for complex protein mixtures such as whole-cell extracts. The maximum capacity for any single protein spot is ~0.5 to 5 μg, depending on the solubility of the protein near its isoelectric point and the separation distance from any near neighbors. A variety of alternative gel sizes, their limits, and their advantages are summarized in Table 3. The lower protein limit for any of the systems is determined strictly by the available detection methods (UNIT 10.5).
Proteins can be detected in 2D gels by the same wide range of techniques used for one-dimensional gels. Autoradiography (UNIT 10.11), silver staining, Sypro Ruby, or other fluorescence tags or stains (UNIT 10.5), and electroblotting to PVDF membranes (UNIT 10.7) followed by colloidal gold or colloidal silver staining (UNIT 10.8) or immunodetection (UNIT 10.10) are among the most sensitive techniques available. If a larger amount of protein is available, Coomassie blue staining of the gel (UNIT 10.5) or amido black staining of a PVDF membrane after electrotransfer (UNIT 10.8) are typically the detection methods of choice. Advantages of using sensitive fluorescent stains such as Sypro Ruby include a wide linear dynamic range for detection (up to 4 orders of magnitude) as well as reduced problems with horizontal streaking caused by reduced solubility near the pI of most proteins because lower total protein loads can be used.
A major technical limitation when comparing multiple samples using 2D PAGE is gel-to-gel variation. The use of commercial precast IPG strip gels has not only greatly increased the reproducibility of 2D gels both within a laboratory and between laboratories, but it has also greatly eased the workload and reduced the hands-on time required to perform experiments. When using gels for the first or second dimension that are cast in the laboratory, maximum reproducibility will be obtained when multiple gels are cast and run simultaneously (Anderson and Anderson, 1978a,b). Even when extreme care is exercised to produce highly reproducible first- and second-dimension gels, some gel-related variability among gels cast at the same time is likely to persist. Another source of variability includes differences in extraction or recovery of proteins during sample solubilization and handling. Maximizing resolution and reproducibility is especially important if computerized comparisons of two-dimensional gels of complex protein mixtures such as cell or tissue extracts are being attempted.
Immobilized pH gradient gels
In IPG gels (Basic Protocol 1), the pH gradient is an integral part of the polyacrylamide matrix (Strahler and Hanash, 1991). Because the pH gradient is covalently associated with the polyacrylamide gel matrix, precise, reproducible, and very high-resolution separations can be achieved. A variety of precast gels and all the necessary equipment are commercially available from Bio-Rad and Thermo Fisher Scientific. Isoelectrofocusing is performed in a horizontal electrophoresis unit in which multiple gel strips may be run simultaneously. As noted above, typically the reproducibility of 2D gels will be highest when the first and second dimensions are each run in single batches.
In addition to ease of use and reproducibility, precast IPG gels are also available in both broad and narrow pH formats and the precise narrow-range gradients can be particularly useful for resolving small charge differences. Because the pH gradient is covalently coupled to the polyacrylamide gel matrix, the pH gradient remains stable and linear during prolonged electrophoresis, thus ensuring reproducibility. This contrasts with conventional soluble ampholyte IEF gels, where gradient drift occurs during prolonged electrophoresis. Furthermore, precast IPG gels are shipped dry, have a long shelf life, and can be rehydrated using a range of solutions tailored to specific applications with additives such as urea, CHAPS or Triton X-100, carrier ampholytes, glycerol, and reducing agents, which may help to increase protein solubility. Additionally, inclusion of the sample during active rehydration may facilitate entry of larger protein loads, use of large sample volumes and better uptake of high molecular weight proteins into the IPG strip, while eliminating some of the sample handling steps.
Precast gel strips are available with pH ranges such as pH 3.0 to 10.0 or pH 4.0 to 7.0 as well as narrower ranges. Table 2 lists types and sizes of commercially available gels. With the Immobiline system, the apparent pI of a given protein of interest may be slightly different from that determined by other methods. Therefore, it is recommended that a broad-range gradient be tried initially, followed by a narrower-range gradient, if needed.
Isoelectrofocusing using soluble ampholytes
Soluble ampholytes are mixtures of low-molecular-weight organic compounds with differing side-chain pKa values that provide buffering capacity. In an IEF gel, the ampholytes migrate to their isoelectric point, where they provide buffering capacity and hence produce stable pH gradients. In theory, any desired pH gradient could be produced by blending ampholytes with appropriate pKa values. In practice, it is relatively easy to produce pH gradients from ~pH 3.0 or 4.0 to pH 8.0, but stable soluble gradients outside this range are usually not technically feasible. Within these pH limits, some manipulation of the gradient shape and pH range is possible by blending different amounts of specific pH range ampholytes. For example, 0.50 ml of pH 5–7 ampholytes plus 0.25 ml of pH 4–8 ampholytes can be used instead of 0.75 ml of pH 4–8 ampholytes alone to increase the separation distance of proteins in the pH 5.0 to 7.0 range.
Basic Protocol 3 is based on use of 3-mm first-dimension isoelectrofocusing gels and 1.5-mm second-dimension gels using the Bio-Rad 2D gel apparatus (Protean II xi 2D). The method can be easily adapted to equipment from other suppliers or to different-sized gels by adjusting the quantities of reagents used. The protocol uses 8 M urea and Triton X-100 as solubilizing agents. Solubilization of the protein sample applied to the gel as well as maintenance of solubility during electrofocusing are the most critical factors influencing the quality of separation in the first dimension. The most common modification to Triton X-100-based procedures is addition of 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) to the gel and solubilizing buffer mixtures. Addition of SDS to complex samples such as tissue or cell extracts can also enhance reproducible solubilization of the largest possible subset of proteins. Although SDS is charged, in the presence of Triton X-100 it is separated from proteins during focusing and migrates to the acidic end of the gel. Regardless of the method used to maintain solubility, some proteins (especially those >100kDa) tend to precipitate at pH values approaching their isoelectric point and thus produce horizontal smears on the final second-dimension gel. Most studies do not attempt to analyze proteins above about 150 kDa due to this poor and variable separation of large proteins;
When using soluble ampholytes several modifications to Basic Protocol 3 are required for successful separation of very acidic (Alternate Protocol 1) or very basic (Alternate Protocol 2) proteins. In both cases, prefocusing of the gels must be avoided, and the isoelectrofocusing time has to be reduced, which for basic separations has the added advantage of reducing modification of lysines by cyanate produced by urea decomposition. A short separation time does not allow the system to reach equilibrium and is used to establish the desired pH gradient. Separation of very acidic proteins requires modifications of gel and electrode solutions. Separation of very basic proteins has to be performed with the positions of electrodes and electrode solutions reversed in the electrophoresis chamber.
The total protein load per gel depends on the complexity of the sample, the solubility of proteins in the sample, and the diameter of the first-dimension gel. Approximately 4 times as much total sample can be applied to a 3-mm gel compared with a 1.5-mm gel. Another advantage of a larger-diameter isoelectrofocusing gel is that extrusion and gel handling are easier owing to improved strength of the gel. If care is exercised in loading the first-dimension gel onto the 1.5-mm second-dimension gel, the final resolution will be similar to that obtained using smaller-diameter IEF gels. The separating or resolving power of a system is dependent on the quality of ampholytes used, the slope of the pH gradient, and the lengths of both first- and second-dimension gels.
Diagonal gel electrophoresis
Diagonal gel electrophoresis is a form of 2D analysis useful for investigating the subunit composition of multisubunit proteins containing interchain disulfide bonds (Goverman and Lewis, 1991). Proteins are electrophoresed in the first-dimension in a tube gel (or a slab gel) under nonreducing conditions. The proteins are then reduced in situ, and the first gel (or a strip from a slab gel) is layered onto a second gel and electrophoresed. In the second gel, the proteins migrate at right angles to the original, first-dimension migration. Most cellular proteins are not disulfide-linked and will fall on the “diagonal” in this system; that is, they migrate approximately equal distances in both directions during electrophoresis and lie approximately on the diagonal line connecting opposite corners of the gel. On reduction, component subunits of proteins connected by disulfide bonds will resolve below the diagonal because the individual subunits migrate faster than the disulfide-linked complex during the second electrophoresis. Some proteins with internal disulfide bonds, but no interchain disulfides, may migrate slightly above the diagonal because internal disulfides can produce a more compact molecular shape (causing faster migration in the first dimension).
Critical Parameters and Troubleshooting
Although no individual steps 2D PAGE are exceptionally difficult, the large number of steps involved increase the likelihood and possible severity of errors or problems. Several steps are especially critical and may require optimization. The first is sample preparation. Proteins applied to IEF gels have to be completely solubilized. Residual precipitate or even soluble aggregates are likely to cause artifacts on the end of the tube gel or position on the IPG strip where the sample is loaded. Precipitated or aggregated proteins may also interact with soluble proteins, causing components with normally good solubility to coprecipitate or migrate anomalously.
Two general rules of thumb apply to sample solubility: (1) the more complex the sample or the more crude the extract, the more likely problems will be encountered with sample solubility, and (2) the higher the protein load applied to the gel, the more likely solubility problems will arise. Care must also be taken to avoid proteolysis during sample preparation, especially when complex samples with high levels of proteases such as cell or tissue extracts are used. If samples are frozen either before or after solubilization, they should be stored at −80°C and should not be subjected to repeated freeze-thawing. Addition of moderate levels of SDS to the solubilization solution can increase solubilization of some proteins; however, SDS should only be used when the IEF gels contain urea and Triton X-100, as these solubilizing agents are required for effective dissociation of the strongly anionic SDS molecules from the proteins during isoelectrofocusing. Heating of samples in urea-containing solutions must be avoided because urea readily decomposes to cyanate, which reacts with amino groups and causes charge heterogeneity. High concentrations of salts and buffers in the sample should be avoided. Ionic compounds increase the conductivity of the sample and can result in localized overheating, especially for IPG gels.
Samples analyzed on IPG gels should not contain precipitates. The concentration of salts and buffer ions in the sample should be kept to a minimum (<50 mM) to avoid local overheating of the gel during electrophoresis. Sample loading during active rehydration is recommended to prevent the formation of aggregates or precipitates at the point of application.
Early decisions that must be made include the size of the gels required and whether an an IPG or a soluble ampholyte system will be used. The major consideration affecting appropriate gel size is the degree of resolution needed. In general, the smallest gel format should be selected that will provide the needed degree of resolution, because smaller gels are easier, less expensive, and faster to run. Therefore, quick screening of samples or analysis of relatively simple samples can easily be accomplished with microgels or minigels. In contrast, if detailed qualitative or quantitative comparisons of cell or tissue extracts are planned, standard-size or large gels are indicated. Similarly, 3-mm or larger first-dimension tube gels followed by 1.5-mm second-dimension gels in the standard or large format should be considered if the 2D gels will be used for preparative isolation of a protein for applications such as raising antibodies. As noted above, IPG gels are usually preferred over soluble ampholyte gels for most applications due to their commercial availability, ease of use, and stable, reproducible pH profiles. The IPG gels are particularly appropriate when a narrow pH range is required.
IPG gels are typically run at 2500 to 3500 V and require a focusing time of 16 to 18 hr. Optimal focusing conditions may be experimentally determined by applying the sample to different positions on the gel and estimating the time for the migration patterns to coincide. Some proteins may require longer run times to reach their pI. Because there is no gradient drift, the potential problems with longer run times are limited to sample denaturation or drying out of the gel. These problems usually can be minimized by including a reducing agent in the rehydration solution and/or coating the top surface of the gel with paraffin oil.
The quality of the first-dimensional separation is strongly dependent on the purity of the reagents used, especially the urea and ampholytes. One fairly commonly encountered frustration, when soluble ampholyte gels are used, is that different batches of ampholytes from the same supplier will sometimes produce markedly different 2D gel patterns. Therefore, it is advisable to purchase an adequate supply of a single lot of ampholytes to meet anticipated needs for an entire study where such an approach is feasible. However, whereas ampholytes usually have a reasonably long shelf life at 4°C (usually up to a year), shelf life as well as total ampholyte requirements often cannot be predicted with much certainty.
When any doubt arises about the purity or quality of ampholytes or any other reagent, the reagent should be replaced immediately. Constant monitoring of the system performance, especially when changing lots of ampholytes, urea, acrylamide, or IPG strips can help minimize potential reagent-associated problems. In most cases, the best standard for a given 2D gel system is an experimental sample or control that is available in sufficient quantity so that many replicate aliquots can be frozen and stored for an extended time at −80°C. Such an experimental standard or reference is more likely to detect subtle, but experimentally important, changes in the 2D gel system than commercially available IEF or SDS gel standard mixes.
Another critical factor is equilibration of the first-dimension gel in the second-dimension equilibration buffer. During this step, urea diffuses out of the IEF gel or IPG strip while SDS and reducing reagent diffuse into the gel. If the gel is inadequately saturated with SDS, vertical streaking will result. However, if the IEF gel or IPG strip is incubated in the equilibration buffer for an extended time, a substantial amount of the protein can rapidly diffuse out of the large-pore IEF gel. Losses arising from diffusion can be critical for any experiment because different proteins will diffuse at different rates, but rigorous control of the incubation step is especially important if quantitative comparisons across multiple gels are planned. The simplest method of controlling the incubation time is to freeze the extruded IEF gel or IPG strip after a carefully controlled 5-min incubation in the equilibration buffer; any additional equilibration incubation time required can then be incorporated and carefully controlled when the sample is thawed for loading onto the second-dimension gel.
Another crucial step is loading of the equilibrated IPG strip or IEF gel onto the top of the second-dimension gel. Any irregularity or obstruction between the two gels, including particles of dirt or air bubbles, will affect the flow of current and disrupt the resolution of proteins in the final gel. Similarly, poor contact or any movement of the IEF gel or IPG strip during electrophoresis of the proteins out of the IEF gel or IPG strip into the second-dimension gel will lead to artifacts. Therefore, it is advisable to embed the IEF gel or IPG strip in a buffered agarose matrix to ensure good electrical contact between the gels and to prevent gel movement after electrophoresis is initiated.
Finally, the choice of second-dimension gel composition and separation conditions can influence the quality of results. A proper percentage of acrylamide should be selected to optimize resolution within the desired molecular mass range. If gradient gels are needed, use of a multiple gel casting stand is the best way to ensure reproducibility among samples within a single experiment. Further details on optimization of 2D gel systems are presented by Hochstrasser et al. (1988).
When no technical, sample-related, or reagent-related problems are encountered, the final stained 2D gels should contain numerous rounded or slightly elliptical spots. Typically, more than 1000 spots can be detected on a standard 16 × 14-cm gel when using a sensitive staining protocol such as Sypro Ruby, silver staining or autoradiography and a whole-cell extract as a sample. Some horizontal streaks for most high-molecular-mass proteins (proteins exceeding ~100– 120 kDa) are common owing to the decreased solubility of larger proteins near their isoelectric points, even in the presence of urea and nonionic detergent. However, excessive horizontal smearing of proteins smaller than 100 kDa indicates poor isoelectrofocusing, which could be related to one or more of the following factors: sample improperly solubilized or contaminated with interfering substances such as large nucleic acid molecules, poor purity of reagents (check the urea first), poor-quality ampholytes, or insufficient isoelectrofocusing (total volt-hours too low). It is important to note that in general the solubility of any protein is the lowest near its isoelectric point, but there are vast differences among proteins in terms of both the minimum concentration where precipitation becomes a problem and the degree to which precipitation can be prevented by adding different solubilization agents. The best conditions for maintaining solubility during isoelectrofocusing for a given sample type must be determined empirically, although the most universal conditions are those described in the protocols herein. In contrast, IEF systems that do not use any detergents or denaturants are limited to that fairly small percentage of proteins which maintain good solubility near their isoelectric point.
If high-molecular-weight proteins are expected but are not present in the final 2D gel, check the sample preparation protocol as well as the sample storage conditions. The most likely problem is proteolysis during sample preparation. Multiple freeze-thawing cycles could contribute to this problem. Vertical smears on the 2D gel suggest (1) insufficient equilibration of the IEF gel (not enough SDS bound to the proteins), (2) poor contact between the IEF and second-dimension gels, or (3) problems related to the stacking gel (too short or wrong buffer). Use of a stacking gel is especially important when large-diameter IEF gels are loaded onto smaller second-dimension gels (Basic Protocol 4).
Omission of Triton X-100 or other nonionic or zwitterionic detergent from the final sample loaded on the gel can yield poor results, especially for samples containing SDS, because the amount of detergent in the IEF gels alone may be insufficient to remove bound SDS from proteins. The presence of Triton X-100 in the sample is especially important when SDS sample buffer is used to solubilize protein samples (i.e., after immunoprecipitation). The amount of Triton X-100 in the lysis buffer is normally sufficient for effective dissociation of SDS from proteins. If poor results are encountered with SDS-containing samples, try decreasing the final SDS concentration and/or increasing the final Triton X-100 concentration in the sample.
If no proteins are detected on the gel, check whether (1) the total protein load is appropriate for detection method used, (2) the orientation of electrical connections is wrong or electrical connection during isoelectrofocusing is poor (all gels from that run will be blank), (3) an air bubble obstructs current in a single IEF tube, or (4) the electrical connection is incorrect or is poor during the second-dimension gel separation. Careful monitoring of current and voltage at the beginning, during, and at the end of electrophoretic separations is strongly recommended in order to detect potential problems that will affect final results. Recording the initial and final current and voltage will also facilitate troubleshooting of future separations. Additional guidelines for troubleshooting and evaluating artifacts in 2D gel electrophoresis are described by Dunbar (1987).
Anticipated Results
A 2D electrophoretic separation of proteins should produce a pattern of round or elliptical spots, with many of the spots separated from one another. The pI range of the separated proteins as well as the observed molecular weight range depend on the first-dimension isoelectrofocusing protocol and the percentage of acrylamide used for the second-dimension gel. A complex protein mixture such as a whole-cell extract should produce more than 1000 silver-stained spots or close to 2000 Sypro Ruby stained spots that are distributed over most of the gel area. Fewer spots will be detected with less sensitive detection techniques such as Coomassie blue staining. On the other hand, separation of radiolabeled proteins and use of multiple exposures permit detection of many low-abundance proteins.
Time Considerations
Time requirements are very dependent on gel size and whether an external cooling unit is used to permit faster separations. Isoelectrofocusing of 18-cm-long IPG gels requires ~16 to 18 hr. Isoelectrofocusing using the standard-size gel format described in Basic Protocol 3 with 3-mm tubes is most conveniently done in an overnight run of ~16 to 18 hr. This separation time can be decreased to ~5 to 6 hr using higher voltages and an external cooling device. Extruding a set of sixteen IEF tube gels and freezing them in equilibration buffer takes 1 to 2 hr, including setup time. Preparing and running SDS gels is described in UNIT 10.1. It takes ~30 min to thaw, equilibrate, and load two second-dimension gels.
Overall, if standard-size gels are used without external cooling, it will take ~3 working days before the results of 2D electrophoresis are obtained. A single person can conveniently run about 16 2D gels in one week, depending on the amount of electrophoresis equipment available. The rate-limiting step in most laboratories is running the second-dimension gels because 16 soluble ampholyte IEF gels or 12 IPG gels can be focused in one run, but loading, running, and detecting results from 12 to 16 second-dimension gels requires substantial operator time and electrophoresis equipment.
Acknowledgements
This work was supported in part by National Institutes of Health (NIH) grants P50 CA174523, P01 CA140043, and R01 CA131582 (D.W.S.).
Footnotes
Key Reference
Hochstrasser et al., 1988. See above.
Discusses methods for improving and troubleshooting two-dimensional separation.
Internet Resources
http://world-2dpage.expasy.org/swiss-2dpage/
A list of links to many two-dimensional electrophoresis gel databases.
Literature Cited
- Anderson NG and Anderson NL 1978a. Two-dimensional analysis of serum and tissue proteins: Multiple isoelectrofocusing. Anal. Biochem 85:331–340. [DOI] [PubMed] [Google Scholar]
- Anderson NL and Anderson NG 1978b. Two-dimensional analysis of serum and tissue proteins: Multiple gradient-slab gel electrophoresis. Anal. Biochem 85:341–354. [DOI] [PubMed] [Google Scholar]
- Celis JE, Rasmussen HH, Leffers H, Madsen P, Honore B, Gesser B, Dejgaard K, and Vandekerckhove J 1991. Human cellular protein patterns and their link to genome DNA sequence data: Usefulness of two-dimensional gel electrophoresis and microsequencing. FASEB J. 5:2200–2208. [DOI] [PubMed] [Google Scholar]
- Dunbar B 1987. Troubleshooting and artifacts in two-dimensional polyacrylamide gel electrophoresis In Two-Dimensional Electrophoresis and Immunological Techniques (Dunbar BS, ed.) pp. 173–195. Plenum, New York. [Google Scholar]
- Garrels JI 1979. Two-dimensional gel electrophoresis and computer analysis of proteins synthesized by cloned cell lines. J. Biol. Chem 54:7961–7977. [PubMed] [Google Scholar]
- Garrels JI 1989. The QUEST system for quantitative analysis of two-dimensional gels. J. Biol. Chem 264:5269–5282. [PubMed] [Google Scholar]
- Garrels JI and Franza BR Jr. 1989. The REF52 protein database: Methods of database construction and analysis using the QUEST system and characterizations of protein patterns from proliferating and quiescent REF52 cells. J. Biol. Chem 264:5283–5298. [PubMed] [Google Scholar]
- Goverman J and Lewis K 1991. Separation of disulfide-bonded polypeptides using two-dimensional diagonal gel electrophoresis. Methods 3:125–127. [Google Scholar]
- Hochstrasser DF, Harrington MC, Hochstrasser AC, Miller MJ, and Merril CR 1988. Methods for increasing the resolution of two-dimensional protein electrophoresis. Anal. Biochem 173:424–435. [DOI] [PubMed] [Google Scholar]
- Meleady P 2018. Two-dimensional gel electrophoresis and 2D-DIGE. Meth. Mol. Biol 1664: 314. [DOI] [PubMed] [Google Scholar]
- O’Farrell PH 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem 250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Rabilloud T, Chevallet M, Luche S, and Lelong C 2010. Two-dimensional gel electrophoresis: Past, Present and future. J. of Protemics 73:2064–2077. [DOI] [PubMed] [Google Scholar]
- Reed PW, Densmore A, and Bloch RJ, 2012. Optimization of large gel 2D electrophoresis for proteomic studies of skeletal muscle. Electrophoresis 33:1263–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogowska-Wrzesinska A, BiHan MC, Thaysan-Andersen M, Roepstorff P 2013. 2D gels still have a niche in proteomics. J of Proteomics. 88:4–13. [DOI] [PubMed] [Google Scholar]
- Strahler JR and Hanash SM 1991. Immobilized pH gradients: Analytical and preparative use. Methods 3:109–114. [Google Scholar]
- Strohkamp S, Gemoll T and Habermann JK 2016. Possibilities and limitations of 2DE-based analysis for identifying low-abundant tumor markers in human serum and plasma. Proteomics. 16:2519–2532. [DOI] [PubMed] [Google Scholar]
- Young DA, Voris BP, Maytin EV, and Colbert RA 1983. Very high resolution two-dimensional electrophoretic separation of proteins on giant gels. Methods Enzymol. 91:190–214. [DOI] [PubMed] [Google Scholar]