Abstract
Mitogen-Activated Protein (MAP) Kinase pathway involves several oncogenic genes which can serve as potential targets for therapy. Therefore, aim of the present study is to analyze mutations in the MAP Kinase pathway in pulmonary adenocarcinoma (ADCA) of Indian patients along with clinico-pathologic correlation and determination of the survival status in patients receiving therapy. Blocks and slides of 125 pulmonary ADCA of last 5 years were retrieved. Histo-morphology and tumor content were determined. EGFR, KRAS, BRAF and MEK1 genes were analyzed using Sanger sequencing and Real-time polymerase chain reaction (PCR). Clinico-pathologic correlation and survival analysis were performed. Fifty-eight (46.4%) patients harbored genetic mutations of which 49 had single somatic mutations, 5 had multiple exonic and 4 showed coexisting EGFR and KRAS mutations. EGFR mutations were seen in 24.8%, KRAS in 19.2% and BRAF (non-V600E) in 2.4% cases. There was no difference in progression-free survival of wild- type/single mutations when compared with multiple/ coexisting mutations (P = 0.09). However, the P value may indicate borderline correlation. To conclude, EGFR and KRAS mutations may coexist in the same patient in lung ADCA. Multiple exonic mutations of KRAS gene formed substantial percentage of our cohort, requiring further exploration. Lung ADCA harbouring BRAF mutations are commonly non-V600E. Testing of all major genetic driver mutations of lung ADCA irrespective of histology and other demographic characteristics is necessary.
Introduction
Constitutive activation of signaling pathways is a common occurrence in human cancers and is often associated with molecular alteration of the key components of the signaling cascade.1,2 One such intracellular signaling pathway playing an important role in lung cancer progression and development is the mitogen-activated protein (MAP) kinase pathway. Aberrantly activated MAP kinase pathway has been associated with tumor development and chemotherapy resistance.1,3 This pathway is evolutionarily conserved and can be activated by several factors such as mitogens, growth factors, hormones, chemokines, and cytokines.1,3,4 It consists of a cascade of signaling molecules which are sequentially activated.1,2,4
Epidermal growth factor receptors (EGFR) are tyrosine kinases receptors which are overexpressed or mutated in various malignancies including lung cancer.5 Binding of epidermal growth factor (EGF) to EGFR activates RAS which is then followed by sequential activation of RAF and finally mitogen-activated protein kinase kinase (MEK).1,2,4
EGFR-MAP kinase pathway has been studied intensively and inhibitors against EGFR, RAS, RAF, and MEK have been developed that target different components of this signaling cascade.1–3 Moreover, it is a commonly known fact that EGFR and other driver mutations of lung cancer are mutually exclusive. However, with the advancement in molecular technology, multiple and coexisting EGFR mutations have been detected, without much exhaustive studies on their clinical significance.6
Moreover, complete and concurrent analysis of EGFR and MAP kinase signaling genes has not been explored widely in nonsmall cell lung carcinoma (NSCLC) from India; except for 1 study where only Kristen Rat Sarcoma Viral oncogene (KRAS) and v-raf murine sarcoma viral oncogene homolog B (BRAF) genes were analyzed.7
Therefore, the aim of the present study is to analyze the mutations in the EGFR-MAP kinase pathway in pulmonary adenocarcinoma (ADCA) in Indian patients, along with its clinicopatho-logic correlation and determination of the survival status in patients receiving targeted therapy. We also intended to compare our findings with the existing world literature.
Material and methods
Sample collection and diagnosis
One hundred and twenty-five cases of pulmonary ADCA diagnosed between January 2012 and December 2017, where adequate material was available, were retrieved from the archives of the Department of Pathology after approval from institute’s ethics committee. The hematoxylin and eosin (H&E) stained slides were analyzed and histologic type of the tumor was determined according to World Health Organization 2015 classification of tumors of the lung, pleura, thymus, and heart.8 Immunohistochemistry for thyroid transcription factor-1 was done in morphologically undifferentiated cases for definite characterization. Cases showing histomorphologically low-grade patterns namely acinar, lepidic, and papillary were grouped together as good prognostic histology (Group 1). Those having high-grade patterns such as solid, micropapillary, or sarcomatoid were grouped together as poor prognostic histology (Group 2) and invasive mucinous carcinomas were kept as a separate group (Group 3).8 Similarly, tumors in stage I and II which are resectable were grouped together and stages III and IV tumors which are unresectable were kept in a separate group. Blocks showing more than 50% tumor component in their respective sections were used for DNA extraction and mutation analysis. All cases of small cell carcinoma, lung metastases, and those showing predominant necrosis were excluded from this study. Treatment and follow-up details were retrieved from case record files. Patients were managed in a multidisciplinary clinic as per stage, Eastern Cooperative Oncology Group (ECOG) performance status, and molecular profile which were available at the time of treatment decision making. Treatment response were assessed radiologically and labeled according to RECIST v 1.1. Progression-free survival was calculated from date of diagnosis till date of disease progression or death.
DNA isolation and quantification
DNA extraction was performed using 50-µm-thick sections of formalin-fixed paraffin embedded tissue samples. Formalin-fixed paraffin embedded DNA tissue extraction kit (cat. No. A2352, Promega) was used for DNA extraction. The isolated DNA was assessed both qualitatively and quantitatively by spectrophotometry (Nanodrop, Biodrop Resolution, Cambridge, UK).
Real-time polymerase chain reaction (PCR)
The EGFR RGQ PCR Kit (cat no. 870111, Therascreen, Qiagen Ltd, Manchester, UK) was used for detecting the presence of 29 EGFR mutations spanning exons 18–21. KRAS PCR Kit (cat no. 870001 Therascreen, Qiagen Ltd, Manchester, UK) was used for detecting 7 KRAS mutations in exon 2 (codons 12 and 13). The analysis was performed according to the manufacturers’ instructions.
Sanger sequencing
PCR was carried out to amplify exons using G2 colorless master mix (cat no. M7422 Promega) on ABI Palm thermal cycler (Applied Biosystem, California), using exon-specific primers. 3 µL of the purified PCR product was used. The sequencing was done using both forward and reverse primers for greater accuracy and the results were analyzed using SeqMan II software (DNASTAR).
Statistical analysis
Data analysis was done using statistical software Stata 14.0 (StataCorp LLC, Texas). Categorical data were expressed as frequency and percentage and quantitative data was expressed as mean ± standard deviation and median (minimum and maximum). Chi-square test and/or Fisher-exact test, independent t test, and rank-sum were used to check the statistical significance of the data. Survival analysis (Kaplan-Meier) was used to check the time to event (recurrence and/or metastasis) relationship. A P value <0.05 was considered significant.
Results
Patient characteristics
Of the 125 samples included in the study, 40 were resections and 85 were small biopsies. There was male predominance with a male: female ratio of 2.6:1. Median age was 58 years (26–85 years). Eighty-six (68.8%) patients were smokers, 72% of which were males. Ninety-five (76%) cases were in advanced clinical stage (stage III/IV). Approximately 90% patients were clinically well preserved with ECOG performance status between 0 and 2. Treatment history and follow-up status were known in 80 cases. Of these 80, 59 (73.7%) patients received chemotherapy as first-line treatment and 21 (26.2%) were subjected to tyrosine kinase inhibitors (TKIs) as the therapy of choice. Demographic characteristics have been shown in Table 1.
Table 1.
Clinicopathological parameters and mutation profile of pulmonary adenocarcinoma patients.
| Total N = 125 | EGFR +ve N = 31 | KRAS +ve N = 24 | BRAF +ve N = 3 | Coexisting mutations N = 4 | Multiple mutations N = 5 | |
|---|---|---|---|---|---|---|
| Median age (years) | 58 | 60 | 65 | 55 | 63 | 60 |
| Male: Female | 2.6:1 | 2.1:1 | 3:1 | 3:0 | 3:1 | 1.5:1 |
| Smoker | 86(68.8%) | 19(61.2%) | 18(75%) | 2 (66.6%) | 2(50%) | 3(60%) |
| Non smoker | 39(31.2%) | 12(38.7%) | 6 (25%) | 1(33.3%) | 2(50%) | 2(40%) |
| Resection | 40(32.0%) | 11(35.4%) | 8(33.3%) | 1(33.3%) | 2(50%) | 0 |
| Biopsy | 85(68.0%) | 20(64.5%) | 16(66.6%) | 2(66.6%) | 2(50%) | 5(100%) |
| Stage1 and 2 | 30(24.0%) | 4(12.9%) | 7(29.1%) | 1(33.3%) | 1(25%) | 0 |
| Stage 3 and 4 | 95(76.0%) | 27(87.0%) | 17(70.8%) | 2(66.6%) | 3(75%) | 5(100%) |
| ECOG 0–2 | 112(89.6%) | 26(83.8%) | 23(95.8%) | 3(100%) | 4(100%) | 5(100%) |
| ECOG 3–4 | 13(10.4%) | 5(16.1%) | 1 (4.1%) | 0 | 0 | 0 |
| Histology patterns | ||||||
| Group 1 low grade | 74 (59.2%) | 24 (77.4%) | 14 (58.3%) | 1(33.3%) | 3(75%) | 5(100%) |
| Group 2 high grade | 43 (34.4%) | 7 (22.5%) | 8(33.3%) | 1(33.3%) | 1(25%) | 0 |
| Group 3 mucinous | 8 (6.4%) | 0 | 2(8.3%) | 1(33.3%) | 0 | 0 |
| Available treatment details | ||||||
| N = 80 | N = 21 | N = 16 | N = 1 | N = 3 | N = 4 | |
| Chemo | 59 (73.7%) | 10 (47.6%) | 14 (87.5%) | 1 (100%) | 2 (66.6%) | 4 (100%) |
| TKI | 21 (26.2%) | 11 (52.3%) | 2 (12.5%) | 0 | 1 (33.3%) | 0 |
| Both | 10 (12.5%) | 7 (33.3%) | 8 (50%) | 0 | 1 (33.3%) | 0 |
| Treatment response | ||||||
| N = 61 | N = 18 | N = 14 | N = 1 | N = 3 | N = 3 | |
| Group 1 (Stable/progressive) | 44 (72.1%) | 15 (83.3%) | 8 (57.1%) | - | 2 (66.6%) | 1(33.3%) |
| Group 2 (Partial/Complete) | 17 (27.8%) | 3 (16.6%) | 6 (42.8%) | 1 (100%) | 1(33.3%) | 2 (66.6%) |
| Median PFS (in months) | 8.3 | 8.1 | 8.3 | - | 7.3 | |
Abbreviations: Chemo, chemotherapy; ECOG, Eastern Cooperative Oncology Group; PFS, progression-free survival; +ve, positive; TKI, tyrosine kinase inhibitors.
Histopathologic examination
The most common histologic pattern of ADCA was acinar (78/125; 62.4%) followed by solid pattern (37/125; 29.6%). Around 10% (12/125) had lepidic morphology and 8.8% (11/125) revealed papillary architecture. Mucinous ADCA formed 6.4% (8/125) of study population; micropapillary and sarcomatoid patterns were seen in 1.6% (2/125) cases each. Ten cases showed undifferentiated morphology all of which were positive for thyroid transcription factor-1. Twenty percent (25/125) tumors showed mixed morphology with varying combinations of acinar, lepidic, solid, papillary and micropapillary patterns. Cases with low-risk patterns were put together in Group 1 (74/125; 59.2%) (Fig 1a-c) and those with even minor components of high-risk patterns were segregated in Group 2 (43/125; 34.4%) (Fig 1d and e). The mucinous pattern was included in Group 3 (8/125; 6.4%) (Fig 1f) (Table 1).
Fig. 1.
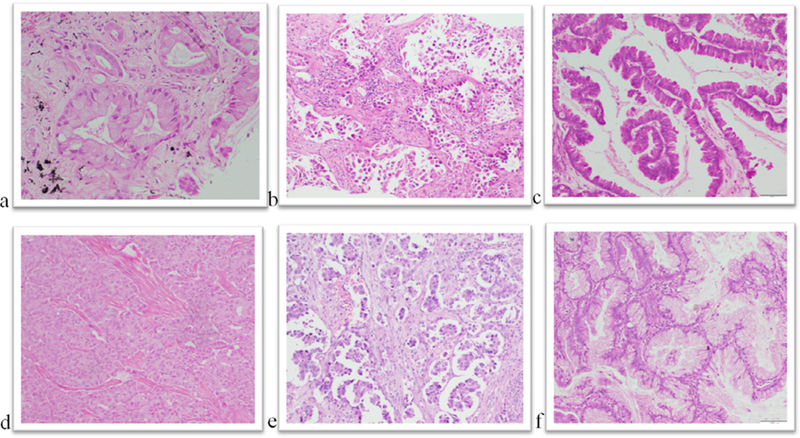
Histomorphologic variants of pulmonary adenocarcinoma. (a) Acinar (Hematoxylin and Eosin (H&E) X 200). (b) Lepidic (H&E X 40). (c) Papillary (H&E X100). (d) Solid (H&E X100). (e) Micropapillary (H&EX100). (f) Mucinous (h&E X100).
All histopathologic groups irrespective of the histology had significantly more number of patients in the higher stage (P = 0.003) (Fisher’s exact test). Correlation of smoking status with histologic subtype also showed statistical significance with smokers being predominant in Groups 1 (48/74; 64.8%) and 2 (36/43; 83.7%), whereas nonsmokers were predominant in Group 3 (6/8;75%) (P = 0.002) (Fisher’s exact test).
Mutation distributions
Fifty-eight of 125 (46.4%) showed genetic mutations. Forty-nine of these had single mutations and 9 patients exhibited multiple mutations (which included multiple exonic mutations of the same gene or presence of multiple gene mutations in the same case). Overall, there were 24.8% (31/125) patients with EGFR mutation, 19.2% (24/125) with KRAS mutation and 2.4% (3/125) with BRAF mutation. The demographic parameters of individual genes have been shown in Table 1.
EGFR mutation
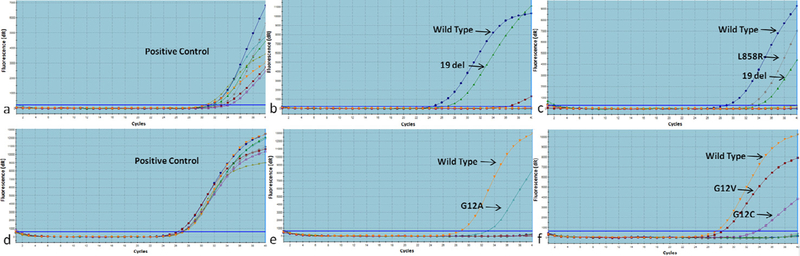
EGFR positivity had statistically significant correlation with histology, where the predominant pattern was acinar (group 1) and none were mucinous (P = 0.036) (Fisher’s exact test). The most commonly detected mutations were exon 19 deletions (20/31) (Fig 2a and b) followed by exon 21 point mutations (5/31). The mutations of exon 21 were L858R (4/5 cases) and L861Q (1/5 case). Exon 20 insertion and T790M mutations were detected in 2 cases each. Exon 18 mutation (G719X) was found positive in 1 case. EGFR dual positivity was seen in 1 case (exon 19 deletion and exon 21 L858R point mutation) (Fig 2c) and had Group 1 histology.
Fig. 2.
Real-time PCR analysis of EGFR and KRAS mutations in pulmonary adenocarcinoma. (a) EGFR control assay. (b) EGFR exon 19 deletion. (c) EGFR exon 19 deletion and exon 21 L858R dual mutation. (d) KRAS control assay. (e) KRAS exon 2 G12A point mutation. (f) KRAS exon 2 G12V and G12C dual mutation.
KRAS mutation
KRAS positivity showed no significant correlation with histologic groups. KRAS mutation was seen mainly in codons 12 and 13 of exon 2. The most commonly detected mutations were G12A (6/24) (Fig 2d and e) and G12C (5/24), followed by G12V (4/24) and G12D (3/24). G12S and G13D mutations were detected in 1 sample each respectively. KRAS dual mutations were seen in 4 cases and revealed combinations of G12A and G12V in 3 cases and G12V and G12C co-positivity in 1 case (Fig 2f), all bearing Group 1 histology.
Coexistence of EGFR and KRAS mutations in the same patients were seen in 4 of 125 cases (3.2%). The combinations noticed were as follow:
EGFR Exon 18 G719X KRAS Exon 2 G12D
EGFR Exon 19 Del KRAS Exon 2 G12A
EGFR Exon 19 Del KRAS Exon 2 G12V
EGFR Exon 21 L858R KRAS Exon 2 G12C
Out of these 4 positive cases, 2 cases were reconfirmed by Sanger sequencing. Histologically, 3 of them showed acinar pattern (Group 1) and 1 was solid ADCA.
BRAF mutation
BRAF mutations consisted of point mutations in exon 11(G442D) (Fig 3a) in 1 case and exon 15 (L597V) in 2 cases. BRAF positivity had no histologic predominance as 1 case each fell into the 3 histologic groups. In addition, BRAF polymorphism was seen in 14.4 % cases (18/125) (Fig 3b). One case showed both BRAF polymorphism as well as L597V point mutation.
Fig. 3.
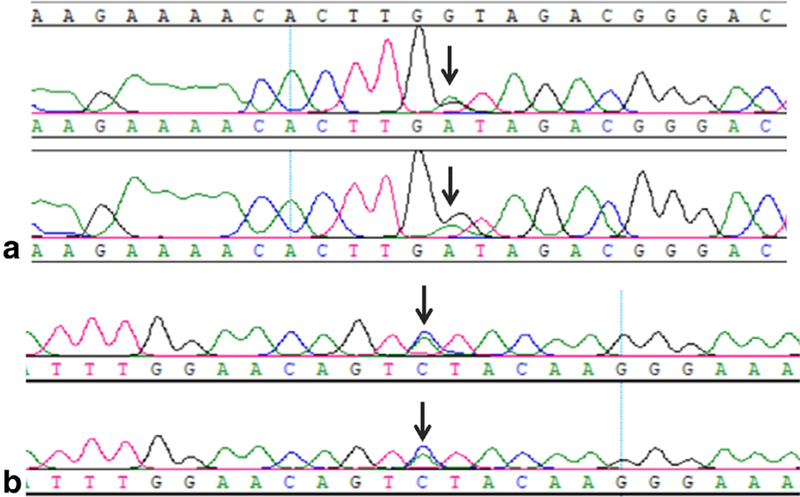
Sanger sequencing analysis of BRAF mutation and polymorphism in pulmonary adenocarcinoma. (a) BRAF exon 11(G442D) point mutation. (b) BRAF polymorphism.
MEK1 mutation
None of the cases exhibited MEK1 (exon1) mutation.
Follow-up
Out of 80 patients whose treatment details were available, 19 patients were lost to follow-up. To evaluate treatment response, we divided patients into 2 groups; response group 1 included cases with either stable disease or progressive disease (44/61; 72.1%) and response group 2 consisted of patients with either partial or complete response (17/61, 27.8%). We had a median follow-up of 5.5 months. The Kaplan-Meir curve analysis and log rank test did not show any significant difference in the median progression-free survival (PFS) of wild type or mutated patients (P = 0.09). We tried correlating progression with the different therapeutic regimens (TKI vs chemotherapy). However, due to limited number of patients with available follow-up in each subgroup, no reproducible statistics could be performed.
Due to small sample size and even smaller subgroup of patients in each mutation type, the power of this study is less than 80%.
Discussion
In NSCLC, the incidence of EGFR mutations varies considerably across different regions of the world.9,10 Various studies from India have shown regional diversity with its frequency ranging from 23% to 51.8%.11,12 One of these studies has compared the frequency of EGFR mutations in the population from North and South India and have found lower incidence of the mutation in people from North (33%) than Southern part of India (65%).13 Our study population being predominantly from North India had a frequency of 25% EGFR positivity which corroborates with their findings. Similar observation was found in previous studies from our institute.14,15
KRAS is the most frequently mutated gene across all cancer types with varying frequency in different regions of the world. The percentage of KRAS mutations in NSCLC observed in Western countries varies from 11% to 38% where Austria shows maximum frequency.16–18 In Asian countries, the range is much lower and constitutes only 3% to 19% in all NSCLCs.9,10,19–23 Two previous studies from India, both from the same center have revealed lower frequencies of KRAS positivity of 6.4% and 1.5% respectively7,24; as compared to 19.5% in the present study. The primary reason for this variation can be attributed to the method of detection of the mutation and higher prevalence of smokers in this cohort. Real-time PCR -based kits used in the present study have better sensitivity and specificity as compared to Sanger sequencing used in other studies.
Biologically, BRAF alterations are associated with increased kinase activity thereby rendering constitutive activation of the MAP kinase pathway. The percentage of BRAF mutation in NSCLC observed across the globe varies from 0.4% to 4.9% in the Western countries16,18 to 0.3% to 1.9% in Asia.7,22–24 Our findings are in concordance with global data. The targetable mutations which have been identified are BRAF L597V and V600E point mutations. Two of our cases had L597V point mutation while the V600E point mutation was absent in our cohort. Other BRAF mutations like G446D, G4469A/L, and Y472C have been identified but lack therapeutic trials. The third BRAF positive case in our study exhibited G442D point mutation. This mutation has earlier been reported in colon cancers25 and proven to be the causative factor.26,27 Additionally, we also observed BRAF polymorphism in 14.4% cases. However, the clinical implications of BRAF G442D point mutation and polymorphism require further exploration in NSCLC.
We had twice the population of males and approximately 70% of patients were smokers. KRAS mutations are known to be common in males and smokers.9,22,28,29 On the other hand, we differed in the gender distribution and smoking status of EGFR and BRAF positive cases.9,30–32 The overall predominance of male smokers in our cohort may be a confounding factor for the above results.
EGFR mutations are strongly associated with acinar, lepidic and papillary subtypes and are rarely found in mucinous.32–36 In contrast, the association of KRAS and BRAF mutations with histologic subtypes is controversial.33 Multiple reports have shown KRAS mutations to be prominent in invasive mucinous ADCA, whereas others have reported them to be associated with the solid subtypes.33,36–40 We found significant correlation of EGFR mutation with acinar, lepidic and papillary patterns; however, KRAS mutations were not associated with mucinous histology (P = 0.036).
The treatment modality offered to our patients solely depended on the clinicians’ discretion keeping in mind the ECOG performance status, stage, and the mutation profile available at the time of treatment decision making. We estimated the PFS in all cases with available follow-up post-treatment. The median PFS of EGFR mutated and wild type patients were almost similar. This may be due to the fact that some patients had not received TKI despite being EGFR mutated. Effective therapy targeting KRAS mutation in lung cancer has not been developed yet. Studies on PFS of KRAS-mutated lung ADCA, irrespective of mode of treatment, also contradict each other.20,41–44 PFS of KRAS wild type and mutated patients in our study differed marginally.
One area requiring further research is the co-existence of multiple exonic mutations in a single gene in both EGFR as well as KRAS genes. We had nearly 4% of patients harboring multiple mutations in single gene. All of them were in advanced clinical stage and had no significant association with smoking. Various clinical trials and studies have reported incidence of EGFR multiple mutations varying from 0.47% to 2.1%9,45 with poorer response rate.45 We had only 1 case with multiple mutations in the EGFR gene whose survival status could not be determined as he was lost to follow-up. A unique finding was the detection of KRAS multiple mutations, which has not been reported in lung cancers. Those conducted in colorectal cancers have shown an incidence of 2.1% and are seen to be associated with advanced clinical stage and metastases.46 We had 4 KRAS mutated cases with multiple mutations. All 4 had received conventional chemotherapy and had shown variable treatment response. Due to the small number of cases with KRAS multiple mutations, it is difficult to draw any conclusion about the treatment response and survival characteristics of such patients, needing further extensive studies to address the issue.
Besides the presence of multiple mutations in the same gene, coexistence of multiple gene mutations is also known with incidence rates up to 5%.47–50 The demographic profile of such patients has not been described earlier. Our patients with such mutational profile were predominantly males in the older age group and all were in the advanced clinical stage. The coexistence of mutations in a patient may be a consequence of intratumoral heterogeneity due to coexisting divergent clones within the same tumor.49,50 Tumors with EGFR and KRAS co-positivity generally show histologic features typical of EGFR positive cases (predominant acinar pattern).49 Similarly, 3 of our 4 cases with dual positivity had acinar histology. Further, it has been hypothesized that presence of coexisting mutations is associated with an aggressive disease profile with suboptimal response to therapy. We had follow-up available in 2 cases with coexisting mutations, both of which had initial short lasting clinical response to TKI and subsequently progressed (approximate PFS was 5 months). One of them was later found to have developed T790M mutation after treatment with EGFR TKI. Presence of co-existing mutations challenges the concept of mutual exclusivity and highlights intratumoral heterogeneity which may be helpful in predicting clinical response to targeted therapy.
On comparing the PFS status of patients with single mutations or wild type status with those having multiple or coexisting mutations, the difference was of only 1 month and was not statistically significant (P = 0.09). However, the P value of 0.09 may indicate a borderline correlation and extensive follow-up of such patients is warranted in further studies.
Also we are unable to comment on the benefits of different therapeutic regimens (TKI vs chemotherapy) in the different mutant groups due to minimal number of cases with satisfactory follow-up in each subgroup.
To conclude, this study examined the genetic alterations of key genes involved in MAP kinase pathway in lung ADCA among Indians. In addition, the study highlights the importance of co-existence of driver mutations in lung cancer which may have clinical implications on disease progression and therapeutic responses. We also found that, not only EGFR but KRAS also exhibited multiple mutations in lung ADCA, a finding which needs further exploration. Lung ADCA commonly exhibits non-V600E BRAF mutations. Finally, we reiterate the importance of testing of all major genetic driver mutations in lung ADCA irrespective of histology, smoking status, and other demographic characteristics.
Acknowledgments
Funding
This work was supported by Lady Tata Memorial Trust, India [Grant number N-1590].
References
- 1.Santarpia L, Lippman SM, El-Naggar AK. Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy. Expert Opin Ther Targets 2012;16:103–119. doi: 10.1517/14728222.2011.645805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen–activated protein kinase cascade for the treatment of cancer. Oncogene 2007;26:3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 3.De Luca A, Maiello MR, D’Alessio A, Pergameno M, Normanno N. The RAS/RAF/MEK/ERK and the PI3K/AKT signalling pathways: role in cancer pathogenesis and implications for therapeutic approaches. Expert Opin Ther Targets 2012;16(Suppl 2):S17–S27. doi: 10.1517/14728222.2011.639361. [DOI] [PubMed] [Google Scholar]
- 4.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene 2007;26:3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 5.Grandis JR, Sok JC. Signaling through the epidermal growth factor receptor during the development of malignancy. Pharmacol Ther 2004;102:37–46. doi: 10.1016/j.pharmthera.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Guibert N, Barlesi F, Descourt R, et al. Characteristics and outcomes of patients with lung cancer harboring multiple molecular alterations: results from the IFCT Study Biomarkers France. J Thorac Oncol 2017;12:963–973. doi: 10.1016/j.jtho.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Bhaumik S, Ahmad F, Das BR. Somatic mutation analysis of KRAS, BRAF, HER2 and PTEN in EGFR mutation-negative non-small cell lung carcinoma: determination of frequency, distribution pattern and identification of novel deletion in HER2 gene from Indian patients. Med Oncol 2016;33:117. doi: 10.1007/s12032-016-0828-7. [DOI] [PubMed] [Google Scholar]
- 8.Travis WD, Brambilla E, Nicholson AG. The 2015 World Health Organization Classification of Lung Tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol 2015;10:1243–1260. doi: 10.1097/jto.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 9.Tam IYS, Chung LP, Suen WS, et al. Distinct epidermal growth factor receptor and KRAS mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res 2006;12:1647–1653. doi: 10.1158/1078-0432.CCR-05-1981. [DOI] [PubMed] [Google Scholar]
- 10.Kim HR, Shim HS, Chung J-H, et al. Distinct clinical features and outcomes in never-smokers with nonsmall cell lung cancer who harbor EGFR or KRAS mutations or ALK rearrangement. Cancer 2012;118:729–739. doi: 10.1002/cncr.26311. [DOI] [PubMed] [Google Scholar]
- 11.Bal A, Singh N, Agarwal P, Das A, Behera D. ALK gene rearranged lung adenocarcinomas: molecular genetics and morphology in cohort of patients from North India. APMIS 2016;124:832–838. doi: 10.1111/apm.12581. [DOI] [PubMed] [Google Scholar]
- 12.Noronha V, Prabhash K, Thavamani A, et al. EGFR mutations in Indian lung cancer patients: clinical correlation and outcome to EGFR targeted therapy. PLoS One 2013;8:e61561. doi: 10.1371/journal.pone.0061561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aggarwal S, Patil S, Minhans S, Pungliya M. A study of EGFR mutation in nonsmoker NSCLC: striking disparity between north and south india patients http://ascopubs.org/doi/abs/10.1200/jco.2012.30.15_suppl.e18041; 2012. Accessed July 7, 2018.
- 14.Jain D, Iqbal S, Walia R, et al. Evaluation of epidermal growth factor receptor mutations based on mutation specific immunohistochemistry in non-small cell lung cancer: a preliminary study. Indian J Med Res 2016;143:308–314. doi: 10.4103/0971-5916.182621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain D, Ramachandrappa VS, Singh V, et al. Use of exfoliative specimens and fine-needle aspiration smears for mutation testing in lung adenocarcinoma. Acta Cytol 2017;61:455–461. doi: 10.1159/000479217. [DOI] [PubMed] [Google Scholar]
- 16.Schmid K, Oehl N, Wrba F, Pirker R, Pirker C, Filipits M. EGFR/KRAS/BRAF mutations in primary lung adenocarcinomas and corresponding locoregional lymph node metastases. Clin Cancer Res 2009;15:4554–4560. doi: 10.1158/1078-0432.CCR-09-0089. [DOI] [PubMed] [Google Scholar]
- 17.Ohtaki Y, Shimizu K, Kakegawa S, et al. Postrecurrence survival of surgically resected pulmonary adenocarcinoma patients according to EGFR and KRAS mutation status. Mol Clin Oncol 2014;2:187–196. doi: 10.3892/mco.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serizawa M, Koh Y, Kenmotsu H, et al. Assessment of mutational profile of Japanese lung adenocarcinoma patients by multitarget assays: a prospective, single-institute study. Cancer 2014;120:1471–1481. doi: 10.1002/cncr.28604. [DOI] [PubMed] [Google Scholar]
- 19.Arrieta O, Cardona AF, Federico Bramuglia G, et al. CLICaP, genotyping non-small cell lung cancer (NSCLC) in Latin America. J Thorac Oncol 2011;6:1955–1959. doi: 10.1097/JTO.0b013e31822f655f. [DOI] [PubMed] [Google Scholar]
- 20.Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer. Cancer Res 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 21.Bacchi CE, Ciol H, Queiroga EM, Benine LC, Silva LH, Ojopi EB. Epidermal growth factor receptor and KRAS mutations in Brazilian lung cancer patients. Clinics (Sao Paulo) 2012;67:419–424. http://www.ncbi.nlm.nih.gov/pubmed/22666783. Accessed July 6, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paik PK, Arcila ME, Fara M, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol 2011;29:2046–2051. doi: 10.1200/JCO.2010.33.12803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim SM, Kim EY, Kim HR, et al. Genomic profiling of lung adenocarcinoma patients reveals therapeutic targets and confers clinical benefit when standard molecular testing is negative. Oncotarget 2016;7:24172–24178. doi: 10.18632/oncotarget.8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das BR, Bhaumik S, Ahmad F, Mandsaurwala A, Satam H. Molecular spectrum of somatic EGFR and KRAS gene mutations in nonsmall cell lung carcinoma: determination of frequency, distribution pattern and identification of novel variations in Indian patients. Pathol Oncol Res 2015;21:675–687. doi: 10.1007/s12253-014-9874-7. [DOI] [PubMed] [Google Scholar]
- 25.Kovaleva V, Geissler A-L, Lutz L, et al. Spatio-temporal mutation profiles of case-matched colorectal carcinomas and their metastases reveal unique de novo mutations in metachronous lung metastases by targeted next generation sequencing. Mol Cancer 2016;15:63. doi: 10.1186/s12943-016-0549-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. http://www.mutationtaster.org/cgi-bin/MutationTaster/MutationTaster69.cgi (AccessedAugust8,2018).
- 27.Mutation overview page BRAF - p.G442D (Substitution - Missense) https://cancer.sanger.ac.uk/cosmic/mutation/overview?id=6196839 (Accessed August 8, 2018).
- 28.Le Calvez F, Mukeria A, Hunt JD, et al. TP53 and KRAS mutation load and types in lung cancers in relation to tobacco smoke: distinct patterns in never, former, and current smokers. Cancer Res 2005;65:5076–5083. doi: 10.1158/0008-5472.CAN-05-0551. [DOI] [PubMed] [Google Scholar]
- 29.Dogan S, Shen R, Ang DC, et al. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res 2012;18:6169–6177. doi: 10.1158/1078-0432.CCR-11-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maturu VN, Singh N, Bal A, Gupta N, Das A, Behera D. Relationship of epidermal growth factor receptor activating mutations with histologic subtyping according to International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society 2011 adenocarcinoma classification and their impact on overall survival. Lung India 2016;33:257–266. doi: 10.4103/0970-2113.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui G, Liu D, Li W, et al. A meta-analysis of the association between BRAF mutation and nonsmall cell lung cancer. Medicine (Baltimore) 2017;96:e6552. doi: 10.1097/MD.0000000000006552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen D, Zhang L-Q, Huang J-F, et al. BRAF mutations in patients with non-small cell lung cancer: a systematic review and meta-analysis. PLoS One 2014;9. doi: 10.1371/journal.pone.0101354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Dong Y, Cai Y, et al. Clinicopathologic characteristics of ALK rearrangements in primary lung adenocarcinoma with identified EGFR and KRAS status. J. Cancer Res Clin Oncol 2014;140:453–460. doi: 10.1007/s00432-014-1584-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Motoi N, Szoke J, Riely GJ, et al. Lung adenocarcinoma: modification of the 2004 WHO mixed subtype to include the major histologic subtype suggests correlations between papillary and micropapillary adenocarcinoma subtypes, EGFR mutations and gene expression analysis. Am J Surg Pathol 2008;32:810–827. doi: 10.1097/PAS.0b013e31815cb162. [DOI] [PubMed] [Google Scholar]
- 35.Sun P-L, Seol H, Lee HJ, et al. High incidence of EGFR mutations in Korean men smokers with no intratumoral heterogeneity of lung adenocarcinomas: correlation with histologic subtypes, EGFR/TTF-1 expressions, and clinical features. J Thorac Oncol 2012;7:323–330. doi: 10.1097/JTO.0b013e3182381515. [DOI] [PubMed] [Google Scholar]
- 36.Yoshizawa A, Sumiyoshi S, Sonobe M, et al. Validation of the IASLC/ATS/ERS lung adenocarcinoma classification for prognosis and association with EGFR and KRAS gene mutations: analysis of 440 Japanese patients. J Thorac Oncol 2013;8:52–61. doi: 10.1097/JTO.0b013e3182769aa8. [DOI] [PubMed] [Google Scholar]
- 37.Dong Y-J, Cai Y-R, Zhou L-J, et al. Association between the histological subtype of lung adenocarcinoma, EGFR/KRAS mutation status and the ALK rearrangement according to the novel IASLC/ATS/ERS classification. Oncol Lett 2016;11:2552–2558. doi: 10.3892/ol.2016.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell PA, Barnett SA, Walkiewicz M, et al. Correlation of mutation status and survival with predominant histologic subtype according to the new IASLC/ATS/ERS lung adenocarcinoma classification in stage III (N2) patients. J Thorac Oncol 2013;8:461–468. doi: 10.1097/JTO.0b013e3182828fb8. [DOI] [PubMed] [Google Scholar]
- 39.Kakegawa S, Shimizu K, Sugano M, et al. Clinicopathological features of lung adenocarcinoma with KRAS mutations. Cancer 2011;117:4257–4266. doi: 10.1002/cncr.26010. [DOI] [PubMed] [Google Scholar]
- 40.Rekhtman N, Ang DC, Riely GJ, Ladanyi M, Moreira AL. KRAS mutations are associated with solid growth pattern and tumor-infiltrating leukocytes in lung adenocarcinoma. Mod Pathol 2013;26:1307–1319. doi: 10.1038/modpathol.2013.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jia Y, Jiang T, Li X, et al. Characterization of distinct types of KRAS mutation and its impact on first-line platinum- based chemotherapy in Chinese patients with advanced non-small cell lung cancer. Oncol Lett 2017;14:6525–6532. doi: 10.3892/ol.2017.7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roman M, Baraibar I, Lopez I, et al. KRAS oncogene in non-small cell lung cancer: clinical perspectives on the treatment of an old target. Mol Cancer 2018;17:33. doi: 10.1186/s12943-018-0789-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ludovini V, Bianconi F, Pistola L, et al. Phosphoinositide-3-kinase catalytic alpha and KRAS mutations are important predictors of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in patients with advanced non-small cell lung cancer. J Thorac Oncol 2011;6:707–715. doi: 10.1097/JTO.0b013e31820a3a6b. [DOI] [PubMed] [Google Scholar]
- 44.Mellema WW, Dingemans A-MC, Thunnissen E, et al. KRAS mutations in advanced nonsquamous non-small-cell lung cancer patients treated with first-line platinum-based chemotherapy have no predictive value. J Thorac Oncol 2013;8:1190–1195. doi: 10.1097/JTO.0b013e318298764e. [DOI] [PubMed] [Google Scholar]
- 45.Wei Z, An T, Wang Z, et al. Patients harboring epidermal growth factor receptor (EGFR) double mutations had a lower objective response rate than those with a single mutation in non-small cell lung cancer when treated with EGFR-tyrosine kinase inhibitors. Thorac Cancer 2014;5:126–132. doi: 10.1111/1759-7714.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macedo MP, Andrade LDB, Coudry R, et al. Multiple mutations in the KRAS gene in colorectal cancer: review of the literature with two case reports. Int J Colorectal Dis 2011;26:1241–1248. doi: 10.1007/s00384-011-1238-0. [DOI] [PubMed] [Google Scholar]
- 47.Han S-W, Kim T-Y, Jeon YK, et al. Optimization of patient selection for gefitinib in non-small cell lung cancer by combined analysis of epidermal growth factor receptor mutation, K-ras mutation, and Akt phosphorylation. Clin Cancer Res 2006;12:2538–2544. doi: 10.1158/1078-0432.CCR-05-2845. [DOI] [PubMed] [Google Scholar]
- 48.Choughule A, Sharma R, Trivedi V, et al. Coexistence of KRAS mutation with mutant but not wild-type EGFR predicts response to tyrosine-kinase inhibitors in human lung cancer. Br. Cancer 2014;111:2203–2204. doi: 10.1038/bjc.2014.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee T, Lee B, Choi Y-L, Han J, Ahn M-J, Um S-W. Non-small cell lung cancer with concomitant EGFR, KRAS, and ALK mutation: clinicopathologic features of 12 cases. J Pathol Transl Med 2016;50:197–203. doi: 10.4132/jptm.2016.03.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benesova L, Minarik M, Jancarikova D, Belsanova B, Pesek M. Multiplicity of EGFR and KRAS mutations in nonsmall cell lung cancer (NSCLC) patients treated with tyrosine kinase inhibitors. Anticancer Res 2010;30:1667–1671. http://www.ncbi.nlm.nih.gov/pubmed/20592359. Accessed July 7, 2018. [PubMed] [Google Scholar]