Abstract
Purpose:
Gene-disease associations implicated in hereditary colorectal cancer and polyposis susceptibility were evaluated using the ClinGen Clinical Validity framework.
Methods:
Forty-two gene-disease pairs were assessed for strength of evidence supporting an association with hereditary colorectal cancer and/or polyposis. Genetic and experimental evidence supporting each gene-disease relationship was curated independently by two trained biocurators. Evidence was reviewed with experts and assigned a final clinical validity classification.
Results:
Of all gene-disease pairs evaluated, 14/42 (33.3%) were Definitive, 1/42 (2.4%) were Strong, 6/42 (14.3%) were Moderate, 18/42 (42.9%) were Limited, and 3/42 (7.1%) were either No Reported Evidence, Disputed, or Refuted. Of panels in the NIH Genetic Testing Registry, 4/26 (~15.4%) contain genes with Limited clinical evidence.
Conclusion:
Clinicians and laboratory diagnosticians should note that <60% of the genes on clinically available panels have Strong or Definitive evidence of association with hereditary colon cancer or polyposis, and >40% have only Moderate, Limited, Disputed, or Refuted evidence. Continuing to expand the structured assessment of the clinical relevance of genes listed on hereditary cancer testing panels will help clinicians and diagnostic laboratories focus the communication of genetic testing results on clinically significant genes.
Keywords: ClinGen, classification, cancer, validity, NGS
Introduction
Colorectal cancer affects roughly 1.3 million individuals in the United States and is estimated to account for approximately 50,000 deaths in 20171. It is estimated that 5% of cases of colorectal cancer are associated with inheritance of a single gene abnormality. About 25% of all cases have a family history suggestive of a genetic influence2 suggesting that discovery of additional complex gene-gene and gene-environment features is likely. Detection of hereditary colorectal cancer susceptibility can focus clinical management, leading to lifesaving interventions that reduce cancer mortality and incidence3,4. Patients presenting at a young age or with other clinical or family history suggesting a hereditary colorectal cancer or polyposis syndrome should be referred for genetic evaluation and possibly germline testing5,6.
With recent advances in next generation sequencing (NGS) technology, we have revolutionized our ability to identify genetic variants implicated in hereditary colorectal cancer susceptibility, polyposis susceptibility, and other diseases7. These advances have fueled a dramatic expansion of genes sequenced on diagnostic NGS panel tests. In May 2018, in the Genetic Testing Registry, there are 33 diagnostic testing panels with roughly 40 genes offered for hereditary colorectal cancer and polyposis susceptibility. Concerns have arisen that not all genes under testing have been rigorously evaluated for their clinical validity8,9. The National Comprehensive Cancer Network (NCCN) has indicated the challenge of limited data and a lack of clear guidance regarding how to evaluate the robustness of clinical evidence supporting the cancer association of some genes on colorectal cancer panels6. In their recent guideline for hereditary colon cancer, the NCCN provides an assessment of clinical relevance for colon cancer for 22 genes. A more complete understanding of the clinical relevance of genes being tested on hereditary colorectal cancer susceptibility panels will aid both laboratories that develop these tests/interpret results and clinicians who communicate these results to patients10,11.
The Clinical Genome Resource (ClinGen) is an NIH-funded program dedicated to creating publicly available data that assesses the clinical relevance of genes and variants within specific diseases10. In an attempt to standardize the evaluation of the clinical effects of disease-associated genes, ClinGen created a Clinical Validity framework that facilitates systematic evaluation of literature evidence to assign the strength of a gene’s association with disease into one of seven clinical validity classifications: Definitive, Strong, Moderate, Limited, No Reported Evidence, Disputed, or Refuted11. The semiquantitative framework assesses and assigns points to genetic evidence (consisting of probands, segregation data, and case-control studies) and experimental evidence (protein interaction data, functional alteration studies, mouse models, etc)12. In this report, the ClinGen Colon Cancer and Polyposis Gene Curation Panel reports our results applying this framework to evaluate the strength of evidence for 40 genes associated with syndromic and isolated hereditary colorectal cancer and polyposis susceptibility, confirming assessments for all of the established colorectal cancer (CRC) genes and clarifying evaluations for many recently described genes putatively associated with CRC or polyposis. This information will benefit the genomic medicine community in evaluating the clinical relevance of genes listed on hereditary colorectal cancer susceptibility panels.
Materials and Methods
The ClinGen Hereditary Colorectal Cancer and Polyposis Susceptibility Gene Curation Panel (GCP) completed the analysis described here. The GCP was composed of seven biocurators (B.A.S, S.A.J., J.L.M., D.I.R., M.E.R., R.J.S., B.A.T), coordinator (K.L.), and experts comprised of a medical geneticist (S.E.P.), three medical oncologists with cancer genetics expertise (K.O., Z.K.S., M.S.G.), and two clinical molecular geneticists (L.Z., M.J.F.). We also benefitted from the informal contributions of other ClinGen experts who were not formal members of the panel. Further details of the gene curation panel structure can be found atwww.clinicalgenome.org. All data were derived from published literature and so no institutional review board review was necessary.
Analysis of gene content on clinically available hereditary colorectal cancer and/or polyposis testing panels was conducted via the NIH Gene Test Registry (GTR). The NIH GTR13 was queried for all genes listed on hereditary colorectal cancer and/or polyposis susceptibility panels using terms pertaining to colorectal cancer and adenomatous polyposis. There were 23 genes found on 3 or more (~10%) hereditary colorectal cancer and/or polyposis susceptibility panels within the NIH GTR. Based on review of recent data, the domain experts of the GCP recommended 17 additional genes including newly published genes for a total of 40 genes (Table 1, Table S1).
Table 1.
Phenotypic Evaluation for polyposis and/or colorectal cancer for syndromic and non-syndromic genes found on clinically available testing panels
| Gene | Disease Entity Curated (Inheritance) |
Disease MIM ID, Orphanet ID |
Phenotype(s) of Curation Focus |
Classification | aNotes |
|---|---|---|---|---|---|
| APC | Familial Adenomatous Polyposis (AD) | MIM:175100, ORPHA:247806 | Adenomatous polyps and Colorectal cancer | Definitive (12 GE, 6 EE, r/t) | |
| ATM | Colorectal cancer Susceptibility (AD) | MIM:N/A, ORPHA: 443909 | Colorectal cancer | Moderate (5 GE, 2 EE) | |
| AXIN2 | Oligodontia-colorectal cancer syndrome (AD) | MIM:608615, ORPHA:401911 | Hyperplastic polyps, Adenomatous polyps, Colorectal cancer | Moderate (8.5 GE, 2.5 EE) | |
| BLM | Colorectal cancer/Polyposis Susceptbility (AD) | MIM:N/A, ORPHA:N/A | Adenomatous polyps, Colorectal cancer | Definitive (8.3 GE, 6 EE, r/t) | |
| BMPR1A | Juvenile Polyposis Syndrome (AD) | MIM:174900, ORPHA:329971 | Hamartomatous polyps | Definitive (12 GE, 4.5 EE, r/t) | |
| CHEK2 | Colorectal cancer Susceptibility (AD) | MIM:604373, ORPHA: 443909 | Colorectal cancer | Limited (3.25 GE, 2.5 EE) | |
| ENG | Colorectal cancer/Polyposis Susceptibility (AD) | MIM:N/A, ORPHA:329971 | Hamartomatous polyps, Hyperplastic polyps, Adenomatous polyps, Colorectal cancer | Limited (0.2 GE, 1 EE) | |
| EPCAM | Lynch Syndrome (AD) | MIM:613244, ORPHA:144 | Colorectal cancer | Definitive (12 GE, 4 EE, r/t) | GE consists of probands with 3’UTR deletions/ exonic deletions at 3’ end of EPCAM, PMID: 19098912, 19177550 |
| GALNT12 | Colorectal cancer Susceptibility (AD) | MIM:608812, ORPHA: 443909 | Colorectal cancer | Limited (4 GE, 0.5 EE) | |
| GREM1 | Hereditary Mixed Polyposis Syndrome (AD) | MIM:601228, ORPHA:157794 | Hamartomatous polyps Adenomatous polyps Colorectal cancer | Strong (8.5 GE, 6 EE) | GE consists of probands with a single tandem duplication upstream of GREM1, PMID: 22561515 |
| MLH1 | Lynch Syndrome (AD) | MIM:609310, ORPHA:144 | Colorectal cancer | Definitive (12 GE, 6 EE, r/t) | |
| MLH3 | Colorectal cancer Susceptibility (AD) | MIM:614385, ORPHA: 443909 | Colorectal cancer | Moderate (5 GE, 5 EE) | |
| MSH2 | Lynch Syndrome (AD) | MIM:120435, ORPHA:144 | Colorectal cancer | Definitive (12 GE, 6 EE, r/t) | |
| MSH6 | Lynch Syndrome (AD) | MIM:614350, ORPHA:144 | Colorectal cancer | Definitive (12 GE, 6 EE, r/t) | |
| MUTYH | Colorectal cancer Susceptibility (AD) | MIM:N/A, ORPHA: 247798 | Colorectal cancer | Moderate (7 GE, 4.5 EE) | GE consists of case-level and case-control data in MUTYH monoallelic carriers |
| MUTYH | MUTYH associated Polyposis (AR) | MIM:608456, ORPHA:247798 | Adenomatous polyps, Colorectal cancer | Definitive (12 GE, 6 EE, r/t) | |
| PALB2 | Colorectal cancer Susceptibility (AD) | MIM:N/A ORPHA: 443909 | Colorectal cancer | Limited (5.2 GE, 0.5 EE) | |
| PMS1 | Lynch Syndrome (AD) | MIM:N/A, ORPHA:144 | Colorectal cancer | Refuted | |
| PMS2 | Lynch Syndrome (AD) | MIM:614337, ORPHA:144 | Colorectal cancer | Definitive (12 GE, 3 EE, r/t) | |
| POLD1 | Colorectal cancer/Polyposis Susceptbility (AD) | MIM:612591, ORPHA:447877 | Adenomatous polyps, Colorectal cancer | Definitive (8.5 GE, 5.5 EE, r/t) | GE consists of probands with missense substitutions in the exonuclease domain, PMID: 23263490 |
| POLE | Colorectal cancer/Polyposis Susceptbility (AD) | MIM:615083, ORPHA:447877 | Adenomatous polyps, Colorectal cancer | Definitive (12 GE, 3 EE, r/t) | GE consists of probands with missense substitutions in the exonuclease domain, PMID: 23263490 |
| PTEN | PTEN Hamartoma Tumor Syndrome (AD) | MIM:601728, ORPHA:306498 | Hamartomatous polyps, Hyperplastic polyps, Adenomatous polyps | Definitive (12 GE, 6 EE, r/t) | |
| SMAD4 | Juvenile Polyposis Syndrome (AD) | MIM:174900, ORPHA:329971 | Hamartomatous polyps | Definitive (12 GE, 6 EE, r/t) | |
| STK11 | Peutz-Jeghers Syndrome (AD) | MIM:175200, ORPHA:2869 | Hamartomatous polyps, Colorectal cancer | Definitive (12 GE, 6 EE, r/t) |
Abbreviations- AD: Autosomal Dominant, AR: Autosomal Recessive, r/t: replicated over time, N/A: Not Applicable, MIM: Mendelian Inheritance in Man, ORPHA:Orphanet identifier. GE: Genetic Evidence, EE: Experimental Evidence, PMID: PubMed identifier.
These notes serve as an explanation for the asterisks in Figure 1.
Two biocurators independently assessed the data for all genes, each assigning a provisional classification for association with either CRC or polyposis. We separately curated biallelic and monoallelic associations for MSH3 and MUTYH, for a total of 42 gene-disease associations. Discussions were held by the entire GCP in which curators summarized the literature evidence supporting a gene-disease association, and the expert-led group assigned a final clinical validity classification by consensus. All final clinical validity classifications were reviewed and approved by the ClinGen Hereditary Cancer Working Group, a large consortium of 22 members and submitted to the ClinGen website14. Clinical validity classifications consisted of Refuted, Disputed, or No Reported Evidence (n=0 total points), Limited (n=0.1–6 points), Moderate (n=7–11 points), Strong (n=12–18 points), and Definitive (n=12–18 points with replication over time). Genetic evidence could receive a maximum of 12 points and experimental evidence a maximum of 6 points, for a combined total of up to 18 points.
Each gene-disease association was curated using the Standard Operating Procedures (SOP), Version 415 according to the methods of Strande et al. (2017)11. Several adjustments were made to the scoring system as the curation process proceeded. Specifically, due to concern for phenocopies because of the high population incidence of CRC, genetic evidence points were adjusted downward from the default suggestions in the SOP as follows. When present, case-control data, which were considered the strongest form of genetic evidence, were used to maximize genetic evidence points prior to the inclusion of case-level data from segregation and single variant carrier evidence (see Figure 2 of Strande, et al.11). With respect to single case-level variant evidence, 0.5 points (instead of the suggested default of 1.5 in the SOP) were assigned to affected probands with either null/predicted null (i.e. loss-of-function or LoF) variants or presumed de novo variants (regardless of confirmation of paternity and maternity). Rare missense variants (MAF<0.001) were assigned 0.5 points/proband when functional evidence supported a damaging effect to the protein by the variant, and 0.1 points/proband assigned to rare missense variants lacking functional evidence. Additionally, for each variant type, points were assigned to probands according to zygosity of the variant (e.g. 0.5 points/proband with a heterozygous LoF variant, 1 point/proband with a homozygous or two compound heterozygous LoF variants in trans). All variants under consideration within probands or case-control studies, unless known to be founder mutations, had to have maximum minor allele frequencies within any subpopulation of ≤0.001 to be counted toward genetic evidence.
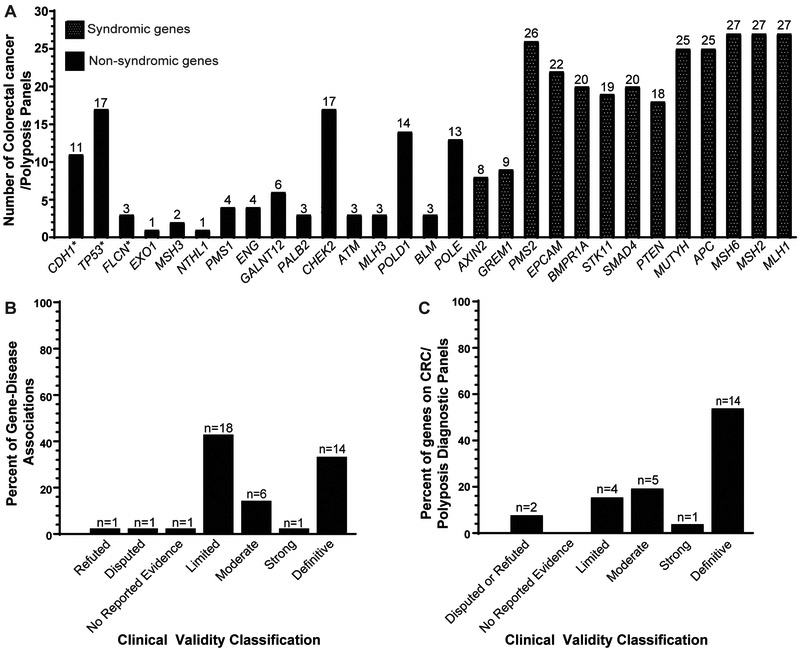
Figure 2.
Distribution of genes and clinical validity classifications on clinically available hereditary colorectal cancer and/or polyposis susceptibility testing panels. A. Quantity of testing panels containing each gene shown in a bar graph of the total number of testing panels for each gene curated as part of a gene-disease pair (See Figure 1, Table 1, and Table S1 for the list of genes). *CDH1, TP53 and FLCN were found on testing panels, but were not assigned a clinical validity classification for either hereditary colorectal cancer or hereditary polyposis susceptibility. B. Percent distribution of each clinical validity classification. The percentages represent the proportion of the 42 gene-disease pairs assigned a certain clinical validity classification. C. Percent distribution of classifications for genes listed on testing panels. The percentages represent the number of genes within a given clinical validity classification out of all genes that (i) were assessed in this manuscript and (ii) exist on testing panels (n=26 genes). For genes curated multiple times for different inheritance patterns, the highest clinical validity classification was used for plotting the graph (e.g. MUTYH was considered a Definitive gene for the graph although it was Definitive for biallelic MUTYH-associated polyposis, but Moderate for monoallelic isolated hereditary colorectal cancer susceptibility).
Four genes were curated for specific mutation types. EPCAM was curated for 3’ deletions and 3’UTR deletions, POLE and POLD1 were curated for variants localized within their exonuclease domains, GREM1 was curated for the single duplication occurring upstream of GREM116 that has previously been reported to be associated with polyposis, and PTPRJ was curated for a single tandem duplication that had been associated with familial colorectal cancer17. Three genes (FLCN, TP53 and CDH1) were curated but not included in our final totals because of their known association with syndromes for which CRC and polyposis are possible rare manifestations, and thus it was difficult to separate out CRC specific risks (see Table 2).
Table 2.
Evidence for TP53, CDH1 and FLCN associations with colorectal cancer in Li-Fraumeni Syndrome, Hereditary Diffuse Gastric Cancer Syndrome and Birt-Hogg-Dubé Syndrome
| Gene | Disease Entity Association (Inheritance) |
Disease MIM ID, Orphanet ID |
Proband Inclusion Criteria |
Phenotype(s) and Diagnostic Age(s) | PMIDs Evaluated |
|---|---|---|---|---|---|
| TP53 | Li-Fraumeni Syndrome (AD) | MIM:151623, ORPHA:524 |
Li-Fraumeni Syndrome Colon Cancer Family Registry lacking known without known hereditary cancer syndrome |
Colon cancer, 21 years Colon cancer, 9 years; Colon and rectum cancer, 41 years Colon cancer, 38 years; Melanoma, 34 years |
12885464 16401470 26086041 |
| CDH1 | Hereditary Diffuse Gastric Cancer Syndrome (AD) | MIM:137215, ORPHA:26106 |
Hereditary Diffuse Gastric Cancer | Colorectal cancer, 30 years Rectal cancer, 30 years Colorectal cancer, 55 years Colon cancer, 59 years |
9537325 10072428 17221870 11968083 |
| FLCN | Birt-Hogg-Dubé Syndrome (AD) | MIM:135150 ORPHA:122 |
Colorectal Cancer Polyposis | Colorectal cancer, 3 probands- no dx. age specified Colorectal cancer, 3 probands- 48 - 64 years Polyps, 59 years Colorectal cancer, 56 years |
27356891 20522427 19785621 18573707 |
The ClinGen Clinical Validity framework was developed to evaluate genes associated with monogenic disorders that are inherited in an autosomal dominant, autosomal recessive, or X-linked manner11. To adjust for confounding factors of potential phenocopies and low penetrance (current diagnostic testing panels include moderate-penetrance genes8), we operated under the assumption that a given variant represented a monogenic etiology significantly raising the risk for the manifested disease phenotype. We did not include Genome-Wide Association Study (GWAS) publications in our curation of each gene-disease association, since the causative variant in most GWAS associations is not known. Diagnostic age of the case(s) were carefully evaluated to attempt to exclude phenocopies18,19. For instance, an individual harboring a genetic variant who was diagnosed with colorectal cancer at 85 years old would have not been counted toward proband evidence as the colorectal cancer was likely a phenocopy rather than being due to a monogenic susceptibility.
Results
Forty genes encompassing 42 gene-disease pairs were curated and classified for an association with a specific disease entity, either syndromic hereditary colorectal cancer and/or polyposis susceptibility, or isolated hereditary colorectal cancer and/or polyposis susceptibility (MUTYH and MSH3 curated for both CRC and polyposis assuming two different phenotypic entities and inheritance patterns) (Table 1, Figure 1). For named syndromes such as Lynch syndrome, curators focused on colorectal cancer susceptibility and/or gastrointestinal polyps as the phenotypic feature(s) of interest when evaluating the literature as these features were within the clinical expertise of our GCP. Generally, this single phenotype was sufficient to reach a definitive conclusion.
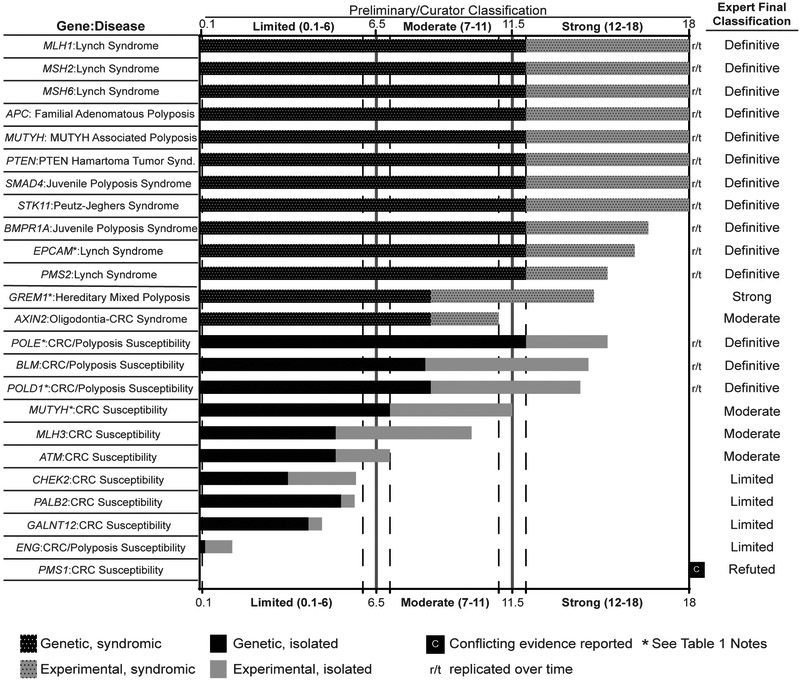
Figure 1.
Clinical validity classifications of 24 gene-disease pairs associated with syndromic or isolated hereditary colorectal cancer and/or polyposis susceptibility. Consensus genetic and experimental evidence scores are depicted for each gene:disease relationship wherein a Limited preliminary classification scored 0.1-6 total points, a Moderate preliminary classification had 7-11 total points, a Strong preliminary classification scored 12-18 total points, and a Definitive classification scored 12-18 total points and achieved replication over time (indicated by r/t).
Gene:disease pairs associated with syndromic hereditary colorectal cancer and/or polyposis are indicated by dotted black and grey bars for genetic and experimental evidence, respectively. Gene:disease pairs associated with isolated hereditary colorectal cancer and/or polyposis are indicated by solid black and grey bars for genetic and experimental evidence, respectively.
Of all gene-disease pairs evaluated, 14/42 (33.3%) were Definitive, 1/42 (2.4%) were Strong, 6/42 (14.3%) were Moderate, 18/42 (42.9%) were Limited, 3/42 (7.1%) were No Reported Evidence, Disputed, or Refuted (Figure 1, Figure 2B, Table 1). Long-established syndromic genes such as MLH1, MSH2, MSH6, PMS2, and EPCAM for Lynch syndrome, and APC, MUTYH, PTEN, and SMAD4 for polyposis were classified Definitive. Of note, in the ClinGen framework there may be no difference in score between Definitive and Strong except for the length of time that the association had been reported. One syndromic gene (GREM1) had a non-Definitive classification. Though the evidence for the role of this gene in syndromic hereditary colorectal cancer and/or polyposis susceptibility was replicated in both experimental and clinical diagnostic settings, the evidence for GREM1:Hereditary Mixed Polyposis Syndrome was due entirely to a single duplication upstream of GREM116, and the association was therefore considered to lack replication over time and granted a Strong classification (Figure 1, Table 1). Additionally, although the association of MUTYH with autosomal recessive MUTYH-associated polyposis was Definitive, the association of MUTYH with autosomal dominant hereditary colorectal cancer susceptibility was only Moderate and the genetic evidence comprised solely of case-control studies (i.e. MUTYH heterozygotes).
Another syndromic gene, POLD1, was initially classified as Moderate for Colorectal cancer/Polyposis Susceptibility. However, based on analysis of a large published pedigree, the experts concluded that obligate carriers within a single polyposis/colorectal cancer family20 should be counted as segregation evidence, raising the classification from Moderate to Definitive (Figure S1).
Classifications of Limited comprised 18/42 (42.9%) of gene-disease associations. Some of them included genes posed as candidates for colorectal cancer/polyposis susceptibility either in the past or more recently, but lacked additional evidence to be classified as Moderate (e.g. GALNT12, SEMA4A, FAN1, MSH3, ENG, XRCC4, BUB1, BUB3, PTPRJ)(Table S1). Others consisted of genes originally associated with cancer or tumor susceptibilities that are unrelated to the colon, such as breast cancer (BARD121) or rhabdoid tumors (SMARCA422). CTNNA1 is not on hereditary colorectal cancer and/or polyposis susceptibility testing panels within the GTR, and was assigned a classification of No Reported Evidence after a PubMed search returned only clinical evidence of somatic variants rather than germline/constitutional variants.
Within the NIH GTR13, we identified 33 diagnostic panels for hereditary colorectal cancer and/or polyposis susceptibility, and 26/40 (65%) genes evaluated by our GCP were found on 1 or more panels (Figure 2A). Although over half of the genes that exist on hereditary colorectal cancer/polyposis testing panels received a Definitive classification (14/26 genes, 53.8%), we also found 4/26 (15.4%) genes on these panels to have Limited evidence. Additionally, we found 2/26 (7.7%) genes to have Disputed (EXO1) or Refuted (PMS1) evidence, identified on 1 and 4 diagnostic testing panels, respectively (Figure 2A). EXO1 had variants previously identified in colorectal cancer probands, however with the advent of large population databases such as ExAC23 and gnomAD24, the maximum minor allele frequencies for the variants were above our threshold of 0.001. Similar to EXO1, one missense variant found in PMS125 was at a minor allele frequency of 0.046 in the African subpopulation, and another proband harbored a PMS1 null variant in addition to a deletion of MSH2 exons 1–726.
Within the Clinical Validity framework, the clinical validity matrix is used to assign points to both to genetic evidence (i.e. probands, segregation evidence, case-control data) and experimental evidence supporting a gene-disease association11. A proband with null/predicted null variant (i.e. loss-of-function) is typically assigned 1.5 points for autosomal dominant genetic disorders11. When curating ATM, we found many examples of frameshift/null/ canonical splicing variants that were found in CRC probands, but no convincing case-control or segregation evidence. Assigning 1.5 points of genetic evidence to each proband would have maximized the genetic evidence based on few case-level associations, and would have allowed a gene with no experimental evidence to reach a Strong classification quickly. To avoid this, and promote a more robust classification, we have modified the semiquantitative system by reducing the number of points per case to 0.5. These changes assure that to become Definitive, a gene-disease association must have both genetic and experimental evidence, and increase the number of cases required to reach the case-level point cap. Using this rule, ATM received 7 points, enough for the lowest score in the Moderate category. The GCP reached consensus that the current level of evidence was consistent with this. We thought that this was reasonable to apply to every cancer gene. For instance, when applying this rule to BLM, we found 6.3 points of case-level evidence, 2 points of case-control evidence, and 6 points of experimental evidence for a total of 14.3 points. Given the association of BLM with hereditary colorectal cancer susceptibility was first established in 2015, the association was considered Definitive (Table 1, Figure 1).
Three genes that we curated for hereditary colorectal cancer, but did not classify, were FLCN, TP53 and CDH1. While we found that these genes are found on 3 (9.1%), 17 (51.5%) and 11 (33.3%) testing panels, respectively (Figure 2A), this is presumably because of isolated reports of early onset colorectal cancer in probands with Birt-Hogg-Dubé (FLCN), Li-Fraumeni Syndrome (TP53) or in families with Hereditary Diffuse Gastric Cancer (CDH1). We reviewed the small amount of evidence supporting hereditary colorectal cancer susceptibility and found that most probands with pathogenic TP53 variants that developed early-onset colorectal cancer existed within Li-Fraumeni Syndrome families (Table 2). However, in a recent case-control study of ~1000 early-onset (age at diagnosis ≤55 years) colorectal cancer cases and genetic ancestry-matched controls27, none of these genes were significantly enriched for null/predicted variants nor nonsynonymous variants in colorectal cancer cases27 suggesting that these genes play small roles in hereditary CRC outside the well-described syndromes. Thus, we did not classify associations of these genes with isolated hereditary colorectal cancer susceptibility.
Discussion
Through the application of ClinGen’s clinical validity framework, this study provides a systematic evaluation of the strength of association of 42 gene-disease pairs implicated in either syndromic hereditary colorectal cancer or polyposis susceptibility, or isolated susceptibility to either disease. The classic genes associated with inherited susceptibilities to either colorectal cancer or polyposis (e.g. APC, MUTYH, MLH1, MSH2, etc.) are well established in the clinical cancer genetics field, and as expected, resulted in a Definitive classification within our framework. We expect that many of the current Strong and Moderate classifications will change over time as evidence accrues. We anticipate the majority to increase to Strong or Definitive, as many of the associations only need a few more points to increase in classification (Figure 1). The Limited genes seemed to lack a clear unifying theme beyond being observed in a few reported probands, and it remains to be determined whether these classifications will increase or decrease in the future. While the rationale for listing Limited genes on genetic testing panels remains unknown, these genes may have been added when the association was first published without being later excluded when the association did not increase in strength. As knowledge of these genes and their variants will undoubtedly advance28,29, we expect classifications to change and new ones to be established as novel candidate genes are identified, enabling Limited genes to potentially increase in strength with the accumulation of more evidence, or perhaps regress to a lower classification.
This semiquantitative approach is novel for gene curation in colon cancer susceptibility and hereditary polyposis. The classifications we derived using the ClinGen framework provide a foundation that can be evaluated over time and could change as more data accumulate. These gene-disease association evaluations are not exhaustive. Since we expect this to be an iterative process, it should not be construed as definitive. We hope that this framework will prove valuable to the broader genetics community and help build a consensus for a generally accepted process for assessing gene-disease associations in cancer.
To safeguard against unintentional bias in the classification system, two separate and independent curators were used for each gene-disease pair. Discussion of the evidence among all curators and experts followed, culminating with approval of the final classifications by the Hereditary Cancer Working Group. To reduce conflicts of interest, none of the curators or experts has been listed as a discoverer of any of the genes curated in this work, and none is currently studying any of the genes that show less than Definitive evidence.
Our assessment of the available evidence was not without challenges. Segregation data could be quantitated and points assigned for most gene-disease pairs. However in the case of some families where only affected relatives were genotyped, the concern arose about potentially missed non-segregations, wherein some of the unaffected older relatives could have been positive for the variant under assessment. In the absence of such data, downgrading segregation points was deemed appropriate. Additionally, not all consensus splice-site variants were functionally verified to impact transcript splicing. Without knowing whether a transcript specific to the colonic epithelium was impacted by a splice-site variant, determining point values for probands with these variants was difficult at times. Another challenge pertained to functional alteration evidence within colorectal cancer cell lines. Unless an assay was specific to a particular biological pathway that is clearly linked to colorectal cancer and/or polyposis biology (e.g. mismatch repair assays or assays focused on the WNT pathway), our GCP applied a conservative approach by either downgrading points, or not scoring the evidence. Future versions of the ClinGen Standard Operating Procedures may include greater detail to address these challenges15.
Defining polyposis susceptibility as a disease in probands and their relatives was particularly challenging for gene-disease pairs not associated with established hereditary polyposis syndromes. Although >20 colorectal adenomas is often used as a clinical indicator of a genetic polyposis syndrome (e.g. FAP and MUTYH-associated polyposis30,31), it is unclear how often polyp numbers between 2 and 20 represent hereditary predisposition or phenocopies. When only 2–10 colorectal adenomas were reported in some carriers, it became difficult to assess whether the proband under evaluation was affected with hereditary polyposis susceptibility, or if the low polyp count was a phenocopy because of the high prevalence of polyps in individuals undergoing screening colonoscopy in the general population32,33. Conservative assessment of the literature with clinical domain experts was crucial to prevent over classification of gene-disease associations where mild polyposis (i.e. ≥2 colorectal adenomas) was evident. Overall, larger genotype-phenotype correlation studies will enable further discrimination between polyp count and disease diagnosis.
Clinical and diagnostic communities utilize current guidelines that guide clinical care of patients according to disease risk6,34, but in contrast to expert consensus guidelines, the approach here allows clinicians to inspect the primary datasets that support gene curation. Thus, the information we provide here aims to augment these communities’ assessments of genes listed on testing panels for hereditary colorectal cancer and/or polyposis susceptibility. Our clinical validity classifications help clarify some issues in interpreting genetic testing results by pointing out which genes have a low versus high prior probability of being associated with these phenotypes. While our classifications do not correlate directly with disease risk, we did include both case-control studies and risk studies in our evaluations (e.g. relative risk, lifetime risk, etc) to optimize interpretation in a transparent manner that is not always apparent in the long lists of genes reported on clinical testing panels.
Although recommendations for gene content on current diagnostic panels is outside the purview of this GCP, our clinical validity assessment highlights that laboratories and physicians receiving these results should be aware that a number of genes included in clinically available hereditary colorectal cancer/polyposis susceptibility testing panels have only Limited evidence for disease association (Figure 2C). The possible harms and costs that come from having genes with Limited evidence present on clinical testing panels may include unnecessary screening and visits, with accompanying costs and potential procedure risks. Unnecessary patient anxiety and increased time spent communicating results to patients are additional harms that may stem from the inclusion of genes with Limited evidence on clinical testing panels. Alternatively, an advantage of including Limited genes on testing panels is the opportunity to accumulate more evidence and potentially raise the classification of these genes in the future. Along these lines, our work highlights some genes such as CDKN1B and FAN1 that have an excess of experimental evidence compared to genetic evidence, indicating that these genes may be good candidates for future clinical studies to further clarify the risk of these diseases.
Ultimately, our evaluation of these gene-disease relationships using the ClinGen Clinical Validity framework is one step in an iterative process toward helping clinicians and diagnostic laboratories in communicating genetic testing results to patients and assessing the clinical relevance of genes listed on their hereditary colorectal cancer and/or polyposis susceptibility testing panels. This important work also augments the work of the International Society for Gastrointestinal Hereditary Tumors (InSiGHT) in variant classification35 and of other groups researching hereditary colorectal cancer and polyposis susceptibility, highlighting areas for focus of future studies to either advance genes from Limited/Moderate to Strong/Definitive or to refute weak evidence.
Supplementary Material
Acknowledgments
We are very grateful to members of the ClinGen Hereditary Cancer Clinical Domain Working Group Executive Committee and the ClinGen Gene Curation Working Group for their expertise and guidance in assessing these gene-disease curation pairs. We would also like to Jonathan S. Berg, MD, PhD and Courtney Thaxton, PhD at the University of North Carolina at Chapel Hill for their thoughtful review and edits of this manuscript. This Gene Curation Panel is funded by National Human Genome Research & National Cancer Institutes (1U41HG006834, 1U01HG007437, 1U01HG007436, HHSN261200800001E, U41HG009650). The following authors have no conflicts of interest to disclose: MSG, SJ, KL, JLM, DIR, BAS, RJS, ZKS, BAT, MT and LZ. The following authors have made extensive contributions to the literature relative to gene curation and genetic testing: KO. The following authors are an employee, trainee or consultant for a commercial laboratory that offers hereditary cancer panel testing: MJF, SAJ, MER and SEP.
Footnotes
Conflict of Interest:
The following authors have no conflicts of interest to disclose: Marc S. Greenblatt, Sharisse Jimenez, Kristy Lee, Jennifer L. McGlaughon, Deborah I. Ritter, Bryce A. Seifert, Ryan J. Schmidt, Zsophia K. Stadler, Bryony A. Thompson, Mackenzie Trapp and Liying Zhang.
The following author has made extensive contributions to the literature relative to gene curation and genetic testing: Kenneth Offit,
The following authors are an employee, trainee or consultant for a commercial laboratory that offers hereditary cancer panel testing: Matthew J. Ferber is employed at clinical molecular lab at Mayo Medical Laboratory. Sarah A. Jackson and Maegan E. Roberts are employed at GeneDx Laboratories that provides hereditary cancer panel testing. Sharon E. Plon who consults for the Baylor College of Medicine Molecular Diagnostic Laboratory.
References
- 1.Cancer Statistics Review, 1975–2014 - SEER Statistics. Available at: https://seer.cancer.gov/csr/1975_2014/. Accessed December 20, 2017.
- 2.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med 2003; 348(10):919–32. DOI: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 3.Järvinen HJ, Aarnio M, Mustonen H, et al. Controlled 15-year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer. Gastroenterology 2000;118(5):829–834. [DOI] [PubMed] [Google Scholar]
- 4.Järvinen HJ, Renkonen-Sinisalo L, Aktán-Collán K, Peltomäki P, Aaltonen LA, Mecklin J-P. Ten years after mutation testing for Lynch syndrome: cancer incidence and outcome in mutation-positive and mutation-negative family members. J Clin Oncol 2009;27(28):4793–4797. [DOI] [PubMed] [Google Scholar]
- 5.American Gastroenterological Association. American Gastroenterological Association medical position statement: hereditary colorectal cancer and genetic testing. Gastroenterology 2001;121(1):195–197. [DOI] [PubMed] [Google Scholar]
- 6.National Comprehensive Cancer Network (NCCN), 2017. Version 3.2017 Genetic/Familial High-Risk Assessment: Colorectal. Available at: https://www.nccn.org/professionals/physician_gls/pdf/genetics_colon.pdf Accessed November 21, 2017.
- 7.Chong JX, Buckingham KJ, Jhangiani SN, et al. The Genetic Basis of Mendelian Phenotypes: Discoveries, Challenges, and Opportunities. Am J Hum Genet 2015;97(2):199–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robson ME, Bradbury AR, Arun B, et al. American Society of Clinical Oncology Policy Statement Update: Genetic and Genomic Testing for Cancer Susceptibility. J Clin Oncol 2015;33(31):3660–3667. [DOI] [PubMed] [Google Scholar]
- 9.Teutsch SM, Bradley LA, Palomaki GE, et al. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Initiative: methods of the EGAPP Working Group. Genet Med 2009;11(1):3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rehm HL, Berg JS, Brooks LD, et al. ClinGen--the Clinical Genome Resource. N Engl J Med 2015;372(23):2235–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strande NT, Riggs ER, Buchanan AH, et al. Evaluating the Clinical Validity of Gene-Disease Associations: An Evidence-Based Framework Developed by the Clinical Genome Resource. Am J Hum Genet 2017;100(6):895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacArthur DG, Manolio TA, Dimmock DP, et al. Guidelines for investigating causality of sequence variants in human disease. Nature 2014;508(7497):469–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubinstein WS, Maglott DR, Lee JM, et al. The NIH genetic testing registry: a new, centralized database of genetic tests to enable access to comprehensive information and improve transparency. Nucleic Acids Res 2013;41(Database issue):D925–935. doi: 10.1093/nar/gks1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Results - ClinGen ∣ Clinical Genome Resource. Available at: https://www.clinicalgenome.org/curation-activities/gene-disease-validity/results/. Accessed December 15, 2017.
- 15.Standard Operating Procedures and Appendices - ClinGen ∣ Clinical Genome Resource. Available at: https://www.clinicalgenome.org/curation-activities/gene-disease-validity/educational-and-training-materials/standard-operating-procedures/. Accessed December 15, 2017.
- 16.Jaeger E, Leedham S, Lewis A, et al. Hereditary mixed polyposis syndrome is caused by a 40-kb upstream duplication that leads to increased and ectopic expression of the BMP antagonist GREM1. Nat Genet 2012;44(6):699–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venkatachalam R, Ligtenberg MJ, Hoogerbrugge N, et al. Germline epigenetic silencing of the tumor suppressor gene PTPRJ in early-onset familial colorectal cancer. Gastroenterology 2010;139(6):2221–2224. [DOI] [PubMed] [Google Scholar]
- 18.Basso G, Bianchi P, Malesci A, Laghi L. Hereditary or sporadic polyposis syndromes. Best Pract Res Clin Gastroenterol 2017;31(4):409–417. [DOI] [PubMed] [Google Scholar]
- 19.Xiang H, Geng X, Ge W, Li H. Meta-analysis of CHEK2 1100delC variant and colorectal cancer susceptibility. Eur J Cancer 2011;47(17):2546–2551. [DOI] [PubMed] [Google Scholar]
- 20.Palles C, Cazier J-B, Howarth KM, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet 2013;45(2):136–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Couch FJ, Shimelis H, Hu C, et al. Associations Between Cancer Predisposition Testing Panel Genes and Breast Cancer. JAMA Oncol 2017;3(9):1190–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneppenheim R, Frühwald MC, Gesk S, et al. Germline nonsense mutation and somatic inactivation of SMARCA4/BRG1 in a family with rhabdoid tumor predisposition syndrome. Am J Hum Genet. 2010;86(2):279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536(7616):285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.gnomAD browser. Available at: http://gnomad.broadinstitute.org/. Accessed December 20, 2017.
- 25.Wang Q, Lasset C, Desseigne F, et al. Prevalence of germline mutations of hMLH1, hMSH2, hPMS1, hPMS2, and hMSH6 genes in 75 French kindreds with nonpolyposis colorectal cancer. Hum Genet 1999;105(1–2):79–85. [DOI] [PubMed] [Google Scholar]
- 26.Liu T, Yan H, Kuismanen S, et al. The role of hPMS1 and hPMS2 in predisposing to colorectal cancer. Cancer Res 2001;61(21):7798–7802. [PubMed] [Google Scholar]
- 27.Chubb D, Broderick P, Dobbins SE, et al. Rare disruptive mutations and their contribution to the heritable risk of colorectal cancer. Nat Commun 2016;7:11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoffel EM, Koeppe E, Everett J, et al. Germline Genetic Features of Young Individuals With Colorectal Cancer. Gastroenterology 2018;154(4):897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung DC. Genetic Testing and Early Onset Colon Cancer. Gastroenterology 2018;154(4):788–789. [DOI] [PubMed] [Google Scholar]
- 30.Perencevich M, Stoffel EM. A multidisciplinary approach to the diagnosis and management of multiple colorectal polyps. Gastroenterol Hepatol (N Y) 2011;7(6):420–423. [PMC free article] [PubMed] [Google Scholar]
- 31.Nielsen M, Lynch H, Infante E, Brand R. MUTYH-Associated Polyposis In: Adam MP, Ardinger HH, Pagon RA, et al. , editors. GeneReviews®. Seattle (WA): University of Washington, Seattle; 1993. Available at: http://www.ncbi.nlm.nih.gov/books/NBK107219/. Accessed December 20, 2017. [Google Scholar]
- 32.Penn E, Garrow D, Romagnuolo J. Influence of race and sex on prevalence and recurrence of colon polyps. Arch Intern Med 2010;170(13):1127–1132. [DOI] [PubMed] [Google Scholar]
- 33.Winawer SJ, Zauber AG, O’Brien MJ, et al. The National Polyp Study. Design, methods, and characteristics of patients with newly diagnosed polyps. The National Polyp Study Workgroup. Cancer 1992;70(5 Suppl):1236–1245. [DOI] [PubMed] [Google Scholar]
- 34.Gupta S, Provenzale D, Regenbogen SE, et al. NCCN Guidelines Insights: Genetic/Familial High-Risk Assessment: Colorectal, Version 3.2017. J Natl Compr Canc Netw 2017;15(12):1465–1475. [DOI] [PubMed] [Google Scholar]
- 35.Thompson BA, Spurdle AB, Plazzer JP, et al. Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat Genet 2014;46(2):107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.