Abstract
Background
Oncologists use patients’ life expectancy to guide decisions and may benefit from a tool that accurately predicts prognosis. Existing prognostic models generally use only a few predictor variables. We used an electronic medical record dataset to train a prognostic model for patients with metastatic cancer.
Methods
The model was trained and tested using 12 588 patients treated for metastatic cancer in the Stanford Health Care system from 2008 to 2017. Data sources included provider note text, labs, vital signs, procedures, medication orders, and diagnosis codes. Patients were divided randomly into a training set used to fit the model coefficients and a test set used to evaluate model performance (80%/20% split). A regularized Cox model with 4126 predictor variables was used. A landmarking approach was used due to the multiple observations per patient, with t0 set to the time of metastatic cancer diagnosis. Performance was also evaluated using 399 palliative radiation courses in test set patients.
Results
The C-index for overall survival was 0.786 in the test set (averaged across landmark times). For palliative radiation courses, the C-index was 0.745 (95% confidence interval [CI] = 0.715 to 0.775) compared with 0.635 (95% CI = 0.601 to 0.669) for a published model using performance status, primary tumor site, and treated site (two-sided P < .001). Our model’s predictions were well-calibrated.
Conclusions
The model showed high predictive performance, which will need to be validated using external data. Because it is fully automated, the model can be used to examine providers’ practice patterns and could be deployed in a decision support tool to help improve quality of care.
Most patients with metastatic solid tumors cannot be cured of their disease. Instead, treatments aim to improve quality and quantity of life. At the end of life, quality of life decreases along with the benefit-to-risk ratio of treatments like systemic therapy (1,2). Oncologists use life expectancy to guide treatment decisions but can be overly optimistic when predicting survival (3–6). Interventions such as intensive care unit stays and systemic therapy administration are commonly performed close to death (7,8). Conversely, palliative care and transition from active treatment are not always discussed early enough (9).
Several research groups have worked to develop prognostic models for patients with metastatic cancer (6,10–16). Deploying a prognostic model in the clinic could help doctors discuss goals of care with their patients at the appropriate time (9). Existing prognostic models have limited accuracy and generally require manual entry of variables such as performance status, which may hinder use.
There has recently been interest in using electronic medical record (EMR) data to create predictive models in oncology (17,18). We propose to use EMR data to train a prognostic model for patients with metastatic cancer. By including provider note text, important features such as performance status and symptom burden can be captured (11). By including thousands of patients, it is feasible to fit high-dimensional models with thousands of predictor variables; the most important variables can be found automatically. The model can be deployed in the clinic in an automated fashion (17).
In this article, we describe the methods used to train the prognostic model and then evaluate its performance in the general setting of metastatic cancer and the specific setting of palliative radiotherapy.
Methods
Patients
The prognostic model was trained on a database of adult patients seen in the Stanford Health Care system from 2008 to 2017. Clinical sites included one hospital, one freestanding cancer center, and several outpatient clinics. The database included data from the EMR (EPIC, Verona, WI), inpatient billing system, and institutional cancer registry. We identified patients with metastatic solid tumors. Visits from date of metastatic cancer diagnosis to death were analyzed. Date of last follow-up or death was determined using the EMR, Social Security Death Index, and cancer registry. For details of database construction and data quality validation, see the Supplementary Methods (available online). This study was approved by the Stanford University Institutional Review Board.
Prognostic Model
We developed a Cox proportional hazards model that takes as input structured and unstructured EMR data and outputs a predicted overall survival curve from the time of a visit. There were 4126 predictor variables, including laboratory values, vital signs, ICD-9 diagnosis codes, CPT procedure codes, medication administrations and prescriptions, and the text of inpatient and outpatient provider notes. Note text was represented using a bag-of-words approach, in which high-level document structure is discarded and the counts of many 1- to 2-word phrases are tallied for each note. This approach was shown to work well for clinical predictive modeling (19).
To predict survival time from a visit, all past data were used, with more recent information weighted more heavily. For details of predictor construction and a list of all predictors, see the Supplementary Information (available online). Each variable was standardized to a mean of 0 and standard deviation of 1. If a patient had no past data for a numeric variable (laboratory value or vital sign), the value was set to 0 (ie, the sample mean).
Patients were divided randomly with an 80%/20% split into a training set used to fit the model coefficients and a test set used to evaluate the model’s performance. A standard Cox model enables prediction of survival time from a single baseline time point, for instance, the time of metastatic cancer diagnosis. Instead, we used a dynamic prediction/landmarking approach to train and evaluate the model (20,21). This approach enables prediction of survival from baseline, but also from later time points using updated data, conditional on the patient having survived up to the later time. The date of the first visit after metastatic cancer diagnosis was designated as time t0. Landmark time points were set at times [t0, t0+ 0.5 years, …, t0+ 5 years]. For each landmark time point tLM, a dataset was constructed using all patients still in view at time tLM (not deceased or lost to follow-up) and their updated predictor data up to time tLM. These datasets were then “stacked” into a combined dataset. A Cox proportional hazards model was fit to the combined dataset. Landmark time point was included as a predictor so the baseline hazard function could vary smoothly with landmark time. Administrative censoring was enforced at thor=tLM+ 5 years. In some versions of the landmarking method (21), survival predictions are made only at thor, but we found that predictions were well calibrated from times tLM to thor, so we generated predicted survival curves spanning this time frame. We investigated allowing the β coefficients to vary depending on tLM but saw no improvement in model performance with this approach so therefore used time-fixed β in the final model. Because of the large number of predictors, L2 regularization was used to avoid overfitting, using the glmnet R package (22). The regularization strength (λ parameter) was chosen using 10-fold cross-validation on the training set. The λ value that maximized mean likelihood for the held-out folds was chosen. Because there were multiple observations per patient, the sandwich estimator could be used to calculate standard errors, but these were not needed in this study. Source code and simulated data are available at https://github.com/MGensheimer/prognosis-model.
Comparison With Existing Models Using Palliative Radiotherapy Courses
For test set patients receiving palliative radiotherapy, we compared the model’s performance with that of two existing prognostic models published by Chow et al. and Jang et al. (10,11) Radiation courses were identified using two databases that contained information including radiation-treated site and provider-rated performance status. Database 1 included 963 courses from 2008 to 2014 with specific fractionation schedules (8 Gy/1 fraction, 20 Gy/5 fractions, 30 Gy/10 fractions, 37.5 Gy/15 fractions) and excluded stereotactic techniques. Database 2 included 1173 courses from 2015 to 2016 and included a variety of fractionation schedules and both conventional and stereotactic techniques.
The two existing prognostic models both used the Cox proportional hazards model, similar to our approach, but both used hand-collected data and evaluated many fewer candidate predictors than we did. The model of Chow et al. (10) uses three predictors: Karnofsky performance status (KPS; >60 vs ≤60), primary tumor site (breast, prostate, lung, or other), and treated site (bone only vs others). In their article, patients were binned into three risk groups, but to create a fair comparison with our model, we used the exact model coefficients listed in their Table 2 to construct a linear predictor. Patients in our database 1 had Eastern Cooperative Oncology Group (ECOG) performance status recorded but not KPS, so we converted from ECOG to KPS using Table 1 in the Chow paper (10). The prognostic model of Jang et al. uses a single predictor: ECOG performance status (11). A few patients in our database 2 had KPS recorded but not ECOG performance status, so for these patients KPS was converted to ECOG performance status using a validated method (23).
Table 2.
Survival model coefficients for selected note text terms
| Term | Coefficient* |
|---|---|
| Symptoms/appearance | |
| Cachectic | 0.020 |
| Fatigued | 0.0059 |
| Ascites | 0.0085 |
| Completely asymptomatic | −0.0054 |
| Anxious | −0.0031 |
| Feel well | −0.0073 |
| Cancer location/response | |
| Disease progression | 0.012 |
| Leptomeningeal | 0.0067 |
| Mixed response | 0.014 |
| Innumerable pulmonary | 0.0046 |
| Minimal progression | −0.0012 |
| Oligometastatic | −0.0066 |
| Systemic therapy agents | |
| Nivolumab | −0.00065 |
| Liposomal doxorubicin† | 0.011 |
| Anastrozole† | −0.00051 |
| Leuprolide† | −0.0037 |
| Tamoxifen | −0.0034 |
A positive coefficient indicates shorter survival.
Brand name converted to generic name for display.
Table 1.
Patient characteristics
| Characteristic | Patients, n (%) |
|---|---|
| Total | 12 588 (100.0) |
| Median age, y (IQR)* | 63.5 (53.2–72.1) |
| Sex | |
| Female | 6384 (50.7) |
| Male | 6204 (49.3) |
| Primary site | |
| Breast | 1362 (10.8) |
| Endocrine | 209 (1.7) |
| Gastrointestinal | 3404 (27.0) |
| Genitourinary | 1407 (11.2) |
| Gynecologic | 799 (6.3) |
| Head and neck | 496 (3.9) |
| Skin | 425 (3.4) |
| Thorax | 2063 (16.4) |
| Other/multiple/unknown | 2449 (19.5) |
Age at diagnosis of metastatic disease.
Statistical Analysis
Our model’s performance was evaluated using Harrell’s C-index and calibration plots, as described in Royston et al., 2013 (24). To calculate C-index for a landmark time point, for each test set patient still at risk at that time point, the Cox model linear predictor was calculated using data from visits prior to the time point. To generate predicted survival curves, the baseline hazard function was estimated using the training set. We also used the model to predict survival from the time of specific radiation treatments. Because the model requires landmark time point to be specified, the landmark time closest to the treatment date was used.
For radiotherapy courses, we compared the model’s performance to two existing prognostic models. To measure difference in discrimination between models and obtain a P value, the rcorrp.cens function in R’s rms package was used. All statistical tests were two-sided and a P value of less than .05 was considered statistically significant. These data were not fully statistically independent because some patients had multiple radiation courses. However, because 224 of 296 patients had a single course, the effect of clustering was expected to be small and we assumed independence. To generate receiver operating characteristic curves at specific follow-up times, the survivalROC R package was used. To calculate 95% confidence intervals for the area under the receiver operating curve, we used bootstrapping with 10 000 samples.
Results
Patient Characteristics
We identified 13 523 adult patients with metastatic cancer seen at Stanford from 2008 to 2017. A total of 935 patients were excluded because of having no follow-up or death information or having died on the day of their only visit. The prognostic model was trained and evaluated using the remaining 12 588 patients. Table 1 lists patient characteristics.
Of the 12 588 analyzed patients, 7629 (60.6%) have died. From the first visit after metastatic cancer diagnosis, median follow-up was 14.5 months and median overall survival was 20.9 months (data not shown). Patients were seen for 384 402 daily visits after metastatic cancer diagnosis. Patients were hospitalized for 94 826 visits (24.7%). There were 1 390 032 provider notes, 12 876 137 lab values (200 most common labs), 1 451 740 vital signs, 357 981 diagnoses (500 most common codes), 1 162 164 procedures (500 most common codes), and 1 834 477 medication orders (500 most common medications).
Model Performance
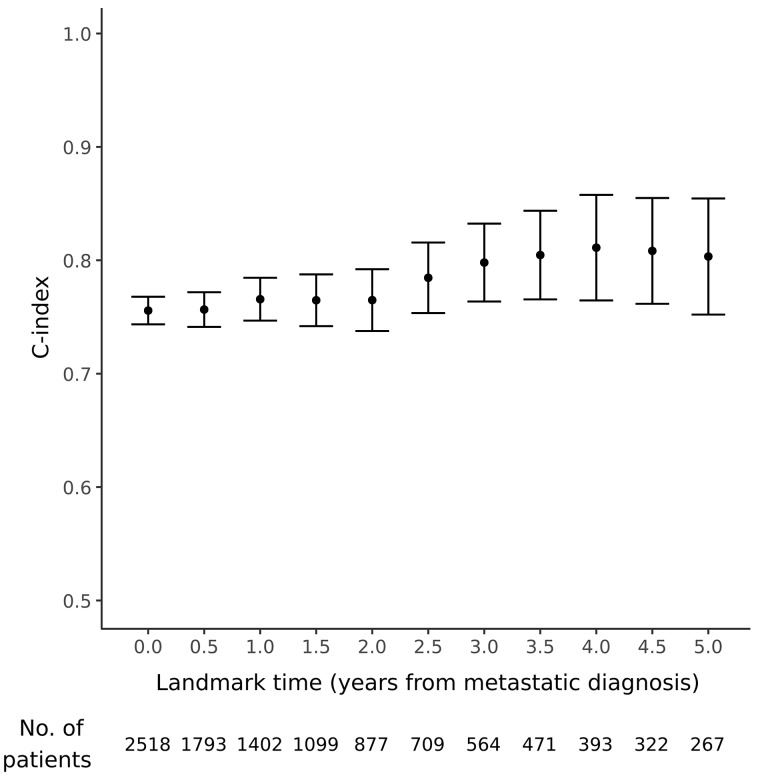
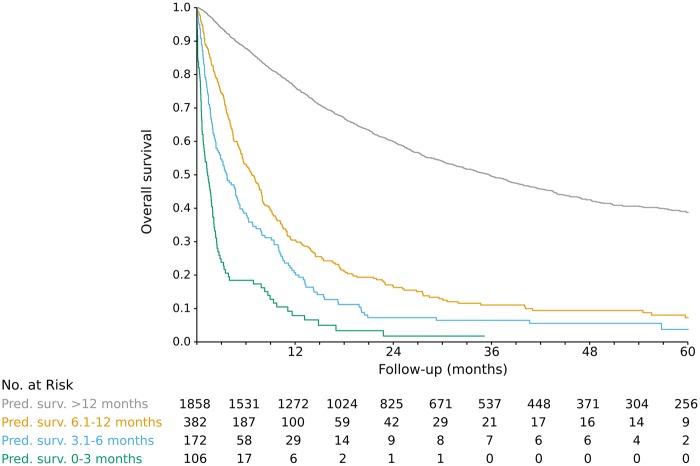
There were 10 070 patients in the training set and 2518 in the test set. Characteristics were well balanced between the two groups; see Supplementary Table 1 (available online). Because the test set patients were not used in model training, they were used to evaluate model performance. For each landmark time point (0–5 years after metastatic cancer diagnosis), the test set patients still in view at that time were ranked by predicted overall survival time using updated predictor data. Then, for each time point, predicted survival was compared with actual survival, with concordance measured using the C-index (0.5 = no better than random chance, 1.0 = perfect prediction). The model had good performance, with C-index ranging from 0.757 to 0.812 (mean 0.786) at various landmark time points (Figure 1). The predicted survival curves were fairly well calibrated (no systematic over- or underestimation of survival), as seen in Figure 2; Supplementary Figures 1 and 2; and Supplementary Table 2 (available online). For visual clarity, the model’s predictions were grouped into four clinically relevant bins, with median predicted survival of 0 to 3 months, 3.1 to 6 months, 6.1 to 12 months, and longer than 12 months. When binned in this way, model predictions were well calibrated. For instance, for test set patients at landmark time 0, the actual median survival for visits in these bins was (95% CI) = 1.3 (0.9 to 2.0), 3.7 (2.5 to 5.2), 6.7 (5.6 to 7.9), and 35.7 (31.8 to 39.1) months, respectively (Figure 2). Discrimination performance was similar when patients were compared only with other patients with the same primary tumor site (mean C-index across landmark time points of 0.729 to 0.866; Supplementary Figure 3, available online).
Figure 1.
Prognostic model performance at various landmark time points. The number of test set patients still at risk at each time point is listed below the figure. Error bars represent 95% confidence intervals.
Figure 2.
Predicted vs actual survival for 2518 test set patients at landmark time t0 (first visit after diagnosis of metastatic cancer). Patients were grouped into four clinically relevant bins by median predicted survival.
We examined which data source was most important to the model’s performance by re-fitting models using different subsets of predictors. As seen in Figure 3, note text was the most useful data source. We examined model coefficients for various note text terms and found that they aligned well with clinical intuition and established prognostic factors (Table 2). Supplementary Table 3 (available online) lists all predictors and their model coefficients.
Figure 3.
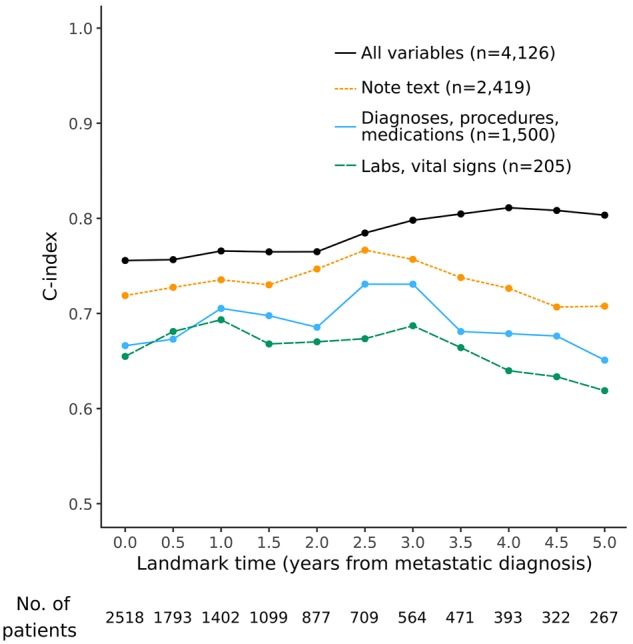
Test set performance of models that incorporate different subsets of the available predictor variables. The model using all variables had the highest performance, followed by the model using only note text. All models included two demographic variables (age and sex).
Comparison With Existing Models
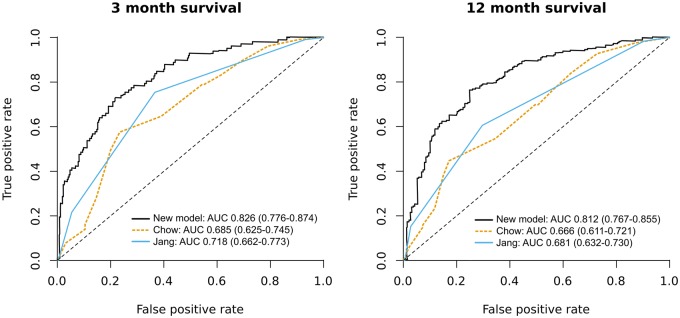
For patients receiving palliative radiotherapy, we compared the model’s performance with that of existing prognostic models. An accurate prognostic model could be useful in this scenario, because providers are known to overestimate survival for such patients and could use life expectancy to tailor factors such as number of radiation fractions (5,10). We used a database of palliative radiotherapy that had details recorded such as provider-rated performance status. Of 2136 radiation courses, 399 were in 296 test set patients and were analyzed. Characteristics of radiation courses are listed in Table 3. Median overall survival from the first day of radiation was 11.3 months. When comparing predicted vs actual survival, our model’s C-index was 0.745 (95% CI = 0.715 to 0.775). The C-index for the validated model of Chow et al. (10) using performance status, primary tumor site, and radiation-treated site was lower at 0.635 (95% CI = 0.601 to 0.669). The difference in discrimination performance between the two models was statistically significant (P < .001). The simple model of Jang et al. (11) using ECOG performance status alone also had inferior performance to our model, with C-index of 0.647 (95% CI = 0.615 to 0.678) (P < .001). Our model had superior discrimination at both short (3 months) and intermediate (12 months) time horizons (Figure 4). For prediction of 3-month survival, area under the receiver operating characteristic curve for our model, the model of Chow et al., and the model of Jang et al. was 0.826 (95% CI = 0.776 to 0.874), 0.685 (95% CI 0.625 to 0.745), and 0.718 (95% CI = 0.662 to 0.773), respectively. For prediction of 12-month survival, the corresponding values were 0.812 (95% CI = 0.767 to 0.855), 0.666 (95% CI = 0.611 to 0.721), and 0.681 (95% CI = 0.632 to 0.730), respectively.
Table 3.
Characteristics of 399 palliative radiation courses in test set patients*
| Characteristic | Patients, n (%) |
|---|---|
| Primary site | |
| Breast | 78 (19.5) |
| Prostate | 34 (8.5) |
| Lung | 125 (31.3) |
| Other | 162 (40.6) |
| Treated site | |
| Bone only | 210 (52.6) |
| Other | 189 (47.4) |
| ECOG performance status | |
| 0 | 22 (5.5) |
| 1 | 194 (48.6) |
| 2 | 146 (36.6) |
| 3 | 36 (9.0) |
| 4 | 1 (0.3) |
| Fractions | |
| 1 | 95 (23.8) |
| 2–5 | 98 (24.6) |
| 6–10 | 151 (37.8) |
| >10 | 55 (13.8) |
| Technique | |
| Conventional | 296 (74.2) |
| Stereotactic | 103 (25.8) |
*ECOG = Eastern Cooperative Oncology Group.
Figure 4.
Receiver operating characteristic curves for palliative radiation survival prediction. For each plot, a positive case was defined as a patient who lived for longer than the specified time. AUC = area under the curve. 95% confidence intervals for AUC are in parentheses.
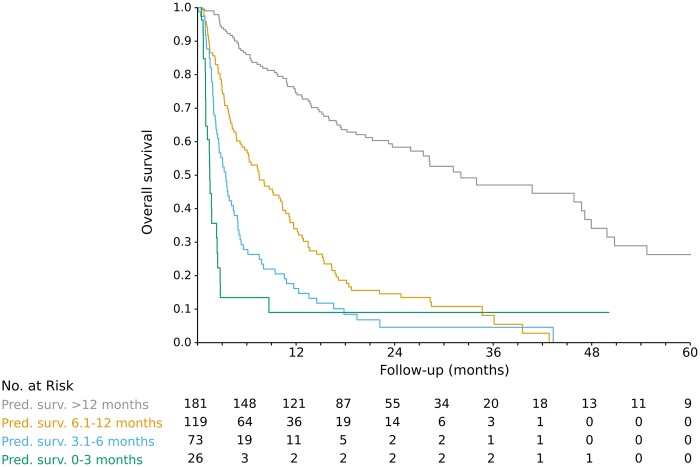
Our model’s predictions for the palliative radiotherapy patients were well calibrated: for the four predicted survival bins (0–3 months, 3.1–6 months, 6.1–12 months, >12 months), actual median survival (95% CI) was 1.5 (1.0 to 2.5), 3.5 (2.6 to 4.9), 7.5 (5.4 to 10.4), and 32.1 (26.0 to 48.0) months, respectively (Figure 5). Of the 26 radiation courses with model-predicted survival of 0 to 3 months, 18 (69.2%) had 10 or more fractions. Of the 73 courses with predicted survival of 3.1 to 6 months, 40 (54.8%) had 10 or more fractions.
Figure 5.
Predicted vs actual survival for 399 palliative radiation courses in test set patients.
Discussion
We used a large sample of patients with metastatic cancer to train a prognostic model with EMR data. The model bridges the gap between simpler models using provider-entered variables, which limits dataset size, and models using population-based data, which lack important information such as patients’ performance status (25). It is notable that the most useful data source was free text provider notes, because most existing prognostic models using large datasets use billing codes instead (25–27), which are often inaccurate (28). The usefulness of note text for predictive modelling has been seen in other recent studies (29–31), though this may be the first example in oncology. The note text features that influenced survival predictions were sensible. For instance, when notes mentioned systemic therapies used as first-line treatment of metastatic breast cancer (anastrozole, tamoxifen), survival was longer than when notes mentioned drugs used as later-line treatment for poorer prognosis cancers (pegylated doxorubicin, nivolumab).
The model showed a statistically significant increase in accuracy compared with prior prognostic models for metastatic cancer patients. C-index on the test set was 0.786 (averaged over landmark time points), and calibration was good. For patients receiving palliative radiotherapy, C-index was higher than that of two highly cited models used for patients receiving palliative treatments: 0.745 vs 0.635 to 0.647. Papers describing other prognostic models have also shown lower discrimination performance than ours. Three models for patients receiving palliative radiotherapy had validation set C-index of 0.59 to 0.72 (10,12,15). Two studies in patients referred to either a palliative care or hospice service showed C-index of 0.62 to 0.73 (11,14). It is impossible to make definitive comparisons between studies, since different groups of patients were used to evaluate the models. Also, we were unable to compare our model with less parsimonious models than those of Chow and Jang (10,11), such as one developed in the multicenter PiPS study (32), because we lacked data such as mental test scores.
The use of EMR data allows the model to be automated and enables several novel applications. The model can be used to examine practice patterns and identify areas of potential improvement. For patients receiving palliative radiotherapy who had model-predicted survival 6 months or less, 59% had at least 10 radiation fractions. Many of these patients could be treated with a single fraction, which provides equivalent results to longer schedules for poor-prognosis patients and has fewer side effects (33). Other areas of care that could be examined include timing of palliative care referral and use of systemic therapy at the end of life (7–9). It can be misleading to identify a group of deceased patients and examine practice patterns during a time period prior to death (34). This ignores the unpredictability of survival, with some healthier patients dying unexpectedly and some sicker patients surviving for years. Because our model enables estimation of the patient’s prognosis using information that was available to the providers at that time, the relationship of practice patterns to prognosis can be examined in a more rigorous way.
The model could also be used as part of a decision support tool, with predictions displayed to providers (17). This could help remind providers to discuss goals of care with their patients and refer to appropriate resources. It is straightforward to understand the reasons for each prediction (in contrast to nonlinear models like neural networks). As each predictor variable was centered before the model was fit, the element-wise product of the model coefficients and a visit’s variables can be interpreted as the influence of each variable on predicted survival. The variables with the most extreme influence values could be displayed to the provider.
This work has several limitations. First, the model was trained using patients seen in one health system, so it likely contains biases due to idiosyncrasies of our patient population and treatment algorithms. Although a mix of patient ages, sex, and cancer type was represented in the training data, the model may generalize poorly to different practice settings such as a rural private practice. Because only care given at Stanford was captured in the structured data, the model performance could drop for patients who mostly received care at other centers. Techniques such as multiple imputation could be used to reduce the effect of such missing data; for example, note text data could be used to impute missing laboratory values (35). It will be important to perform external validation using data from other centers. Better infrastructure for sharing data in common formats between institutions is needed.
Second, patients were identified by a mix of manual and automated methods. Patients who have initially localized disease, then develop metastatic disease and are too frail to receive any cancer treatment, may not be captured well (36). This may cause the model to perform poorly for older patients who do not have initially metastatic disease.
Finally, the model ignores interactions between variables. For instance, the current method of note text analysis discards sentence- and document-level structure. Therefore, sequencing of therapies and changes in patient condition are poorly represented. Neural networks can capture more complicated structure and may improve performance (30,37–39).
In conclusion, using EMR data, it was possible to train a high-performance automated prognostic model for metastatic cancer patients. We plan to deploy the model in clinical trials as part of a decision support tool to help physicians and patients choose treatments and decide on goals of care.
Funding
This work was supported in part by the National Cancer Institute (Cancer Center Support Grant no. 5P30CA124435); and National Institutes of Health/National Center for Research Resources (CTSA award no. UL1 RR025744).
Notes
Affiliations of authors: Department of Radiation Oncology (MFG, SA, SAD, PP, EP, ACK, DTC) and Department of Biomedical Data Science (ASH, DJW, TJH, DLR) and Department of Statistics (TJH) and Department of Radiology (IB, DLR), Stanford University, Stanford, CA; Genentech, South San Francisco, CA (EC); Department of Medicine, Stanford University, Stanford, CA (KR); Department of Radiation Oncology, The University of Texas MD Anderson Cancer Center, Houston, TX (ACK).
The funders had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
MFG reports research funding from Varian Medical Systems and Philips Healthcare. DLR reports research funding from Philips Healthcare. DTC reports research funding and honoraria from Varian Medical Systems, and stock ownership in ViewRay.
Supplementary Material
References
- 1. Costantini M, Beccaro M, Higginson IJ.. Cancer trajectories at the end of life: is there an effect of age and gender? BMC Cancer. 2008;8:127.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mccarthy EP, Phillips RS, Zhong Z, Drews RE, Lynn J.. Dying with cancer: patients’ function, symptoms, and care preferences as death approaches. J Am Geriatr Soc. 2000;48(5 suppl):S110–S121. [DOI] [PubMed] [Google Scholar]
- 3. Fairchild A, Barnes E, Ghosh S, et al. International patterns of practice in palliative radiotherapy for painful bone metastases: evidence-based practice? Int J Radiat Oncol Biol Phys. 2009;755:1501–1510. [DOI] [PubMed] [Google Scholar]
- 4. Chow E, Davis L, Panzarella T, et al. Accuracy of survival prediction by palliative radiation oncologists. Int J Radiat Oncol Biol Phys. 2005;613:870–873. [DOI] [PubMed] [Google Scholar]
- 5. Hartsell WF, Desilvio M, Bruner DW, et al. Can physicians accurately predict survival time in patients with metastatic cancer? Analysis of RTOG 97-14. J Palliat Med. 2008;115:723–728. [DOI] [PubMed] [Google Scholar]
- 6. Krishnan M, Temel JS, Wright AA, Bernacki R, Selvaggi K, Balboni T.. Predicting life expectancy in patients with advanced incurable cancer: a review. J Support Oncol. 2013;112:68–74. [DOI] [PubMed] [Google Scholar]
- 7. Bekelman JE, Halpern SD, Blankart CR, et al. Comparison of site of death, health care utilization, and hospital expenditures for patients dying with cancer in 7 developed countries. JAMA. 2016;3153:272–283. [DOI] [PubMed] [Google Scholar]
- 8. Earle CC, Neville BA, Landrum MB, Ayanian JZ, Block SD, Weeks JC.. Trends in the aggressiveness of cancer care near the end of life. J Clin Oncol. 2004;222:315–321. [DOI] [PubMed] [Google Scholar]
- 9. Bernacki R, Obermeyer Z.. A need for more, better, and earlier conversations with cancer patients about goals of care. Am J Managed Care. 2015;21(s6):166–167. [Google Scholar]
- 10. Chow E, Abdolell M, Panzarella T, et al. Predictive model for survival in patients with advanced cancer. J Clin Oncol. 2008;2636:5863–5869. [DOI] [PubMed] [Google Scholar]
- 11. Jang RW, Caraiscos VB, Swami N, et al. Simple prognostic model for patients with advanced cancer based on performance status. J Oncol Pract. 2014;105:e335–e341. [DOI] [PubMed] [Google Scholar]
- 12. Westhoff PG, De Graeff A, Monninkhof EM, et al. An easy tool to predict survival in patients receiving radiation therapy for painful bone metastases. Int J Radiat Oncol Biol Phys. 2014;904:739–747. [DOI] [PubMed] [Google Scholar]
- 13. Wallington M, Saxon EB, Bomb M, et al. 30-day mortality after systemic anticancer treatment for breast and lung cancer in England: a population-based, observational study. Lancet Oncol. 2016;179:1203–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maltoni M, Scarpi E, Pittureri C, et al. Prospective comparison of prognostic scores in palliative care cancer populations. Oncologist. 2012;173:446–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krishnan MS, Epstein-Peterson Z, Chen YH, et al. Predicting life expectancy in patients with metastatic cancer receiving palliative radiotherapy: the TEACHH model. Cancer. 2014;1201:134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ramchandran KJ, Shega JW, Von Roenn J, et al. A predictive model to identify hospitalized cancer patients at risk for 30-day mortality based on admission criteria via the electronic medical record. Cancer. 2013;11911:2074–2080. [DOI] [PubMed] [Google Scholar]
- 17. Sledge GW, Hudis CA, Swain SM, et al. ASCO’s approach to a learning health care system in oncology. J Oncol Pract. 2013;93:145–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miller RS. CancerLinQ update. J Oncol Pract. 2016;1210:835–837. [DOI] [PubMed] [Google Scholar]
- 19. Boag W, Doss D, Naumann T, Szolovits P.. What’s in a note? Unpacking predictive value in clinical note representations. AMIA Jt Summits Transl Sci Proc. 2018;2017:26–34. [PMC free article] [PubMed] [Google Scholar]
- 20. Van Houwelingen HC. Dynamic prediction by landmarking in event history analysis. Scand J Stat. 2007;341:70–85. [Google Scholar]
- 21. Putter H. The landmark approach: an introduction and application to dynamic prediction in competing risks. Talk presented at Dynamic Prediction Workshop; October 10, 2013; Bordeaux, France. http://www.canceropole-gso.org/download/fichiers/2777/4_Putter.pdf. Accessed April 25, 2018.
- 22. Simon N, Friedman J, Hastie T, Tibshirani R.. Regularization paths for Cox’s proportional hazards model via coordinate descent. J Stat Soft. 2011;395:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ma C, Bandukwala S, Burman D, et al. Interconversion of three measures of performance status: an empirical analysis. Eur J Cancer. 2010;4618:3175–3183. [DOI] [PubMed] [Google Scholar]
- 24. Royston P, Altman DG.. External validation of a Cox prognostic model: principles and methods. BMC Med Res Methodol. 2013;13:33.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pfister DG, Rubin DM, Elkin EB, et al. Risk adjusting survival outcomes in hospitals that treat patients with cancer without information on cancer stage. JAMA Oncol. 2015;19:1303–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Makar M, Ghassemi M, Cutler DM, Obermeyer Z.. Short-term mortality prediction for elderly patients using medicare claims data. Int J Mach Learn Comput. 2015;53:192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nguyen P, Tran T, Wickramasinghe N, Venkatesh S.. Deepr: a convolutional net for medical records. IEEE J Biomed Health Inform. 2017;211:22–30. [DOI] [PubMed] [Google Scholar]
- 28. Jollis JG, Ancukiewicz M, Delong ER, Pryor DB, Muhlbaier LH, Mark DB.. Discordance of databases designed for claims payment versus clinical information systems. Implications for outcomes research. Ann Intern Med. 1993;1198:844–850. [DOI] [PubMed] [Google Scholar]
- 29. Weissman GE, Hubbard RA, Ungar LH, et al. Inclusion of unstructured clinical text improves early prediction of death or prolonged ICU stay. Crit Care Med. 2018;467:1125–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rajkomar A, Oren E, Chen K, et al. Scalable and accurate deep learning with electronic health records. Npj Digital Med. 2018;11:18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kwong C, Ling AY, Crawford MH, Zhao SX, Shah NH.. A clinical score for predicting atrial fibrillation in patients with cryptogenic stroke or transient ischemic attack. Cardiology. 2017;1383:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gwilliam B, Keeley V, Todd C, et al. Development of prognosis in palliative care study (PiPS) predictor models to improve prognostication in advanced cancer: prospective cohort study. BMJ. 2011;343:d4920.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hartsell WF, Scott CB, Bruner DW, et al. Randomized trial of short- versus long-course radiotherapy for palliation of painful bone metastases. J Natl Cancer Inst. 2005;9711:798–804. [DOI] [PubMed] [Google Scholar]
- 34. Bach PB, Schrag D, Begg CB.. Resurrecting treatment histories of dead patients: a study design that should be laid to rest. JAMA. 2004;29222:2765–2770. [DOI] [PubMed] [Google Scholar]
- 35. Spratt M, Carpenter J, Sterne JA, et al. Strategies for multiple imputation in longitudinal studies. Am J Epidemiol. 2010;1724:478–487. [DOI] [PubMed] [Google Scholar]
- 36. Warren JL, Mariotto A, Melbert D, et al. Sensitivity of medicare claims to identify cancer recurrence in elderly colorectal and breast cancer patients. Med Care. 2016;548:e47–e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Graves A. Generating sequences with recurrent neural networks. arXiv. 2013;1308.0850. [Google Scholar]
- 38. Banerjee I, Gensheimer MF, Wood DJ, et al. Probabilistic prognostic estimates of survival in metastatic cancer patients (PPES-Met) utilizing free-text clinical narratives. Sci Rep. 2018;81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gensheimer MF, Narasimhan BA.. Simple discrete-time survival model for neural networks. arXiv. 2018;1805.00917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.