Abstract Abstract
DNA sequences from the nuclear LSU and ITS regions were used for phylogenetic analyses of Thelephorales with a focus on the stipitate hydnoid genera Hydnellum and Sarcodon. Analyses showed that Hydnellum and Sarcodon are distinct genera but that the current division, based on basidioma texture, makes Sarcodon paraphyletic with respect to Hydnellum. In order to make genera monophyletic several species are moved from Sarcodon to Hydnellum and the following new combinations are made: Hydnellumamygdaliolens, H.fennicum, H.fuligineoviolaceum, H.fuscoindicum, H.glaucopus, H.joeides, H.lepidum, H.lundellii, H.martioflavum, H.scabrosum, H.underwoodii, and H.versipelle. Basidiospore size seems to separate the genera in most cases. Hydnellum species have basidiospore lengths in the range 4.45−6.95 µm while the corresponding range for Sarcodon is 7.4−9 µm. S.quercinofibulatus deviates from this pattern with an average spore length around 6 µm. Neotropical Sarcodon species represent a separate evolutionary lineage.
Keywords: Phylogeny, stipitate hydnoid, taxonomy, Thelephorales , tooth fungi
Introduction
The order Thelephorales is a distinctive lineage of Agaricomycetes, well-known for its almost ubiquitous ectomycorrhizal life style (Tedersoo et al. 2010). Several species have stipitate hydnoid basidiomata (Fig. 1). They have traditionally been divided into four genera, Phellodon and Bankera with hyaline basidiospores, and Hydnellum and Sarcodon with yellow to brown tinted basidiospores (Maas Geesteranus 1975). In both cases the genera within each pair differ in basidioma structure, with Phellodon and Hydnellum being hard and dry, and Bankera and Sarcodon forming softer, fleshier basidiomata. This difference in texture is, however, difficult to assess and a series of recent molecular phylogenetic analyses, as outlined below, have indicated that the traditional, morphology-based generic limits are equivocal.
Figure 1.
Fruiting bodies of Hydnellum and SarcodonAHydnellumsuaveolensBH.aurantiacumCH.ferrugineumDSarcodonimbricatus.
In a recent comprehensive study of stipitate hydnoid species from south-eastern North America, Baird et al. (2013) found that Bankera could not be separated from Phellodon and the genera were hence combined into a more comprehensive Phellodon. The same study suggested that the generic limits of Sarcodon and Hydnellum need reassessment.
Nitare and Högberg (2012) examined the Nordic species of Sarcodon and included a preliminary molecular phylogeny for the species accepted in Sarcodon. Hydnellum species were also included in non-published test runs and found to be nested among Sarcodon species. They concluded that revisions of limits of both genera were probably necessary. Miscevic (2013) expanded on the results in Nitare and Högberg (2012) by including more sequences for each species and by including a selection of Hydnellum species in published phylogenies. The results were in congruence with Baird et al. (2013) with regard to overall tree topology and again the conclusion was that the limits of Sarcodon and Hydnellum need further study. A recent phylogenetic overview of Thelephorales (Vizzini et al. 2016) and a study of Hydnellum from the Mediterranean region (Loizides et al. 2016) came to similar conclusions, although Vizzini et al. (2016) did not include sequences from several Neotropical Sarcodon species described by Grupe et al. (2015, 2016).
In this paper we analyse ITS and nuclear LSU sequences from a wide selection of Thelephorales species with a focus on Hydnellum and Sarcodon in order to resolve the relationship between these two genera. We also make some nomenclatural changes that follow from the revision of genus circumscriptions. We demonstrate that Neotropical Sarcodon species do not cluster with temperate and boreal species and may be warranted as one or more new genera with more data.
Methods
For the phylogenetic analyses we compiled two datasets. The first dataset consists of nuclear LSU sequences from most genera in Thelephorales and from a majority of the Hydnellum and Sarcodon species occurring in Europe. For our two target genera we chose only sequences generated for this study from recently collected basidiomata. We deliberately excluded sequences from specimens identified as H.concrescens or H.scrobiculatum since these names seem to cover more than just two species and it is currently unclear how the names should be applied (Ainsworth et al. 2010). Since this study is positioned as a revision of the genus limits we were more interested in sequence quality control than a complete coverage of all species reported from Europe.
For our second dataset we chose a different strategy. Here we included ITS sequences from all Hydnellum and Sarcodon species represented among our own sequences and in GenBank as of December 1, 2018. The reason is that many species, and especially the recently described species from tropical regions, are only available as ITS sequences. However, we made no attempt to verify the identifications given in GenBank and do not endorse them as correct.
DNA was extracted from recent dried collections of basidiomata from North Europe. Voucher numbers, herbarium location, and GenBank numbers are given in Table 1. DNA extraction and PCR protocols follow Larsson et al (2018). Sequencing was either done in-house at University of Oslo, or as a commercial service by Macrogen Inc., South Korea. Assembly of chromatograms was done with Sequencher 5.2.4 (Gene Codes Co., Ann Arbor). Aligning was performed either manually using the editor in PAUP* 4.0a (Swofford 2002) or the software ALIVIEW 1.18 (Larsson 2014), or automatically utilising the L-INS-i strategy as implemented in MAFFT v. 7.017 (Katoh and Standley 2013), followed by manual adjustment.
Table 1.
Specimens sequenced or downloaded from GenBank. Herbarium acronyms follow Thiers. Sequences generated for this study are marked in bold.
| Species | Voucher | Herb. | GenBank number | |
|---|---|---|---|---|
| ITS | LSU | |||
| Amaurodonaquicoeruleus Agerer | Agerer & Bougher | M | AM490944 | AM490944 |
| Amaurodonviridis (Alb. & Schwein.:Fr.) J.Schröt | KH Larsson 14947b | O | MK602707 | MK602707 |
| Bankerafuligineoalba (J.C.Schmidt:Fr.) Pouzar | E Larsson 400-13 | GB | MK602708 | MK602708 |
| Bankeraviolascens (Alb. & Schwein.:Fr.) Pouzar | MV 130902 | GB | MK602709 | MK602709 |
| Boletopsisleucomelaena (Pers.:Fr.) Fayod | M Krikorev 140912 | GB | MK602710 | MK602710 |
| Hydnellumaurantiacum (Batsch:Fr.) P.Karst. | RG Carlsson 08-105 | GB | MK602711 | MK602711 |
| Hydnellum aurantiacum | E Bendiksen 177-07 | O | MK602712 | MK602712 |
| Hydnellum aurantiacum | O-F-295029 | O | MK602713 | MK602713 |
| Hydnellumauratile (Britzelm.) Maas Geest. | O-F-294095 | O | MK602714 | MK602714 |
| Hydnellum auratile | O-F-242763 | O | MK602715 | MK602715 |
| Hydnellum auratile | J Nitare 110926 | GB | MK602716 | MK602716 |
| Hydnellumcaeruleum (Hornem.:Fr.) P.Karst. | O-F-291490 | O | MK602717 | MK602717 |
| Hydnellum caeruleum | E Bendiksen 575-11 | O | MK602718 | MK602718 |
| Hydnellum caeruleum | E Bendiksen 584-11 | O | MK602719 | MK602719 |
| Hydnellumcomplicatum Banker | REB 71 | KC571711 | ||
| Hydnellumconcrescens (Pers.) Banker | K(M)134463 | K | EU784267 | |
| Hydnellumcristatum (G.F.Atk.) Stalpers | REB 169 | TENN | JN135174 | |
| Hydnellumcumulatum K.A.Harrison | SE Westmoreland 69 | AY569026 | ||
| Hydnellumcyanopodium K.A.Harrison | SE Westmoreland 85 | AY569027 | ||
| Hydnellumdiabolus Banker | KAH 13873 | MICH | AF351863 | |
| Hydnellumdianthifolium Loizides, Arnolds & P.-A.Moreau | ML61211HY | KX619419 | ||
| Hydnellumearlianum Banker | REB 375 | TENN | JN135179 | |
| Hydnellumferrugineum (Fr.:Fr.) P.Karst. | O-F-297319 | O | MK602720 | MK602720 |
| Hydnellum ferrugineum | E Larsson 356-16 | GB | MK602721 | MK602721 |
| Hydnellum ferrugineum | E Larsson 197-14 | GB | MK602722 | MK602722 |
| Hydnellumferrugipes Coker | REB 176 | KC571727 | ||
| Hydnellumgeogenium (Fr.) Banker | O-F-66379 | O | MK602723 | MK602723 |
| Hydnellum geogenium | O-F-296213 | O | MK602724 | MK602724 |
| Hydnellum geogenium | E Bendiksen 526-11 | O | MK602725 | MK602725 |
| Hydnellumgracilipes (P.Karst.) P.Karst. | E Larsson 219-11 | GB | MK602726 | MK602726 |
| Hydnellum gracilipes | GB-0113779 | GB | MK602727 | MK602727 |
| Hydnellummirabile (Fr.) P.Karst. | RG Carlsson 11-119 | GB | MK602728 | MK602728 |
| Hydnellum mirabile | E Larsson 170-14 | GB | MK602729 | MK602729 |
| Hydnellum mirabile | S Lund 140912 | GB | MK602730 | MK602730 |
| Hydnellumpeckii Banker | S Svantesson 328 | GB | MK602731 | MK602731 |
| Hydnellum peckii | E Larsson 174-14 | GB | MK602732 | MK602732 |
| Hydnellum peckii | E Bendiksen 567-11 | O | MK602733 | MK602733 |
| Hydnellumpineticola K.A.Harrison | RB 94 | KC571734 | ||
| Hydnellumpiperatum Maas Geest. | REB 322 | TENN | JN135173 | |
| Hydnellumregium K.A.Harrison | SE Westmoreland 93 | AY569031 | ||
| Hydnellumscleropodium K.A.Harrison | REB 3 | TENN | JN135186 | |
| Hydnellumscrobiculatum (Fr.) P.Karst. | REB 78 | TENN | JN135181 | |
| Hydnellumspongiosipes (Peck) Pouzar | REB 52 | TENN | JN135184 | |
| Hydnellumsuaveolens (Scop.:Fr.) P.Karst. | E Larsson 139-09 | GB | MK602734 | MK602734 |
| Hydnellum suaveolens | E Larsson 8-14 | GB | MK602735 | MK602735 |
| Hydnellum suaveolens | S Svantesson 877 | GB | MK602736 | MK602736 |
| Hydnellumsubsuccosum K.A.Harrison | REB 10 | TENN | JN135178 | |
| Lenzitopsisdaii L.W.Zhou & Kõljalg | Yuan 2959 | IFP | JN169799 | JN169793 |
| Lenzitopsisoxycedri Malençon & Bertault | KH Larsson 15304 | GB | MK602774 | MK602774 |
| Odontiafibrosa (Berk. & M.A.Curtis) Kõljalg | TU115028 | TU | MK602775 | MK602775 |
| Phellodon cf niger | E Larsson 35-14 | GB | MK602782 | MK602782 |
| Phellodontomentosus (L.:Fr.) Banker | E Bendiksen 118-10 | O | MK602781 | MK602781 |
| Pseudotomentellaflavovirens (Höhn. & Litsch.) Svrček | KH Larsson 16190 | O | MK602780 | MK602780 |
| Sarcodonamygdaliolens Rubio Casas, Rubio Roldán & Català | SC 2011 | JN376763 | ||
| Sarcodonaspratus (Berk.) S.Ito | DQ448877 | |||
| Sarcodonatroviridis (Morgan) Banker | REB 104 | TENN | JN135190 | |
| Sarcodon atroviridis | REB 61 | KC571768 | ||
| Sarcodonbairdii A.C.Grupe & Vasco-Pal. | Vasco 990 | HUA | KR698938 | |
| Sarcodoncolombiensis A.C.Grupe & Vasco-Pal. | Vasco 2084 | HUA | KP972654 | |
| Sarcodonfennicus (P.Karst.) P.Karst. | S Westerberg 110909 | GB | MK602739 | MK602739 |
| Sarcodon fennicus | O-F-242833 | O | MK602738 | MK602738 |
| Sarcodon fennicus | O-F-204087 | O | MK602737 | MK602737 |
| Sarcodonfuligineoviolaceus (Kalchbr.) Pat. | LA 120818 | GB | MK602740 | MK602740 |
| Sarcodon fuligineoviolaceus | B Nylén 130918 | GB | MK602741 | MK602741 |
| Sarcodon fuligineoviolaceus | A Molia 160-2011 | O | MK602742 | MK602742 |
| Sarcodonfuscoindicus (K.A.Harrison) Maas Geest. | OSC 113622 | OSC | EU669228 | |
| Sarcodonglaucopus Maas Geest. & Nannf. | RG Carlsson 13-060 | GB | MK602743 | MK602743 |
| Sarcodon glaucopus | J Nitare 060916 | GB | MK602744 | MK602744 |
| Sarcodon glaucopus | Å Edvinson 110926 | GB | MK602745 | MK602745 |
| Sarcodonimbricatus (L.:Fr.) P.Karst. | S Svantesson 355 | GB | MK602748 | MK602748 |
| Sarcodon imbricatus | J Rova 140829-2 | GB | MK602746 | MK602746 |
| Sarcodon imbricatus | E Larsson 384-10 | GB | MK602747 | MK602747 |
| Sarcodonjoeides (Pass.) Bataille | RG Carlsson 11-090 | GB | MK602749 | MK602749 |
| Sarcodon joeides | K Hjortstam 17589 | GB | MK602750 | MK602750 |
| Sarcodon joeides | J Nitare 110829 | GB | MK602751 | MK602751 |
| Sarcodon joeides | REB 270 | KC571772 | ||
| Sarcodonlepidus Maas Geest. | E Grundel 110916 | GB | MK602753 | MK602753 |
| Sarcodon lepidus | RG Carlsson 10-065 | GB | MK602752 | MK602752 |
| Sarcodon lepidus | J Nitare 110829 | GB | MK602754 | MK602754 |
| Sarcodonleucopus (Pers.) Maas Geest. & Nannf. | O-F-296944 | O | MK602756 | MK602756 |
| Sarcodon leucopus | O-F-296099 | O | MK602755 | MK602755 |
| Sarcodon leucopus | P Hedberg 080811 | GB | MK602757 | MK602757 |
| Sarcodonlundellii Maas Geest. & Nannf. | L&A Stridvall 06-049 | GB | MK602758 | MK602758 |
| Sarcodon lundellii | O-F-242639 | O | MK602759 | MK602759 |
| Sarcodon lundellii | O-F-295814 | O | MK602760 | MK602760 |
| Sarcodonmartioflavus (Snell, K.A.Harrison & H.A.C.Jacks.) Maas Geest. | A Delin 110804 | GB | MK602763 | MK602763 |
| Sarcodon martioflavus | O-F-242435 | O | MK602762 | MK602762 |
| Sarcodon martioflavus | O-F-242872 | O | MK602761 | MK602761 |
| Sarcodonpakaraimensis A.C.Grupe & T.W.Henkel | T Henkel 9554 | BRG | KM668103 | |
| Sarcodonpallidogriseus A.C.Grupe & Vasco-Pal. | Vasco 989 | HUA | KR698939 | |
| Sarcodonportoricensis A.C.Grupe & T.J.Baroni | TG Baroni 8776 | NY | KM668100 | |
| Sarcodonquercophilus A.C.Grupe & Lodge | CFMR-BZ-3833 | NY | KM668101 | |
| Sarcodonquercinofibulatus Pérez-De-Greg., Macau & J.Carbó | JC 20090718-2 | JX271818 | MK602773 | |
| Sarcodonrufobrunneus A.C.Grupe & Vasco-Pal. | Vasco 1989 | HUA | KR698937 | |
| Sarcodonscabripes (Peck.) Banker | REB 351 | TENN | JN135191 | |
| Sarcodonscabrosus (Fr.) P.Karst. | O-F-295824 | O | MK602764 | MK602764 |
| Sarcodon scabrosus | O-F-292320 | O | MK602766 | MK602766 |
| Sarcodon scabrosus | O-F-360777 | O | MK602765 | MK602765 |
| Sarcodonsquamosus (Schaeff.) Quél. | O-F-177452 | O | MK602768 | MK602768 |
| Sarcodon squamosus | E Larsson 248-12 | GB | MK602767 | MK602767 |
| Sarcodon squamosus | O-F-295554 | O | MK602769 | MK602769 |
| Sarcodonumbilicatus A.C.Grupe, T.J.Baroni & Lodge | TJ Baroni 10201 | NY | KM668102 | |
| Sarcodonunderwoodii Banker | REB 50 | KC571781 | ||
| Sarcodonversipellis (Fr.) Nikol. | RG Carlsson 13-057 | GB | MK602771 | MK602771 |
| Sarcodon versipellis | RG Carlsson 11-085 | GB | MK602772 | MK602772 |
| Sarcodon versipellis | E Bendiksen 164-07 | O | MK602770 | MK602770 |
| Sistotremabrinkmannii (Bres.) J.Erikss. | KH Larsson 14078 | GB | KF218967 | KF218967 |
| Steccherinumochraceum (J.F.Gmel.:Fr.) Gray | KH Larsson 11902 | GB | JQ031130 | JQ031130 |
| Thelephoracaryophyllea (Schaeff.:Fr.) Pers. | E Larsson 89-09S | GB | MK602776 | MK602776 |
| Thelephoraterrestris Ehrh.:Fr. | E Larsson 295-13 | GB | MK602777 | MK602777 |
| Tomentellastuposa (Link) Stalpers | Th-0764 | O | MK602778 | MK602778 |
| Tomentellopsispulchella Kõljalg & Bernicchia | KH Larsson 16366 | O | MK602779 | MK602779 |
In the phylogenetic analyses we assumed the following minimal partitions for the nrDNA region: ITS1, 5.8S, ITS2 and LSU (approximately 1200 bases of the 5’ end). Two datasets were analysed separately: an LSU dataset only including the LSU region, and an ITS dataset including ITS1, 5.8S and ITS2. We used the automated best-fit tests implemented in PAUP* 4.0a (Swofford 2002) to select optimal substitution models for each complete, non-partitioned dataset (PHYML) and optimal substitution model partitions for each minimal partition (BEAST). Models and partitions were chosen based on BIC score for the BEAST analysis and AICc score for the PHYML analysis. All tests were conducted using three substitution schemes and evaluated substitution models with equal and gamma-distributed among-site rate variation. The tests for the PHYML analysis also evaluated substitution models with invariant sites. The following partitions and models had the highest ranking, according to BIC: ITS1+ITS2 (GTR+G), 5.8S (K80+G), LSU (GTR+G). According to AICc the GTR+I+G model provided the best fit for both the ITS and the LSU datasets.
To generate Bayesian phylogenetic trees (BI) from the alignments we used BEAST 2.4.7 (Bouckaert et al. 2014). We prepared the xml-files for the BEAST 2 runs in BEAUTI 2.4.7 (Bouckaert et al. 2014). We set the substitution model to GTR+G for the LSU run. In the ITS run we set it to HKY+G for 5.8S, since it is the most similar model to K80+G available in the program. Test runs revealed convergence problems due to insufficient data for some substitution rates in the GTR+G model initially used for the ITS1+ITS2 partition, and it was hence changed to HKY+G. In the ITS run the substitution rate of both partitions were estimated independently. We set the trees of the minimal nrDNA partitions as linked in this analysis and the clock models as unlinked. A lognormal, relaxed clock model was assumed for each partition, as test runs had shown that all partitions had a coefficient of variation well above 0.1 (i.e. implying a relatively high rate variation among branches). The clock rate of each partition was estimated in the runs, using a lognormal prior with a mean set to one in real space. We set the growth rate prior to lognormal, with a mean of 5 and a standard deviation of 2. We ran the Markov Chain Monte Carlo (MCMC) chains of both datasets for 20 million generations with tree and parameter files sampled every 1,000 generations. The analyses all converged well in advance of the 10 % burn-in threshold, had ESS values well above 200 for all parameters, and chain mixing was found to be satisfactory as assessed in TRACER 1.6.0 (Rambaut et al. 2014). After discarding the burn-in trees, maximum clade credibility trees were identified by TREEANNOTATOR 2.4.7 (Bouckaert et al. 2014).
To generate Maximum Likelihood (ML) gene trees we used PHYML 3.1 (Guindon et al. 2010). We set the substitution model to GTR+I+G for both the ITS and LSU datasets. Tree topology search was conducted using NNI+SPR, with ten random starting trees. Non-parametric bootstrap analyses with 1000 replicates were performed on the resulting trees.
Results
Seventy-five Thelephorales specimens from the genera Amaurodon, Bankera, Boletopsis, Hydnellum, Lenzitopsis, Phellodon, Pseudotomentella, Sarcodon, Thelephora, Tomentella, and Tomentellopsis, were sequenced for this study. In addition, 39 sequences were downloaded from public databases (GenBank, UNITE) including outgroup sequences of Steccherinumochraceum (Polyporales) and Sistotremabrinkmannii (Cantharellales) included in the LSU dataset. The ITS analyses were rooted by the default method (BEAST) or left unrooted (PHYML).
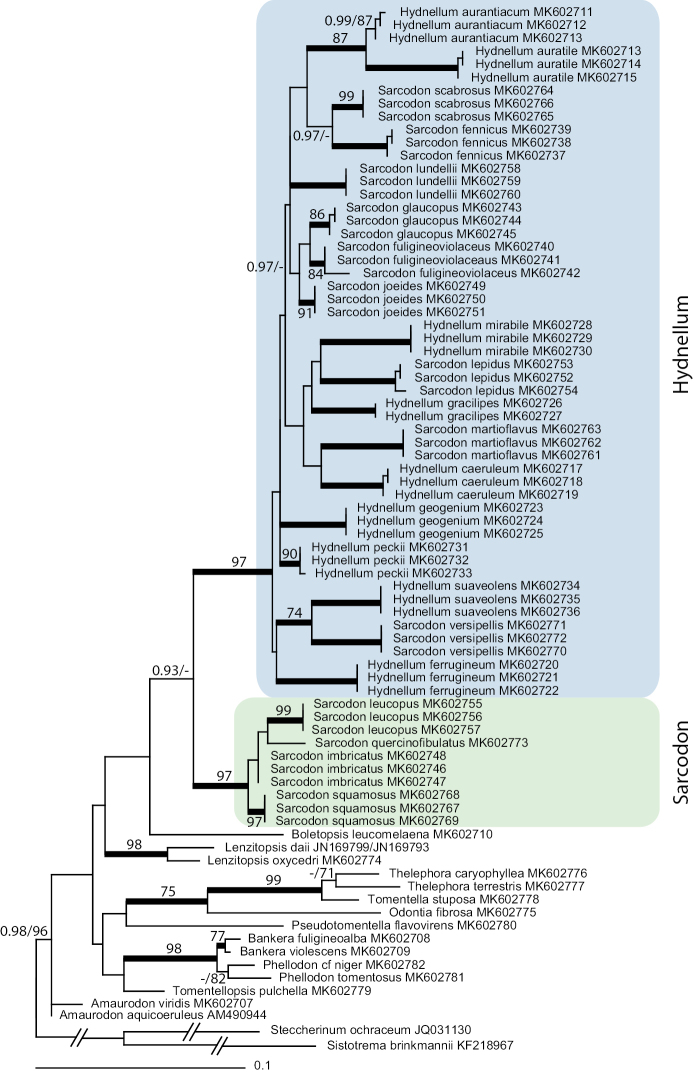
The aligned LSU dataset consisted of 1443 nucleotide positions. After exclusion of ambiguous regions 1377 positions remained for the analyses. BI returned a tree where the focus genera Hydnellum and Sarcodon are distributed over two strongly supported clades. The larger of these clades includes the type of Hydnellum, H.suaveolens, and an additional 17 species, all except one forming strongly supported terminal clades. Nine of these taxa are currently placed in Sarcodon. With a few exceptions the relationships within Hydnellum are not resolved. H.aurantiacum and H.auratile are recovered as a strongly supported group; Sarcodonscabrosus and S.fennicus are grouped with 0.97 posterior probability support; S.fuligineoviolaceus, S.glaucopus, and S.joeides form a subclade with 0.97 posterior probability support; and finally H.suaveolens and S.versipellis form a strongly supported clade. The type of Sarcodon, S.imbricatus, and three other species form the second main clade. The three sequences of S.imbricatus cluster together but the clade is unsupported. Hydnellum and Sarcodon are recovered as sister clades but the support for this arrangement is weak.
For target taxa the ML tree is essentially similar to the BI tree with strong support for the similarly composed Hydnellum and Sarcodon clades (Fig. 2). As for the BI analysis the relationships among species within Hydnellum and Sarcodon are not resolved except for a weak to moderate support for grouping H.aurantiacum with H.auratile and H.suaveolens with S.versipellis. S.fuligineoviolaceus, S.glaucopus, and S.joeides also group together in the ML tree but without support. Again S.imbricatus does not get support and is not separated from S.quercinofibulatus.
Figure 2.
Maximum likelihood analyses of LSU dataset for Thelephorales. Branches in bold have a posterior probability value of 1 in Bayesian inference and 100% bootstrap support in ML analysis, if not otherwise indicated by a figure. Lower support values on other branches are indicated by figures. Steccherinumochraceum and Sistotremabrinkmannii are used as outgroup (branch lengths shortened).
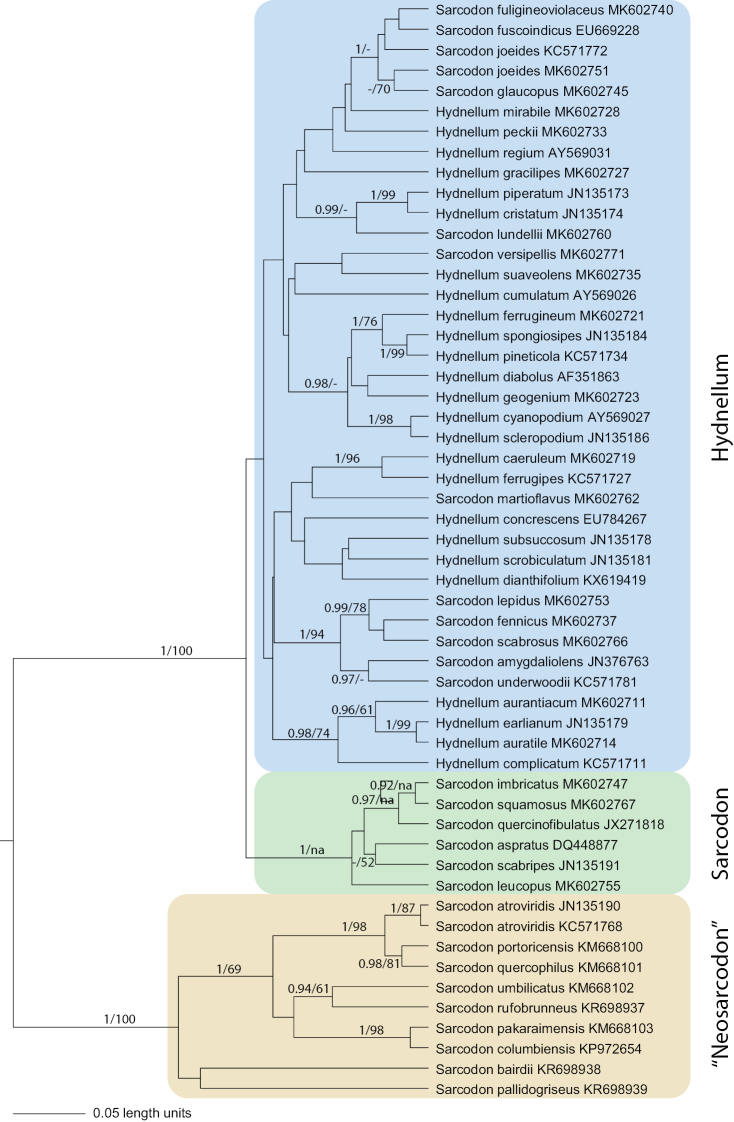
The aligned ITS dataset consisted of 1068 nucleotide positions of which 505 remained for the analyses after removal of ambiguous regions. Bayesian inference produced a tree with two strongly supported clades (Fig. 3). The smaller one, which we here informally call “Neosarcodon”, contains nine Sarcodon species, all with a distribution in the tropical and subtropical Americas. Remaining Hydnellum and Sarcodon taxa, including both type species, formed the other clade. Within the latter clade two subclades are visible, corresponding to the genera Hydnellum and Sarcodon, and with the same delimitation as in the LSU trees. Only the Sarcodon subclade has strong support. Within each larger clade several groups of taxa received moderate to strong support. The reader is referred to Fig. 2 for further details.
Figure 3.
Ultrametric default rooted BEAST tree of ITS dataset for Hydnellum and Sarcodon. Posterior probability values and bootstrap percent support from ML analysis are indicated by figures; na = not applicable.
The ML tree recovered the same two main clades with strong support but could not resolve the relationships within the larger Hydnellum/Sarcodon clade. In the ML tree the clade corresponding to Hydnellum in the LSU tree is correctly identified but not supported while the clade corresponding to Sarcodon appears polyphyletic.
Based on these results we hereby revise the limits of the two genera by moving a number of species from Sarcodon to Hydnellum. Consequently the genus description for Hydnellum must be emended while the genus description for Sarcodon can remain unaltered.
Taxonomy
Hydnellum
P.Karst., Meddn Soc. Fauna Flora fenn. 5: 41 (1879).
Type species.
Hydnellumsuaveolens (Scop.:Fr.) P.Karst. (1879)
Basionym.
Hydnumsuaveolens Scop.:Fr. (1772)
Basidiomata with pileus and stipe, single or concrescent; pileus thin to thick, at first smooth and velutinous, when mature felted, fibrillose, scaly, ridged, or irregularly pitted and scrupose, mostly brownish but also with white, olive yellowish, orange, purplish or bluish colours, often concentrically zonate; stipe narrow to thick, solid, mostly short; hymenophore hydnoid, usually strongly decurrent; context from soft and brittle to corky or woody; hyphal system monomitic, septa with or without clamps, context hyphae inflated or not; cystidia lacking; basidia narrowly clavate, producing four sterigmata; basidiospores with irregular outline, more or less lobed, verrucose, brownish. Terrestrial, forming ectomycorrhiza with forest trees.
Hydnellum amygdaliolens
(Rubio Casas, Rubio Roldán & Català) E.Larss., K.H.Larss. & Kõljalg comb. nov.
830570
Basionym.
Sarcodonamygdaliolens Rubio Casas, Rubio Roldán & Català, Boln Soc. Micol. Madrid 35: 44−45. 2011. Holotype: Spain, Tamajón, Barranco la Jara. L. Rubio-Casas & L. Rubio-Roldán, AH 42113.
Hydnellum fennicum
(P.Karst.) E.Larss., K.H.Larss. & Kõljalg comb. nov.
830571
Basionym.
Sarcodonscabrosusvar.fennicus P.Karst., Bidr. Känn. Finl. Nat. Folk 37: 104. 1882. Type: not indicated (neotype: H, designated by Maas Geesteranus & Nannfeldt 1969: 406)
Hydnellum fuligineoviolaceum
(Kalchbr.) E.Larss., K.H.Larss. & Kõljalg comb. nov.
830572
Basionym.
Hydnumfuligineoviolaceum Kalchbr., in Fries, Hymenomyc. eur. (Upsaliae): 602. 1874. Holotype: Slovakia, Presovsky kraj, Olaszi. C. Kalchbrenner, UPS F-173546.
Hydnellum fuscoindicum
(K.A.Harrison) E.Larss., K.H.Larss. & Kõljalg comb. nov.
830573
Basionym.
Hydnumfuscoindicum K.A.Harrison, Can. J. Bot. 42: 1213. 1964. Holotype: USA, Washington, Olympic Nat. Park, A.H. Smith. MICH 10847.
Hydnellum glaucopus
(Maas Geest. & Nannf.) E.Larss., K.H.Larss. & Kõljalg comb. nov.
830574
Basionym.
Sarcodonglaucopus Maas Geest. & Nannf., Svensk bot. Tidskr. 63: 407. 1969. Holotype: Sweden, Uppland, Börje par., J. Eriksson. UPS F-013955.
Hydnellum joeides
(Pass.) E.Larss., K.H.Larss. & Kõljalg comb. nov.
830575
Basionym.
Hydnumjoeides Pass., Nuovo G. bot. ital. 4: 157. 1872. Holotype: Italy, Emilia-Romagna, Collecchio, G. Passerini. PAD.
Hydnellum lepidum
(Maas Geest.) E. Larss., K.H.Larss. & Kõljalg comb. nov.
830576
Basionym.
Sarcodonlepidus Maas Geest., Verh. K. ned. Akad. Wet., tweede sect. 65: 105. 1975. Holotype: The Netherlands, Lochem, Ampsen, G. & H. Piepenbroek. L.
Hydnellum lundellii
(Maas Geest. & Nannf.) E.Larss., K.H.Larss. & Kõljalg comb. nov.
830577
Basionym.
Sarcodonlundellii Maas Geest. & Nannf., Svensk bot. Tidskr. 63: 421. 1969. Type: Sweden, Uppland, Storvreta, S. Lundell & J.A. Nannfeldt, distributed in S. Lundell & J.A. Nannfeldt Fungi exs. suec. as number 252 (lectotype, designated here, UPS F-010975; MycoBank No.: MBT387081). The UPS herbarium has two copies of the exsiccate and the specimens of H.lundellii are registered as F-010975 and F-013956, respectively. From F-010975 an ITS2 sequence has been generated [GenBank MK753037] and this specimen is here selected as lectotype).
Hydnellum martioflavum
(Snell, K.A.Harrison & H.A.C.Jacks.) E.Larss., K.H.Larss. & Kõljalg comb. nov.
830578
Basionym.
Hydnummartioflavum Snell, K.A.Harrison & H.A.C.Jacks., Lloydia 25: 161. 1962. Holotype: Canada, Quebec, Ste Anne de la Pocatière, H.A.C. Jackson & W.H. Snell 13 Sep. 1954, BPI 259438.
Hydnellum scabrosum
(Fr.) E.Larss., K.H.Larss. & Kõljalg comb. nov.
830579
Basionym.
Hydnumscabrosum Fr., Anteckn. Sver. Ätl. Svamp.: 62. 1836. Type: not indicated (neotype: Sweden, Småland, Femsjö, S. Lundell, UPS F-013954, designated by Maas Geesteranus & Nannfeldt 1969: 426)
Hydnellum underwoodii
(Banker) E.Larss., K.H.Larss. & Kõljalg comb. nov.
830580
Basionym.
Sarcodonunderwoodii Banker, Mem. Torrey bot. Club 12: 147. 1906. Holotype: USA, Connecticut, NY 776131.
Hydnellum versipelle
(Fr.) E.Larss., K.H.Larss. & Kõljalg comb. nov.
830581
Basionym.
Hydnumversipelle Fr., Öfvers. K. Svensk. Vetensk.-Akad. Förhandl. 18(1): 31. 1861. Type: not indicated (neotype: Sweden, Uppland, Danmark par., J. Eriksson & H. Nilsson, UPS F-013958, designated by Maas Geesteranus & Nannfeldt 1969: 430)
Sarcodon
Quél. ex P.Karst., Revue mycol., Toulouse 3 (no. 9): 20 (1881).
Type species.
Sarcodonimbricatus (L.:Fr.) P.Karst. (1881)
Basionym.
Hydnumimbricatum L.:Fr. (1753).
Basidiomata with pileus and stipe, single or concrescent; pileus thin to thick, at first smooth and velutinous, when mature smooth or scaly, brownish; stipe thick, solid, mostly short; hymenophore hydnoid, usually strongly decurrent; context soft and brittle; hyphal system monomitic, septa with clamps, context hyphae inflated; cystidia lacking; basidia narrowly clavate, producing four sterigmata; basidiospores with irregular outline, more or less lobed, verrucose, brownish. Terrestrial, forming ectomycorrhiza with forest trees.
Discussion
In this paper we show that the current morphology-based concepts of Sarcodon and Hydnellum do not correspond to monophyletic subgroups within the Thelephorales. The characters traditionally used to separate the two genera do not reflect true relationships. These characters, however, are vague and open to subjectivity; hence it is not surprising that they have now been shown to be unreliable. Maas Geesteranus (1975) pointed to the context structure and consistency as the main differentiating character. For Hydnellum he describes the context as “... fibrillose, soft or tough, corky to woody, more or less duplex, zoned, ...” and hyphae are said to be “...usually not inflating ...”. In Sarcodon the same structures are described as “... fleshy, brittle, soft or firm (never corky or woody), not duplex, not zoned ...” and “...hyphae inflating ...”. While these morphological characteristics remain true for Sarcodon, the corresponding descriptions for Hydnellum had to be emended.
Instead of context structure it seems that average basidiospore size may in most cases offer a possibility to separate a Sarcodon species from one belonging to Hydnellum. Table 2 summarizes basidiospore measurements from the literature. Average basidiospore lengths in Hydnellum fall between 4.45 and 6.95 µm while the same figures for Sarcodon are 7.4 and 9 µm, ornamentation excluded. However, S.quercinofibulatus clearly deviates from this pattern. According to measurements in the protologue (Pérez-de-Gregorio et al. 2011) and in Vizzini et al. (2013) average basidiospore length was measured to 6.95 and 7.0, respectively, but then included the ornamentation. Measurements excluding ornamentation would be approximately 1 µm less. Clearly, for S.quercinofibulatus basidiospore length alone will not be decisive for genus placement.
Table 2.
Basidiospore measurements for Hydnellum and Sarcodon from the literature. Sources: B = Baird et al. (2013), M = Maas Geesteranus (1975), J = Johannesson et al. (1999). All measurements exclude ornamentation. For species treated in this paper names follow our new classification. For other species names are according to cited authors.
| Species | Measurements | Mean length |
| Hydnellumaurantiacum (M) | (5.8−)6−6.7 × (4−)4.3−4.9 | 6.35 |
| Hydnellumauratile (M) | 4.9−5.8 × 3.6−4.5 | 5.35 |
| Hydnellumcaeruleum (M) | 5.4−6(−6.3) × 3.4−4.3 | 5.70 |
| Hydnellumcompactum (Pers.:Fr.) P.Karst. (M) | 5.4−6.3 × 3.6−4.5 | 5.85 |
| Hydnellumcomplicatum (B) | 4−5 × 3−5 | 4.50 |
| Hydnellumconcrescens (M) | 5.4−6.1 × (3.6−)4−4.5 | 5.75 |
| Hydnellumcristatum (B) | 5−6 × 4−5 | 5.50 |
| Hydnellumcruentum K.A.Harrison (B) | 4−5 × 3−4 | 4.50 |
| Hydnellumcumulatum (M) | 4.3−5.6 × 3.6−4.3 | 4,95 |
| Hydnellumdiabolus (B) | 6−7 × 5−6 | 6.50 |
| Hydnellumearlianum (B) | 5−6 × 4−5 | 5.50 |
| Hydnellumfennicum (M) | 6.3−7.6 × 4.5−5.2 | 6.95 |
| Hydnellumferrugineum (M) | (5.4−)5.8−6.3 × 3.6−4.5 | 6.05 |
| Hydnellumferrugipes (B) | 5−7 × 5−6 | 6.00 |
| Hydnellumfuligineoviolaceum (M) | 5.4−6.5 × 4−4.7(−5.4) | 5.95 |
| Hydnellumgeogenium (M) | 4.5−5.2 × 3.1−3.6 | 4.85 |
| Hydnellumglaucopus (M) | (5−)5.4−5.8(−6.3) × (3.6−)4−4.5 | 5.60 |
| Hydnellumgracilipes (M) | 4.3−4.6 × 2.7−3.6 | 4.45 |
| Hydnellumjoeides (M) | 5.4−5.8 × 3.6−4.2 | 5.60 |
| Hydnellumlepidum (M) | 5.8−6.3 × 3.6−4.3 | 6.05 |
| Hydnellumlundellii (M) | 4.9−5.8 × 3.6−4.2 | 5.35 |
| Hydnellummartioflavum (M) | 5−6.3 × 3.6−4.5 | 5.65 |
| Hydnellumpeckii (M) | 4.9−5.4 × 3.8−4 | 5.15 |
| Hydnellumpineticola (B) | 5−7 × 4−6 | 6.00 |
| Hydnellumpiperatum (B) | 4−6 × 4−5 | 5.00 |
| Hydnellumscabrosum (M) | (5.4−)6.3−7.3 × (3.6−)4−5 | 6.80 |
| Hydnellumscleropodium (B) | 4−6 × 3−4 | 5.00 |
| Hydnellumspongiosipes (B) | 6−7 × 5−6 | 6.50 |
| Hydnellumsuaveolens (M) | 4−5 × 3−3.6 | 4.50 |
| Hydnellumsubsuccosum (B) | 5−6 × 4−6 | 5.50 |
| Hydnellumversipelle (M) | 4.5−5.5 × 3.5−4.5 | 5.00 |
| Hydnellumunderwoodii (B) | 5−7 × 5−6 | 6.00 |
| Sarcodonatroviridis (B) | 8−9 × 7−8 | 8.50 |
| Sarcodonexcentricus R.E.Baird (B) | 8−9 × 6−8 | 8.50 |
| Sarcodonharrisonii R.E.Baird (B) | 7−9 × 6−8 | 8.00 |
| Sarcodonleucopus (M) | (6.7−)7.2−7.6(−9) × 4.5−5.6 | 7.40 |
| Sarcodonimbricatus (M) | 7.2−8.2 × 4.9−5.4 | 7.70 |
| Sarcodonscabripes (B) | 8−10 × 7−9 | 9.00 |
| Sarcodonsquamosus (J) | 7.2−8.2 × 4.9−5.4 | 7.70 |
Not all sequences from species described as Sarcodon spp. were recovered within either Sarcodon or Hydnellum. In our ITS-only analyses nine species formed a well-supported clade of their own, separated from Sarcodon sensu stricto and Hydnellum (Fig. 3). This clade, here informally called “Neosarcodon”, contains species collected in tropical and subtropical regions of the Western Hemisphere and may represent one or several distinct genera. However, further analyses based on an expanded dataset using more conservative molecular markers would be required to definitely identify any new higher taxa in the group.
The failure to generate support for Sarcodon and Hydnellum in the ITS-only analyses reflects the large genetical distances present among the species within this marker. Our general experience with the ITS region for thelephoralean target genera is that species are extremely well separated and the internal variation surprisingly low, even when a large number of specimens from both Europe and America are considered. On the other hand, the genetical difference among species is moderate to high, making alignments difficult and prone to ambiguities. In our ITS analyses we chose to remove ambiguous regions, thus halving the number of nucleotide positions suggested by automatic alignment through MAFFT. This seems to have affected the ML analyses most. However, the ITS analyses only served to position neotropical Sarcodon species and the results clearly show that they belong to a separate lineage.
Otto (1997) suggested that Hydnumauratile is a later synonym of Hydnumaurantiacum and that the species we now call Hydnellumaurantiacum should be named Hydnellumfloriforme (Schaeff.) Banker. The name change is based on a reinterpretation of Batsch’s original illustration, which, according to Otto, clearly shows the same species as Hydnumauratile. In phylogenetic analyses H.aurantiacum and H.auratile are sister taxa and during our study we have sequenced several specimens identified as H.auratile that turned out to be H.aurantiacum. Thus separating these species can be hazardous and to interpret illustrations must be even harder. We currently do not accept this unfortunate name change.
The present study will serve as the basis for further exploration of species limits within Hydnellum and Sarcodon. As has been demonstrated for the genera, many species interpretations are in need of revision. Over the years we have found numerous specimen misidentifications as well as specimens that could not be assigned to pre-existing names. A closer inspection of the ITS tree in Fig. 3, where we let the terminals retain the identifications given in GenBank, shows some examples. The American sequence of Sarcodonjoeides (KC571772) does not cluster with the European representative of the same species (MK602751) and the American sequence named Hydnellumearlianum seems to be identical to what is in Europe called H.auratile. Considering that many stipitate hydnoid species are red-listed and used as indicators of forests in need of conservation (Ainsworth 2005, Nitare 2019), it is of utmost importance to sort out the taxonomy of these species.
Supplementary Material
Acknowledgements
This study was supported by grants from ArtsDatabanken, Norway, to KH Larsson (ADB54-09), from Artdatabanken, Sweden, to E Larsson (2014-152 4.3), and from Estonian Research Council to U Kõljalg (IUT20-30). We also acknowledge support to S Svantesson from Kungliga Vetenskaps- och Vitterhetssamhället i Göteborg and from Kapten Carl Stenholms donatationsfond. We are grateful to many dedicated mycologists in Norway, Sweden and Finland for sending valuable collections. We are especially grateful to Johan Nitare for sharing with us his outstanding knowledge of stipitate hydnoid fungi and for duplicates from his herbarium. We also thank Martyn Ainsworth and Terry Henkel whose thorough reviews improved this paper considerably.
Citation
Larsson K-H, Svantesson S, Miscevic D, Kõljalg U, Larsson E (2019) Reassessment of the generic limits for Hydnellum and Sarcodon (Thelephorales, Basidiomycota) MycoKeys 54: 31–47. https://doi.org/10.3897/mycokeys.54.35386
References
- Ainsworth AM. (2005) BAP Fungi Handbook. English Nature Research Reports 600, English Nature, Peterborough, 115 pp. [Google Scholar]
- Ainsworth AM, Parfitt D, Rogers HJ, Boddy L. (2010) Cryptic taxa within European species of Hydnellum and Phellodon revealed by combined molecular and morphological analysis. Fungal Ecology 3: 65−80. 10.1016/j.funeco.2009.07.001 [DOI]
- Baird R, Wallace LE, Baker G, Scruggs M. (2013) Stipitate hydnoid fungi of the temperate southeastern United States. Fungal Diversity 62: 41−114. 10.1007/s13225-013-0261-6 [DOI]
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ. (2014) BEAST 2: A software platform for Bayesian evolutionary analysis. PLOS Computational Biology 10(4). 10.1371/journal.pcbi.1003537 [DOI] [PMC free article] [PubMed]
- Grupe A, Baker A, Uehling JK, Smith ME, Baroni TJ, Lodge DJ, Henkel TW. (2015) Sarcodon in the Neotropics I: new species from Guyana, Puerto Rico and Belize. Mycologia 107: 591–606, 10.3852/14-185 [DOI] [PubMed]
- Grupe A, Vasco-Palacios AM, Smith ME, Boekhout T, Henkel TW. (2016) Sarcodon in the Neotropics II: four new species from Colombia and a key to the regional species. Mycologia 108: 791–805. 10.3852/15-254 [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59: 307−321. 10.1093/sysbio/syq010 [DOI] [PubMed]
- Johannesson H, Ryman S, Lundmark H, Danell E. (1999) Sarcodonimbricatus and S.squamosus - two confused species. Mycological Research 103: 1447−1452. 10.1017/S0953756299008709 [DOI]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7, improvements in performance and usability. Molecular Biology and Evolution 30: 772−780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed]
- Larsson A. (2014) AliView: a fast and lightweight alignment viewer and editor for large data sets. Bioinformatics 30: 3276−3278. 10.1093/bioinformatics/btu531 [DOI] [PMC free article] [PubMed]
- Larsson E, Vauras J, Cripps CL. (2018) Inocybepraetervisa group - a clade of four closely related species with partly different geographical distribution ranges in Europe. Mycoscience 59: 277−287. 10.1016/j.myc.2017.11.002 [DOI]
- Loizides M, Alvarado P, Assoyov B, Arnolds E, Moreau P-A. (2016) Hydnellumdianthifolium sp. nov. (Basidiomycota, Thelephorales, a new tooth-fungus from southern Europe with notes on H.concrescens and H.scrobiculatum Phytotaxa 280: 23−35. 10.11646/phytotaxa.280.1.2 [DOI]
- Maas Geesteranus RA. (1975) Die Terrestrischen Stachelpilze Europas (The Terrestrial Hydnums of Europe). North-Holland Publishing, 1−127.
- Maas Geesteranus RA, Nannfeldt JA. (1969) The genus Sarcodon in Sweden in the light of recent investigations. Svensk Botanisk Tidskrift 63: 401−440.
- Miscevic D. (2013) A phylogenetic study of the genus Sarcodon. MSc degree project, Department of Biological and Environmental Sciences, University of Gothenburg, Sweden.
- Nitare J. (2019) Skyddsvärd skog. Naturvårdsarter och andra kriterier för naturvärdesbedömning. Skogsstyrelsens förlag, Jönköping.
- Nitare J, Högberg N. (2012) Svenska arter av fjälltaggsvampar (Sarcodon) – en preliminär rapport. Svensk Mykologisk Tidskrift 33(3): 2−49.
- Otto P. (1997) Kommentierter Bestimmungsschlüssel der terrestrischen Stachelpilze Deutschlands mit taxonomischen and nomenklatorischen Anmerkungen. Boletus 21(1): 1−21.
- Pérez-de-Gregorio MÁ, Macau N, Carbó J. (2011) Sarcodonquercinofibulatum, una nueva especie del género con hifas fibulíferas. Revista Catalana de Micologia 33: 25−30.
- Rambaut A, Suchard MA, Xie D, Drummond AJ. (2014) Tracer v1.6. http://tree.bio.ed.ac.uk/software/tracer
- Swofford DL. (2002) PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4.0a, build 159. Sinauer Associates, Sunderland, MA.
- Tedersoo L, May TW, Smith ME. (2010) Ectomycorrhizal lifestyle in fungi: global diversity, distribution, and evolution of phylogenetic lineages. Mycorrhiza 20: 217–263. 10.1007/s00572-009-0274-x [DOI] [PubMed] [Google Scholar]
- Thiers B (continuously updated) Index Herbariorum: A global directory of public herbaria and associated staff. New York Botanical Garden’s Virtual Herbarium. http://sweetgum.nybg.org/science/ih [accessed 1 December 2018]
- Vizzini A, Carbone M, Boccardo F, Ercole E. (2013) Molecular validation of Sarcodonquercinofibulatus, a species of the S.imbricatus complex associated with Fagaceae, and notes on Sarcodon. Mycological Progress 12: 465–474. 10.1007/s11557-012-0851-9 [DOI] [Google Scholar]
- Vizzini A, Angelini C, Los C, Ercole E. (2016) Thelephoradominicana (Basidiomycota, Thelephorales), a new species from the Dominican Republic, and preliminary notes on thelephoroid genera. Phytotaxa 265: 27–38. 10.11646/phytotaxa.265.1.2 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.