Abstract
Introduction
An increasing number of studies are using magnetoencephalography (MEG) to study dementia. Here we define a common methodological framework for MEG resting-state acquisition and analysis to facilitate the pooling of data from different sites.
Methods
Two groups of patients with mild cognitive impairment (MCI, n = 84) and healthy controls (n = 84) were combined from three sites, and site and group differences inspected in terms of power spectra and functional connectivity. Classification accuracy for MCI versus controls was compared across three different types of MEG analyses, and compared with classification based on structural MRI.
Results
The spectral analyses confirmed frequency-specific differences in patients with MCI, both in power and connectivity patterns, with highest classification accuracy from connectivity. Critically, site acquisition differences did not dominate the results.
Discussion
This work provides detailed protocols and analyses that are sensitive to cognitive impairment, and that will enable standardized data sharing to facilitate large-scale collaborative projects.
Keywords: Magnetoencephalography, Multi-site, Mild cognitive impairment, Spectral analysis, Functional connectivity, Harmonization
Highlights
-
•
Magnetoencephalography is sensitive to mild cognitive impairment.
-
•
Magnetoencephalography data sets can be usefully harmonized across sites.
-
•
Site differences remain important factors to consider.
-
•
Different analyses reveal robust frequency-specific changes in power and connectivity.
-
•
Our database of mild cognitive impairment and matched healthy controls is available for research.
1. Introduction
A major challenge to the development of new mechanistic therapies for dementia is to establish robust biomarkers that can detect and monitor early stages of illness, and which directly reflect the consequences of underlying pathology on neurophysiology and function. Magnetoencephalography (MEG) is a promising tool to study neurodegeneration, not only by its proven sensitivity to dementia and safety as a noninvasive test, but also by its direct representation of network and synaptic physiology [1], [2], [3]. MEG provides rich, high-dimensional data on neuronal activity, oscillatory dynamics and connectivity at a millisecond time scale. Critically, the evoked and oscillatory signals can be used to estimate neural interactions between brain regions, which are key to establishing reliable biomarkers of the macroscopic sequelae of neurodegeneration.
Capitalizing on the rich data provided by MEG an increasing number of studies use resting-state paradigms to examine dementia. However, variability in acquisition parameters and use of diverse analytical approaches can hinder direct comparisons between studies at a time when it is critically important to understand the common brain responses to damage versus those that are disease-specific and potential targets for therapeutic treatment [4]. MEG data are characterized along numerous dimensions with many different methodological techniques for preprocessing and analysis, with a variety of endpoints used as indices for clinical features [5]. A standardized approach to pool data from different sites will facilitate data sharing and provide a core data set for validation benchmarks in future studies. Larger data sets can benefit from advanced analyses, utilizing approaches taken with “big data” to identify features relevant to dementia [5], and from multisite collaborations to enable faster throughput of data. A clear example in the MRI field is the Alzheimer's Disease Neuroimaging Initiative [6], the success of which is promising for a similar MEG database. There are already MEG guidelines for acquisition, reporting, and analysis in the general population [7], and more specific frameworks for examining Alzheimer's disease (AD) with MEG [2]. Here we extend these reports, in a multicenter approach to harmonizing data across sites, to examine mild cognitive impairment (MCI). We report three different sets of analyses, each performed by a different research group (Oxford, Cambridge, and Madrid), which all examined MEG power spectra using a variety of techniques.
More specifically, our primary aims were to (1) examine potential differences across sites in the data, (2) examine potential differences across sites in the results from the different analyses, and (3) report preliminary classification accuracy for distinguishing patients with MCI versus controls based on different features of MEG data (e.g., peak frequency, power distributions over sensors, power distribution over sources, and connectivity between sources) and in comparison with that based on MRI data.
2. Methods
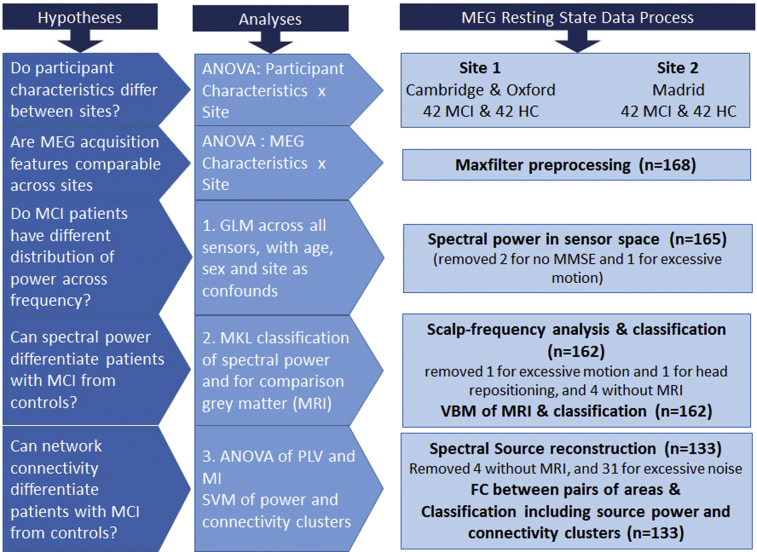
The data set was created by pooling data from three sites: the Medical Research Council's Cognition and Brain Sciences Unit in Cambridge, UK, the Oxford Centre for Human Brain Activity in Oxford, UK, and the Centre for Biomedical Technology in Madrid, Spain. A description of the steps taken to create the shared data set is in Appendix 1, with a detailed description of preprocessing steps in Appendix 2. The raw data set and preprocessing scripts are available on request from the FTP site “https://biofind-archive.mrc-cbu.cam.ac.uk”, following permission from the corresponding author. Fig. 1 shows a flow diagram of the analyses.
Fig. 1.
Flow chart of analyses undertaken in this study. Abbreviations: MCI, mild cognitive impairment; MMSE, Mini-Mental State Examination; VBM, voxel-based morphometry; FC, functional connectivity.
2.1. Participant details
Cohorts of patients and controls were created from existing data sets in two sites: the cohort of 42 patients scanned at Cambridge and Oxford were recruited with an MCI diagnosis from tertiary clinics at Cambridge University Hospitals NHS Trust. Three of these were scanned in Oxford, and the remaining 39 in Cambridge. The patients were recruited for one of several MEG projects but all underwent the same MEG resting-state paradigm. The 42 controls were selected from the population-representative CamCAN database (http://camcan-archive.mrc-cbu.cam.ac.uk). The controls were randomly selected from the database to match the Cambridge/Oxford patients in terms of the number of males/females, and within four years of patient ages. These controls all underwent the same resting-state MEG paradigm in Cambridge.
The cohorts of 42 patients and 42 controls from Madrid were recruited from the Neurology and Geriatric Departments of the University Hospital San Carlos, and from the Centre for Prevention of Cognitive Impairment and the Seniors Center of Chamartin District. They were matched by number of males/females, and the mean and standard deviation of the age range. For all patients, the MCI diagnosis was established in a memory clinic according to the National Institute on Aging–Alzheimer Association criteria [8]. Details of the data are presented in Table 1. The studies were approved by the local Research Ethics Committees. All participants gave written informed consent before participation according to the 1991 Declaration of Helsinki.
Table 1.
Participant characteristics as a function of recruitment site (patients scanned in Oxford were recruited via Cambridge)
| Data Characteristic | Madrid |
Cambridge |
All |
ANOVA |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Controls | MCI | Controls | MCI | Controls | MCI | Site | Group | Interaction | |
| N | 42 | 42 | 42 | 42 | 84 | 84 | - | - | - |
| Sex (M/F) | 19/23 | 19/23 | 28/14 | 28/14 | 47/37 | 47/37 | - | - | - |
| Age | 72.3 (2.7) | 72.2 (3.3) | 69.0 (8) | 69.0 (8) | 70.8 (6.1) | 70.8 (6.2) | Sig∗ | ns | ns |
| MMSE | 29.0 (1.1) | 26.9† (2.8) | 28.8 (1.2) | 25.1 (3.1) | 28.9 (1.1) | 26.0 (3.1) | Sig∗ | Sig∗ | Sig∗ |
Means have standard deviation in parentheses.
Abbreviations: MMSE, Mini-Mental State Examination; MCI, mild cognitive impairment.
P < .05.
Two patients had missing MMSE scores.
2.2. Procedure: Resting-state eyes-closed paradigm
During the five-minute MEG recording, all participants were asked to close their eyes and stay still for the duration of the recording. What participants had been doing before the eyes-closed recording varied depending on the project that they were recruited for, and is a potential source of unmodeled variance, contributing to, for example, tiredness. Drowsiness was also monitored before and after the recording (by self-report) to ensure they stayed awake.
2.3. MEG and MRI data acquisition
MEG data from all three sites were recorded using the same model of scanner: the 306-channel Vectorview system (Elekta Neuromag, Helsinki), which contains two orthogonal planar gradiometers and one magnetometer at each of 102 positions. Data were acquired continuously at 1 kHz in a magnetically shielded room. Head position indicator coils were used to monitor head position. Electro-oculograms were recorded using paired EOG electrodes in a bipolar montage. The three-dimensional locations of the head position indicator coils, over 100 “head points” across the scalp, and three anatomical fiducials (the nasion and left and right preauricular points), were recorded using a 3D Fastrak digitizer (Polhemus Inc., Colchester, VA).
T1-weighted MRIs were acquired for all participants, except 4 patients. Acquisition sequences are detailed in Appendix 2. The MRIs were used for coregistration for the source-space analyses, and also used in a voxel-based morphometry (VBM) analysis (standard procedures were applied, detailed in Appendix 2).
2.4. Analyses of participant and acquisition differences
To validate combining data sets from different sites (scanners), we compared participant characteristics and acquisition variables for Site (Madrid x Cambridge) and Group (MCI x Controls). We predicted that site differences would be apparent, as often found with different brain scanners, but we were more interested in whether Site interacted with Group, that is, whether differences between patients with MCI and Controls depended on the Site.
2.5. MEG analyses (also see Appendix 2 for details)
MEG data were preprocessed with tSSS MaxFilter and corrected for head motion at least every 1s. Bad channels (those channels with artifacts and excessive noise) were identified either visually or by MaxFilter's 'autobad' option. Specific details are in Appendix 2.
To examine frequency-specific changes in MCI three different analyses were performed (by separate groups of co-authors): (1) spectral power in sensor space, (2) classification of scalp-frequency images of power in sensor space, and (3) functional connectivity in source-space. A description of each analysis is in the following sections, with extra details in Appendix 2.
2.5.1. Analysis 1: Spectra averaged across sensors
Analysis 1 was performed in Oxford. Power at each frequency 1-45 Hz was examined to test the hypothesis that patients with MCI have slowed dynamics, as reflected for example by lower peak frequency for the alpha rhythm. For this analysis, 165 participants were included (one participant was removed for excessive head movement, and two for not having Mini-Mental State Examination [MMSE] scores). MEG data were preprocessed following a previously published pipeline [9]. The data recorded from planar gradiometers were downsampled to 250 Hz and a 1–45 Hz bandpass filter applied. Bad segments were rejected using an automatic algorithm and excluded from subsequent preprocessing and analysis. ICA was used to detect and remove oculographic and cardiographic artifacts. Data were z-transformed to normalize total spectral power across sensors and participants, before being converted into time-frequency data using a 5-cycle wavelet transform. The mean power for each frequency was averaged across time and entered into a general linear model (GLM) to test for differences in power spectra across participants. The GLM regressors coded the two Groups (MCI Patients and Controls), MMSE scores (for each Group separately), as well as Site, Sex, and Age (across both Groups).
Contrast of parameter estimates were used to compare the two Groups and two Sites. Permutation stats were computed with 5000 permutations of rows of the design matrix. A null distribution was built for each frequency using the maximum statistic across sensors of each permutation. The observed statistics were compared with this distribution to establish significance. Finally, the peak of the dominant alpha frequency peak was identified for each participant and compared across Groups.
2.5.2. Analysis 2: Power-frequency topographies
Analysis 2 was performed in Cambridge. The power at each frequency was further divided according to spatial location across the scalp, by interpolating the sensor locations across a 2D grid. MEG data were preprocessed using MaxFilter with the additional step of transforming the data from participant head space into common device space, which helps aligned sensor-level analyses. 166 participants were included, after excluding two patients, one because of a large distance from center of helmet (>30 mm), for which the transformation to a common device space breaks down, and one because of excessive motion (>6 mm). The data were converted to SPM12 format and segmented into 4-second epochs, and epochs with artifacts (based on visual inspection) were removed. A fast Fourier transform with multiple Hanning tapers was applied for frequencies from 1 to 92 Hz in steps of 0.25 Hz. The frequency resolution increased approximately exponentially with mean frequency. The resulting power spectra were averaged over each epoch and the (base 10) logarithm taken. The frequencies were subsampled approximately logarithmically leaving 94 frequencies in total. The sensor locations were projected onto a 32 × 32 grid to produce a scalp-frequency image (for gradiometers, the power was averaged across each pair of gradiometer directions). These topographic images were smoothed in the frequency dimension with a 2-pixel full width half maximum Gaussian kernel. The smoothed images were entered into a GLM, with regressors for Group, Site, Age, and Sex. Random field theory was used to estimate the corrected P value for the extent of scalp-frequency clusters of voxels surviving P < .001 uncorrected when comparing controls and patients with MCI.
To test the sensitivity of MEG to MCI, and compare it to the sensitivity of sMRI, both MEG and VBM images were used to assess classification performance of MCI versus controls using multikernel learning (MKL) within the PRoNTO v2.0 toolbox (http://www.mlnl.cs.ucl.ac.uk/pronto), with leave-one-out cross-validation with additional covariates of Site, Age, and Sex.
2.5.3. Analysis 3: Source-level power analyses and functional connectivity
Analysis 3 was performed in Madrid. For this analysis, 133 participants were included, removing those without an MRI, and those whose data were deemed excessively noisy, (reflected by the absence of the characteristic oscillatory activity of the eyes-closed resting state when visually reviewing the individual power spectra). MEG data were preprocessed using MaxFilter and left in participant head space. Oculographic, cardiographic, muscular, and jump artifacts were detected by the automatic artifact detection in the FieldTrip toolbox [10] confirmed by visual inspection, or by using a second-order blind identification separation algorithm. Artifact-related components were removed. MEG data were segmented into 4-second epochs and filtered using a 2000th order FIR band-pass filter with a Hanning window into five bands (with 2 seconds of real data padding added either side): for source analysis: broad band (2–45 Hz), and for connectivity analysis: theta (4–8 Hz), alpha (8–12 Hz), beta (12–30 Hz), and gamma (30–45 Hz). The data were coregistered to the T1-weighted MRI and the forward model calculated using a realistic single shell head [11].
For source-space analyses, a volumetric regular grid was generated from the MNI (MNI coordinate system from the Montreal Neurological Institute) template resulting in a model with 2459 sources. Cortical (including hippocampal and parahippocampal) anatomical labels were assigned according to the Automated Anatomical Labeling atlas [12] and a reduced version of the Harvard-Oxford atlas [13] leaving 1467 and 1489 sources, respectively. Using the T1-weighted MRI and a binary mask, the MNI defined grid was transformed to subject space. A mesh surface was generated from the mask. Both grid and brain surface were realigned to Neuromag coordinate system.
Source reconstruction used linearly constrained minimum variance beamformer [14] for each band. The source-space time series were grouped according to atlas, obtaining one representative time series for each area using [1] the principal component analysis (PCA) of all the sources in the area and [2] the source closest to the centroid of the area.
To localize frequency band power changes, relative power in frequency bands (delta-beta) was compared using a 2 × 2 ANOVA with group (Controls vs. MCI) as the main factor and the site as a covariate. Multiple comparisons were controlled using cluster-based permutation test with α = 0.05 for cluster thresholding.
Functional connectivity analyses were performed using phase-locking value (PLV) and mutual information (MI). The functional connectivity between pairs of areas was estimated between [1] the sources closest to their respective centroids and [2] the PCA of all sources in each area using the Automated Anatomical Labeling atlas. Statistical comparisons were performed using 2 × 2 ANOVA, with group and site as factors. Nonparametric statistics were calculated with 100,000 permutations. False discovery rate of 10% (Q = 0.10) was applied to the results.
Classification of the source-space results included power and connectivity clusters using a support vector machine with a simple linear kernel, (MATLAB 2018a, MathWorks) [15], [16]. A 5-fold cross-validation method was used to estimate the accuracy of the classifier [17]. The significant values obtained from the analyses of in the source domain of spectral power and functional connectivity were used as features to train a classification model. In the case of power measures, the average power value for each subject in each of the four significant clusters was used as features. In the case of functional connectivity measures, the average connectivity for each one of the two largest connected clusters (motifs) in each of the six metrics was selected. Altogether, four power-related features and twelve connectivity-related features were included. Note that this represents a drastic dimensionality reduction approach. Although this may not be the optimal approach in this context (cf. [18]), it has the advantage of selecting a small set of features with a clear-cut procedure.
3. Results
3.1. Participant characteristics
Participant characteristics including sex, age, and MMSE score are summarized in Table 1. The numbers of men and women differed between sites, and therefore sex was used as a covariate, where appropriate, in MEG analyses in the following. Age and MMSE scores were analyzed using a 2 × 2 (Site: Cambridge vs. Madrid x Group: Control vs. MCI), between-subject ANOVA.
For the ANOVA on age, although there was a significant main effect of Site, F(3,164) = 10.3, P = .002, with the Madrid group being older, there was no significant main effect of Group, or interaction between Group and Site, Fs < 1. Age was also used as a covariate to clarify any Site effects.
For MMSE (excluding the two missing scores), there was the expected main effect of Group, F(3,162) = 70.9, P < .001, with patients scoring lower than controls. There was also a main effect of Site, F(3,162) = 7.44, P = .007, with Cambridge site having lower scores, and an interaction, F(3,162) = 5.67, P = .018, with the control-patient difference being larger from the Cambridge site.
3.2. MEG acquisition characteristics
Acquisition characteristics are summarized in Table 2. Each measure was analyzed using a 2 × 2 (Site x Group), between-subject ANOVA (excluding three patients scanned at the Oxford Centre for Human Brain Activity).
Table 2.
MEG data characteristics
| Data characteristic | Madrid |
Cambridge |
Oxford |
ANOVA |
||||
|---|---|---|---|---|---|---|---|---|
| Controls | MCI | Controls | MCI | MCI | Site | Group | Interaction | |
| N | 42 | 42 | 42 | 39 | 3 | - | - | - |
| Bad channels | 7.38 | 6.93 | 4.64 | 6.07 | 4.33 | Sig∗ | ns | ns |
| M move (mm)† | 0.80 | 1.10 | 0.61 | 0.94 | 3.15 | ns | Sig∗ | ns |
| SD move (mm)† | 0.18 | 0.30 | 0.06 | 0.24 | 3.04 | ns | ns | ns |
| Position (mm) | 10.5 | 12.0 | 10.0 | 12.9 | 13.9 | ns | Sig∗ | ns |
| Time of day (24h) | 12.0 | 11.6 | 14.2 | 14.0 | 13.5 | Sig∗ | ns | ns |
| Data onset (s) | 250 | 234 | 125 | 132 | 124 | - | - | - |
| Data offset (s) | 419 | 404 | 298 | 306 | 294 | - | - | - |
| No. of epochs | 38.2 | 39.7 | 39.8 | 37.8 | 33.0 | ns | ns | Sig∗ |
Abbreviations: M, mean; MEG, magnetoencephalography; SD, standard deviation.
P < .05.
Head motion excluded 12 participants for whom head motion could not be estimated.
Bad channels for present purposes were those defined by the MaxFilter “autobad” option. The number of bad channels showed no difference between Groups, F < 1, although there were more bad channels for Madrid than Cambridge, F(1,163) = 13.5, P < .001 (and no significant interaction, F(1,163) = 1.11, P = .29).
Head motion was summarized by the mean (M) and the standard deviation (SD) of translations over the recording. Excluding the 12 participants for whom head motion could not be estimated, there was significantly more mean movement (F(1,151) = 4.96, P < .05) in the MCI group, but no difference between sites (F(1,151) = 1.52, P = .22), nor interaction (F < 1). A similar pattern was seen for the SD of translations, with a borderline group difference (F(1,151) = 3.36, P = .07), but no difference between sites (F(1,151) = 1.20, P = .27), nor interaction (F < 1). The translation between the head center and device center (“Position” in Table 2) was significantly greater for the MCI group (F(1,163) = 5.10, P < .05), but did not differ by, or interact with, site (F < 1). The preprocessing “movement compensation” step in MaxFilter corrects for head movements at least every 1 second, but, where appropriate, participants whose overall motion was much higher than other participants were also removed from the spectral analyses (described in sections in the following).
The time of day in which the MEG data were acquired did not differ between Groups, (F(1,163) = 1.07, P = .3), but was later for Cambridge than Madrid sites (F(1,163) = 66.1, P < .001) with no interaction (F < 1). The number of valid 4s epochs (as defined manually) did not show main effects of Group or Site (Fs < 1), although there was an interaction between these factors (F(1,163) = 4.22, P < .05), with numerically more valid epochs for the MCI group than controls at Madrid site, but vice versa at the Cambridge site.
3.3. Analysis 1: Spectral power averaged over sensors
Spectral power for both patients and controls declined with frequency apart from a notable peak at alpha frequency (around 10 Hz; Fig. 2A). The most obvious feature distinguishing groups, as we predicted, was a reduction in the peak frequency of the alpha oscillation, which averaged at 9–10 Hz for controls and 8–9 Hz for patients, and occurred regardless of any effects of sex or age. The contrast between the two groups confirmed higher spectral power for patients in low-frequency bands (<∼8 Hz) and for controls in higher frequencies (>∼8 Hz), Fig. 2E. Alpha power was more prominent in posterior sensors while the frontal sensors were dominated by the overall decline of power with frequency, and showed little group difference. In the MCI group, there was a positive linear parametric relationship between MMSE and alpha power, significant over posterior sensors, showing reduced alpha power with a lower MMSE score. The contrast between sites revealed more posterior alpha power in the Cambridge data but more frontal theta in the Madrid data (Fig. 2F–H).
Fig. 2.
Power spectrum in sensor space. (A) The mean power spectrum across participants for each sensor. (B) Sensors are color-coded by location. (C) To aid visualization, the spectrum is split into 5 bands defined by clustering the diagonal of a frequency-by-frequency correlation matrix. These bands are indicated by gray bars in panel A, together with the average sensor, topology is shown above each band. (E) The result of the MCI-Control contrast for each sensor and frequency (frequency-by-frequency correlation for this differential contrast in panel D). Statistical significance limits are indicated by black lines. Panels F, G, and H show corresponding data for the Site contrast (Cambridge–Madrid). Panel I shows the ROC for the Alpha peak frequency. Abbreviation: MCI, mild cognitive impairment.
3.3.1. Classification based on peak frequency
A t-test on peak frequencies showed a significant reduction in patients relative to controls T(164) = 2.86, P = .0049. To provide a reference point for later MEG analyses, a receiver operating characteristic curve was plotted for distinguishing controls from patients (Fig. 2I) on the basis of the peak alpha frequency (which has been shown to decrease in MCI, [19]). This revealed an area under the curve (AUC) of 63% (a balanced accuracy of 61%), which only dropped to 62% when first adjusting for age, sex, and site.
3.4. Analysis 2: Scalp-frequency power images in sensor-space
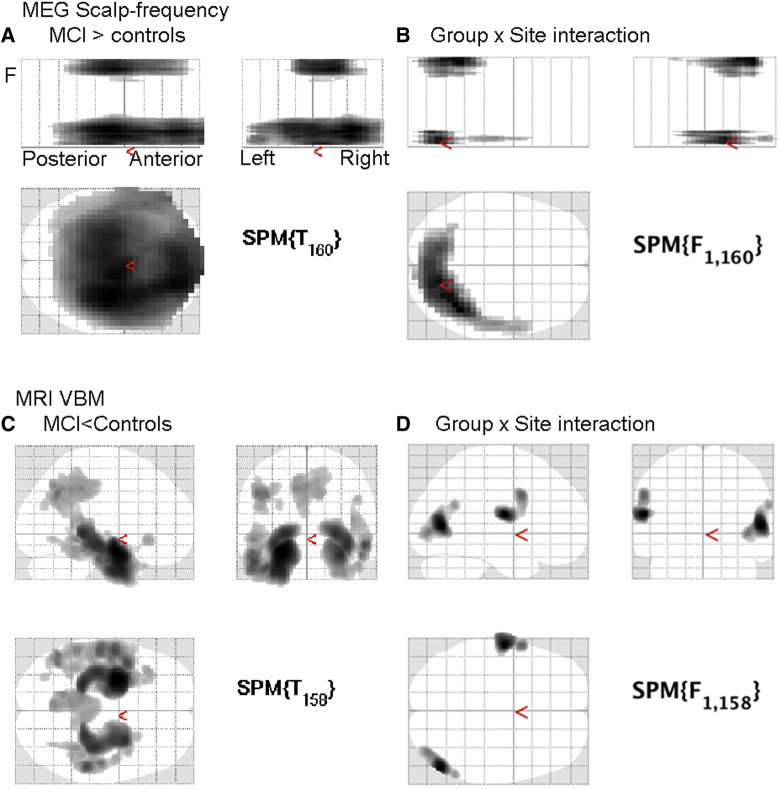
Comparison of the gradiometer scalp-frequency images revealed patients with MCI had greater power in two clusters that survived correction for their extent. One cluster was in low frequencies from 1 to 6.5 Hz and spread over most of the scalp (Fig. 3A), consistent with Analysis 1. There was a second cluster from 27 to 92 Hz, maximal more centrally (beyond the frequency range examined in Analysis 1). Controls did not show significantly greater power than patients anywhere (unlike the increase from 15 to 25 Hz in Analysis 1, possibly reflecting the more stringent statistical thresholds for this whole-image search). The results for magnetometers were similar.
Fig. 3.
Group and Site analyses for MEG and MRI. (A) Scalp-frequency results for gradiometers for regions showing greater power for patients than controls (P < .001 uncorrected height threshold, P < .05 corrected for cluster extent). (B) Scalp-frequency interaction of group by site. (C) MRI VBM results showing where local grey-matter volume is greater from Controls than Patients (P < .001 uncorrected height threshold, P < .05 corrected for cluster extent). (D) VBM group-by-site interaction. Abbreviations: MEG, magnetoencephalography; VBM, voxel-based morphometry; MCI, mild cognitive impairment.
There were also significant main effects of Site over much of the space, particularly higher frequencies over frontal scalp regions (not shown). More importantly, there were two small clusters that showed a significant Group-by-Site interactions (Fig. 3B), one at low and one at high frequencies, but both over more posterior sensors than the main effect of Group aforementioned.
3.4.1. Classification based on MRI images
Using the same mass univariate approach as on the aforementioned MEG scalp-frequency power images, a standard VBM of grey-matter volume at each voxel from the segmented MRI images revealed the expected reductions in grey-matter volume in patients, most prominently in bilateral medial temporal regions, as well as medial parietal and some lateral temporal regions (Fig. 3C). There were also large effect of Site across the whole brain, as often found with different MRI scanners, and two clusters showing Site by Group interactions (in right superior occipital and left inferior frontal cortices) but these were not in regions associated with dementia (Fig. 3D).
Grey-matter images were used to assess classification performance using MKL (see Appendix 2 for details) in 162 participants (to match those used in the MEG classification, in the following). The AUC for the MRI was 75% (with a balanced accuracy of 68%), as shown in Fig. 4.
Fig. 4.
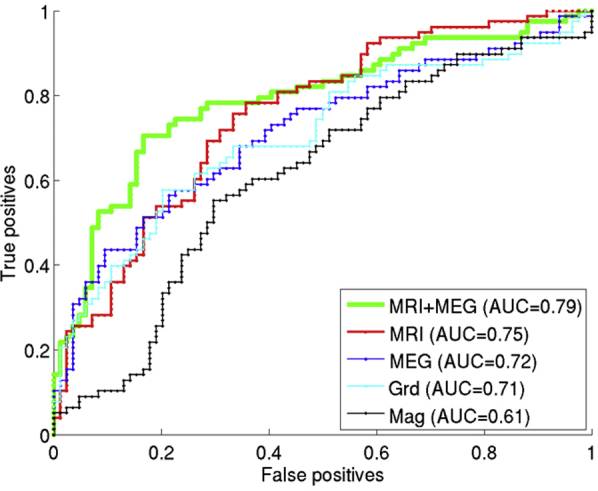
ROCs from MKL image-based classification. MEG power spectra (gradiometers + magnetometers), MRI, and combined MEG-power spectra and MRI. Abbreviations: MKL, multikernel learning; ROC, receiver operating characteristic; MEG, magnetoencephalography.
3.4.2. Classification based on MEG images
Smoothed topographic scalp-frequency images were also used to assess classification performance using MKL in 162 participants (to match those used in the MRI classification). The AUC was 61% for magnetometers (balanced accuracy of 59%) and 71% for gradiometers (balanced accuracy of 66%). When combining both types of sensor, the AUC increased slightly to 72% (Fig. 4).
3.4.3. Combined multivariate classification (MKL) of MRI and MEG
The MRI grey-matter images were combined with the magnetometer and gradiometer scalp-frequency images in an MKL classifier with 3 kernels. The AUC for this multimodal classification was 79% with balanced accuracy of 76%, better (numerically at least) than for any one modality alone (Fig. 4).
3.5. Analysis 3: Spectral power and connectivity in source space
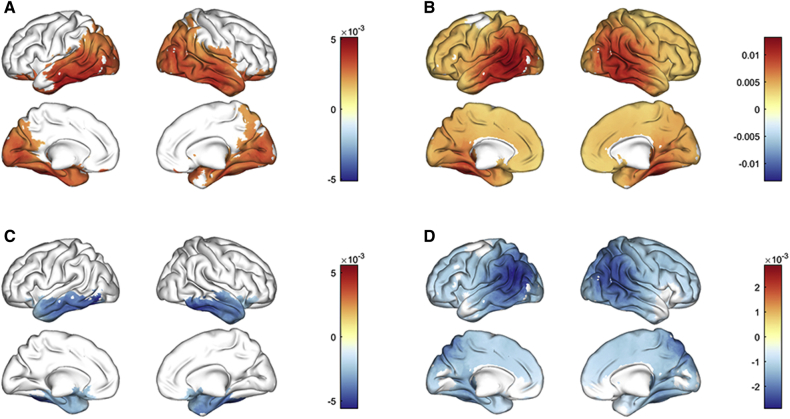
The spectral analysis from the source reconstruction provided relative power in each frequency band that was compared with a 2-way ANOVA using group (MCI vs. Control) as the main factor and the site as a covariate. Fig. 5 shows differences in relative power across frequency bands. Patients with MCI showed increased relative delta power (2–4 Hz) in a cluster comprising bilateral regions over posterior areas such as cuneus, precuneus, and calcarine; widespread bilateral temporal structures including hippocampus and parahippocampal cortices among others and extending into bilateral orbitofrontal areas (P = .01). Relative theta band power (4–8 Hz) was significantly increased in MCI (P = 9.9 · 10−5). Areas affected by theta power increase were distributed across the entire cortical surface, but were more intense over bilateral middle temporal gyri, commensurate with the sensor space analysis in section 3.4. Patients with MCI exhibited a significant decrease in relative alpha power (8–12 Hz) (P = .027), although this could reflect the shift in the peak of their Alpha power (in Analysis 1). Alpha disruption affected bilateral temporal cortices, but also extended into orbitofrontal regions. Relative power in beta band (12 - 30 Hz) was significantly decreased in MCI (P = 9.9 · 10−5), as in Analysis 1, although the significant cluster included widespread regions affecting most cortical regions, the peak was localized over bilateral temporoparietal areas such as angular gyrus, inferior parietal lobe, and middle temporal gyri.
Fig. 5.
Differences in relative power across frequency bands between MCI and controls. (A) Delta band (2–4 Hz); (B) Theta band (4–8 Hz); (C) Alpha band (8–12 Hz); (D) Beta band (12–30 Hz). Abbreviation: MCI, mild cognitive impairment.
3.5.1. Functional connectivity analysis
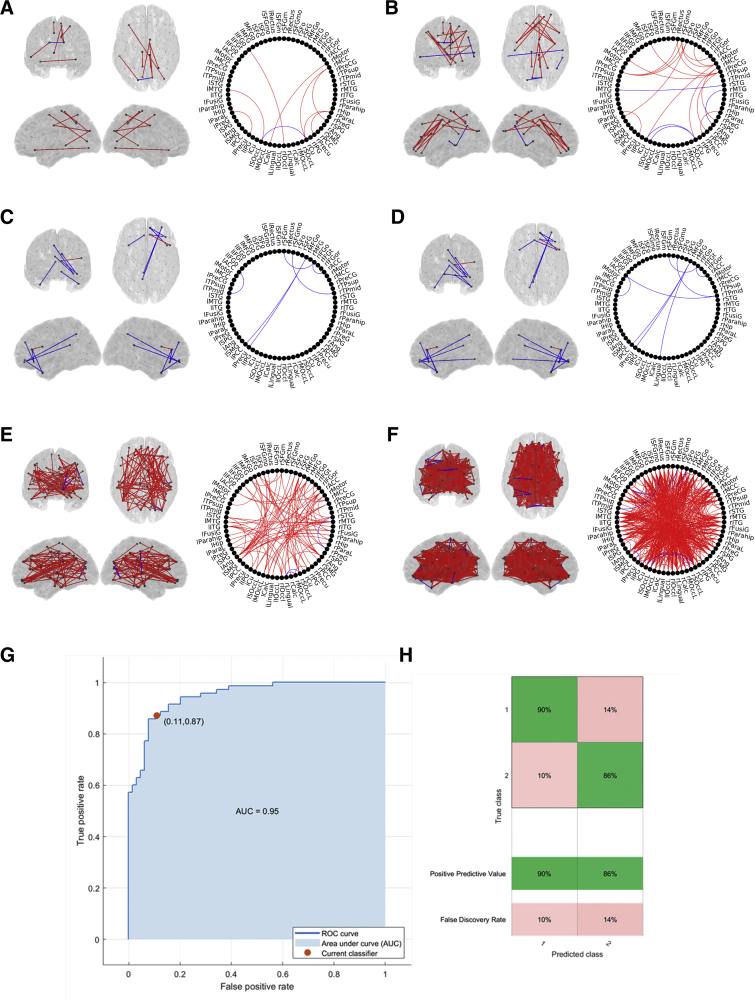
The functional connectivity analyses revealed several networks with an abnormal pattern in the MCI group (Fig. 6). In theta band, both PLV and MI showed differences between the patients with MCI and the controls. Several motifs could be identified. In essence, two main results were found: (1) PLV and MI measurements, using PCA and centroid methods, detected a parieto-occipital increased connectivity in the MCI group when compared with the control group; (2) in addition, the MI values, using the centroid method, showed a frontofrontal enhanced connectivity along with a diminished parietotemporal connectivity in the MCI group when compared with controls. Alpha band showed the same significant motif for both PLV (up) and MI (down) using the PCA method. The motif showed a frontofrontal and frontoparietal decreased connectivity in the MCI group when compared with the healthy control group. In broadband MI analyses showed an increase in connectivity in MCI at a global level, when compared with the healthy controls. These results appeared both using both the PCA-based and the centroid-based connectivity. Analysis of Site effects revealed distributed brain-wide differences, but there were no significant differences in the Site by Group interaction, suggesting that the group differences are not driven by acquisition differences.
Fig. 6.
Differences in functional connectivity (PLV and MI) between MCI and controls within frequency bands. (A) Theta band PLV based on centroid areas. (B) Theta band MI based on centroids areas; (C) Alpha band PLV based on PCA areas; (D) Alpha band MI based on PCA areas; (E) Broadband MI based on PCA areas; (F) Broadband MI based on areas centroids. Red lines: MCI > Controls; Blue Lines: Controls > MCI. (G) Receiving operator curve (ROC) of the support vector machine classifier trained using six FC-related variables. (H) Confusion matrix showing the positive predictive values and false discovery rate of the classifier. Abbreviations: MCI, mild cognitive impairment; PCA, principal component analysis; FC, functional connectivity; MI, mutual information; PLV, phase-locking value.
3.5.2. Classification based on source power and connectivity
The classifier was first trained using four power-related features, calculated as the mean value for each subject in the significant clusters in the source level power analysis, reaching an accuracy of 62.7 % (AUC of 0.69), comparable with Analysis 2 that used the distribution of power over all sensors. However, using functional connectivity features produced much better classification. The best trade-off between a reduced subset of features and a high accuracy was obtained by selecting the MI-derived subnetworks in theta, alpha, and broadband (6 features). In this case, the classifier reached an accuracy of 88% (AUC of 0.95), as shown in Fig. 6. Therefore, we conclude that using a few MEG functional connectivity-related features in the source domain from two frequency bands and the broadband signal allows us to distinguish between control and MCI subjects with high accuracy in this multisite study.
4. Discussion
We have explored MEG data acquisition, preprocessing, and analyses for multisite resting-state data, where data were pooled a posteriori (i.e., after individual studies were designed). We show that it is feasible to estimate differences between patients with MCI and controls in MEG signals, without variations in acquisition dominating the outcome. Nonetheless, we recommend that when combing existing data sets, the raw data characteristics are compared (for example as in Tables 1 and 2), and potential site differences in the MEG data included in the statistical models. We found MCI/control differences according to three different analyses, confirming that MEG is sensitive to changes in the underlying neurophysiology in patients with MCI, and report cross-validated classification accuracies for future reference for properly planned trials and stratification studies.
In general, we confirmed the slowing in oscillatory power and reduction in the peak frequency of the alpha band in patients, which are robust features of MCI and AD [19], [20], [21]. This pattern, particularly the increase in low-frequency power in the MCI group, was common across all three analyses, despite their differences in using normalized versus raw power, sensor versus source-space and different spectral estimators. When examining a single data feature—the frequency of the peak in the alpha band—patients could be classified with an accuracy of 61%. When considering the distribution of power over all (gradiometer) sensors and a large range of frequencies, an AUC of 66% could be achieved. This was less than that achieved (using the same classification algorithm) based on the gray-matter estimated from an MRI scan (accuracy of 68%). However, it should be noted that the MCI label is often influenced by examination of a comparable clinical MRI scan, which may bias the present MRI-based accuracy. More importantly, when combining the MEG and MRI data, the classification accuracy increased to 76%, suggesting that MEG power changes provide complementary information above MRI. When going further and estimating the cortical sources of the MEG power, and then the functional connectivity between those sources, accuracy of 88% was achieved. Although this figure may be somewhat inflated by virtue of the fact that the connectivity features used for classification were first selected by using all of the data (when comparing groups), it suggests that alterations to brain functional connectivity are the most sensitive index of MCI.
We also showed differences in power spectra across the Cambridge and Madrid sites. These were quite marked in some cases. This could reflect differences in the MEG scanners, or in the characteristics of the controls and/or patients recruited at each site (beyond differences in age and sex, which were included as covariates of no interest). Interestingly, site differences were also seen in the MRI VBM analyses, across many voxels throughout the brain, which probably reflects well-known scanner differences (e.g., imaging gradient nonlinearity [22]) rather than participant characteristics. There were also smaller interactions between site and group, that is, situations where the differences between controls and patients depended on site. Importantly, however, all of the differences between patients and controls, for example, in the above classification results, were after adjusting for any site effects. In other words, these frequency-specific abnormalities appear robust to differences in acquisition, as well as type of analysis.
Patients and controls, as well as sites, also differed in other acquisition details, such as number of bad channels, head motion in the MEG scanner, mean head position in the MEG scanner, time of day of scan and the tasks performed before the resting-state scan. We did not explore the effects of all these variables on the MEG data, and some of these differences (such as head motion and position) should have been corrected by the data preprocessing. Nonetheless, these factors will be important to measure and consider in future, more controlled studies/trials.
Validation of these measures on an independent data set would indicate the utility in obtaining a clinically valid biomarker. Brain network connectivity measures are key markers of disease; neural networks are particularly vulnerable to neural dysfunction even in preclinical disease [23] and abnormalities can index disease severity [24], [25] and predict conversion from MCI to AD [26], [27]. We show patterns of functional connectivity within frequency bands, comparable with previous research: In frontal regions, connectivity was enhanced in the theta band, but diminished in the alpha band, and connectivity to posterior regions was reduced in both bands (broadband enhancement was probably dominated by slow frequency bands). Hyperconnectivity is a recurrent finding observed in MCI [28], [29], which typically diminishes with advancing disease [30], [31]. Although changes in connectivity are common to other dementia syndromes (e.g. [32]), networks are selectively vulnerable depending on the region affected and pathology of disease [33], [34] and a pool of connectivity measures can be used to discriminate between illnesses [33], [35].
Further work to validate reliability and reproducibility of MEG features for dementia classification, as well work to inform disease biology, would be facilitated by multisite studies that can provide faster and larger throughput of data from diverse populations. Pooled data could be used to benchmark future studies, including validation of new experiments, increasing specificity by including other types of neurodegeneration, examining prodromal genetically susceptible individuals and assessing of therapeutic treatments. Using a standardized framework mutually agreed across scientific groups for a short protocol such as resting state, would reduce methods variance to allow for direct comparisons of the data, but without limiting scientific exploration of novel methods and paradigms. It could also, of course, be applied to task-based analyses. These outputs, with open-source code and accessible data, will support academic and pharma initiatives in dementia research, potentially providing crucial tools for stratifying patients and identifying disease modifying targets.
Research in context.
-
1.
Systematic review: The literature on magnetoencephalography methods and analyses for dementia and other disorders was examined using PubMED as well as in depth discussions as part of the BioFIND meetings.
-
2.
Interpretation: It is evident that magnetoencephalography can identify sensitive and robust features of neurodegeneration in dementia. With an increasing number of studies using similar protocols, harmonizing data sets across multiple sites will be beneficial for collaborative large-scale studies. Here we pooled resting-state data from three sites, and illustrate three different sets of analyses. Importantly, our results show clear differences between patients with mild cognitive impairment and healthy controls, over and above differences in site acquisition and analysis techniques.
-
3.
Future directions: We recommend a strategy for harmonizing data acquisition for resting state and, for existing data sets, methods to harmonize pooled data from different sites. We created a database of patients with mild cognitive impairment and matched healthy controls, with scripts for potential analyses, which are available for research purposes.
Acknowledgments
The authors would like to thank all the BioFIND members for their input, including Arjan Hillebrand, Alida Gouw, and Jyrki Makela. They thank Christophe Phillips for his help implementing MKL within the PRONTO toolbox. The study was funded by JPND “Harmonisation and Alignment in Brain Imaging Methods for Neurodegeneration” (MR/P502017/1). R.N.H. was additionally supported by MRC grant SUAG/010-RG91365; L.E.H. and J.B.R. were additionally supported by MRC grant SUAG/004-RG91365, the Cambridge NIHR Biomedical Research Centre and Wellcome Trust (103838) and the Dementias Platform UK. Cam-CAN data collection was supported by the Biotechnology and Biological Sciences Research Council (BB/H008217/1). A.J.Q., M.W.W., and A.C.N. are additionally supported by a NIHR Oxford Health Biomedical Research Centre and Wellcome Investigator Awards to MWW (106183/Z/14/Z) and ACN (104571/Z/14/Z), The Wellcome Centre for Integrative Neuroimaging is supported by core funding from the Wellcome Trust (203139/Z/16/Z).
Footnotes
A full list of contributors can be found in: http://www.neurodegenerationresearch.eu/wp-content/uploads/2016/06/JPND_Project-Fact-Sheet_BioFIND.pdf
Supplementary data related to this article can be found at https://doi.org/10.1016/j.dadm.2019.04.009.
Supplementary Data
References
- 1.Lopez-Sanz D., Serrano N., Maestu F. The Role of Magnetoencephalography in the Early Stages of Alzheimer's Disease. Front Neurosci. 2018;12:572. doi: 10.3389/fnins.2018.00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zamrini E., Maestu F., Pekkonen E., Funke M., Makela J., Riley M. Magnetoencephalography as a putative biomarker for Alzheimer's disease. Int J Alzheimers Dis. 2011;2011:280289. doi: 10.4061/2011/280289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsvetanov K.A., Henson R.N., Tyler L.K., Davis S.W., Shafto M.A., Taylor J.R. The effect of ageing on fMRI: Correction for the confounding effects of vascular reactivity evaluated by joint fMRI and MEG in 335 adults. Hum Brain Mapp. 2015;36:2248–2269. doi: 10.1002/hbm.22768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hillary F.G., Roman C.A., Venkatesan U., Rajtmajer S.M., Bajo R., Castellanos N.D. Hyperconnectivity is a fundamental response to neurological disruption. Neuropsychology. 2015;29:59–75. doi: 10.1037/neu0000110. [DOI] [PubMed] [Google Scholar]
- 5.Hillebrand A., Gaetz W., Furlong P.L., Gouw A.A., Stam C.J. Practical guidelines for clinical magnetoencephalography - Another step towards best practice. Clin Neurophysiol. 2018;129:1709–1711. doi: 10.1016/j.clinph.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Weiner M.W., Veitch D.P., Aisen P.S., Beckett L.A., Cairns N.J., Cedarbaum J. Impact of the Alzheimer's Disease Neuroimaging Initiative, 2004 to 2014. Alzheimers Dement. 2015;11:865–884. doi: 10.1016/j.jalz.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hari R., Baillet S., Barnes G., Burgess R., Forss N., Gross J. IFCN-endorsed practical guidelines for clinical magnetoencephalography (MEG) Clin Neurophysiol. 2018;129:1720–1747. doi: 10.1016/j.clinph.2018.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albert M.S., DeKosky S.T., Dickson D., Dubois B., Feldman H.H., Fox N.C. The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quinn A.J., Vidaurre D., Abeysuriya R., Becker R., Nobre A.C., Woolrich M.W. Task-Evoked Dynamic Network Analysis Through Hidden Markov Modeling. Front Neurosci. 2018;12:603. doi: 10.3389/fnins.2018.00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oostenveld R., Fries P., Maris E., Schoffelen J.M. FieldTrip: Open source software for advanced analysis of MEG, EEG, and invasive electrophysiological data. Comput Intell Neurosci. 2011;2011:156869. doi: 10.1155/2011/156869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nolte G. The magnetic lead field theorem in the quasi-static approximation and its use for magnetoencephalography forward calculation in realistic volume conductors. Phys Med Biol. 2003;48:3637–3652. doi: 10.1088/0031-9155/48/22/002. [DOI] [PubMed] [Google Scholar]
- 12.Tzourio-Mazoyer N., Landeau B., Papathanassiou D., Crivello F., Etard O., Delcroix N. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- 13.Desikan R.S., Segonne F., Fischl B., Quinn B.T., Dickerson B.C., Blacker D. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 14.Van Veen B.D., Van Drongelen W., M Y., Suzuki A. Localization of brain electrical activity via linearly constrained minimum variance spatial filtering. IEEE Trans Biomed Eng. 1997;44:867–880. doi: 10.1109/10.623056. [DOI] [PubMed] [Google Scholar]
- 15.Li Y., Qin Y., Chen X., Li W. Exploring the functional brain network of Alzheimer's disease: based on the computational experiment. PLoS One. 2013;8:e73186. doi: 10.1371/journal.pone.0073186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui X., Xiang J., Guo H., Yin G., Zhang H., Lan F. Classification of Alzheimer's disease, mild cognitive impairment, and normal controls with subnetwork selection and graph Kernel principal component analysis based on minimum spanning tree brain functional network. Front Comput Neurosci. 2018;12:31. doi: 10.3389/fncom.2018.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varoquaux G., Raamana P.R., Engemann D.A., Hoyos-Idrobo A., Schwartz Y., Thirion B. Assessing and tuning brain decoders: Cross-validation, caveats, and guidelines. Neuroimage. 2017;145:166–179. doi: 10.1016/j.neuroimage.2016.10.038. [DOI] [PubMed] [Google Scholar]
- 18.Pereda E., Garcia-Torres M., Melian-Batista B., Manas S., Mendez L., Gonzalez J.J. The blessing of Dimensionality: Feature Selection outperforms functional connectivity-based feature transformation to classify ADHD subjects from EEG patterns of phase synchronisation. PLoS One. 2018;13:e0201660. doi: 10.1371/journal.pone.0201660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garces P., Vicente R., Wibral M., Pineda-Pardo J.A., Lopez M.E., Aurtenetxe S. Brain-wide slowing of spontaneous alpha rhythms in mild cognitive impairment. Front Aging Neurosci. 2013;5:100. doi: 10.3389/fnagi.2013.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engels M.M., Hillebrand A., van der Flier W.M., Stam C.J., Scheltens P., van Straaten E.C. Slowing of Hippocampal activity correlates with cognitive decline in early onset Alzheimer's disease. An MEG study with virtual electrodes. Front Hum Neurosci. 2016;10:238. doi: 10.3389/fnhum.2016.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez-Sanz D., Bruna R., Garces P., Camara C., Serrano N., Rodriguez-Rojo I.C. Alpha band disruption in the AD-continuum starts in the Subjective Cognitive Decline stage: a MEG study. Sci Rep. 2016;6:37685. doi: 10.1038/srep37685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jovicich J., Czanner S., Greve D., Haley E., van der Kouwe A., Gollub R. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. Neuroimage. 2006;30:436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- 23.Maestu F., Pena J.M., Garces P., Gonzalez S., Bajo R., Bagic A. A multicenter study of the early detection of synaptic dysfunction in Mild Cognitive Impairment using Magnetoencephalography-derived functional connectivity. Neuroimage Clin. 2015;9:103–109. doi: 10.1016/j.nicl.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez M.E., Cuesta P., Garces P., Castellanos P.N., Aurtenetxe S., Bajo R. MEG spectral analysis in subtypes of mild cognitive impairment. Age (Dordr) 2014;36:9624. doi: 10.1007/s11357-014-9624-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stam C.J., Jones B.F., Manshanden I., van Cappellen van Walsum A.M., Montez T., Verbunt J.P. Magnetoencephalographic evaluation of resting-state functional connectivity in Alzheimer's disease. Neuroimage. 2006;32:1335–1344. doi: 10.1016/j.neuroimage.2006.05.033. [DOI] [PubMed] [Google Scholar]
- 26.Bajo R., Castellanos N.P., Cuesta P., Aurtenetxe S., Garcia-Prieto J., Gil-Gregorio P. Differential patterns of connectivity in progressive mild cognitive impairment. Brain Connect. 2012;2:21–24. doi: 10.1089/brain.2011.0069. [DOI] [PubMed] [Google Scholar]
- 27.Lopez M.E., Turrero A., Cuesta P., Lopez-Sanz D., Bruna R., Marcos A. Searching for Primary Predictors of Conversion from Mild Cognitive Impairment to Alzheimer's Disease: A Multivariate Follow-Up Study. J Alzheimers Dis. 2016;52:133–143. doi: 10.3233/JAD-151034. [DOI] [PubMed] [Google Scholar]
- 28.Bajo R., Maestu F., Nevado A., Sancho M., Gutierrez R., Campo P. Functional connectivity in mild cognitive impairment during a memory task: implications for the disconnection hypothesis. J Alzheimers Dis. 2010;22:183–193. doi: 10.3233/JAD-2010-100177. [DOI] [PubMed] [Google Scholar]
- 29.Lopez-Sanz D., Bruna R., Garces P., Martin-Buro M.C., Walter S., Delgado M.L. Functional connectivity disruption in subjective cognitive decline and mild cognitive impairment: a common pattern of alterations. Front Aging Neurosci. 2017;9:109. doi: 10.3389/fnagi.2017.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engels M.M.A., van der Flier W.M., Stam C.J., Hillebrand A., Scheltens P., van Straaten E.C.W. Alzheimer's disease: The state of the art in resting-state magnetoencephalography. Clin Neurophysiol. 2017;128:1426–1437. doi: 10.1016/j.clinph.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 31.Stam C.J., de Haan W., Daffertshofer A., Jones B.F., Manshanden I., van Cappellen van Walsum A.M. Graph theoretical analysis of magnetoencephalographic functional connectivity in Alzheimer's disease. Brain. 2009;132:213–224. doi: 10.1093/brain/awn262. [DOI] [PubMed] [Google Scholar]
- 32.Hughes L.E., Ghosh B.C., Rowe J.B. Reorganisation of brain networks in frontotemporal dementia and progressive supranuclear palsy. Neuroimage Clin. 2013;2:459–468. doi: 10.1016/j.nicl.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sami S., Williams N., Hughes L.E., Cope T.E., Rittman T., Coyle-Gilchrist I.T.S. Neurophysiological signatures of Alzheimer's disease and frontotemporal lobar degeneration: pathology versus phenotype. Brain. 2018;141:2500–2510. doi: 10.1093/brain/awy180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu M., Gouw A.A., Hillebrand A., Tijms B.M., Stam C.J., van Straaten E.C. Different functional connectivity and network topology in behavioral variant of frontotemporal dementia and Alzheimer's disease: an EEG study. Neurobiol Aging. 2016;42:150–162. doi: 10.1016/j.neurobiolaging.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 35.de Haan W., Pijnenburg Y.A., Strijers R.L., van der Made Y., van der Flier W.M., Scheltens P. Functional neural network analysis in frontotemporal dementia and Alzheimer's disease using EEG and graph theory. BMC Neurosci. 2009;10:101. doi: 10.1186/1471-2202-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.