Abstract
Colorectal cancer (CRC) is the third most common cancer worldwide, and liver metastasis presents a major cause of CRC-associated death. Extensive genomic analysis has provided valuable insight into the pathogenesis and progression of CRC; however, a comprehensive proteogenomic characterization of CRC liver metastasis (CLM) has yet to be reported. Here, we analyzed the proteomes of 44 paired normal colorectal tissues and CRC tissues with or without liver metastasis, as well as analyzed genomics of CRC characterized previously by The Cancer Genome Atlas (TCGA) to conduct integrated proteogenomic analyses. We identified a total of 2,170 significantly deregulated proteins associated with CLM, 14.88% of which were involved in metabolic pathways. The mutated peptide number was found to have potential prognosis value, and somatic variants revealed two metabolism-related genes UQCR5 and FDFT1 that frequently mutated only in the liver metastatic cohort and displayed dysregulated protein abundance with biological function and clinical significance in CLM. Proteogenomic characterization and integrative and comparative genomic analysis provides functional context and prognostic value to annotate genomic abnormalities and affords a new paradigm for understanding human colon and rectal cancer liver metastasis.
Keywords: CRC, liver metastasis, proteomics, genomics, integrated analysis
Introduction
Colorectal cancer (CRC) is a significant contributor of cancer morbidity and mortality.1, 2, 3 Almost half of CRC patients die within 5 years of diagnosis due to the development of recurrent disease and metastasis.4, 5, 6, 7, 8 Therefore, it is important to illuminate the molecular basis of CRC liver metastasis (CLM) in hopes of developing new effective treatment modalities. The Cancer Genome Atlas (TCGA) has characterized the genomic features of many types of human cancers, including CRC,9, 10, 11 and the Clinical Proteomic Tumor Analysis Consortium has also performed CRC-integrated proteomic analyses.12, 13, 14, 15 However, the primary genetic basis of CLM has not been fully elucidated. Understanding the genetic and proteogenomic differences between primary colon cancer and associated metastases to the liver is essential for discovering metastasis-specific molecular biomarkers and for devising a better therapeutic approach for this disease.
Results and Discussion
To identify protein variants associated with CLM, we collected tumor samples and paired normal colorectal tissues (PNs) from 44 CRC tumor samples with (MT, n = 23) or without liver metastasis (NM, n = 21) in comparison with PNs (Table S1) to perform liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based shotgun proteomics profiling (Figure S1). A total of 12,177,738 spectra were used in the Andromeda engine search, and 63,720 unique peptides were identified in an assembly of 5,758 protein groups with a protein-level false discovery rate (FDR) of 1.0%. Ingenuity pathway analysis with all 5,758 identified proteins showed that about 55% of the proteins were from the cytoplasm, 28% were from the nucleus, 7% were from the plasma membrane, and 2% were from the extracellular space, whereas 8% of proteins remained unclassified (Figure S2A). The random predicted cellular distribution of the proteins supports the quality of the sample preparation. A scatterplot of protein abundance between CRC tissues and PNs showed that there was a great variation between the NM or MT tumors and PN (Figures S2B and S2C). However, the protein expression between the MT and NM group was positively correlated (R2 = 0.86) (Figure S2D). These results suggest that metastatic and non-metastatic CRCs share similar protein profiles and that there are common molecular alterations at each stage of tumor development.16, 17, 18
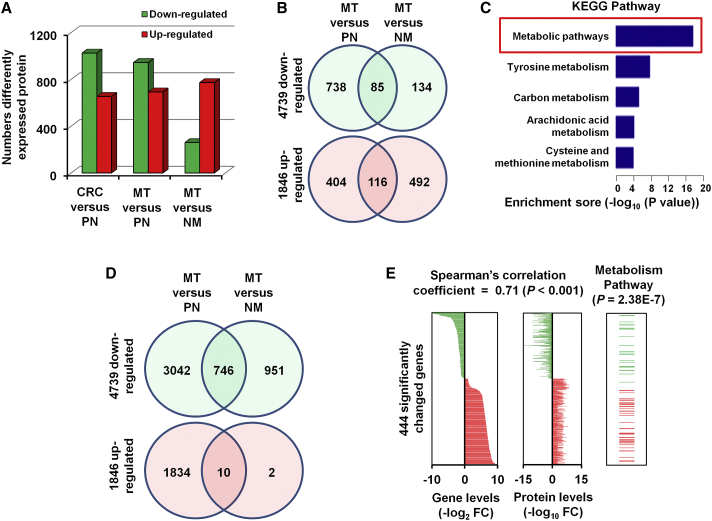
Among the 5,758 proteins, a total of 1,679 proteins were significantly altered between CRC tissue and normal colorectal tissues (Figure 1A, left bars). There were 2,170 proteins with significant differences in MT tissues when compared with PNs or NM tissues (Figures 1A, middle and right bars, and 1B). To explore the functions of proteins that are dysregulated in CLM, we used DAVID (Database for Annotation, Visualization and Integrated Discovery) analysis software to classify the gene ontology of the 2,170 significantly altered proteins in MT tissues19 (Figures S3A–S3C). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of differentially expressed proteins20 revealed that metabolic pathways were altered in the MT samples, with 323 significantly differentially expressed proteins (14.88%) involved in metabolic pathways (Figure S3D), which suggests that metabolism-related pathways may play important roles in the liver metastasis of CRC.21, 22, 23, 24
Figure 1.
Differently Expressed Protein and mRNA-Protein Correlation Analysis
(A) Numbers of differently expressed proteins (≥2-fold difference; p < 0.05 with a FDR q value < 0.05) between the 44 CRC and paired PN samples, and the 23 MT and 21 NM samples. (B) Significantly changed proteins among three groups from LC-MS/MS data. (C) KEGG pathway analysis of the 2,170 differentially expressed proteins. (D) Significantly changed genes among three groups from RNA-sequencing data. (E) 444 significantly changed genes showed significant mRNA-protein correlation, with a mean Spearman’s correlation coefficient of 0.71. Among these, 57 genes were enriched in metabolism pathways.
We performed unsupervised hierarchical clustering of the expression data from RNA-sequencing data to evaluate the genetic diversity between the primary and metastatic CRCs, based on the assumption that genetically similar CRCs and their matched metastatic foci would be closely related.25, 26, 27 As expected, the results showed that the MT and LM tissues had closely related expression profiles, indicating clonal and genetic similarity for these pairs. However, the NM samples were more distantly related, suggesting distinct genetic relationships between patients with and without metastasis (Figure S4A). A total of 2,299 genes from the RNA-sequencing were significantly changed in CLM (Figure S4B). KEGG pathway classification enrichment analysis indicated that 915 genes (39.8%) were enriched in metabolism pathways (Figure 1C), which was similar to the findings of the protein analysis (Figure S3D). These results provide additional confirmation that metabolism pathways may play a role in the carcinogenesis and development of CRC.28
For further confirmation of these results, we analyzed RNA-sequencing data from TCGA portal. A total of 6,585 significantly changed genes in CRC tissues were identified (Figure 1D). When compared with the 2,170 significantly changed CLM-associated proteins identified by LC-MS/MS, 444 significantly changed genes showed significant positive mRNA-protein correlation (Figure 1E, left two panels). To determine whether the concordance between protein and mRNA variation is related to the biological function of the gene product, we performed KEGG enrichment analysis, which indicated that among the 444 significantly deregulated genes or proteins, 57 are enriched in metabolic pathways (p = 2.38E−7) (Figure 1E, right panel). These findings further verify the role of metabolic pathway genes in CLM.
To examine the clinical significance of our findings, we evaluated the clinical characters data of 374 CRC patients from TCGA database for the 444 significantly deregulated genes or proteins (Table S2). Our results showed that 17 dysregulated genes were significantly associated with overall survival (OS) and progression-free survival (PFS) of CRC patients. Among them, high expression of four upregulated genes, BGN, EHD2, UQCR5, or COL1A2, was associated with poor survival of CRC patients.29, 30, 31, 32 Furthermore, high expression of FDFT1 significantly prolongs the OS and PFS of CRC patients relative to low expression of FDFT1 (Table S3).
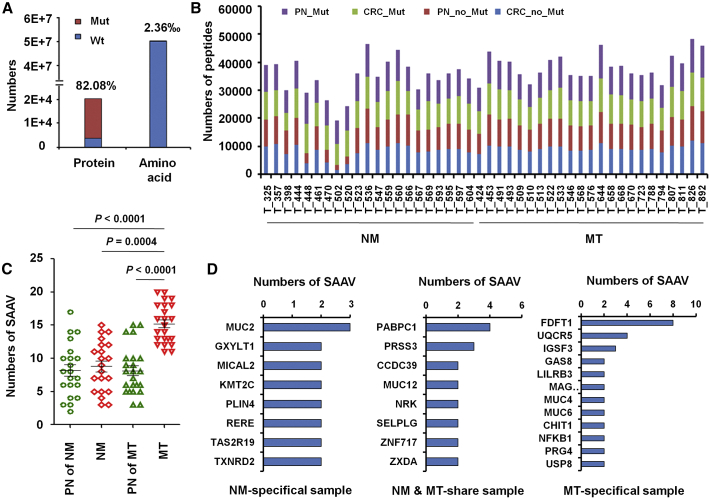
A fundamental goal of proteogenomics is to identify protein-coding alterations that are expressed at the protein level.33, 34, 35 However, standard database search approaches cannot identify variant peptides from MS/MS data.36, 37, 38 Therefore, we created a customized mutation database to search for single amino acid variants (SAAVs) in CRC. A SAAV library was prepared using 113,844 mutated sites in CRC tissues from cBioport, and 16,581 mutated proteins were identified, which constitutes 82.08% of 20,201 proteins in the human protein library (Figure 2A). We determined the total numbers of mutated and non-mutated peptides and tumor-specific mutant peptides (Figure 2B) and found that mutated peptide numbers in MT samples were significantly increased (Figure 2C), which indicates that the mutated peptide number has potential predictive value for CLM.
Figure 2.
Numbers of SAAVs in Paired PN, NM, or MT Samples
(A) The proportion of mutated proteins and amino acids in CRC samples were calculated by comparing LC-MS/MS data for the standard protein library and SAAV library. (B) The mutated and non-mutated peptides numbers of 44 paired CRC tissues. (C) Numbers of SAAVs in 21 NM, 23 MT, and their PNs. (D) Numbers of NM-specific, MT-specific, and NM- and MT-shared SAAVs. The mutated peptides were identified by comparing LC-MS/MS data for the standard protein library and SAAV library.
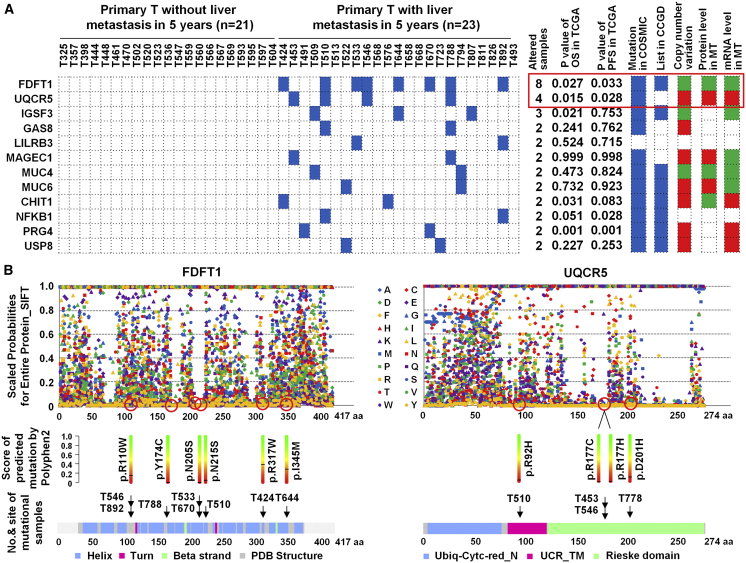
Among those, 140 SAAVs in 131 proteins occurred only in NM patients (Figure 2D; Table S4), and 223 proteins in 18 MT patients had 256 SAAVs, of which 110 SAAVs in 100 proteins occurred in both NM and MT samples (Figure 2D; Table S5), and 203 SAAVs in 184 proteins only occurred in MT samples (Figure 2D; Table S6). Similarly to single nucleotide variants (SNVs), some mutations reported previously, such as those in TP53, antigen-presenting cells (APCs), vascular endothelial growth factor (VEGF), and SMAD4 were found.39, 40, 41, 42, 43 Among these, two mRNA-protein positively changed and metabolism-related proteins, FDFT1, a 47-kDa membrane-associated enzyme located at a branch point in the mevalonate pathway involved in the replication stage of the hepatitis virus C (HCV) life cycle44 and paclitaxel sensitivity in hypopharynx cancer cell,45 and UQCR5, a component of the ubiquinol-cytochrome c reductase complex, amplifying in primary breast cancer core biopsy samples46, 47, 48 and overexpressing in gastric cancer,49 were found to be an occurrence of somatic alterations, which was validated with Sanger sequencing, in the 18.2% (8/44) and 9.1% (4/44) of CRC, and are specific to CLM (Figure 3A). Mutation in FDFT1 was predicted to lose the function; however, mutation in UQCR5 was predicted to gain the function, and increased copy number enhanced UQCR5 gene expression and lead to increased protein expression when scale-invariant feature transform (SIFT) and PolyPhen2 were used for predicted function of FDFT1 and UQCR5 mutations (Figure 3B).
Figure 3.
The Frequency and Distribution of Mutational Proteins Specifically Altered in MT for 44 CRC
(A) Heatmap comparing the frequency and distribution of 12 mutational proteins in 44 CRC (including 23 MT and 21 NM). Red, amplification or high expression; green, deletion or low expression. (B) Scaled probabilities for entire protein score of predicted mutation using SIFT and PolyPhen2 for FDFT1 and UQCR5 mutations.
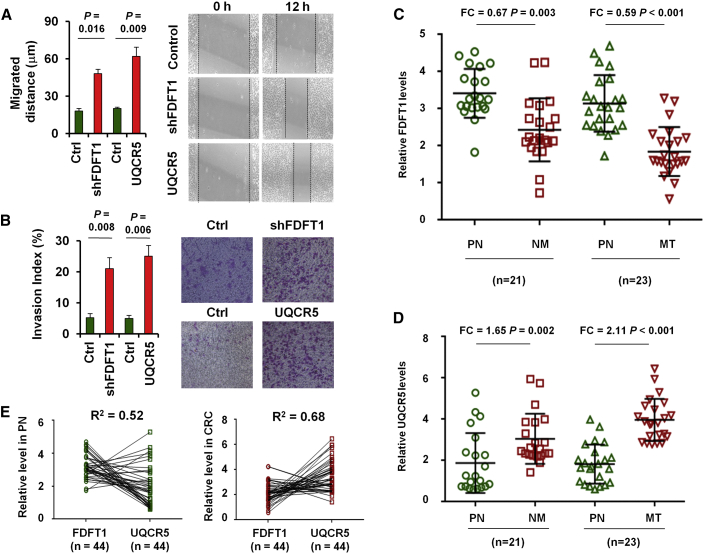
Moreover, FDFT1 knockdown (Figures S5A and S5B) or UQCR5 overexpression (Figures S5C and S5D) led to a significant increase of migrated distance (Figure 4A) and an obvious increase in matrigel invasion (Figure 4B). FDFT1 was found to downregulate in CRC tissues when compared with normal colorectal tissues (Figure 4C). Determination of UQCR5 gene expression identified overexpression (Figure 4D) and negatively correlated with FDFT1 expression in normal colorectal tissues and CRC tissues (Figure 4E).
Figure 4.
The Biological Function and Clinical Significance of FDFT1 and UQCR5 in CRC
Wound-healing assay (A) and migration abilities (B) of the parental and shFDFT1 or UQCR5 overexpressed SW480 cells. LC-MS/MS to quantify FDFT1 (C) and UQCR5 (D) levels in 44 paired CRC (including 23 MT and 21 NM) and normal colorectal tissues. (E) The expression correlation between FDFT1 and UQCR5 expression levels in 44 adjacently normal colorectal tissues or CRC tissues.
Patients with lower FDFT1 expression were found to have shorter median OS (Figures S6A and S7A) and PFS (Figures S6B and S7B), and high UQCR5 expression was an important risk factor for OS (Figure S6A and S7A) and PFS (Figures S6B and S7B). Moreover, concomitant low expression of FDFT1 and high expression of UQCR5 correlated with a shorter median OS (Figures S6A and S7A) and PFS (Figures S6B and S7B) in CRC patients. These studies provide direct evidence for contribution of metabolism-related genes.
To the best of our knowledge, this is the first comprehensive study to use proteogenomic profiling of primary CRCs from patients with or without liver metastasis to define the dominant events of metastatic lesions in terms of their expression and mutation. Our comprehensive integrative analysis of 44 colorectal tumors and normal pairs provides a number of insights into the biology of CLM and identifies potential therapeutic targets. Moreover, our characterization of the annotated metastatic CRC proteome clarifies the power of integrating genomics (SNVs) and proteomics (SAAVs). This approach provides new insights into the roles of these protein alterations in CLM, which can be broadly extended to understand the roles of protein mutation in other cancers.
Materials and Methods
Details of sample preparation and analysis are described in the Supplemental Materials and Methods. The study was examined and approved by the Ethics Committee of the Shanghai Tenth People’s Hospital, Tongji University School of Medicine. This study was registered with ClinicalTrials.gov: NCT02917707). Forty-four paired fresh primary tumors from the colorectal and PNs were used for proteogenomic analysis. The reliability of the exome analysis and somatic variant identification strategies was assessed using PCR and Sanger sequencing. Nine specimens from three CRC patients with metastasis and three specimens from three CRC patients without liver metastasis were obtained for RNA-sequencing analysis. Human CRC cell line SW480 was used for scratch-wound and in vitro invasion assays. All calculations were performed with SPSS 20.0 software.
Whole-exome sequencing data from this study are available for download through NCBI Sequence Read Archive: PRJNA358865. All RNA-sequencing data have been deposited in the Gene Expression Omnibus: GSE92914. All of the MS proteomics data have been deposited to iProX (https://www.iprox.org/index): IPX00083203 and IPX00083210.
Author Contributions
Y.-S.M., J.-B.L., X.-M.W., and D.F. designed and supervised the research and experiments. Y.-S.M., Z.-J.W., B.C., H.-W.Z., T.H., H.-D.L., H.X., Y.-Z.Z., Y.-Z.Y., R.-T.X., L.L.T., and D.F. analyzed the genetic data. Y.-S.M., Z.-J.W., B.C., H.-W.Z., T.H., S.-B.X., L.L., C.-L.L., J.-B.L., X.-M.W., and D.F. performed the proteogenomic experiments and analyzed the data. Y.-S.M., Z.-J.W., B.C., H.-W.Z., T.H., S.-B.X., L.L., R.-T.X., J.-B.L., and X.-M.W. performed tissue acquisition and clinical data collection. Y.-S.M., Z.-J.W., B.C., H.-W.Z., S.-B.X., L.L., R.-T.X., J.B.L., and D.F. performed functional validation experiments. All authors contributed to the final version of the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We would like to thank Dr. Wei Zhang for data analysis and critical discussion of the manuscript. This study was supported partly by grants from the National Natural Science Foundation of China (81772932, 81472202, 81201535, 81302065, 81671716, 81301993, 81702243, 81372175, and 81472209), The Fundamental Research Funds for the Central Universities (22120170212 and 22120170117), the Shanghai Natural Science Foundation (12ZR1436000 and 16ZR1428900), the Shanghai Municipal Commission of Health and Family Planning (201540228 and 201440398), the Construction of Clinical Medical Center for Tumor Biological Samples in Nantong (HS2016004), and the Jiangsu 333 Program (BRA2017205).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2019.04.008.
Contributor Information
Ji-Bin Liu, Email: tians2008@163.com.
Xu-Ming Wu, Email: ntzlyyywk@163.com.
Da Fu, Email: fu800da900@126.com.
Supplemental Information
References
- 1.Laissue P. The forkhead-box family of transcription factors: key molecular players in colorectal cancer pathogenesis. Mol. Cancer. 2019;18:5. doi: 10.1186/s12943-019-0938-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cristóbal I., Sanz-Alvarez M., Torrejón B., Santos A., Luque M., Rojo F., García-Foncillas J. Potential therapeutic impact of miR-145 deregulation in colorectal cancer. Mol. Ther. 2018;26:1399–1400. doi: 10.1016/j.ymthe.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X., Sun D., Tai J., Chen S., Hong S., Wang L. ZNF280A promotes proliferation and tumorigenicity via inactivating the hippo-signaling pathway in colorectal cancer. Mol. Ther. Oncolytics. 2019;12:204–213. doi: 10.1016/j.omto.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X.N., Wang Z.J., Ye C.X., Zhao B.C., Li Z.L., Yang Y. RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J. Exp. Clin. Cancer Res. 2018;37:325. doi: 10.1186/s13046-018-1006-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu Y., Wang C., Becker S.A., Hurst K., Nogueira L.M., Findlay V.J., Camp E.R. miR-145 antagonizes SNAI1-mediated stemness and radiation resistance in colorectal cancer. Mol. Ther. 2018;26:744–754. doi: 10.1016/j.ymthe.2017.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ibarrola-Villava M., Cervantes A., Bardelli A. Preclinical models for precision oncology. Biochim Biophys Acta Rev Cancer. 2018;1870:239–246. doi: 10.1016/j.bbcan.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 7.O’Leary M.P., Warner S.G., Kim S.I., Chaurasiya S., Lu J., Choi A.H., Park A.K., Woo Y., Fong Y., Chen N.G. A novel oncolytic chimeric orthopoxvirus encoding luciferase enables real-time view of colorectal cancer cell infection. Mol. Ther. Oncolytics. 2018;9:13–21. doi: 10.1016/j.omto.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bleau A.M., Redrado M., Nistal-Villan E., Villalba M., Exposito F., Redin E., de Aberasturi A.L., Larzabal L., Freire J., Gomez-Roman J., Calvo A. miR-146a targets c-met and abolishes colorectal cancer liver metastasis. Cancer Lett. 2018;414:257–267. doi: 10.1016/j.canlet.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu H., Wang C., Song H., Xu Y., Ji G. RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers. Mol. Cancer. 2019;18:8. doi: 10.1186/s12943-018-0932-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toiyama Y., Okugawa Y., Fleshman J., Richard Boland C., Goel A. MicroRNAs as potential liquid biopsy biomarkers in colorectal cancer: A systematic review. Biochim Biophys Acta Rev Cancer. 2018;1870:274–282. doi: 10.1016/j.bbcan.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kryeziu K., Bruun J., Guren T.K., Sveen A., Lothe R.A. Combination therapies with HSP90 inhibitors against colorectal cancer. Biochim Biophys Acta Rev Cancer. 2019;1871:240–247. doi: 10.1016/j.bbcan.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Zhang B., Wang J., Wang X., Zhu J., Liu Q., Shi Z., Chambers M.C., Zimmerman L.J., Shaddox K.F., Kim S., NCI CPTAC Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:382–387. doi: 10.1038/nature13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrera M., Llorens C., Rodríguez M., Herrera A., Ramos R., Gil B., Candia A., Larriba M.J., Garre P., Earl J. Differential distribution and enrichment of non-coding RNAs in exosomes from normal and Cancer-associated fibroblasts in colorectal cancer. Mol. Cancer. 2018;17:114. doi: 10.1186/s12943-018-0863-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J., Song P., Jiang T., Dai D., Wang H., Sun J., Zhu L., Xu W., Feng L., Shin V.Y. Heat shock factor 1 epigenetically stimulates glutaminase-1-dependent mTOR activation to promote colorectal carcinogenesis. Mol. Ther. 2018;26:1828–1839. doi: 10.1016/j.ymthe.2018.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhartiya D., Patel H., Ganguly R., Shaikh A., Shukla Y., Sharma D., Singh P. Novel insights into adult and cancer stem cell biology. Stem Cells Dev. 2018;27:1527–1539. doi: 10.1089/scd.2018.0118. [DOI] [PubMed] [Google Scholar]
- 17.Ribatti D. An historical note on the cell theory. Exp. Cell Res. 2018;364:1–4. doi: 10.1016/j.yexcr.2018.01.038. [DOI] [PubMed] [Google Scholar]
- 18.Shastri A., Choudhary G., Teixeira M., Gordon-Mitchell S., Ramachandra N., Bernard L., Bhattacharyya S., Lopez R., Pradhan K., Giricz O. Antisense STAT3 inhibitor decreases viability of myelodysplastic and leukemic stem cells. J. Clin. Invest. 2018;128:5479–5488. doi: 10.1172/JCI120156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma Y.S., Yu F., Zhong X.M., Lu G.X., Cong X.L., Xue S.B., Xie W.T., Hou L.K., Pang L.J., Wu W. miR-30 family reduction maintains self-renewal and promotes tumorigenesis in NSCLC-initiating cells by targeting oncogene TM4SF1. Mol. Ther. 2018;26:2751–2765. doi: 10.1016/j.ymthe.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y.J., Ma Y.S., Xia Q., Yu F., Lv Z.W., Jia C.Y., Jiang X.X., Zhang L., Shao Y.C., Xie W.T. MicroRNA-mRNA integrated analysis based on a case of well-differentiated thyroid cancer with both metastasis and metastatic recurrence. Oncol. Rep. 2018;40:3803–3811. doi: 10.3892/or.2018.6739. [DOI] [PubMed] [Google Scholar]
- 21.Xing F., Wang S., Zhou J. The expression of microRNA-598 inhibits ovarian cancer cell proliferation and metastasis by targeting URI. Mol. Ther. Oncolytics. 2018;12:9–15. doi: 10.1016/j.omto.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Jin F., Yang R., Wei Y., Wang D., Zhu Y., Wang X., Lu Y., Wang Y., Zen K., Li L. HIF-1α-induced miR-23a∼27a∼24 cluster promotes colorectal cancer progression via reprogramming metabolism. Cancer Lett. 2019;440-441:211–222. doi: 10.1016/j.canlet.2018.10.025. [DOI] [PubMed] [Google Scholar]
- 23.Pang R., Law W.L., Chu A.C., Poon J.T., Lam C.S., Chow A.K., Ng L., Cheung L.W., Lan X.R., Lan H.Y. A subpopulation of CD26+ cancer stem cells with metastatic capacity in human colorectal cancer. Cell Stem Cell. 2010;6:603–615. doi: 10.1016/j.stem.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Yue B., Liu C., Sun H., Liu M., Song C., Cui R., Qiu S., Zhong M. A positive feed-forward loop between lncRNA-CYTOR and Wnt/β-catenin signaling promotes metastasis of colon cancer. Mol. Ther. 2018;26:1287–1298. doi: 10.1016/j.ymthe.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu H.M., Yi W.W., Ma Y.S., Wu W., Yu F., Fan H.W., Lv Z.W., Yang H.Q., Chang Z.Y., Zhang C. Prognostic implications of decreased microRNA-101-3p expression in patients with non-small cell lung cancer. Oncol. Lett. 2018;16:7048–7056. doi: 10.3892/ol.2018.9559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu F., Liu J.B., Wu Z.J., Xie W.T., Zhong X.J., Hou L.K., Wu W., Lu H.M., Jiang X.H., Jiang J.J. Tumor suppressive microRNA-124a inhibits stemness and enhances gefitinib sensitivity of non-small cell lung cancer cells by targeting ubiquitin-specific protease 14. Cancer Lett. 2018;427:74–84. doi: 10.1016/j.canlet.2018.04.022. [DOI] [PubMed] [Google Scholar]
- 27.Ma Y.S., Wu Z.J., Bai R.Z., Dong H., Xie B.X., Wu X.H., Hang X.S., Liu A.N., Jiang X.H., Wang G.R. DRR1 promotes glioblastoma cell invasion and epithelial-mesenchymal transition via regulating AKT activation. Cancer Lett. 2018;423:86–94. doi: 10.1016/j.canlet.2018.03.015. [DOI] [PubMed] [Google Scholar]
- 28.Bhullar K.S., Lagarón N.O., McGowan E.M., Parmar I., Jha A., Hubbard B.P., Rupasinghe H.P.V. Kinase-targeted cancer therapies: progress, challenges and future directions. Mol. Cancer. 2018;17:48. doi: 10.1186/s12943-018-0804-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwon J.J., Factora T.D., Dey S., Kota J. A systematic review of miR-29 in cancer. Mol. Ther. Oncolytics. 2018;12:173–194. doi: 10.1016/j.omto.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma Y.S., Huang T., Zhong X.M., Zhang H.W., Cong X.L., Xu H., Lu G.X., Yu F., Xue S.B., Lv Z.W., Fu D. Proteogenomic characterization and comprehensive integrative genomic analysis of human colorectal cancer liver metastasis. Mol. Cancer. 2018;17:139. doi: 10.1186/s12943-018-0890-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nimmakayala R.K., Batra S.K., Ponnusamy M.P. Unraveling the journey of cancer stem cells from origin to metastasis. Biochim Biophys Acta Rev Cancer. 2019;1871:50–63. doi: 10.1016/j.bbcan.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ylä-Herttuala S. The pharmacology of gene therapy. Mol. Ther. 2017;25:1731–1732. doi: 10.1016/j.ymthe.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bach D.H., Park H.J., Lee S.K. The dual role of bone morphogenetic proteins in cancer. Mol. Ther. Oncolytics. 2017;8:1–13. doi: 10.1016/j.omto.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hou Z., Guo K., Sun X., Hu F., Chen Q., Luo X., Wang G., Hu J., Sun L. TRIB2 functions as novel oncogene in colorectal cancer by blocking cellular senescence through AP4/p21 signaling. Mol. Cancer. 2018;17:172. doi: 10.1186/s12943-018-0922-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krishnamurthy N., Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treat. Rev. 2018;62:50–60. doi: 10.1016/j.ctrv.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu S., Yue Y., Zhang S., Zhou C., Cheng X., Xie X., Wang X., Lu W. STON2 negatively modulates stem-like properties in ovarian cancer cells via DNMT1/MUC1 pathway. J. Exp. Clin. Cancer Res. 2018;37:305. doi: 10.1186/s13046-018-0977-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Duijnhoven F.J.B., Jenab M., Hveem K., Siersema P.D., Fedirko V., Duell E.J., Kampman E., Halfweeg A., van Kranen H.J., van den Ouweland J.M.W. Circulating concentrations of vitamin D in relation to pancreatic cancer risk in European populations. Int. J. Cancer. 2018;142:1189–1201. doi: 10.1002/ijc.31146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H.P., Peng C.C., Wu C.C., Chen C.H., Shih M.J., Huang M.Y., Lai Y.R., Chen Y.L., Chen T.W., Tang P. Inactivation of the tight junction gene CLDN11 by aberrant hypermethylation modulates tubulins polymerization and promotes cell migration in nasopharyngeal carcinoma. J. Exp. Clin. Cancer Res. 2018;37:102. doi: 10.1186/s13046-018-0754-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X., Guan H., Liu X.D., Xie D.F., Wang Y., Ma T., Huang B., Zhou P.K. p53 positively regulates the expression of cancer stem cell marker CD133 in HCT116 colon cancer cells. Oncol. Lett. 2018;16:431–438. doi: 10.3892/ol.2018.8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bressy C., Hastie E., Grdzelishvili V.Z. Combining oncolytic virotherapy with p53 tumor suppressor gene therapy. Mol. Ther. Oncolytics. 2017;5:20–40. doi: 10.1016/j.omto.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y., Lv J., Liu J., Liang X., Jin X., Xie J., Zhang L., Chen D., Fiskesund R., Tang K. STAT3/p53 pathway activation disrupts IFN-β-induced dormancy in tumor-repopulating cells. J. Clin. Invest. 2018;128:1057–1073. doi: 10.1172/JCI96329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tzchori I., Falah M., Shteynberg D., Levin Ashkenazi D., Loberman Z., Perry L., Flugelman M.Y. Improved patency of ePTFE grafts as a hemodialysis access site by seeding autologous endothelial cells expressing Fibulin-5 and VEGF. Mol. Ther. 2018;26:1660–1668. doi: 10.1016/j.ymthe.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang T., Ye L., Han Z., Liu Y., Yang Y., Peng Z., Fan J. miR-19b-3p promotes colon cancer proliferation and oxaliplatin-based chemoresistance by targeting SMAD4: validation by bioinformatics and experimental analyses. J. Exp. Clin. Cancer Res. 2017;36:131. doi: 10.1186/s13046-017-0602-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park E.M., Nguyen L.N., Lim Y.S., Hwang S.B. Farnesyl-diphosphate farnesyltransferase 1 regulates hepatitis C virus propagation. FEBS Lett. 2014;588:1813–1820. doi: 10.1016/j.febslet.2014.03.043. [DOI] [PubMed] [Google Scholar]
- 45.Xu C.Z., Shi R.J., Chen D., Sun Y.Y., Wu Q.W., Wang T., Wang P.H. Potential biomarkers for paclitaxel sensitivity in hypopharynx cancer cell. Int. J. Clin. Exp. Pathol. 2013;6:2745–2756. [PMC free article] [PubMed] [Google Scholar]
- 46.Natrajan R., Mackay A., Wilkerson P.M., Lambros M.B., Wetterskog D., Arnedos M., Shiu K.K., Geyer F.C., Langerød A., Kreike B. Functional characterization of the 19q12 amplicon in grade III breast cancers. Breast Cancer Res. 2012;14:R53. doi: 10.1186/bcr3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato T., Chang H.C., Bayeva M., Shapiro J.S., Ramos-Alonso L., Kouzu H., Jiang X., Liu T., Yar S., Sawicki K.T. mRNA-binding protein tristetraprolin is essential for cardiac response to iron deficiency by regulating mitochondrial function. Proc. Natl. Acad. Sci. USA. 2018;115:E6291–E6300. doi: 10.1073/pnas.1804701115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim H.C., Chang J., Lee H.S., Kwon H.J. Mitochondrial UQCRB as a new molecular prognostic biomarker of human colorectal cancer. Exp. Mol. Med. 2017;49:e391. doi: 10.1038/emm.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jun K.H., Kim S.Y., Yoon J.H., Song J.H., Park W.S. Amplification of the UQCRFS1 gene in gastric cancers. J. Gastric Cancer. 2012;12:73–80. doi: 10.5230/jgc.2012.12.2.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.