Abstract
Bordetella pertussis (B. pertussis) is the causative agent of pertussis, also referenced as whooping cough. Although pertussis has been appropriately controlled by routine immunization of infants, it has experienced a resurgence since the beginning of the 21st century. Given that elucidating the immune response to pertussis is a crucial factor to improve therapeutic and preventive treatments, we re-analyzed a time course microarray dataset of B. pertussis infection by applying a newly developed dynamic data analysis pipeline. Our results indicate that the immune response to B. pertussis is highly dynamic and heterologous across different organs during infection. Th1 and Th17 cells, which are two critical types of T helper cell populations in the immune response to B. pertussis, and follicular T helper cells (TFHs), which are also essential for generating antibodies, might be generated at different time points and distinct locations after infection. This phenomenon may indicate that different lymphoid organs may have their unique functions during infection. These findings provide a better understanding of the basic immunology of bacterial infection, which may provide valuable insights for the improvement of pertussis vaccine design in the future.
1. Introduction
1.1. Bordetella pertussis infection
B. pertussis is a Gram-negative coccobacillus responsible for pertussis in humans, also known as whooping cough (Parkhill et al., 2003). This pathogen acts by colonizing in the respiratory tract of its host resulting in a constellation of symptoms that affect the respiratory tract and other organs (J D Cherry, 1996). Every year, pertussis is responsible for 195,000 human deaths around the world, despite abundant efforts in preventing and treating the infection (Baxter, Bartlett, Rowhani-Rahbar, Fireman, & Klein, 2013; Celentano, Massari, Paramatti, Salmaso, & Tozzi, 2005; James D Cherry, 2012; Kretzschmar, Teunis, & Pebody, 2010; Mutangadura, 2004). Immunity to pertussis has been shown to last longer following natural pertussis infections compared to the immunity obtained through vaccination (Jason M Warfel, Zimmerman, & Merkel, 2014; Wendelboe, Van Rie, Salmaso, & Englund, 2005). The reemerging of pertussis is temporally relate to the shift of vaccine usage from whole-cell (wP) vaccine and acellular (aP) vaccine. The aP vaccine causes less side effect such as inflammation at the site of injection, which also means that weaker responses were generated at the invasion site. Thus, the local immune response at the invasion site might be essential for an optional immune response. Therefore, understanding the immune response to pertussis is critical to improving therapeutic and preventive measures.
1.2. Immunology of Bordetella pertussis infection
After B. pertussis infection, pulmonary antigen-presenting cells (APCs) will migrate and deliver bacterial antigens to local draining lymph nodes (DLN) via lymphatics (Hamilton-Easton & Eichelberger, 1995; Legge & Braciale, 2003). Also, inducible bronchus-associated lymphoid tissue (iBALT) may be induced in the lung and participate in the initiation of the immune response during the pulmonary infection (Fleige & Förster, 2017; Fleige et al., 2014). Besides these locally primed immunities, the spleen is the major organ that monitors the antigens in blood and provides a large pool of leukocytes during infection. Leukocytes will be activated in the spleen and migrate to the site of infection after activation and differentiation. CD4+ T helper cells are important immune response regulators, which can differentiate into different lineages, such as Th1, Th2, Th9, Th17, TFH, and Treg cells (Rebhahn et al., 2008). Each of these linages shapes the overall immune response in different ways to protect the host from various infections. For example, the Th1/Th17 combined response was shown to play a crucial protection role and lead to the optimal immunity against B. pertussis infection (Charlotte Andreasen & Carbonetti, 2009; Miller et al., 2008; Ross et al., 2013; J M Warfel & Merkel, 2013). However, it is unclear how Th1 and Th17 responses are generated, and it is also unclear how these can achieve the maximal level against B. pertussis infection (Raeven et al., 2014). On the other hand, follicular helper T cells (TFHs) are essential in helping B cells' antibody affinity maturation and memory B cell response in general (Cannons, Lu, & Schwartzberg, 2013; Tangye, Ma, Brink, & Deenick, 2013), but the specific role of TFH cells in immune responses against B. pertussis infection is not yet clear (Brummelman, Wilk, Han, van Els, & Mills, 2015).
1.3. Data resource
To investigate immunological signatures in different tissues during B. pertussis infection, Raeven et al. (Raeven et al., 2014). Conducted an elegant and extensive system biology study in the mouse model. By collecting and analyzing microarray data, flow cytometry data, and multiplex immunoassays data from different tissues at multiple time points after B. pertussis infection, Raeven et al. systematically elucidated a series of subsequent molecular and cellular events related with the innate and adaptive immune response to B. pertussis infection. In addition to these novel findings, we believe that this time-course gene expression dataset may also provide more valuable information regarding location and timing of immune responses against B. pertussis infection, which is worth more detailed reanalyses.
1.4. Modeling work and aims
In this paper, we re-analyzed the microarray data from Raeven et al. using a novel time course gene expression data analysis pipeline (Carey, Ramírez, Wu, & Wu, 2018), recently developed by Carey et al. In particular, we first identified dynamic response genes (DRGs), i.e., genes that exhibited significant changes in expression levels over time, and then clustered the DRGs with similar temporal expression patterns into groups of genes called gene response modules (GRMs) using the new analytic pipeline. Compared to the ANOVA analysis used in Raeven's original paper (Raeven et al., 2014), in our pipeline, we considered the time patterns and temporal information more efficiently so that intricate behavioral patterns of genetic response over time can be identified. Also, because genes are grouped by their expression patterns (GRMs), a more thorough investigation of these patterns could be conducted separately to determine their aggregated role in response to infection. This approach assumes that genes displaying similar expression behaviors over time are likely to play a similar role within the cell. Also, our study focused on the adaptive immune response and showed that different types of immune responses were initiated in the lung and the spleen, respectively. In particular, we found that Th17 cell response was significant only in the lung instead of spleen, the TFH cell response was contributed mainly from the spleen while the GC reactions appeared in the lung, which suggested that the B cells' affinity maturation and generation of memory B cells might happen only locally in the lung. Our analysis also showed the importance of the immune responses generated in the antigen invasion sit, which might explain the reduced effectiveness of the current B. Pertussis vaccine.
2. Materials and methods
2.1. Experiments and microarray data
In this study, we reanalyzed the microarray dataset GSE53294 from Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse53294). The metadata and detailed experiments were described in the original publication by Raeven et al. (Raeven et al., 2014). Briefly, a group of 8-weeks-old female BALB/c mice was intranasally infected with B. pertussis B1817 at 2 × 105 cfu, and infected mice were euthanized at different time points (three mice at each time point) until 28 days post infection (p.i.). RNA samples were extracted from lungs at 9 different time points, including naïve samples before infection (time 0) as a control, 4 h, and 2, 4, 7, 10, 14, 21 and 28 days p.i., while RNA samples were extracted from spleens at 10 different time points, including naïve samples before infection (time 0), 4 h, 1, 2, 4, 7, 10, 14, 21 and 28 days p.i. The RNA samples were amplified and labeled with Cy3. Also, equimolar amounts of amplified RNA samples from lung or spleen tissues were pooled together and labeled with Cy5 as standard reference samples for lung or spleen tissues, respectively. Then mixtures (1:1) of RNA samples and standard reference were hybridized on NimbleGen 12 × 135 k Mus musculus microarrays and read by an Agilent DNA microarray scanner G2565CA. The raw microarray data were normalized and summarized using a four-step approach: (1) natural log-transformation; (2) quantile normalization of all scans; (3) correcting the sample spot signal for the corresponding reference spot signal; and (4) averaging data from replicate probe spots.
2.2. Data analysis pipeline
We applied our newly developed analysis pipeline to the time course microarray data collected by Raeven et al. (Raeven et al., 2014). For the two types of tissue samples (i.e., ‘lung’ and ‘spleen’), the microarray gene expression data from three replicates were combined by taking the median of all probes' expression level at each time point. Then we applied the data analysis pipeline (Carey et al., 2018) to the combined data. Our data analysis pipeline consists of several steps of statistical and modeling approaches.
2.2.1. Detect dynamic response genes (DRGs)
We applied a modified F-test to the normalized gene expression data at probe levels to identify the genes (or probes) with significant expression level changes over time, which were then referred to as dynamic response genes (DRGs).
We assume the expression level of the ith gene (xi) in a sample is a smooth function over time, and the measurement of this gene (yi) in microarray assay is a discrete observation of this smooth function with error ().
For i = 1, …,N, and k = 1, …,K, where N is the number of genes, K is the number of times points. The noise is assumed to be an independently identically distributed (i.i.d.) Gaussian random viable with mean 0 and variance σ2.
For each gene, a K × 1 vector of the estimated measurement of expression level at each time point was generated by spline smoothing (SS). xi was approximated by a linear combination of L independent basis functions, , where the K × L matrix Bi denotes the basis function evaluated at time point t. The corresponding coefficients ci can be estimated by minimizing, where the first term defines the squared discrepancy between the observed centered gene expression and the estimated gene expression level . In the second term, the L × L matrix Ri is the integral of the squared second derivative of Bi. This term penalizes the curvature of so that the parameter λ controls the trade of the fit to the data and the smoothness of the curve. All the genes in one subject are assumed to have the same λ, which is estimated by the conventional method of minimizing the prediction error with generalized cross-validation (GCV). To identify DRGs after smoothing, we perform an F-test, which compare the goodness-of-fit of the null hypothesis vs. the alternative hypothesis . The F-statistic is given by
where are the degrees of freedom of the estimated curve, and are the residual sum of the square under the null model and alternative model. The total number of DRGs discovered might reflect the strength of stimulations or effect of interventions under each experimental condition, which is B. pertussis infection in this study. To make the results comparable for different experimental conditions, we focused on the top 10,000 DRGs with the highest F-scores for each of the experimental conditions (lung or spleen tissues in this study).
2.2.2. Cluster the DRGs into gene response modules (GRMs)
The infection progress is occupied by multiple asynchronous biological events which are controlled or regulated by a different set of genes. Thus, we assume that genes having temporally similar expression pattern may tend to participate in similar biological events. To have a better understanding biological events during the infection, we clustered the top 10,000 DRGs into gene response modules (GRMs) based on their temporal expression patterns using the iterative hierarchical clustering method (Carey, Wu, Gan, & Wu, 2016). A parameter α is needed to set the threshold of clustering. The average within-cluster correlation will be approximate α, while the between-cluster correlation will be below α. Briefly, this algorithm has four major steps: (1) Initialization: Selected DRGs firstly clustered by the hierarchical agglomerative clustering approach, where Spearman rank correlation used as distance metric with a threshold of α and the average of the gene expression level in the cluster is used in the linkage criteria; (2) Merge: Each cluster is treated as “new genes” and used the same rule as the initialization step to merge similar clusters. (3) Prune: New center of the cluster is calculated. Any gene, whose correlation to the center of the cluster is smaller than the threshold of α, is removed from this cluster and form a single gene cluster. (4) Converge: The merge and prune steps are repeated until the index of clusters converges, or between-cluster correlation is less than α.
2.2.3. Annotation
Also, the genes in the GRMs were annotated by gene ontology (GO) enrichment analysis, which is achieved by using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (Huang, Sherman, & Lempicki, 2008).
3. Results
3.1. Identification of dynamic response genes (DRGs)
We applied the analysis pipeline, as described above, to the time-course gene expression data set from the study of B. pertussis infection. We identified 11255 and 10273 significant DRGs out of 44,170 probes from the lung and spleen samples, respectively. For fair comparisons between the two tissues (lung vs. spleen), we focused on the top 10,000 DRGs and clustered them into gene response modules (GRMs) for each of the two tissue samples, from which the lung tissue data produced 61 GRMs while the spleen tissue data produced 88 GRMs.
The identified top 10,000 significant DRGs at the probe level corresponds to 7,645 and 7,495 unique genes in the lung and spleen, respectively. In both lung and spleen tissues, our analysis pipeline identified a considerably higher number of significant response genes after B. pertussis infection compared to those reported by Raeven et al., in which only 558 and 798 differentially expressed genes were identified for the lung tissue and spleen tissue, respectively. Because we considered the temporal gene expression data as the functional data and the time course information was efficiently used to identify DRGs with more power in our analysis pipeline, we can identify more significant genes.
To examine the general functions of these DRGs, we performed functional enrichment analyses on the identified DRGs, 460 biological process ontology terms were found significant (p < 0.05) in the spleen samples (Table S1) and 558 ontology terms were significant in the lung samples (Table S2). For the spleen samples, we found that most significant gene ontology (GO) terms were related to general biological cell behaviors, such as cellular metabolic process (GO:0044237) and single-organism cellular process (GO:0044763). In contrast, many genes (354 genes) were significantly related to regulations of the immune system process (GO: 0002682) in the lung samples. Although many of these gene ontology terms are vague and general, some of them are related to bacterial infection.
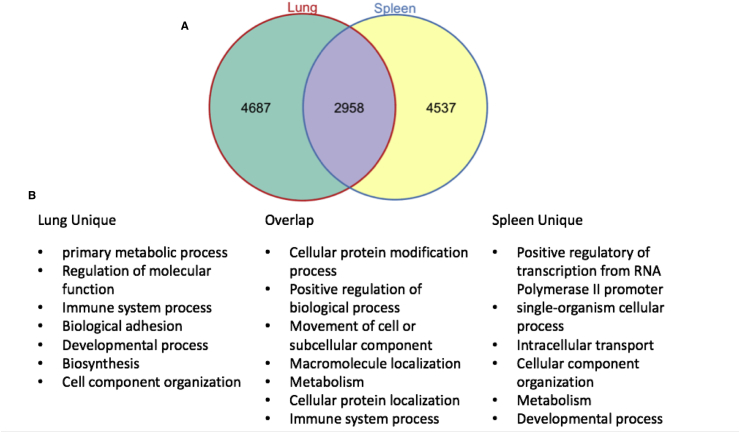
Among the top 10,000 significant probes, 2958 significant DRGs were shared between the lung and spleen, 4687 DRGs were only significant in the lung, and 4537 DRGs were only significant in the spleen (Fig. 1). Remarkably, among the 4687 DRGs uniquely identified in the lung, 419 of them were related to the immune system process (GO:0002376, p = 2.20E-07) via functional enrichment analysis. Similarly, the immune system process was also significant for the shared DRGs between the lung and spleen. In contrast, the immune response related genes were not significantly enriched among the significant 4537 DRGs uniquely identified in the spleen (Fig. 1). In particular, some significant immune response-related genes in the lung were related to effector functions of immune cells, such as IFNɣ, IL17a, and IL17f, which were consistent with the finding of a Th1/Th17 biased immune response against the pertussis infection identified by Raeven et al. Also, we also found CXCR5 and ICOS among unique significant DRGs in the lung and these genes are essential markers for follicular T helper cells (TFHs), which are critical to the germinal center formation, B cell proliferation, memory cell differentiation and affinity maturation (Crotty, 2011; Morita et al., 2011; Tangye et al., 2013).
Fig. 1.
The differences of the gene ontology enrichment of DRGs between spleen and lung. A. The Venn diagram for the number of significant DRGs in the lung and spleen. B. The significant gene ontology terms for each group of genes.
Among the shared 2958 significant DRGs between the lung and spleen, the enrichment analysis identified a total of 359 ontology terms. Despite lots of general biology process terms, we did find that the immune system process term (GO:0002376) was significant. We also identified different vital immune response processes or pathways (child terms of GO:0002376) such as innate immune (GO:0045087), antigen processing and presentation (KEGG:04612), leukocyte aggregation (GO:0070486), leukocyte activation (GO:0045321), and T cell proliferation (GO:0042098), which are necessary components for general immune responses to infections.
3.2. Gene response modules (GRMs)
To have a better understanding of the dynamic response genes (DRGs), we clustered them into gene response modules (GRMs) based on their expression patterns over time. Then we annotated each module and identified the critical biological functions or related biological processes for each module (Linel, Wu, Deng, & Wu, 2014; S.; Wu & Wu, 2013; Zou & Conzen, 2005). The top 10,000 most significant DRGs (probes) from spleen and lung samples were selected for clustering analysis, respectively.
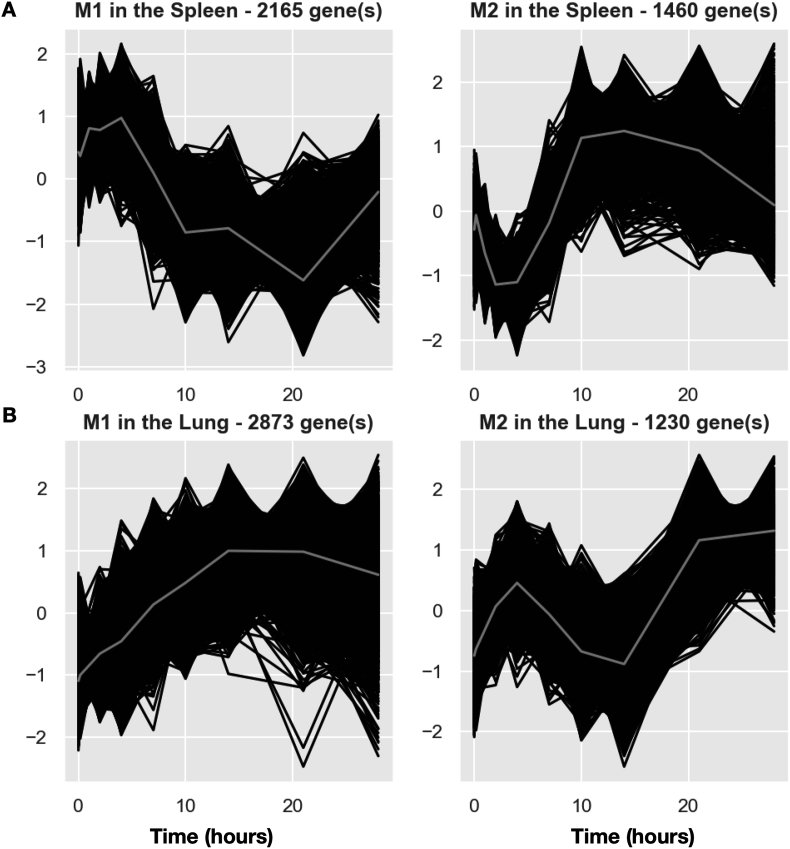
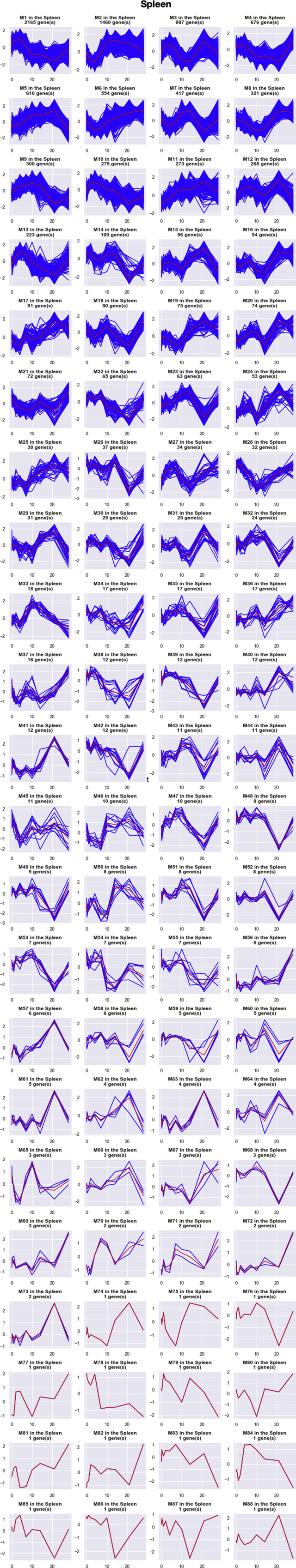
For spleen, the top 10,000 DRGs were clustered into 88 GRMs (i.e., clusters of genes with similar expression patterns over time) (Fig. 2A, S1 and S6 Table). Even though the top 10,000 DRGs from spleen cells were grouped into 88 GRMs, we found that immune-related genes were only enriched in the two largest modules (named splenic M1 and M2). Splenic M1 with 2165 genes had a gradual increase from day 0 to day 4, followed by a decrease from day 4 to day 21, and then recovered back almost to the baseline level at day 28. A important annotation of splenic M1 is the adaptive immune (GO:0002250) (S8 Table). After infection, the antigen-presenting cells (APCs) uptook the antigen from the lung and delivered the antigens to the spleen so that the adaptive immune cells were activated. Thus, the gene expression of the adaptive immune cells started to increase slowly in the spleen as the splenic M1's mean curve exhibited (Fig. 2A). After 4–5 days of infection, the activated adaptive immune cells began to migrate out of the spleen and were recruited to the lung rapidly. When the migration of adaptive immune cells from the spleen to the lung could not compensate for the generation or activation of new adaptive immune cells, it resulted in a decrease of gene expression levels for splenic M1 from 4 to 21 days after infection. At the same time, the immune cells still proliferated fast in the spleen and compensated the loss of splenic immune cells due to emigration. Then, the expression levels of M1 started to increase from day 21 to the pre-infection level at day 28 after the infection was controlled. The pattern of splenic M2, enriched as the general immune response, started to decrease rapidly from day 1 to day 4, followed by a recovery and overshoot to a peak at day 14 and then dropped back to the baseline level at day 28 (Fig. 2A). Although no specific immune response was significantly enriched for splenic M2, we suspect that M2 might be related to the innate immune response and we identified several innate immune genes such as PRDX2, ZBTB3, and CXCL1 in splenic M2. After infection, the innate immune cells usually were immediately recruited and migrated from the spleen to the lung. After day 4 of infection, the adaptive immune response started to kick in, and the innate immune cell recruitment from the spleen to the lung might slow down, that is why the M2 pattern started to recover quickly after a 4-day dip, and then it increased rapidly to a peak at day 14, possibly because the activation of innate immune cells continued even though these innate immune cells slowed down to migrate to the lung. Finally, the gene expression dropped back to the baseline level after the pathogen was controlled and cleared at day 28.
Fig. 2.
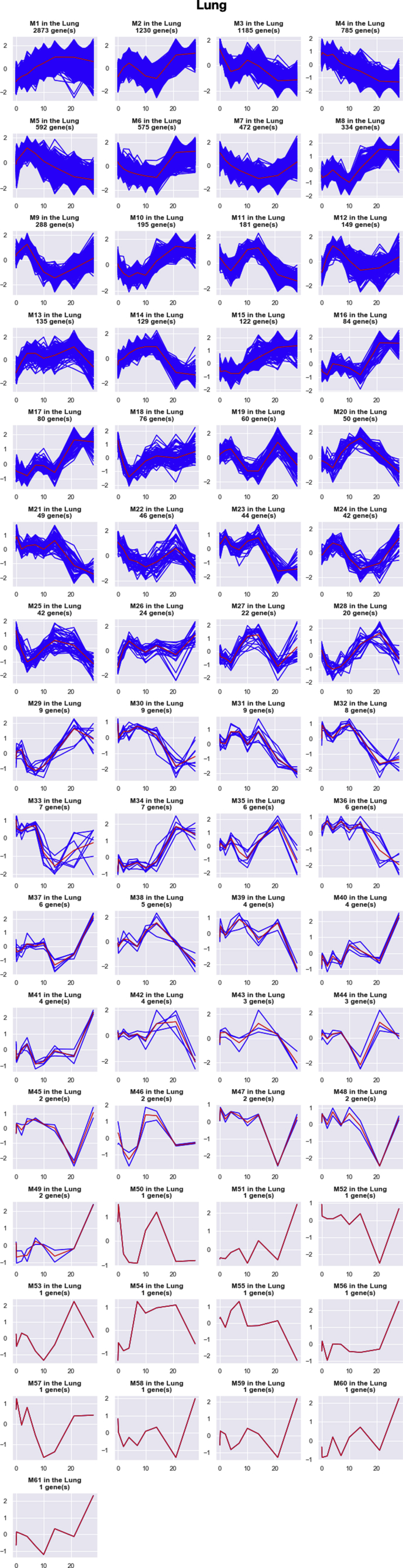
Pipeline analysis results in spleen and lung. A. Expression patterns of top 2 GRMs in the spleen. B. Expression patterns of top 2 GRMs in the lung. The total number of DRGs in each GRM is marked at the top of each plot. Black curves represent the expression of each DRGs in the cluster. Grey curves represent the mean expression of the GRM.
Similarly, we clustered the top 10,000 DRGs in the lung into 61 GRMs (Fig. 2B, S2 and S7 Table). In these GRMs, only the largest module, named pulmonary M1, was significant in immune response processes in gene ontology enrichment analysis (S9 Table). The mean curve of pulmonary M1 gradually and continuously increased from day 1 to day 21, and it started to decline when the pathogen was controlled after day 21. Correspondingly, the bacteria peaked at day 7–10 p.i. and decreased after that (see Raeven et al.). This GRM might reflect the overall immune response pattern, including both innate and adaptive immune responses in the lung against B. pertussis infection.
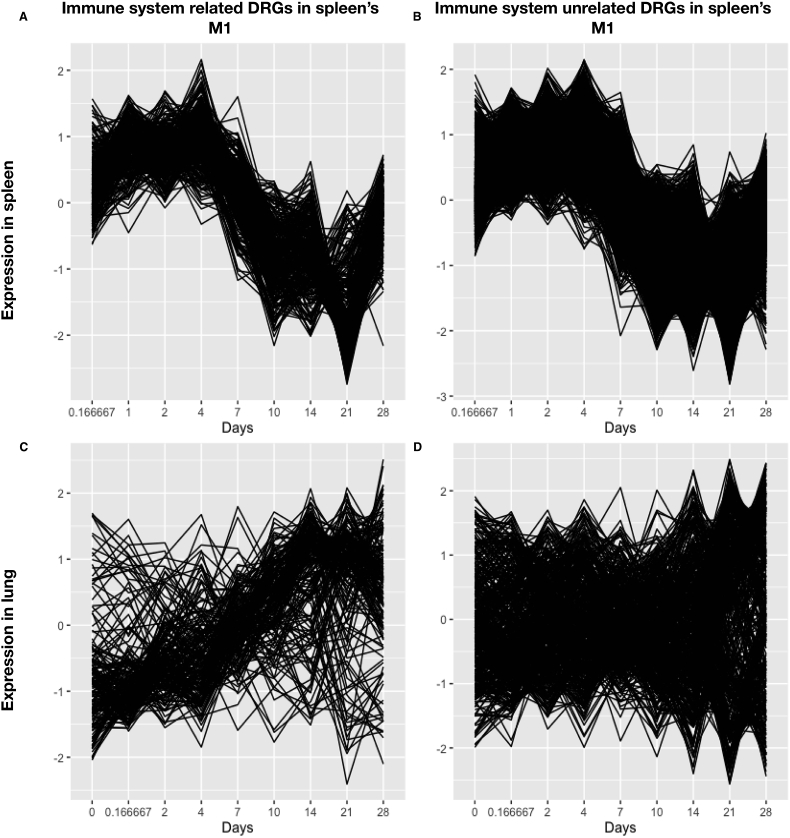
The spleen is the largest secondary lymphoid organ. It maintains a vast repertoire of immune cells and monitors the antigens in the blood. After infection, immune cells in the spleen will be activated and migrate to the infection site. We assume that this process can be monitored by time-course microarray assay, and we can get details of the development and migration of immune cells from microarray data. If a group of immune cells migrates for the spleen to lung, the ratio of those cells will decrease in the spleen and increase in the lung. Thus, the expression level of signature genes of this group of immune cells, i.e., immune-related genes, will decrease in the spleen and increase in the lung. The migrated cells have similar signature genes. After they migrated into the lung, the expression of those signature genes would be highly correlated with each other in the lung. We divide the genes in splenic M1 into immune-related genes (GO:0002376) and other non-immune related genes by their gene ontology terms, and we compared the expression patterns of the genes in these two groups in the spleen (Fig. 3A and B) with those in the lung (Fig. 3C and D). We observed that, for the immune response related DRGs identified in splenic M1, a large proportion of them either also belong to pulmonary M1 or expressed a similar temporal pattern to pulmonary M1 (Fig. 3C). In matter of fact, 34.7% immune response related DRGs identified in splenic M1 belong to pulmonary M1, and 61.4% of them were significantly (p < 0.05, Spearman's correlation test) correlated with the mean expression curve of pulmonary M1, which means that a large proportion of pulmonary M1 may be contributed by immune cells for spleen.
Fig. 3.
Expression of immune system-related genes and unrelated genes in the spleen and the lung, respectively. Immune-related genes (A, C) and unrelated (B, D) DRGs in the spleen M1 (A, B) and the lung (C, D) are plotted.
In contrast, for those genes unrelated to the immune response (Fig. 3 right panel), the temporal expression patterns in the lung were more chaotic (Fig. 3D). It supports our hypothesis that mainly immune cells migrating from the spleen to lung. We did similar analyses for other splenic GRMs, which are not significantly related to immune response, such as M2 and M3 (summarized in S10 table), this phenomenon was not observed. We carefully examined and compared the temporal patterns of splenic M1 and pulmonary M1 (Fig. 2) and found that the expression patterns of the splenic M1 and pulmonary M1 are similar for the first four days (slowly increased), and then become the opposite pattern from day 4 to day 28. During the initial stage of infection, the adaptive immune cells might not be fully differentiated in the spleen and might not migrate out of spleen to the lung. As aforementioned, some immune-related DRGs were significant uniquely in the lung, but not in the spleen (Fig. 1). We found that these immune-related DRGs, significantly expressed in the lung alone (under the GO term GO:0002376), were only enriched in pulmonary M1 (the odds ratio = 1.69, p = 3.54e-9 based on the hypergeometric test). The initial increase of the immune-related gene expression in the lung might suggest that some immune responses were initiated independently in the lung. After day 4 of infection, the expression of these immune-related DRGs in splenic M1 was reversed and started to decline (Fig. 2), which might reflect that the splenic immune cells were activated, recruited and migrated to the lung. It is warranted to tease out the relative contributions of the immune response between the spleen and local lymphoid tissues in the lung, which is beyond the scope of this study.
3.3. Different CD4 populations might be generated from separate compartments
It is important to dissect the origins and timing of different immune responses. CD4 T cells play an essential role in regulating the adaptive immune response. The characteristic of an immune response can be determined mainly by which CD4 T cell lineage was developed during the response (Jeong et al., 2014). In the original paper, Raeven et al. suggested that the immune response against B. pertussis infection was a Th1/Th17-bias response. By carefully examining the temporal response patterns of genetic markers for different types of immune cells, we may be able to extract more information regarding the origins and timing of different types of immune responses, including Th1, Th17, TFH and follicular dendritic cells (FDCs), so that a more detailed picture of immune response processes against B. pertussis infection can be established.
In general, all the key transcription factors such as TBX21, STAT4, RORC, and PD1 of Th1, Th2, Th17, and TFH were not in the top 10,000 significant DRGs (S11, S12 Table), which is not surprising since the transcription factors are usually expressed at extremely low levels and may not be detected using the gene expression microarray. However, we may use IFNɣ as a marker for Th1 cells, IL-17a as a marker for Th17 cells, and CXCR5 and ICOS as markers for TFH cells (Chevalier et al., 2011; Crotty, 2011; Rasheed, Rahn, Sallusto, Lipp, & Müller, 2006; Zhu, Yamane, & Paul, 2010). We also examined other types of immune responses such as Th2 cells, but their corresponding genetic markers, including IL-4, IL-5, and IL-13 were not in the top 10,000 significant DRGs in the lung or spleen (S11 and S12 Table), which might suggest that Th2 response was not significant for B. pertussis infection.
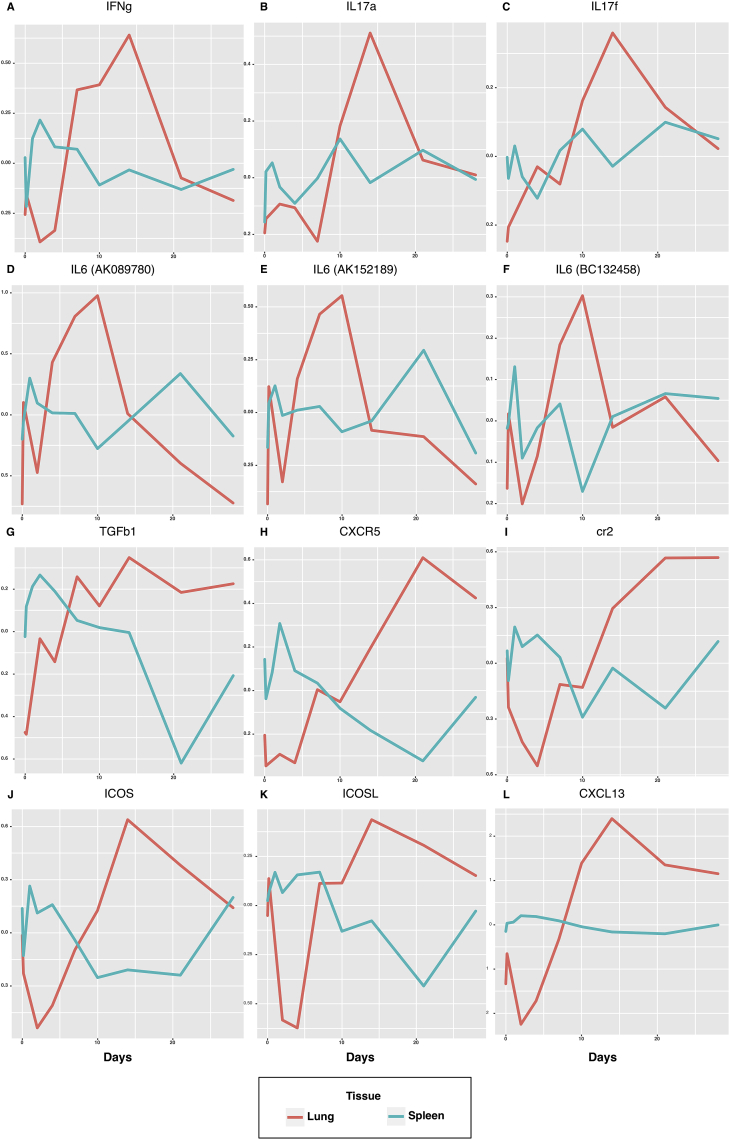
Th1 cells can magnify the macrophages' phagocytosis activity to eliminate extracellular bacterial infection (Ross et al., 2013). In the spleen, IFNG, coding the key cytokine of Th1 cell IFNɣ, was upregulated since day 1, and then decreased after day two p.i.(Fig. 4A). At the same time, the IFNɣ significantly increased in the lung after an initial dip, suggesting that the spleen might be an essential source of Th1 cells. Also, before the migration of IFNɣ expressing cells from the spleen to the lung on day 2–3 p.i., there was no IFNɣ increase in the lung (Fig. 4A). Thus, it was possible that all the Th1 response was initiated in the spleen and migrated to the lung after day two p.i. As early as day 7, the Th1 response reached a relatively high level and peaked at day 14 p.i. in the lung. After day 14 p.i., the expression of IFNɣ began to decrease to the baseline level after the infection was controlled.
Fig. 4.
Expression patterns ofbiomarker genesof CD4+ T-cell subpopulationsin the lung (red curve) and spleen (blue curve).
On the other hand, Th17 cells play a crucial function in mucosal immunity against fungi and extracellular bacteria (Stockinger, Veldhoen, & Martin, 2007). We found that the Th17's effecter cytokines IL-17a and IL-17f's expression level was only in the top 10,000 significant genes of the lung (Fig. 4B–C, S11, and S12 Table), and similar to Th1 cell's marker, Th17 cells' key cytokines in the lung started to increase after infection, peaked on day 14 p.i., and then declined after the infection was controlled. Also, IL6 and TGFβ are essential inducers of Th17 lineage (Ghoreschi et al., 2010; Jacobo, Pérez, Theas, Guazzone, & Lustig, 2011; Kobayashi et al., 2008). TGFβ was significant in both spleen and lung, but it is upregulated in the lung and downregulated in the spleen after infection (Fig. 4G). All the three IL6 probes were also upregulated in the lung and peaked at day ten p.i (Fig. 4D–F), which might indicate that IL-6 induced differentiation of Th17 and Th17 began to express IL-17a and IL-17f in the lung. This phenomenon was not observed in the spleen.
TFH cells' primary function is to support the germinal center (GC) formation and to help B cells' isotype switching, somatic mutation, and proliferation in GCs to generate high-affinity antibody-producing B cells and memory B cells. CXCR5 and ICOS are two significant markers for TFH cells (Bauquet et al., 2009; Chevalier et al., 2011), whose expression level was mildly elevated in the spleen in the early stage of infection and substantially decreased to a lower level with recovery close to the baseline at the end of the infection. In contrast, the expression level of CXCR5 and ICOS in the lung mirrored the patterns of those in the spleen (Fig. 4H, J). Thus, we hypothesize that TFH cells might be activated in the spleen, and then migrate to the lung before the follicles were formed in the spleen. That is why the expression of TFH cell marker genes (CXCR5 and ICO) showed opposite dynamic patterns in the spleen and lung. To further validate this hypothesis, we also examined the expression patterns of genetic markers for other two types of cells that also participated in the GC reaction, i.e., CR2 (Fig. 4I) as the marker for follicle dendritic cell and ICOSL (Fig. 4K) as the marker for follicle B cells which is the ICOS's ligand and plays an essential role for the T-B cell interaction in follicle forming. We found that both of these markers also followed a similar pattern as that of CXCR5 and ICOS. Thus, the three major types of cells (T, B, and dendritic cells) composing follicles for the GC reaction might all be initiated in the spleen initially, and then migrate into the lung where the germinal centers were formed. This migration can also be evidenced by examining the expression pattern of gene CXCL13, the ligand of CXCR5, which is a significant recruiter of T and B cells to form GCs/follicles. The expression pattern of CXCL13 was almost a flat line in the spleen but significantly increased in the lung after a small initial dip (Fig 4L). Follicles might only be formed in the lung instead of the spleen. We also expect that experimental scientists may design prospective studies to confirm our findings shortly further.
4. Discussion
There are more and more temporal genetics data with multiple time points available to allow systematical investigation of dynamic responses of a biological system with intervention at the genetic level. In particular, the temporal microarray gene expression data that were generated in both lung and spleen of mice in the response of Bordetella pertussis infection from day 0–28 days by Raeven et al. (Raeven et al., 2014), allow us to gain insight of dynamic immune responses at different locations. The B.pertusiss infection affects the behavior of infected cells, such as proliferation, survival, and apoptosis, as well as induces the immune response. A viral infection can cause these general biological processes. Also, the development of immune cells also involves similar general biological processes. Unfortunately, the samples in this study were unsorted so that we can't separate that two effect. Thus, we focused on the immune-related genes, which is specific to the immune response. The B.pertusiss also can inhibit the immune response by complement evasion (Jongerius, Schuijt, Mooi, & Pinelli, 2015), reducing chemokines production (C. Andreasen & Carbonetti, 2008), and inhibition of phagocyte (Schaeffer & Weiss, 2001). However, the gene expression of B.pertusiss was not included in this dataset. We can only descriptively illustrate the immune response during infection, but we can't determine the details of host-pathogen interaction in this study.
Based on the modeling and analysis pipeline proposed by Carey et al., we analyzed the genetic response differences between local and systematic immunities during B. pertussis infection. In particular, the new analytic pipeline also allows us to explore the gene response data in more details by examining the temporal response of the genes, grouping their responses by their temporal behaviors over time from which we expect to discover more results from the rich and detailed data generated in this study by Raeven et al. Comparing with the original analysis, we also reached the same conclusion that a Th1 and Th17 mixed response dominates the immune response during B.pertussis infection. Thanks to our pipeline, which has a better ability to analyze the dynamics, we found more details of the development of Th1 and Th17 in a different organ.
Th1 and Th17's responses against B. pertussis infection have been documented in the literature (Charlotte Andreasen, Powell, & Carbonetti, 2009; Brummelman et al., 2015; J M Warfel & Merkel, 2013). However, few reports detail the dynamic features and locations of Th1 and Th17 responses. Wu et al.(V. Wu, Smith, You, & Nguyen, 2016) collected the data of pulmonary Th1 and Th17 cell counts at 0, 5, 15, 35 and 50 days after B. pertussis infection in a mouse model, which suggested that the initial Th1-bias response was converted to a Th1/Th17 mixed response later during B. pertussis infection. But no details on the dynamic patterns and locations of Th1 and Th17 responses were investigated. Our analysis results based on time course gene expression response data showed that the Th1/Th17 mixed response in B. pertussis infection might be initiated at different locations.
Th1/Th17 combined response was observed, and this coupled response provided better protection than Th1-alone or Th17-alone response (Charlotte Andreasen et al., 2009; Baxter et al., 2013; Ross et al., 2013; J M Warfel & Merkel, 2013). Th1 can enhance the macrophage activation to engulf the bacteria. Although B. pertussis is considered as an extracellular pathogen and CD8+ cells are not essential for the protection (Leef, Elkins, Barbic, & Shahin, 2000), there was evidence that B. pertussis could be found intracellularly during transmission (Lamberti, Hayes, Perez Vidakovics, Harvill, & Rodriguez, 2010). The Th1 response might also inhibit transmissions by killing intracellularly infected cells. Th17 cells mediate mucosal immunity such as activating and maintaining the neutrophils. Notably, it seems that Th1 and Th17 might need different conditions or environments for their activation and differentiation (Hirahara et al., 2011; Stockinger et al., 2007; Zhu et al., 2010). Our new findings can explain this discord that Th17 cells might be primed in the lung only while Th1 cells might mainly be activated in the spleen. The spatial heterogeneity might ensure that the two lineages for Th1 and Th17 cells could have optimal differentiation conditions and environments at different locations. Wu et al. proposed that the plasmacytoid dendritic cell (pDC) might inhibit Th17 cells via secreting IFNα in the lung and caused a delayed Th17 response. We found that the expression of many IFNα family members did upregulate initially after infection but decreased back to the baseline level before day ten p.i. in the lung (S1 Fig). This observation agreed with Wu et al.'s conclusion (V. Wu et al., 2016). In some circumstances, a committed CD4 population could express key cytokines of other CD4 populations. It was reported that Th17 exhibited plasticity and could be converted to Th1 cells or express IFNɣ(Bending et al., 2011; Muranski et al., 2011), but this reversed process has not been directly observed. Thus, the switch from Th17 response to Th1/Th17 response in the lung was unlikely to be explained by the T cell plasticity.
TFH cells are required for the optimized antibody response and play an essential role in the GC formation, from which B cells develop affinity maturation (Crotty, 2011; Tangye et al., 2013). In LCMV and HIV infection, TFH cells were shown to play an essential role in generating long-term broadly neutralizing antibodies (Greczmiel et al., 2017; Tangye et al., 2013; Thornhill, Fidler, Klenerman, Frater, & Phetsouphanh, 2017). However, the presence and function of TFH cells in bacterial infection, especially for B. pertussis infection, are not well investigated before (Brummelman et al., 2015). Our results suggested that TFH cells might be generated initially in the spleen during B. pertussis infection, and then migrate to the lung to participate in the GC reaction.
In summary, our results derived from the temporal gene expression data from both spleen and lung under a mouse infection model suggested that the local lymphoid tissues around the lung/antigen evasion site and spleen might play distinct roles in the adaptive immune response against B. pertussis infection. Systematic biological research by Cave in 2016 also emphasized the vital part of the pulmonary local immune response. To some extent, it can be a validation of our analysis. However, they more focused on innate immune response, but we focused on the adaptive immune response directed by CD4 cell populations.
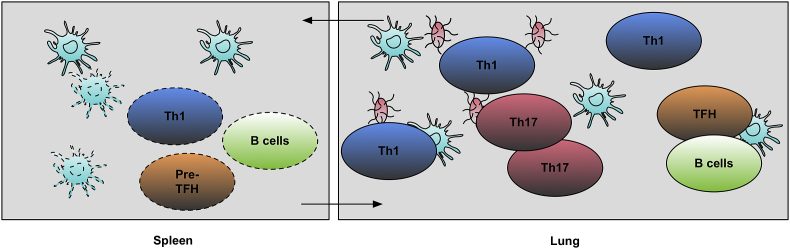
We summarize our findings for this new adaptive immune response mechanism against B. pertussis infection in Fig. 5. After infection, APCs delivered antigens to the spleen where Th1 cells, TFH cells, B cells, and follicle DC were activated. After T and B cells' differentiation in the spleen (3–4 days post-infection), activated Th1 cells and B cells started to migrate to the lung. The Th1 response could also activate CTLs and macrophages in the lung. Then, the Th17 response was initiated in the lung, and this Th1/Th17 mixed response was initiated and peaked on day 14 p.i. Th17 cells could induce an inflammation condition and activate the neutrophils to clean extracellular pathogens. At the same time, the DCs, TFH cells, and B cells started to migrate to the lung and form germinal centers in the lung under the guidance of CXCL13. B cells started to be matured and produce high-affinity antibodies. Memory B cells were also generated under TFH cells' help in follicles. Our findings and conclusions are promising and worth to be further verified by future prospective experimental studies. Our results also suggest that it will be essential to induce both systematic and local immunity responses simultaneously for B. pertussis vaccine design.
Fig. 5.
Adaptive Immune Response Model for B. Pertussis Infection. After infection, APC up-took B. Pertussis antigens and delivered them to the spleen. In the spleen, Th1 and TFH cells began differentiation and then migrated to the lung later. Th17 cells only differentiated in the lung. Although spleen might provide a big pool of TFH cells and the TFH cells might be activated in the spleen, but they only form follicles in the lung.
Conflicts of interest
We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
Acknowledgments
This work is partially supported by NIH/NIAID grant RO1 AI087135. CAA is supported by NIH/NIAID grant K24 AI114818.
Handling Editor: J. Wu
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.idm.2019.06.001.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Gene ontology enrichment analysis of the spleen.
Gene ontology enrichment analysis of the lung.
GO enrichment analysis of the spleen's unique DRGs.
GO enrichment analysis of the lung’s unique DRGs.
GO enrichment analysis of overlapped DRGs.
The clustering result of the spleen’s DRGs.
The clustering result of the lung’s DRGs.
GO enrichment analysis of the spleen’s GRMs.
GO enrichment analysis of the lung's GRMs.
The summarization correlations of pulmonary/splenic DRGs’ expression in the spleen/lung.
The significant tests of gene expression in the spleen.
The significant tests of gene expression in the lung.
figs1.
Expression patterns of all GRMs in the spleen. The total number of DRGs in each GRM is marked at the top of each plot. Blue curves represent the expression of each DRGs in the cluster. Red curves represent the mean expression of the GRM.
figs2.
Expression patterns of all GRMs in the lung. The total number of DRGs in each GRM is marked at the top of each plot. Blue curves represent the expression of each DRGs in the cluster. Red curves represent the mean expression of the GRM.
References
- Andreasen C., Carbonetti N.H. Pertussis toxin inhibits early chemokine production to delay neutrophil recruitment in response to Bordetella pertussis respiratory tract infection in mice. Infection and Immunity. 2008;76(11):5139–5148. doi: 10.1128/IAI.00895-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen C., Carbonetti N.H. Role of neutrophils in response to Bordetella pertussis infection in mice. Infection and Immunity. 2009;77(3):1182–1188. doi: 10.1128/IAI.01150-08. http://iai.asm.org/cgi/doi/10.1128/IAI.01150-08 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen C., Powell D.A., Carbonetti N.H. Pertussis toxin stimulates IL-17 production in response to Bordetella pertussis infection in mice. PLoS One. 2009;4(9) doi: 10.1371/journal.pone.0007079. http://dx.plos.org/10.1371/journal.pone.0007079 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauquet A.T., Jin H., Paterson A.M., Mitsdoerffer M., Ho I.-C., Sharpe A.H. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nature Immunology. 2009;10(2):167–175. doi: 10.1038/ni.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter R., Bartlett J., Rowhani-Rahbar A., Fireman B., Klein N.P. Vol. 347. Clinical Research Ed.; 2013. Effectiveness of pertussis vaccines for adolescents and adults: Case-control study; p. f4249.http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=23873919&retmode=ref&cmd=prlinks Retrieved from. BMJ. [DOI] [PubMed] [Google Scholar]
- Bending D., Newland S., Krejcí A., Phillips J.M., Bray S., Cooke A. Epigenetic changes at Il12rb2 and Tbx21 in relation to plasticity behavior of Th17 cells. Journal of Immunology (Baltimore, Md. : 1950) 2011;186(6):3373–3382. doi: 10.4049/jimmunol.1003216. [DOI] [PubMed] [Google Scholar]
- Brummelman J., Wilk M.M., Han W.G.H., van Els C.A.C.M., Mills K.H.G. Roads to the development of improved pertussis vaccines paved by immunology. Pathogens and Disease. 2015;73(8):ftv067. doi: 10.1093/femspd/ftv067. https://academic.oup.com/femspd/article-lookup/doi/10.1093/femspd/ftv067 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannons J.L., Lu K.T., Schwartzberg P.L. T follicular helper cell diversity and plasticity. Trends in Immunology. 2013;34(5):200–207. doi: 10.1016/j.it.2013.01.001. http://www.ncbi.nlm.nih.gov/pubmed/23395212 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M., Ramírez J.C., Wu S., Wu H. A big data pipeline: Identifying dynamic gene regulatory networks from time-course Gene Expression Omnibus data with applications to influenza infection. Statistical Methods in Medical Research. 2018;27(7):1930–1955. doi: 10.1177/0962280217746719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M., Wu S., Gan G., Wu H. Correlation-based iterative clustering methods for time course data: The identification of temporal gene response modules for influenza infection in humans. Infectious Disease Modelling. 2016;1(1):28–39. doi: 10.1016/j.idm.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celentano L.P., Massari M., Paramatti D., Salmaso S., Tozzi A.E. Resurgence of pertussis in Europe. The Pediatric Infectious Disease Journal. 2005;24(9):761–765. doi: 10.1097/01.inf.0000177282.53500.77. http://content.wkhealth.com/linkback/openurl?sid=WKPTLP:landingpage&an=00006454-200509000-00003 Retrieved from. [DOI] [PubMed] [Google Scholar]
- Cherry J.D. Historical review of pertussis and the classical vaccine. The Journal of Infectious Diseases. 1996;174(Supplement 3):S259–S263. doi: 10.1093/infdis/174.supplement_3.s259. https://academic.oup.com/jid/article-lookup/doi/10.1093/infdis/174.Supplement_3.S259 Retrieved from. [DOI] [PubMed] [Google Scholar]
- Cherry J.D. Epidemic pertussis in 2012 — the resurgence of a vaccine-preventable disease. New England Journal of Medicine. 2012;367(9):785–787. doi: 10.1056/NEJMp1209051. http://www.nejm.org/doi/abs/10.1056/NEJMp1209051 Retrieved from. [DOI] [PubMed] [Google Scholar]
- Chevalier N., Jarrossay D., Ho E., Avery D.T., Ma C.S., Yu D. CXCR5 expressing human central memory CD4 T cells and their relevance for humoral immune responses. Journal of Immunology (Baltimore, Md. : 1950) 2011;186(10):5556–5568. doi: 10.4049/jimmunol.1002828. [DOI] [PubMed] [Google Scholar]
- Crotty S. Follicular helper CD4 T cells (TFH) Annual Review of Immunology. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- Fleige H., Förster R. Induction and analysis of bronchus-associated lymphoid tissue. Methods in Molecular Biology (Clifton, N.J.) 2017;1559:185–198. doi: 10.1007/978-1-4939-6786-5_13. http://link.springer.com/10.1007/978-1-4939-6786-5_13 Chapter 13. Retrieved from. [DOI] [PubMed] [Google Scholar]
- Fleige H., Ravens S., Moschovakis G.L., Bölter J., Willenzon S., Sutter G. IL-17-induced CXCL12 recruits B cells and induces follicle formation in BALT in the absence of differentiated FDCs. The Journal of Experimental Medicine. 2014;211(4):643–651. doi: 10.1084/jem.20131737. http://www.jem.org/lookup/doi/10.1084/jem.20131737 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoreschi K., Laurence A., Yang X.-P., Tato C.M., McGeachy M.J., Konkel J.E. Generation of pathogenic T(H)17 cells in the absence of TGF-β signalling. Nature. 2010;467(7318):967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greczmiel U., Kräutler N.J., Pedrioli A., Bartsch I., Agnellini P., Bedenikovic G. Sustained T follicular helper cell response is essential for control of chronic viral infection. Science Immunology. 2017;2(18) doi: 10.1126/sciimmunol.aam8686. http://immunology.sciencemag.org/lookup/doi/10.1126/sciimmunol.aam8686 eaam8686. Retrieved from. [DOI] [PubMed] [Google Scholar]
- Hamilton-Easton A., Eichelberger M. Virus-specific antigen presentation by different subsets of cells from lung and mediastinal lymph node tissues of influenza virus-infected mice. Journal of Virology. 1995;69(10):6359–6366. doi: 10.1128/jvi.69.10.6359-6366.1995. //pmc/articles/PMC189535/?report=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirahara K., Vahedi G., Ghoreschi K., Yang X.-P., Nakayamada S., Kanno Y. Helper T-cell differentiation and plasticity: Insights from epigenetics. Immunology. 2011;134(3):235–245. doi: 10.1111/j.1365-2567.2011.03483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2008;4(1):44–57. doi: 10.1038/nprot.2008.211. http://www.nature.com/doifinder/10.1038/nprot.2008.211 Retrieved from. [DOI] [PubMed] [Google Scholar]
- Jacobo P., Pérez C.V., Theas M.S., Guazzone V.A., Lustig L. CD4+ and CD8+ T cells producing Th1 and Th17 cytokines are involved in the pathogenesis of autoimmune orchitis. Reproduction (Cambridge, England) 2011;141(2):249–258. doi: 10.1530/REP-10-0362. [DOI] [PubMed] [Google Scholar]
- Jeong Y.H., Jeon B.-Y., Gu S.-H., Cho S.-N., Shin S.J., Chang J. Differentiation of antigen-specific T cells with limited functional capacity during Mycobacterium tuberculosis infection. Infection and Immunity. 2014;82(1):132–139. doi: 10.1128/IAI.00480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongerius I., Schuijt T.J., Mooi F.R., Pinelli E. Complement evasion by Bordetella pertussis: Implications for improving current vaccines. Journal of Molecular Medicine (Berlin, Germany) 2015;93(4):395. doi: 10.1007/S00109-015-1259-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Okamoto S., Hisamatsu T., Kamada N., Chinen H., Saito R. IL23 differentially regulates the Th1/Th17 balance in ulcerative colitis and Crohn's disease. Gut. 2008;57(12):1682–1689. doi: 10.1136/gut.2007.135053. [DOI] [PubMed] [Google Scholar]
- Kretzschmar M., Teunis P.F.M., Pebody R.G. Incidence and reproduction numbers of pertussis: Estimates from serological and social contact data in five European countries. PLoS Medicine. 2010;7(6) doi: 10.1371/journal.pmed.1000291. http://dx.plos.org/10.1371/journal.pmed.1000291 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamberti Y.A., Hayes J.A., Perez Vidakovics M.L., Harvill E.T., Rodriguez M.E. Intracellular trafficking of Bordetella pertussis in human macrophages. Infection and Immunity. 2010;78(3):907–913. doi: 10.1128/IAI.01031-09. http://iai.asm.org/cgi/doi/10.1128/IAI.01031-09 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leef M., Elkins K.L., Barbic J., Shahin R.D. Protective immunity to Bordetella pertussis requires both B cells and CD4(+) T cells for key functions other than specific antibody production. Journal of Experimental Medicine. 2000;191(11):1841–1852. doi: 10.1084/jem.191.11.1841. http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=10839801&retmode=ref&cmd=prlinks Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legge K.L., Braciale T.J. Accelerated migration of respiratory dendritic cells to the regional lymph nodes is limited to the early phase of pulmonary infection. Immunity. 2003;18(2):265–277. doi: 10.1016/s1074-7613(03)00023-2. http://linkinghub.elsevier.com/retrieve/pii/S1074761303000232 Retrieved from. [DOI] [PubMed] [Google Scholar]
- Linel P., Wu S., Deng N., Wu H. Dynamic transcriptional signatures and network responses for clinical symptoms in influenza-infected human subjects using systems biology approaches. Journal of Pharmacokinetics and Pharmacodynamics. 2014;41(5):509–521. doi: 10.1007/s10928-014-9365-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.D., van der Most R.G., Akondy R.S., Glidewell J.T., Albott S., Masopust D. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity. 2008;28(5):710–722. doi: 10.1016/j.immuni.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Morita R., Schmitt N., Bentebibel S.-E., Ranganathan R., Bourdery L., Zurawski G. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34(1):108–121. doi: 10.1016/j.immuni.2010.12.012. Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranski P., Borman Z.A., Kerkar S.P., Klebanoff C.A., Ji Y., Sanchez-Perez L. Th17 cells are long lived and retain a stem cell-like molecular signature. Immunity. 2011;35(6):972–985. doi: 10.1016/j.immuni.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutangadura G. Vol. 2002. Promoting Healthy Life World Health Organization; Geneva: 2004. p. 250.http://doi.wiley.com/10.1016/j.agecon.2003.11.006 (World health report 2002: Reducing risks). US$ 13.50, ISBN 9-2415-6207-2. Agricultural Economics, 30(2), 170–172. Retrieved from. [Google Scholar]
- Parkhill J., Sebaihia M., Preston A., Murphy L.D., Thomson N., Harris D.E. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nature Genetics. 2003;35(1):32–40. doi: 10.1038/ng1227. http://www.nature.com/doifinder/10.1038/ng1227 Retrieved from. [DOI] [PubMed] [Google Scholar]
- Raeven R.H.M., Brummelman J., Pennings J.L.A., Nijst O.E.M., Kuipers B., Blok L.E.R. Molecular signatures of the evolving immune response in mice following a Bordetella pertussis infection. PLoS One. 2014;9(8) doi: 10.1371/journal.pone.0104548. http://dx.plos.org/10.1371/journal.pone.0104548 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed A.-U., Rahn H.-P., Sallusto F., Lipp M., Müller G. Follicular B helper T cell activity is confined to CXCR5(hi)ICOS(hi) CD4 T cells and is independent of CD57 expression. European Journal of Immunology. 2006;36(7):1892–1903. doi: 10.1002/eji.200636136. [DOI] [PubMed] [Google Scholar]
- Rebhahn J.A., Bishop C., Divekar A.A., Jiminez-Garcia K., Kobie J.J., Lee F.E.-H. Automated analysis of two- and three-color fluorescent Elispot (Fluorospot) assays for cytokine secretion. Computer Methods and Programs in Biomedicine. 2008;92(1):54–65. doi: 10.1016/j.cmpb.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P.J., Sutton C.E., Higgins S., Allen A.C., Walsh K., Misiak A. Relative contribution of Th1 and Th17 cells in adaptive immunity to Bordetella pertussis: Towards the rational design of an improved acellular pertussis vaccine. PLoS Pathogens. 2013;9(4) doi: 10.1371/journal.ppat.1003264. http://dx.plos.org/10.1371/journal.ppat.1003264 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer L.M., Weiss A.A. Pertussis toxin and lipopolysaccharide influence phagocytosis of Bordetella pertussis by human monocytes. Infection and Immunity. 2001;69(12):7635. doi: 10.1128/IAI.69.12.7635-7641.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger B., Veldhoen M., Martin B. Th17 T cells: Linking innate and adaptive immunity. Seminars in Immunology. 2007;19(6):353–361. doi: 10.1016/j.smim.2007.10.008. http://linkinghub.elsevier.com/retrieve/pii/S1044532307000851 Retrieved from. [DOI] [PubMed] [Google Scholar]
- Tangye S.G., Ma C.S., Brink R., Deenick E.K. The good, the bad and the ugly - TFH cells in human health and disease. Nature Reviews Immunology. 2013;13(6):412–426. doi: 10.1038/nri3447. [DOI] [PubMed] [Google Scholar]
- Thornhill J.P., Fidler S., Klenerman P., Frater J., Phetsouphanh C. The role of CD4+ T follicular helper cells in HIV infection: From the germinal center to the periphery. Frontiers in Immunology. 2017;8(3):46. doi: 10.3389/fimmu.2017.00046. http://journal.frontiersin.org/article/10.3389/fimmu.2017.00046/full Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfel J.M., Merkel T.J. Bordetella pertussis infection induces a mucosal IL-17 response and long-lived Th17 and Th1 immune memory cells in nonhuman primates. Mucosal Immunology. 2013;6(4):787–796. doi: 10.1038/mi.2012.117. http://www.nature.com/doifinder/10.1038/mi.2012.117 Retrieved from. [DOI] [PubMed] [Google Scholar]
- Warfel J.M., Zimmerman L.I., Merkel T.J. Acellular pertussis vaccines protect against disease but fail to prevent infection and transmission in a nonhuman primate model. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(2):787–792. doi: 10.1073/pnas.1314688110. http://www.pnas.org/cgi/doi/10.1073/pnas.1314688110 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendelboe A.M., Van Rie A., Salmaso S., Englund J.A. Duration of immunity against pertussis after natural infection or vaccination. The Pediatric Infectious Disease Journal. 2005;24(Supplement):S58–S61. doi: 10.1097/01.inf.0000160914.59160.41. http://content.wkhealth.com/linkback/openurl?sid=WKPTLP:landingpage&an=00006454-200505001-00011 Retrieved from. [DOI] [PubMed] [Google Scholar]
- Wu V., Smith A.A., You H., Nguyen T.A. Plasmacytoid dendritic cell-derived IFN[alpha] modulates Th17 differentiation during early Bordetella pertussis infection in mice - ProQuest. Immunology. 2016 doi: 10.1038/mi.2015.101. http://search.proquest.com/openview/565ae7665f323a2f9e83eca6d28a89d5/1?pq-origsite=gscholar&cbl=2041940 Retrieved from. [DOI] [PubMed] [Google Scholar]
- Wu S., Wu H. More powerful significant testing for time course gene expression data using functional principal component analysis approaches. BMC Bioinformatics. 2013;14(1):6. doi: 10.1186/1471-2105-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Yamane H., Paul W.E. Differentiation of effector CD4 T cell populations (*) Annual Review of Immunology. 2010;28(1):445–489. doi: 10.1146/annurev-immunol-030409-101212. http://www.annualreviews.org/doi/10.1146/annurev-immunol-030409-101212 Retrieved from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou M., Conzen S.D. A new dynamic Bayesian network (DBN) approach for identifying gene regulatory networks from time course microarray data. Bioinformatics. 2005;21(1):71–79. doi: 10.1093/bioinformatics/bth463. http://bioinformatics.oxfordjournals.org/cgi/doi/10.1093/bioinformatics/bth463 Retrieved from. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene ontology enrichment analysis of the spleen.
Gene ontology enrichment analysis of the lung.
GO enrichment analysis of the spleen's unique DRGs.
GO enrichment analysis of the lung’s unique DRGs.
GO enrichment analysis of overlapped DRGs.
The clustering result of the spleen’s DRGs.
The clustering result of the lung’s DRGs.
GO enrichment analysis of the spleen’s GRMs.
GO enrichment analysis of the lung's GRMs.
The summarization correlations of pulmonary/splenic DRGs’ expression in the spleen/lung.
The significant tests of gene expression in the spleen.
The significant tests of gene expression in the lung.