Abstract
Circulating microRNAs (miRNAs) have been suggested as non-invasive biomarkers for the diagnosis of several autoimmune diseases. However, to the best of our knowledge, no studies have yet examined the miRNA expression profiles in autoimmune inner ear disease (AIED). The present study aimed to use an miRNA sequencing assay to detect the miRNA expression profiles of serum samples from 3 control mice and 3 antigen-induced AIED model mice. Differentially expressed miRNAs (DE-miRNAs) were screened using a t-test. miRNA target prediction was performed using TargetScan Mouse. Then, the miRNA-target gene interaction network was constructed and visualized using Cytoscape software. The underlying functions of the target genes of the DE-miRNAs were predicted using the clusterProfiler package. As a result, 22 miRNAs were identified as DE-miRNAs between AIED and control mice, including 10 upregulated and 12 downregulated genes. Based on the TargetScan Mouse prediction, 1,958 genes were identified as the targets for the 22 DE-miRNAs. Functional analysis indicated that only the target genes of 8 miRNAs were respectively enriched for Gene Ontology terms and Kyoto Encyclopedia of Genes and Genomes pathways, among which miR-10b-3p, let-7j and miR-8112 were shared between the two pathway analyses. These 3 miRNAs may be involved in AIED by affecting inflammatory chemokine (miR-10b-3p-C-C motif chemokine 12), Wnt signaling (miR-8112-Wnt9b/Wnt 3a/Wnt2b) and Mucin type O-glycan biosynthesis pathways (let-7j-Galnt2/Galnt12). In conclusion, miR-10b-3p, miR-8112 and let-7j may be underlying biomarkers for diagnosing AIED.
Keywords: hearing loss, autoimmune diseases, inflammation, microRNAs
Introduction
Autoimmune inner ear disease (AIED) is a rare, but rapidly progressive, bilateral form of sensorineural hearing loss (SNHL) (1). Patients with AIED exhibit a loss of hearing over a period of weeks to months, often accompanied by tinnitus and vertigo, which may cause depressive thoughts and emotions, and disruption to daily activities and personal relationships for patients, ultimately affecting their quality of life (2,3). However, this disease is reversible, and early diagnosis of AIED with timely administration of glucocorticoids may prevent damage to the inner ear structures and result in hearing preservation. Therefore, the investigation of rapid and reliable diagnostic biomarkers for AIED is an important field of clinical study.
Although the pathogenesis of AIED remains unclear, accumulating evidence suggests that activation of the innate immune system may be the primary contributor (4). When an antigen enters the inner ear, it may be initially processed by the immunocompetent cells present in and around the endolymphatic sac. Through the secretion of various cytokines, these immunocompetent cells additionally stimulate migration of the inflammatory cells in the systemic circulation to the inner ear, subsequently amplifying the immune response and inducing more severe inner ear injuries (4–6). Therefore, several inflammatory cytokines, including tumor necrosis factor-α (TNF-α) (7), interleukin-1β (IL-1β), Il-6, IL-17 (8), C-X-C motif chemokine 10 (CXCL10) (9) and interferon-γ (IFN-γ) (10) have been suggested as biomarkers for the diagnosis of AIED. However, the implications of these biomarkers in a clinical setting remains limited and additional exploration of the key genes involved in the development of AIED is essential.
MicroRNAs (miRNAs) are endogenous, noncoding RNAs that have been demonstrated to serve an important role in regulating autoimmunity via binding to the 3′-untranslated region of their mRNA targets and thereby affecting the development of a number of autoimmune diseases (11). For example, Ishida et al (12) identified that the levels of miR-142-5p and miR-21 were significantly increased, whereas the level of miR-182 was significantly decreased, in experimental autoimmune uveoretinitis, and suggested that these miRNAs may participate in immunity by affecting the expression of IL-17. Fang et al (13) demonstrated that the increased expression of miR-30a was associated with the development and progression of autoimmune encephalomyelitis. miR-30a increased the expression of pro-inflammatory IL-1β and inducible nitric oxide synthase in primary cultured mouse microglia. These data suggested that cytokine production and associated AIED may also be controlled by miRNAs. This hypothesis has been preliminarily supported by Rudnicki et al (14) who demonstrated that inflammatory stimuli in the inner ear induced activation of the innate immune system via miR-224 and its target pentraxin 3. Furthermore, Xin et al (15) also identified that compared with Fas(lpr/lpr) mice, a spontaneous AIED model accompanied by systemic lupus erythematosus, miR-155(−/-)Fas(lpr/lpr) mice exhibited decreased levels of total immunoglobulin (Ig)A, IgM and IgG and less infiltration of inflammatory cells in the kidney. In addition, the serum levels of IL-4 and IL-17a, secreted by T helper (Th)2 and Th17 cells, were decreased, and the cluster of differentiation (CD)4(+)/CD8(+) T cell ratio was restored in the miR-155(−/-)Fas(lpr/lpr) mice. These data implied that miR-155 may also be an important miRNA in the regulation of inflammation in AIED. However, the specific miRNAs that are involved in the development of AIED remain unclear. Therefore, the aim of the present study was to screen crucial serum miRNAs in an antigen-induced AIED mouse model.
Materials and methods
Experimental animals
A total of 10 male adult (4 weeks) white guinea pigs weighing ~270 g and 54 female C57BL/6 4-week old mice weighing ~15 g (specific pathogen-free) were purchased from the Shanghai Laboratory Animal Center. Animals were housed in a temperature- (20–25°C) and humidity-controlled environment (40–70%). The animal procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals (16) and were approved by the Institutional Animal Care and Use Committees of Shandong University (Jinan, China).
Preparation of purified inner ear antigen
Following ether anesthesia, which was verified by the observation of muscle relaxation and loss of the pedal reflex with the presence of a normal heartbeat, guinea pigs were sacrificed by cardiac perfusion using cold PBS followed by cold 4% paraformaldehyde (17). Animal death was defined as mydriasis and respiratory arrest. The temporal bones were obtained following decapitation, which were then placed in 10 mM sterile TBS supplemented with 1 µg/ml aprotinin to isolate the inner ear tissue. Following homogenization and centrifugation at 100,000 × g for 1 h at 4°C, the supernatant was separated and inner ear tissue antigen was prepared. The protein content was indirectly estimated by using a UV–Vis spectrophotometer at excitation and emission wavelengths of 260 and 280 nm, respectively (protein concentration =1.45 A280-0.74 A260) following sedimentation in TBS supplemented with 0.5% SDS, 0.5% mercaptoethanol, 1% glycerinum, 2 mM phenylmethylsulfonyl fluoride and 10 mM decyl maleic dimethylamine and centrifugation at 1,000 × g for 10 min at 4°C.
Construction of an AIED animal model
The C57BL/6 mice were randomly assigned into three groups, including the control (n=18), immunization for 1 week (AIED-1; n=18) and immunization for 2 weeks (AIED-2; n=18) groups. Inner ear tissue antigen was emulsified with an equal volume of complete Freund's adjuvant (Sigma-Aldrich; Merck KGaA), which was subcutaneously injected (200 µl, containing 100 µg antigen) into multiple points in the neck and back of animals to induce AIED. In the control group, only complete Freund's adjuvant was injected. Injection sites were examined daily and reactogenicity was observed macroscopically (18). Pain was evaluated by torsion observation and topical opioids (19) were applied on the skin area if pain was present. All ulceration was observed macroscopically, and was spontaneously resolved without the requirement for treatment. The AIED model was confirmed by histopathological analysis of the cochlea via standard hematoxylin and eosin (H&E) staining: The cochlea tissue was fixed in 4% paraformaldehyde at 4°C for 3–5 days, dehydrated in graded ethanol (80, 90, 95, 100% ethanol I, 100% ethanol II and 100% ethanol III, 5 min each, at room temperature), cleared in xylene (I, II, each 30 min, at room temperature), embedded in paraffin (I, 1 h; II, 6 h; at 65°C) and sectioned to a thickness of 4 µm. Following deparaffinization and rehydration (xylene I 15 min, xylene II 15 min, 100% ethanol 5 min, 100% ethanol II 5 min, 95% ethanol 5 min, 80% ethanol 5 min, running water 1 min, at room temperature), the sections were stained with 0.5% hematoxylin and eosin for 5 min each at room temperature and then washed with distilled water. The slices were viewed under a light microscope (Olympus BX43; Olympus Corporation; magnification, ×200).
Small RNAseq library preparation and sequencing
The serum samples of 3 AIED-1 and 3 control mice were obtained and subjected to miRNA deep sequencing analysis (LC Sciences). Briefly, total RNA was extracted from serum using TRIzol (Thermo Fisher Scientific, Inc.) and ligated to 3′ and RNA 5′ adapters of RNA at 70°C for 2 min each. The total RNA was reverse transcribed into cDNA using the SuperrScript II Reverse Transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.) at 50°C for 1 h. PCR amplification was run with 25 µl reaction mixture, including 11.5 µl cDNA, 12.5 µl Phusion® Hot Start Flex 2X Master Mix (New England Biolabs, Inc.) and 0.5 µl primers (each, 5′-GATCGGAAGAGCACACGTCTGAACTCCAGTCACAAGACGGAATCTCGTATGCCGTCTTCTGCTTG-3′; 5′-AGATCGGAAGAGCGTCGTGTAGGGAAAGA-3′). PCR amplification was carried out under conditions of 98°C for 3 sec followed by 12 cycles of 98°C for 10 sec, 60°C for 30 sec, and 72°C for 15 sec, and 72°C for 10 min. Small cDNA fractions were isolated by using 6% Tris-borate-EDTA/polyacrylamide gel electrophoresis (TBE-PAGE). Subsequently, the cDNA constructs were purified and the library was validated. Finally, the purified PCR products were used for sequencing analysis via the Illumina Solexa Sequencer (Illumina, Inc.). Raw sequencing reads were obtained using Illumina's Sequencing Control Studio software (v2.8; Illumina, Inc.).
Data preprocessing
Preliminary quality control analysis of the FASTQ files was performed with FASTQC software (v0.10.0; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Cutadapt (v1.1) (20) was then used to trim 5′ (TGGAATTCTCGGGTGCCAAGG) and 3′ (GTTCAGAGTTCTACAGTCCGACG) adaptor sequences. Reads that were <17 nucleotides following trimming were discarded. Trimmed reads were then additionally filtered using the FASTQC software.
Quantification of miRNA gene expression
miRDeep software (v 2.0) (21) was used to perform the qualitative and quantitative analysis for miRNAs. Briefly, the filtered ‘clean reads’ were aligned to mouse genome reference sequence (University of California Santa Cruz mm10) (22) using the Mapper module in miRDeep2 software, which uses the bowtie algorithm. Then, the Quantifier module from the miRDeep2 package was used to generate miRNAs alignment files against known miRNAs from an miRNA database (miRBASE release 20) (23). The novel miRNAs were predicted by the ViennaRNA (https://www.tbi.univie.ac.at/RNA/) (24) and RNAFold (25) algorithms from miRDeep2 package. The principal-component analysis (PCA) was performed using the ‘Factoextra’ package in R (v1.0.3) (26) to examine the associations and variation between samples.
Identification of differentially expressed miRNAs (DE-miRNAs)
The quantitative miRNAs data were normalized to read per million (RPM), which was calculated as RPM = miRNA counts / total counts of each sample × 1,000,000, and then log 2-transformed. The DE-miRNAs between the control and AIED groups were identified using a paired Student's t-test. miRNAs were considered to be differentially expressed at P<0.05, fold change (FC) >1.5 or <1/1.5 and average expression >(log2) 5 RPM. To determine whether the DE-miRNAs were able to differentiate AIED from control mice, clustering analysis (27) was performed to generate a heat map using the hclust function implemented in the R package (v2.14.1; http://www.R-project.org), which uses the Euclidean distance and Ward's method.
Target genes prediction of DE-miRNAs
miRNA target prediction was performed using the online software TargetScan Mouse (v7.1) (28). The threshold value was set at Context Score <-0.4. Then, the miRNA-target gene interaction network was constructed and visualized using Cytoscape software (v3.4; www.cytoscape.org/) (29).
Functional enrichment analysis
To examine the underlying functions of target genes of DE-miRNAs, Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed using the clusterProfiler package in R (v3.0.1) (30) with the mouse annotations from package org.Mm.eg.db (v3.3.0) (31) and KEGG.db (v2.2.11) (32). P<0.05 and multiple-adjusted P<0.05 were set as the cut-off values.
Statistical analysis
The data are expressed as mean ± standard deviation, and were analyzed by GraphPad Prism software (v6; GraphPad Software, Inc.). The DE-miRNAs between the control and AIED groups were identified using a paired Student's t-test. Comparisons between multiple groups were performed by one-way analysis of variance followed by Duncan's post-hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
AIED model construction
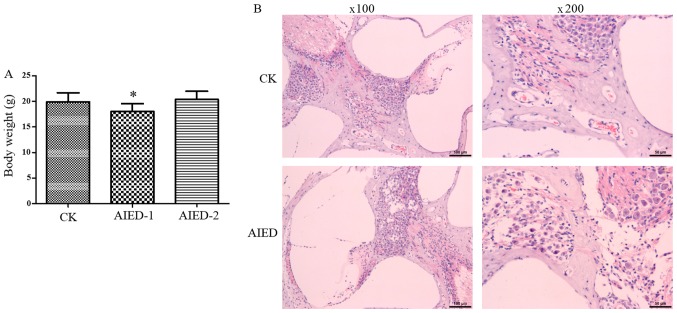
In the control group, the hair of mice was smooth and glossy, and their body weight increased over time. However, in the inner ear tissue antigen immunization group, the activity of mice was decreased, with significantly decreased body weight after 1 week (Fig. 1A). In addition, slight ulceration, but no evidence of pain, was evident in the skin injected with the antigen. However, the ulcer scar was formed and then sloughed after 2 weeks, leading to a restored increase in the body weight in the AIED-2 group (Fig. 1A). These data suggest that the immune response was marked in the groups with inner ear tissue antigen immunization for 1 week and may have been sufficient to cause serious inner ears injuries. Subsequently, H&E staining was performed to observe the histopathological changes of the cochlea. As hypothesized, the inflammatory reactions in the inner ears of animals were evident in groups following immunization. Infiltration of inflammatory cells, primarily lymphocytes, was observed in the cochlear axis. The number of spiral ganglion cells was decreased and their distribution disordered. Shrinkage occurred in the organ of Corti, with a small level of necrosis observed in the outer hair cells. Vacuolar degeneration was occasionally visible in the hair cells of the cristae ampullaris (Fig. 1B). These changes were not present in the control mice, indicating that the AIED model was successfully established.
Figure 1.
Construction of autoimmune inner ear disease model mice. (A) Measurement of body weight in each group. (B) Histopathological analysis of the cochlea via standard hematoxylin and eosin staining. 2, for two weeks. *P<0.05 vs. CK group. CK, control; AIED, autoimmune inner ear disease; AIED-1, 1 week AIED model; AIED-2, 2 week AIED model.
Identification of miRNAs
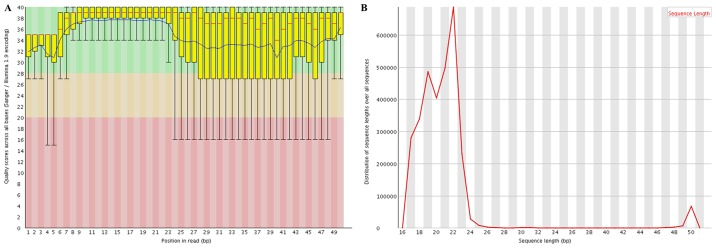
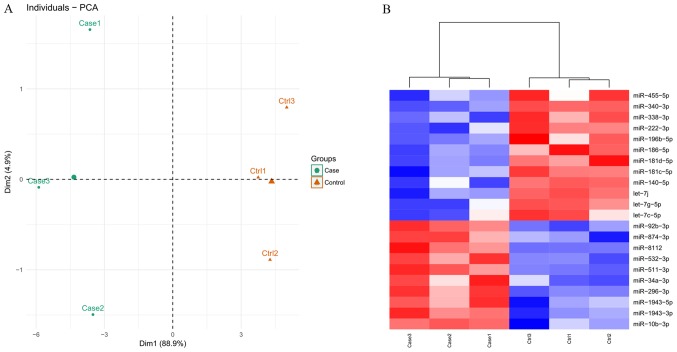
Following quality control analysis, 3,060,225–13,477,582 clean reads were obtained for the small RNA library. The length distribution analysis indicated that the majority of the reads were between 19–24 nucleotides (nt) long, with the most common length being 22 nt (Fig. 2). The trimmed reads were aligned against the mouse genome reference sequence. As a result, 67.7–80.7% of reads were mapped perfectly to the mouse genome (Table I), which were subsequently screened to identify known and novel miRNAs. Consequently, a total of 995 mature mouse miRNAs were identified and 337 novel miRNAs were annotated. PCA analysis (Fig. 3A) indicated that the known miRNAs clearly distinguished each AIED from each control sample, indicating there was there was a high level of consistency between samples.
Figure 2.
Quality control analysis. (A) Quality scores. (B) Length distribution. Bp, base pairs.
Table I.
Quality control and alignment results.
| Sample | Raw reads | Following QC reads | Mapped reads | Mapped reads. % |
|---|---|---|---|---|
| Case 1 | 8,827,017 | 3,060,225 | 2,070,930 | 67.7 |
| Case 2 | 12,375,221 | 9,365,942 | 6,493,558 | 69.3 |
| Case 3 | 11,975,356 | 8,136,614 | 5,522,291 | 67.9 |
| Ctrl 1 | 12,655,012 | 8,128,777 | 6,558,442 | 80.7 |
| Ctrl 2 | 17,988,328 | 13,477,582 | 10,106,224 | 75.0 |
| Ctrl 3 | 10,252,143 | 6,396,838 | 4,773,578 | 74.6 |
QC, quality control; Ctrl, control.
Figure 3.
Cluster analysis. (A) Principal-component analysis for samples. (B) Heat map of differentially expressed miRNAs between autoimmune inner ear disease model mice and control groups. Red, upregulated; blue, downregulated. miR, microRNA, ctrl, control.
Screening of DE-miRNAs and prediction of their target genes
A total of 22 miRNAs were identified as DE-miRNAs between AIED and control mice based on the thresholds, including 10 upregulated and 12 downregulated genes (Table II). The identified DE-miRNAs also significantly differentiated AIED from control samples according to the heat map presented in Fig. 3B.
Table II.
Differentially expressed miRNAs screened.
| miRNA | P-value | Fold change | Average expression (log2 RPM) |
|---|---|---|---|
| mmu-let-7j | 0.018 | −0.42 | 9.73 |
| mmu-miR-181c-5p | 0.025 | −0.45 | 8.12 |
| mmu-miR-340-3p | 0.003 | −0.48 | 3.14 |
| mmu-miR-181d-5p | 0.007 | −0.48 | 8.73 |
| mmu-miR-186-5p | 0.013 | −0.50 | 9.04 |
| mmu-miR-196b-5p | 0.038 | −0.55 | 6.05 |
| mmu-miR-455-5p | 0.042 | −0.56 | 7.58 |
| mmu-miR-338-3p | 0.007 | −0.60 | 6.42 |
| mmu-let-7g-5p | 0.034 | −0.62 | 13.41 |
| mmu-miR-140-5p | 0.043 | −0.62 | 9.82 |
| mmu-let-7c-5p | 0.045 | −0.63 | 11.67 |
| mmu-miR-222-3p | 0.018 | −0.65 | 11.19 |
| mmu-miR-10b-3p | 0.044 | 1.54 | 3.83 |
| mmu-miR-34a-3p | 0.024 | 1.57 | 2.56 |
| mmu-miR-92b-3p | 0.001 | 1.62 | 7.53 |
| mmu-miR-1943-5p | 0.020 | 1.71 | 4.11 |
| mmu-miR-532-3p | 0.005 | 1.74 | 6.53 |
| mmu-miR-874-3p | 0.011 | 1.76 | 4.74 |
| mmu-miR-8112 | 0.017 | 1.82 | 6.14 |
| mmu-miR-296-3p | 0.014 | 2.17 | 5.59 |
| mmu-miR-1943-3p | 0.006 | 2.41 | 2.51 |
| mmu-miR-511-3p | 0.007 | 2.43 | 8.44 |
mmu, Mus musculus; miR, microRNA; RPM, read per million.
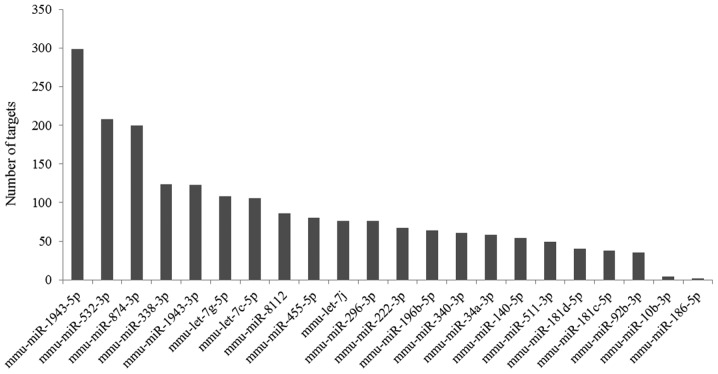
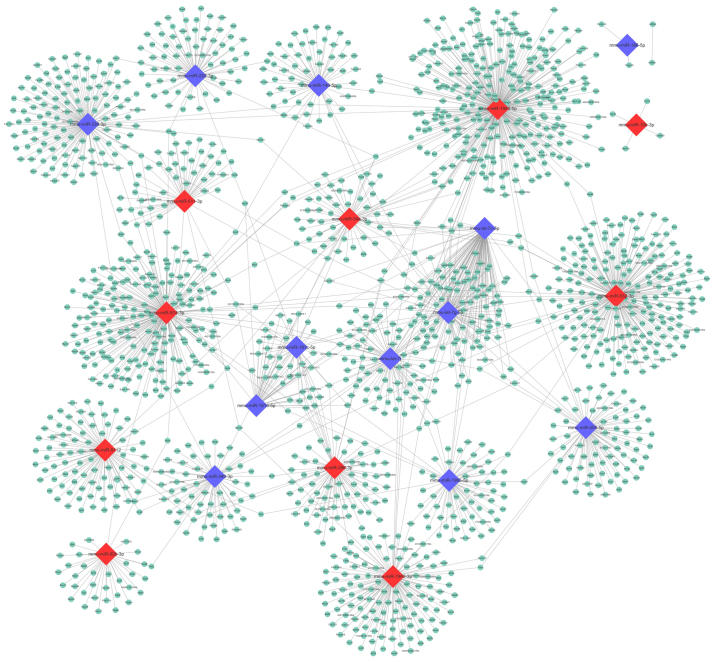
Based on the TargetScan Mouse prediction, 1,958 genes were identified as the targets for the 22 DE-miRNAs. The number of target genes of each miRNA is demonstrated in Fig. 4. Then, a miRNA-mRNA interaction network was constructed, in which 1,696 genes and 22 miRNAs were included (Fig. 5).
Figure 4.
Number of target genes of differentially expressed miRNAs. miR, microRNA; mmu, Mus musculus.
Figure 5.
miRNA-mRNA interaction network. Diamond shapes represent miRNAs, and circles represent target genes. Red color denotes upregulated, and blue denotes downregulated. miR, microRNA.
Functional enrichment analysis of target genes
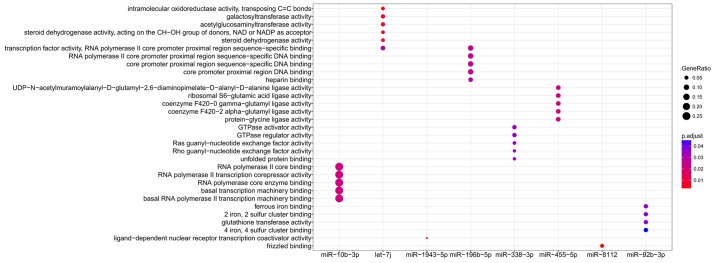
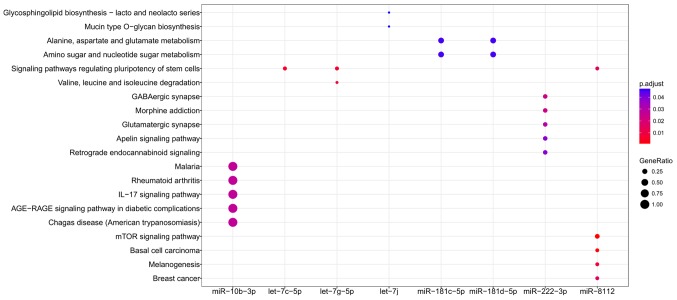
Functional enrichment analysis was performed for all target genes to reveal their underlying functions. As a result, only the target genes of 8 miRNAs were enriched for GO terms (miR-10b-3p, let-7j, miR-1943-5p, miR-196b-5p, miR-338-3p, miR-455-5p, miR-8112 and miR-92b-3p; Fig. 6) and KEGG pathways (miR-10b-3p, let-7c-5p, let-7g-5p, let-7j, miR-181c-5p, miR-181d-5p, miR-222-3p and miR-8112; Fig. 7), amongst which miR-10b-3p, let-7j and miR-8112 were common in each group. The results (Tables III and IV) indicated that miR-10b-3p may be particularly important for AIED as its target genes included inflammatory chemokines [including C-C motif chemokine (CCL) 12]. miR-8112 may be involved in AIED by affecting factors within the Wnt signaling pathway, including Wnt9b, Wnt 3a and Wnt2b (Tables III and IV). The target genes of let-7j were enriched in Glycosphingolipid biosynthesis - lacto and neolacto series and Mucin type O-glycan biosynthesis pathways, including polypeptide N-acetylgalactosaminyltransferase (Galnt) 2 and Galnt12 (Tables III and IV); these 2 pathways are associated with glycosylation, and induce the activation of the immune response.
Figure 6.
Gene ontology enrichment terms for differentially expressed miRNAs. miR, microRNA.
Figure 7.
Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses for differentially expressed miRNAs. miR, microRNA.
Table III.
GO enrichment for the target genes of differentially expressed miRNAs.
| Cluster | ID | Description | P-value | Adjusted P-value | Gene ID |
|---|---|---|---|---|---|
| let-7j | GO:0016863 | Intramolecular oxidoreductase activity, transposing C=C bonds | 1.07×105 | 2.99×103 | Hsd3b6, Hsd3b3, Hsd3b2 |
| let-7j | GO:0008378 | Galactosyltransferase activity | 1.20×104 | 7.81×103 | B3gnt5, Galnt2, Fut9, Galnt12 |
| let-7j | GO:0008375 | Acetylglucosaminyltransferase activity | 2.42×104 | 7.81×103 | B3gnt5, Galnt2, Fut9, Galnt12 |
| let-7j | GO:0033764 | Steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | 3.63×104 | 7.81×103 | Hsd3b6, Hsd3b3, Hsd3b2 |
| let-7j | GO:0016229 | Steroid dehydrogenase activity | 4.99×104 | 7.81×103 | Hsd3b6, Hsd3b3, Hsd3b2 |
| miR-196b-5p | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding | 2.53×104 | 2.42×102 | Nr6a1, Hoxa7, Otx1, Hoxa5, Hand1, Sarnp |
| miR-196b-5p | GO:0000987 | Core promoter proximal region sequence-specific DNA binding | 3.44×104 | 2.42×102 | Nr6a1, Hoxa7, Otx1, Hoxa5, Hand1, Sarnp |
| miR-196b-5p | GO:0001159 | Core promoter proximal region DNA binding | 3.60×104 | 2.42×102 | Nr6a1, Hoxa7, Otx1, Hoxa5, Hand1, Sarnp |
| miR-196b-5p | GO:0000982 | Transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding | 3.76×104 | 2.42×102 | Nr6a1, Hoxa7, Otx1, Hoxa5, Hand1, Sarnp |
| miR-196b-5p | GO:0008201 | Heparin binding | 5.62×104 | 2.90×102 | Fgfbp3, Lxn, Prss57, Ecm2 |
| miR-455-5p | GO:0008766 | UDP-N-acetylmuramoylalanyl-D-glutamyl-2, 6-diaminopimelate-D-alanyl-D-alanine ligase activity | 9.11×104 | 2.14×102 | Nr6a1, Hoxa7, Otx1, Hoxa5, Hand1, Sarnp |
| miR-455-5p | GO:0018169 | Ribosomal S6-glutamic acid ligase activity | 9.11×104 | 2.14×102 | Nr6a1, Hoxa7, Otx1, Hoxa5, Hand1, Sarnp |
| miR-455-5p | GO:0043773 | Coenzyme F420-0 gamma-glutamyl ligase activity | 9.11×104 | 2.14×102 | Nr6a1, Hoxa7, Otx1, Hoxa5, Hand1, Sarnp |
| miR-455-5p | GO:0043774 | Coenzyme F420-2 alpha-glutamyl ligase activity | 9.11×104 | 2.14×102 | Nr6a1, Hoxa7, Otx1, Hoxa5, Hand1, Sarnp |
| miR-455-5p | GO:0070735 | Protein-glycine ligase activity | 9.11×104 | 2.14×102 | Fgfbp3, Lxn, Prss57, Ecm2 |
| miR-338-3p | GO:0005096 | GTPase activator activity | 1.76×104 | 1.76×104 | Nr6a1, Hoxa7, Otx1, Hoxa5, Hand1, Sarnp |
| miR-338-3p | GO:0030695 | GTPase regulator activity | 2.94×104 | 2.94×104 | Nr6a1, Hoxa7, Otx1, Hoxa5, Hand1, Sarnp |
| miR-338-3p | GO:0005088 | Ras guanyl-nucleotide exchange factor activity | 3.44×104 | 3.44×104 | Nr6a1, Hoxa7, Otx1, Hoxa5, Hand1, Sarnp |
| miR-338-3p | GO:0005089 | Rho guanyl-nucleotide exchange factor activity | 4.38×104 | 4.38×104 | Nr6a1, Hoxa7, Otx1, Hoxa5, Hand1, Sarnp |
| miR-338-3p | GO:0051082 | Unfolded protein binding | 4.88×104 | 4.88×104 | Fgfbp3, Lxn, Prss57, Ecm2 |
| miR-10b-3p | GO:0001106 | RNA polymerase II transcription corepressor activity | 4.20×103 | 3.67×103 | 76365 |
| miR-10b-3p | GO:0043175 | RNA polymerase core enzyme binding | 4.20×103 | 4.20×103 | Rprd1b |
| miR-10b-3p | GO:0048020 | CCR chemokine receptor binding | 5.59×103 | 5.59×103 | Ccl12 |
| miR-10b-3p | GO:0008009 | Chemokine activity | 6.81×103 | 6.81×103 | Ccl12 |
| miR-10b-3p | GO:0042379 | Chemokine receptor binding | 8.73×103 | 8.73×103 | Ccl12 |
| miR-92b-3p | GO:0008198 | Ferrous iron binding | 5.34×104 | 5.34×104 | Isca1, 432732 |
| miR-92b-3p | GO:0051537 | 2 iron, 2 sulfur cluster binding | 6.84×104 | 6.84×104 | Isca1, 432732 |
| miR-92b-3p | GO:0004364 | Glutathione transferase activity | 9.74×104 | 9.74×104 | Gsta2, Gm10639 |
| miR-92b-3p | GO:0051539 | 4 iron, 4 sulfur cluster binding | 1.62×103 | 1.62×103 | Isca1, 432732 |
| miR-1943-5p | GO:0030374 | Ligand-dependent nuclear receptor transcription coactivator activity | 1.57×105 | 1.57×105 | Atxn7l3, Usp22, 15361, Actn2; Hmga1-rs1, Rbm14 |
| miR-8112 | GO:0005109 | Frizzled binding | 1.14×105 | 1.14×105 | Ror2, Wnt9b, Wnt3a, Wnt2b |
GO, Gene ontology.
Table IV.
Kyoto Encyclopedia of Genes and Genomes pathway enrichment for the target genes of differentially expressed miRNAs..
| Cluster | ID | Description | P-value | Adjusted P-value | Gene ID |
|---|---|---|---|---|---|
| let-7j | mmu00601 | Glycosphingolipid biosynthesis - lacto and neolacto series | 2.85×103 | 4.75×102 | B3gnt5, Fut9 |
| let-7j | mmu00512 | Mucin type O-glycan biosynthesis | 3.06×103 | 4.75×102 | Galnt2, Galnt12 |
| miR-181c-5p | mmu00250 | Alanine, aspartate and glutamate metabolism | 1.37×102 | 4.53×102 | Gfpt1 |
| miR-181c-5p | mmu00520 | Amino sugar and nucleotide sugar metabolism | 1.81×102 | 4.53×102 | Gfpt1 |
| miR-181d-5p | mmu00250 | Alanine, aspartate and glutamate metabolism | 1.37×102 | 4.53×102 | Gfpt1 |
| miR-181d-5p | mmu00520 | Amino sugar and nucleotide sugar metabolism | 1.81×102 | 4.53×102 | Gfpt1 |
| let-7g-5p | mmu04550 | Signaling pathways regulating pluripotency of stem cells | 6.84×105 | 6.57×103 | Smarcad1, Skil, Wnt16, Hand1, Hoxb1, Acvr1c |
| let-7g-5p | mmu00280 | Valine, leucine and isoleucine degradation | 1.92×104 | 9.22×103 | Agxt2, Bcat1, Ehhadh, Acat1 |
| let-7c-5p | mmu04550 | Signaling pathways regulating pluripotency of stem cells | 5.95×105 | 5.59×103 | Smarcad1, Skil, Wnt16, Hand1, Hoxb1, Acvr1c |
| miR-222-3p | mmu04727 | GABAergic synapse | 6.35×104 | 2.35×102 | Gng5, Gabra1, Gnb3 |
| miR-222-3p | mmu05032 | Morphine addiction | 7.23×104 | 2.35×102 | Gng5, Gabra1, Gnb3 |
| miR-222-3p | mmu04724 | Glutamatergic synapse | 1.35×103 | 2.92×102 | Gng5, Ppp3r1, Gnb3 |
| miR-222-3p | mmu04371 | Apelin signaling pathway | 2.38×103 | 3.85×102 | Agtr1a, Gng5, Gnb3 |
| miR-222-3p | mmu04723 | Retrograde endocannabinoid signaling | 2.96×103 | 3.85×102 | Gng5, Gabra1, Gnb3 |
| miR-222-3p | mmu04151 | PI3K-Akt signaling pathway | 4.02×103 | 4.36×102 | Ddit4, Gng5, Gnb3, Kit |
| miR-10b-3p | mmu05144 | Malaria | 6.08×103 | 2.68×102 | Ccl12 |
| miR-10b-3p | mmu05323 | Rheumatoid arthritis | 1.03×102 | 2.68×102 | Ccl12 |
| miR-10b-3p | mmu04657 | IL-17 signaling pathway | 1.13×102 | 2.68×1024 | Ccl12 |
| miR-10b-3p | mmu04933 | AGE-RAGE signaling pathway in diabetic complications | 1.24×102 | 2.68×102 | Ccl12 |
| miR-10b-3p | mmu05142 | Chagas disease (American trypanosomiasis) | 1.26×102 | 2.68×102 | Ccl12 |
| miR-10b-3p | mmu04668 | TNF signaling pathway | 1.34×102 | 2.68×102 | Ccl12 |
| miR-10b-3p | mmu05418 | Fluid shear stress and atherosclerosis | 1.76×102 | 2.78×102 | Ccl12 |
| miR-10b-3p | mmu04621 | NOD-like receptor signaling pathway | 2.08×102 | 2.78×102 | Ccl12 |
| miR-10b-3p | mmu05164 | Influenza A | 2.08×102 | 2.78×102 | Ccl12 |
| miR-10b-3p | mmu04062 | Chemokine signaling pathway | 2.42×102 | 2.90×102 | Ccl12 |
| miR-10b-3p | mmu05168 | Herpes simplex infection | 2.67×102 | 2.91×102 | Ccl12 |
| miR-10b-3p | mmu04060 | Cytokine-cytokine receptor interaction | 3.36×102 | 3.36×102 | Ccl12 |
| miR-8112 | mmu04150 | mTOR signaling pathway | 1.40×102 | 8.55×104 | Fzd8, Clip1, Wnt9b, Wnt3a, Wnt2b, Lpin1 |
| miR-8112 | mmu05217 | Basal cell carcinoma | 4.06×105 | 1.24×103 | Fzd8, Wnt9b, Wnt3a, Wnt2b |
| miR-8112 | mmu04916 | Melanogenesis | 4.17×104 | 8.48×103 | Fzd8, Wnt9b, Wnt3a, Wnt2b |
| miR-8112 | mmu04550 | Signaling pathways regulating pluripotency of stem cells | 1.36×103 | 1.60×102 | Fzd8, Wnt9b, Wnt3a, Wnt2b |
| miR-8112 | mmu05224 | Breast cancer | 1.64×103 | 1.60×102 | Fzd8, Wnt9b, Wnt3a, Wnt2b |
| miR-8112 | mmu04310 | Wnt signaling pathway | 1.72×103 | 1.60×102 | Fzd8, Wnt9b, Wnt3a, Wnt2b |
| miR-8112 | mmu04390 | Hippo signaling pathway | 2.10×103 | 1.60×102 | Fzd8, Wnt9b, Wnt3a, Wnt2b |
| miR-8112 | mmu00532 | Glycosaminoglycan biosynthesis - chondroitin sulfate/dermatan sulfate | 2.28×103 | 1.60×102 | Chst15, B3galt6 |
| miR-8112 | mmu05200 | Pathways in cancer | 2.36×103 | 1.60×102 | Fzd8, Wnt9b, Lpar1, Wnt3a, Wnt2b, Bcr |
| miR-8112 | mmu04974 | Protein digestion and absorption | 3.98×103 | 2.43×102 | Col2a1, Atp1a2, Col11a1 |
| miR-8112 | mmu05205 | Proteoglycans in cancer | 5.96×103 | 3.30×102 | Fzd8; Wnt9b, Wnt3a, Wnt2b |
miR, microRNA; mmu, Mus musculus.
Discussion
Previous studies have indicated that there are miRNA profiles associated with noise-induced hearing loss (33,34), but the AIED-associated miRNAs remain undetermined. To the best of our knowledge, the present study investigated, for the first time, the serum miRNA profiles in AIED by constructing an AIED mouse model. The results identified 22 DE-miRNAs in the AIED mice compared with the control mice. The functions of 13 of these miRNAs were enriched, among which miR-10b-3p, miR-8112 and let-7j may be particularly important for the development of AIED, as suggested by the GO and KEGG pathway enrichment analysis of their target genes, in which they were predicted to affect chemokine signaling (miR-10b-3p-Ccl12), Wnt signaling (miR-8112-Wnt9b; miR-8112-wnt 3a and miR-8112-Wnt2b) and O-glycan biosynthesis pathways (let-7j-Galnt2a and let-7j-Galnt12). This appears to be in concordance with the previously identified inflammatory mechanisms of AIED (4).
It has been suggested that miR-10b was significantly upregulated in ankylosing spondylitis (35), which is a common autoimmune disease associated with the induction of SNHL (36–38). Therefore, miR-10b may also be hypothesized to be highly expressed in AIED, which was demonstrated in the present study (FC =1.54; P=0.04). However, Chen et al (35) identified that miR-10b overexpression inhibited the production of IL-17A by total CD4 and differentiating Th17 cells, suggesting that miR-10b may likely constitute part of a negative feedback loop that restricts the enhanced inflammatory Th17 responses. Although these results appeared to be inconsistent with our data, they may be reasonable as the present study similarly predicted that miR-10b may downregulate CCL12, a known inflammatory chemokine (39). Furthermore, it has been demonstrated that the use of the C-C chemokine receptor type 2 agonists, CCL2 and CCL12, elicited migration in Th17-polarized cells and additionally stimulated a modest enrichment of IL-17(+) cells (40). Therefore, miR-10b may serve crucial roles in AIED by regulating CCL-12 mediated Th17 responses. However, additional exploration of the roles of miR-10b-3p in AIED is required, as the study conducted by Mojsilovic-Petrovic et al (39) only examined the role of miR-10b-5p, although it may be inferred that miR-10b-3p may have a similar effect to miR-10b-5p (41).
miR-8112 is a rarely studied miRNA involved in disease, with the exception of one study by Wang et al (42), which indicated that miR-8112 was highly expressed in MC3T3-E1 cells exposed to high-dose fluoride and may induce the formation of impaired bone diseases, including skeletal fluorosis. Patients with skeletal fluorosis have also been demonstrated to exhibit varying degrees of hearing loss (43). Therefore, miR-8112 was hypothesized to be upregulated in AIED, which was verified in the results of the present study (FC=1.82; P=0.02). Notably, the present study suggested that inhibition of Wnt signaling pathway genes (Wnt9b, Wnt 3a and Wnt2b) may be an underlying mechanism of miR-8112, which has not been demonstrated previously, to the best of our knowledge. This prediction may be indirectly supported by the observation that the loss of Wnt/β-catenin made hair cells more vulnerable to neomycin-induced injury and caused hearing loss (44), while activation of Wnt/β-catenin markedly promoted the mitotic regeneration of new hair cells and attenuated neurodegeneration in the auditory cortex (45,46). The association between the Wnt signaling pathway and AIED may also be associated with the inflammatory and oxidative stress, as the study by Liu et al (44) indicated that loss of β-catenin in hair cells led to decreased expression of antioxidant enzymes.
The Let-7 miRNA family has been suggested to exert inhibitory effects on the inflammation. For example, Brennan et al (47) observed that ectopic overexpression of let-7 inhibited the inflammatory responses including proliferation, migration, monocyte adhesion and NF-κB activation in vascular smooth muscle cells. In addition, Kumar et al (48) demonstrated that exogenous administration of let-7 mimics decreased IL-13 levels and relieved allergic airway inflammation. In concordance with these studies, the present study identified that let-7j, let-7g-5p and let-7c-5p were also downregulated in AIED mice. In addition, subsequent analysis also revealed that let-7j may be involved in AIED by regulating Galnts. Galnts are enzymes responsible for initiating the cascade of mucin-type O-linked glycosylation for certain inflammation-associated proteins, including selectin ligands. Block et al (49) demonstrated that compared with wild-type mice, L-selectin-dependent leukocyte rolling was completely abolished in Galnt1(−/-) mice, as its ligand was not able to be glycosylated by Galnt1. Although Galnt1 and Galnt12 have not been demonstrated in AIED, we hypothesized they may be upregulated by let-7j; however, this requires additional investigation.
In conclusion, the present study preliminarily revealed several important inflammatory-associated miRNAs, miR-10b-3p, miR-8112 and let-7j, in an AIED mouse model. They may be potential biomarkers and their abnormal expression may be conducive to early diagnosis of AIED and timely treatment with glucocorticoids. However, additional studies are required to confirm their diagnostic values using clinical samples and to investigate the direct interaction between these miRNAs and their target genes, including miR-10b-3p-Ccl12, miR-8112-Wnt9b/Wnt 3a/Wnt2b and let-7j-Galnt2/Galnt12.
Acknowledgements
Not applicable.
Funding
The present study was supported by National Natural Science Foundation of China (grant nos., 81371084 and 81570924).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JZ and AX participated in the design of this study. JZ and NW performed the animal model experiments. JZ performed the bioinformatics analyses. NW and AX contributed to the acquisition and interpretation of data. JZ was involved in drafting the manuscript. AX was involved in the manuscript revision. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The animal procedures were approved by the Institutional Animal Care and Use Committees at Shandong University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Matsuoka AJ, Harris JP. Autoimmune inner ear disease: A retrospective review of forty-seven patients. Audiol Neurotol. 2013;18:228–239. doi: 10.1159/000351289. [DOI] [PubMed] [Google Scholar]
- 2.Chen J, Liang J, Ou J, Cai W. Mental health in adults with sudden sensorineural hearing loss: An assessment of depressive symptoms and its correlates. J Psychosom Res. 2013;75:72–74. doi: 10.1016/j.jpsychores.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 3.Carlsson PI, Hall M, Lind KJ, Danermark B. Quality of life, psychosocial consequences, and audiological rehabilitation after sudden sensorineural hearing loss. Int J Audiol. 2011;50:139–144. doi: 10.3109/14992027.2010.533705. [DOI] [PubMed] [Google Scholar]
- 4.Goodall AF, Siddiq MA. Current understanding of the pathogenesis of autoimmune inner ear disease: A review. Clin Otolaryngol. 2015;40:412–419. doi: 10.1111/coa.12432. [DOI] [PubMed] [Google Scholar]
- 5.Gopen Q, Keithley EM, Harris JP. Mechanisms underlying autoimmune inner ear disease. Drug Discov Today Dis Mech. 2006;3:137–142. doi: 10.1016/j.ddmec.2006.02.006. [DOI] [Google Scholar]
- 6.Satoh H, Firestein GS, Billings PB, Harris JP, Keithley EM. Proinflammatory cytokine expression in the endolymphatic sac during inner ear inflammation. J Assoc Res Otolaryngol. 2003;4:139–147. doi: 10.1007/s10162-002-3025-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Svrakic M, Pathak S, Goldofsky E, Hoffman R, Chandrasekhar SS, Sperling N, Alexiades G, Ashbach M, Vambutas A. Diagnostic and prognostic utility of measuring tumor necrosis factor in the peripheral circulation of patients with immune-mediated sensorineural hearing loss. Arch Otolaryngol Head Neck Surg. 2012;138:1052–1058. doi: 10.1001/2013.jamaoto.76. [DOI] [PubMed] [Google Scholar]
- 8.Pathak S, Hatam LJ, Bonagura V, Vambutas A. Innate immune recognition of molds and homology to the inner ear protein, cochlin, in patients with autoimmune inner ear disease. J Clin Immunol. 2013;33:1204–1215. doi: 10.1007/s10875-013-9926-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JM, Kim JY, Bok J, Kim KS, Choi JY, Kim SH. Identification of evidence for autoimmune pathology of bilateral sudden sensorineural hearing loss using proteomic analysis. Clin Immunol. 2017;183:24–35. doi: 10.1016/j.clim.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Lorenz RR, Solares CA, Williams P, Sikora J, Pelfrey CM, Hughes GB, Tuohy VK. Interferon-γ production to inner ear antigens by T cells from patients with autoimmune sensorineural hearing loss. J Neuroimmunol. 2002;130:173–178. doi: 10.1016/S0165-5728(02)00190-X. [DOI] [PubMed] [Google Scholar]
- 11.Chen JQ, Papp G, Szodoray P, Zeher M. The role of microRNAs in the pathogenesis of autoimmune diseases. Autoimmun Rev. 2016;15:1171–1180. doi: 10.1016/j.autrev.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Ishida W, Fukuda K, Higuchi T, Kajisako M, Sakamoto S, Fukushima A. Dynamic changes of microRNAs in the eye during the development of experimental autoimmune uveoretinitis. Invest Ophthalmol Vis Sci. 2011;52:611–617. doi: 10.1167/iovs.10-6115. [DOI] [PubMed] [Google Scholar]
- 13.Fang X, Sun D, Wang Z, Yu Z, Liu W, Pu Y, Wang D, Huang A, Liu M, Xiang Z, et al. miR-30a positively regulates the inflammatory response of microglia in experimental autoimmune encephalomyelitis. Neurosci Bull. 2017;33:603–615. doi: 10.1007/s12264-017-0153-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rudnicki A, Shivatzki S, Beyer LA, Takada Y, Raphael Y, Avraham KB. MicroRNA-224 regulates Pentraxin 3, a component of the humoral arm of innate immunity, in inner ear inflammation. Hum Mol Genet. 2014;23:3138–3146. doi: 10.1093/hmg/ddu023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xin Q, Li J, Dang J, Bian X, Shan S, Yuan J, Qian Y, Liu Z, Liu G, Yuan Q, et al. miR-155 deficiency ameliorates autoimmune inflammation of systemic lupus erythematosus by targeting S1pr1 in Faslpr/lpr mice. J Immunol. 2015;194:5437–5445. doi: 10.4049/jimmunol.1403028. [DOI] [PubMed] [Google Scholar]
- 16.National Research Council (US) Institute for Laboratory Animal Research, corp-author. National Academies Press (US); Washingto, DC: 1996. Guide for the Care and Use of Laboratory Animals. [PubMed] [Google Scholar]
- 17.Watanabe K, Inai S, Jinnouchi K, Baba S, Yagi T. Expression of caspase-activated deoxyribonuclease (CAD) and caspase 3 (CPP32) in the cochlea of cisplatin (CDDP)-treated guinea pigs. Auris Nasus Larynx. 2003;30:219–225. doi: 10.1016/S0385-8146(03)00049-X. [DOI] [PubMed] [Google Scholar]
- 18.Silva-Gomes R, Marcq E, Trigo G, Gonçalves CM, Longatto-Filho A, Castro AG, Pedrosa J, Fraga AG. Spontaneous healing of Mycobacterium ulcerans lesions in the guinea pig model. PLoS Negl Trop Dis. 2015;9:e0004265. doi: 10.1371/journal.pntd.0004265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poonawala T, Levay-Young BK, Hebbel RP, Gupta K. Opioids heal ischemic wounds in the rat. Wound Repair Regen. 2005;13:165–174. doi: 10.1111/j.1067-1927.2005.130207.x. [DOI] [PubMed] [Google Scholar]
- 20.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 21.Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haeussler M, Zweig AS, Tyner C, Speir ML, Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Hinrichs AS, Gonzalez JN, et al. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 2019;47(D1):D853–D858. doi: 10.1093/nar/gky1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozomara A, Griffiths-Jones S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(D1):D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonnet E, Wuyts J, Rouzé P, Van de Peer Y. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics. 2004;20:2911–2917. doi: 10.1093/bioinformatics/bth374. [DOI] [PubMed] [Google Scholar]
- 26.Kassambara A, Mundt F. factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R package version 1.0.3, 2017. https://rpkgs.datanovia.com/factoextra/index.html
- 27.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 28.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:4. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carlson M. org.Mm.eg.db: Genome wide annotation for Mouse. R package version 3.7.0, 2018. https://bioconductor.org/packages/release/data/annotation/html/org.Mm.eg.db.html
- 32.Carlson M., KEGG.db A set of annotation maps for KEGG. R package version 3.2.3, 2016. https://bioconductor.org/packages/release/data/annotation/html/KEGG.db.html
- 33.Ding L, Liu J, Shen HX, Pan LP, Liu QD, Zhang HD, Han L, Shuai LG, Ding EM, Zhao QN, et al. Analysis of plasma microRNA expression profiles in male textile workers with noise-induced hearing loss. Hear Res. 2016;333:275–282. doi: 10.1016/j.heares.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 34.Li YH, Yang Y, Yan YT, Xu LW, Ma HY, Shao YX, Cao CJ, Wu X, Qi MJ, Wu YY, et al. Analysis of serum microRNA expression in male workers with occupational noise-induced hearing loss. Braz J Med Biol Res. 2018;51:e6426. doi: 10.1590/1414-431x20176426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L, Al-Mossawi MH, Ridley A, Sekine T, Hammitzsch A, de Wit J, Simone D, Shi H, Penkava F, Kurowska-Stolarska M, et al. miR-10b-5p is a novel Th17 regulator present in Th17 cells from ankylosing spondylitis. Ann Rheum Dis. 2017;76:620–625. doi: 10.1136/annrheumdis-2016-210175. [DOI] [PubMed] [Google Scholar]
- 36.Eryilmaz A, Dagli M, Karabulut H, Sivas Acar F, Erkol Inal E, Gocer C. Evaluation of hearing loss in patients with ankylosing spondylitis. J Laryngol Otol. 2007;121:845–849. doi: 10.1017/S0022215106004488. [DOI] [PubMed] [Google Scholar]
- 37.Kahveci OK, Demirdal US, Duran A, Altuntas A, Kavuncu V, Okur E. Hearing and cochlear function of patients with ankylosing spondylitis. Clin Rheumatol. 2012;31:1103–1108. doi: 10.1007/s10067-012-1984-6. [DOI] [PubMed] [Google Scholar]
- 38.Sugahara K, Hashimoto M, Hirose Y, Shimogori H, Yamashita H. Autoimmune inner ear disease associated with ankylosing spondylitis. Egypt J Otolaryngol. 2014;30:176–179. doi: 10.4103/1012-5574.133224. [DOI] [Google Scholar]
- 39.Mojsilovic-Petrovic J, Callaghan D, Cui H, Dean C, Stanimirovic DB, Zhang W. Hypoxia-inducible factor-1 (HIF-1) is involved in the regulation of hypoxia-stimulated expression of monocyte chemoattractant protein-1 (MCP-1/CCL2) and MCP-5 (Ccl12) in astrocytes. J Neuroinflammation. 2007;4:12. doi: 10.1186/1742-2094-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Webb A, Johnson A, Fortunato M, Platt A, Crabbe T, Christie MI, Watt GF, Ward SG, Jopling LA. Evidence for PI-3K-dependent migration of Th17-polarized cells in response to CCR2 and CCR6 agonists. J Leukoc Biol. 2008;84:1202–1212. doi: 10.1189/jlb.0408234. [DOI] [PubMed] [Google Scholar]
- 41.Yoon EL, Yeon JE, Ko E, Lee HJ, Je JH, Yoo YJ, Kang SH, Suh SJ, Kim JH, Seo YS, et al. An explorative analysis for the role of serum miR-10b-3p levels in predicting response to sorafenib in patients with advanced hepatocellular carcinoma. J Korean Med Sci. 2017;32:212–220. doi: 10.3346/jkms.2017.32.2.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Zhang X, Zhao Z, Xu H. Preliminary analysis of microRNAs expression profiling in MC3T3-E1 cells exposed to fluoride. Biol Trace Elem Res. 2017;176:367–373. doi: 10.1007/s12011-016-0887-9. [DOI] [PubMed] [Google Scholar]
- 43.Haimanot RT. Neurological complications of endemic skeletal fluorosis, with special emphasis on radiculo-myelopathy. Paraplegia. 1990;28:244–251. doi: 10.1038/sc.1990.31. [DOI] [PubMed] [Google Scholar]
- 44.Liu L, Chen Y, Qi J, Zhang Y, He Y, Ni W, Li W, Zhang S, Sun S, Taketo MM, et al. Wnt activation protects against neomycin-induced hair cell damage in the mouse cochlea. Cell Death Dis. 2016;7:e2136. doi: 10.1038/cddis.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xia MY, Zhao XY, Huang QL, Sun HY, Sun C, Yuan J, He C, Sun Y, Huang X, Kong W, et al. Activation of Wnt/β-catenin signaling by lithium chloride attenuates d-galactose-induced neurodegeneration in the auditory cortex of a rat model of aging. FEBS Open Bio. 2017;7:759–776. doi: 10.1002/2211-5463.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ni W, Zeng S, Li W, Chen Y, Zhang S, Tang M, Sun S, Chai R, Li H. Wnt activation followed by Notch inhibition promotes mitotic hair cell regeneration in the postnatal mouse cochlea. Oncotarget. 2016;7:66754–66768. doi: 10.18632/oncotarget.11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brennan E, Wang B, McClelland A, Mohan M, Marai M, Beuscart O, Derouiche S, Gray S, Pickering R, Tikellis C, et al. Protective effect of let-7 miRNA family in regulating inflammation in diabetes-associated atherosclerosis. Diabetes. 2017;66:2266–2277. doi: 10.2337/db16-1405. [DOI] [PubMed] [Google Scholar]
- 48.Kumar M, Ahmad T, Sharma A, Mabalirajan U, Kulshreshtha A, Agrawal A, Ghosh B. Let-7 microRNA-mediated regulation of IL-13 and allergic airway inflammation. J Allergy Clin Immunol. 2011;128:1077–85.e1, 10. doi: 10.1016/j.jaci.2011.04.034. [DOI] [PubMed] [Google Scholar]
- 49.Block H, Ley K, Zarbock A. Severe impairment of leukocyte recruitment in ppGalNAcT-1-deficient mice. J Immunol. 2012;188:5674–5681. doi: 10.4049/jimmunol.1200392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.