Abstract
It has been demonstrated that microRNAs (miRNAs) serve important roles in various biological processes, such as tumorigenesis. In the present study, the role of miR-30a-3p in the pathogenesis of esophageal carcinoma (EC) was investigated. Reverse transcription-quantitative polymerase chain reaction was performed to determine the levels of miR-30a-3p expression in EC tissues and cell lines. Then, the effects of miR-30a-3p on the migration, invasion and radiosensitivity of EC cells were investigated using scratch-wound, Transwell and radiosensitivity assays, respectively. A dual-luciferase reporter assay was performed to determine potential interactions between miR-30a-3p and the 3′-untranslated region (3′-UTR) of insulin-like growth factor 1 receptor (IGF-1R). The results demonstrated that the levels of miR-30a-3p expression in EC tissues and cell lines were significantly decreased compared with those in paired healthy tissues and a human esophageal epithelial cell line. Upregulation of miR-30a-3p expression significantly suppressed migration, invasion and epithelial-mesenchymal transition (EMT), and enhanced radiosensitivity in EC cells. Analysis of luciferase activity demonstrated that miR-30a-3p interacted with the 3′-UTR of IGF-1R, and knockdown of IGF-1R induced similar effects on the migration, invasion, EMT and radiosensitivity of EC cells. The results indicated that miR-30a-3p suppressed metastasis and enhanced the radiosensitivity of EC cells via downregulation IGF-1R, suggesting that miR-30a-3p may be a potential therapeutic target in the treatment of EC.
Keywords: microRNA-30a-3p, esophageal carcinoma, insulin-like growth factor 1 receptor, metastasis, radiosensitivity
Introduction
Esophageal carcinoma (EC) is one of the most common types of malignant tumor of the digestive tract, with high morbidity and mortality (1,2). In developed countries, the morbidity and mortality of EC ranks 20 and 11th, respectively, whereas in developing countries, the morbidity and mortality rank eighth and fifth, respectively (3). China accounts for >50% of the world's annual incidence (4). Esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma are the main subtypes, with ESCC most commonly reported, particularly in China (5). As of the lack of clear symptoms during the early stages of EC, the majority of patients are first diagnosed with advanced EC (6,7). Tumor metastasis and recurrence are the leading causes of poor prognosis of EC; the 5-year survival rate of patients with EC is 9.4–35% (8,9). Radiotherapy is an important therapeutic strategy for the treatment of EC to decrease the risk of recurrence, and improve quality of life and survival; however, numerous factors affect the efficacy of radiotherapy, with resistance to radiotherapy the main reason for the failure of treatment of patients with EC (10–12). Therefore, improved understanding of the mechanisms underlying the metastasis and radiotherapy resistance of EC is required.
MicroRNAs (miRNAs/miRs) are a class of small (17–24 nucleotides) noncoding RNAs that regulate gene expression at the post-transcriptional level via interactions with the 3′-untranslated region (3′-UTR) of target mRNAs (13). Increasing evidence suggests that miRNAs are involved in the differentiation, proliferation, invasion, metastasis and apoptosis of cells, and may represent novel diagnostic biomarkers and therapeutic targets for various cancers, such as EC (14–18). For example, miRNA-377 inhibits the initiation and progression of esophageal cancer via suppression of cluster of differentiation 133 and vascular endothelial growth factor (19). miRNA-200c enhanced the radiosensitivity of esophageal cancer by arresting the cell cycle and targeting p21 (20).
Analysis of miRNA expression profiles using Affymetrix GeneChip® technology revealed the miR-30 family as downregulated in numerous tumors, including lung, prostate, thyroid and liver cancers (21,22). A member of this family, miR-30a-3p, exhibited reduced expression in various tumors (23,24). Insulin-like growth factor type 1 receptor (IGF-1R) is a heterotetrameric tyrosine kinase belonging to the receptor tyrosine kinase superfamily. In the present study, putative miR-30a-3p targets were identified using TargetScan, suggesting that IGF-1R may be a potential target gene of miR-30a-3p. It was demonstrated that miR-30a-3p was downregulated in EC tissues and cell lines. Additionally, the role of miR-30a-3p in the migration, invasion and radiosensitivity of EC cells was investigated in vitro. Furthermore, it was revealed that IGF-1R was a direct functional target of miR-30a-3p. The findings of the present study suggested the regulation of IGF-1R by miR-30a-3p as a potential mechanism underlying cellular migration and invasion in EC, and also demonstrated that miR-30a-3p may be a novel biomarker to determine the radiosensitivity of tumors in patients with EC.
Materials and methods
Patients and tissue specimens
A total of 30 patients (17 female and 13 male patients), who were histopathologically and clinically diagnosed at Jiangsu Cancer Hospital (Nanjing, China) were included in the present study. Patients with EC were diagnosed via a combination of esophageal X-ray barium meal examination, three-dimensional computerized tomography imaging and histopathological examination. Clinicopathological data were collected, including gender, age, tumor location, differentiation grade, tumor size and tumor, necrosis and metastasis stage (25) (Table I). EC tissues and paired normal tissues were obtained from patients (age, 51±12 years) that underwent esophagectomy or endoscopic submucosal dissection without radiotherapy or chemotherapy in Jiangsu Cancer Hospital (Nanjing, China) from August 2016 to September 2017. The present study was approved by the Ethics Committee of Nanjing Medical University (Nanjing, China), and each patient provided written informed consent. All specimens were snap-frozen in liquid nitrogen within 2 h and stored at −80°C until use.
Table I.
Associations between levels of'miR-30a-3p expression and the clinicopathological data of patients with esophageal carcinoma.
| Clinical characteristics | Nο. of patients | Relative expression | P-value |
|---|---|---|---|
| Age (years) | 0.572 | ||
| ≤60 | 18 | 0.425 | |
| >60 | 12 | 0.581 | |
| Sex | 0.653 | ||
| Male | 20 | 0.366 | |
| Female | 10 | 0.418 | |
| Drinking | 0.544 | ||
| Yes | 11 | 0.545 | |
| No | 19 | 0.411 | |
| Smoking | 0.661 | ||
| Yes | 14 | 0.622 | |
| No | 16 | 0.541 | |
| pT stage | 0.526 | ||
| T1 + T2 | 13 | 0.366 | |
| T3 + T4 | 17 | 0.521 | |
| TNM stage | 0.492 | ||
| ≤II | 10 | 0.512 | |
| >II | 20 | 0.478 | |
| Lymphatic metastasis | 0.517 | ||
| Positive | 11 | 0.362 | |
| Negative | 19 | 0.428 |
miR, microRNA; pT, pathologic T stage; TNM, tumor, node and metastasis.
Cell culture and transfection
Human EC cell lines (EC9706 and EC109) and a human esophageal epithelial cell line (HET-1A) were purchased from the Cell Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Human cell lines were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 µg/ml streptomycin and 100 U/ml penicillin (Gibco; Thermo Fisher Scientific, Inc.), and maintained at 37°C in a humidified incubator with 5% CO2 for 48 h prior to further experiments.
miR-30a-3p mimics, miRNA mimics negative control (mimics NC), miR-30a-3p inhibitors, miRNA inhibitors negative control (inhibitors NC), small interfering RNA (siRNA) targeted against IGF-1R (siRNA IGF-1R) and negative control siRNA (siRNA NC) were chemically synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The sequences were as follows: miR-30 mimics forward, 5′-ACUGGUAGUAAGUUGUAAUGCU-3′ and reverse, 5′-UCAUAUAACUUCAUACCUGCCU-3′; and miR-30 inhibitors, 5′-CGTGGCACCAATAGAATTGAGA-3′; NC forward, 5′-GGCTGCATTGGCTGGCGAAACCCGUC-3′ and reverse, 5′-ATGCGUGCCCTGCTGTTGCTCCATGTCG-3′. Cells were seeded into 6-well plates at 60–70% confluence 1 day prior to transfection. Cell transfection was conducted using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. The RNA was transfected at a concentration of 50 nM.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from EC tissues, paired normal tissues, EC cells and normal cells using TRIzol® solution (Invitrogen; Thermo Fisher Scientific, Inc.). For miRNA expression, RT reactions were performed with a One Step PrimeScript miRNA cDNA Synthesis kit (Takara Biotechnology Co., Ltd., Dalian, China) at 37°C for 30 min according to the manufacturer's instructions, followed by qPCR with SYBR® Premix Ex Taq (Takara Biotechnology Co., Ltd.). For mRNA expression, cDNA was synthesized from total RNA using a PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd.). For miRNA and mRNA amplifications, analysis was performed with an ABI 7500 Fast Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc). qPCR was performed using the SYBR Premix ExTaq (Takara Bio, Inc., Otsu, Japan) according to the manufacturer's protocols. The thermocycling conditions comprised one cycle at 95°C for 30 sec, followed by 40 cycles of amplification (95°C for 3 sec and 60°C for 30 sec). All alterations in the expression of miR and mRNA were calculated via the 2−ΔΔcq method using U6 or GAPDH for normalization, respectively (26). The primer sequences used were as follows: miR-30a-3p, forward 5′-CCCTGCTCTGGCTGGTCAAACGGAAC-3′, reverse, 5′-TTGCCAGCCCTGCTGTAGCTGGTTGAAG-3′; U6, forward 5′-CTCGCTTCGGCAGCACA-3′, reverse, 5′-AACGCTTCACGAATTTGCGT-3′; IGF-1R, forward 5′-CAACGGCAACCTGAGTTAC-3′, reverse, 5′-GCACGAAGATGGAGTTGTG-3′; and GAPDH, forward 5′-ACAACTTTGGTATCGTGGAAGG-3′ and reverse, 5′-GCCATCACGCCACAGTTTC-3′. Each experiment was performed in triplicate.
Scratch-wound assay
EC9706 and EC109 cells were seeded into a 6-well plate following transfection at a density of 5×105 cells/well. When 90% confluence was attained, a sterilized plastic scraper was used to scratch the well axis, and the floating cells were removed with PBS. The medium was subsequently replaced with fresh culture medium and incubated at 37°C for 48 h. The cells were then washed twice with PBS and were photographed at 0 and 48 h under a light microscope (Nikon Corporation, Tokyo, Japan) in five random fields of view (magnification, ×200). Then, the distance of cell migration was determined.
Transwell assay
The migratory and invasive abilities of transfected EC cells were evaluated using Transwell inserts (Costar; Corning, Inc., Corning, NY, USA) with polycarbonate membrane filters (8-µm pores). For the Transwell migration assay, EC9706 and EC109 cells at density of 5×104 cells/well were plated in the upper chambers of Transwell plates in fresh culture media. For the Transwell invasion assay, Matrigel (BD Biosciences, San Jose, CA, USA) was dissolved at 4°C overnight, diluted with fresh culture media (1:3), added to the upper chambers (30 µl/well) and incubated for 30 min at 37°C prior to the addition of cells. Culture medium containing 20% FBS was added to the lower chambers. Following incubation with RPMI-1640 medium supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) for 48 h at 37°C, filter inserts were removed from the wells and the cells on the upper surface were removed with a cotton swab. The filters were fixed in 4% paraformaldehyde for 30 min at room temperature and then stained with 0.1% crystal violet for 30 min at room temperature. The migratory and invasive cells were observed and counted in six fields with a light microscope (magnification, ×200).
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was determined using a CCK-8 assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan). EC9706 and EC109 cells were cultured in a 96-well plate at a density of 3×103 cells/well, and transfected with miR-30a-3p mimics, miR-30a-3p mimics NC, miR-30a-3p inhibitors or miR-30a-3p inhibitors NC. Following incubation for 24, 48 or 72 h at 37°C with 5% CO2, CCK-8 solution (10 µl) was added to each well, followed by incubation for a further 4 h at room temperature. The optical density values were measured at 450 nm using a microplate reader (Shanghai, China).
Western blotting
Tissues and transfected cells were lysed using radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology, Haimen, China). Total protein was quantified using a bicinchoninic acid protein assay kit (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Supernatant samples (20 µg) were heated at 99°C for 5 min prior to loading, separated via 10% SDS-PAGE (Bio-Rad Laboratories, Inc., Hercules, CA, USA), and transferred to polyvinylidene difluoride membranes (Merck KGaA) according to the manufacturer's protocols. The membranes were then blocked with 5% non-fat milk for 1 h at room temperature prior to incubation overnight with primary antibodies at 4°C. The specific primary antibodies from Cell Signaling Technology, Inc. (Danvers, MA, USA) used during the study were as follows: IGF-1R (1:1,000; cat. no. 3027); E-cadherin (1:1,000, cat. no. 4A2); N-cadherin (1:1,000; cat. no. 13A9); and vimentin (1:1,000; cat. no. 49636). Following three washes with 0.1% TBS-Tween 20 (TBS-T). Subsequently, the membranes were incubated with the horseradish peroxidase-conjugated anti-rabbit secondary antibody (cat. no. ab6721; 1:2,000; Abcam) for 1 h at room temperature. The samples were washed three times using TBS-T and agitated for 5 min at room temperature, and the proteins were then visualized using an enhanced chemiluminescence system (Pierce; Thermo Fisher Scientific, Inc.). Protein expression was quantified using ImageJ software (version 1.48; National Institutes of Health, Bethesda, MD, USA). GAPDH were used as a loading control (ab9485; 1:1,000 dilution; Abcam, Cambridge, MA, USA). The following antibodies were purchased from Abcam: IGF-1R (ab39675; 1:1,000 dilution); matrix metalloproteinase-2 (MMP-2; ab37150; 1:1,000 dilution); MMP-9 (ab73734; 1:1,000 dilution); E-cadherin (ab1416; 1:1,000 dilution); vimentin (ab8978; 1:1,000 dilution); N-cadherin (ab18203; 1:1,000 dilution) and GAPDH (ab8245; 1:1,000 dilution).
Flow cytometry analysis
A total of 5×104 cells were seeded in 6-well plates, cultured for 48 h at 37°C and subsequently cells were collected. Cells were fixed with precooled 70% ethanol for 30 min at room temperature, and collected following centrifugation at 12,000 × g for 5 min at room temperature. The cells were resuspended in PBS containing 50 mg/ml propidium iodide and 50 mg/ml RNaseA (cat. no. 40711ES10; Shanghai Yeasen Biotechnology, Co., Ltd., Shanghai, China) for 30 min at room temperature. Cells were incubated for 1 h at 37°C in the dark, and analyzed using flow cytometry (FACSCalibur; BD Biosciences) and analyzed using FlowJo 10.06 software (FlowJo LLC, Ashland, OR, USA). The flow cytometry analyses were repeated three times.
Immunohistochemical analysis
The surgical EC tissues and paired adjacent tissues were fixed in 10% neutral buffered formalin at room temperature for 20 min and embedded in paraffin wax. Tissue sections with a thickness of 4 µm were mounted onto slides, then the slides were deparaffinized with xylene at room temperature, rehydrated with a graded alcohol series, and incubated with H2O2 at 37°C for 10 min. Following blocking using 1.5% normal goat serum (Shanghai Yeasen Biotechnology Co., Ltd.) at 37°C for 20 min, the primary IGF-1R antibody (CST; Danvers, MA, USA; 1:1,000; cat. no. 3027) was incubated on the slides at 4°C overnight. The slides were washed with PBS three times, then incubated with a secondary antibody (horseradish peroxidase-conjugated; cat. no. ab6721; 1:2,000; Abcam) and stained with 3,3′-diaminobenzidine. Images were obtained using a fluorescence microscope in five randomly-selected fields of view (magnification, ×200) (FSX100; Olympus Corporation, Tokyo, Japan).
Radiosensitivity assay
Transfected EC9706 and EC109 cells were seeded at a density of 3×103 cells/well in a 96-well plate, and irradiated with various doses of radiation (0, 2, 4, 6 and 8 Gy) using an X-RAD 320 X-ray radiator (Softex Co., Ltd., Tokyo, Japan) at a dose rate of 2 Gy/min. A CCK-8 assay was subsequently performed as previously described to determine cell proliferation.
Luciferase assay
A search for putative targets of miR-30a-3p was performed with TargetScan Human 7.2 (www.targetscan.org/vert_72/) and miRanda software (www.microrna.org/). The 3′-untranslated region (3′-UTR) of IGF-1R was cloned into an miRNA Expression Reporter Vector psiCheck-2 (Promega Corporation, Madison, WI, USA). Cells were plated in 48-well plates for 24 h, psiCheck-2 with the wild-type (WT) or mutant (MUT) 3′-UTR of IGF-1R was cotransfected with miR-30a-3p mimics or miR-NC using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.). Then, 48 h following transfection, cells were collected and subjected to a luciferase assay using a Dual Luciferase Reporter Assay kit (Promega Corporation). Luciferase activity was normalized using Renilla luciferase activity (Promega Corporation).
Statistical analysis
Data are expressed as the mean ± standard deviation and analyzed with SPSS 19.0 (IBM Corp., Armonk, NY, USA). Comparisons between two groups were performed using Student's t-tests. Comparisons across three or more groups were performed using one-way analyses of variance and a Tukey's post-hoc test. P<0.05 was considered to indicate a statistically significant difference. All experiments were repeated a minimum of three times.
Results
miR-30a-3p is downregulated in EC tissues and cell lines
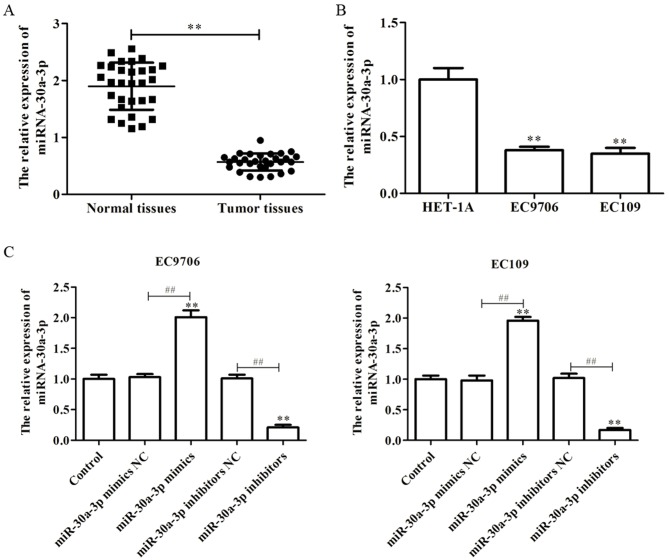
miR-30a-3p, a member of the miR-30 family, has been reported as downregulated in numerous tissues (24,27). In the present study, to investigate the role of miR-30a-3p in the development of EC, the expression of miR-30a-3p in EC tissues and cell lines was determined via RT-qPCR. As presented in Fig. 1A, it was observed that the levels of miR-30a-3p expression were significantly downregulated in EC tissues compared with in paired normal tissues. Furthermore, it was demonstrated that the levels of miR-30a-3p expression in the EC cell lines, EC9706 and EC109, were significantly decreased compared with in a human esophageal epithelial cell line, HET-1A (Fig. 1B). The findings suggested that miR-30a-3p is downregulated in EC tissues and cell lines.
Figure 1.
miR-30a-3p is downregulated in EC tissues and cell lines. (A) miR-30a-3p expression in EC tissues and paired normal tissues as determined by RT-qPCR. (B) miR-30a-3p expression in EC cell lines (EC9706 and EC109) and a human esophageal epithelial cell line (HET-1A) as determined by RT-qPCR. (C) Following transfection with miR-30a-3p mimics or miR-30a-3p inhibitors in EC9706 and EC109 cells, the efficiency of transfection was evaluated by RT-qPCR. Data are presented as the mean ± standard deviation of three independent experiments, each performed in triplicate. **P<0.01 vs. HET-1A or control. ##P<0.01 vs. the NC group. EC, esophageal carcinoma; miR, microRNA; NC, negative control; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
In order to investigate the effects of miR-30a-3p on EC, miR-30a-3p mimics, mimics NC, miR-30a-3p inhibitors or inhibitors NC were transfected into EC9706 and EC109 cells. RT-qPCR was performed to determine the efficiency of transfection, and the results revealed that miR-30a-3p mimics could significantly promote the expression of miR-30a-3p compared with the control, while miR-30a-3p inhibitors significantly reduced the expression of miR-30a-3p in EC9706 and EC109 cells (Fig. 1C).
miR-30a-3p suppresses the migration and invasion of EC cells
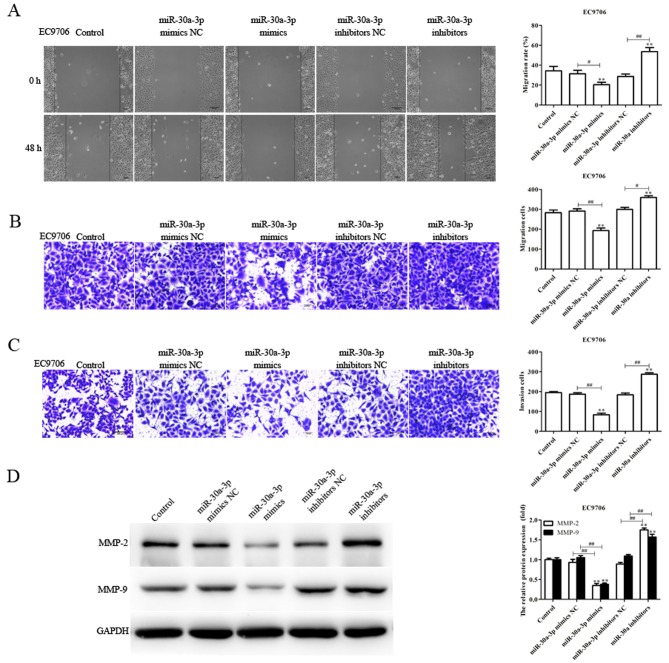
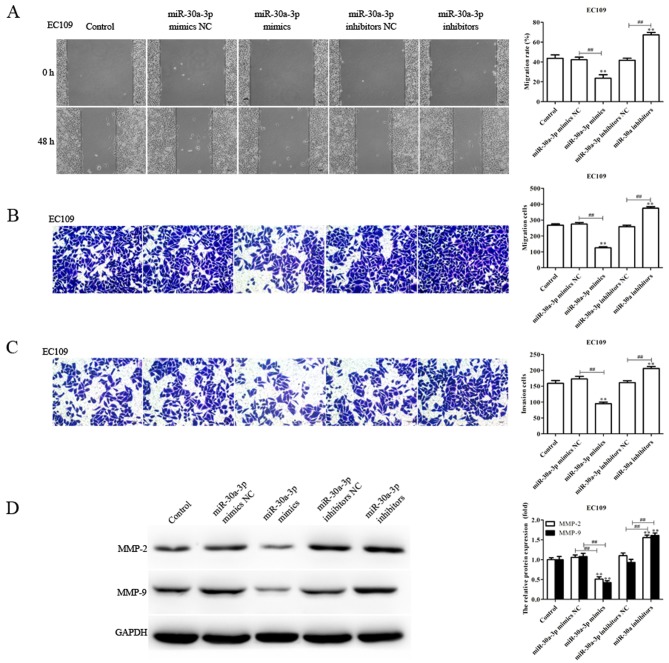
As the levels of miR-30a-3p expression are associated with lymph node metastasis (28), the migratory and invasive abilities of EC9706 and EC109 cells transfected with miR-30a-3p mimics, mimics NC, miR-30a-3p inhibitors and inhibitors NC were determined via a scratch-wound, and Transwell migration and invasion assays, respectively. Compared with the control, the migration and invasion of EC9706 and EC109 cells transfected with miR-30a-3p mimics were significantly reduced, whereas miR-30a-3p inhibitors significantly promoted the migratory and invasive abilities of EC9706 and EC109 cells (Figs. 2 and 3). The results indicated that miR-30a-3p inhibits EC cell migration and invasion in vitro.
Figure 2.
miR-30a-3p suppresses the migration and invasion of EC9706 cells. The migratory abilities of EC9706 cells transfected with miR-30a-3p mimics or miR-30a-3p inhibitors for 48 h were investigated using (A) scratch-wound and (B) Transwell assays. (C) Invasive abilities of transfected EC9706 cells were evaluated via a Transwell invasion assay. Cells were counted in each well under an inverted microscope at ×200 magnification. (D) Expression levels of MMP-2 and MMP-9 proteins were determined by western blot analysis. Data are presented as the mean ± standard deviation of three independent experiments; each experiment was performed in triplicate. **P<0.01 vs. the control group. #P<0.05, ##P<0.01 vs. the NC group. miR, microRNA; MMP, matrix metalloproteinase; NC, negative control.
Figure 3.
miR-30a-3p suppresses the migration and invasion of EC109 cells. The migratory abilities of EC109 cells transfected with miR-30a-3p mimics or miR-30a-3p inhibitors for 48 h were determined using (A) scratch-wound and (B) Transwell assays. (C) Invasive abilities of transfected EC109 cells were evaluated via a Transwell invasion assay. Cells were counted in each well under an inverted microscope at ×200 magnification. (D) Expression levels of MMP-2 and MMP-9 protein were determined by western blot analysis. Data are presented as the mean ± standard deviation of three independent experiments; each was performed in triplicate. **P<0.01 vs. the control group. ##P<0.01 vs. the NC group. miR, microRNA; MMP, matrix metalloproteinase; NC, negative control.
Tumor metastasis is a sequential process involving interactions between tumor cells, host cells and the tissue microenvironment, in which MMPs serve an essential role; MMP-2 and MMP-9 have been reported to be associated with the migration and invasion of various tumors (29,30). In the present study, the levels of MMP-2 and MMP-9 protein expression in transfected EC9706 and EC109 cells were determined. The expression levels of MMP-2 and MMP-9 protein were significantly decreased in EC9706 and EC109 cells transfected with miR-30a-3p mimics compared with the control; however, the expression levels were significantly increased in EC9706 and EC109 cells transfected with miR-30a-3p inhibitors (Figs. 2D and 3D). The results suggested that miR-30a-3p serves an inhibitory role in EC cell metastasis.
Effects of miR-30a-30p on the apoptosis and cell cycle of EC cells
To determine whether miR-30a-3p is able to affect the cell cycle and apoptosis, flow cytometry was performed. The results demonstrated that overexpression of miR-30a-3p markedly increased the number of cells arrested in the G1 phase compared with the control group (Fig. 4A). Furthermore, the number of cells in the G1 phase was notably reduced following transfection with miR-30a-3p inhibitors compared with the control group. Additionally, it was revealed that the number of apoptotic cells was markedly decreased following transfection with miR-30a-3p inhibitors, whereas, overexpression of miR-30a-3p notably increased the number of apoptotic cells compared with the control group (Fig. 4B). The results indicated that miR-30a-3p may suppress cell cycle progression and induce the apoptosis of EC cells.
Figure 4.
miR-30a-3p regulates the cell cycle and apoptosis. The effects of miR-30a-3p on (A) the cell cycle and (B) apoptosis were determined by flow cytometry in EC9706 and EC109 cells. Data are presented as the mean ± standard error of the mean of three independent experiments. *P<0.05, **P<0.01 vs. the control group. #P<0.05, ##P<0.01 vs. the NC group. AV, annexin V; miR, microRNA; NC, negative control; PI, propidium iodide.
miR-30a-3p inhibits epithelial-mesenchymal transition (EMT) in EC cells
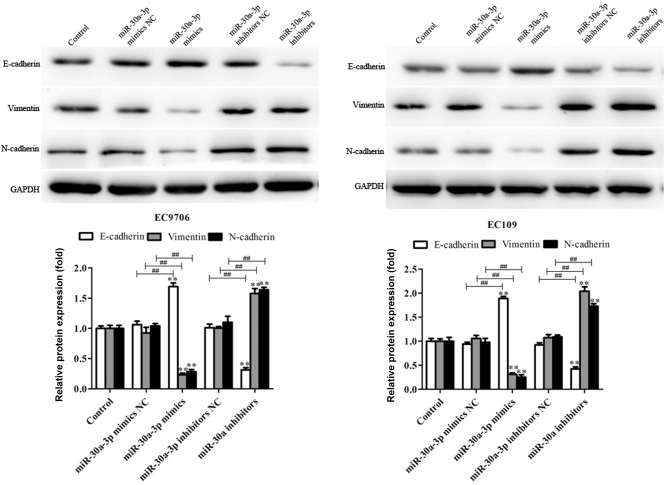
EMT is characterized by the loss of differentiation of epithelial cells. Important features of EMT include loss of the epithelial cell phenotype and the acquisition of interstitial properties, represented by the downregulation of E-cadherin, and upregulated expression of N-cadherin and vimentin. This results in reduced adhesion between cells, and enhanced migration and invasion (31–33). In the present study, the effects of miR-30a-3p on the EMT of EC9706 and EC109 cells transfected with miR-30a-3p mimics, mimics NC, miR-30a-3p inhibitors and inhibitors NC were investigated. As presented in Fig. 5, miR-30a-3p mimics significantly upregulated the levels of E-cadherin protein expression, and decreased those of N-cadherin and vimentin compared with the control; opposing effects were observed in EC9706 and EC109 cells transfected with miR-30a-3p inhibitors. The results suggested that miR-30a-3p inhibits EC cell metastasis via the regulation of EMT.
Figure 5.
miR-30a-3p inhibits EMT in esophageal carcinoma cells. Expression levels of EMT-associated proteins (E-cadherin, vimentin and N-cadherin) were determined in EC9706 and EC109 cells transfected with miR-30a-3p mimics or miR-30a-3p inhibitors for 48 h via western blot analysis. Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01 vs. the control group. ##P<0.01 vs. the NC group. EMT, epithelial-mesenchymal transition; miR, microRNA; NC, negative control.
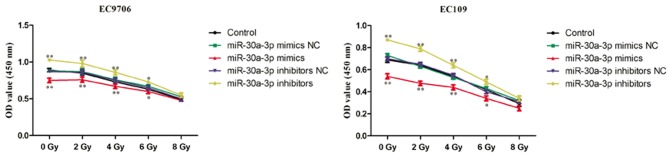
miR-30a-3p enhances the radiosensitivity of EC cells
Radiotherapy is commonly used in the treatment of EC; however, patients may exhibit recurrence of EC following radiotherapy due to low radiosensitivity. It is important to identify strategies that decrease the resistance of EC to radiotherapy to improve the therapeutic effects (11). Therefore, the role of miR-30a-3p in the radiosensitivity of EC cells was investigated. EC9706 and EC109 cells were transfected with miR-30a-3p mimics, mimics NC, miR-30a-3p inhibitors and inhibitors NC, and irradiated with various radiation doses (0, 2, 4, 6 and 8 Gy) for 48 h. The CCK-8 assay was subsequently performed to evaluate the proliferation of cells. As presented in Fig. 6, miR-30a-3p mimics significantly decreased the proliferation of EC9706 and EC109 cells compared with the control group indicating an increased radiosensitivity, whereas miR-30a-3p inhibitors significantly promoted the proliferation of EC9706 and EC109 cells. The results suggested that miR-30a-3p may act as a radiosensitizer.
Figure 6.
miR-30a-3p enhances the radiosensitivity of esophageal carcinoma cells. EC9706 and EC109 cells were transfected with miR-30a-3p mimics or miR-30a-3p inhibitors and irradiated with various radiation doses (0, 2, 4, 6 and 8 Gy) for 48 h. A Cell Counting Kit-8 assay was subsequently performed to evaluate the proliferation of cells. Data are presented as the mean ± standard deviation of three independent experiments, each performed in triplicate. *P<0.05, **P<0.01 vs. the control group. miR, microRNA; NC, negative control; OD, optical density.
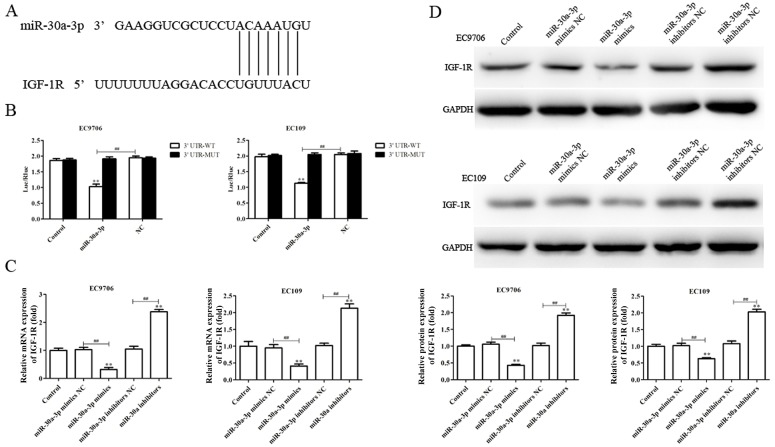
IGF-1R is a direct target of miR-30a-3p
To investigate the molecular mechanisms by which miR-30a-3p may suppress metastasis and EMT, and enhance radiosensitivity in EC cells, putative miR-30a-3p targets were determined using TargetScan. The software analysis suggested that IGF-1R may present a potential candidate for regulation by miR-30a-3p (Fig. 7A). Therefore, dual luciferase reporter gene analysis was performed to investigate whether miR-30a-3p directly targets the 3′-UTR of IGF-1R. As presented in Fig. 7B, miR-30a-3p mimics significantly reduced the luciferase activity of cells containing the IGF-1R 3′-UTR-WT compared with the control, but not 3′-UTR-MUT, indicating that miR-30a-3p directly binds to the 3′UTR of IGF-1R.
Figure 7.
IGF-1R is a direct target of miR-30a-3p. (A) Prediction of interaction between miR-30a-3p and IGF-1R was determined by TargetScan. (B) Luciferase reporter assays were performed to verify the binding of miR-30a-3p to the 3′-UTR of IGF-1R. (C) Reverse transcription-quantitative polymerase chain reaction and (D) western blotting were performed to evaluate the levels of IGF-1R mRNA and protein expression, respectively, following transfection with miR-30a-3p mimics or miR-30a-3p inhibitors. Band intensity was quantified using ImageJ software. Data are presented as the mean ± standard deviation of three independent experiments; each was performed in triplicate. **P<0.01 vs. the control group. ##P<0.01 vs. the NC group. IGF-1R, insulin-like growth factor 1 receptor; miR, microRNA; MUT, mutant; NC, negative control; 3′-UTR, 3′-untranslated region; WT, wild-type.
RT-qPCR and western blotting were utilized to investigate the effects of miR-30a-3p on endogenous IGF-1R expression in EC cells. As presented in Fig. 7C and D, the levels of IGF-1R mRNA and protein expression were significantly downregulated in EC9706 and EC109 cells transfected with miR-30a-3p mimics compared with the control, whereas miR-30a-3p inhibitors promoted IGF-1R expression (Fig. 7C and D). The results suggested that IGF-1R is a direct target gene of miR-30a-3p in EC.
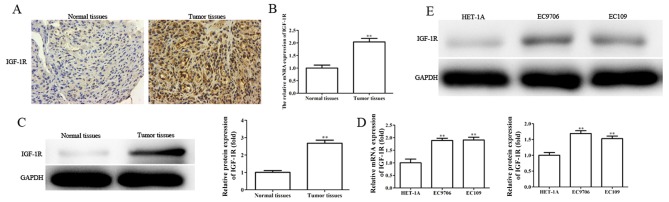
IGF-1R is upregulated in EC tissues and cell lines, and silencing IGF-1R suppresses the migration, invasion and EMT, and enhances the radiosensitivity of EC cells
Providing IGF-1R is a direct target gene of miR-30a-3p, the role of IGF-1R in EC was investigated. The levels of IGF-1R mRNA and protein expression were determined in EC tissues and cells by RT-qPCR, western blotting and immunohistochemical analysis. It was revealed that the levels of IGF-1R mRNA and protein expression were significantly upregulated in EC tissues and cell lines (Fig. 8A-E).
Figure 8.
IGF-1R is upregulated in EC tissues and cell lines. The levels of IGF-1R expression in EC tissues and paired normal tissues were evaluated by (A) immunohistochemistry, (B) RT-qPCR and (C) western blot analysis. The levels of IGF-1R expression in HET-1A, EC9706 and EC109 were determined by (D) RT-qPCR and (E) western blot analysis. The band intensity was quantified using ImageJ software. Data are presented as the mean ± standard deviation of three independent experiments; each was performed in triplicate. **P<0.01 vs. the control group. EC, esophageal carcinoma; IGF-1R, insulin-like growth factor 1 receptor; NC, negative control; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
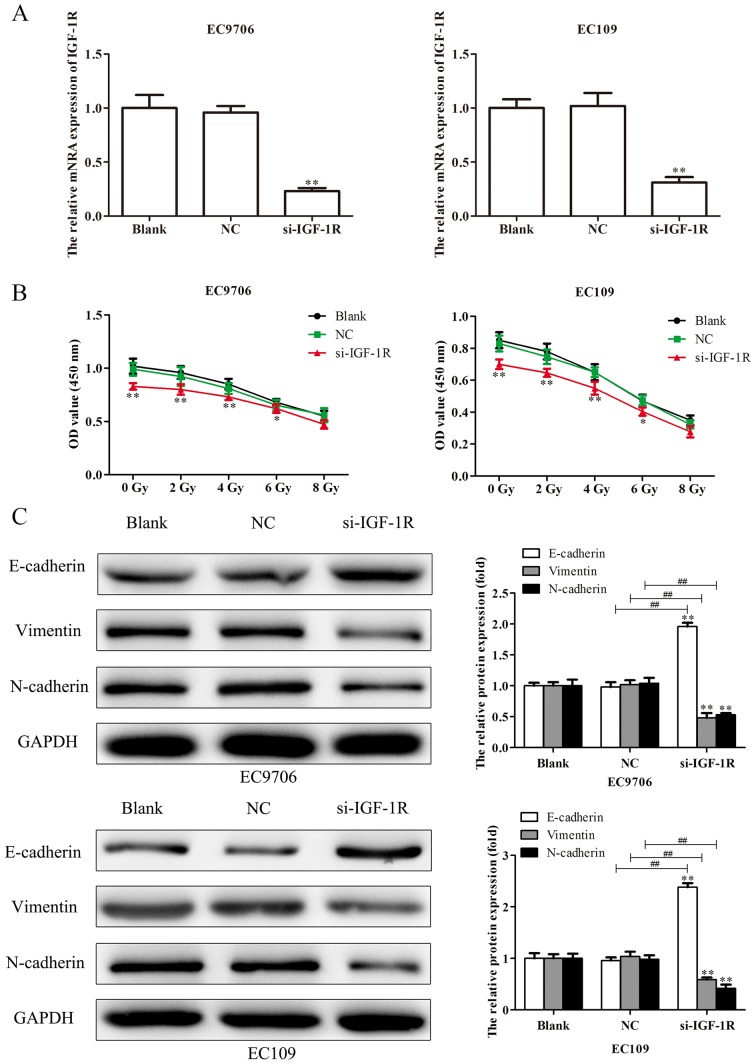
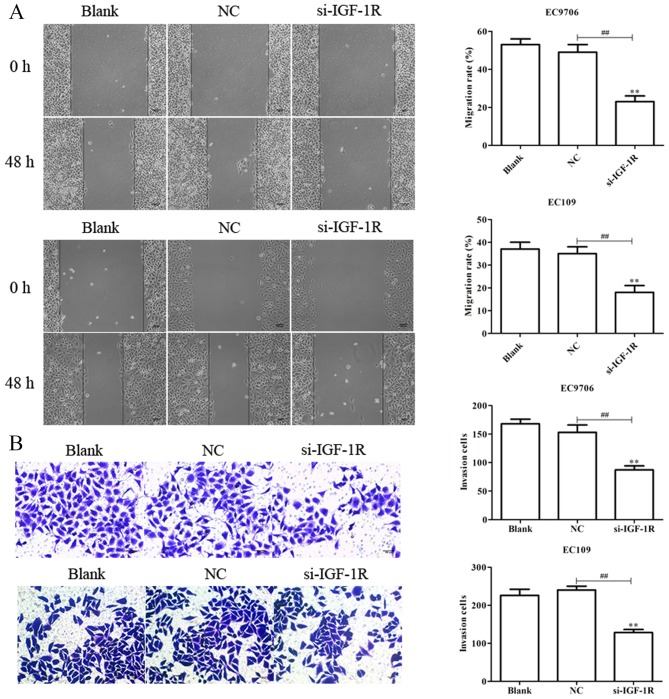
To investigate the effects of IGF-1R on the migration, invasion, EMT and radiosensitivity of EC cells, EC9706 and EC109 cells were transfected with si-IGF-1R, and RT-qPCR analysis was performed to determine the transfection efficiency. As presented in Fig. 9A, the levels of IGF-1R mRNA expression were significantly reduced in EC9706 and EC109 cells following transfection with si-IGF-1R. EC9706 and EC109 cells transfected with si-IGF-1R were irradiated with various radiation doses (0, 2, 4, 6 and 8 Gy) for 48 h, and a CCK-8 assay was performed to evaluate the proliferation of cells. As presented in Fig. 9B, transfection with si-IGF-1R significantly decreased the proliferation indicating increased radiosensitivity of EC9706 and EC109 cells. Additionally, western blotting was performed to evaluate the levels of expression of EMT-associated proteins. It was demonstrated that, compared with the control, E-cadherin protein was significantly upregulated, and vimentin and N-cadherin proteins were significantly downregulated following transfection with si-IGF-1R (Fig. 9C). Scratch-wound, and Transwell migration and invasion assays were subsequently performed to determine the effects of IGF-1R on the migration and invasion of EC9706 and EC109 cells. It was demonstrated that si-IGF-1R significantly suppressed the migration and invasion of EC9706 and EC109 cells compared with the control (Fig. 10A and B). The results indicated that IGF-1R, a candidate target of miR-30a-3p, serves important roles in the migration, invasion, EMT and radiosensitivity of EC cells.
Figure 9.
Silencing IGF-1R suppresses the proliferation and EMT of esophageal carcinoma cells. (A) Following transfection of EC9706 and EC109 cells with si-IGF-1R, the efficiency of transfection was determined via reverse transcription-quantitative polymerase chain reaction analysis. (B) EC9706 and EC109 cells transfected with si-IGF-1R were irradiated with various radiation doses (0, 2, 4, 6 and 8 Gy) for 48 h, then a Cell Counting Kit-8 assay was performed to evaluate the proliferation of cells. (C) Expression levels of EMT-associated proteins (E-cadherin, vimentin and N-cadherin) were determined in EC9706 and EC109 cells transfected with si-IGF-1R by western blot analysis. Band intensity was quantified using ImageJ software. Data are presented as the mean ± standard deviation of three independent experiments; each was performed in triplicate. *P<0.05 and **P<0.01 vs. the blank group. ##P<0.01 vs. the NC group. EMT, epithelial-mesenchymal transition; IGF-1R, insulin-like growth factor 1 receptor; NC, negative control; si, small interfering RNA.
Figure 10.
Silencing IGF-1R suppresses the migration and invasion of esophageal carcinoma cells. (A) Migratory abilities of EC9706 and EC109 cells transfected with si-IGF-1R for 48 h were determined via a scratch-wound assay. (B) Invasive abilities of EC9706 and EC109 cells transfected with si-IGF-1R for 48 h were evaluated using a Transwell invasion assay. Cells were counted in each well under an inverted microscope at ×200 magnification. Data are presented as the mean ± standard deviation of three independent experiments; each was performed in triplicate. **P<0.01 vs. the blank group. ##P<0.01 vs. the NC group. IGF-1R, insulin-like growth factor 1 receptor; NC, negative control; si, small interfering RNA.
Discussion
Studies investigating the association between miRNAs and tumor metastasis have focused on tumors located in various regions and systems, including the head and neck, as well as the respiratory, digestive and urinary systems (34–37). For example, numerous miRNAs have been reported to contribute to the regulation of breast cancer metastasis. miR-21 promoted the invasion of breast cancer cells via negative regulation of its target genes tropomyosin 1, programmed cell death 4 and maspin (38); a mutual feedback loop between zinc-finger E-box binding homeobox 1, miR-141 and miR-200c in regulating the invasion and metastasis of breast, pancreatic and colorectal cancer cells via EMT was reported (39).
miRNAs are not only involved in physiological processes, including growth and development of the body, but also serve important roles in various pathological processes, including the proliferation, apoptosis, invasion, angiogenesis and metastasis of various malignancies via downstream target genes (40–42). miR-30a-3p has been reported as downregulated, and involved in the progression and development of a number of tumors (28–30). For example, miRNA-30a-3p is downregulated in hepatocellular carcinoma, and inhibits the proliferation, invasiveness and metastasis of tumors (43). In the present study, it was demonstrated that the levels of miRNA-30a-3p expression were decreased in EC tissues and cell lines, and contributed to the migration and invasion of EC cells.
The role of EMT in tumor metastasis has received increasing focus (44,45). EMT is a complex process in which epithelial cells develop a mesenchymal-like phenotype. E-cadherin downregulation is an important feature of EMT and has been considered to be the most reliable indicator of EMT occurrence. E-cadherin serves an important role in the homeostasis of epithelial cells; its reduced expression during EMT leads to the downregulation of epithelial cell-associated proteins or reconstruction of complexes (including desmosome-associated proteins, tight junction proteins and cell polarity complex components), and upregulated expression of proteins associated with mesenchymal cells (including vimentin and N-cadherin), which is accompanied with reconstruction of the actin cytoskeleton (46–50). Therefore, the expression levels of EMT-associated proteins were evaluated in EC cells following transfection via western blotting, and it was revealed that miR-30a-3p significantly altered the expression of EMT-associated proteins.
Radiotherapy is an important therapy in the treatment of EC, reducing the recurrence risk of EC, and improving quality of life and survival; however, numerous factors affect the efficacy of radiotherapy, including the degree of hypoxia, glutathione content and individual differences in the sensitivity to radiation, which may result in an unsuccessful response to radiotherapy, and even induce severe radiation resistance (11,12,51,52). Radiation resistance of tumors is a complex phenomenon, involving complex molecular mechanisms and genetic alterations. DNA damage is the main mechanism by which radiotherapy-induced cell death occurs, including single and double strand breaks, and base damage (50,53–55). Ionizing radiation can provide sufficient time to repair damaged DNA and enable cells to survive via cell cycle arrest; however, when DNA repair fails, apoptosis is induced, an important defense and protection mechanism for the maintenance of genomic stability in normal cells. In tumor cells, cell cycle arrest and the suppression of apoptosis are important factors associated with the development of radiation resistance (51). Increasing evidence suggests that miRNAs are involved in these processes (56,57). Thus, CCK-8 assays were performed in the present study to evaluate the role of miR-30a-3p in transfected EC9706 and EC109 cells irradiated with various radiation doses. The results suggested that miR-30a-3p may act to enhance the radiosensitivity of EC cells.
IGF-1R belongs to the insulin receptor (IR) family, which also includes IR, IGF-1R and IGF-2R. Previous studies reported that the IGF-1R signaling pathway serves a pivotal role in the growth and progression of cancer, and resistance to anticancer therapies (58,59). It was revealed that IGF-1R promoted the growth of whole-body tissues and organs via the induction of protein synthesis. Autocrine or paracrine IGF-2 from tumor cells activated IGF-1R, and regulated the proliferation and differentiation of tumor cells (60–63). Nussbaum et al (63) observed that autocrine IGF-2 release from hepatocytes signaled via IGF-1R, interacting with hepatocyte growth factors to inhibit cell apoptosis, and promote cell growth and metastasis. Kim et al (64) reported that bronchial epithelial cells lacking p53 or expressing mutations in v-Ki-Ras2 Kirsten rat sarcoma viral oncogene homolog exhibited upregulation of IGF-1 and IGF-2; the transformed characteristics of these cells could be suppressed by IGF-1R inactivation, or enhanced by overexpression of IGF-1R. Furthermore, activated IGF-1R induced cisplatin resistance in numerous ovarian cancer cell lines via its downstream target gene phosphatidylinositol-3-kinase (65). IGF-1R has also been reported to contribute to radiosensitivity in oral squamous cell carcinoma (66). In the present study, the luciferase assay indicated that miR-30a-3p binds to the 3′-UTR of IGF-1R mRNA. Furthermore, silencing of IGF-1R affected the migration, invasion and radiosensitivity of EC cells; however, whether miR-30a-3p exerts its effects on EC via targeting IGF-1R requires further investigation.
The present study is a preliminary study into the role of miR-30a-3p in EC. Based on the findings, future work will aim to further investigate the role and mechanisms of miR-30a-3p in the progression and development of cancer in vitro and in vivo. Collectively, it was demonstrated that miR-30a-3p expression was upregulated in EC tissues and cell lines, and that miR-30a-3p may function as a potential tumor suppressor in EC, inhibiting metastasis and enhancing radiosensitivity via downregulation of IGF-1R. Therefore, miR-30a-3p may represent a potential therapeutic target in the treatment of EC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YF and QZ conceived and designed the study. PQ, PY and JWe performed the experiments. YL, JWu and XB wrote the manuscript and contibuted to the analysis or interpretation of the data. All authors have read and approved the final manuscript and agreed to be accountable for all aspects of the research in ensuring that the accuracy and integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The present study was approved by The Ethics Committee of Nanjing Medical University (Nanjing, China), and each patient provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Demeester SR. Epidemiology and biology of esophageal cancer. Gastrointest Cancer Res. 2009;32(Suppl):S2–S5. [PMC free article] [PubMed] [Google Scholar]
- 4.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 5.Zhang F, Yang Z, Cao M, Xu Y, Li J, Chen X, Gao Z, Xin J, Zhou S, Zhou Z, et al. MiR-203 suppresses tumor growth and invasion and down-regulates MiR-21 expression through repressing Ran in esophageal cancer. Cancer Lett. 2014;342:121–129. doi: 10.1016/j.canlet.2013.08.037. [DOI] [PubMed] [Google Scholar]
- 6.Iyer RB, Silverman PM, Tamm EP, Dunnington JS, DuBrow RA. Diagnosis, staging, and follow-up of esophageal cancer. AJR Am J Roentgenol. 2003;181:785–793. doi: 10.2214/ajr.181.3.1810785. [DOI] [PubMed] [Google Scholar]
- 7.Subasinghe D, Samarasekera DN. Delay in the diagnosis of esophageal carcinoma: Experience of a single unit from a developing country. Indian J Cancer. 2010;47:151–155. doi: 10.4103/0019-509X.63009. [DOI] [PubMed] [Google Scholar]
- 8.Lv XB, Lian GY, Wang HR, Song E, Yao H, Wang MH. Long noncoding RNA HOTAIR is a prognostic marker for esophageal squamous cell carcinoma progression and survival. PLoS One. 2013;8:e63516. doi: 10.1371/journal.pone.0063516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson SK, Ruszkiewicz AR, Jamieson GG, Esterman A, Watson DI, Wijnhoven BP, Lamb PJ, Devitt PG. Improving the accuracy of TNM staging in esophageal cancer: A pathological review of resected specimens. Ann Surg Oncol. 2008;15:3447–3458. doi: 10.1245/s10434-008-0155-0. [DOI] [PubMed] [Google Scholar]
- 10.Li CC, Chen CY, Chien CR. Comparative effectiveness of image-guided radiotherapy for non-operated localized esophageal squamous cell carcinoma patients receiving concurrent chemoradiotherapy: A population-based propensity score matched analysis. Oncotarget. 2016;7:71548–71555. doi: 10.18632/oncotarget.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kondo S, Tajika M, Tanaka T, Kodaira T, Mizuno N, Hara K, Hijioka S, Imaoka H, Goto H, Yamao K, Niwa Y. Prognostic factors for salvage endoscopic resection for esophageal squamous cell carcinoma after chemoradiotherapy or radiotherapy alone. Endosc Int Open. 2016;4:E841–E848. doi: 10.1055/s-0042-109609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roeder F, Nicolay NH, Nguyen T, Saleh-Ebrahimi L, Askoxylakis V, Bostel T, Zwicker F, Debus J, Timke C, Huber PE. Intensity modulated radiotherapy (IMRT) with concurrent chemotherapy as definitive treatment of locally advanced esophageal cancer. Radiat Oncol. 2014;9:191. doi: 10.1186/1748-717X-9-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 14.Li S, Li Z, Guo F, Qin X, Liu B, Lei Z, Song Z, Sun L, Zhang HT, You J, Zhou Q. miR-223 regulates migration and invasion by targeting Artemin in human esophageal carcinoma. J Biomed Sci. 2011;18:24. doi: 10.1186/1423-0127-18-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagman Z, Larne O, Edsjö A, Bjartell A, Ehrnström RA, Ulmert D, Lilja H, Ceder Y. miR-34c is downregulated in prostate cancer and exerts tumor suppressive functions. Int J Cancer. 2010;127:2768–2776. doi: 10.1002/ijc.25269. [DOI] [PubMed] [Google Scholar]
- 16.Lin RJ, Xiao DW, Liao LD, Chen T, Xie ZF, Huang WZ, Wang WS, Jiang TF, Wu BL, Li EM, Xu LY. MiR-142-3p as a potential prognostic biomarker for esophageal squamous cell carcinoma. J Surg Oncol. 2012;105:175–182. doi: 10.1002/jso.22066. [DOI] [PubMed] [Google Scholar]
- 17.Yokobori T, Suzuki S, Tanaka N, Inose T, Sohda M, Sano A, Sakai M, Nakajima M, Miyazaki T, Kato H, Kuwano H. MiR-150 is associated with poor prognosis in esophageal squamous cell carcinoma via targeting the EMT inducer ZEB1. Cancer Sci. 2013;104:48–54. doi: 10.1111/cas.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong L, Han Y, Zhang H, Zhao Q, Qiao Y. miR-210: A therapeutic target in cancer. Expert Opin Ther Targets. 2013;17:21–28. doi: 10.1517/14728222.2013.819853. [DOI] [PubMed] [Google Scholar]
- 19.Li B, Xu WW, Han L, Chan KT, Tsao SW, Lee NPY, Law S, Xu LY, Li EM, Chan KW, et al. MicroRNA-377 suppresses initiation and progression of esophageal cancer by inhibiting CD133 and VEGF. Oncogene. 2017;36:3986–4000. doi: 10.1038/onc.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng R, Liu Y, Zhang X, Zhao P, Deng Q. miRNA-200c enhances radiosensitivity of esophageal cancer by cell cycle arrest and targeting P21. Biomed Pharmacother. 2017;90:517–523. doi: 10.1016/j.biopha.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 21.Shao C, Yu Y, Yu L, Pei Y, Feng Q, Chu F, Fang Z, Zhou Y. Amplification and up-regulation of microRNA-30b in oral squamous cell cancers. Arch Oral Biol. 2012;57:1012–1017. doi: 10.1016/j.archoralbio.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Jiang BY, Zhang XC, Su J, Meng W, Yang XN, Yang JJ, Zhou Q, Chen ZY, Chen ZH, Xie Z, et al. BCL11A overexpression predicts survival and relapse in non-small cell lung cancer and is modulated by microRNA-30a and gene amplification. Mol Cancer. 2013;12:61. doi: 10.1186/1476-4598-12-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi B, Wang Y, Chen ZJ, Li XN, Qi Y, Yang Y, Cui GH, Guo HZ, Li WH, Zhao S. Down-regulation of miR-30a-3p/5p promotes esophageal squamous cell carcinoma cell proliferation by activating the Wnt signaling pathway. World J Gastroenterol. 2017;23:7965–7977. doi: 10.3748/wjg.v23.i45.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Ji Q, Zhang C, Liu X, Liu Y, Liu N, Sui H, Zhou L, Wang S, Li Q. miR-30a acts as a tumor suppressor by double-targeting COX-2 and BCL9 in H. pylori gastric cancer models. Sci Rep. 2017;7:7113. doi: 10.1038/s41598-017-07193-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas W. Staging of esophageal cancer: TNM and beyond. Esophagus. 2010;7:189–195. doi: 10.1007/s10388-010-0246-4. [DOI] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Wen XP, Ma HL, Zhao LY, Zhang W, Dang CX. MiR-30a suppresses non-small cell lung cancer progression through AKT signaling pathway by targeting IGF1R. Cell Mol Biol (Noisy-le-Grand) 2015;61:78–85. [PubMed] [Google Scholar]
- 28.Wang X, Qiu H, Tang R, Song H, Pan H, Feng Z, Chen L. miR-30a inhibits epithelial-mesenchymal transition and metastasis in triple-negative breast cancer by targeting ROR1. Oncol Rep. 2018;39:2635–2643. doi: 10.3892/or.2018.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiang AC, Massague J. Molecular basis of metastasis. N Engl J Med. 2008;359:2814–2823. doi: 10.1056/NEJMra0805239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen DX, Bos PD, Massague J. Metastasis: From dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 31.Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions the importance of changing cell state in development and disease. J Clin Invest. 2009;19:1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Weinberg RA. Epithelial mesenchymal transition: At the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 34.Tie J, Pan Y, Zhao L, Wu K, Liu J, Sun S, Guo X, Wang B, Gang Y, Zhang Y, et al. MiR-218 inhibits invasion and metastasis of gastric cancer by targeting the robo1 receptor. PLoS Genet. 2010;6:e1000879. doi: 10.1371/journal.pgen.1000879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu ZL, Wang H, Liu J, Wang ZX. MicroRNA-21 (miR-21) expression promotes growth, metastasis, and chemo- or radioresistance in non-small cell lung cancer cells by targeting PTEN. Mol Cell Biochem. 2013;372:35–45. doi: 10.1007/s11010-012-1443-3. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Z, Liu S, Shi R, Zhao G. miR-27 promotes human gastric cancer cell metastasis by inducing epithelial-to-mesenchymal transition. Cancer Genet. 2011;204:486–491. doi: 10.1016/j.cancergen.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 37.Sachdeva M, Mo YY. miR-145-mediated suppression of cell growth, invasion and metastasis. Am J Transl Res. 2010;2:170–180. [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 39.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEBl and membem of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 41.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 42.Liu H, Zhao J, Lv J. Inhibitory effects of miR-101 overexpression on cervical cancer Siha cells. Eur J Gynaecol Oncol. 2017;38:236–240. [PubMed] [Google Scholar]
- 43.Wang W, Lin H, Zhou L, Zhu Q, Gao S, Xie H, Liu Z, Xu Z, Wei J, Huang X, Zheng S. MicroRNA-30a-3p inhibits tumor proliferation, invasiveness and metastasis and is downregulated in hepatocellular carcinoma. Eur J Surg Oncol. 2014;40:1586–1594. doi: 10.1016/j.ejso.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 44.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 45.Vandewalle C, Comijn J, De Craene B, Vermassen P, Bruyneel E, Andersen H, Tulchinsky E, Van Roy F, Berx G. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res. 2005;33:6566–6578. doi: 10.1093/nar/gki965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eades G, Yuan Y, Yang M, Zhang Y, Chumsri S, Zhou Q. MiR-200a regulates SIRT1 and EMT-like transformation in mammary epithelial cells. J Biol Chem. 2011;286:25992–6002. doi: 10.1074/jbc.M111.229401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiang X, Zhuang X, Jiang H, Zhang S, Jiang H, Mu J, Zhang L, Miller D, Grizzle W, Zhang HG. miR-155 promotes macroscopic tumor formation yet inhibits tumor dissemination from mammary fat pads to the lung by preventing EMT. Oncogene. 2011;30:3440–3453. doi: 10.1038/onc.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ke Y, Zhao W, Xiong J, Cao R. miR-149 inhibits non-small-cell lung cancer cells EMT by targeting FOXM1. Biochem Res Int. 2013;2013:506731. doi: 10.1155/2013/506731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin Z, Zhou B, He Q, Li M, Guan P, Li X, Cui Z, Xue X, Su M, Ma R, et al. Association between polymorphisms in DNA repair genes and survival of non-smoking female patients with lung adenocarcinoma. BMC Cancer. 2009;9:439. doi: 10.1186/1471-2407-9-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Connell PP, Kron SJ, Weichselbaum RR. Relevance and irrelevance of DNA damage response to radiotherapy. DNA Repair (Amst) 2004;3:1245–1251. doi: 10.1016/j.dnarep.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 51.Woodward WA, Chen MS, Behbod F, Alfaro MP, Buchholz TA, Rosen JM. WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc Natl Acad Sci USA. 2007;104:618–623. doi: 10.1073/pnas.0606599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anastasov N, Höfig I, Vasconcellos IG, Rappl K, Braselmann H, Ludyga N, Auer G, Aubele M, Atkinson MJ. Radiation resistance due to high expression of miR-21 and G2/M checkpoint arrest in breast cancer cells. Radiat Oncol. 2012;7:206. doi: 10.1186/1748-717X-7-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Naidu MD, Mason JM, Pica RV, Fung H, Peña LA. Radiation resistance in glioma cells determined by DNA damage repair activity of Ape1/Ref-1. J Radiat Res. 2010;51:393–404. doi: 10.1269/jrr.09077. [DOI] [PubMed] [Google Scholar]
- 54.Cho S, Cinghu S, Yu JR, Park WY. Helicase-like transcription factor confers radiation resistance in cervical cancer through enhancing the DNA damage repair capacity. J Cancer Res Clin Oncol. 2011;137:629–637. doi: 10.1007/s00432-010-0925-5. [DOI] [PubMed] [Google Scholar]
- 55.Hazawa M, Hosokawa Y, Monzen S, Yoshino H, Kashiwakura I. Regulation of DNA damage response and cell cycle in radiation-resistant HL60 myeloid leukemia cells. Oncol Rep. 2012;28:55–61. doi: 10.3892/or.2012.1771. [DOI] [PubMed] [Google Scholar]
- 56.Liamina D, Sibirnyj W, Khokhlova A, Saenko V, Rastorgueva E, Fomin A, Saenko Y. Radiation-induced changes of microRNA expression profiles in radiosensitive and radioresistant leukemia cell lines with different levels of chromosome abnormalities. Cancers. 2017;9(pii):E136. doi: 10.3390/cancers9100136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu J, Li M, Wang Y, Luo J. Curcumin sensitizes prostate cancer cells to radiation partly via epigenetic activation of miR-143 and miR-143 mediated autophagy inhibition. J Drug Target. 2017;25:645–652. doi: 10.1080/1061186X.2017.1315686. [DOI] [PubMed] [Google Scholar]
- 58.Hartog H, Wesseling J, Boezen HM, van der Graaf WT. The insulin-like growth factor 1 receptor in cancer: Old focus, new future. Eur J Cancer. 2007;43:1895–1904. doi: 10.1016/j.ejca.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 59.Bähr C, Groner B. The insulin like growth factor-1 receptor (IGF-1R) as a drug target: Novel approaches to cancer therapy. Growth Horm IGF Res. 2004;14:287–295. doi: 10.1016/j.ghir.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 60.Lovly CM, McDonald NT, Chen H, Ortiz-Cuaran S, Heukamp LC, Yan Y, Florin A, Ozretić L, Lim D, Wang L, et al. Rationale for co-targeting IGF-1R and ALK in ALK fusion positive lung cancer. Nat Med. 2014;20:1027–1034. doi: 10.1038/nm.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buck E, Mulvihill M. Small molecule inhibitors of the IGF-1R/IR axis for the treatment of cancer. Expert Opin Investig Drugs. 2011;20:605–621. doi: 10.1517/13543784.2011.558501. [DOI] [PubMed] [Google Scholar]
- 62.Shiratsuchi I, Akagi Y, Kawahara A, Kinugasa T, Romeo K, Yoshida T, Ryu Y, Gotanda Y, Kage M, Shirouzu K. Expression of IGF-1 and IGF-1R and their relation to clinicopathological factors in colorectal cancer. Anticancer Res. 2011;31:2541–2545. [PubMed] [Google Scholar]
- 63.Nussbaum T, Samarin J, Ehemann V, Bissinger M, Ryschich E, Khamidjanov A, Yu X, Gretz N, Schirmacher P, Breuhahn K. Autocrine insulin-like growth factor-II stimulation of tumor cell migration is a progression step in human hepatocarcinogenesis. Hepatology. 2008;48:146–156. doi: 10.1002/hep.22297. [DOI] [PubMed] [Google Scholar]
- 64.Kim WY, Jin Q, Oh SH, Kim ES, Yang YJ, Lee DH, Feng L, Behrens C, Prudkin L, Miller YE, et al. Elevated epithelial insulin-like growth factor expression is a risk factor for lung cancer development. Cancer Res. 2009;69:7439–7448. doi: 10.1158/0008-5472.CAN-08-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eckstein N, Servan K, Hildebrandt B, Pölitz A, von Jonquières G, Wolf-Kümmeth S, Napierski I, Hamacher A, Kassack MU, Budczies J, et al. Hyperactivation of the insulin-like growth factor receptor I signaling pathway is an essential event for cisplatin resistance of ovarian cancer cells. Cancer Res. 2009;69:2996–3003. doi: 10.1158/0008-5472.CAN-08-3153. [DOI] [PubMed] [Google Scholar]
- 66.Zhang B, Li Y, Hou D, Shi Q, Yang S, Li Q. MicroRNA-375 inhibits growth and enhances radiosensitivity in oral squamous cell carcinoma by targeting insulin like growth factor 1 receptor. Cell Physiol Biochem. 2017;42:2105–2117. doi: 10.1159/000479913. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.