Abstract
Multiple sclerosis (MS) is a chronic inflammatory disease of the central nervous system of autoimmune etiopathogenesis, and is characterized by various neurological symptoms. Glatiramer acetate and interferon-β are administered as first-line treatments for this disease. In non-responsive patients, several second-line therapies are available, including natalizumab; however, a percentage of MS patients does not respond, or respond partially. Therefore, it is of the utmost importance to develop a diagnostic test for the prediction of drug response in patients suffering from complex diseases, such as MS, where several therapeutic options are already available. By a machine learning approach, the UnCorrelated Shrunken Centroid algorithm was applied to identify a subset of genes of CD4+ T cells that may predict the pharmacological response of relapsing-remitting MS patients to natalizumab, before the initiation of therapy. The results from the present study may provide a basis for the design of personalized therapeutic strategies for patients with MS.
Keywords: multiple sclerosis, natalizumab, biomarkers, machine learning, UnCorrelated Shrunken Centroid, personalized medicine
Introduction
Multiple sclerosis (MS) is a chronic inflammatory disease of the central nervous system of autoimmune etiopathogenesis, and is characterized by neurological symptoms (1). A total of ~90% of patients with MS are diagnosed with relapsing-remitting disease, that involve acute periods of neurological dysfunctions followed by a period of recovery (2). Glatiramer acetate and interferon-β (IFN-β) are administered as first-line therapies. In patients non-responsive to treatment, several second-line therapies are available, including natalizumab and fingolimod (3). Natalizumab is a monoclonal antibody against the α4 subunit (CD49d) of α4 integrins [α4β1 (VLA-4) and α4β7), that prevents the transmigration of leukocytes across the blood-brain barrier by inhibiting interactions between α4β1 integrin/vascular cell adhesion molecule-1 and mucosal vascular addressin cell adhesion molecule-1 (4). In 2012, a longitudinal study assessing the effects of natalizumab on 333 patients with MS revealed that 69–88% of patients exhibited a positive outcome in all Patient-Reported Outcomes measures assessed (5). In the Natalizumab Safety and Efficacy in Relapsing Remitting Multiple Sclerosis (AFFIRM) trial (ClinicalTrials.gov Identifier: NCT00027300), of 942 patients, 627 were randomly selected for treatment with natalizumab and 315 were administered a placebo. The results revealed that natalizumab reduced the risk of sustained disability progression by 42% in a 2-year time-frame and decreased the rate of clinical relapse in 1 year by 68%, leading to an 83% reduction in the accumulation of new or enlarging T-2 hyperintense lesions (6). Additionally, the results of a SENTINEL trial indicated that 67% of patients receiving natalizumab plus IFN-β-1a remained free of new or enlarging T2-lesions compared with 30% of patients receiving IFN-β-1a alone (7). These findings indicate that despite the high efficacy, a percentage of patients with MS do not respond, or respond partially to natalizumab. Therefore, it is of the utmost importance to develop a diagnostic test to predict drug response in patients suffering from complex diseases, such as MS, in which several therapeutic options are readily available. This could lead to a double-fold advantage: Patients would benefit by avoiding ineffective therapies and healthcare costs would be notably reduced. In the present study, a machine learning approach was utilized to identify a subset of genes that may predict the response of patients with MS to natalizumab prior to the initiation of therapy.
Materials and methods
Molecular patterns of pharmacological resistance to natalizumab
For the identification of the molecular patterns underlying the pharmacological resistance to natalizumab in MS, we selected the GSE44964 microarray dataset, available from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/). The dataset comprised whole-genome expression data from CD4+ T cells isolated from patients with MS and stimulated in vitro with precoated anti-CD3/-CD28 monoclonal antibodies for 48 h. The Agilent Sureprint G3 Human Gene Expression 8×60k platform was used to generate the dataset (Agilent Technologies, Inc., Santa Clara, CA, USA). The raw data were quantile normalized and batch effect-corrected using ComBat v2 (8). The patients were of Swedish origin and had relapsing-remitting disease. The GSE44964 dataset comprises data generated from two different microarray platforms. To avoid obtaining biased results, the data of two platforms were not combined; thus, analysis was conducted using the largest set of samples only. Patients were diagnosed with MS according to the McDonald criteria (9) and prospectively classified as low responders (LRs, n=6), if at least one period of relapse occurred during the follow-up period (3 years) and as high responders (HRs, n=6), providing no relapse was observed. Other parameters, such as magnetic resonance imaging could be used for the classification of LRs and HRs; however, the relapse rate is considered as a primary endpoint of several phase 2/3 clinical trials (6), and classifying patients as responsive and non-responsive on the basis of whether relapse had occurred or not is appropriate for a preliminary transcriptomic analysis. All samples were collected for gene expression analysis prior to the initiation of natalizumab treatment. The LR and HR groups were matched for sex, age, Expanded Disability Status Scale score (10) and disease duration (11). A total of 5/6 patients in the groups were males; the age of patients was 36±6.3 and 33.7±7.1 years old for LRs and HRs, respectively. Statistical differences between HR and LR patients were assessed using LIMMA version 3.26.8 (Linear models for microarray data) in R version 3.2.3 (12). P<0.01 was considered to indicate a statistically significant difference. Statistical analysis and principal component analysis (PCA) were performed using MultiExperiment Viewer software (http://mev.tm4.org/). Gene Ontology (GO) analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 web-based tool (13,14). Functional annotation provided by DAVID comprises >40 annotation categories, including GO terms, protein-protein interactions, protein domains, disease associations, pathways, homology, gene function, gene tissue expression and literature. Network analysis was performed using the GeneMania utility (15).
Identification of biomarkers for natalizumab responsiveness
In order to identify a specific gene expression signature for predicting patient responsiveness to natalizumab treatment, the UnCorrelated Shrunken Centroid (UCSC; http://home.cc.umanitoba.ca/~psgendb/birchhomedir/BIRCHDEV/doc/MeV/manual/usc.html) algorithm was used. UCSC analysis was performed with the probes that were determined to be significantly modulated in HRs compared with the LR group. Cross-validation was conducted using the following parameters: 5-fold and 10-fold cross-validation; each cross-validation run was divided five-fold and therefore, a total 10 cross-validation runs were performed. Δ-(shrinkage threshold) and ρ-(correlation threshold) values were empirically selected so that the smallest number of classification errors were obtained using the fewest genes. Subsequently, PCA and Hierarchical Clustering (HCL) was performed using only the set of the identified predictors. For HCL, Euclidean distance and average linkage degree were used.
Results
HR and LR patients have different transcriptomic patterns
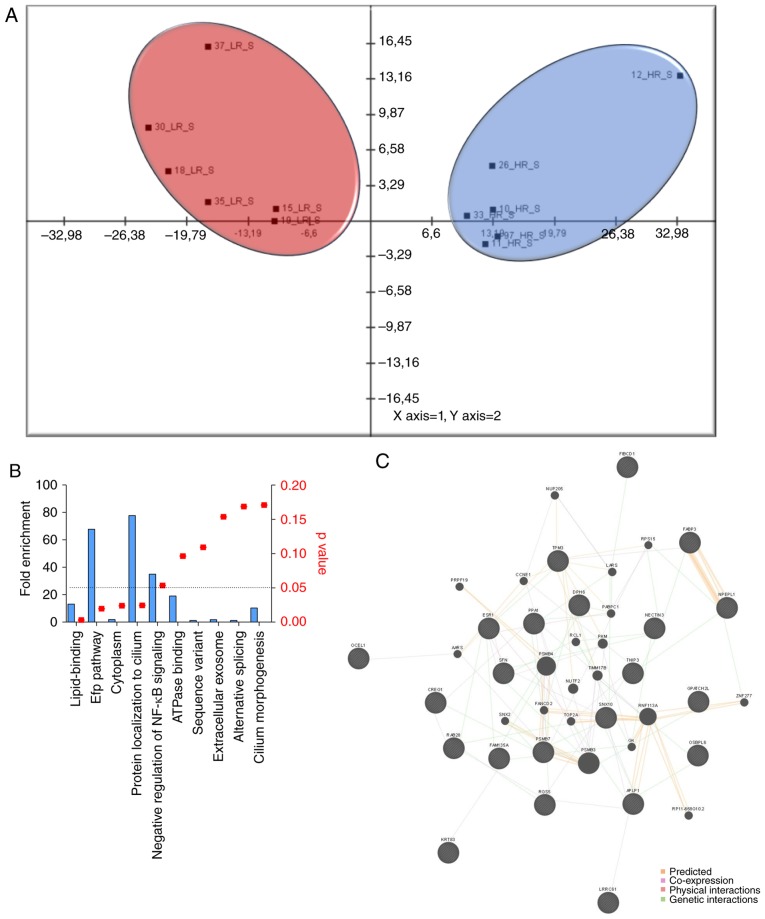
Statistical analysis of the transcriptomic differences between CD4+ T cells from patients of the HR and LR groups revealed 45 significant probes (Table I). PCA (Fig. 1A) produced two main clusters that contained HRs and LRs, respectively. The results indicated that the mRNA expression levels of several genes notably differed between natalizumab-responsive and non-responsive patients, and that a distinct pattern of gene expression could be associated with natalizumab resistance. Functional annotation revealed that the most enriched categories and their associated genes were: ‘Lipid-binding’ [oxysterol binding protein like 6 (OSBPL6), fatty acid binding protein 3 (FABP3), estrogen receptor 1 (ESR1) and sorting nexin 10 (SNX10)], ‘estrogen-responsive protein Efp controls cell cycle and breast tumors growth’ [ESR1 and stratifin (SFN)], ‘cytoplasm’ [OSBPL6, ESR1, SFN, amyloid β precursor-like protein 1 (APLP1), inorganic pyrophosphatase PPA1), tropomyosin 3 (TPM3), 20S proteasome subunit β-2 (PSMB7), RAB28, member RAS oncogene family (RAB28), regulator of G-protein signaling 5, FABP3, TBC1 domain family member 32 (TBC1D32), SNX10 and coiled-coil domain-containing protein 8] and ‘protein localization to cilium’ (SNX10 and TBC1D32) (Fig. 1B). A regulatory network comprising the significant genes and the top 20 related genes, was presented as Fig. 1C. The computational gene network prediction tool GeneMania identified the DNA topoisomerase II α gene to interact with PPA1, TNFAIP3 interacting protein 3, PSMB3, RAB28, APLP1, ESR1 and ring finger protein 113A. Other nodes were represented by PSMB7, alanyl-tRNA synthetase, APLP1 and ESR1 (Fig. 1C).
Table I.
List of genes significantly modulated between LRs and HRs CD4+ T cells.
| Probe | Gene accession no. | Gene symbol | Gene name | P-value | Log fold change |
|---|---|---|---|---|---|
| A_19_P00317412 | XLOC_000787 | 0.0048 | −0.4088 | ||
| A_19_P00317731 | XLOC_002473 | 0.0078 | 0.6845 | ||
| A_19_P00319311 | 0.0046 | 0.6016 | |||
| A_19_P00321466 | 0.0079 | 0.5516 | |||
| A_19_P00805263 | XLOC_001851 | 0.0088 | 0.3698 | ||
| A_19_P00807336 | 0.0030 | −0.5004 | |||
| A_23_P108823 | NM_032523 | OSBPL6 | Oxysterol binding protein-like 6 | 0.0054 | −0.3303 |
| A_23_P112187 | NM_032843 | FIBCD1 | Fibrinogen C domain containing 1 | 0.0059 | −1.1805 |
| A_23_P164958 | NM_032040 | CCDC8 | Coiled-coil domain containing 8 | 0.0044 | 0.6849 |
| A_23_P27983 | NM_005166 | APLP1 | Amyloid β (A4) precursor-like protein 1 | 0.0077 | −0.7417 |
| A_23_P381017 | NM_152559 | WBSCR27 | Williams Beuren syndrome chromosome region 27 | 0.0051 | 0.8273 |
| A_23_P386478 | NM_024873 | TNIP3 | TNFAIP3 interacting protein 3 | 0.0065 | −0.9948 |
| A_23_P398172 | NM_020819 | FAM135A | Family with sequence similarity 135, member A | 0.0099 | 0.7579 |
| A_23_P401547 | NM_015480 | PVRL3 | Poliovirus receptor-related 3 | 0.0076 | 0.4645 |
| A_23_P54205 | NM_017926 | C14orf118 | Chromosome 14 open reading frame 118 | 0.0067 | 0.6402 |
| A_23_P77135 | NM_080650 | ATPBD4 | ATP binding domain 4 | 0.0084 | 1.0763 |
| A_23_P90523 | NM_024578 | OCEL1 | Occludin/ELL domain containing 1 | 0.0025 | −0.3799 |
| A_24_P62783 | NM_004102 | FABP3 | Fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) | 0.0016 | −0.6532 |
| A_24_P920048 | AK092807 | LOC100127972 | Uncharacterized LOC100127972 | 0.0043 | 0.9088 |
| A_32_P204376 | NM_001012421 | ANKRD20A2 | Ankyrin repeat domain 20 family, member A2 | 0.0084 | 0.3258 |
| A_32_P7204 | NM_004249 | RAB28 | RAB28, member RAS oncogene family | 0.0057 | 0.8683 |
| A_32_P797019 | XM_003403482 | NPEPL1 | Aminopeptidase-like 1 | 0.0060 | 0.6953 |
| A_32_P80245 | NM_001109809 | ZFP57 | Zinc finger protein 57 homolog (mouse) | 0.0042 | 1.0614 |
| A_33_P3221925 | NM_152730 | C6orf170 | Chromosome 6 open reading frame 170 | 0.0018 | 0.6414 |
| A_33_P3227269 | NR_024256 | FLJ45983 | Uncharacterized LOC399717 | 0.0047 | 0.9719 |
| A_33_P3231297 | NM_003851 | CREG1 | Cellular repressor of E1A-stimulated genes 1 | 0.0073 | −0.3014 |
| A_33_P3233273 | NM_001142928 | LRRC61 | Leucine rich repeat containing 61 | 0.0067 | 0.4877 |
| A_33_P3243093 | NM_003617 | RGS5 | Regulator of G-protein signaling 5 | 0.0042 | −0.7003 |
| A_33_P3271241 | NM_021129 | PPA1 | Pyrophosphatase (inorganic) 1 | 0.0079 | −0.3602 |
| A_33_P3273552 | NM_002282 | KRT83 | Keratin 83 | 0.0004 | −0.9603 |
| A_33_P3289536 | NM_001199835 | SNX10 | Sorting nexin 10 | 0.0082 | 0.9177 |
| A_33_P3292724 | BC017576 | 0.0042 | 0.6890 | ||
| A_33_P3333600 | LOC400950 | Uncharacterized LOC400950 | 0.0078 | −0.6855 | |
| A_33_P3338674 | NR_033298 | CCDC163P | Coiled-coil domain containing 163, pseudogene | 0.0006 | −1.1794 |
| A_33_P3341474 | NM_001080412 | ZBTB38 | Zinc finger and BTB domain containing 38 | 0.0067 | 0.7430 |
| A_33_P3346193 | NM_001043351 | TPM3 | Tropomyosin 3 | 0.0022 | 0.8160 |
| A_33_P3354514 | BC047507 | SLC2A13 | Solute carrier family 2 (facilitated glucose transporter), member 13 | 0.0004 | 0.4126 |
| A_33_P3379356 | NM_001122742 | ESR1 | Estrogen receptor 1 | 0.0062 | 0.5516 |
| A_33_P3382489 | 0.0056 | 0.6209 | |||
| A_33_P3387110 | NR_015419 | LOC145783 | Uncharacterized LOC145783 | 0.0081 | −0.5672 |
| A_33_P3389286 | NM_006142 | SFN | Stratifin | 0.0059 | 0.7813 |
| A_33_P3400292 | 0.0094 | −0.5263 | |||
| A_33_P3407429 | PSMB7 | Proteasome (prosome, macropain) subunit, β type, 7 | 0.0052 | −0.4364 | |
| A_33_P3541279 | AF116649 | 0.0021 | 0.8729 | ||
| A_33_P3876414 | AJ272176 | C17orf6 | Chromosome 17 open reading frame 6 | 0.0090 | 0.5759 |
Figure 1.
Molecular patterns of pharmacological resistance to natalizumab. For the identification of the molecular patterns underlying the pharmacological resistance to natalizumab in MS, we selected the GSE44964 microarray dataset that included expression data from CD4+ T cells isolated from patients with MS, which were stimulated in vitro with anti-CD3/-CD28 monoclonal antibodies for 48 h. Statistical differences between HR (n=6) and LR (n=6) patients were assessed using LIMMA. (A) Principal component analysis was performed using MultiExperiment Viewer software and the genes significantly modulated between HR and LR patients. The distribution of the samples of the first two components was presented. (B) Gene Ontology analysis for the genes significantly modulated between HR and LR patients was performed using the Database for Annotation, Visualization and Integrated Discovery v6.8 web-based tool. (C) Network analysis for genes significantly modulated between HR and LR patients was performed using the GeneMania. HR, high responder; LR, low responder; NF-κB, nuclear factor-κB.
Machine learning-identified genes for predicting natalizumab responsiveness
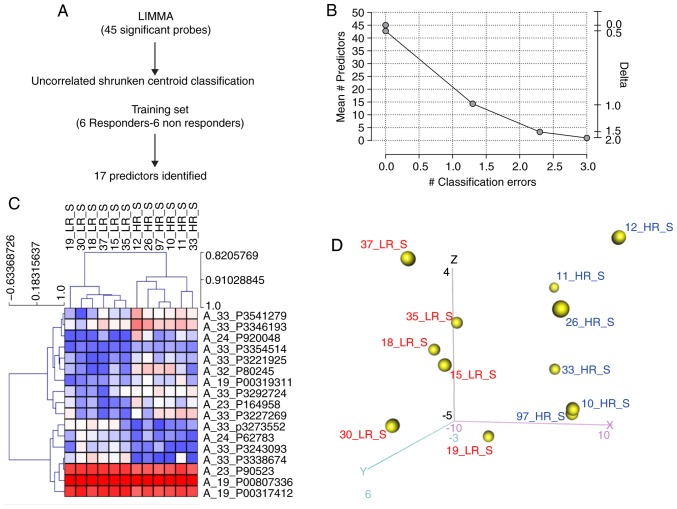
To identify a specific gene signature for natalizumab responsiveness, the UCSC algorithm was applied to the significant probes identified. The following parameters were selected: Δ=1 and ρ=1 for UCSC analysis. A total of 17 predictors of the 45 probes (Fig. 2A) were identified from UCSC analysis that were able to classify HR and LR samples with 89.2% agreement with the clinical data (Fig. 2B). Consistent with these findings, HCL and PCA based on the 17 markers were able to accurately separate HR and LR patients (Fig. 2C and D). The 17 identified predictors were presented in Table II.
Figure 2.
Prediction of pharmacological response to natalizumab. In order to determine a specific gene expression signature for predicting responsiveness to natalizumab treatment, the UCSC algorithm was applied to the genes significantly modulated between HR and LR patients. (A) The experimental procedure. (B) The number of classification errors obtained from UCSC analysis using different Δ-values (C) Hierarchical clustering of the 17 predictor genes obtained via UCSC analysis. (D) Principal component analysis using the 17 predictor genes obtained via UCSC analysis. The distribution of the samples on the first three components. HR, high responder; LR, low responder; UCSC, UnCorrelated Shrunken Centroid.
Table II.
Predictors of natalizumab responsiveness.
| Probe | Gene accession no. | Gene symbol | Description |
|---|---|---|---|
| A_33_P3273552 | NM_002282 | KRT83 | Keratin 83 |
| A_33_P3338674 | NR_033298 | CCDC163P | Coiled-coil domain containing 163, pseudogene |
| A_24_P62783 | NM_004102 | FABP3 | Fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| A_33_P3221925 | NM_152730 | C6orf170 | Chromosome 6 open reading frame 170 |
| A_33_P3541279 | AF116649 | ||
| A_33_P3346193 | NM_001043351 | TPM3 | Tropomyosin 3 |
| A_33_P3243093 | NM_003617 | RGS5 | Regulator of G-protein signaling 5 |
| A_32_P80245 | NM_001109809 | ZFP57 | Zinc finger protein 57 homolog (mouse) |
| A_24_P920048 | AK092807 | LOC100127972 | Uncharacterized LOC100127972 |
| A_33_P3227269 | NR_024256 | FLJ45983 | Uncharacterized LOC399717 |
| A_23_P381017 | NM_152559 | WBSCR27 | Williams Beuren syndrome chromosome region 27 |
| A_32_P7204 | NM_004249 | RAB28 | RAB28, member RAS oncogene family |
| A_23_P112187 | NM_032843 | FIBCD1 | Fibrinogen C domain containing 1 |
| A_33_P3389286 | NM_006142 | SFN | Stratifin |
| A_23_P386478 | NM_024873 | TNIP3 | TNFAIP3 interacting protein 3 |
| A_33_P3289536 | NM_001199835 | SNX10 | Sorting nexin 10 |
| A_23_P77135 | NM_080650 | ATPBD4 | ATP binding domain 4 |
Discussion
Several studies have investigated expression array phenotyping as a means for predicting drug response and clinical prognosis (16–18), and for the classification of diseases (18–25)the most common lymphoid malignancy in adults, is curable in less than 50% of patients. Prognostic models based on pre-treatment characteristics, such as the International Prognostic Index (IPI. Predicting diagnostic classes based on a sample using its expression profile is known as supervised learning or classification. The use of microarray data, although practical, poses the problem of predicting diagnostic classes using a number of genes that is notably higher than the number of sample types available. Therefore, it is necessary to select subsets of genes that are relevant for the characterization of the different diagnostic classes. In addition, the identification of specific subsets of genes may improve the classification accuracy, allow the development of cost-effective diagnostic tests and may provide novel biological insight into certain diseases. Classification can be defined as a supervised learning approach, in which the classes of a series of samples are inputted to an algorithm. This is distinct from unsupervised clustering, in which no prior knowledge of the samples is available. The aim of classification is to identify the smallest possible subgroup of genes highly associated with the known sample classes. The UCSC algorithm is based on the ‘Shrunken Centroid’ algorithm reported by Tibshirani et al (25). Briefly, genes are considered one at a time and the difference between the class centroid (the mean expression in a class) of a gene and the overall centroid (the mean expression level across all classes) of a gene is compared with the within-class standard deviation plus a Δ-value, which is determined by cross-validation, in order to minimize classification errors. In the present study, the UCSC algorithm was applied for the identification of a subset of genes that could predict the pharmacological response to natalizumab treatment among patients with relapsing-remitting MS. Natalizumab is a disease-modifying drug that can effectively reduce the frequency of relapse and short-term disability progression in relapsing-remitting MS, and it is often used as second-line treatment in patients exhibiting active disease, despite treatment with glatiramer acetate or IFN-β (26).
To the best of our knowledge, the present study is the first to identify, at the whole-genome level, the genes that were significantly modulated in HR patients compared with the LR group. Our findings suggest that a specific gene expression profile of CD4+ T cells may characterize the pharmacological responsiveness to natalizumab in patients with MS. Interestingly, no significant differences in the transcription levels of CD49d and CD29, which encode the target of natalizumab comprising the α4 and β1 subunits of VLA-4, were observed between the HR and LR groups of patients (data not shown). Additionally, we applied machine learning to select the minimum number of genes able to predict the response to natalizumab. The results indicate the genes that may be relevant for P4 medicine, which constitutes predictive, preventative, personalized and participatory medicine (27). At present, the mechanisms of resistance to natalizumab remains largely unknown. Recently, Cavaliere et al (28) applied molecular dynamics simulation to determine whether a polymorphism could induce conformational changes in VLA-4, affecting the binding affinity with natalizumab; expression profiling of circulating blood cells should be conducted for the identification of biomarkers of natalizumab resistance. The role of the genes identified in our study requires further investigation; however, certain genes may be involved in immunity and the pathology of MS. In particular, the Ras-related protein, Rab-28, has been detected in the serum of Alzheimer's disease and MS patients (29). TPM3 was determined to be phosphorylated following T cell costimulation, resulting in downregulated interleukin-2-stimulated T cells (30). The locus 3 kb upstream of the zinc finger 57, that encodes a protein likely to act as a transcriptional repressor, has been reported to be hypomethylated in CD4+ T cells from patients with MS compared with healthy controls (31). Increasing efforts are required to validate our findings determine of the role of the genes involved.
The use of biomarkers to predict natalizumab resistance in MS would lead to notable therapeutic and economic benefits; however, our study has several limitations. The number of patients is limited and no external validation could be performed. In addition, it has not been disclosed by the original authors of the microarray datasets whether natalizumab treatment was administered as first-line therapy or after failure with other medications, such as glatiramer acetate or IFN-β. Furthermore, a comparison with other second-line drugs, such as fingolimod, should be conducted. Finally, the expression profiles of unsorted and unstimulated circulating cell populations, such as whole blood cells or peripheral blood mononuclear cells should be determined for the development of simple and economically viable diagnostic tests. Despite these limitations of the present study, findings may serve as a basis for the design of personalized therapeutic options for patients with MS.
Acknowledgements
Not applicable.
Funding
The present study was supported by current research funds 2018 of IRCCS ‘Centro Neurolesi ‘Bonino Pulejo’, Messina-Italy.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus repository (https://www.ncbi.nlm.nih.gov/geo/), with accession number GSE44964.
Authors' contributions
PF, EM, FN and KM made substantial contributions to the conception and design of the study. SM, FS, MSB and MCP analyzed the data and prepared the figures. PF, RDM, PB and KM interpreted the data and drafted the manuscript. FN, EM, RDM and PB critically revised the final manuscript. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Compston A, Coles A. Multiple sclerosis. Lancet. 2008;372:1502–1517. doi: 10.1016/S0140-6736(08)61620-7. [DOI] [PubMed] [Google Scholar]
- 2.Tullman MJ. Overview of the epidemiology, diagnosis, and disease progression associated with multiple sclerosis. Am J Manag Care 19 (2 Suppl) 2013:S15–S20. [PubMed] [Google Scholar]
- 3.Garg N, Smith TW. An update on immunopathogenesis, diagnosis, and treatment of multiple sclerosis. Brain Behav. 2015;5:e00362. doi: 10.1002/brb3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clerico M, Artusi CA, Liberto AD, Rolla S, Bardina V, Barbero P, Mercanti SF, Durelli L. Natalizumab in multiple sclerosis: Long-term management. Int J Mol Sci. 2017;18(pii):E940. doi: 10.3390/ijms18050940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sospedra M, Planas R, Martin R. Long-term safety and efficacy of natalizumab in relapsing-remitting multiple sclerosis: Impact on quality of life. Patient Relat Outcome Meas. 2014;5:25–33. doi: 10.2147/PROM.S41768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polman CH, O'Connor PW, Havrdova E, Hutchinson M, Kappos L, Miller DH, Phillips JT, Lublin FD, Giovannoni G, Wajgt A, et al. A randomized, placebo-controlled trial of natalizumab for relapsing multiple sclerosis. N Engl J Med. 2006;354:899–910. doi: 10.1056/NEJMoa044397. [DOI] [PubMed] [Google Scholar]
- 7.Radue EW, Stuart WH, Calabresi PA, Confavreux C, Galetta SL, Rudick RA, Lublin FD, Weinstock-Guttman B, Wynn DR, Fisher E, et al. Natalizumab plus interferon beta-1a reduces lesion formation in relapsing multiple sclerosis. J Neurol Sci. 2010;292:28–35. doi: 10.1016/j.jns.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 8.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 9.Polman CH, Reingold SC, Banwell B, Clanet M, Cohen JA, Filippi M, Fujihara K, Havrdova E, Hutchinson M, Kappos L, et al. Diagnostic criteria for multiple sclerosis: 2010 Revisions to the McDonald criteria. Ann Neurol. 2011;69:292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saccà F, Costabile T, Carotenuto A, Lanzillo R, Moccia M, Pane C, Russo CV, Barbarulo AM, Casertano S, Rossi F, et al. The EDSS integration with the brief international cognitive assessment for multiple sclerosis and orientation tests. Mult Scler. 2017;23:1289–1296. doi: 10.1177/1352458516677592. [DOI] [PubMed] [Google Scholar]
- 11.Gustafsson M, Edström M, Gawel D, Nestor CE, Wang H, Zhang H, Barrenäs F, Tojo J, Kockum I, Olsson T, et al. Integrated genomic and prospective clinical studies show the importance of modular pleiotropy for disease susceptibility, diagnosis and treatment. Genome Med. 2014;6:17. doi: 10.1186/gm534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 15.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38 (Web Server Issue) 2010:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 17.Nutt CL, Mani DR, Betensky RA, Tamayo P, Cairncross JG, Ladd C, Pohl U, Hartmann C, McLaughlin ME, Batchelor TT, et al. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63:1602–1607. [PubMed] [Google Scholar]
- 18.Shipp MA, Ross KN, Tamayo P, Weng AP, Kutok JL, Aguiar RC, Gaasenbeek M, Angelo M, Reich M, Pinkus GS, et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. 2002;8:68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- 19.Alon U, Barkai N, Notterman DA, Gish K, Ybarra S, Mack D, Levine AJ. Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc Natl Acad Sci USA. 1999;96:6745–6750. doi: 10.1073/pnas.96.12.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schummer M, Ng WV, Bumgarner RE, Nelson PS, Schummer B, Bednarski DW, Hassell L, Baldwin RL, Karlan BY, Hood L. Comparative hybridization of an array of 21,500 ovarian cDNAs for the discovery of genes overexpressed in ovarian carcinomas. Gene. 1999;238:375–385. doi: 10.1016/S0378-1119(99)00342-X. [DOI] [PubMed] [Google Scholar]
- 21.Ramaswamy S, Tamayo P, Rifkin R, Mukherjee S, Yeang CH, Angelo M, Ladd C, Reich M, Latulippe E, Mesirov JP, et al. Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci USA. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 23.Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 24.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorscheider J, Benkert P, Lienert C, Hänni P, Derfuss T, Kuhle J, Kappos L, Yaldizli Ö. Comparative analysis of natalizumab versus fingolimod as second-line treatment in relapsing-remitting multiple sclerosis. Mult Scler J. 2018;24:777–785. doi: 10.1177/1352458518768433. [DOI] [PubMed] [Google Scholar]
- 27.Bousquet J, Anto JM, Sterk PJ, Adcock IM, Chung KF, Roca J, Agusti A, Brightling C, Cambon-Thomsen A, Cesario A, et al. Systems medicine and integrated care to combat chronic noncommunicable diseases. Genome Med. 2011;3:43. doi: 10.1186/gm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cavaliere F, Montanari E, Emerson A, Buschini A, Cozzini P. In silico pharmacogenetic approach: The natalizumab case study. Toxicol Appl Pharmacol. 2017;330:93–99. doi: 10.1016/j.taap.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 29.Hajipour MJ, Ghasemi F, Aghaverdi H, Raoufi M, Linne U, Atyabi F, Nabipour I, Azhdarzadeh M, Derakhshankhah H, Lotfabadi A, et al. Sensing of Alzheimer's disease and multiple sclerosis using nano-bio interfaces. J Alzheimers Dis. 2017;59:1187–1202. doi: 10.3233/JAD-160206. [DOI] [PubMed] [Google Scholar]
- 30.Lichtenfels R, Rappl G, Hombach AA, Recktenwald CV, Dressler SP, Abken H, Seliger B. A proteomic view at T cell costimulation. PLoS One. 2012;7:e32994. doi: 10.1371/journal.pone.0032994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhead B, Brorson IS, Berge T, Adams C, Quach H, Moen SM, Berg-Hansen P, Celius EG, Sangurdekar DP, Bronson PG, et al. Increased DNA methylation of SLFN12 in CD4+ and CD8+ T cells from multiple sclerosis patients. PLoS One. 2018;13:e0206511. doi: 10.1371/journal.pone.0206511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus repository (https://www.ncbi.nlm.nih.gov/geo/), with accession number GSE44964.