Abstract
Long non-coding RNAs (lncRNAs) have been proven to be critical gene regulators of development and disease. The main aim of the present study was to elucidate the lncRNA-mRNA regulation network in ischemic stroke induced by middle cerebral artery occlusion (MCAO) using RNA sequencing (RNA-seq) in rats. lncRNA expression profiles were screened in brain tissues to identify a number of differentially expressed lncRNAs (DELs) and genes (DEGs) by RNA-seq. Reverse transcription-quantitative polymerase chain reaction was performed to further confirm the lncRNA expression data. Furthermore, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were used to mine mRNA functions, and a lncRNA-mRNA network was constructed. Additionally, cis- and trans-regulatory gene analyses of DELs were predicted. A total of 134 DELs (fold change >2, false discovery rate <0.05) and 1,006 DEGs (fold change >2 and P<0.05) were identified. Eighteen lncRNAs were predicted to regulate heme oxygenase 1, mitotic checkpoint serine/threonine kinase B, chemokine ligand 2 and DNA Topoisomerase IIα, amongst other genes. These genes are all associated with a cellular response to inorganic substances, alkaloids, estradiol, reactive oxygen species, metal ions, oxidative stress, and are associated with metabolic pathways, chemokine signaling pathways, malaria, Parkinson's disease, the cell cycle and other GO and KEGG pathway enrichments. The present study identifies novel DELs and an lncRNA-mRNA regulatory network that may allow for an improved understanding of the molecular mechanism of ischemic stroke induced by MCAO.
Keywords: middle cerebral artery occlusion, long non-coding RNA
Introduction
Stroke, universally acknowledged as a cerebrovascular accident, may result in lasting brain damage, long-term disability or even mortality (1,2). A multitude of biological processes are implicated in ischemic stroke, including oxygen deprivation, neuronal necrosis and an intense inflammatory response (3,4). MicroRNAs (miRNAs), long non-coding RNAs (lncRNAs) and even circular RNAs (circRNAs) contribute to RNA-mediated networks (5–8) that regulate notable cellular events through a variety of complicated mechanisms (9,10). These networks have been implicated in ischemic stroke in previous studies (5–10); however, there remain gaps in current knowledge in this regard, and novel ncRNAs need be mined in order to provide a better understanding of the precise molecular mechanisms involved in ischemic stroke.
lncRNAs have been proven to be critical gene regulators of development and disease (11–13). lncRNAs may also perform functions through competitively binding to miRNAs known as competitive endogenous RNAs (14). Washietl et al (15) systematically analyzed the conservatism of human lncRNA and other six mammalian lncRNA and identified that ~54% human lncRNA loci may be mapped to that of a rat. A previous study has demonstrated that significantly differentially expressed lncRNAs (DELs) may contribute to the stabilization of mRNA expressions in stroke (7). Stroke-induced lncRNAs may also interact with chromatin-modifying proteins and modulate genes associated with ischemic brain damage (16,17). Furthermore, lncRNA BC088414 was revealed to be involved with apoptosis-associated genes following hypoxic-ischemic brain damage (8). Similarly, another study suggested that lncRNA C2dat1 may modulate calcium/calmodulin-dependent protein kinase II expression to promote neuronal survival following cerebral ischemia (10). Although a host of lncRNAs have been identified by massive parallel sequencing, to date, little is known on functional RNA molecules and RNA-mediated regulation networks in ischemic stroke.
The main aim of the present study is to elucidate the lncRNA-mRNA regulation networks in ischemic stroke induced by middle cerebral artery occlusion (MCAO) using RNA sequencing (RNA-seq) in rats.
Materials and methods
MCAO model and tissue preparation
A focal cerebral ischemia model induced by MCAO, prepared as previously described (18), was prepared using 20 7-week-old male Sprague-Dawley rats of a specific pathogen-free grade (weighing 200±20 g), purchased from the experimental animal center of Anhui Medical University (Anhui, China). The study protocol was ethically approved by the Committee on the Ethics of Animal Experiments of Anhui University of Chinese Medicine (approval no. 2012AH-036-03). In brief, the animals were fasted overnight but allowed ad libitum access to water. They were then anesthetized with chloral hydrate (350 mg/kg, intraperitoneal injection). A 4-0 silicon-coated monofilament nylon suture with a round tip was inserted through an arteriectomy in the common carotid artery just below the carotid bifurcation and then advanced into the internal carotid artery ~18 mm distal to the carotid bifurcation until a mild resistance was felt. Following 2 h of MCAO, the filament was removed to allow reperfusion. As a control, control-operated rats underwent identical surgery but did not have the suture inserted. Four days subsequent to MCAO, the left hemispheres were collected and immediately frozen in liquid nitrogen.
RNA-seq
RNA-seq was performed by Ao-Ji Bio-Tech (Shanghai, China). Briefly, total RNA was extracted using an RNeasy Mini kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer's protocol. The RNA quality control was performed using Nanodrop 2000 and Agilent 2100, and mainly depended on the concentration, purity and integrity of the RNA. Ribosomal RNA was removed from total RNA using Ribo-Zero rRNA removal beads (Illumina, Inc., San Diego, CA, USA). Libraries were constructed according to the standard TruSeq protocol (19). Purified cDNA libraries were prepared for cluster generation and sequencing on an Illumina HiSeq 2500 (Illumina, Inc.) according to the manufacturer's protocol. Subsequently, data analyses were performed in silico.
lncRNA annotation
Quality control of the RNA-Seq reads was conducted using FastQC (v0.11.3) (The Babraham Institute, Cambridge, UK). Reads were trimmed using the software seqtk (github.com/lh3/seqtk) for known Illumina TruSeq adapter sequences, poor reads and ribosome RNA reads. Trimmed reads were aligned to the rat genome (Rn6) using Hisat2 (version 2.0.4) (20). Transcripts were assembled using Stringtie (v1.3.0) (20,21). Transcripts constructed from Stringtie were compiled together by gffcompare (v0.9.8) (20,21). Transcripts detected in at least five samples (half of the total number) were considered to be bona fide transcripts. Transcripts, with the exception of those with just one exon and shorter than 200 base pairs, were further analyzed for the identification of lncRNAs. Transcripts with class codes ‘i,’ ‘u,’ and ‘x,’ were considered to be potential novel long transcripts. Pfam (22), Coding Potential Calculator (CPC) (23) and Coding-Non-Coding Index (CNCI) (24) were used to estimate the coding potential of each novel transcript. Transcripts with a Pfam score <0, CNCI <0 and CPC non-significant were considered to lack coding potential. Transcripts were compared with annotation databases, including NONCODE (v4) (http://www.noncode.org) and Ensembl (25). The matched transcripts were considered to be known lncRNAs, and others were considered to be novel lncRNAs. All lncRNAs were quantified using Stringtie. According to the positional association between lncRNA and mRNA in the genome, lncRNA may be classified into six types: Bidirectional, exonic_antisense, exonic_sense, intergenic, intronic_antisense and intronic_sense (26).
The lncRNA-mRNA coexpression network
Initially, the DELs and differentially expressed genes (DEGs) were analyzed using EdgeR (27). For DEGs, log2| [fold change (FC)] |>1 and P<0.05 were used as the cutoff values. Meanwhile, log2| (FC) |>1 and false discovery rate (FDR) <0.05 were used as the threshold for DELs. Hierarchical clustering of DELs was performed based on mean signals using a Euclidean distance function. In addition, a volcano plot was generated. The Pearson's correlation coefficient (PCC) between lncRNAs and mRNAs was calculated (cutoff value, PCC>0.9, P<0.05) and the lncRNA-mRNA regulatory network was structured using Cytoscape 2.8.3 (28).
Prediction of target genes and enrichment analysis
cis- or trans-acting algorithms were used to predict the potential targets of lncRNAs. The first algorithm predicted potential target genes of cis-acting lncRNAs that were physically located within 10 kb upstream or 20 kb downstream of lncRNAs using liftOver genome browser (genome.ucsc.edu/cgi-bin/hgLiftOver). The second algorithm predicted potential target genes of trans-acting lncRNAs based on the lncRNA-mRNA complementary sequences, and predicted lncRNA-mRNA duplex energy. First, BLASTN (29) was performed to detect potential target mRNA sequences with >95% identity and E value <1×10−5 (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Then, RNAplex (30) was used to calculate the complementary energy between lncRNAs and their potential trans-regulated target genes with RNAplex-10−30. Gene Ontology (GO) (31) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (32) enrichment analyses of the identified potential target genes were performed using the Database for Annotation, Visualization and Integrated Discovery (33); and P<0.05 was considered to indicate a statistically significant difference.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from left hemisphere samples using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and reverse-transcribed using a Thermo Fisher Scientific RevertAid First Strand cDNA Synthesis kit (cat. no. K1622; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol at 42°C for 60 min. To further confirm the expression data from RNA-seq, a cutoff value (FC>2, P<0.05) was randomly selected for qPCR verification. The expression levels of six randomly DELs (NONRATT027551.2, MSTRG.1836.1, MSTRG.4344.10, MSTRG.7720.11, NONRATT005132.2 and MSTRG.20633.3) were assayed using a SYBRGreen flurophore (Applied Biosystems; Thermo Fisher Scientific, Inc.) using the PikoReal real-time PCR system (Thermo Fisher Scientific, Inc.) under the following conditions: Initial denaturation at 95°C for 30 sec, followed by 40 cycles at 95°C for 30 sec and 60°C for 30 sec, and a final extension step at 4°C for 20 min. FC was determined using the 2−ΔΔCq method (34). GAPDH mRNA was used as an internal control. The primers used are listed in Table I.
Table I.
Primer sequences.
| Gene | Sequence | Polymerase chain reaction product length (base pairs) |
|---|---|---|
| GAPDH | F: 5′-CCTGGTATGACAACGAATTTG-3′ | 131 |
| R: 5′-CAGTGAGGGTCTCTCTCTTCC-3′ | ||
| NONRATT027551.2 | F: 5′- GGACCTGGAAGGTGAACAGG-3′ | 118 |
| R: 5′-TGAATGGGTGACCAACAGGG-3′ | ||
| MSTRG.1836.1 | F: 5′-CCATTGTCCTTCCATCCCCC-3′ | 85 |
| R: 5′-CCACCCTACCAAACTTCCCC-3′ | ||
| MSTRG.4344.10 | F: 5′-GACTTAGGCACAGTGGGTGG-3′ | 119 |
| R: 5′-ATGGCAGAGAGCGAATGGAG-3′ | ||
| MSTRG.7720.11 | F: 5′-TCCCTAGAGCAGTCCTCACC-3′ | 97 |
| R: 5′- ATCTCGGGTTCGCCTTTTGT-3′ | ||
| NONRATT005132.2 | F: 5′-CCTGACTATGGCACGTCCTC-3′ | 152 |
| R: 5′-CTGAGTCCAGTGTGCCTGTT-3′ | ||
| MSTRG.20633.3 | F: 5′-CTTTCACTCCGAGAACCCCC-3′ | 117 |
| R: 5′-GCAAGCAGGTTGGTTCCTTG-3′ |
F, forward; R, reverse.
Statistical analysis
The comparisons between the MCAO group and the control group were determined using a Student's t-test for the RT-qPCR results by SPSS 22.0 statistical software (IBM Corp., Armonk, NY, USA). P<0.05 was considered to indicate a statistically significant difference. The PCC between lncRNAs and mRNAs was calculated using the Hmisc package in R based on the expression determined using RNA-seq (PCC>0.9, P<0.05). The correlation analysis between the RT-qPCR results and RNA-seq results was calculated in Excel 2013 (Microsoft Corporation, Redmond, WA, USA) with the function of CORR.
Results
lncRNA-sequencing data analysis
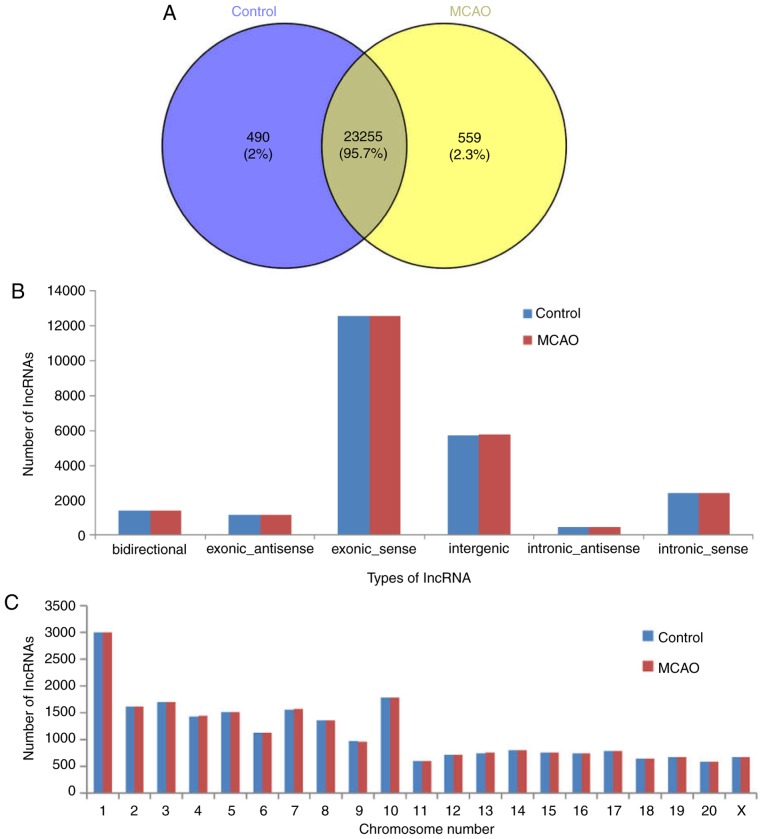
The present study characterized the lncRNA landscape and expression by performing deep RNA-seq experiments on three control and three MCAO tissue samples. Subsequent to the seqtk quality assessment of sequencing, >33 million total original reads for each sample were obtained, and the proportion of bases with quality values >20 was >94%. These results indicated that the quality of the sequencing results was acceptable (Table II). Subsequent to filtering out the adaptor sequence and low quality reads, the percentage of clean reads within the raw reads accounted for 94% of the total sequences in two groups. Hisat2 software was used to map the obtained clean reads to the Rattus norvegicus reference genome. As presented in Table II, ~97% of the trimmed reads were mapped onto the reference genome. In total, 24,304 lncRNAs were screened from six samples, and there were 23,255 shared lncRNAs detected in the MCAO and control groups (Fig. 1A). The majority of the identified lncRNAs were transcribed from protein-coding exons; others were from introns and intergenic regions (Fig. 1B). In addition, the present study analyzed the distribution of the identified lncRNAs on the rat chromosomes; 24,304 lncRNA transcripts were identified in all chromosomes, and chromosome 1 included the most lncRNAs (Fig. 1C).
Table II.
Results of the RNA sequencing.
| Sample ID | Raw reads | Clean reads | Clean ratio (%) | rRNA trimmed | Mapped reads | Mapped ratio |
|---|---|---|---|---|---|---|
| MCAO 1 | 155190870 | 147282901 | 94.90 | 147214274 | 131290423 | 0.824026529 |
| MCAO 2 | 144450130 | 136930064 | 94.79 | 136768708 | 120311195 | 0.812346697 |
| MCAO 3 | 151411986 | 142862439 | 94.35 | 142724911 | 125540743 | 0.804354762 |
| Control 1 | 169303916 | 160544960 | 94.83 | 160466568 | 142289885 | 0.819832697 |
| Control 2 | 136533930 | 129672892 | 94.97 | 129609605 | 116072061 | 0.828973282 |
| Control 3 | 124878376 | 118432971 | 94.84 | 118349299 | 105506286 | 0.821661742 |
MCAO, middle cerebral artery occlusion.
Figure 1.
Class type and chromosome distribution of lncRNAs identified in the control and MCAO group. (A) Venn diagram of lncRNA in the control and MCAO groups. (B) According to the positional association between lncRNA and mRNA in the genome, lncRNAs may be classified into six types: Bidirectional, exonic antisense, exonic sense, intergenic, intronic antisense and intronic sense. (C) Number of lncRNAs on each chromosome in the MCAO and control groups. MCAO, Middle cerebral artery occlusion; lncRNA, long noncoding RNA.
Identification of DEGs and DELs
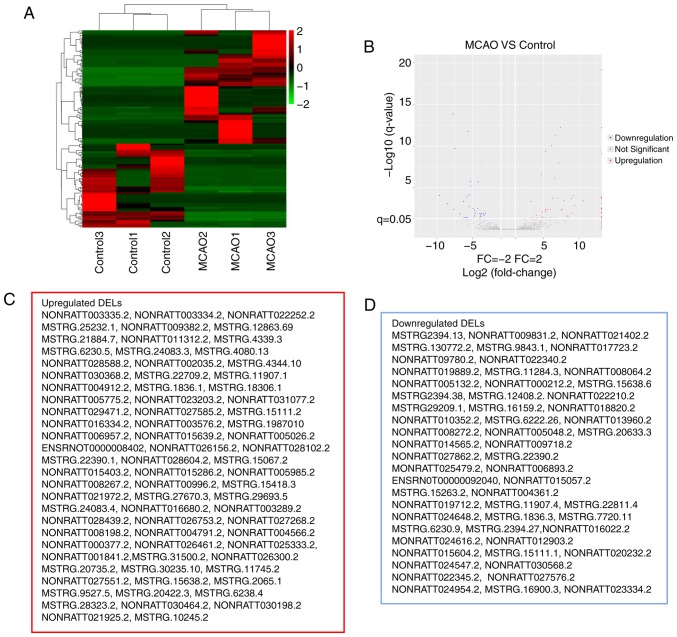
EdgeR was used to filter DEGs and DELs and differentiate their expression between the control and MCAO groups. A total of 1,007 DEGs (|FC|>2, P<0.05) were identified, including 785 upregulated genes and 222 downregulated genes. Similarly, as presented in Fig. 2, 134 DELs (|FC|>2, FDR<0.05) were identified in the MCAO group (Fig. 2A and B), including 77 upregulated and 57 downregulated DELs (Fig. 2C and D). In the present study, it was revealed that the FC values of certain DELs were equal to positive infinity and negative infinity, meaning that these lncRNAs are switched-on or off with MCAO. Essentially, positive or negative infinity indicates zero expression of the lncRNA in normal or MCAO groups. It was speculated that this may be associated with the abundance of lncRNAs and the sensitivity to RNA-seq. The top five upregulated DELs were NONRATT027551.2, MSTRG.1836.1, MSTRG.4344.10, NONRATT028102.2 and MSTRG.31500.2; the top five downregulated DELs were MSTRG.7720.11, NONRATT005132.2, MSTRG.20633.3, NONRATT020232.2 and MSTRG.1836.3.
Figure 2.
RNA-seq data on the differentially expressed lncRNAs between the model and control groups. (A) Hierarchical cluster of DELs between the MCAO and control groups. The color code in each heat map is linear, with green indicating the least and red indicating the greatest differentiation. The mean signals of the altered lncRNAs in each of the two groups were clustered using a Euclidean distance function. The lncRNAs with the most similar expression patterns were placed next to each other (n=3 per group). (B) A volcano plot of the RNA-seq FC and P-value of MCAO group compared with the control group. Blue and red points stand for DELs. Gray points represent lncRNAs which are not differentially expressed. (C) Upregulated and (D) downregulated DELs as exhibited in the red and blue boxes, respectively, represent the results. MCAO, Middle cerebral artery occlusion; lncRNA, long noncoding RNA; RNA-seq, RNA sequencing; FC, fold change; DEL, differentially expressed lncRNAs.
lncRNA-mRNA network
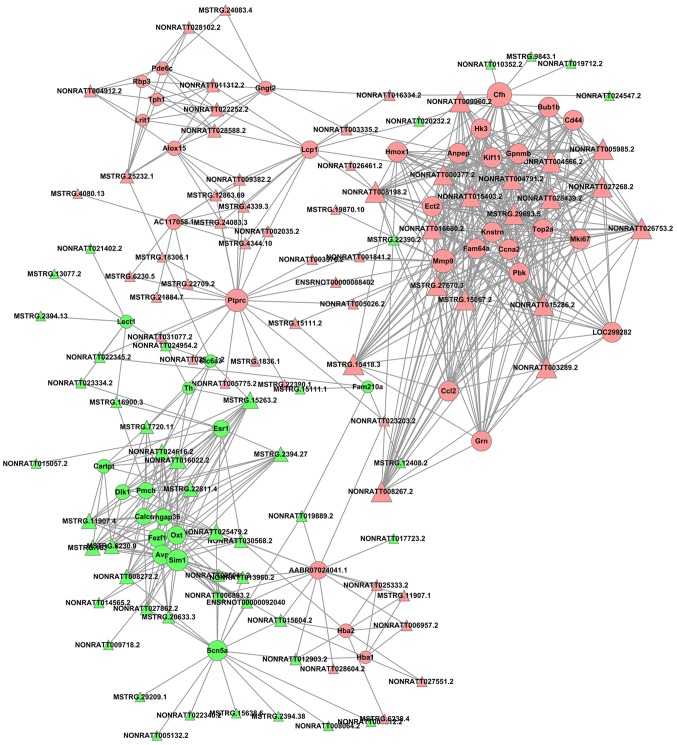
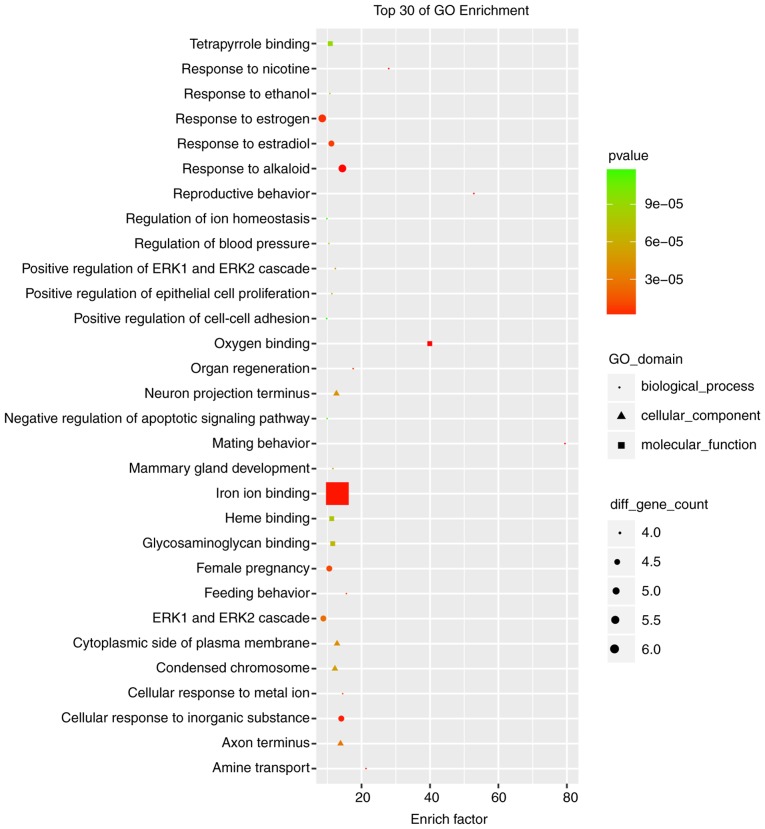
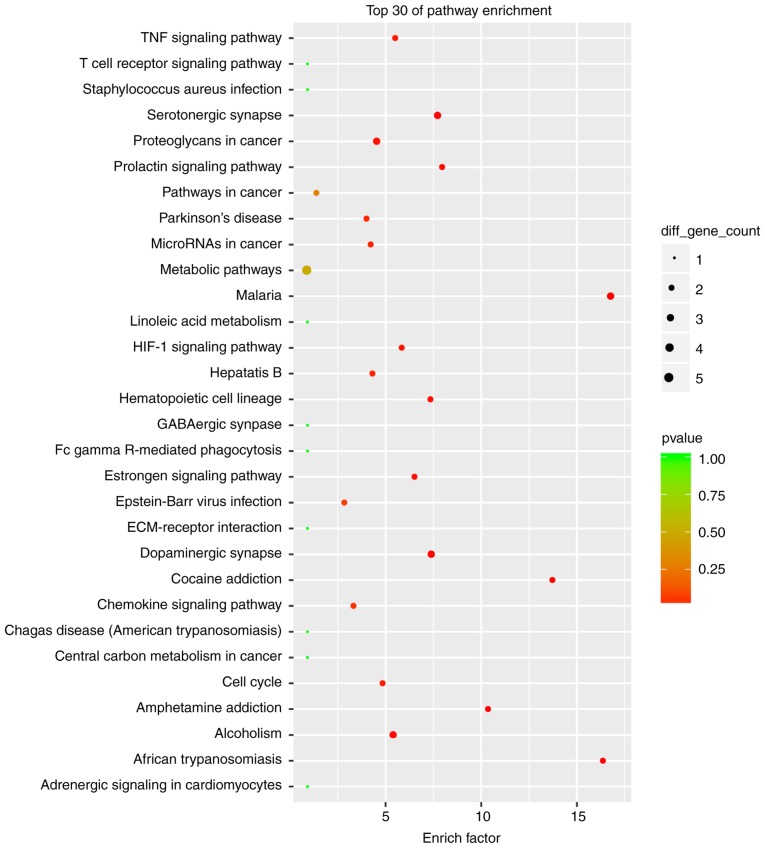
The cutoff correlation r-values (|PCC|>0.9) and P-values (P<0.05) were selected to structure a lncRNA-mRNA co-expression network between DEGs and DELs. As Fig. 3 presents, 46 DEGs, 104 DELs and 664 edges were filtered out using Cytoscape to construct the co-expression network. The co-expression-associated top 30 GO terms and pathway terms enrichment analyses presented in Figs. 4 and 5 suggest that these DELs were associated with the cellular response to inorganic substances, alkaloids, estradiols, reactive oxygen species, metal ions and oxidative stress. In particular, the heme oxygenase 1 (HO-1) gene participates in many of these functions. A multitude of pathways were implicated, including metabolic pathways, chemokine signaling pathways, malaria, Parkinson's disease and the cell cycle. Notably, the BUB1 mitotic checkpoint serine/threonine kinase B (BUB1B) and C-C motif chemokine ligand 2 (CCL2) genes were associated with the cell cycle.
Figure 3.
An lncRNA-gene-network based on Pearson's correlation coefficient. Pink nodes indicate the upregulated mRNAs or lncRNAs, and green nodes indicate the downregulated mRNAs or lncRNAs. lncRNA, long noncoding RNA.
Figure 4.
Top 30 significant enrichment of GO terms in the long noncoding RNA-mRNA network. GO, gene ontology; ERK, extracellular regulated kinase.
Figure 5.
Top 30 significant enrichment of KEGG pathway terms in the long noncoding RNA-mRNA network. KEGG, Kyoto Encyclopedia of Genes and Genomes; TNF, tumor necrosis factor; HIF-1, hypoxia-inducible factor 1; ECM, extracellular matrix.
Regulatory analysis of DELs
A total of 91 cis-regulatory genes of 94 DELs, including 55 upregulated lncRNAs in the MCAO group were identified; 14 of the 91 cis-regulatory genes exhibited differential expression. A total of 13 of the DEL/cis-regulatory gene pairs had positive correlations as follows: NONRATT021925.2 (Rho GDP dissociation inhibitor β), NONRATT004791.2 (G protein subunit γ transducing 2), NONRATT015286.2 (periostin), MSTRG.30235.10 (LRR binding FLII interacting protein 1), NONRATT015403.2 (IQ motif containing GTPase activating protein 3), NONRATT008267.2 (kinesin family member 14), NONRATT009960.2 (non-SMC condensin I complex subunit G), NONRATT011312.2 (retinol binding protein 3), NONRATT005985.2 (DNA topoisomerase IIα), NONRATT016680.2 (ENSRNOG00000053081), NONRATT013960.2 (galanin receptor 1), NONRATT016022.2 (CART prepropeptide) and NONRATT024616.2 (δ like non-canonical Notch ligand 1) (Table III). Additionally, 90 trans-regulatory genes of lncRNAs were filtered by BLASTN and RNAplex, with a negative correlation identified between ENSRNOT00000092040 and Ccl9 (Table IV).
Table III.
Differentially expressed lncRNAs mechanisms involved in cis-regulatory elements.
| lncRNA ID | log2FC | Q value | Up/downregulated | Type | Gene ID | Gene name | log2FC | P-value | Up/downregulated |
|---|---|---|---|---|---|---|---|---|---|
| NONRATT017723.2 | −3.68 | 0.0125 | Down | cis | ENSRNOG00000000633 | Rhobtb1 | 0.85 | 0.1148 | – |
| NONRATT008064.2 | −6.73 | 0.0145 | Down | cis | ENSRNOG00000001183 | Hnf1a | 1.25 | 0.7907 | – |
| NONRATT000377.2 | +∞ | 0.0492 | Up | cis | ENSRNOG00000001492 | Slc8a2 | 0.13 | 0.9091 | – |
| NONRATT006957.2 | 6.69 | 0.0000 | Up | cis | ENSRNOG00000001982 | Cblb | 0.60 | 0.1402 | – |
| NONRATT009831.2 | −5.72 | 0.0425 | Down | cis | ENSRNOG00000002137 | Aasdh | 0.21 | 0.7048 | – |
| NONRATT009718.2 | −5.48 | 0.0020 | Down | cis | ENSRNOG00000002146 | Pkd2 | 0.29 | 0.5035 | – |
| NONRATT009780.2 | −3.94 | 0.0473 | Down | cis | ENSRNOG00000002208 | Shroom3 | 0.83 | 0.1301 | – |
| NONRATT005026.2 | 6.14 | 0.0000 | Up | cis | ENSRNOG00000002607 | Sox9 | 0.25 | 0.5566 | – |
| NONRATT014565.2 | −2.91 | 0.0492 | Down | cis | ENSRNOG00000002864 | Nacc1 | 0.26 | 0.5744 | – |
| NONRATT005048.2 | -∞ | 0.0432 | Down | cis | ENSRNOG00000003144 | Gprc5c | 0.48 | 0.5195 | – |
| NONRATT024547.2 | -∞ | 0.0082 | Down | cis | ENSRNOG00000003955 | Spata7 | 0.26 | 0.6325 | – |
| NONRATT026753.2 | 3.08 | 0.0450 | Up | cis | ENSRNOG00000003993 | Thap2 | 0.26 | 0.7629 | – |
| NONRATT005132.2 | −8.71 | 0.0471 | Down | cis | ENSRNOG00000004049 | Baiap2 | −0.04 | 0.8211 | – |
| NONRATT026300.2 | +∞ | 0.0469 | Up | cis | ENSRNOG00000004628 | Dazap2 | 0.13 | 0.8554 | – |
| NONRATT026156.2 | 7.44 | 0.0000 | Up | cis | ENSRNOG00000005332 | Csdc2 | −0.58 | 0.1263 | – |
| NONRATT005775.2 | 3.69 | 0.0453 | Up | cis | ENSRNOG00000005538 | Psmd11 | −0.03 | 0.7616 | – |
| NONRATT023334.2 | −4.85 | 0.0448 | Down | cis | ENSRNOG00000005711 | Ptprd | −0.21 | 0.4658 | – |
| NONRATT021925.2 | +∞ | 0.0000 | Up | cis | ENSRNOG00000005809 | Arhgdib | 1.81 | 0.0074 | Up |
| NONRATT004791.2 | 4.02 | 0.0218 | Up | cis | ENSRNOG00000006108 | Gngt2 | 4.95 | 0.0000 | Up |
| MSTRG.22811.4 | -∞ | 0.0388 | Down | cis | ENSRNOG00000006966 | Nfia | 0.21 | 0.6590 | – |
| NONRATT025479.2 | −4.14 | 0.0000 | Down | cis | ENSRNOG00000007610 | Gdf11 | 0.08 | 0.9402 | – |
| NONRATT020232.2 | −7.72 | 0.0000 | Down | cis | ENSRNOG00000007804 | C1galt1 | 0.45 | 0.2729 | – |
| NONRATT008198.2 | 4.83 | 0.0305 | Up | cis | ENSRNOG00000007887 | Elk4 | 0.76 | 0.2762 | – |
| NONRATT022252.2 | +∞ | 0.0305 | Up | cis | ENSRNOG00000008099 | Galnt12 | 0.85 | 0.2496 | – |
| NONRATT024954.2 | −3.98 | 0.0471 | Down | cis | ENSRNOG00000008155 | Dus4l | −0.17 | 0.7340 | – |
| NONRATT028439.2 | 5.06 | 0.0430 | Up | cis | ENSRNOG00000008187 | Ubash3b | −0.21 | 0.4764 | – |
| NONRATT022210.2 | −4.24 | 0.0492 | Down | cis | ENSRNOG00000008237 | Unc13b | 0.30 | 0.6027 | – |
| NONRATT027551.2 | 9.08 | 0.0119 | Up | cis | ENSRNOG00000008709 | Arhgap32 | −0.24 | 0.5090 | – |
| NONRATT027576.2 | -∞ | 0.0061 | Down | cis | ENSRNOG00000008757 | Tmem218 | 0.16 | 0.8511 | – |
| NONRATT021402.2 | -∞ | 0.0291 | Down | cis | ENSRNOG00000009156 | Tra2a | 0.47 | 0.2267 | – |
| NONRATT021972.2 | +∞ | 0.0335 | Up | cis | ENSRNOG00000009338 | Kras | −0.11 | 0.6201 | – |
| NONRATT022345.2 | −5.23 | 0.0000 | Down | cis | ENSRNOG00000009795 | Nfib | 0.04 | 0.9239 | – |
| MSTRG.16900.3 | −4.70 | 0.0248 | Down | cis | ENSRNOG00000009882 | Ppp3ca | 0.07 | 1.0000 | – |
| MSTRG.19870.10 | +∞ | 0.0000 | Up | cis | ENSRNOG00000010993 | Dpm1 | −0.14 | 0.5966 | – |
| NONRATT008272.2 | −3.96 | 0.0431 | Down | cis | ENSRNOG00000011063 | Dennd1b | 0.20 | 0.7240 | – |
| MSTRG.10245.2 | 6.31 | 0.0002 | Up | cis | ENSRNOG00000011704 | Fbxo34 | −0.37 | 0.2745 | – |
| NONRATT001841.2 | 3.41 | 0.0430 | Up | cis | ENSRNOG00000012110 | Col17a1 | −1.52 | 0.0483 | Down |
| NONRATT002035.2 | 7.89 | 0.0003 | Up | cis | ENSRNOG00000012324 | Soga3 | −0.04 | 0.7913 | – |
| MSTRG.22390.2 | -∞ | 0.0000 | Down | cis | ENSRNOG00000012634 | Fbxo10 | −0.41 | 0.1989 | – |
| MSTRG.22390.1 | +∞ | 0.0000 | Up | cis | ENSRNOG00000012634 | Fbxo10 | −0.41 | 0.1989 | – |
| NONRATT015286.2 | 5.30 | 0.0425 | Up | cis | ENSRNOG00000012660 | Postn | 3.66 | 0.0004 | Up |
| MSTRG.1836.3 | −7.60 | 0.0041 | Down | cis | ENSRNOG00000012716 | Chd2 | 0.06 | 0.9929 | – |
| MSTRG.1836.1 | 9.24 | 0.0001 | Up | cis | ENSRNOG00000012716 | Chd2 | 0.06 | 0.9929 | – |
| NONRATT016334.2 | +∞ | 0.0000 | Up | cis | ENSRNOG00000012734 | Dcun1d1 | −0.20 | 0.5140 | – |
| NONRATT015057.2 | −4.70 | 0.0049 | Down | cis | ENSRNOG00000012799 | Prkaa1 | −0.28 | 0.3193 | – |
| NONRATT000212.2 | −6.12 | 0.0378 | Down | cis | ENSRNOG00000013194 | Rps6ka2 | −0.14 | 0.5704 | – |
| NONRATT030198.2 | 5.34 | 0.0000 | Up | cis | ENSRNOG00000013213 | Epha4 | 0.35 | 0.5640 | – |
| NONRATT010352.2 | −6.23 | 0.0004 | Down | cis | ENSRNOG00000013353 | Tmem260 | 0.07 | 0.9952 | – |
| NONRATT029471.2 | 4.91 | 0.0082 | Up | cis | ENSRNOG00000013557 | Lancl1 | −0.17 | 0.5373 | – |
| NONRATT028588.2 | 6.05 | 0.0378 | Up | cis | ENSRNOG00000013829 | Chrna3 | 1.17 | 0.2523 | – |
| NONRATT023203.2 | +∞ | 0.0007 | Up | cis | ENSRNOG00000013956 | Rnf38 | 0.17 | 0.7841 | – |
| MSTRG.29693.5 | +∞ | 0.0484 | Up | cis | ENSRNOG00000013991 | Creg2 | −0.69 | 0.2339 | – |
| MSTRG.12408.2 | -∞ | 0.0041 | Down | cis | ENSRNOG00000014007 | Gfod1 | 0.04 | 0.9704 | – |
| NONRATT004566.2 | 3.44 | 0.0440 | Up | cis | ENSRNOG00000015002 | Abhd15 | 0.76 | 0.3266 | – |
| MSTRG.15067.2 | +∞ | 0.0090 | Up | cis | ENSRNOG00000015334 | Fcho2 | 0.49 | 0.2280 | – |
| NONRATT003289.2 | 7.56 | 0.0052 | Up | cis | ENSRNOG00000015717 | Ptpre | 0.21 | 0.6977 | – |
| NONRATT013960.2 | −3.81 | 0.0248 | Down | cis | ENSRNOG00000016654 | Galr1 | −3.38 | 0.0001 | Down |
| NONRATT028604.2 | 4.71 | 0.0000 | Up | cis | ENSRNOG00000017193 | Lingo1 | −0.52 | 0.2454 | – |
| NONRATT016022.2 | −5.74 | 0.0001 | Down | cis | ENSRNOG00000017712 | Cartpt | −3.83 | 0.0001 | Down |
| NONRATT015604.2 | −5.60 | 0.0000 | Down | cis | ENSRNOG00000018166 | Prkab2 | 0.00 | 0.8495 | – |
| NONRATT024616.2 | −3.79 | 0.0143 | Down | cis | ENSRNOG00000019584 | Dlk1 | −4.21 | 0.0000 | Down |
| MSTRG.30235.10 | 8.50 | 0.0275 | Up | cis | ENSRNOG00000019892 | Lrrfip1 | 1.14 | 0.0479 | Up |
| NONRATT026461.2 | +∞ | 0.0118 | Up | cis | ENSRNOG00000020230 | Pias4 | 0.17 | 0.8050 | – |
| NONRATT018820.2 | -∞ | 0.0002 | Down | cis | ENSRNOG00000020337 | Sla2 | 0.52 | 0.5953 | – |
| NONRATT004912.2 | 6.00 | 0.0041 | Up | cis | ENSRNOG00000020658 | Aarsd1 | 0.05 | 0.9927 | – |
| NONRATT019712.2 | −6.20 | 0.0446 | Down | cis | ENSRNOG00000021262 | Slc23a2 | 0.09 | 0.9742 | – |
| NONRATT027268.2 | 5.27 | 0.0071 | Up | cis | ENSRNOG00000022570 | Pus7l | 0.51 | 0.5096 | – |
| MSTRG.12863.69 | +∞ | 0.0304 | Up | cis | ENSRNOG00000023661 | Celf2 | 0.30 | 0.5495 | – |
| NONRATT030368.2 | 5.55 | 0.0043 | Up | cis | ENSRNOG00000025527 | Mtcl1 | 0.16 | 0.8136 | – |
| NONRATT027862.2 | −3.75 | 0.0304 | Down | cis | ENSRNOG00000027145 | Rora | −0.16 | 0.5963 | – |
| NONRATT015403.2 | 4.83 | 0.0409 | Up | cis | ENSRNOG00000027894 | Iqgap3 | 2.03 | 0.0137 | Up |
| NONRATT003576.2 | +∞ | 0.0028 | Up | cis | ENSRNOG00000028017 | Tmem109 | 0.36 | 0.4097 | – |
| NONRATT004361.2 | −5.86 | 0.0409 | Down | cis | ENSRNOG00000028341 | Alkbh5 | 0.00 | 0.8591 | – |
| NONRATT012903.2 | −5.26 | 0.0000 | Down | cis | ENSRNOG00000031706 | AABR07027388.1 | −0.85 | 0.1761 | – |
| MSTRG.15111.2 | +∞ | 0.0000 | Up | cis | ENSRNOG00000032735 | Srek1 | 0.18 | 0.7203 | – |
| MSTRG.15111.1 | −7.42 | 0.0000 | Down | cis | ENSRNOG00000032735 | Srek1 | 0.18 | 0.7203 | – |
| NONRATT028102.2 | 10.21 | 0.0000 | Up | cis | ENSRNOG00000033809 | Mlh1 | 0.76 | 0.1272 | – |
| NONRATT019889.2 | −4.60 | 0.0113 | Down | cis | ENSRNOG00000034031 | Vstm2l | −0.45 | 0.3771 | – |
| NONRATT008267.2 | +∞ | 0.0039 | Up | cis | ENSRNOG00000037211 | Kif14 | 5.80 | 0.0005 | Up |
| NONRATT009960.2 | 4.06 | 0.0430 | Up | cis | ENSRNOG00000038572 | Ncapg | 3.28 | 0.0054 | Up |
| MSTRG.21884.7 | +∞ | 0.0492 | Up | cis | ENSRNOG00000042458 | Stau2 | −0.20 | 0.5104 | – |
| NONRATT030464.2 | 5.30 | 0.0042 | Up | cis | ENSRNOG00000046053 | Nudt10 | −0.59 | 0.1573 | – |
| MSTRG.15418.3 | +∞ | 0.0448 | Up | cis | ENSRNOG00000048993 | Metazoa_SRP | 0.03 | 0.9881 | – |
| MSTRG.28323.2 | +∞ | 0.0492 | Up | cis | ENSRNOG00000049584 | AABR07070555.1 | −0.13 | 0.8591 | – |
| MSTRG.4080.13 | +∞ | 0.0002 | Up | cis | ENSRNOG00000049768 | Adcy9 | −0.04 | 0.7770 | – |
| NONRATT025333.2 | 6.61 | 0.0000 | Up | cis | ENSRNOG00000051719 | AABR07065498.1 | 0.11 | 0.9304 | – |
| NONRATT011312.2 | +∞ | 0.0479 | Up | cis | ENSRNOG00000051911 | Rbp3 | 9.84 | 0.0000 | Up |
| NONRATT024648.2 | -∞ | 0.0492 | Down | cis | ENSRNOG00000052540 | SNORD113 | 0.45 | 0.6631 | – |
| NONRATT005985.2 | 4.53 | 0.0490 | Up | cis | ENSRNOG00000053047 | Top2a | 5.28 | 0.0000 | Up |
| NONRATT016680.2 | 7.62 | 0.0041 | Up | cis | ENSRNOG00000053081 | 3.59 | 0.0011 | Up | |
| NONRATT009382.2 | 8.35 | 0.0257 | Up | cis | ENSRNOG00000056826 | Arap2 | 0.21 | 0.6941 | – |
| MSTRG.15263.2 | -∞ | 0.0041 | Down | cis | ENSRNOG00000060329 | Emb | −0.06 | 0.7939 | – |
| NONRATT015639.2 | 3.25 | 0.0005 | Up | cis | ENSRNOG00000061058 | Csde1 | 0.02 | 0.8739 | – |
| NONRATT027585.2 | 7.19 | 0.0000 | Up | cis | ENSRNOG00000061656 | SNORD14 | 0.17 | 0.9160 | – |
lncRNA, long noncoding RNA; FC, fold change.
Table IV.
Differentially expressed lncRNA mechanisms involved in trans-regulatory elements.
| lncRNA ID | log2FC | Q value | Up/downregulated | Type | Gene name | log2FC | P-value | Up/downregulated |
|---|---|---|---|---|---|---|---|---|
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Ccl9 | 5.19 | 0.0017 | Up |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Homer3 | 1.00 | 0.0611 | – |
| ENSRNOT00000088402 | +∞ | 0.0471 | Up | Trans | Aurkb | 2.35 | 0.0723 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Galns | 1.03 | 0.0724 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Slc39a1 | 0.92 | 0.0739 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | AC099384.2 | +∞ | 0.1052 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Hsd3b7 | 0.83 | 0.1369 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Klrb1b | 1.98 | 0.1413 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Me2 | 0.63 | 0.1558 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Pnma2 | −0.51 | 0.1564 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Cass4 | 1.24 | 0.1599 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Klhl23 | −0.44 | 0.2192 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Elac1 | −0.49 | 0.2260 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Clcf1 | 1.26 | 0.2519 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Fzd6 | 0.80 | 0.2590 | – |
| NONRATT009960.2 | 4.06 | 0.0430 | Up | Trans | Lcorl | −0.44 | 0.3199 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Gpr37l1 | 0.40 | 0.3297 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Mmab | −0.35 | 0.3391 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Spon1 | −0.28 | 0.3414 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Ifngr2 | 0.44 | 0.3660 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Foxred2 | −0.35 | 0.3732 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Prr22 | −0.67 | 0.3889 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Sppl2a | 0.40 | 0.3894 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Haus5 | 0.55 | 0.4019 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Cpvl | 0.80 | 0.4041 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Fam120b | −0.24 | 0.4061 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Fbxo3 | −0.21 | 0.4206 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Ypel4 | −0.24 | 0.4477 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Klhl26 | −0.20 | 0.4480 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Sync | 0.43 | 0.5076 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Lhfpl5 | −0.31 | 0.5231 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Mrpl52 | 0.44 | 0.5306 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Cldn15 | −0.39 | 0.5315 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Mrps35 | −0.18 | 0.5443 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Zfp382 | −0.17 | 0.5640 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Ube2k | −0.14 | 0.5697 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Drg1 | −0.12 | 0.5755 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | P2rx5 | −0.36 | 0.5770 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Xpnpep3 | 0.25 | 0.5933 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | RGD1311345 | 0.28 | 0.5950 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Acer2 | 0.32 | 0.6034 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Urgcp | 0.28 | 0.6182 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Megf8 | −0.10 | 0.6223 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Lrtm2 | −0.14 | 0.6301 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | RGD1562299 | 0.26 | 0.6325 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Rpl9 | 0.38 | 0.6347 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Slc25a44 | −0.10 | 0.6348 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Ssmem1 | −0.28 | 0.6350 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Stt3a | 0.24 | 0.6468 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Spire1 | −0.09 | 0.6631 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | RGD1561777 | −0.13 | 0.6846 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Luzp1 | −0.09 | 0.6941 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Acot2 | 0.26 | 0.7000 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Fitm2 | −0.07 | 0.7111 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Aox4 | 0.32 | 0.7374 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Golga1 | −0.04 | 0.7775 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Lefty2 | 1.23 | 0.7977 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Zdhhc24 | −0.04 | 0.8075 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Zfp329 | −0.03 | 0.8205 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Tmem101 | −0.03 | 0.8236 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Dhh | 0.28 | 0.8353 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Ncr3 | 0.57 | 0.8354 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Ppp1r15b | 0.14 | 0.8423 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Zfyve27 | −0.01 | 0.8439 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Ints7 | 0.15 | 0.8457 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Dus1l | 0.14 | 0.8545 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Ahsa2 | 0.15 | 0.8576 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Aif1l | 0.15 | 0.8579 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Rsg1 | 0.16 | 0.8645 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Fuom | 0.15 | 0.8883 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Cwf19l1 | 0.12 | 0.8894 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Paqr7 | 0.01 | 0.8942 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Coa5 | 0.02 | 0.8985 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Cdc14b | 0.13 | 0.8989 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Crebl2 | −0.01 | 0.9081 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | RGD1564541 | 0.10 | 0.9210 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Slc15a1 | −0.03 | 0.9211 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Ppm1k | 0.03 | 0.9299 | – |
| NONRATT025479.2 | −4.14 | 0.0000 | Down | Cis and trans | Gdf11 | 0.08 | 0.9402 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Fam20b | 0.04 | 0.9436 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Anapc11 | 0.09 | 0.9536 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Tmem79 | −0.04 | 0.9595 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Cwc25 | 0.06 | 0.9692 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Blvrb | 0.07 | 0.9859 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Stk4 | 0.08 | 0.9880 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Rbm20 | 0.08 | 0.9896 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Tbc1d10b | 0.06 | 0.9927 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Mapkbp1 | 0.07 | 0.9986 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Psma8 | 1.00 | 1.0000 | – |
| ENSRNOT00000092040 | −3.35 | 0.0204 | Down | Trans | Rbbp8nl | 0.37 | 1.0000 | – |
lncRNA, long non-coding RNA; FC, fold change.
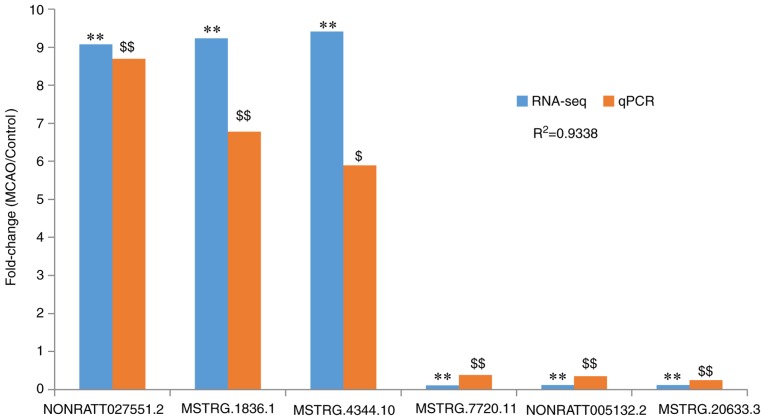
Validation of expression of DELs by RT-qPCR
From the data in Fig. 6, NONRATT027551.2, MSTRG.1836.1 and MSTRG.4344.10 were identified to be significantly upregulated in the MCAO group compared with the control (P<0.05), consistent with the RNA-seq data, while MSTRG.7720.11, NONRATT005132.2 and MSTRG.20633.3 were significantly downregulated in the MCAO group compared with the control (P<0.01), also consistent with the RNA-seq data. These results, revealing that the RNA-seq results were consistent with the RT-qPCR results, verified that the RNA-seq results were reliable (Fig. 6).
Figure 6.
Validation of lncRNA RNA-seq data by RT-qPCR. Fold changes represent the comparison of the MCAO group with the control group. Blue bars indicate the fold change were detected with RNA-seq. **P<0.01 vs. the control group. The orange bars indicate the fold change detected using RT-qPCR. $P<0.05 and $$P<0.01 vs. the control group. Comparison of the results obtained from RT-qPCR and RNA-seq revealed satisfactory consistency (R2=0.9338). MCAO, Middle cerebral artery occlusion; lncRNA, long noncoding RNA; RNA-seq, RNA sequencing; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Discussion
A host of lncRNAs have been indicated to be involved in ischemic stroke by microarray or RNA-seq studies (35,36). Metastasis associated lung adenocarcinoma transcript 1 was identified to have a function in ischemic stroke through inhibiting endothelial cell death and inflammation (36,37). Additionally, the upregulation of H19 imprinted maternally expressed transcript may induce apoptosis and necrosis in cerebral ischemic reperfusion injury (38–40). In the present study, a total of 77 upregulated and 57 downregulated DELs (|FC|>2, P<0.05) were identified through reliable RNA-seq and validated using RT-qPCR in an ischemic stroke group induced by MCAO compared with a control group.
HO-1-mediated neurogenesis has been demonstrated to be enhanced in ischemic stroke in mice (41). HO-1 has been revealed to promote angiogenesis following cerebral ischemic reperfusion injury in rats (42). GO enrichment analysis suggested that HO-1 was associated with responses to alkaloids, cellular responses to oxidative stress and responses to reactive oxygen species. BUB1B has been reported to promote tumor proliferation in glioblastoma (43,44). Similarly, BUB1B has been implicated in tumor growth and the progression of prostate cancer (45), and overexpressed BUB1B has been demonstrated to be involved in lung adenocarcinoma in humans (46). The KEGG enrichment analysis in the present study indicated that BUB1B was associated with the cell cycle. It has previously been reported that upregulated CCL2 is associated with protection from stroke induced by hypoxic preconditioning (47), and the knockdown of CCL2 was used to successfully reverse the drug resistance of tumor cells in gastric cancer (48). In the KEGG enrichment analysis performed in the present study, CCL2 was additionally associated with the cell cycle. Furthermore, based on the data presented in Fig. 3, HO-1, BUB1B and CCL2 may be regulated by a number of novel lncRNAs, including NONRATT008267.2, NONRATT015286.2, NONRATT004791.2, MSTRG.15067.2, NONRATT003289.2, NONRATT004566.2, NONRATT005985.2, NONRATT008198.2, NONRATT028439.2, NONRATT026753.2, NONRATT027268.2, MSTRG.15418.3, NONRATT016680.2, NONRATT015403.2, MSTRG.29693.5, NONRATT009960.2, MSTRG.27670.3 and NONRATT000377.2. A previous study has suggested that the knockdown of DNA topoisomerase IIα (Top2a) may suppress proliferation and invasion of colon cancer cells (49); based on the present regulatory analysis of DELs, Top2a, as a cis-regulatory gene of NONRATT005985.2, may have a vital function in ischemic stroke. Overall, the analyzed data provide novel DELs and an lncRNA-mRNA regulatory network that may provide a better understanding of ischemic stroke induced by MCAO.
Acknowledgements
The authors would like to thank Mr. Qiang Fan (Ao-Ji Bio-tech Co., Ltd., Shanghai, China) for help with data analysis.
Funding
The present study was financially supported by the National Key Research and Development Plan (grant nos. 2017YFC1701600 and 2017YFC1701601), the National Natural Science Foundation of China (grant nos. 81473387, 81503291 and 81703805), the Anhui Provincial Natural Science Foundation of China (grant no. 1508085QH191) and the Key Project of the National Science Fund of Anhui Province (grant no. KJ2013A169).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XD and DP conceived and designed the study. XD and QB performed the experiments. LH, CP, LX and HP analyzed the data and drafted the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by The Animal Experiments Ethics Committee of The Anhui University of Chinese Medicine (Hefei, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Writing Group Members, Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, Das SR, de Ferranti S, Després JP, et al. Heart disease and stroke statistics-2016 update: A report from the american heart association. Circulation. 2016;133:e38–e360. doi: 10.1161/CIR.0000000000000350. [DOI] [PubMed] [Google Scholar]
- 2.Tewari D, Majumdar D, Vallabhaneni S, Bera AK. Aspirin induces cell death by directly modulating mitochondrial voltage-dependent anion channel (VDAC) Sci Rep. 2017;7:45184. doi: 10.1038/srep45184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitsios N, Gaffney J, Kumar P, Krupinski J, Kumar S, Slevin M. Pathophysiology of acute ischaemic stroke: An analysis of common signalling mechanisms and identification of new molecular targets. Pathobiology. 2006;73:159–175. doi: 10.1159/000096017. [DOI] [PubMed] [Google Scholar]
- 4.Deb P, Sharma S, Hassan KM. Pathophysiologic mechanisms of acute ischemic stroke: An overview with emphasis on therapeutic significance beyond thrombolysis. Pathophysiology. 2010;17:197–218. doi: 10.1016/j.pathophys.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Dharap A, Bowen K, Place R, Li LC, Vemuganti R. Transient focal ischemia induces extensive temporal changes in rat cerebral microRNAome. J Cereb Blood Flow Metab. 2009;29:675–687. doi: 10.1038/jcbfm.2008.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeyaseelan K, Lim KY, Armugam A. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke. 2008;39:959–966. doi: 10.1161/STROKEAHA.107.500736. [DOI] [PubMed] [Google Scholar]
- 7.Dharap A, Nakka VP, Vemuganti R. Effect of focal ischemia on long noncoding RNAs. Stroke. 2012;43:2800–2802. doi: 10.1161/STROKEAHA.112.669465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao F, Qu Y, Liu J, Liu H, Zhang L, Feng Y, Wang H, Gan J, Lu R, Mu D. Microarray profiling and co-expression network analysis of lncRNAs and mRNAs in neonatal rats following hypoxic-ischemic brain damage. Sci Rep. 2015;5:13850. doi: 10.1038/srep13850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei N, Xiao L, Xue R, Zhang D, Zhou J, Ren H, Guo S, Xu J. MicroRNA-9 mediates the cell apoptosis by targeting Bcl2l11 in ischemic stroke. Mol Neurobiol. 2016;53:6809–6817. doi: 10.1007/s12035-015-9605-4. [DOI] [PubMed] [Google Scholar]
- 10.Xu Q, Deng F, Xing Z, Wu Z, Cen B, Xu S, Zhao Z, Nepomuceno R, Bhuiyan MI, Sun D, et al. Long non-coding RNA C2dat1 regulates CaMKIIδ expression to promote neuronal survival through the NF-κB signaling pathway following cerebral ischemia. Cell Death Dis. 2016;7:e2173. doi: 10.1038/cddis.2016.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci. 2012;13:528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaukowitch K, Kim TK. Emerging epigenetic mechanisms of long non-coding RNAs. Neuroscience. 2014;264:25–38. doi: 10.1016/j.neuroscience.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Briggs JA, Wolvetang EJ, Mattick JS, Rinn JL, Barry G. Mechanisms of long non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution. Neuron. 2015;88:861–877. doi: 10.1016/j.neuron.2015.09.045. [DOI] [PubMed] [Google Scholar]
- 14.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Washietl S, Kellis M, Garber M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. 2014;24:616–628. doi: 10.1101/gr.165035.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dharap A, Pokrzywa C, Vemuganti R. Increased binding of stroke-induced long non-coding RNAs to the transcriptional corepressors Sin3A and coREST. ASN Neuro. 2013;5:283–289. doi: 10.1042/AN20130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehta SL, Kim T, Vemuganti R. Long noncoding RNA FosDT promotes ischemic brain injury by interacting with REST-associated chromatin-modifying proteins. J Neurosci. 2015;35:16443–16449. doi: 10.1523/JNEUROSCI.2943-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han L, Ji Z, Chen W, Yin D, Xu F, Li S, Chen F, Zhu G, Peng D. Protective effects of tao-Hong-si-wu decoction on memory impairment and hippocampal damage in animal model of vascular dementia. Evid Based Complement Alternat Med. 2015;2015:195835. doi: 10.1155/2015/195835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duan X, Han L, Peng D, Chen W, Peng C, Xiao L, Bao Q. High throughput mRNA sequencing reveals potential therapeutic targets of Tao-Hong-Si-Wu decoction in experimental middle cerebral artery occlusion. Front Pharmacol. 2019;9:1570. doi: 10.3389/fphar.2018.01570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun L, Zhang Z, Bailey TL, Perkins AC, Tallack MR, Xu Z, Liu H. Prediction of novel long non-coding RNAs based on RNA-Seq data of mouse Klf1 knockout study. BMC Bioinformatics. 2012;13:331. doi: 10.1186/1471-2105-13-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35 (Web Server Issue) 2007:W345–D349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C, Liu Y, Chen R, Zhao Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013;41:e166. doi: 10.1093/nar/gkt646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG, et al. Ensembl 2018. Nucleic Acids Res 46 D. 2018:D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knauss JL, Sun T. Regulatory mechanisms of long noncoding RNAs in vertebrate central nervous system development and function. Neuroscience. 2013;235:200–214. doi: 10.1016/j.neuroscience.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikolayeva O, Robinson MD. edgeR for differential RNA-seq and ChIP-seq analysis: An application to stem cell biology. Methods Mol Biol. 2014;1150:45–79. doi: 10.1007/978-1-4939-0512-6_3. [DOI] [PubMed] [Google Scholar]
- 28.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.Tafer H, Hofacker IL. RNAplex: A fast tool for RNA-RNA interaction search. Bioinformatics. 2008;24:2657–2663. doi: 10.1093/bioinformatics/btn193. [DOI] [PubMed] [Google Scholar]
- 31.Gene Ontology Consortium, Blake JA, Dolan M, Drabkin H, Hill DP, Li N, Sitnikov D, Bridges S, Burgess S, Buza T, et al. Gene ontology annotations and resources. Nucleic Acids Res 41 (Database Issue) 2013:D530–D535. doi: 10.1093/nar/gks1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Dykstra-Aiello C, Jickling GC, Ander BP, Shroff N, Zhan X, Liu D, Hull H, Orantia M, Stamova BS, Sharp FR. Altered expression of long noncoding RNAs in blood after ischemic stroke and proximity to putative stroke risk loci. Stroke. 2016;47:2896–2903. doi: 10.1161/STROKEAHA.116.013869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, Yuan L, Zhang X, Hamblin MH, Zhu T, Meng F, Li Y, Chen YE, Yin KJ. Altered long non-coding RNA transcriptomic profiles in brain microvascular endothelium after cerebral ischemia. Exp Neurol. 2016;277:162–170. doi: 10.1016/j.expneurol.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X, Tang X, Liu K, Hamblin MH, Yin KJ. Long noncoding RNA malat1 regulates cerebrovascular pathologies in ischemic stroke. J Neurosci. 2017;37:1797–1806. doi: 10.1523/JNEUROSCI.3389-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Cao B, Han D, Sun M, Feng J. Long non-coding RNA H19 induces cerebral ischemia reperfusion injury via activation of autophagy. Aging Dis. 2017;8:71–84. doi: 10.14336/AD.2016.0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tao H, Cao W, Yang JJ, Shi KH, Zhou X, Liu LP, Li J. Long noncoding RNA H19 controls DUSP5/ERK1/2 axis in cardiac fibroblast proliferation and fibrosis. Cardiovasc Pathol. 2016;25:381–389. doi: 10.1016/j.carpath.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 40.Puyal J, Clarke PG. Targeting autophagy to prevent neonatal stroke damage. Autophagy. 2009;5:1060–1061. doi: 10.4161/auto.5.7.9728. [DOI] [PubMed] [Google Scholar]
- 41.Nada SE, Tulsulkar J, Shah ZA. Heme oxygenase 1-mediated neurogenesis is enhanced by Ginkgo biloba (EGb 761®) after permanent ischemic stroke in mice. Mol Neurobiol. 2014;49:945–956. doi: 10.1007/s12035-013-8572-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong B, Zhang Z, Xie K, Yang Y, Shi Y, Wang C, Yu Y. Hemopexin promotes angiogenesis via up-regulating HO-1 in rats after cerebral ischemia-reperfusion injury. BMC Anesthesiol. 2018;18:2. doi: 10.1186/s12871-017-0466-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma Q, Liu Y, Shang L, Yu J, Qu Q. The FOXM1/BUB1B signaling pathway is essential for the tumorigenicity and radioresistance of glioblastoma. Oncol Rep. 2017;38:3367–3375. doi: 10.3892/or.2017.6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee E, Pain M, Wang H, Herman JA, Toledo CM, DeLuca JG, Yong RL, Paddison P, Zhu J. Sensitivity to BUB1B inhibition defines an alternative classification of glioblastoma. Cancer Res. 2017;77:5518–5529. doi: 10.1158/0008-5472.CAN-17-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu X, Chen G, Cai ZD, Wang C, Liu ZZ, Lin ZY, Wu YD, Liang YX, Han ZD, Liu JC, Zhong WD. Overexpression of BUB1B contributes to progression of prostate cancer and predicts poor outcome in patients with prostate cancer. Onco Targets Ther. 2016;9:2211–2220. doi: 10.2147/OTT.S101994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen H, Lee J, Kljavin NM, Haley B, Daemen A, Johnson L, Liang Y. Requirement for BUB1B/BUBR1 in tumor progression of lung adenocarcinoma. Genes Cancer. 2015;6:106–118. doi: 10.18632/genesandcancer.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stowe AM, Wacker BK, Cravens PD, Perfater JL, Li MK, Hu R, Freie AB, Stüve O, Gidday JM. CCL2 upregulation triggers hypoxic preconditioning-induced protection from stroke. J Neuroinflammation. 2012;9:33. doi: 10.1186/1742-2094-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu W, Wei Q, Han M, Zhou B, Wang H, Zhang J, Wang Q, Sun J, Feng L, Wang S, et al. CCL2-SQSTM1 positive feedback loop suppresses autophagy to promote chemoresistance in gastric cancer. Int J Biol Sci. 2018;14:1054–1066. doi: 10.7150/ijbs.25349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang R, Xu J, Zhao J, Bai JH. Proliferation and invasion of colon cancer cells are suppressed by knockdown of TOP2A. J Cell Biochem. 2018;119:7256–7263. doi: 10.1002/jcb.26916. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.