Abstract
Intermuscular adipose tissue (IMAT) is negatively related to insulin sensitivity, but a causal role of IMAT in the development of insulin resistance is unknown. IMAT was sampled in humans to test for the ability to induce insulin resistance in vitro and characterize gene expression to uncover how IMAT may promote skeletal muscle insulin resistance. Human primary muscle cells were incubated with conditioned media from IMAT, visceral (VAT), or subcutaneous adipose tissue (SAT) to evaluate changes in insulin sensitivity. RNAseq analysis was performed on IMAT with gene expression compared with skeletal muscle and SAT, and relationships to insulin sensitivity were determined in men and women spanning a wide range of insulin sensitivity measured by hyperinsulinemic-euglycemic clamp. Conditioned media from IMAT and VAT decreased insulin sensitivity similarly compared with SAT. Multidimensional scaling analysis revealed distinct gene expression patterns in IMAT compared with SAT and muscle. Pathway analysis revealed that IMAT expression of genes in insulin signaling, oxidative phosphorylation, and peroxisomal metabolism related positively to donor insulin sensitivity, whereas expression of macrophage markers, inflammatory cytokines, and secreted extracellular matrix proteins were negatively related to insulin sensitivity. Perilipin 5 gene expression suggested greater IMAT lipolysis in insulin-resistant individuals. Combined, these data show that factors secreted from IMAT modulate muscle insulin sensitivity, possibly via secretion of inflammatory cytokines and extracellular matrix proteins, and by increasing local FFA concentration in humans. These data suggest IMAT may be an important regulator of skeletal muscle insulin sensitivity and could be a novel therapeutic target for skeletal muscle insulin resistance.
Keywords: adipose tissue distribution, EMCL, insulin sensitivity, myosteatosis
INTRODUCTION
Intermuscular adipose tissue (IMAT) is marbled within skeletal muscle and accounts for <5% of total thigh fat, but directly relates to insulin resistance, subclinical atherosclerosis, and metabolic syndrome (18, 20, 49). The first report showing IMAT content was negatively related to insulin sensitivity was published in 2000 (20) and has been reinforced by nearly every investigation in this area across a wide range of muscle groups and individuals (6, 15, 18, 19, 35, 46, 48). IMAT content is negatively related to insulin sensitivity in many populations, including in men compared with women (6), blacks compared with Caucasians (15), and older compared with younger individuals (48). Unlike intramuscular triglyceride, IMAT is low in insulin-sensitive, endurance-trained athletes (26), consistent with a negative relationship of IMAT to insulin sensitivity. Lifestyle interventions that increase insulin sensitivity either prevent IMAT accretion or reduce IMAT content, whereas detraining increases IMAT content (11, 14, 21, 27, 37, 40). Combined, these data suggest IMAT may influence muscle insulin sensitivity. Although IMAT is uniquely positioned to influence muscle metabolism and insulin sensitivity by bathing muscle with hormones, adipokines, proteins, and free fatty acids, little is known about potential mechanisms by which IMAT may influence adjacent tissues due to the difficulty of accessing it.
Signaling and secretory properties of other adipose tissue depots such as subcutaneous and visceral fat play key roles in the induction of insulin resistance, inflammation, and tissue dysfunction (22, 30). To date, we know adipose tissue secretes nearly 300 proteins, which act as endocrine, paracrine, and autocrine signals (30). Because of its physical proximity to skeletal muscle, IMAT may be a key factor regulating muscle insulin resistance and metabolic dysfunction, without measurable changes in systemic blood concentrations. Such proximity also means that therapeutic interventions directed at IMAT may powerfully alter muscle insulin sensitivity and metabolic dysfunction. To date, IMAT has not been characterized in detail, so how interventions could be directed at IMAT are not known.
The purpose of this study was to directly sample IMAT in humans with a wide range of insulin sensitivities to directly test if IMAT induces insulin resistance in vitro, and to characterize IMAT gene expression to gain insight into how IMAT may promote skeletal muscle insulin resistance.
METHODS
Subjects.
Seven lean endurance trained athletes (athletes), 7 lean sedentary controls (lean), 21 sedentary obese individuals (obese), and 6 individuals with type 2 diabetes (T2D) were included in this study for IMAT-conditioned media and RNAseq analysis. Skeletal muscle biopsies were also used for RNAseq analysis from 12 lean individuals in this study. In a separate cohort of 10 lean men and women that has been previously described (2), gluteal subcutaneous adipose tissue (SAT) was biopsied using needle aspiration after a 12-h fast and used for RNAseq analysis (4). Conditioned media was generated from visceral (VAT) and SAT biopsies obtained from 7 obese individuals who were undergoing gastric bypass surgery. Subjects gave written informed consent and were excluded if they had a body mass index (BMI) < 20 kg/m2 or > 25 kg/m2 for lean and athletes, and BMI < 30 kg/m2 for obese and T2D individuals; or had fasting triglycerides >150 mg/dl, liver, kidney, thyroid, or lung disease. Sedentary subjects were engaged in planned physical activity <2 h/wk. Endurance athletes were masters athletes training for cycling and triathlon competitions. Individuals with type 2 diabetes were excluded from the study if they used insulin and/or thiazolidinediones. All other medications were permissible, but washed out for 2 wk before metabolic testing. Volunteers in other groups were not taking medications. Subjects were weight stable in the 6 mo before the study. This study was approved by the Colorado Multiple Institution Review Board at the University of Colorado.
Preliminary testing.
Subjects reported to the Clinical Translational Research Center (CTRC) for screening procedures following a 12-h overnight fast, where they were given a health and physical examination, followed by a fasting blood draw. Volunteers underwent a standard 75-g oral glucose tolerance test to verify glucose tolerance. Percent body fat and leg lean mass were determined using DEXA analysis (Lunar DPX-IQ, Lunar, Madison, WI).
Exercise control.
Subjects were asked to refrain from planned physical activity for 48 h before the metabolic study.
Insulin clamp study.
Volunteers spent the night on the CTRC to ensure compliance with the overnight fast. After a 12-h overnight fast, an antecubital vein was cannulated in one arm for infusions of insulin, [6,6-2H2]glucose, and dextrose, and a retrograde dorsal hand vein was catheterized in the contralateral side for blood sampling via the heated hand technique. A primed continuous infusion of [6,6-2H2]glucose was initiated at 0.04 mg·kg−1·min−1 and continued throughout a 2-h equilibrium period and the 3-h insulin clamp. After 2 h of tracer equilibration, a percutaneous needle biopsy was taken from midway between the greater trochanter of the femur and the patella. In a subset of obese individuals, IMAT sampled from the vastus lateralis was immediately washed in PBS and saved to generate conditioned media. In all other subjects, muscle was immediately flash frozen in liquid nitrogen and stored at −80°C until IMAT dissection described below. A hyperinsulinemic-euglycemic clamp was then initiated and continued for the next 3 h using the method of DeFronzo et al. (10) as previously described for this cohort (39). Briefly, a primed continuous infusion of insulin was administered at 40 mU·m−2·min−1 for 3 h. A variable infusion of 20% dextrose was infused to maintain blood glucose ~90 mg/dl. The dextrose infusion used to maintain euglycemia was labeled with [6,6-2H2]glucose (Cambridge Isotope Laboratories, Tewksbury, MA) to maintain stable enrichment of plasma glucose. Glucose rate of disappearance (glucose Rd) was calculated as previously described (12).
Conditioned media experiments.
IMAT was sampled from the vastus lateralis while obtaining muscle biopsies in seven obese individuals, immediately dissected away from skeletal muscle on ice using a dissecting microscope, washed in PBS, and cultured in DMEM (1 ml media/50 mg IMAT) for 48 h at 37°C in 5% CO2 to generate conditioned media. Conditioned media was generated in the same way from VAT and SAT collected from individuals undergoing gastric bypass surgery. For this comparison, patients with similar fasting glucose, HbA1c, and age were chosen to minimize potential confounding variables. IMAT, VAT, and SAT conditioned media was administered to primary myotubes from a separate group of insulin-resistant obese donors (n = 5; age 36.2 ± 2.1 yr; BMI 37.0 ± 1.7 kg/m2; sex: 3 men and 2 women; 3.8 ± 0.9 mg·kg−1·min−1 glucose infusion rate (GIR) during a 40 mU·m−2·min−1 insulin clamp) during days 5–7 of differentiation at 5% of total media volume for 3.5 h. Myotubes were grown using standard techniques (16), while insulin sensitivity was determined using an insulin-stimulated glycogen synthesis assay using a final insulin concentration of 100 nM (45), and myotube lipid accumulation and conditioned media FFA content were measured as previously described (23). Data were normalized to the control condition to account for differences in donor insulin sensitivity.
Biopsy processing.
Flash-frozen muscle biopsies were cut into smaller 20- to 30-mg pieces and dissected on ice for 1–1.5 min to separate IMAT from skeletal muscle using a dissection microscope. IMAT and skeletal muscle samples were not contaminated by other tissues as observed under a dissecting scope before RNA extraction described below. No attempt was made to separate adipocytes from stromal vascular cells in the isolated IMAT as our goal was to examine IMAT as a complete tissue.
RNAseq.
Total RNA was prepared from IMAT, skeletal muscle, and subcutaneous adipose tissue biopsies using the RNeasy Lipid Tissue Kit (QIAGEN) according to the manufacturer’s instructions. The quality of the isolated RNA was determined with the Agilent 2100 Bioanalyzer (RNA 6000 Nano Kit, Agilent). All samples had a RNA integrity number (RIN) value > 7. Samples were handled in a blinded fashion during the library preparation and sequencing process.
For library preparation, 300 μg of total RNA per sample was used. RNA molecules were poly(A) selected, fragmented, and reverse transcribed with the Elute, Prime, Fragment Mix (Illumina, San Diego, CA). End-repair, A-tailing, adaptor ligation, and library enrichment were performed as described in the Low-Throughput Protocol of the TruSeq RNA Sample Prep Guide (Illumina) using the Bravo Automated Liquid Handling Platform (Agilent Technologies, Santa Clara, CA). RNA libraries were assessed for quality and quantity with the Agilent 2100 Bioanalyzer and the Quant-iT PicoGreen dsDNA Assay Kit (Thermo Fisher, Waltham, MA). RNA libraries were sequenced as 100-bp paired-end runs on an Illumina HiSeq2500 platform. Primary analysis for base calling and quality scorning was performed using the Real-Time Analysis software (Illumina). Sequences were aligned against the hg19 genome and UCSC knownGene annotation assembly (GTF file) with GEM mapper (version 1.7.1) using standard parameters (except mismatches = 0.04 and min-decoded-strata = 2). Read counts were calculated using HTSeq-count (version 0.6.0) with the same annotation files as for mapping.
Statistical analysis.
Differences between groups and tissues were analyzed using a 1-way ANOVA (SPSS, Chicago, IL). When significant differences were detected, individual means were compared using Student’s t-tests to determine differences between groups. For RNAseq analysis, we followed the recently published workflow described by Law et al. to analyze data including the “edgeR” and “limma” package in R. Specifically, for the analysis of RNAseq data comparing SAT, muscle, and IMAT, raw counts were filtered using edgeR (raw counts ≥ 25 in ≥8 samples). The raw counts of the remaining 15,550 genes were converted to counts per million (CPM) and log2-counts per million (log-CPM) values using edgeR (43). Values were normalized by the method of trimmed mean of M-values (TMM) (44). Limma was used to make multidimensional (MDS) plots (42).
To correlate gene expression and glucose Rd during the clamp for the IMAT RNAseq data, one sample was excluded as no glucose Rd was available. We used genes with more than 25 counts ≥ 70% of the IMAT samples; 13,476 genes passed the criteria and were used for downstream analyses. All genes significantly correlated to glucose Rd were evaluated within the GAGE package in R (34). We used the “pathview” package to visualize the correlation coefficients within the pathways in R (33). A priori hypotheses for genes representing macrophages, cytokines, extracellular matrix, and perilipins were corrected for multiple comparisons using the Benjamini-Hochberg method. An α level of 0.05 was used for statistical significance.
RESULTS
Demographic information for obese donors used in conditioned media experiments for IMAT, VAT, and SAT are shown in Table 1. Individuals in the SAT/VAT group were significantly more obese than the IMAT group (P < 0.05). Demographics for lean individuals used to compare gene expression patterns of IMAT, skeletal muscle, and SAT are shown in Table 2. All of the female and male subjects in this comparison were lean with similar BMI and percent body fat between groups. The age of individuals from which SAT biopsies were obtained was significantly lower than the other groups (P < 0.05). Demographic information for individuals used to compare differences in IMAT gene expression across a range of insulin sensitivities are shown in Table 3. There were a roughly equal number of men and women in this study (17/18), with a similar proportion of each sex in all groups. The mean age was not significantly different between groups, with BMI and body fat significantly lower in lean controls and athletes by design. Glucose Rd during the insulin clamp was significantly different between each group except for the comparison between lean individuals and endurance-trained athletes which was of borderline significance (P = 0.06).
Table 1.
Subject demographics for conditioned media analysis
| Variable | IMAT | SAT/VAT |
|---|---|---|
| Sex, M/W | 2/5 | 1/6 |
| n | 7 | 7 |
| BMI, kg/m2 | 34.8 ± 1.3 | 43.6 ± 2.5* |
| Age, yr | 43.3 ± 2.1 | 47.6 ± 3.8 |
| HbA1c, % | 5.9 ± 0.4 | 6.2 ± 0.4 |
| Fasting glucose, mg/dl | 93 ± 12 | 101 ± 13 |
Values are means ± SE. M, men; W, women; BMI, body mass index; HbA1c, hemoglobin A1c; IMAT, intermuscular adipose tissue; SAT, subcutaneous adipose tissue; VAT, visceral adipose tissue.
Significantly different from IMAT conditioned media group.
Table 2.
Subject demographics for skeletal muscle versus IMAT versus SAT RNAseq comparison
| Skeletal Muscle |
IMAT |
SAT |
||||
|---|---|---|---|---|---|---|
| Variable | Men | Women | Men | Women | Men | Women |
| n | 6 | 6 | 3 | 5 | 5 | 4 |
| BMI, kg/m2 | 22.7 ± 1.4 | 21.7 ± 0.5 | 21.0 ± 2.1 | 21.4 ± 0.4 | 23.4 ± 0.7 | 22.6 ± 0.9 |
| Age, yr | 43.2 ± 4.1 | 42.5 ± 2.5 | 44.7 ± 4.7 | 39.6 ± 2.6 | 28.4 ± 3.1*† | 27.5 ± 3.5*† |
| % Fat | 15.1 ± 2.6 | 29.0 ± 2.1 | 18.1 ± 4.0 | 29.7 ± 2.6 | 18.8 ± 3.6 | 29.0 ± 4.5 |
Values are means ± SE. BMI, body mass index; OGTT, oral glucose tolerance test; IMAT, intermuscular adipose tissue; SAT, subcutaneous adipose tissue; VAT, visceral adipose tissue.
Significantly different from skeletal muscle group,
significantly different from IMAT group: P < 0.05.
Table 3.
Subject demographics for IMAT RNAseq comparisons to insulin sensitivity
| Variable | Athletes | Lean Controls | Obese Controls | Type 2 Diabetes |
|---|---|---|---|---|
| n (W/M) | 7 (3/4) | 7 (4/3) | 14 (6/8) | 6 (3/3) |
| Age, yr | 43.3 ± 2.3 | 41.5 ± 2.4 | 40.1 ± 1.9 | 45.7 ± 2.5 |
| BMI, kg/m2 | 22.0 ± 0.7 | 21.2 ± 0.7 | 36.4 ± 1.4†‡ | 34.8 ± 1.7†‡ |
| Body fat, % | 17.0 ± 3.1‡ | 25.3 ± 2.9 | 37.5 ± 2.5†‡ | 35.4 ± 2.5†‡ |
| Fasting glucose, mg/dl | 84 ± 2 | 84 ± 3 | 89 ± 3 | 177 ± 15†‡§ |
| 2-h OGTT, mg/dl | 77 ± 10 | 95 ± 5 | 99 ± 6 | 329 ± 26†‡§ |
| HbA1c, % | 5.4 ± 0.07 | 5.2 ± 0.06†§ | 5.4 ± 0.07 | 8.0 ± 0.5†‡§ |
| Glucose rate of disappearance, mg·kg−1·min−1 | 12.2 ± 0.7*§ | 9.9 ± 0.7*§ | 5.1 ± 0.7*†‡ | 2.3 ± 0.5†‡§ |
| Proportion of parent group with sampled IMAT, % | 62 | 73 | 71 | 83 |
| IMAT weight, mg | 27 ± 5 | 56 ± 13 | 71 ± 17 | 63 ± 20 |
Values are means ± SE. W, women; M, men; BMI, body mass index; OGTT, oral glucose tolerance test; IMAT, intermuscular adipose tissue; SAT, subcutaneous adipose tissue; VAT, visceral adipose tissue.
Significantly different from Type 2 diabetes;
significantly different from athletes;
significantly different from lean;
significantly different from obese: P < 0.05.
IMAT conditioned media decreases myotube insulin sensitivity.
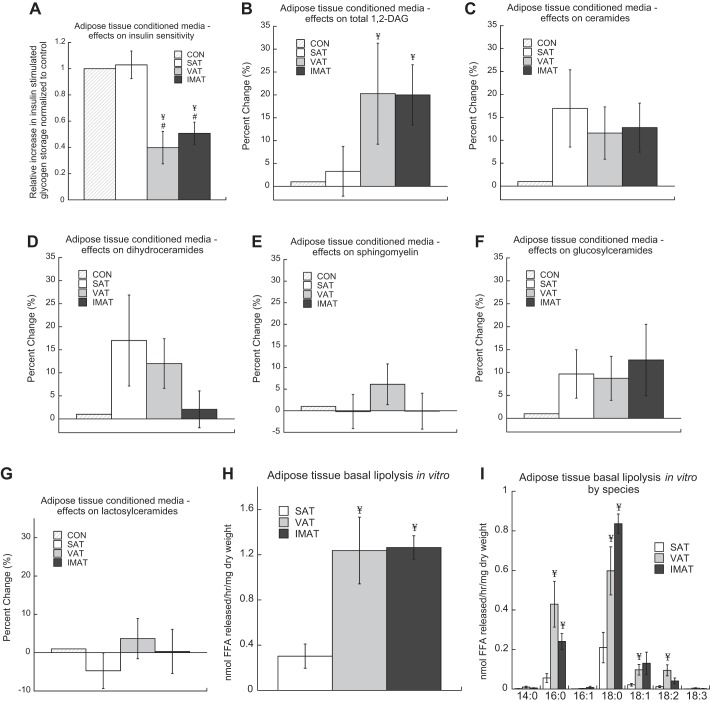
Conditioned media from IMAT, VAT, and SAT of obese patients was administered separately to primary myotubes from obese donors to evaluate if IMAT influences insulin sensitivity in skeletal muscle through secreted factors. In the control condition 100 nM of insulin for 1 h resulted in a 104 ± 15% increase in insulin-stimulated glycogen storage. IMAT and VAT conditioned media significantly decreased insulin sensitivity compared with conditioned media from SAT (P = 0.002) and DMEM control (P = 0.003, Fig. 1A). The decrease in insulin sensitivity was not significantly different between IMAT and VAT. Conditioned media from SAT did not change insulin sensitivity compared with DMEM control. Effects of conditioned media on insulin sensitivity in vitro are not due to a “non-self” immune response because there are no immune cells in this primary culture model. These data indicate that factors secreted from IMAT decrease insulin sensitivity in vitro with a potency similar to VAT.
Fig. 1.
Intermuscular adipose tissue (IMAT) conditioned media decreases insulin sensitivity. Conditioned media was generated from subcutaneous adipose tissue (SAT), visceral adipose tissue (VAT), and IMAT, and administered along with DMEM-only control to primary myotubes from obese donors for 3.5 h at 5% of total media volume. Insulin sensitivity was measured (A), along with changes in the content of 1,2-diacyl glycerol (1,2-DAG) (B), ceramides (C), dihydroceramides (D), sphingomyelin (E), glucosylceramides (F), and lactosylceramides (G). The free fatty acid (FFA) content of conditioned media was measured to calculate basal lipolytic rates (H) as well as the composition of individual FFA species (I) from SAT, VAT, and IMAT in culture. Values are means ± SE. ¥Significantly different from SAT, #significantly different from control: P < 0.05.
IMAT conditioned media increases myotube 1,2-diacylglycerol concentration.
To evaluate mechanisms explaining the induction of insulin resistance in vitro, we administered conditioned media to myotubes to evaluate if SAT, VAT, and IMAT conditioned media altered bioactive lipid accumulation. We found that VAT and IMAT conditioned media significantly increased myotube 1,2-diacylglycerol (DAG) content, but did not result in significant changes in ceramides, dihydroceramides, sphingomyelins, glucosylceramides, or lactosylceramides (Fig. 1, B–G). These data suggest that the VAT and IMAT secretome promote insulin resistance by inducing 1,2-DAG accumulation in skeletal muscle.
Basal lipolytic rates of SAT, VAT, and IMAT.
Conditioned media from SAT, VAT, and IMAT were lipid extracted and analyzed for free fatty acids (FFA) using lipidomics, with FFA release rates normalized to dry tissue weight. We found similar rates of basal lipolysis in VAT and IMAT, which were both significantly greater than SAT (Fig. 1H). Composition analysis revealed the majority of the increased rates of lipolysis in VAT and IMAT could be explained by release of 16:0 and 18:0, with significantly greater rates of 18:1 and 18:2 release also found in VAT compared with SAT (Fig. 1I).
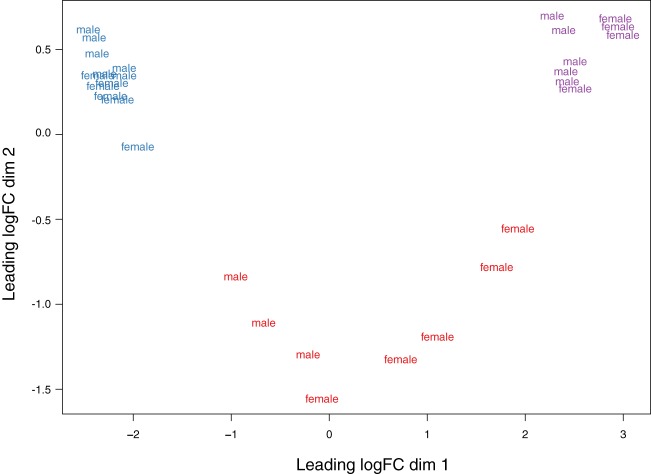
IMAT has a gene expression pattern distinct from skeletal muscle and subcutaneous fat.
A multidimensional scaling (MDS) plot in Fig. 2 shows differences in gene expression between IMAT, muscle, and SAT based on the 1,500 most divergent genes. All three tissues showed clearly separated expression profiles with all samples from one depot clustering together, with further grouping within adipose tissues based on sex. These results indicate that IMAT has a unique transcriptome, which is different from that of muscle and subcutaneous fat.
Fig. 2.
Intermuscular adipose tissue (IMAT) has a unique transcriptome compared with muscle and subcutaneous adipose tissue. Multidimensional scaling (MDS) plot of logCPM for IMAT, skeletal muscle, and subcutaneous adipose tissue (SAT) samples from RNAseq analyses over dimensions 1 and 2 based on the 1,500 most divergent genes. Red = IMAT; blue = muscle; purple = SAT.
IMAT gene expression correlates with insulin sensitivity.
The schematic of the workflow is depicted in Fig. 3. Within the IMAT transcriptome, 988 genes correlated negatively and 1,234 genes correlated positively with glucose Rd. We used significantly correlated genes to find enriched pathways using the KEGG database (24a). Fifty-four pathways were identified containing at least 10 significantly enriched correlated genes (Table 4). Among these pathways, there were eight pathways directly linked to the immune system and four pathways related to the extracellular matrix and local fibrosis.
Fig. 3.
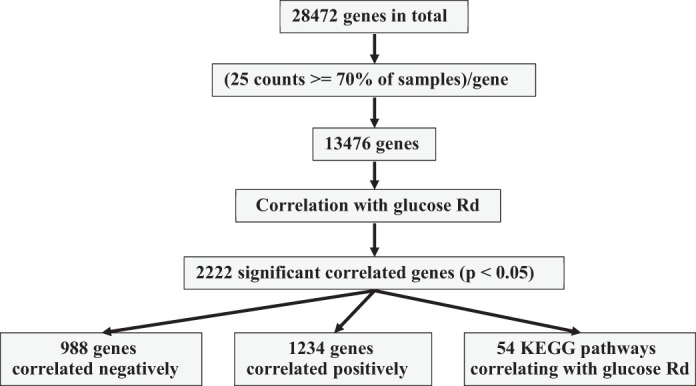
Schematic workflow of correlating gene expression with glucose rate of disappearance (Rd). From the initial 28,472 genes we only used genes with at least 25 counts in 70% of the intermuscular adipose tissue (IMAT) samples. This left 13,476 genes, from which 2,222 genes were significantly correlated with glucose Rd (P < 0.05). Of these genes, 988 genes correlated negatively and 1,234 genes correlated positively with glucose Rd. Additionally, we found 54 KEGG pathways which showed a correlation with glucose Rd. For more details, see text and methods.
Table 4.
Complete list of the 54 identified enriched KEGG pathways using all 2,222 genes significantly correlated to Rd with a P value < 0.05
| KEGG ID | Pathway | Gene Set Level Change | Gene (n) |
|---|---|---|---|
| hsa03010 | Ribosome | 5.541 | 52 |
| hsa00190 | Oxidative phosphorylation | 4.295 | 31 |
| hsa00280 | Valine. leucine and isoleucine degradation | 3.689 | 18 |
| hsa00240 | Pyrimidine metabolism | 2.702 | 15 |
| hsa00640 | Propanoate metabolism | 2.537 | 10 |
| hsa00230 | Purine metabolism | 2.113 | 17 |
| hsa04146 | Peroxisome | 1.807 | 13 |
| hsa04910 | Insulin signaling pathway | 1.304 | 25 |
| hsa03040 | Spliceosome | 1.291 | 22 |
| hsa04150 | mTOR signaling pathway | 1.168 | 15 |
| hsa04141 | Protein processing in endoplasmic reticulum | 0.838 | 19 |
| hsa04920 | Adipocytokine signaling pathway | 0.637 | 11 |
| hsa04012 | ErbB signaling pathway | 0.605 | 11 |
| hsa04142 | Lysosome | 0.529 | 14 |
| hsa04530 | Tight junction | 0.418 | 14 |
| hsa03008 | Ribosome biogenesis in eukaryotes | 0.332 | 13 |
| hsa04114 | Oocyte meiosis | 0.315 | 11 |
| hsa00564 | Glycerophospholipid metabolism | 0.167 | 14 |
| hsa04914 | Progesterone-mediated oocyte maturation | 0.146 | 15 |
| hsa03013 | RNA transport | −0.118 | 22 |
| hsa04960 | Aldosterone-regulated sodium reabsorption | −0.202 | 10 |
| hsa04722 | Neurotrophin signaling pathway | −0.222 | 16 |
| hsa04110 | Cell cycle | −0.278 | 17 |
| hsa04144 | Endocytosis | −0.311 | 30 |
| hsa04370 | VEGF signaling pathway | −0.374 | 13 |
| hsa04120 | Ubiquitin mediated proteolysis | −0.465 | 24 |
| hsa04912 | GnRH signaling pathway | −0.483 | 11 |
| hsa04620 | Toll-like receptor signaling pathway | −0.532 | 19 |
| hsa04730 | Long-term depression | −0.567 | 10 |
| hsa03015 | mRNA surveillance pathway | −0.619 | 10 |
| hsa04010 | MAPK signaling pathway | −0.766 | 33 |
| hsa04916 | Melanogenesis | −0.907 | 10 |
| hsa04310 | Wnt signaling pathway | −0.955 | 23 |
| hsa04510 | Focal adhesion | −0.986 | 18 |
| hsa04210 | Apoptosis | −1.068 | 16 |
| hsa04630 | Jak-STAT signaling pathway | −1.140 | 17 |
| hsa04360 | Axon guidance | −1.225 | 13 |
| hsa04115 | p53 signaling pathway | −1.457 | 12 |
| hsa04070 | Phosphatidylinositol signaling system | −1.488 | 11 |
| hsa04660 | T cell receptor signaling pathway | −1.535 | 20 |
| hsa04664 | Fc epsilon RI signaling pathway | −1.759 | 18 |
| hsa04810 | Regulation of actin cytoskeleton | −2.065 | 28 |
| hsa04020 | Calcium signaling pathway | −2.393 | 16 |
| hsa04621 | NOD-like receptor signaling pathway | −2.457 | 12 |
| hsa04662 | B cell receptor signaling pathway | −2.388 | 22 |
| hsa04062 | Chemokine signaling pathway | −2.398 | 35 |
| hsa04514 | Cell adhesion molecules (CAMs) | −2.618 | 18 |
| hsa04145 | Phagosome | −2.707 | 18 |
| hsa04640 | Hematopoietic cell lineage | −3.747 | 11 |
| hsa04666 | Fc gamma R-mediated phagocytosis | −3.125 | 21 |
| hsa04610 | Complement and coagulation cascades | −3.828 | 12 |
| hsa04670 | Leukocyte transendothelial migration | −3.453 | 15 |
| hsa04650 | Natural killer cell mediated cytotoxicity | −3.269 | 23 |
| hsa04380 | Osteoclast differentiation | −3.335 | 32 |
A gene set level change > 0 implies a positive change of genes within the pathway to insulin sensitivity and vice versa. Gene (n) indicates the number of genes used in the gene set test by gage().
Analysis of IMAT signaling pathways associated with insulin sensitivity.
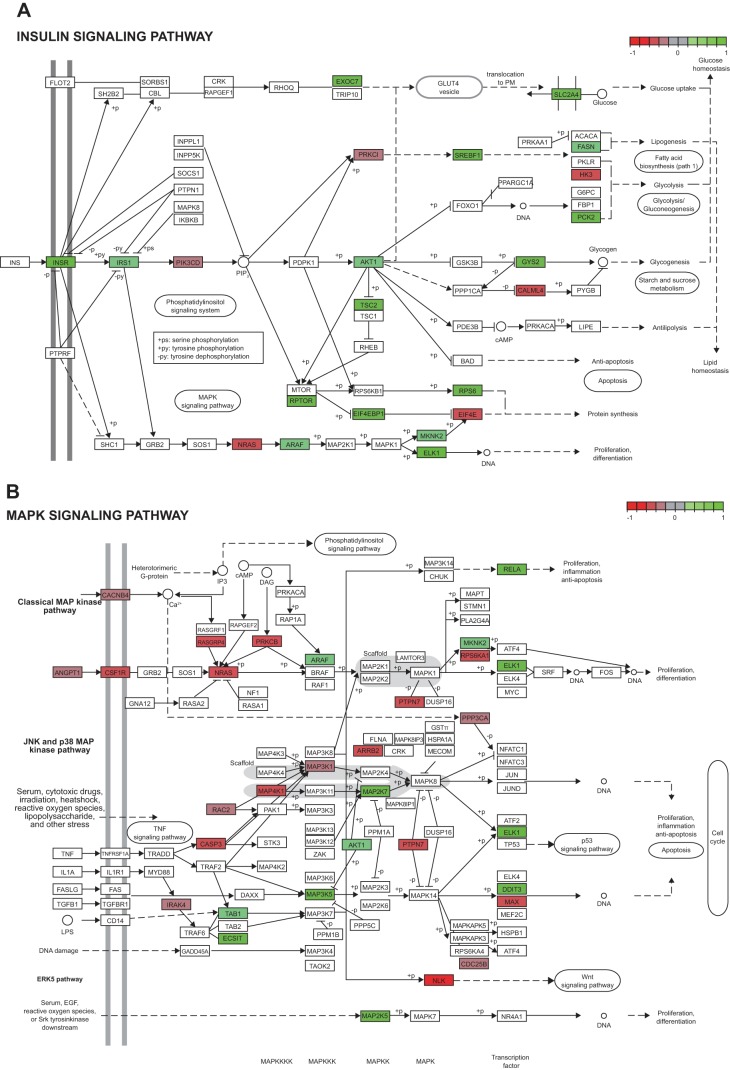
Pathway analysis showed the insulin signaling pathway for IMAT was positively related to insulin sensitivity, with relationship of individual genes to glucose Rd shown in Fig. 4A. The JAK/STAT and MAPK signaling pathways in IMAT were negatively related to insulin sensitivity with the relationship of individual genes to glucose Rd shown in Fig. 4, B and C. These data suggest that IMAT insulin signaling parallels whole body insulin sensitivity, and could be negatively influenced by inflammation through the JAK/STAT and MAPK pathways. Insulin normally inhibits lipolysis in adipose tissue, so insulin-resistant IMAT could increase local release of free fatty acids into interstitial fluid surrounding muscle. Pathway analysis revealed a positive relationship of IMAT oxidative phosphorylation to insulin sensitivity (Fig. 4D). These data suggest insulin-resistant IMAT may have decreased mitochondrial function and lipid oxidation, which could also contribute to enhanced release of free fatty acids into interstitial fluid surrounding muscle.
Fig. 4.
Continued
Fig. 4.
Pathway analysis of human intermuscular adipose tissue (IMAT) revealed a downregulation of key pathways associated with insulin resistance. Green indicates a positive correlation whereas red indicates a negative correlation between the patient’s gene expression and insulin sensitivity. Key genes like the insulin receptor (INSR), the serine-threonine protein kinase (AKT), and the glucose transporter GLUT4 (SLC2A4) were positively associated with insulin sensitivity, and therefore were downregulated in patients with low insulin sensitivity (A). The MAPK (B) and JAK/STAT (C) signaling pathways were inversely related to insulin sensitivity. Oxidative phosphorylation (D) was associated with insulin sensitivity, and therefore is consistent with mitochondrial dysfunction in insulin-resistant individuals. [KEGG pathways (24a) in panels A–D redrawn with permission. Copyright 2018 Kanehisa Laboratories.]
Macrophage marker and inflammatory cytokine gene expression.
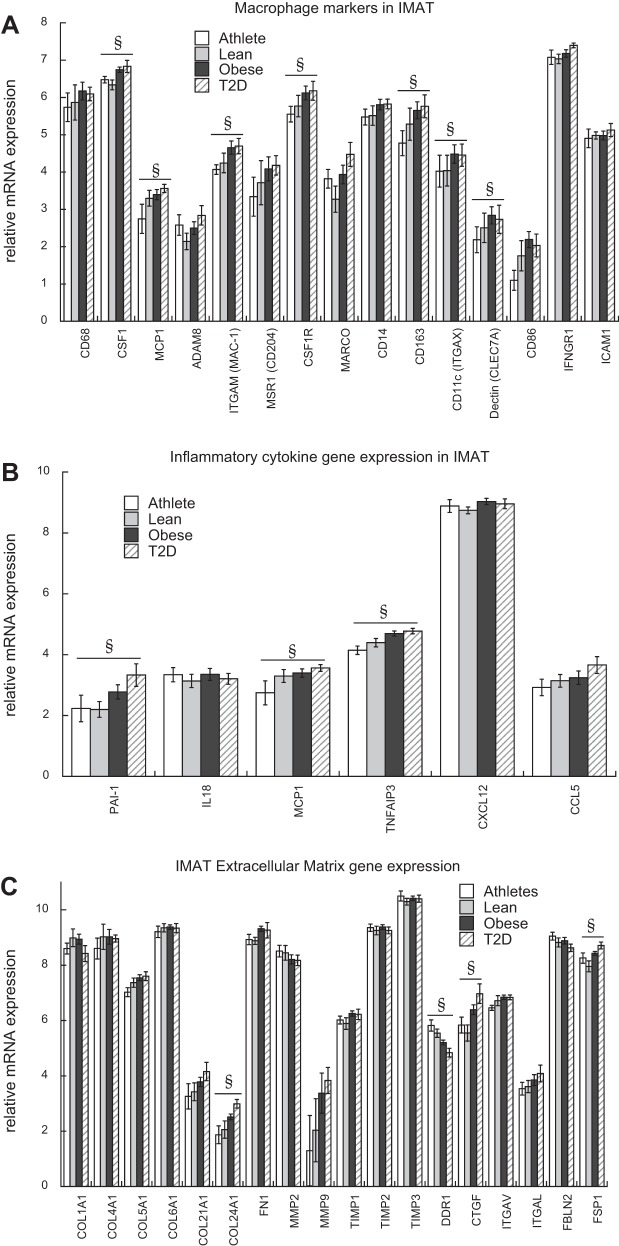
Common macrophage marker genes were evaluated in IMAT, and we found colony stimulating factor 1 (CSF1), cluster of differentiation 163 (CD163), monocyte chemotactic protein 1 (MCP1), integrin subunit alpha M (ITGAM), colony stimulating Factor 1 Receptor (CSF1R), integrin subunit alpha X (ITGAX), and C-type lectin domain family 7 member A (CLEC7A) were negatively correlated to insulin sensitivity (Fig. 5A). We found significant negative relationships between insulin sensitivity and several inflammatory cytokines such as plasminogen activator inhibitor 1 (PAI-1) and MCP1 (Fig. 5B), whereas there were no significant relationships between IMAT anti-inflammatory gene expression and insulin sensitivity. TNF alpha-induced protein 3 (TNFAIP3) codes for a protein that limits TNFα-induced NFκB inflammation (9), and was also significantly related to glucose Rd. These data not only indicate that IMAT contains macrophages proportional to insulin sensitivity, but also suggests that macrophage cytokine secretion within IMAT is negatively related to insulin sensitivity.
Fig. 5.
Intermuscular adipose tissue (IMAT) macrophage, cytokine, and extracellular matrix gene expression correlates to insulin sensitivity. IMAT mRNA expression of macrophage markers (A), inflammatory cytokines (B), and extracellular matrix proteins (C). Values are means ± SE. CD68, cluster of differentiation 68; CSF1, colony stimulating factor 1; MCP1, monocyte chemotactic protein 1; ADAM8, ADAM metallopeptidase domain 8; ITGAM, integrin subunit alpha M; MSR1, macrophage scavenger receptor 1; CSF1R, colony stimulating factor 1 receptor; MARCO, Macrophage Receptor With Collagenous Structure; CD14, CD14 molecule; CD163, Cluster of Differentiation 163; ITGAX, integrin subunit alpha X; CLEC7A, C-type lectin domain family 7 member A; IFNGR1, interferon gamma receptor 1; SIGLEC1, sialic acid binding Ig like lectin 1; PAI-1, plasminogen activator inhibitor type 1; IL18, interleukin 18; TNFAIP3, TNF alpha induced protein 3; CXCL12, stromal cell-derived factor 1; CCL5, C-C motif chemokine ligand 5. COL1A1, collagen type I alpha 1; COL4A1, collagen type IV alpha 1; COL5A1, collagen type V alpha 1; COL6A1, collagen type VI alpha 1; COL21A1, collagen type XXI alpha 1; COL24A1, collagen type XXIV alpha 1; FN1, fibronectin 1; MMP2 and MMP9, matrix metalloproteinases 2 and 9, respectively; TIMP1–3, tissue inhibitor of metalloproteinases 1–3; DDR1, discoidin domain receptor family, member 1; CTGF, connective tissue growth factor; ITGAV, integrin alpha chain 5; ITGAL, integrin alpha L; FBLN2, fibulin 2; FSP1, fibroblast specific protein. §Significantly correlated to insulin sensitivity.
Extracellular matrix gene expression.
Among the IMAT genes that strongly correlated with glucose Rd, the expression of extracellular matrix genes collagen type XXIV alpha 1 (COL24A1), discoidin domain receptor family, member 1 (DDR1), and connective tissue growth factor (CTGF) scaled to insulin sensitivity (Fig. 5C). There are many cell types in adipose tissue, including fibroblasts, that may also contribute to extracellular matrix protein secretion (13). Therefore, we evaluated the relationship of fibroblast markers fibroblast specific protein 1 (FSP1), prolyl 4-hydroxylase subunit alpha 2 (P4HA2), fibroblast activation protein alpha (FAP), prolyl 4-hydroxylase subunit alpha 1 (P4HA1), and prolyl 4-hydroxylase subunit beta (P4HB) to insulin sensitivity and only found FSP1 was significantly related to insulin sensitivity after correcting for multiple comparisons (Fig. 5C).
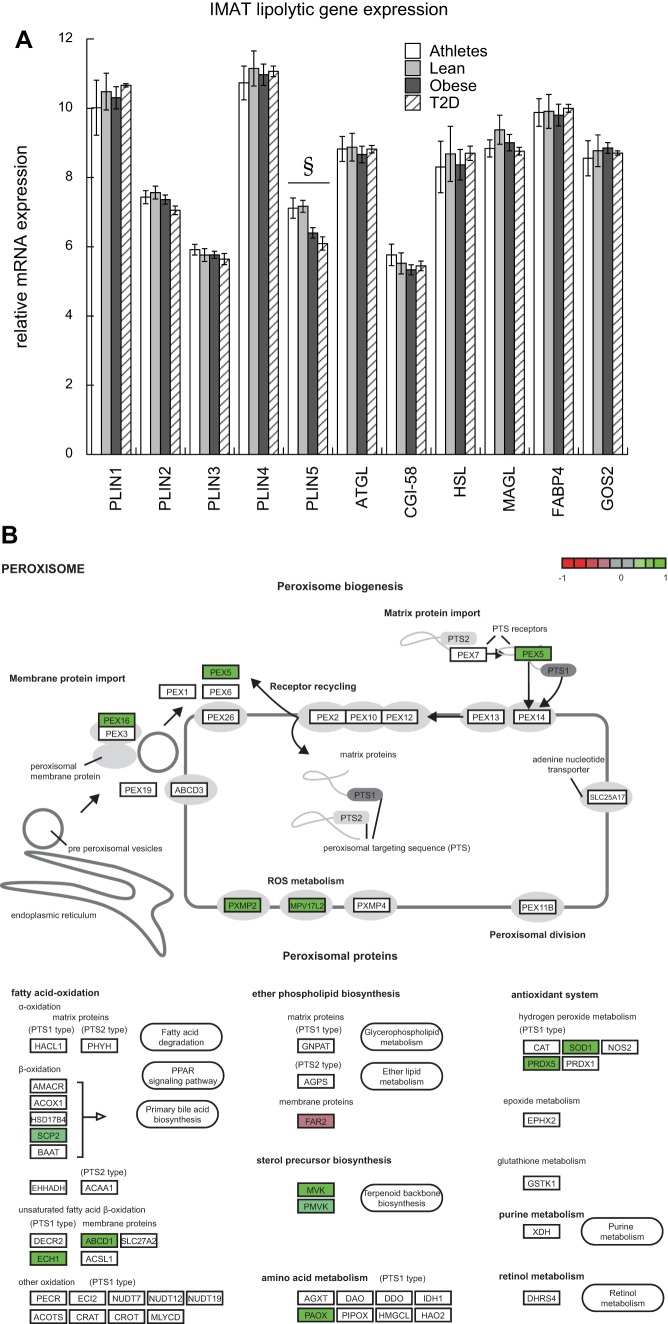
Lipolytic and peroxisome gene expression.
Expression of genes involved in regulating lipolysis was analyzed to evaluate potential regulation of lipid droplet lipolysis in IMAT. Other than a significant relationship between PLIN5 and glucose Rd, there were no other significant relationships to insulin sensitivity for genes regulating lipolysis (Fig. 6A). Additionally, there was a positive relationship of the KEGG “Peroxisome” pathway to insulin sensitivity (Fig. 6B). We did not find significant differences between groups in gene expression for pathways of beta-oxidation or triglyceride reesterification. Although we cannot exclude alterations in protein content or activity to these pathways, these data suggest reduced peroxisome pathway expression may contribute to greater IMAT FFA release in insulin-resistant individuals.
Fig. 6.
Intermuscular adipose tissue (IMAT) may influence interstitial free fatty acid (FFA) concentration promote muscle lipid accumulation. IMAT mRNA expression of genes involved in lipolytic regulation and their relationship to insulin sensitivity (A), and a positive relationship between the KEGG peroxisome pathway and insulin sensitivity (B). PLIN1 encodes perilipin 1, PLIN2, perilipin 2; PLIN3, perilipin 3; PLIN4, perilipin 4, PLIN5, perilipin 5, ATGL, adipose triglyceride lipase, CGI-58, comparative gene identification-58, HSL, hormone sensitive lipase, MAGL, monacylglycerol lipase, FABP4, fatty acid binding protein 4, GOS2, G0/G1 switch 2. §Significantly correlated to insulin sensitivity. Green indicates a positive correlation between the gene expression and insulin sensitivity. Key genes like the sterol carrier protein 2 (SCP2), enoyl-CoA hydratase 1 (ECH1), and ATP binding cassette subfamily D member 1 (ABCD1) were downregulated in insulin-resistant patients. [KEGG pathway (24a) in panel B redrawn with permission. Copyright 2018 Kanehisa Laboratories.]
Figure 7 is a summary figure showing potential mechanisms by which IMAT may promote decreased skeletal muscle insulin sensitivity implicated in this study.
Fig. 7.
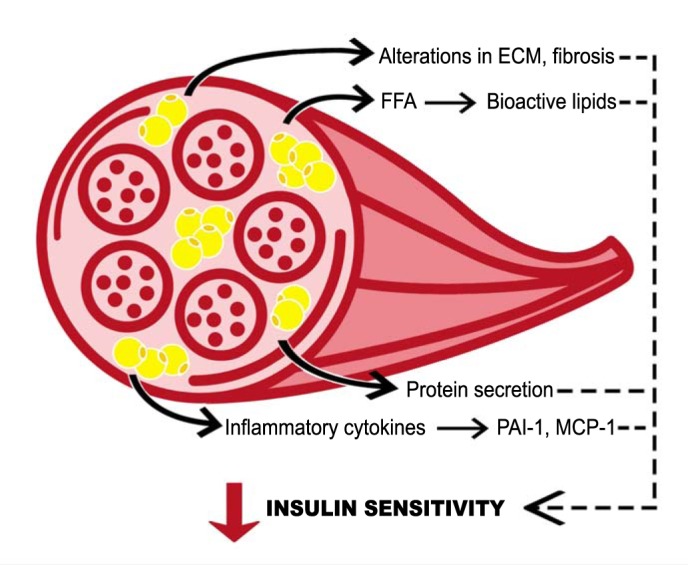
Conceptual model for intermuscular adipose tissue (IMAT)-induced insulin resistance. Potential mechanisms by which IMAT decreases insulin sensitivity in neighboring skeletal muscle. FFA, free fatty acids; PAI-1, plasminogen activator inhibitor 1; MCP-1, monocyte chemotactic protein 1.
DISCUSSION
Due to its physical proximity, secreted FFA, proteins, and cytokines from IMAT are likely particularly potent for regulating muscle insulin sensitivity and metabolic function. Such proximity also means that therapeutic interventions directed at IMAT may powerfully alter muscle insulin sensitivity and metabolic dysfunction. However, specific components of IMAT signaling in humans that modify muscle metabolism are not known and, therefore, cannot be targeted by a therapeutic intervention. To our knowledge, this is the first publication to directly sample IMAT in humans to reveal how IMAT may impact muscle metabolism and insulin resistance. Key findings from this study include: conditioned media from IMAT decreased insulin sensitivity in primary human myotubes with a potency similar to VAT; administration of conditioned media from IMAT and VAT increased myotube 1,2-DAG content; VAT and IMAT have greater rates of basal lipolysis compared with SAT; expression of macrophage markers, inflammatory cytokines, and connective tissue markers in IMAT were significantly greater in insulin-resistant individuals; and, finally, IMAT RNA expression in insulin-resistant individuals was consistent with increased rates of IMAT lipolysis and interstitial FFA concentration leading to muscle lipid accumulation. These data provide the first direct evidence that factors secreted from IMAT contribute to the development of insulin resistance.
To date, studies in humans have been limited to associations between IMAT accumulation and metabolic outcomes due to the difficulty in sampling this tissue depot. To determine if skeletal muscle or SAT was collected in our IMAT samples, we compared gene expression patterns in skeletal muscle, IMAT, and SAT from lean individuals. Our analyses revealed distinct gene expression patterns between these tissue depots, suggesting contamination between tissues was unlikely. Similar to what has been previously reported in livestock (8, 29), we interpret these data to suggest that IMAT is a separate tissue depot with a unique gene expression signature that differs markedly from skeletal muscle and subcutaneous adipose tissue in humans.
Our functional in vitro experiments show IMAT and VAT secreted factors that decreased muscle insulin sensitivity relative to SAT. The components of IMAT conditioned media causing insulin resistance are not known. However, the transcriptomic signature of IMAT suggests it may contain macrophages that secrete inflammatory cytokines and could promote local inflammation in skeletal muscle. These data also support the idea that IMAT may secrete proteins that influence the extracellular matrix, and could increase local FFA concentration, both of which are known to impact muscle insulin sensitivity (50). Our data show that IMAT and VAT conditioned media increased muscle cell accumulation of 1,2-DAG, which is known to decrease insulin signaling and sensitivity through activation of novel PKC isoforms, and may help explain the insulin resistance observed in vitro. These ideas are supported by a recent study showing conditioned media from fibroadipogenic precursors differentiated into adipocytes decreased insulin sensitivity and signaling in primary muscle cell cultures (28). Together, these data indicate that secretions from IMAT can decrease muscle insulin sensitivity similar to VAT, and suggest that the IMAT secretome bathes the muscle in factors that attenuate insulin sensitivity.
Although exact mechanisms by which IMAT promotes insulin resistance are unknown, the initial report by Goodpaster et al. (20) suggested IMAT may induce insulin resistance by impairing muscle blood flow or insulin diffusion capacity, or increasing local FFA concentration. Our data support the concept that IMAT triglyceride lipolysis may increase interstitial FFA concentration, leading to insulin resistance. This idea is supported by human microdialysis studies showing that obese compared with lean individuals have greater interstitial glycerol concentrations (5, 47), and that individuals with type 2 diabetes compared with normal glucose-tolerant controls have less inhibition of muscle and IMAT lipolysis in response to insulin (24). Additionally, when compared with SAT, lipolysis in muscle and IMAT is less inhibited in response to insulin (7, 36, 47) and increases more in response to fasting (17). IMAT mRNA expression of genes controlling lipolysis did not reveal coordinated changes suggestive of increased capacity for IMAT lipolysis in insulin-resistant individuals. However, basal lipolytic rates were significantly greater in VAT and IMAT compared with SAT. These data suggest that regulation of lipolysis, rather than capacity for lipolysis, may explain differences in basal lipolytic rates measured between tissues in vitro. IMAT’s high lipolytic activity and close proximity to muscle mean that it likely plays an important role in regulating local FFA concentration, and the increase in muscle interstitial glycerol found in insulin-resistant individuals.
Similar to a report on fibroadipogenic precursors from human skeletal muscle (1), our data suggest IMAT insulin sensitivity parallels whole body insulin sensitivity. Similar to reports of adipose tissue insulin resistance in other studies (3, 24), decreased IMAT insulin sensitivity would attenuate insulin inhibition of lipolysis and also contribute to increased interstitial FFA concentration. Moreover, we found key genes of the peroxisomal beta-oxidation pathway to be downregulated in obesity and T2D. If there were no adaptive increases in mitochondrial beta-oxidation or reesterification, this could contribute to increased interstitial FFA concentration (32). Together, these findings suggest IMAT may explain increased muscle lipolytic rates found in obese individuals that promote increased interstitial FFA concentration, leading to muscle lipid accumulation, and ultimately decreased muscle insulin sensitivity.
Visceral adipose tissue is thought to be uniquely deleterious toward insulin sensitivity (41). Importantly, the relationship between IMAT and insulin resistance is almost equally as strong as for VAT (51), and our conditioned media data revealed that the secretome of IMAT and VAT have a similar ability to decrease insulin sensitivity. Animal studies found low expression of mRNA for oxidative metabolism, and high expression of inflammatory cytokines in IMAT (29), as well as similar DNA methylation in VAT and IMAT with increased expression of IL-6, TNFα, and PAI-1 compared with subcutaneous adipose tissue (31). Therefore, it appears that IMAT is a uniquely regulated tissue that is functionally similar to VAT, and is capable of secreting cytokines known to influence tissue inflammation and function. We found that PAI-1 and MCP1 gene expression scaled to insulin resistance, as did TNFAIP3, which codes for a protein that limits TNFα-induced NFκB inflammation (9). Therefore, IMAT may be both a target and a source of inflammation in insulin-resistant humans.
The ability of IMAT to expand is limited by the morphology of muscle, and we reasoned may also be limited by the extracellular matrix (ECM), which is known to influence insulin sensitivity in adipose tissue, liver, and skeletal muscle (25, 50). It is possible that IMAT, being contained with the muscle fascia, may also be an example of adipose tissue with limitations for expansion with negative effects on insulin sensitivity. Our data indicate IMAT may secrete proteins that modify muscle ECM and decrease insulin sensitivity in humans. Therefore, IMAT may increase muscle fibrosis, and could influence the development of sarcopenia, age-related metabolic dysfunction, as well as the greater declines in these parameters in individuals with type 2 diabetes (38).
There are several limitations to this work. Individuals in our SAT group were significantly younger compared with the IMAT and skeletal muscle cohort, making it possible that the differences shown in Fig. 2 could be influenced by age. We do not have any measurements of total IMAT content in these volunteers and therefore cannot determine if IMAT gene expression was related to IMAT content to understand the biology of this tissue. Flash-frozen skeletal muscle biopsies from which IMAT was isolated for RNAseq analysis were thawed and quickly dissected on ice, which may have influenced the gene expression profile. We did not separate IMAT into various cell types as we wanted to study the tissue as a whole. It would have been ideal to measure stimulated IMAT lipolysis to evaluate the potential impact on muscle lipid delivery. Unfortunately, fresh tissue was not available for these measurements. Every attempt was made to prevent muscle contamination of IMAT, but it is possible that muscle was also measured along with IMAT in these analyses.
These data represent the first direct sampling and analysis of IMAT in humans. IMAT conditioned media decreased insulin sensitivity and caused accumulation of 1,2-DAG in primary myotubes with a potency similar to VAT. These data indicate that factors secreted by IMAT can contribute to the development of muscle insulin resistance. We found IMAT mRNA expression consistent with macrophage infiltration, cytokine, and extracellular matrix secretion, and decreased insulin signaling. Collectively, these data indicate that IMAT may be an important regulator of skeletal muscle insulin sensitivity whose importance we are just starting to appreciate.
GRANTS
This work was partially supported by the National Institutes of Health General Clinical Research Center Grant RR-00036; National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Grant R01-DK-089170 to B. C. Bergman and NIDDK Grant T32-DK-07658 to N. Krebs; Colorado Nutrition Obesity Research Center Grant P30-DK-048520; the Helmholtz Alliance ICEMED (Imaging and Curing Environmental Metabolic Diseases); the Network Fund of the Helmholtz Association; and the German Center for Diabetes Research (DZD).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
L.P. and B.C.B. conceived and designed research; S.S., S.Z., D.E.K., K.A.H., L.P., T.P., S.A.N., A.S., A.K., J.A.S., D.H.B., J.K., and B.C.B. performed experiments; S.S., S.Z., D.E.K., K.A.H., T.P., S.A.N., A.S., A.K., D.H.B., T.S., E.G., D.L., J.K., S.H., and B.C.B. analyzed data; S.S., S.Z., D.E.K., K.A.H., L.P., T.P., S.A.N., A.K., T.S., E.G., D.L., J.K., S.H., and B.C.B. interpreted results of experiments; S.S., S.H., and B.C.B. prepared figures; S.S., D.E.K., and B.C.B. drafted manuscript; S.S., S.Z., D.E.K., K.A.H., L.P., T.P., S.A.N., A.S., A.K., J.A.S., D.H.B., T.S., E.G., D.L., J.K., S.H., and B.C.B. edited and revised manuscript; S.S., S.Z., D.E.K., K.A.H., L.P., T.P., S.A.N., A.S., A.K., J.A.S., D.H.B., T.S., E.G., D.L., J.K., S.H., and B.C.B. approved final version of manuscript.
ACKNOWLEDGMENTS
Some of these data were presented as an abstract at the American Diabetes Association 77th Scientific Sessions in San Diego, CA, June 9–13, 2017.
REFERENCES
- 1.Arrighi N, Moratal C, Clément N, Giorgetti-Peraldi S, Peraldi P, Loubat A, Kurzenne JY, Dani C, Chopard A, Dechesne CA. Characterization of adipocytes derived from fibro/adipogenic progenitors resident in human skeletal muscle. Cell Death Dis 6: e1733, 2015. doi: 10.1038/cddis.2015.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergman BC, Cornier MA, Horton TJ, Bessesen DH. Effects of fasting on insulin action and glucose kinetics in lean and obese men and women. Am J Physiol Endocrinol Metab 293: E1103–E1111, 2007. doi: 10.1152/ajpendo.00613.2006. [DOI] [PubMed] [Google Scholar]
- 3.Bergman BC, Howard D, Schauer IE, Maahs DM, Snell-Bergeon JK, Clement TW, Eckel RH, Perreault L, Rewers M. The importance of palmitoleic acid to adipocyte insulin resistance and whole-body insulin sensitivity in type 1 diabetes. J Clin Endocrinol Metab 98: E40–E50, 2013. doi: 10.1210/jc.2012-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bessesen DH, Cox-York KA, Hernandez TL, Erickson CB, Wang H, Jackman MR, Van Pelt RE. Postprandial triglycerides and adipose tissue storage of dietary fatty acids: impact of menopause and estradiol. Obesity (Silver Spring) 23: 145–153, 2015. doi: 10.1002/oby.20935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaak EE, Schiffelers SL, Saris WH, Mensink M, Kooi ME. Impaired beta-adrenergically mediated lipolysis in skeletal muscle of obese subjects. Diabetologia 47: 1462–1468, 2004. doi: 10.1007/s00125-004-1471-y. [DOI] [PubMed] [Google Scholar]
- 6.Boettcher M, Machann J, Stefan N, Thamer C, Häring HU, Claussen CD, Fritsche A, Schick F. Intermuscular adipose tissue (IMAT): association with other adipose tissue compartments and insulin sensitivity. J Magn Reson Imaging 29: 1340–1345, 2009. doi: 10.1002/jmri.21754. [DOI] [PubMed] [Google Scholar]
- 7.Bolinder J, Kerckhoffs DA, Moberg E, Hagström-Toft E, Arner P. Rates of skeletal muscle and adipose tissue glycerol release in nonobese and obese subjects. Diabetes 49: 797–802, 2000. doi: 10.2337/diabetes.49.5.797. [DOI] [PubMed] [Google Scholar]
- 8.Bong JJ, Cho KK, Baik M. Comparison of gene expression profiling between bovine subcutaneous and intramuscular adipose tissues by serial analysis of gene expression. Cell Biol Int 34: 125–133, 2009. [DOI] [PubMed] [Google Scholar]
- 9.Coornaert B, Carpentier I, Beyaert R. A20: central gatekeeper in inflammation and immunity. J Biol Chem 284: 8217–8221, 2009. doi: 10.1074/jbc.R800032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeFronzo RA, Tobin JD, Andres R. Glucose clamp technique: a method for quantifying insulin secretion and resistance. Am J Physiol Endocrinol Metab 237: E214–E223, 1979. doi: 10.1152/ajpendo.1979.237.3.E214. [DOI] [PubMed] [Google Scholar]
- 11.Durheim MT, Slentz CA, Bateman LA, Mabe SK, Kraus WE. Relationships between exercise-induced reductions in thigh intermuscular adipose tissue, changes in lipoprotein particle size, and visceral adiposity. Am J Physiol Endocrinol Metab 295: E407–E412, 2008. doi: 10.1152/ajpendo.90397.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finegood DT, Bergman RN, Vranic M. Estimation of endogenous glucose production during hyperinsulinemic-euglycemic glucose clamps. Comparison of unlabeled and labeled exogenous glucose infusates. Diabetes 36: 914–924, 1987. doi: 10.2337/diab.36.8.914. [DOI] [PubMed] [Google Scholar]
- 13.Frantz C, Stewart KM, Weaver VM. The extracellular matrix at a glance. J Cell Sci 123: 4195–4200, 2010. doi: 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallagher D, Heshka S, Kelley DE, Thornton J, Boxt L, Pi-Sunyer FX, Patricio J, Mancino J, Clark JM; MRI Ancillary Study Group of Look AHEAD Research Group . Changes in adipose tissue depots and metabolic markers following a 1-year diet and exercise intervention in overweight and obese patients with type 2 diabetes. Diabetes Care 37: 3325–3332, 2014. doi: 10.2337/dc14-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallagher D, Kuznia P, Heshka S, Albu J, Heymsfield SB, Goodpaster B, Visser M, Harris TB. Adipose tissue in muscle: a novel depot similar in size to visceral adipose tissue. Am J Clin Nutr 81: 903–910, 2005. doi: 10.1093/ajcn/81.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaster M, Kristensen SR, Beck-Nielsen H, Schrøder HD. A cellular model system of differentiated human myotubes. APMIS 109: 735–744, 2001. doi: 10.1034/j.1600-0463.2001.d01-140.x. [DOI] [PubMed] [Google Scholar]
- 17.Gjedsted J, Gormsen LC, Nielsen S, Schmitz O, Djurhuus CB, Keiding S, Ørskov H, Tønnesen E, Møller N. Effects of a 3-day fast on regional lipid and glucose metabolism in human skeletal muscle and adipose tissue. Acta Physiol (Oxf) 191: 205–216, 2007. doi: 10.1111/j.1748-1716.2007.01740.x. [DOI] [PubMed] [Google Scholar]
- 18.Goodpaster BH, Krishnaswami S, Harris TB, Katsiaras A, Kritchevsky SB, Simonsick EM, Nevitt M, Holvoet P, Newman AB. Obesity, regional body fat distribution, and the metabolic syndrome in older men and women. Arch Intern Med 165: 777–783, 2005. doi: 10.1001/archinte.165.7.777. [DOI] [PubMed] [Google Scholar]
- 19.Goodpaster BH, Krishnaswami S, Resnick H, Kelley DE, Haggerty C, Harris TB, Schwartz AV, Kritchevsky S, Newman AB. Association between regional adipose tissue distribution and both type 2 diabetes and impaired glucose tolerance in elderly men and women. Diabetes Care 26: 372–379, 2003. doi: 10.2337/diacare.26.2.372. [DOI] [PubMed] [Google Scholar]
- 20.Goodpaster BH, Thaete FL, Kelley DE. Thigh adipose tissue distribution is associated with insulin resistance in obesity and in type 2 diabetes mellitus. Am J Clin Nutr 71: 885–892, 2000. doi: 10.1093/ajcn/71.4.885. [DOI] [PubMed] [Google Scholar]
- 21.Greco AV, Mingrone G, Giancaterini A, Manco M, Morroni M, Cinti S, Granzotto M, Vettor R, Camastra S, Ferrannini E. Insulin resistance in morbid obesity: reversal with intramyocellular fat depletion. Diabetes 51: 144–151, 2002. doi: 10.2337/diabetes.51.1.144. [DOI] [PubMed] [Google Scholar]
- 22.Hardin BJ, Campbell KS, Smith JD, Arbogast S, Smith J, Moylan JS, Reid MB. TNF-alpha acts via TNFR1 and muscle-derived oxidants to depress myofibrillar force in murine skeletal muscle. J Appl Physiol (1985) 104: 694–699, 2008. doi: 10.1152/japplphysiol.00898.2007. [DOI] [PubMed] [Google Scholar]
- 23.Harrison KA, Bergman BC. HPLC-MS/MS methods for diacylglycerol and sphingolipid molecular species in skeletal muscle. In: High Throughput Metabolomics and Protocols, edited by D’Alessandro A. New York: Springer Nature, 2019. [Google Scholar]
- 24.Jocken JW, Goossens GH, Boon H, Mason RR, Essers Y, Havekes B, Watt MJ, van Loon LJ, Blaak EE. Insulin-mediated suppression of lipolysis in adipose tissue and skeletal muscle of obese type 2 diabetic men and men with normal glucose tolerance. Diabetologia 56: 2255–2265, 2013. doi: 10.1007/s00125-013-2995-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45: D353–D361, 2017. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan T, Muise ES, Iyengar P, Wang ZV, Chandalia M, Abate N, Zhang BB, Bonaldo P, Chua S, Scherer PE. Metabolic dysregulation and adipose tissue fibrosis: role of collagen VI. Mol Cell Biol 29: 1575–1591, 2009. doi: 10.1128/MCB.01300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J, Heshka S, Gallagher D, Kotler DP, Mayer L, Albu J, Shen W, Freda PU, Heymsfield SB. Intermuscular adipose tissue-free skeletal muscle mass: estimation by dual-energy X-ray absorptiometry in adults. J Appl Physiol (1985) 97: 655–660, 2004. doi: 10.1152/japplphysiol.00260.2004. [DOI] [PubMed] [Google Scholar]
- 27.Konopka AR, Wolff CA, Suer MK, Harber MP. Relationship between intermuscular adipose tissue infiltration and myostatin before and after aerobic exercise training. Am J Physiol Regul Integr Comp Physiol 315: R461–R468, 2018. doi: 10.1152/ajpregu.00030.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laurens C, Louche K, Sengenes C, Coué M, Langin D, Moro C, Bourlier V. Adipogenic progenitors from obese human skeletal muscle give rise to functional white adipocytes that contribute to insulin resistance. Int J Obes 40: 497–506, 2016. doi: 10.1038/ijo.2015.193. [DOI] [PubMed] [Google Scholar]
- 29.Lee HJ, Park HS, Kim W, Yoon D, Seo S. Comparison of metabolic network between muscle and intramuscular adipose tissues in hanwoo beef cattle using a systems biology approach. Int J Genomics 2014: 679437, 2014. doi: 10.1155/2014/679437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehr S, Hartwig S, Lamers D, Famulla S, Müller S, Hanisch FG, Cuvelier C, Ruige J, Eckardt K, Ouwens DM, Sell H, Eckel J. Identification and validation of novel adipokines released from primary human adipocytes. Mol Cell Proteomics 11: M111.010504, 2012. doi: 10.1074/mcp.M111.010504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li M, Wu H, Wang T, Xia Y, Jin L, Jiang A, Zhu L, Chen L, Li R, Li X. Co-methylated genes in different adipose depots of pig are associated with metabolic, inflammatory and immune processes. Int J Biol Sci 8: 831–837, 2012. doi: 10.7150/ijbs.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lodhi IJ, Semenkovich CF. Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metab 19: 380–392, 2014. doi: 10.1016/j.cmet.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo W, Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29: 1830–1831, 2013. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics 10: 161, 2009. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miljkovic-Gacic I, Gordon CL, Goodpaster BH, Bunker CH, Patrick AL, Kuller LH, Wheeler VW, Evans RW, Zmuda JM. Adipose tissue infiltration in skeletal muscle: age patterns and association with diabetes among men of African ancestry. Am J Clin Nutr 87: 1590–1595, 2008. doi: 10.1093/ajcn/87.6.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moberg E, Sjöberg S, Hagström-Toft E, Bolinder J. No apparent suppression by insulin of in vivo skeletal muscle lipolysis in nonobese women. Am J Physiol Endocrinol Metab 283: E295–E301, 2002. doi: 10.1152/ajpendo.00339.2001. [DOI] [PubMed] [Google Scholar]
- 37.Murphy JC, McDaniel JL, Mora K, Villareal DT, Fontana L, Weiss EP. Preferential reductions in intermuscular and visceral adipose tissue with exercise-induced weight loss compared with calorie restriction. J Appl Physiol (1985) 112: 79–85, 2012. doi: 10.1152/japplphysiol.00355.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park SW, Goodpaster BH, Strotmeyer ES, de Rekeneire N, Harris TB, Schwartz AV, Tylavsky FA, Newman AB. Decreased muscle strength and quality in older adults with type 2 diabetes: the health, aging, and body composition study. Diabetes 55: 1813–1818, 2006. doi: 10.2337/db05-1183. [DOI] [PubMed] [Google Scholar]
- 39.Perreault L, Newsom SA, Strauss A, Kerege A, Kahn DE, Harrison KA, Snell-Bergeon JK, Nemkov T, D’Alessandro A, Jackman MR, MacLean PS, Bergman BC. Intracellular localization of diacylglycerols and sphingolipids influences insulin sensitivity and mitochondrial function in human skeletal muscle. JCI Insight 3: e96805, 2018. doi: 10.1172/jci.insight.96805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Popadic Gacesa JZ, Kozic DB, Grujic NG. Triceps brachii strength and regional body composition changes after detraining quantified by MRI. J Magn Reson Imaging 33: 1114–1120, 2011. doi: 10.1002/jmri.22548. [DOI] [PubMed] [Google Scholar]
- 41.Pouliot MC, Després JP, Nadeau A, Moorjani S, Prud’Homme D, Lupien PJ, Tremblay A, Bouchard C. Visceral obesity in men. Associations with glucose tolerance, plasma insulin, and lipoprotein levels. Diabetes 41: 826–834, 1992. doi: 10.2337/diab.41.7.826. [DOI] [PubMed] [Google Scholar]
- 42.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47, 2015. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140, 2010. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25, 2010. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitz-Peiffer C, Craig DL, Biden TJ. Ceramide generation is sufficient to account for the inhibition of the insulin-stimulated PKB pathway in C2C12 skeletal muscle cells pretreated with palmitate. J Biol Chem 274: 24202–24210, 1999. doi: 10.1074/jbc.274.34.24202. [DOI] [PubMed] [Google Scholar]
- 46.Sinha R, Dufour S, Petersen KF, LeBon V, Enoksson S, Ma YZ, Savoye M, Rothman DL, Shulman GI, Caprio S. Assessment of skeletal muscle triglyceride content by (1)H nuclear magnetic resonance spectroscopy in lean and obese adolescents: relationships to insulin sensitivity, total body fat, and central adiposity. Diabetes 51: 1022–1027, 2002. doi: 10.2337/diabetes.51.4.1022. [DOI] [PubMed] [Google Scholar]
- 47.Sjöstrand M, Gudbjörnsdottir S, Holmäng A, Strindberg L, Ekberg K, Lönnroth P. Measurements of interstitial muscle glycerol in normal and insulin-resistant subjects. J Clin Endocrinol Metab 87: 2206–2211, 2002. doi: 10.1210/jcem.87.5.8495. [DOI] [PubMed] [Google Scholar]
- 48.Song MY, Ruts E, Kim J, Janumala I, Heymsfield S, Gallagher D. Sarcopenia and increased adipose tissue infiltration of muscle in elderly African American women. Am J Clin Nutr 79: 874–880, 2004. doi: 10.1093/ajcn/79.5.874. [DOI] [PubMed] [Google Scholar]
- 49.Terry JG, Shay CM, Schreiner PJ, Jacobs DR Jr, Sanchez OA, Reis JP, Goff DC Jr, Gidding SS, Steffen LM, Carr JJ. Intermuscular Adipose Tissue and Subclinical Coronary Artery Calcification in Midlife: The CARDIA Study (Coronary Artery Risk Development in Young Adults). Arterioscler Thromb Vasc Biol 37: 2370–2378, 2017. doi: 10.1161/ATVBAHA.117.309633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams AS, Kang L, Wasserman DH. The extracellular matrix and insulin resistance. Trends Endocrinol Metab 26: 357–366, 2015. doi: 10.1016/j.tem.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yim JE, Heshka S, Albu J, Heymsfield S, Kuznia P, Harris T, Gallagher D. Intermuscular adipose tissue rivals visceral adipose tissue in independent associations with cardiovascular risk. Int J Obes 31: 1400–1405, 2007. doi: 10.1038/sj.ijo.0803621. [DOI] [PMC free article] [PubMed] [Google Scholar]