Abstract
Lung cancer has the lowest survival rate spread globally resulting in a large number of deaths. This is attributed to insufficient measures such as lack of early detection and chemoresistance in the patients. It can be subdivided into two histological groups: Non-Small-Cell Lung Cancer (NSCLC), which is most prevalent (85% of all lung cancers) but less destructive; and Small-Cell Lung Cancer (SCLC), which is intermittently metastatic and less prevalent (15% of all lung cancers). The present study deals with the analysis of gene expression of two subtypes to identify the Differentially Expressed Genes (DEGs). For this study, we selected two datasets from the Omnibus database, which included 50 non-small cell lung cancer samples, 31 small cell lung cancer samples, and 48 samples from normal lung tissue. After DEGs identification using the meta-analysis approach, they were then subjected to further analysis following p-value adjustment via the Benjamini-Hochberg method. We identified 440 overexpressed and 489 underexpressed genes in NSCLC, and 489 overexpressed and 525 underexpressed genes in SCLC, compared with normal lung tissues. Furthermore, we identified 3 overlapping genes between upregulated DEGs in NSCLC and downregulated DEGs in SCLC; and 8 overlapping genes between upregulated DEGs in SCLC and downregulated DEGs in NSCLC. Accordingly, a Protein-Protein Interaction (PPI) network of the overlapping genes was generated, which contained a total of 261 genes, of which the top five were TRIM29, ANK3, CSTA, FGG, and AGR2. These five candidate genes reported herein may prove to be potential therapeutic targets.
Keywords: Systems biology, Bioinformatics, Biological science, Gene expression
1. Introduction
Smoking, pollution and unhealthy toxic environment are the most prominent causes of lung cancer causing deaths world over [1]. Despite glaring advancements in the area of lung cancer-related treatment settings, its rate of cure remains low. This is attributed to several factors such as delayed diagnosis, impoverished prognosis, and enhanced drug resistance. Based on histology, lung cancer is classified into Small-Cell Lung Cancer (SCLC) and Non-Small Cell Lung Cancer (NSCLC) [2]. SCLC is a fatal tumor of epithelial cells. The cellular morphology of SCLC comprises cramped cells including inconspicuous nucleoli, insufficient cytoplasm, obscured cellular margins and extreme granular nuclear chromatin [3]. SCLC has an expeditious growth rate, exaggerated initial response rates, early metastases and a strong association with smoking [4, 5]. NSCLC consists of adenocarcinoma, large cell carcinoma and squamous cell (epidermoid) carcinoma. Adenocarcinoma and large cell carcinoma are peripheral tumors originating from the obscured bronchi, bronchioles, or alveoli of lungs, whereas Squamous cell carcinoma possesses a central origin [6]. Squamous cell carcinoma displays delayed development of distant metastases and is described by hemoptysis or obstructive pneumonia and lobar collapse. On the other hand, few primary tumors of adenocarcinoma are peripheral lesions without any symptoms related to primordial metastases development. Large cell carcinoma exhibits populous peripheral masses, along with occasional cavitation [7]. Lung cancer is the result of augmentation of several genetic and epigenetic modifications, which could be due to multiple reasons [5]. Protracted exposure to carcinogens such as tobacco smoke or asbestos is the customarily identified reason for such alterations. Identification and determination of new diagnostic or prognostic biomarkers, along with the evolution of innovative therapeutic approaches for lung cancer is a foremost upcoming area of translational cancer research. Nonetheless, there are certain limitations to the above owing to the scarcity of complete understanding of the heterogeneous nature of the tumor and involvement of multiple factors in the process of lung carcinogenesis. Lucrative methods of molecular testing for early stage diagnosis also require an extensive understanding of molecular events involved in tumorigenesis and monitoring the expression of one or few genes would not be much helpful [8]. A comprehensive genetic analysis would thus, be more beneficial in elucidating the complex disease. High throughput gene expression analysis has recently come into the limelight in this direction and has enhanced the likelihood of identifying molecular events associated with lung carcinogenesis. A large number of studies have reported multiple plausible biomarkers of cancer and the classification of lung carcinomas on the basis of their gene expression profile. Therefore, the biological connotation of extensive microarray data seems to be a great challenge at this point in time. Microarray technology is a high throughput and highly cost-effective technique as it measures the mRNA levels of several thousands of genes simultaneously [9]. Certain highly significant molecular signature has been identified by microarray technology, and they are currently evaluated in prospective randomized clinical trials. Despite the advantages of microarray technology, there are several studies reporting irreproducibility and non-robustness of the technique, even with moderate alterations. Incompetent reporting of methods, inadequate control of false positives and improper analysis or validation are the most common causes of irreproducibility of the technique. Moreover, gene expression profiling experiments are customarily scrutinized in solitude and are barred by a small number of samples [10, 11, 12]. Thus, the widespread application requires a pre-assessment of generalizability across broad studies. This is where meta-analysis comes into action. A meta-analysis is a combinatorial approach for bringing together the information from multiple extant studies to aggravate the authenticity and generalizability of results from separate analogous studies [12]. However, meta-analysis is not just statistical technique, but a wide description of the whole study process. It increases the statistical competency, leading to the generation of a higher explicit assessment of differential gene expression and evaluation of diversification of the long-term projection. Comprehensive utilization of already available data makes it a relatively inexpensive approach. A scrupulous meta-analysis combining multiple data of a large number of patient populations from several platforms, institutions and numerous methods of data procurement would unravel functionally relevant genes that may have been differently neglected by the secluded analysis of analogous studies of gene expression [13]. It has been customarily observed that there are compelling genetic and molecular perturbations in cells during the process of carcinogenesis. These changes can be dynamic, permanent or irreversible and may cause momentous changes in gene expression. Thus, the detection of these genetic changes would contribute to providing diagnosis markers. Most of the contemporary studies are based on classifying NSCLC or identifying diagnostic/prognostic biomarkers depending upon the gene expression profile [14, 15, 16, 17, 18]. Here, the objective is to select gene expression differentials between the two subtypes (NSCLC and SCLC) of lung cancer through meta-analysis of mRNA expression profiles from different studies to overcome the limitations of individual studies. These gene expression differentials were further functionally enriched to identify the perturbations in regulating pathways, which would further enhance the understanding of the relationship between SCLC and NSCLC besides providing a new context to the research. A Protein-Protein Interaction (PPI) network was constructed to further understand and predict the biological activity of the selected gene expression differentials at the molecular level.
2. Materials and methods
2.1. Gene expression dataset selection for meta-analysis
Gene Expression Omnibus (GEO) [19] databases were exhaustively searched for microarray datasets. The selection process was based on the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines published in 2009 [20]. Inclusion criteria set for the selection of datasets: control studies involving human cases include both lung cancer subtypes (NSCLC and SCLC), comparable conditions, data squeak (both raw and processed data), datasets with greater than 20 samples. Review articles, non-human studies and combined analysis of expression profiles were precluded.
2.2. Data pre-processing
Data pre-processing operations including normalization and probe ID mapping were performed preceding the meta-analysis. The unprocessed CEL info was redressed and graded via Robust Multichip Average using the Affy [21] and Oligo [22] packages in R statistical software to procure the respective probe expression data. After retrieving the data, we created a tab-delimited input text file of normalized intensities for further analysis. The gene probe IDs from each study were mapped to Ensembl gene IDs using different tools such as Synergizer [23], gProfiler id converter [24], DB2DB conversion [25] and GEO2R [19]. It is customarily found that numerous probes map to a specific Gene Symbols for the genes with multifarious splice variants. Thus, the average expression value of such probes was used for gene mapping.
2.3. Meta-analysis of microarray datasets
R statistical software (http://www.r-project.org/) using metaMA [26], limma [27] and MAMA [28] packages were used to conduct a meta-analysis of the gene expression profiles obtained from the selected datasets. This software followed the t-test and p-value combining method for performing meta-analysis. We used the False discovery rate (FDR) approach, as given in Benjamini-Hochberg (BH) method [29] to adjust the p-values. The genes with p-value, less than 0.05 and fold change greater than 2 were selected as differentially expressed genes (DEGs) between normal and cancerous tissue samples.
2.4. Functional enrichment analysis
DEGs identified after meta-analysis were subjected to functional enrichment analysis, in order to understand their biological implications. The gene ontology (GO) [30, 31] function and the Kyoto Encyclopedia of Gene and Genomes (KEGG) [32, 33, 34] pathway enrichment analysis was carried out using the contrivances available in the Database for Annotation, Visualization and Integrated Discovery (DAVID; David.abcc.ncifcrf.gov) [35]. A BH-corrected p-value of less than 0.05 was used as the parameter to select the significantly enriched KEGG pathway.
2.5. PPI network analysis
The identified DEGs were subjected to create protein-protein interaction network using Cytoscape (www.cytoscape.org) [36]. The proteins encoded by DEGs and their interactions with other proteins were computed from the Biological General Repository for Interacting Datasets (BioGRID, http://thebiogrid.org/) [37, 38].
3. Results
3.1. Selection and normalization of microarray datasets
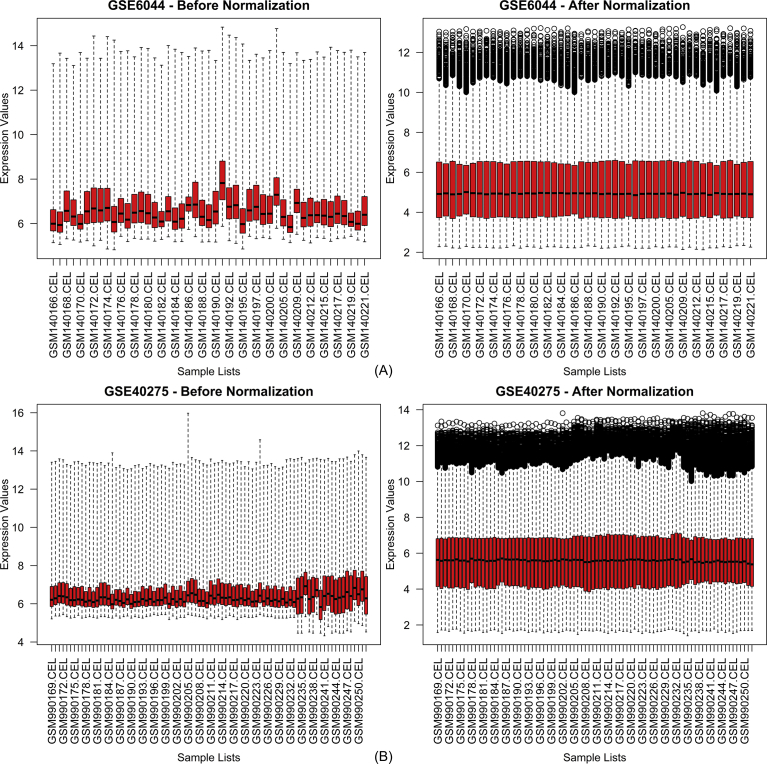
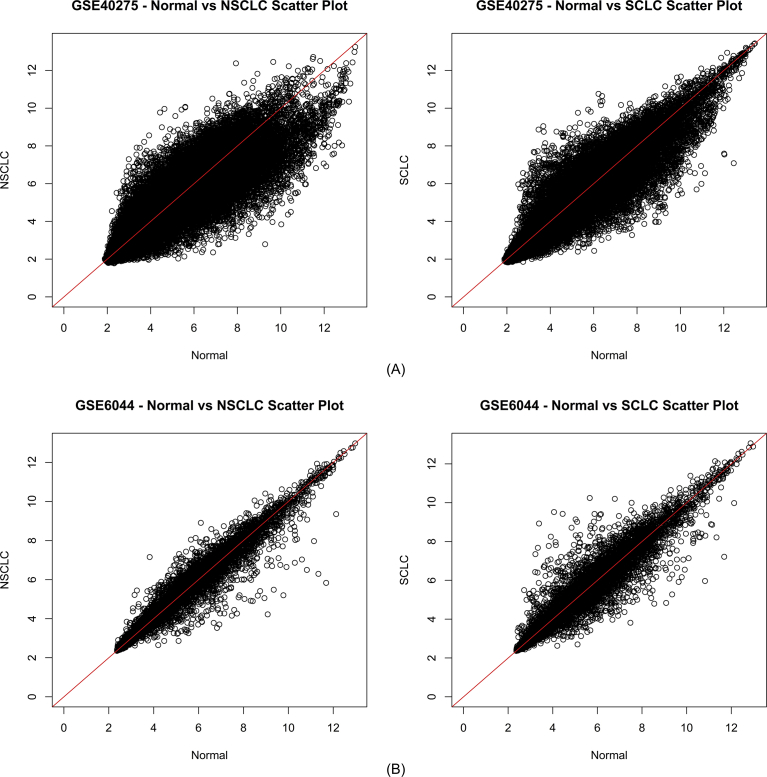
We selected two datasets with accession numbers GSE6044 and GSE40275. The two datasets consisted of a total of 131 samples; 50 of which were non-small cell lung cancer, 31 were small cell lung cancer, 48 were normal tissues, which include 5 samples from different control patients without tumor and rest had been purchased from OriGene technologies and 2 samples were from carcinoma tissues, but their disease type was not defined, so we excluded them, and the total samples we used in this study were 129. The required dossiers were extricated from exclusive studies: platform, type of samples, GEO accession number, number of case and controls, and gene expression profile (Table 1). Significant differential distribution of data was observed before and after normalization of the datasets. They were demonstrated by a box plot of intensity sample CEL files of the two datasets (Fig. 1). Relative changes in differential expression were clearly identified by intensity scatter plots of normal vs NSCLC and normal vs SCLC pertaining to the two datasets (Fig. 2).
Table 1.
Characteristics of individual studies retrieved from GEO Database included in meta-analysis.
Fig. 1.
Distribution of Expression Data Before and After Normalization in (A) GSE6044 and (B) GSE40275 . X-axis represents the sample lists; Y-axis represents expression values. Before normalization box plot shows median of data at different levels whereas after n normalization median is adjusted.
Fig. 2.
Intensity Scattered Plot showing the relationship between expression values of Normal vs NSCLC and Normal vs SCLC in datasets (A) GSE40275 and (B) GSE6044. (A) represents the comparison of expression value of genes in dataset GSE40275 between normal and NSCLC samples and between normal and SCLC samples. (B) represents the comparison of expression value of genes in dataset GSE6044 between normal and NSCLC samples and between normal and SCLC samples. x-axis represents the expression value of normal samples whereas y-axis represents the expression values of diseased samples.
3.2. A set of genes differentially expressed in SCLC and NSCLC
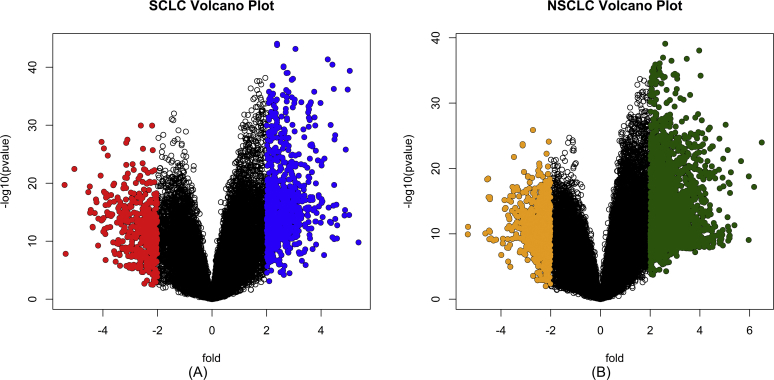
A total of 1,943 genes were identified as differentially expressed ones in both the datasets, including 1,014 DEGs in SCLC samples and 929 DEGs in NSCLC samples. DEGs were identified following more than 2.0-fold enrichment (fold change, biological significance) over random expectation (P < 0.05, statistical significance). Using the same criteria for screening - BH-corrected p-value, less than 0.05 and FC (fold change) of more than 2, an integral 489 DEGs were endowed as up-regulated and 525 DEGs were down-regulated in SCLC samples, whereas 440 DEGs were up-regulated and 489 DEGs were down-regulated in NSCLC samples. Our data reports a momentous negative regulation of several genes in both NSCLC and SCLC groups. However, SCLC group had highly deregulated genes as compared to the NSCLC one (Fig. 3). The leading 10 upregulated and downregulated genes in both NSCLC and SCLC are listed in Table 2. Genes were stacked acceding the fold change, superseded by adjustment of the corresponding p-values using the Benjamini-Hochberg procedure, positioning the false discovery rate.
Fig. 3.
Volcano plot highlighting DEGs: (A) A fold vs -log10(p-value) plot, highlighting DEGs in SCLC, indicates that the down-regulated DEGs highlighted with blue color are more in number than the up-regulated DEGs highlighted with red color. The second fold vs -log10(p-value) plot (B) highlights DEGs in NSCLC, up-regulated DEGs are highlighted with orange color and down-regulated DEGs are highlighted with green color. X-axis represents the fold change (log2 scale) and Y-axis represents the p-value (-log10 scale).
Table 2.
Top 10 upregulated and downregulated DEGs in NSCLC and SCLC.
| Genes |
BH-p-value |
Fold change |
Genes |
BH-p-value |
Fold change |
|---|---|---|---|---|---|
| Upregulated DEGs (SCLC) | Downregulated DEGs (SCLC) | ||||
| AGER | 8.40E-29 | 5.070 | TMSB15B | 0.000902695 | −5.462 |
| SFTPC | 1.77E-07 | 4.982 | BEX1 | 4.07E-14 | −5.418 |
| GSTA1 | 0.0016756 | 4.858 | DLK1 | 0.0076076 | −4.984 |
| CYP2B7P | 2.49E-22 | 4.732 | TOP2A | 3.80E-16 | −4.926 |
| AQP4 | 8.46E-11 | 4.712 | ASCL1 | 6.08E-06 | −4.883 |
| CLDN18 | 3.94E-17 | 4.682 | HIST1H3C | 4.49E-10 | −4.721 |
| C4BPA | 1.87E-11 | 4.658 | HIST1H3B | 4.35E-17 | −4.671 |
| ADH1B | 1.03E-12 | 4.643 | DCX | 2.89E-10 | −4.557 |
| LRRK2 | 2.53E-10 | 4.582 | TUBB2B | 4.59E-14 | −4.472 |
| MSMB |
0.00387654 |
4.473 |
HIST1H3I |
2.63E-18 |
−4.415 |
| Upregulated DEGs (NSCLC) |
Downregulated DEGs (NSCLC) |
||||
| MSMB | 1.95E-10 | 5.855 | TOP2A | 1.63E-17 | −4.515 |
| SFTPC | 3.66E-09 | 4.866 | SPP1 | 8.22E-15 | −4.464 |
| SCARNA17 | 1.58E-08 | 4.550 | RPS27 | 3.86E-10 | −4.444 |
| GSTA1 | 1.35E-09 | 4.326 | LOC727900 | 6.24E-09 | −4.302 |
| AQP1 | 9.81E-13 | 4.278 | ANLN | 1.65E-14 | −3.858 |
| AGER | 1.39E-23 | 4.235 | TPX2 | 2.89E-13 | −3.847 |
| TCF21 | 3.50E-22 | 4.226 | LOC727929 | 1.29E-10 | −3.744 |
| ADH1B | 2.37E-11 | 4.130 | CKS2 | 2.24E-12 | −3.713 |
| SLC6A4 | 1.53E-31 | 4.028 | LOC732426 | 5.52E-06 | −3.711 |
| CENPVL3 | 2.01E-09 | 3.991 | DLGAP5 | 4.55E-12 | −3.669 |
3.3. Overlapping DEGs of NSCLC and SCLC
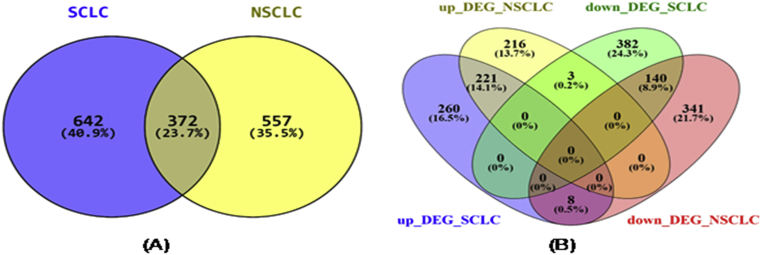
To find out how many of the genes differentially expressed are specific to each group, we checked for the overlap between a differentially expressed gene in non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). For this purpose, we used the Venny 2.1.0 (http://bioinfogp.cnb.csic.es/tools/venny/). 642 genes were included exclusively in “SCLC”, 557 genes were included exclusively in “NSCLC” and 372 genes were common in “NSCLC” and “SCLC”. 221 genes were commonly up-regulated in NSCLC and SCLC whereas 140 genes were commonly down-regulated in NSCLC and SCLC (Fig. 4A). 3 genes (GUSBP8, CHL1, CXCL1) were common between up-regulated DEGs in NSCLC and down-regulated DEGs in SCLC and 8 genes (FGG, IL33, AGR2, ANK3, CSTA, FABP6, S100P, TRIM29) were common between up-regulated DEGs in SCLC and down-regulated DEGs in NSCLC (Fig. 4B).
Fig. 4.
(A) Venn diagram showing overlap in the number of genes identified as differentially expressed in SCLC and NSCLC. As shown in figure (A), 642 genes were included exclusively in “SCLC”, 557 genes were included exclusively in “NSCLC” and 372 genes were common in “NSCLC” and “SCLC”. Blue circle denotes the number of DEGs in SCLC group and yellow circle denotes number of DEGs in NSCLC group. (B) Venn diagram showing overlap between the up-regulated and down-regulated differentially expressed genes in SCLC and NSCLC. As shown in Figure (B), 221 genes were commonly up-regulated in NSCLC group and SCLC group whereas 140 genes were commonly down-regulated in NSCLC group and SCLC group and also 3 genes (GUSBP8, CHL1, CXCL1) were common between up-regulated DEGs in NSCLC and down-regulated DEGs in SCLC and 8 genes (FGG, IL33, AGR2, ANK3, CSTA, FABP6, S100P, TRIM29) were common between up-regulated DEGs in SCLC and down-regulated DEGs in NSCLC. The four ovals highlighted with different colors represent the type of differential expression pattern. Blue color represents up-regulated DEGs in SCLC, yellow color represents up-regulated DEGs in NSCLC, green denotes down-regulated DEGs in SCLC and red shows down-regulated DEGs in NSCLC.
3.4. Characterization of DEGs
We classified the DEGs into different functional categories of cellular integrals, biological systems, and molecular functions with a compelling threshold of <0.05, based on the GO hierarchy and KEGG pathway. The DEGs in the small cell lung cancer group were significantly enriched in the following GO terms (most significant) under the biological process's category (descending order): ‘DNA replication’ (GO:0006260), ‘cell division’ (GO:0051301) and ‘DNA replication initiation’ (GO:0006270). ‘Protein binding (GO:0005515) and ‘extracellular space’ (GO:0005615) were highly enriched GO terms under the molecular functions and cellular components categories. The most enriched KEGG pathway terms in which the DEGs in the small cell lung cancer group were convincingly enriched (in descending order): ‘Systemic lupus erythematosus’ (hsa05322), ‘DNA replication’ (hsa03030) and ‘Cell cycle’ (hsa04110) (Table 3). On the other hand, the DEGs in the non-small cell lung cancer group were highly important for the following GO terms (most significant) under the biological processes such as ‘cell division’ (GO:0051301) ‘mitotic nuclear division’ (GO:0007067) and ‘xenobiotic glucuronidation’ (GO:0052697). The most convincing GO terms under the molecular functions and cellular component categories were ‘protein binding’ (GO:0005515) and ‘extracellular exosome’ (GO:0070062). The most enriched KEGG pathway terms in which the DEGs in the non-small cell lung cancer group were significantly enriched (in descending order): ‘Ascorbate and aldarate metabolism, (has: 00053) ‘Drug metabolism-cytochrome P450’ (hsa00982) and ‘Pentose and glucuronate interconversions’ (hsa00040) (Table 4).
Table 3.
Functional enrichment analysis representing top 10 GO terms and pathways of DEGs in the SCLC group. Enriched terms were ranked based on the BH-adjusted p-value.
| GO ID | GO term | No. of Genes | BH-p-value |
|---|---|---|---|
| Biological Process | |||
| GO:0006260 | DNA replication | 38 | 2.82E-11 |
| GO:0051301 | Cell division | 59 | 1.96E-11 |
| GO:0006270 | DNA replication initiation | 17 | 1.33E-09 |
| Molecular Functions | |||
| GO:0005515 | Protein binding | 559 | 3.30E-09 |
| GO:0046982 | Protein heterodimerization activity | 60 | 1.12E-07 |
| GO:0042393 | Histone binding | 22 | 4.28E-04 |
| Cellular Components | |||
| GO:0005615 | Extracellular space | 144 | 7.73E-15 |
| GO:0070062 | Extracellular exosome | 242 | 5.36E-15 |
| GO:0000786 |
Nucleosome |
30 |
2.94E-13 |
| KEGG ID |
KEGG pathway |
No. of Genes |
BH-p-value |
| hsa05322 | Systemic lupus erythematosus | 37 | 1.17E-11 |
| hsa03030 | DNA replication | 18 | 1.33E-09 |
| hsa04110 | Cell cycle | 29 | 2.23E-07 |
| hsa04640 | Hematopoietic cell lineage | 20 | 4.25E-05 |
| hsa04512 | ECM-receptor interaction | 16 | 0.0113025 |
Table 4.
Functional enrichment analysis representing the top 10 GO terms and pathways of DEGs in the NSCLC group. Enriched terms were ranked based on the BH-adjusted p-value.
| GO ID | GO term | No. of Genes | BH-p-value |
|---|---|---|---|
| Biological Processes | |||
| GO:0051301 | Cell division | 55 | 3.55E-13 |
| GO:0007067 | Mitotic nuclear division | 42 | 1.28E-10 |
| GO:0052697 | Xenobiotic glucuronidation | 9 | 1.01E-07 |
| Molecular Functions | |||
| GO:0005515 | Protein binding | 459 | 6.17E-10 |
| GO:0044822 | Poly(A) RNA binding | 87 | 1.49E-05 |
| GO:0005178 | Integrin binding | 19 | 9.27E-05 |
| Cellular Components | |||
| GO:0070062 | Extracellular exosome | 215 | 5.72E-18 |
| GO:0005654 | Nucleoplasm | 188 | 7.68E-10 |
| GO:0005615 |
Extracellular space |
108 |
6.86E-09 |
| KEGG ID |
KEGG pathway |
No. of Genes |
BH-p-value |
| hsa00053 | Ascorbate and aldarate metabolism | 11 | 1.25E-04 |
| hsa00982 | Drug metabolism – cytochrome P450 | 16 | 1.92E-04 |
| hsa00040 | Pentose and glucuronate interconversions | 11 | 8.21E-04 |
| hsa05150 | Staphylococcus aureus infection | 13 | 9.92E-04 |
| hsa04110 | Cell cycle | 20 | 9.99E-04 |
3.5. PPI network analysis
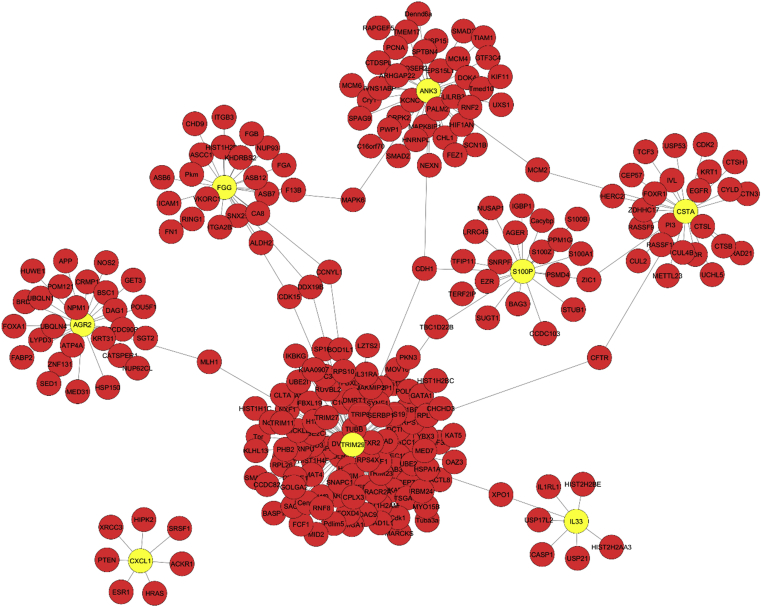
The PPI network generated for 8 DEGs (CXCL1, FGG, IL33, AGR2, ANK3, CSTA, S100P, TRIM29) by Cytoscape software included 261 nodes and 265 edges as shown in Fig. 5. Nodes represent proteins, edges represent the interaction between two proteins. The higher the node shape, the greater the degree of connection. The significant hub proteins containing TRIM29 (Tripartite Motif-Containing Protein 29, Degree = 119), ANK3 (Ankyrin 3, Degree = 42) and CSTA (Cystatin A, Degree = 31). Top five genes having a higher number of interacting partners were identified (Table 5).
Fig. 5.
Protein-protein interaction (PPI) network: This PPI-interaction network was constructed from the 8 overlapping differentially expressed genes (DEGs). A total of 261 proteins participated in this network. Yellow nodes represent 8 DEGs.
Table 5.
Degree of the top 10 genes in the protein-protein interaction network.
| Gene Symbol | Degree |
|---|---|
| TRIM29 | 119 |
| ANK3 | 42 |
| CSTA | 31 |
| FGG | 27 |
| AGR2 | 27 |
| S100P | 24 |
| IL33 | 10 |
| CXCL1 | 9 |
| CDH1 | 3 |
| SPTBN4 | 2 |
4. Discussion and conclusion
Lung cancer is a foremost cause of deaths globally. Currently available options of treatment are restricted due to the chemoresistance and resurgence of recurrence. The two different subtypes of cancers (SCLC and NSCLC) are distinctly different with respect to their characteristic features. These characteristics can be explored to understand the magnitude of the disease. This would serve as a tool for identifying effective therapeutic strategies for the disease. Differential expression analysis is the most commonly used method for identification of aberrantly expressed genes in disease. This analysis utilizes several statistical approaches such as t-tests of cohorts to identify differences in the level of expression between diseased and normal individuals. These differentially expressed genes are then disintegrated into specific dysregulated pathways. Despite the clear expediency of approach, it is limited by a high level of noise in the gene expression data, reproducibility of the results and individual differences due to factors such as age, gender, genotype, and disease stage. Moreover, different stages of treatment, differences in cohort and experimental methods may also result in disparities between studies. Thus, meta-analysis of statistically combining multiple studies is a more powerful tool to abode these issues and elicitate the relevant information from multiple datasets. This would result in identifying disease signatures that are largely consistent across several studies, enhancing the potential of the technique. In order to evaluate the unique contribution of meta-analysis in identifying significant differences in two subtypes of lung cancer in this study, we first applied inclusion criteria to identify datasets available in each subtype specific dataset. All the required information was extracted, and data was normalized before processing it by meta-analysis. We then performed meta-analysis using R-statistical software and identified 1,943 differentially expressed genes, among which 1,014 were differentially expressed in SCLC and 929 were differentially expressed in NSCLC. These genes were then evaluated for GO and KEGG pathway functional enrichment with Padj. < 0.05 for each method. The DEGs found in SCLC group were enriched for DNA replication, cell division, and DNA replication initiation by GO enrichment and; in Systemic lupus erythematosus, DNA replication and cell cycle by KEGG pathway enrichment. DEGs found in NSCLC group were exigently enriched for cell division, mitotic nuclear division, and xenobiotic glucuronidation by GO enrichment and cytochrome p450 and Pentose and glucuronate interconversions by KEGG pathway enrichment. Notably, GO functional enrichment results in a considerably smaller set of enriched GO terms, mostly falling into metabolic, cell division, cell cycle and DNA replication categories. We identified selective biological signals associated with different subtypes of lung cancer. On further examining the differential expression of genes in detail, we found that 3 genes (GUSBP8, CHL1, CXCL1) were common between upregulated DEGs in NSCLC and downregulated DEGs in SCLC and 8 genes (FGG, IL33, AGR2, ANK3, CSTA, FABP6, S100P, and TRIM29) were common between upregulated DEGs in SCLC and downregulated DEGs in NSCLC. These are important findings as they provide an opportunity to look for consensus of the genes affected in two different types of lung cancer. Besides using these genes for diagnostic purposes, they may prove to be authentic targets for therapeutics and for eventual management of lung cancer.
CHL1 is a cell adhesion molecule which acts as a helicase protein during the interphase stage of mitosis during cell cycle. It is known to promote invasion and metastasis in other types of cancers such as breast, colon and ovary. It acts as tumor suppressive or oncogenic factor depending upon the stage and types of cancer [39]. Thus, it might act as a diagnostic biomarker for NSCLC differentiating the stages of tumor growth. GUSBP8, a marker of mature β cells and upregulation of GUSBP8 suggests its role in immune evasion by cancer cells. CXCL1, a small cytokine associated with processes of arteriogenesis, angiogenesis, inflammation and tumorigenesis [40]. Upregulation of CXCL1 might be correlated with aggressiveness of the tumor as infiltration of cytokines at the tumor site promotes tumorigenesis. They also direct the tumor cells to metastatic sites by enhancing angiogenesis of tumor cells. Thus, their expression levels might indicate the aggressiveness of NSCLCs.
On the other hand, FGG, a gene encoding γ component of fibrinogen, has vasonstriction and chemotactic activities that regulates cell adhesion and spreading [41]. Upregulation of the gene in SCLC can be correlated with high metastatic capability of the subtype. IL-33 is another cytokine upregulated in SCLC, can be associated with high initial response rates, as it signals inflammatory cascades by acting upon macrophages, neutrophils and B cells which are the early responding cells of the immune system [42]. Upregulation of AGR2 (anterior gradient 2, also called adenocarcinoma antigen) can be associated with survival, as it is correlated with reduced p53 response and enhanced cell migration and transformation [43]. ANK3, a proteoglycan plays a key role in cellular motility, proliferation, activation, contact and maintenance of specialized membrane domains [44]. Upregulation of ANK3 can be associated with typical cellular morphology of SCLC tumors. Upregulation of CSTA (cystatin A), a stefin that forms tight complexes with papain and cathepsin B, H and L, acts as a cysteine protease inhibitor [45] and can be associated with neoplastic changes in squamous cell epithelium. It might serve as a plausible biomarker for differentiation of the two subtypes based on cellular morphology. FABP6, a gene involved in fatty acid metabolism [46], is associated with nutrient availability for cancer cells. S100P increases cancer cell migration, invasion, and metastasis [47] and is envisaged to be associated with metastasis of SCLC. Similarly, TRIM29 promotes cellular proliferation by reducing acetylation of p53, thereby affecting DNA damage responses, UV resistance, cell adhesion, invasion, and differentiation [48]. Thus, it can be associated with resistance in case of SCLC. From the above, we could find that DEGs associated with the two subtypes are implicated in several aspects of tumorigenesis such as cell cycle, cell division, and DNA replication. But there are other genes to which the DEGs are closely connected. These genes may also undergo several alterations at numerous levels, such as the post-transcriptional level, which might contribute to the process of carcinogenesis. Thus, an accompanying network analysis of 8 DEGs (CXCL1, FGG, IL33, AGR2, ANK3, CSTA, S100P, TRIM29) genes was conducted. The protein information for the rest of the 3 overlapping genes was not available, due to which they were not used for PPI network analysis. Five genes (TRIM29, ANK3, CSTA, FGG, and AGR2) with a higher number of interacting protein partners were identified. TRIM29 is a co-transcriptional regulatory factor which encodes a 588 amino acid protein. It is intricated into differentiation or carcinogenesis of several types of cancers. It is upregulated in bladder, ovarian, endometrial, colon and colorectal cancer but downregulated in prostate and breast cancer [49, 50, 51, 52, 53, 54]. The changes in expression are suggestive of the involvement of particular cellular specificities or connections and conglomerate pathways of signaling. The upregulation of TRIM29 in SCLC and downregulation in NSCLC corresponds with the above findings indicating the importance of a gene in differentiation of the disease. The higher degree of protein interactions is indicative of their roles in several pathways related to cell adhesion, invasion, radioresistance and DNA damage responses. Moreover, it also plays an important role in activating macrophages in response to bacterial or viral infections. An exaggerated polarization of macrophage leads to more conspicuous inflammation, thereby enhancing the severity of the disease. Thus, it might serve as an impressive therapeutic target of lung cancer. Similarly, ANK3 is an integral membrane protein convoluted in the processes of proliferation, activation, and motility. It has been found to be associated with poor prognosis and metastasis. As seen in the PPI network, it has a higher degree of protein interactions and is associated with several proteins such as TIAM1, where it promotes Rac1 signaling and migration in breast SP cells [55]. Thus, upregulation of ANK3 might be responsible for high cell motility and poor prognosis in case of lung cancer. CSTA, another DEG associated with lung cancer has anti-apoptotic properties associated with neoplastic changes in squamous cell epithelium. It has been reported that its expression is regulated by several factors such as smoking, COPD and genetic variability [45]. These findings can be correlated with higher degree of protein interactions in the PPI network constructed in our study. FGG, as discussed before is a gene encoding γ component of fibrinogen, which is involved in the process of blood clotting. Any perturbations in these genes are associated with the homeostatic imbalance between coagulation and anticoagulation [41]. Thus, differentially expressed FGG might contribute to the processes of pathologic thrombosis and angiogenesis associated with cancer cells. Similarly, AGR2 is also a multi-faceted protein known to affect several aspects of tumorigenesis. This is clear from the interactions observed in the PPI network. Thus, this study has identified the differentially expressed genes in NSCLC and SCLC. We have also tried to find out the interactions of their protein products with other proteins. When once this part of the study is complete, that would provide deeper insight into the mechanistic dynamics of lung cancer.
We have identified differential expression of genes in NSCLC and SCLC. However, gene expression changes are driven by alterations in regulatory pathways. Therefore, understanding the regulatory network would lead us to the development of the competent approach in building predictive models of the disease. Notwithstanding this challenge, we used meta-analysis to identify certain gene signatures associated with NSCLC and SCLC. Meticulous statistical analysis using heterogeneous data from multiple studies has enabled us to uncover biologically significant elements for modeling transitional relationships between genotype and phenotype of the lung cancer.
Declarations
Author contribution statement
Nitesh Shriwash: Conceived and designed the experiments; Wrote the paper.
Prithvi Singh: Contributed reagents, materials, analysis tools or data.
Shweta Arora, Sher Ali: Analyzed and interpreted the data; Wrote the paper.
Syed Mansoor Ali: Conceived and designed the experiments.
Ravins Dohare: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was partially supported by the Science and Engineering Research Board (Grant ID: EEQ/2016/000509), Government of India, India and also supported by the Department of Biotechnology (Grant ID: DBT/PR12828/AAQ/1/622/2015), Government of India, India.
Competing interest statement
The authors declare no conflict of interest.
Additional information
Data associated with this study has been deposited at Gene Expression Omnibus under the accession numbers GSE6044 and GSE40275.
References
- 1.Stewart B.W., Wild C. IARC Press; Lyon, France: 2014. World Cancer Report 2014. [Google Scholar]
- 2.Hoffman P.C., Mauer A.M., Vokes E.E. Lung cancer. The Lancet. 2000;355(9202):479–485. doi: 10.1016/S0140-6736(00)82038-3. [DOI] [PubMed] [Google Scholar]
- 3.Travis W.D. World Health Organization Classification of Tumours. IARC Press Oxford University Press (distributor); Lyon Oxford: 2004. Pathology and genetics of tumours of the lung, pleura, thymus and heart; p. 344. [Google Scholar]
- 4.van Meerbeeck J.P., Fennell D.A., De Ruysscher D.K.M. Small-cell lung cancer. The Lancet. 2011;378(9804):1741–1755. doi: 10.1016/S0140-6736(11)60165-7. [DOI] [PubMed] [Google Scholar]
- 5.Loiselle J.J., Roy J.G., Sutherland L.C. RBM5 reduces small cell lung cancer growth, increases cisplatin sensitivity and regulates key transformation-associated pathways. Heliyon. 2016;2(11) doi: 10.1016/j.heliyon.2016.e00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wistuba I. Genetics of preneoplasia: lessons from lung cancer. Curr. Mol. Med. 2007;7(1):3–14. doi: 10.2174/156652407779940468. [DOI] [PubMed] [Google Scholar]
- 7.Long F. Identification of gene biomarkers for distinguishing small-cell lung cancer from non-small-cell lung cancer using a network-based approach. BioMed Res. Int. 2015;2015 doi: 10.1155/2015/685303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu C. Identification of differentially expressed genes between lung adenocarcinoma and lung squamous cell carcinoma by gene expression profiling. Mol. Med. Rep. 2016;14(2):1483–1490. doi: 10.3892/mmr.2016.5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarca A.L., Romero R., Draghici S. Analysis of microarray experiments of gene expression profiling. Am. J. Obstet. Gynecol. 2006;195(2):373–388. doi: 10.1016/j.ajog.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakobs T.C. Differential gene expression in glaucoma. Cold Spring Harb. Perspect. Med. 2014;4(7):a020636. doi: 10.1101/cshperspect.a020636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makashir S.B., Kottyan L.C., Weirauch M.T. Pacific Symposium on Biocomputing Co-chairs. World Scientific; 2014. Meta-analysis of differential gene co-expression: application to lupus. [PubMed] [Google Scholar]
- 12.Ramasamy A. Key issues in conducting a meta-analysis of gene expression microarray datasets. PLoS Med. 2008;5(9):e184. doi: 10.1371/journal.pmed.0050184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen R. A meta-analysis of lung cancer gene expression identifies PTK7 as a survival gene in lung adenocarcinoma. Cancer Res. 2014;74(10):2892–2902. doi: 10.1158/0008-5472.CAN-13-2775. canres. 2775.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillis J., Pavlidis P. A methodology for the analysis of differential coexpression across the human lifespan. BMC Bioinf. 2009;10(1):306. doi: 10.1186/1471-2105-10-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee H.K. Coexpression analysis of human genes across many microarray data sets. Genome Res. 2004;14(6):1085–1094. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dudley J.T. Disease signatures are robust across tissues and experiments. Mol. Syst. Biol. 2009;5(1):307. doi: 10.1038/msb.2009.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guan Y. A genomewide functional network for the laboratory mouse. PLoS Comput. Biol. 2008;4(9) doi: 10.1371/journal.pcbi.1000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He X. Meta-analysis of mRNA expression profiles to identify differentially expressed genes in lung adenocarcinoma tissue from smokers and non-smokers. Oncol. Rep. 2018;39(3):929–938. doi: 10.3892/or.2018.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett T. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2012;41(D1):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moher D. Guidance for developers of health research reporting guidelines. PLoS Med. 2010;7(2) doi: 10.1371/journal.pmed.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gautier L. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 22.Carvalho B.S., Irizarry R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26(19):2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berriz G.F., Roth F.P. The Synergizer service for translating gene, protein and other biological identifiers. Bioinformatics. 2008;24(19):2272–2273. doi: 10.1093/bioinformatics/btn424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reimand J. g:Profiler—a web server for functional interpretation of gene lists (2016 update) Nucleic Acids Res. 2016;44(W1):W83–W89. doi: 10.1093/nar/gkw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mudunuri U. bioDBnet: the biological database network. Bioinformatics. 2009;25(4):555–556. doi: 10.1093/bioinformatics/btn654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marot G. Moderated effect size and P-value combinations for microarray meta-analyses. Bioinformatics. 2009;25(20):2692–2699. doi: 10.1093/bioinformatics/btp444. [DOI] [PubMed] [Google Scholar]
- 27.Ritchie M.E. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7) doi: 10.1093/nar/gkv007. e47-e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ihnatova I. 2013. MAMA : an R Package for Meta-Analysis of MicroArray. [Google Scholar]
- 29.Haynes W. Encyclopedia of Systems Biology. Springer; 2013. Benjamini–hochberg method. 78-78. [Google Scholar]
- 30.Ashburner M. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47(D1):D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanehisa M. KEGG: Kyoto Encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47(D1):D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2008;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shannon P. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chatr-aryamontri A. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017;45:D369–D379. doi: 10.1093/nar/gkw1102. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stark C. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoek K.S. Novel MITF targets identified using a two-step DNA microarray strategy. Pigm. cell & Melanoma Res. 2008;21(6):665–676. doi: 10.1111/j.1755-148X.2008.00505.x. [DOI] [PubMed] [Google Scholar]
- 40.Vries M.H. CXCL1 promotes arteriogenesis through enhanced monocyte recruitment into the peri-collateral space. Angiogenesis. 2015;18(2):163–171. doi: 10.1007/s10456-014-9454-1. [DOI] [PubMed] [Google Scholar]
- 41.Jacquemin M. The amplitude of coagulation curves from thrombin time tests allows dysfibrinogenemia caused by the common mutation FGG-Arg301 to be distinguished from hypofibrinogenemia. Int J Lab Hematol. 2017;39(3):301–307. doi: 10.1111/ijlh.12625. [DOI] [PubMed] [Google Scholar]
- 42.Yagami A. IL-33 mediates inflammatory responses in human lung tissue cells. J. Immunol. 2010;185(10):5743–5750. doi: 10.4049/jimmunol.0903818. 0903818. [DOI] [PubMed] [Google Scholar]
- 43.Alavi M. High expression of AGR2 in lung cancer is predictive of poor survival. BMC Canc. 2015;15(1):655. doi: 10.1186/s12885-015-1658-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Borek10 D., Jaskólski M. Sequence analysis of enzymes with asparaginase activity. Acta Biochim. Pol. 2001;48:893–902. [PubMed] [Google Scholar]
- 45.Butler M.W. Cancer research; 2011. Modulation of Cystatin A Expression in the Human Small Airway Epithelium by Genotype, Smoking and COPD. canres. 2046.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fagerberg L. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. 2014;13(2):397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsu Y.-L. S100P interacts with integrin α7 and increases cancer cell migration and invasion in lung cancer. Oncotarget. 2015;6(30):29585. doi: 10.18632/oncotarget.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hatakeyama S. Taylor & Francis; 2016. Early Evidence for the Role of TRIM29 in Multiple Cancer Models. [DOI] [PubMed] [Google Scholar]
- 49.Savitsky K. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268(5218):1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 50.Dyrskjot L. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 2004;64(11):4040–4048. doi: 10.1158/0008-5472.CAN-03-3620. [DOI] [PubMed] [Google Scholar]
- 51.Ohmachi T. Clinical significance of TROP2 expression in colorectal cancer. Clin. Cancer Res. 2006;12(10):3057–3063. doi: 10.1158/1078-0432.CCR-05-1961. [DOI] [PubMed] [Google Scholar]
- 52.Bignotti E. Differential gene expression profiles between tumor biopsies and short-term primary cultures of ovarian serous carcinomas: identification of novel molecular biomarkers for early diagnosis and therapy. Gynecol. Oncol. 2006;103(2):405–416. doi: 10.1016/j.ygyno.2006.03.056. [DOI] [PubMed] [Google Scholar]
- 53.Nacht M. Combining serial analysis of gene expression and array technologies to identify genes differentially expressed in breast cancer. Cancer Res. 1999;59(21):5464–5470. [PubMed] [Google Scholar]
- 54.Yu Y.P. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J. Clin. Oncol. 2004;22(14):2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 55.Bourguignon L.Y. Ankyrin-Tiam1 interaction promotes Rac1 signaling and metastatic breast tumor cell invasion and migration. J. Cell Biol. 2000;150(1):177–191. doi: 10.1083/jcb.150.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]