Abstract
BACKGROUND
Aberrant expression of stanniocalcin 2 (STC2) is implicated in colon adenocarcinoma (COAD). A previous study identified that STC2 functions as a tumor promoter to drive development of some cancers, but the role of its overexpression in the development of COAD remains unclear.
AIM
To evaluate the regulation mechanism of STC2 overexpression in COAD.
METHODS
The expression of STC2 in COAD was assessed by TCGA COAD database and GEO (GSE50760). Methylation level of the STC2 promoter was evaluated with beta value in UALCAN platform, and the correlation between STC2 expression and survival rate was investigated with TCGA COAD. Transcription binding site prediction was conducted by TRANSFAC and LASAGNA, and a luciferase reporter system was used to identify STC2 promoter activity in several cell lines, including HEK293T, NCM460, HT29, SW480, and HCT116. Western blotting was performed to evaluate the role of Sp1 on the expression of STC2.
RESULTS
The central finding of this work is that STC2 is overexpressed in COAD tissues and positively correlated with poor prognosis. Importantly, the binding site of the transcription factor Sp1 is widely located in the promoter region of STC2. A luciferase reporter system was successfully constructed to analyze the transcription activity of STC2, and knocking down the expression of Sp1 significantly inhibited the transcription activity of STC2. Furthermore, inhibition of Sp1 remarkably decreased protein levels of STC2.
CONCLUSION
Our data provide evidence that the transcription factor Sp1 is essential for the overexpression of STC2 in COAD through activation of promoter activity. Taken together, our finding provides new insights into the mechanism of oncogenic function of COAD by STC2.
Keywords: Transcription factor Sp1, Stanniocalcin 2, Overexpression, Promoter activity, Colon adenocarcinoma
Core tip: This study demonstrated that stanniocalcin 2 (STC2) is overexpressed in colon adenocarcinoma (COAD) and that high expression of STC2 predicts poor prognosis. The promoter of STC2 in COAD was hypermethylated, and the transcription factor Sp1 was essential for STC2 expression. These findings provide new insights into the mechanism of oncogenic function of STC2 in COAD.
INTRODUCTION
Colon cancer is the third most common cancer and the third most common cause of tumor-related death[1]. A digestive system tumor, colon cancer is highly aggressive and malignant and has high mortality and recurrence rates[2,3]. Research on the pathogenesis of colon cancer has identified not only genetic factors, such as gene mutations in the APC/KRAS/p53 pathways and familial adenomatous polyposis (FAP), but also lifestyle factors, such as a high-fat diet and excessive alcohol use[4,5]. Current therapy for colon cancer is mainly surgery, chemotherapy, and targeting therapy. If cancer diagnosis is during the end-stage, drug therapy becomes the most important therapy in the clinical practice. Drug resistance has become a restriction factor during chemotherapy and targeting drug therapy[6,7], revealing the importance of searching for novel drug targets in colon cancer.
Stanniocalcin 2 (STC2) is a glycoprotein that was first identified in bony fish[8], and it has been reported that STC2 is widely expressed in human tissues. Organs with high levels of the transcript are muscle, kidney, brain, bone, and lung[9,10]. A recent study implicated a role of STC2 in phosphorus and calcium homeostasis[11]. Several reports have shown that STC2 is involved in the growth of bone and adult height[12,13]. In addition, the expression of STC2 plays an important role in the development of some cancers, including hepatocellular carcinoma (HCC), head and neck squamous cell carcinoma, lung cancer, laryngeal squamous cell cancer, breast cancer, nasopharyngeal carcinomas, cervical cancer, and so on[14-18]. The expression of STC2 correlates with clinical and pathological parameters in nasopharyngeal carcinomas, laryngeal squamous cell cancer, lung cancer, and other cancers[18-21]. STC2 is overexpressed in some cancers, indicating that it may be a potential cancer biomarker for diagnosis and prognosis analysis[15,21-23]. Also, it was shown that serum STC2 combined with TIMP metallopeptidase inhibitor 1 and kinesin II-associated protein KAP1 could be poor prognostic factors for recurrence of gastric cancer[20]. The mechanistic investigation demonstrated that the expression of STC2 could regulate PI3K/AKT/ERK signaling in head and neck squamous cell carcinoma and HCC[16,24]. In the progression of colorectal cancer, ovarian cancer, and HCC, STC2 contributed to metastasis through promoting epithelial-mesenchymal transition[22,25,26]. STC2 was also identified as a target gene of hypoxia inducible factor 1 alpha in hypoxia and aryl hydrocarbon receptor[27,28]. It was recently shown that STC2 was closely related with chemotherapeutic drug resistance in cervical cancer and other cancers[17,29-31], suggesting that STC2 plays an important role in the regulation of proliferation and apoptosis of cancer cells.
However, the significance of STC2 in colon adenocarcinoma (COAD) remains unclear, especially regarding the mechanism of STC2 overexpression in some cancers. In this study, we showed that STC2 was overexpressed in COAD, and that its expression was closely related with the progression of COAD. We revealed that STC2 is a tumor promoter by analyzing the survival rate of COAD patients. More importantly, we identified the core region promoter of STC2 and showed that the promoter of STC2 in colon cancer tissues was hypermethylated. The transcription factor Sp1 contributed to promoter activity and expression of STC2. Our findings provide evidence of the mechanism for the overexpression of STC2 in COAD, revealing STC2 as a feasible therapeutic target for colon cancer therapy.
MATERIALS AND METHODS
Reagents
Dulbecco’s Modified Eagle’s Medium (DMEM), RMPI1640, fetal bovine serum (FBS), and penicillin-streptomycin were purchased from Gibco (Gaithersburg, MD, United States); RNA extraction reagent, 1st Strand cDNA Synthesis mixture, quantitative PCR SYBR Green kit, lentivirus concentration solution kit, and enhanced ECL chemiluminescent substrate kit were obtained from Yesen Biotechnology (Shanghai, China); Anti-GAPDH (5174S) was purchased from Cell Signaling Technology (Beverly, MA, United States); anti-STC2 antibody was obtained from Santa Cruz (Dallas, TX, United States; horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin G, HRP-conjugated goat anti-rabbit immunoglobulin G were from ProteinTech (Chicago, IL, United States); Dual luciferase reporter kit (Cat. RG028) was obtained from Beyotime Biotechnology (Shanghai, China).
Cell culture
Human colon cancer cell lines HT29, SW480, and HCT116 and normal colonic epithelial cells NCM460 were obtained from America Type Culture Collection (Manassas, VA, United States). Human embryonic kidney cells HEK293T were purchased from Shanghai Institute of Biochemistry and Cell biology, Chinese Academy of Sciences (Shanghai, China). HEK293T cells were cultured in high-glucose DMEM supplemented with 10% FBS. HT29, SW480, HCT116, and NCM460 cell lines were maintained in Roswell Park Memorial Institute 1640 medium with 10% FBS. All cell lines in this project were cultured with 100 U/mL penicillin and 100 µg/mL streptomycin in a humidified 5% CO2 cell incubator at 37 °C. The cell passage was achieved with trypsin with 0.25% EDTA, and the maximum cell passage was 10 times.
Plasmid construction and transfection
The full length of Sp1 was cloned into pCMV-Myc vector to produce overexpression in cells. The pSuper-neo system was used to knockdown Sp1 expression. The oligo sequences for Sp1 lentiviral shRNA clones were as follows: 5’-CCG GGC TGG TGG TGA TGG AAT ACA TCT CGA GAT GTA TTC CAT CAC CAC CAG CTT TTT-3’[32]. All plasmids were verified by sequencing.
Before transfection, cell culture medium was replaced with fresh medium. Transfections were carried out using Lipofectamine 2000, and the cells were incubated with transfection solution for 6 h. The total medium was replaced with fresh medium with 10% FBS. The subsequent steps were conducted after additional 24 h in culture.
TCGA and UALCAN platform analysis
The expression of STC2 in adjacent normal colonic tissues and colon tumor tissues was analyzed using the TCGA COAD database (https://cancergenome.nih.gov/). Briefly, the expression and clinical information of COAD were downloaded with the GDC Data Portal (https://portal.gdc.cancer.gov/). The 41 normal colonic tissue samples and 286 colon tumor tissue samples were included in the analysis of STC2 expression.
The promoter methylation level of STC2 was evaluated with beta value in UALCAN platform (http://ualcan.path.uab.edu/analysis.html), indicating the levels of DNA methylation ranging from 0 (unmethylated) to 1 (fully methylated). Different beta value cut-off has been considered as hypermethylation (Beta calue: 0.7-0.5) or hypo-methylation (Beta value: 0.3-0.25).
The cumulative survival rate of TCGA COAD is shown as a Kaplan-Meier plot. The top 50% expression of STC2 was considered the high expression group, and the bottom 50% expression of STC2 was considered the low expression group. These groups were compared by the log-rank test.
Promoter activity analysis
The potential promoter sequence of STC2 was analyzed by the GeneCopoeia promoter reporter clone platform (www.genecopoeia.com). The promoter sequence was obtained from the total genomic DNA of HT29 cells by polymerase chain reaction (PCR) method. The full length of 1530 nt of promoter was cloned to the Nhe I/Xho I of pGL3-basic vector, and the sequences of the primer were as follows: Forward primer, 5’-CTA GCT AGC AGG CTG GGC AAA GCA GG-3’, reverse primer, 5’-CCG CTC GAG GCG GAG CAT CGC GTG-3’. The full length of the luciferase reporter plasmid was used as a template to clone other truncated reporter plasmids with the same method.
Luciferase reporter assay
The cells were seeded at 80000 cells/well in 24-well plates and transfected with different pGL3-STC2 reporter plasmids and the pRL-Rellina vector. Cells were cultured for an additional 24 h in complete medium, and the activity of reporter activity was measured with Dual-luciferase assay kit (Beyotime).
Western blotting assay
The cells were washed twice with cold phosphate buffer saline and harvested with radioimmunoprecipitation assay lysis buffer containing 150 mmol/L NaCl, 50 mmol/L Tris (pH 7.4), 1 mmol/L EDTA, sodium deoxycholate, 1% (v/v) Triton X-100, 0.1% (w/v) SDS. Equal protein was separated on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membrane (Roche, Rotkreuz, Switzerland). Subsequently, membranes were blocked in 5% non-fat milk and washed with Tris-buffered saline (TBS, 10 mmol/L Tris, 150 mmol/L NaCl) containing 0.05% Tween-20 (TBST) for three times. The membranes were incubated with the indicated antibody at 4 °C overnight, rinsed, and then incubated with HRP-conjugated anti-mouse or anti-rabbit antibody for another 1 h at room temperature. The blots were detected using an electrochemiluminescence system.
RNA extraction and mRNA level analysis
Total RNAs were isolated by TRIeasy total RNA extraction reagent (Yesen Biotech) and reverse transcribed by Hifair 1st strand cDNA synthesis kit (Yesen Biotech). quantitative PCR analysis was carried out using SYBR Green reverse transcription-PCR kits (TaKaRa, Tokyo, Japan). Relative mRNA levels of the target genes were normalized to β-actin levels. The primers were synthesized by Invitrogen (Shanghai Branch), which included: STC2 (forward: 5'-TTG AAA TGT AAG GCC CAC GC -3'; reverse: 5’-CAG GTC AGC AGC AAG TTC AC-3') and β-actin (forward: 5'- CAT CCG CAA AGA CCT GTA CG-3'; reverse: 5’-CCT GCT TGC TGA TCC ACA TC -3'). The data were calculated by the 2- △△CT method.
Statistical analysis
All experiments were performed three times or more. The data are presented as mean ± standard deviation. Statistics analysis was performed by Graphpad 7.0 software (La Jolla, CA, United States) using a two-tailed student's t test. P value less than 0.05 was considered statically significant.
RESULTS
STC2 is overexpressed in human COAD tissues
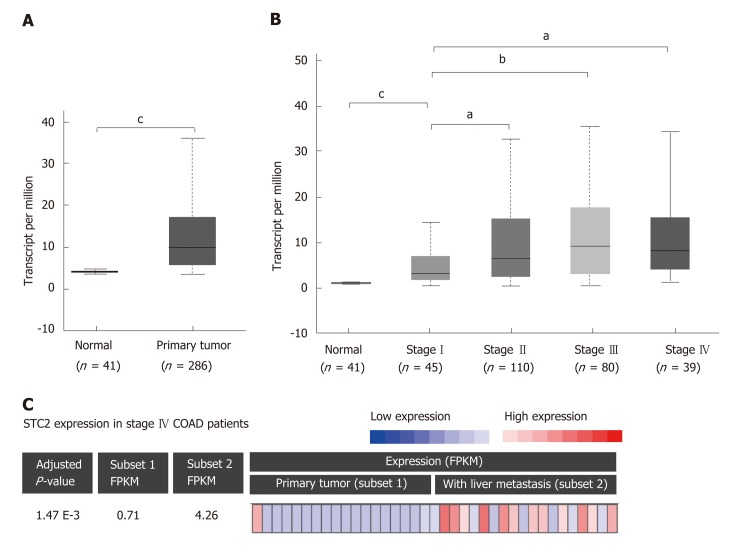
To determine the role of STC2 in COAD, the TCGA COAD database was used to analyze the transcript levels of STC2 in 286 cases COAD tissues and 41 cases normal colonic tissues. The transcript levels of STC2 were significantly higher in COAD tissues than those from normal colonic tissues (Figure 1A). To analyze the potential role of STC2 in the development of COAD, the expression of STC2 in different stages was determined. In the early stage (tumor stage I), the expression of STC2 was higher in COAD tissues than in normal colonic tissues. The expression of STC2 was higher in the more advanced stages than in stage I (Figure 1B). STC2 levels in primary colon tumor and colon tumor with liver metastasis were compared, and the expression of STC2 in the 18 cases of colon cancer with liver metastasis was increased 6-fold compared to 19 cases of primary COAD (Figure 1C). These results suggest that STC2 might play an important role in COAD progression.
Figure 1.
STC2 was overexpressed in colon tumor tissues and contributed to cancer development. A: The relative transcript expression of STC2 was conducted with normal (adjacent colorectal tissues, 41 cases) and colon tumor tissues (286 cases). b), cP < 0.001 vs normal; B: The relative transcript expression of STC2 was evaluated in 274 cases colon tumor tissues with different disease condition, aP < 0.05 vs normal group, cP < 0.001 vs normal group; C: With RNA-seq analysis of STC2 expression in primary colon cancer and liver metastatic colon cancer tissues of stage IV patients (GSE50760). Subset 1: Primary colon cancer; Subset 2: Colon cancer with liver metastasis. STC2: Stanniocalcin 2; FPKM: Fragments per kilobase million.
STC2 is hypermethylated in colon cancer tissues and high expression shows lower survival rate
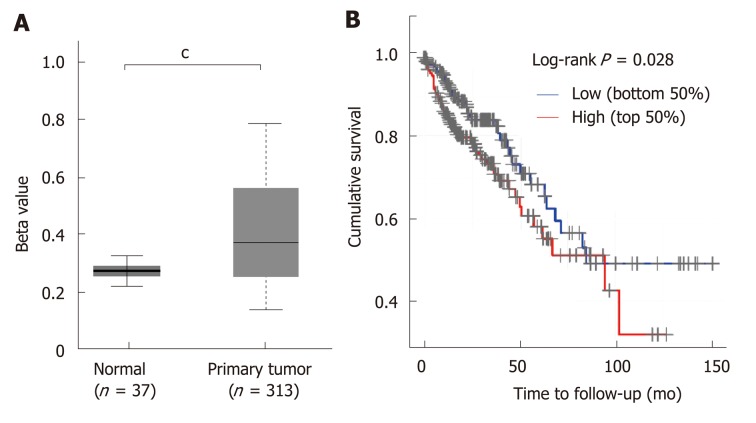
STC2 was overexpressed in COAD, and its expression was positively correlated with the disease progression. Since many studies have shown that aberrant DNA methylation has a significant impact on gene expression and prognosis in some cancers[33,34], and a previous study reported that reducing the expression of DNA methyltransferase 1 could stimulate STC2 expression[35], we hypothesized that methylation might contribute to the overexpression of STC2. To verify this, the promoter DNA methylation data from TCGA Infinium HumanMethylation450K BeadChip arrays for COAD[36] were subjected to methylation analysis of the STC2 promoter. As shown in Figure 2A, promoter methylation levels of STC2 in colon tumors were remarkably upregulated compared with normal colonic tissues. This result suggests that hypermethylation of the STC2 promoter might contribute to the overexpression of STC2 in colon tumor through regulating its transcriptional activity.
Figure 2.
STC2 was hypermethylated in colon cancer tissues and the high expression of STC2 indicated poor prognosis. A: Comparison of the promoter methylation level of STC2 in 37 cases normal colon and 313 cases tumor tissues; B: The cumulative survival rate was conducted between high and low expression of STC2. cP < 0.001 vs normal. STC2: Stanniocalcin 2.
Since STC2 is overexpressed in COAD, we determined the role of STC2 on the tumorigenesis of COAD and the survival rate curve of TCGA COAD was performed. Compared with the low expression group of STC2 patients, the high expression group showed a notable reduction in survival rate. As shown in Figure 1, STC2 was highly overexpressed in colon cancer tissues and its expression was closely related to the development of the disease. Thus, these results indicate that increased expression of STC2 might promote the development of colon cancer and that STC2 is a potential prognostic biomarker and therapeutic target for colon cancer.
Identification of the human STC2 promoter core region
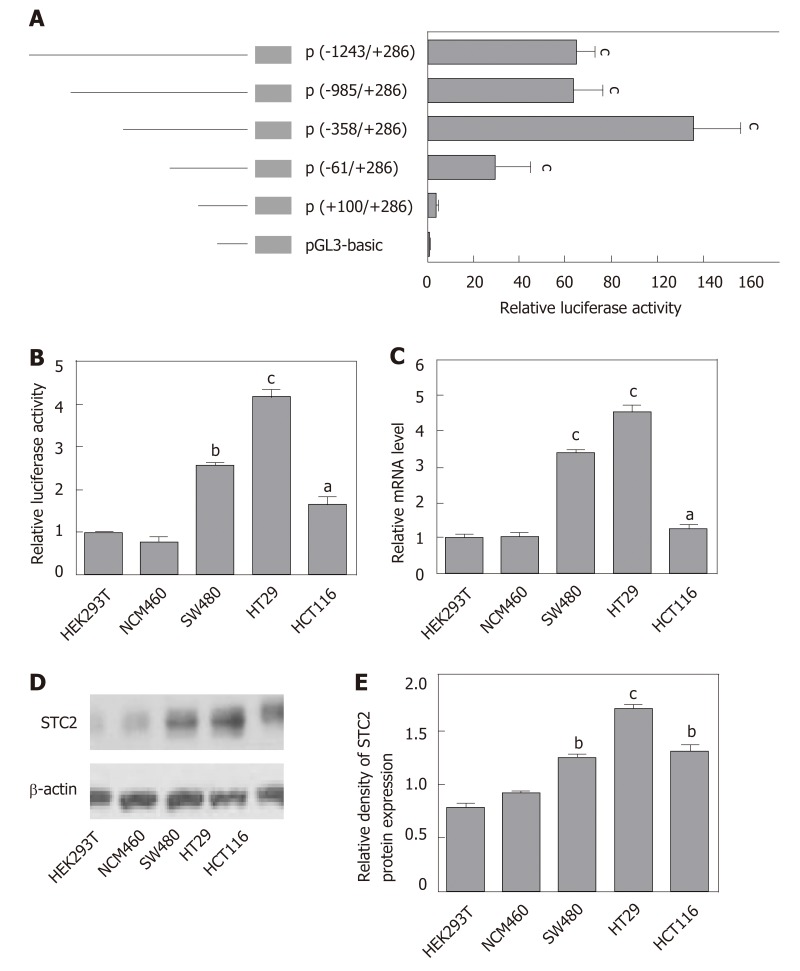
To study the mechanism of STC2 overexpression in colon cancer, we cloned the constructs of the STC2 promoter region, including -1243/+286 (pGL3-STC2-p5), -985/+286 (pGL3-STC2-p4), -358/+286 (pGL3-STC2-p3), -61/+286 (pGL3-STC2-p2), and +100/+286 (pGL3-STC2-p1). A dual luciferase reporter assay was performed to identify the promoter core region of STC2. As shown in Figure 3A, pGL3-STC2-p3 inserted with the -358/+286 promoter fragment exhibited the maximum luciferase reporter activity, indicating that the -358/+286 fragment is the core region of the STC2 promoter. Then, we examined the core region promoter activity in some cell lines, including HEK293T cells, normal colonic epithelial cells NCM460, SW480 cancer cells, HT29 cancer cells, and HCT116 cancer cells. We found that the promoter activity of STC2 in colon cells was significantly higher than that in the normal colonic cells NCM460 and HEK293T cells. In addition, HT29 exhibited higher luciferase promoter activity than SW480 cancer cells and HCT116 cells (Figure 3B). HT29 cells were the most aggressive with high-grade differentiated, SW480 cells were moderately differentiated, and HCT116 cells were poorly differentiated[37,38]. The promoter activity of STC2 was positively correlated with the progression of colon cancer disease. The mRNA levels of STC2 were measured in these cells, and the results showed that the order of mRNA expression levels in the three colon cancer cell lines was HT29, SW480, and HCT116 (Figure 3C). In addition, protein levels were assessed by western blotting, and the protein levels of STC2 in HEK293T, NCM460, HT29, SW480, and HCT116 cells were consistent with the mRNA levels (Figure 3D and E). Based on analysis of promoter activity and mRNA and protein levels, our results indicate that STC2 expression is increased in colon cancer cells and this increase was related to malignant level.
Figure 3.
The core region of the STC2 promoter was -358/+286 nt and correlated with the protein levels. A: The relative promoter activity of different regions of the STC2 promoter was evaluated by luciferase reporter assay in HT29 cells, cP < 0.001 vs pGL3-Basic; B: The relative promoter activity of core region -358/+286 nt and (C: mRNA expression levels were examined in different cell lines, aP < 0.05, bP < 0.01, cP < 0.001 vs HEK293T; D: Western blotting was performed to examine the protein expression of STC2 in different cell lines; E: The relative protein density of panel D was conducted by Quantity One software. bP < 0.01, cP < 0.001 vs HEK293T. STC2: Stanniocalcin 2.
Transcription factor binding sites prediction
Our results showed the STC2 was highly expressed in colon cancer, and the promoter activity of STC2 was closely related with the mRNA and protein levels. Since transcription factors play important roles in the gene regulation, we postulated that some transcription factors might be required for the overexpression of STC2 through binding its promoter region. To test this, the promoter core region of STC2 was analyzed using TRANSFAC and LASAGNA tools. The STC2 promoter core region, along with some putative binding sites for transcriptional regulator, were used. As shown in Figure 4, the core region was GC rich (GC box), which has been reported to be a potential binding site for the Sp1 transcription factor.
Figure 4.
The potential transcription factor Sp1 binding site in core region of STC2 promoter. The prediction of transcription factor binding sites by conducting matrices on the fly from TRANSFAC 4.0 and LASAGNA 2.0 sites. The Min mat conservation was set at 80% (high) and the Sim. of seq to mat. was set at 100% (matrix = seq). STC2: Stanniocalcin 2.
Sp1 transcription factor is required for the promoter activity of STC2
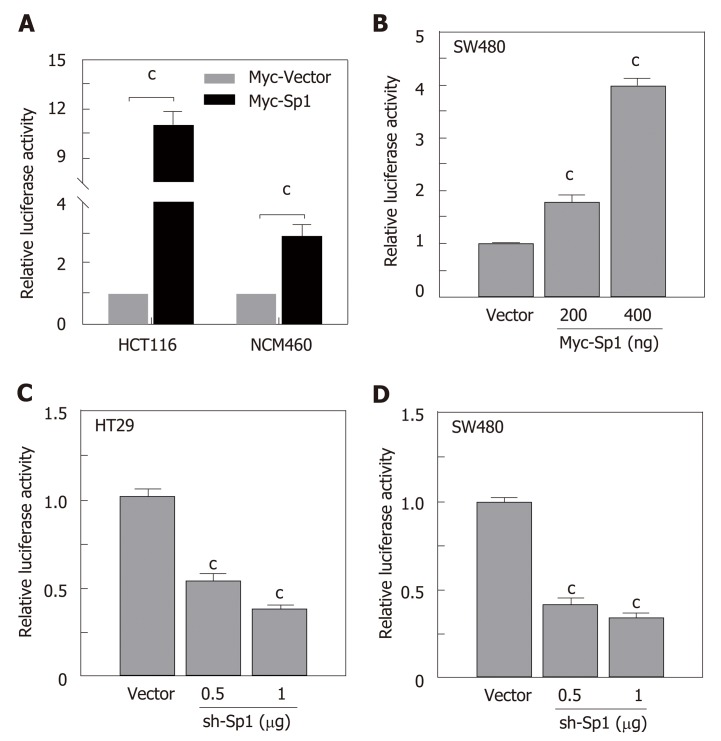
Due to the potential Sp1 binding sites in the core region of the STC2 promoter, we hypothesized that Sp1 has an important role in the regulation of STC2 promoter activity. To evaluate the contribution of Sp1, we constructed an overexpressing Sp1 plasmid and transfected it into HCT116 and NCM460 cells. As shown in Figure 5A, overexpression of Sp1 remarkably increased the promoter reporter activity in both HCT116 cancer cells and normal colonic NCM460 cells. To confirm this result, SW480 colon cancer cells were similarly treated, and the data showed that overexpression of Sp1 in SW480 could upregulate the promoter activity of STC2 in a dose dependent manner (Figure 5B). Moreover, knockdown of Sp1 in HT29 and SW480 cells revealed that silencing of Sp1 could significantly downregulate the promoter activity (Figure 5C and D). These results confirmed that Sp1 transcription factor contributes to the promoter regulation of STC2.
Figure 5.
The transcription factor Sp1 was involved in the promoter activity of STC2. A: Overexpression of Sp1 and control vector in HCT116 and NCM460 cells and the promoter activity were tested by luciferase reporter assay, cP < 0.001 vs Myc-Vector of individual cell group; B: Different Sp1 overexpression plasmids were transfected in SW480 cells, the promoter activity was examined 24 h after transfection, cP < 0.001 vs Myc-Vector group; C: Knockdown of expression of Sp1 in HT29 and D: SW480 cell lines with different dose, and reporter activity was conducted after 48 h transfection, cP < 0.001 vs sh-Vector. STC2: Stanniocalcin 2.
Sp1 transcription factor is required for the expression of STC2
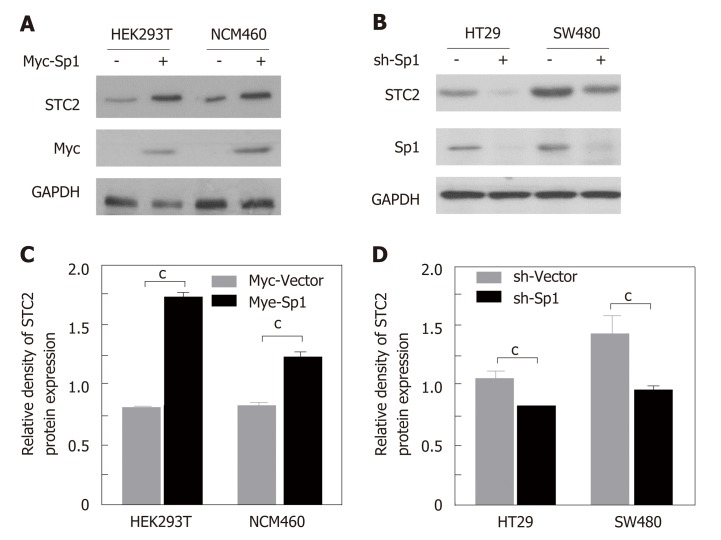
Since transcription factor Sp1 contributes to the promoter activity of STC2, we confirmed the role of Sp1 in the expression of STC2. In STC2 low expressing HEK293T and NCM460 cells, overexpression of Sp1 could enhance the protein expression of STC2 (Figure 6A and B). In STC2 high expressing HT29 and SW480 cells, knocking down the expression of Sp1 decreased the expression of STC2 (Figure 6C and D). These results are consistent with results on promoter regulation shown in Figure 5.
Figure 6.
The transcription factor Sp1 contributed to the overexpression of STC2 in colon cancer cells. A: Overexpression of Sp1 in HEK293T and NCM460 cells. Western blotting was used to evaluate the protein expression of STC2; B: The quantity of panel A protein density with Quantity One software, cP < 0.001 vs Myc-Vector of individual cell group; C: Knocking down the expression of Sp1 with pSuper knockdown system in HT29 and SW480 cell lines. The protein levels of STC2 were evaluated with western blotting assay; D: The quantity of panel C protein density with Quantity One software. cP < 0.001 vs sh-Vector of individual cell group. STC2: Stanniocalcin 2.
DISSCUSION
Previous studies have shown that STC2 was closely associated with some cancers. On the one hand, some reports showed STC2 was an oncogene in gastric, esophageal, liver, colon cancers, and so on[20,22,24,29], but on the other hand, STC2 was reported as a tumor suppressor in breast cancer[39]. Thus, STC2 might play different regulatory functions in different diseases due to the heterogeneity of tumors. In order to fully study the expression of STC2 in colon cancer, the TCGA cancer database was searched to identify the overexpression of STC2 in tumor tissues. The data for survival rate indicated that high expression of STC2 predicted poor prognosis. Importantly, in this study, we fully evaluated the regulatory manner for the overexpression of STC2 in colon cancer. Methylation regulation plays an important role in the expression of genes[40], and we found in the colon cancer tissues that the promoter of STC2 was hypermethylated compared to normal colonic tissues. Thus, regulation of the methylation of the promoter might contribute to the overexpression of STC2 in colon cancer.
Because of the hypermethylated STC2 promoter, we analyzed the promoter region of STC2 and identified that the core region of the STC2 promoter was the -358/+286 fragment. Remarkably, the activity of the STC2 promoter was correlated with the protein and mRNA expression levels of STC2, suggesting that transcriptional regulation is essential to the expression of STC2. Upon analysis of the core region of STC2 promoter, a GC-rich region was identified. Previously, GC box was reported to be a potential binding site of the transcription factor Sp1[41,42]. Thus, we hypothesized that Sp1 might be involved in the regulation of STC2 expression. The overexpression of Sp1 could upregulate the promoter activity of STC2 and enhance its expression levels. To confirm the role of Sp1 in the overexpression of STC2 in colon tumor tissues, knockdown of Sp1 was performed. The results showed that silence of Sp1 expression decreased the expression and promoter activity of STC2. Based on these results, we concluded that the transcription factor Sp1 played an important role in the overexpression of STC2 in COAD.
In this study, we reported that STC2 expression was high in COAD and that the expression levels of STC2 was closely associated with the development of cancer. In addition, high expression of STC2 is linked to low survival rate, suggesting that STC2 might be a novel biomarker for prognosis of COAD. Our study is the first to report that the Sp1 transcription factor can regulate the expression of STC2 at the transcription level. Thus, our findings provide new insights into the overexpression of STC2 in COAD.
ARTICLE HIGHLIGHTS
Research background
To date, growing evidence has shown that stanniocalcin 2 (STC2) might be a cancer-promoter in several cancer types. Some reports demonstrated that the expression of STC2 was higher in colon adenocarcinoma (COAD), but its significance needs further study as the effect of the overexpression of STC2 in COAD remains unclear.
Research motivation
COAD is an aggressive cancer and is linked with high mortality rate. A previous study demonstrated that STC2 might be an oncogenic factor in COAD. This study confirmed that STC2 is overexpressed in COAD and, for the first time, showed that transcription factor Sp1 was essential to the expression of STC2. These findings provide new insight into the oncogenic function of STC2 in COAD.
Research objectives
We confirmed the expression of STC2 and its clinical significance in COAD and evaluated the potential regulation mechanism for the overexpression of STC2.
Research methods
The expression of STC2 in COAD was confirmed with the TCGA database, and expression of STC2 in COAD with liver metastasis or not was confirmed by the GEO database. The methylation levels of the STC2 promoter was analyzed with the UALCAN tool. Survival of the high expression STC2 group and the low expression STC2 group was determined using the Kaplan Meier method. The pGL3 luciferase reporter system to analyze the activity of STC2 promoter. Western blotting was performed to identify the expression of STC2 in several cell lines and evaluate the effect of Sp1 on STC2 expression.
Research results
We confirmed that STC2 was overexpressed in COAD and that the expression of STC2 was positively correlated with the stage of COAD patients and liver metastasis, where high expression of STC2 predicted low survival. Interestingly, the promoter of STC2 was hypermethylated. Through construction of several luciferase reporter systems of STC2 to evaluate its promoter activity, we confirmed the core region of the STC2 promoter. Also, we evaluated if Sp1 was involved in the overexpression of STC2 in COAD.
Research conclusions
Our findings revealed for the first time that transcription factor Sp1 was essential to the overexpression of STC2 in the COAD. Sp1 mediated the expression of STC2 through regulation of its promoter activity. Furthermore, the promoter of STC2 was hypermethylation in COAD tumor tissues, suggesting that hypermethylation of the STC2 promoter might be another factor contributing to the overexpression of STC2. These data provide novel insight into the tumor-promoter function of STC2 in COAD.
Research perspectives
In this study, we showed that Sp1 contributed to the overexpression of STC2 in COAD and that the promoter of STC2 was hypermethylated. In the future, efforts should focus on the interaction between Sp1 and STC2 and the methylation of the promoter for the regulation of STC2.
Footnotes
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
Institutional review board statement: This study was reviewed and approved by the Institutional Review Board of Cancer Hospital of China Medical University, Liaoning Cancer Hospital & Institute.
Conflict-of-interest statement: The authors declare no conflict of interest.
Data sharing statement: No additional data are available.
Peer-review started: March 27, 2019
First decision: April 17, 2019
Article in press: April 29, 2019
P-Reviewer: Maric I, Shin T, Tanabe S S-Editor: Gong ZM L-Editor: Filipodia E-Editor: Zhang YL
Contributor Information
Ji-Bin Li, Department of Colorectal Surgery, Cancer Hospital of China Medical University, Liaoning Cancer Hospital & Institute, Shenyang 110042, Liaoning Province, China.
Zhe-Xian Liu, Department of Colorectal Surgery, Cancer Hospital of China Medical University, Liaoning Cancer Hospital & Institute, Shenyang 110042, Liaoning Province, China.
Rui Zhang, Department of Colorectal Surgery, Cancer Hospital of China Medical University, Liaoning Cancer Hospital & Institute, Shenyang 110042, Liaoning Province, China.
Si-Ping Ma, Department of Colorectal Surgery, Cancer Hospital of China Medical University, Liaoning Cancer Hospital & Institute, Shenyang 110042, Liaoning Province, China.
Tao Lin, Department of Colorectal Surgery, Cancer Hospital of China Medical University, Liaoning Cancer Hospital & Institute, Shenyang 110042, Liaoning Province, China.
Yan-Xi Li, Department of Colorectal Surgery, Cancer Hospital of China Medical University, Liaoning Cancer Hospital & Institute, Shenyang 110042, Liaoning Province, China.
Shi-Hua Yang, Department of Colorectal Surgery, Cancer Hospital of China Medical University, Liaoning Cancer Hospital & Institute, Shenyang 110042, Liaoning Province, China; China Medical University, Shenyang 110000, Liaoning Province, China.
Wan-Chuan Zhang, Department of Colorectal Surgery, Cancer Hospital of China Medical University, Liaoning Cancer Hospital & Institute, Shenyang 110042, Liaoning Province, China; China Medical University, Shenyang 110000, Liaoning Province, China.
Yong-Peng Wang, Department of Colorectal Surgery, Cancer Hospital of China Medical University, Liaoning Cancer Hospital & Institute, Shenyang 110042, Liaoning Province, China. zzzrrr1234@sina.com.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Sobrero A, Grothey A, Iveson T, Labianca R, Yoshino T, Taieb J, Maughan T, Buyse M, André T, Meyerhardt J, Shields AF, Souglakos I, Douillard JY, Cervantes A. The hard road to data interpretation: 3 or 6 months of adjuvant chemotherapy for patients with stage III colon cancer? Ann Oncol. 2018;29:1099–1107. doi: 10.1093/annonc/mdy064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vietti Violi N, Duran R, Demartines N, Sempoux C, Guiu B, Bize PE, Sala N, Halkic N, Knebel JF, Denys A. Local recurrence rate in patients with colorectal cancer liver metastasis after wedge resection or percutaneous radiofrequency ablation. Int J Hyperthermia. 2018;34:1020–1028. doi: 10.1080/02656736.2017.1372644. [DOI] [PubMed] [Google Scholar]
- 4.Suehiro Y, Wong CW, Chirieac LR, Kondo Y, Shen L, Webb CR, Chan YW, Chan AS, Chan TL, Wu TT, Rashid A, Hamanaka Y, Hinoda Y, Shannon RL, Wang X, Morris J, Issa JP, Yuen ST, Leung SY, Hamilton SR. Epigenetic-genetic interactions in the APC/WNT, RAS/RAF, and P53 pathways in colorectal carcinoma. Clin Cancer Res. 2008;14:2560–2569. doi: 10.1158/1078-0432.CCR-07-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith G, Carey FA, Beattie J, Wilkie MJ, Lightfoot TJ, Coxhead J, Garner RC, Steele RJ, Wolf CR. Mutations in APC, Kirsten-ras, and p53--alternative genetic pathways to colorectal cancer. Proc Natl Acad Sci U S A. 2002;99:9433–9438. doi: 10.1073/pnas.122612899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colak S, Medema JP. Human colonic fibroblasts regulate stemness and chemotherapy resistance of colon cancer stem cells. Cell Cycle. 2016;15:1531–1537. doi: 10.4161/15384101.2014.973321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buzzelli JN, Ouaret D, Brown G, Allen PD, Muschel RJ. Colorectal cancer liver metastases organoids retain characteristics of original tumor and acquire chemotherapy resistance. Stem Cell Res. 2018;27:109–120. doi: 10.1016/j.scr.2018.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo CW, Pisarska MD, Hsueh AJ. Identification of a stanniocalcin paralog, stanniocalcin-2, in fish and the paracrine actions of stanniocalcin-2 in the mammalian ovary. Endocrinology. 2005;146:469–476. doi: 10.1210/en.2004-1197. [DOI] [PubMed] [Google Scholar]
- 9.Ishibashi K, Miyamoto K, Taketani Y, Morita K, Takeda E, Sasaki S, Imai M. Molecular cloning of a second human stanniocalcin homologue (STC2) Biochem Biophys Res Commun. 1998;250:252–258. doi: 10.1006/bbrc.1998.9300. [DOI] [PubMed] [Google Scholar]
- 10.Takei Y, Yamamoto H, Masuda M, Sato T, Taketani Y, Takeda E. Stanniocalcin 2 is positively and negatively controlled by 1,25(OH)(2)D(3) and PTH in renal proximal tubular cells. J Mol Endocrinol. 2009;42:261–268. doi: 10.1677/JME-08-0161. [DOI] [PubMed] [Google Scholar]
- 11.Yahata K, Mori K, Mukoyama M, Sugawara A, Suganami T, Makino H, Nagae T, Fujinaga Y, Nabeshima Y, Nakao K. Regulation of stanniocalcin 1 and 2 expression in the kidney by klotho gene. Biochem Biophys Res Commun. 2003;310:128–134. doi: 10.1016/j.bbrc.2003.08.131. [DOI] [PubMed] [Google Scholar]
- 12.Marouli E, Graff M, Medina-Gomez C, Lo KS, Wood AR, Kjaer TR, Fine RS, Lu Y, Schurmann C, Highland HM, Rüeger S, Thorleifsson G, Justice AE, Lamparter D, Stirrups KE, Turcot V, Young KL, Winkler TW, Esko T, Karaderi T, Locke AE, Masca NG, Ng MC, Mudgal P, Rivas MA, Vedantam S, Mahajan A, Guo X, Abecasis G, Aben KK, Adair LS, Alam DS, Albrecht E, Allin KH, Allison M, Amouyel P, Appel EV, Arveiler D, Asselbergs FW, Auer PL, Balkau B, Banas B, Bang LE, Benn M, Bergmann S, Bielak LF, Blüher M, Boeing H, Boerwinkle E, Böger CA, Bonnycastle LL, Bots ML, Bottinger EP, Bowden DW, Brandslund I, Breen G, Brilliant MH, Broer L, Burt AA, Butterworth AS, Carey DJ, Caulfield MJ, Chambers JC, Chasman DI, Chen YI, Chowdhury R, Christensen C, Chu AY, Cocca M, Collins FS, Cook JP, Corley J, Galbany JC, Cox AJ, Danesh J, Davies G, Deary IJ, Dedoussis G, Demerath EW, Dennis JG, Drenos F, Du M, Dunning AM, Easton DF, Ebeling T, Edwards TL, Ellinor PT, Elliott P, Evangelou E, Farmaki AE, Faul JD, Feitosa MF, Feng S, Ferrannini E, Ferrario MM, Ferrieres J, Florez JC, Ford I, Fornage M, Franks PW, Galesloot TE, Gan W, Gandin I, Gasparini P, Giedraitis V, Giri A, Girotto G, Gordon SD, Gorski M, Grarup N, Grove ML, Gudnason V, Gustafsson S, Hansen T, Harris KM, Harris TB, Hattersley AT, Hayward C, He L, Heid IM, Heikkilä K, Helgeland Ø, Hernesniemi J, Hewitt AW, Hocking LJ, Hollensted M, Holmen OL, Hovingh GK, Howson JM, Hoyng CB, Huang PL, Hveem K, Ikram MA, Ingelsson E, Jackson AU, Jansson JH, Jarvik GP, Jensen GB, Jhun MA, Jia Y, Jiang X, Johansson S, Jørgensen ME, Jørgensen T, Jousilahti P, Jukema JW, Kahali B, Kahn RS, Kähönen M, Kamstrup PR, Kanoni S, Kaprio J, Karaleftheri M, Kardia SL, Karpe F, Kee F, Keeman R, Kiemeney LA, Kitajima H, Kluivers KB, Kocher T, Komulainen P, Kontto J, Kooner JS, Kooperberg C, Kovacs P, Kriebel J, Kuivaniemi H, Küry S, Kuusisto J, La Bianca M, Laakso M, Lakka TA, Lange EM, Lange LA, Langefeld CD, Langenberg C, Larson EB, Lee IT, Lehtimäki T, Lewis CE, Li H, Li J, Lin H, Lin LA, Lin X, Lind L, Lindström J, Linneberg A, Liu Y, Liu Y, Lophatananon A, Luan J, Lubitz SA, Lyytikäinen LP, Mackey DA, Madden PA, Manning AK, Männistö S, Marenne G, Marten J, Martin NG, Mazul AL, Meidtner K, Metspalu A, Mitchell P, Mohlke KL, Morgan A, Morris AD, Morris AP, Munroe PB, Nalls MA, Nauck M, Nelson CP, Neville M, Nielsen SF, Nikus K, Njølstad PR, Nordestgaard BG, Ntalla I, O'Connel JR, Oksa H, Loohuis LM, Ophoff RA, Owen KR, Packard CJ, Padmanabhan S, Palmer CN, Pasterkamp G, Patel AP, Pattie A, Pedersen O, Peissig PL, Peloso GM, Pennell CE, Perola M, Perry JA, Perry JR, Person TN, Pirie A, Polasek O, Posthuma D, Raitakari OT, Rasheed A, Rauramaa R, Reilly DF, Reiner AP, Renström F, Ridker PM, Rioux JD, Robertson N, Robino A, Rolandsson O, Rudan I, Ruth KS, Saleheen D, Salomaa V, Samani NJ, Sandow K, Sapkota Y, Sattar N, Schmidt MK, Schreiner PJ, McCarthy MI, Willer CJ, Stefansson K, Borecki IB, Liu DJ, North KE, Pers TH, Lindgren CM, Oxvig C, Kutalik Z, Rivadeneira F, Loos RJ, Frayling TM, Hirschhorn JN, Deloukas P, Lettre G, Vozzi D, Walker M, Wang F, Wang CA, Wang S, Wang Y, Wareham NJ, Warren HR, Wessel J, Willems SM, Wilson JG, Witte DR, Woods MO, Wu Y, Yaghootkar H, Yao J, Yao P, Young R, Zeggini E, Zhan X, Zhang W, Zhao JH, Zhao W, Zhao W, Zheng H, Zhou W, Rotter JI, Boehnke M, Kathiresan S, Schulze MB, Scott RA, Shah S, Sim X, Sivapalaratnam S, Small KS, Smith AV, Smith JA, Southam L, Spector TD, Speliotes EK, Starr JM, Steinthorsdottir V, Stringham HM, Stumvoll M, Surendran P, Tansey KE, Tardif JC, Taylor KD, Teumer A, Thompson DJ, Thorsteinsdottir U, Thuesen BH, Tönjes A, Tromp G, Trompet S, Tsafantakis E, Tuomilehto J, Tyrer JP, Uher R, Uitterlinden AG, Ulivi S, Varbo A, Varga TV, Varma R, Edwards DR, Vermeulen SH, Vestergaard H, Vitart V, Vogt TF, Bork-Jensen J, Frikke-Schmidt R, Gordon-Larsen P, Li-Gao R, Mook-Kanamori DO, Müller-Nurasyid M, 't Hart LM, Yerges-Armstrong LM, Heard-Costa NL, de Bakker PI, de Borst GJ, Segura-Lepe MP, Cuellar-Partida G, Tybjaerg-Hansen A, de Denus S, de Groot MC, de Mutsert R, den Hollander AI, Di Angelantonio E, van der Laan SW, Van Der Leij AR, van Duijn CM, van Schoor NM, van Setten J, 't Hart LM EPIC-InterAct Consortium; CHD Exome+ Consortium; ExomeBP Consortium; T2D-Genes Consortium; GoT2D Genes Consortium; Global Lipids Genetics Consortium; ReproGen Consortium; MAGIC Investigators. Rare and low-frequency coding variants alter human adult height. Nature. 2017;542:186–190. doi: 10.1038/nature21039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jepsen MR, Kløverpris S, Mikkelsen JH, Pedersen JH, Füchtbauer EM, Laursen LS, Oxvig C. Stanniocalcin-2 inhibits mammalian growth by proteolytic inhibition of the insulin-like growth factor axis. J Biol Chem. 2015;290:3430–3439. doi: 10.1074/jbc.M114.611665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu F, Li TY, Su SC, Yu JS, Zhang HL, Tan GQ, Liu JW, Wang BL. STC2 as a novel mediator for Mus81-dependent proliferation and survival in hepatocellular carcinoma. Cancer Lett. 2017;388:177–186. doi: 10.1016/j.canlet.2016.11.039. [DOI] [PubMed] [Google Scholar]
- 15.Na SS, Aldonza MB, Sung HJ, Kim YI, Son YS, Cho S, Cho JY. Stanniocalcin-2 (STC2): A potential lung cancer biomarker promotes lung cancer metastasis and progression. Biochim Biophys Acta. 2015;1854:668–676. doi: 10.1016/j.bbapap.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Yang S, Ji Q, Chang B, Wang Y, Zhu Y, Li D, Huang C, Wang Y, Sun G, Zhang L, Guan Q, Xiang J, Wei W, Lu Z, Liao T, Meng J, Wang Z, Ma B, Zhou L, Wang Y, Yang G. STC2 promotes head and neck squamous cell carcinoma metastasis through modulating the PI3K/AKT/Snail signaling. Oncotarget. 2017;8:5976–5991. doi: 10.18632/oncotarget.13355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jansen MP, Sas L, Sieuwerts AM, Van Cauwenberghe C, Ramirez-Ardila D, Look M, Ruigrok-Ritstier K, Finetti P, Bertucci F, Timmermans MM, van Deurzen CH, Martens JW, Simon I, Roepman P, Linn SC, van Dam P, Kok M, Lardon F, Vermeulen PB, Foekens JA, Dirix L, Berns EM, Van Laere S. Decreased expression of ABAT and STC2 hallmarks ER-positive inflammatory breast cancer and endocrine therapy resistance in advanced disease. Mol Oncol. 2015;9:1218–1233. doi: 10.1016/j.molonc.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou H, Li YY, Zhang WQ, Lin D, Zhang WM, Dong WD. Expression of stanniocalcin-1 and stanniocalcin-2 in laryngeal squamous cell carcinoma and correlations with clinical and pathological parameters. PLoS One. 2014;9:e95466. doi: 10.1371/journal.pone.0095466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esseghir S, Kennedy A, Seedhar P, Nerurkar A, Poulsom R, Reis-Filho JS, Isacke CM. Identification of NTN4, TRA1, and STC2 as prognostic markers in breast cancer in a screen for signal sequence encoding proteins. Clin Cancer Res. 2007;13:3164–3173. doi: 10.1158/1078-0432.CCR-07-0224. [DOI] [PubMed] [Google Scholar]
- 20.Wang YY, Li L, Zhao ZS, Wang HJ. Clinical utility of measuring expression levels of KAP1, TIMP1 and STC2 in peripheral blood of patients with gastric cancer. World J Surg Oncol. 2013;11:81. doi: 10.1186/1477-7819-11-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin S, Guo Q, Wen J, Li C, Lin J, Cui X, Sang N, Pan J. Survival analyses correlate stanniocalcin 2 overexpression to poor prognosis of nasopharyngeal carcinomas. J Exp Clin Cancer Res. 2014;33:26. doi: 10.1186/1756-9966-33-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu J, Lai M, Shao C, Wang J, Wei JJ. STC2 overexpression mediated by HMGA2 is a biomarker for aggressiveness of high-grade serous ovarian cancer. Oncol Rep. 2015;34:1494–1502. doi: 10.3892/or.2015.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang ZH, Wu YG, Qin CK, Rong ZH, Su ZX, Xian GZ. Stanniocalcin 2 expression predicts poor prognosis of hepatocellular carcinoma. Oncol Lett. 2014;8:2160–2164. doi: 10.3892/ol.2014.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, Wu K, Sun Y, Li Y, Wu M, Qiao Q, Wei Y, Han ZG, Cai B. STC2 is upregulated in hepatocellular carcinoma and promotes cell proliferation and migration in vitro. BMB Rep. 2012;45:629–634. doi: 10.5483/BMBRep.2012.45.11.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Law AY, Wong CK. Stanniocalcin-2 is a HIF-1 target gene that promotes cell proliferation in hypoxia. Exp Cell Res. 2010;316:466–476. doi: 10.1016/j.yexcr.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 26.Chen B, Zeng X, He Y, Wang X, Liang Z, Liu J, Zhang P, Zhu H, Xu N, Liang S. STC2 promotes the epithelial-mesenchymal transition of colorectal cancer cells through AKT-ERK signaling pathways. Oncotarget. 2016;7:71400–71416. doi: 10.18632/oncotarget.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Law AY, Wong CK. Stanniocalcin-2 promotes epithelial-mesenchymal transition and invasiveness in hypoxic human ovarian cancer cells. Exp Cell Res. 2010;316:3425–3434. doi: 10.1016/j.yexcr.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 28.Harper TA, Jr, Joshi AD, Elferink CJ. Identification of stanniocalcin 2 as a novel aryl hydrocarbon receptor target gene. J Pharmacol Exp Ther. 2013;344:579–588. doi: 10.1124/jpet.112.201111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Gao Y, Cheng H, Yang G, Tan W. Stanniocalcin 2 promotes cell proliferation and cisplatin resistance in cervical cancer. Biochem Biophys Res Commun. 2015;466:362–368. doi: 10.1016/j.bbrc.2015.09.029. [DOI] [PubMed] [Google Scholar]
- 30.Yuan Q, Zhan L, Zhang LL, Wang Q, Liu J, Jiang ZY, Hu XM, Yuan XC. Stanniocalcin 2 induces oxaliplatin resistance in colorectal cancer cells by upregulating P-glycoprotein. Can J Physiol Pharmacol. 2016;94:929–935. doi: 10.1139/cjpp-2015-0530. [DOI] [PubMed] [Google Scholar]
- 31.Cheng H, Wu Z, Wu C, Wang X, Liow SS, Li Z, Wu YL. Overcoming STC2 mediated drug resistance through drug and gene co-delivery by PHB-PDMAEMA cationic polyester in liver cancer cells. Mater Sci Eng C Mater Biol Appl. 2018;83:210–217. doi: 10.1016/j.msec.2017.08.075. [DOI] [PubMed] [Google Scholar]
- 32.Cheng D, Zhao Y, Wang S, Jia W, Kang J, Zhu J. Human Telomerase Reverse Transcriptase (hTERT) Transcription Requires Sp1/Sp3 Binding to the Promoter and a Permissive Chromatin Environment. J Biol Chem. 2015;290:30193–30203. doi: 10.1074/jbc.M115.662221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strand SH, Orntoft TF, Sorensen KD. Prognostic DNA methylation markers for prostate cancer. Int J Mol Sci. 2014;15:16544–16576. doi: 10.3390/ijms150916544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kordowski F, Kolarova J, Schafmayer C, Buch S, Goldmann T, Marwitz S, Kugler C, Scheufele S, Gassling V, Németh CG, Brosch M, Hampe J, Lucius R, Röder C, Kalthoff H, Siebert R, Ammerpohl O, Reiss K. Aberrant DNA methylation of ADAMTS16 in colorectal and other epithelial cancers. BMC Cancer. 2018;18:796. doi: 10.1186/s12885-018-4701-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Law AY, Lai KP, Ip CK, Wong AS, Wagner GF, Wong CK. Epigenetic and HIF-1 regulation of stanniocalcin-2 expression in human cancer cells. Exp Cell Res. 2008;314:1823–1830. doi: 10.1016/j.yexcr.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Wang Z, Wu X, Wang Y. A framework for analyzing DNA methylation data from Illumina Infinium HumanMethylation450 BeadChip. BMC Bioinformatics. 2018;19:115. doi: 10.1186/s12859-018-2096-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Z, Qi L, Li Y, Zhao X, Sun B. VEGFR2 regulates endothelial differentiation of colon cancer cells. BMC Cancer. 2017;17:593. doi: 10.1186/s12885-017-3578-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo Y, Ye GY, Qin SL, Yu MH, Mu YF, Zhong M. ATAD2 Overexpression Identifies Colorectal Cancer Patients with Poor Prognosis and Drives Proliferation of Cancer Cells. Gastroenterol Res Pract. 2015;2015:936564. doi: 10.1155/2015/936564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hou J, Wang Z, Xu H, Yang L, Yu X, Yang Z, Deng Y, Meng J, Feng Y, Guo X, Yang G. Stanniocalicin 2 suppresses breast cancer cell migration and invasion via the PKC/claudin-1-mediated signaling. PLoS One. 2015;10:e0122179. doi: 10.1371/journal.pone.0122179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang WY, Hsu SD, Huang HY, Sun YM, Chou CH, Weng SL, Huang HD. MethHC: a database of DNA methylation and gene expression in human cancer. Nucleic Acids Res. 2015;43:D856–D861. doi: 10.1093/nar/gku1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacLeod MC, Powell KL, Kuzmin VA, Kolbanovskiy A, Geacintov NE. Interference of benzo[a]pyrene diol epoxide-deoxyguanosine adducts in a GC box with binding of the transcription factor Sp1. Mol Carcinog. 1996;16:44–52. doi: 10.1002/(SICI)1098-2744(199605)16:1<44::AID-MC6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 42.Jitrapakdee S, Slawik M, Medina-Gomez G, Campbell M, Wallace JC, Sethi JK, O'rahilly S, Vidal-Puig AJ. The peroxisome proliferator-activated receptor-gamma regulates murine pyruvate carboxylase gene expression in vivo and in vitro. J Biol Chem. 2005;280:27466–27476. doi: 10.1074/jbc.M503836200. [DOI] [PMC free article] [PubMed] [Google Scholar]