Abstract
A Disintegrin and Metalloproteinase (ADAM) is a family of proteolytic enzymes that possess sheddase function and regulate shedding of membrane-bound proteins, growth factors, cytokines, ligands and receptors. Typically, ADAMs have a pro-domain, and a metalloproteinase, disintegrin, cysteine-rich and a characteristic transmembrane domain. Most ADAMs are activated by proprotein convertases, but can also be regulated by G-protein coupled receptor agonists, Ca2+ ionophores and protein kinase C activators. A Disintegrin and Metalloproteinase with Thrombospondin Motifs (ADAMTS) is a family of secreted enzymes closely related to ADAMs. Like ADAMs, ADAMTS members have a pro-domain, and a metalloproteinase, disintegrin, and cysteine-rich domain, but they lack a transmembrane domain and instead have characteristic thrombospondin motifs. Activated ADAMs perform several functions and participate in multiple cardiovascular processes including vascular smooth muscle cell proliferation and migration, angiogenesis, vascular cell apoptosis, cell survival, tissue repair, and wound healing. ADAMs may also be involved in pathological conditions and cardiovascular diseases such as atherosclerosis, hypertension, aneurysm, coronary artery disease, myocardial infarction and heart failure. Like ADAMs, ADAMTS have a wide-spectrum role in vascular biology and cardiovascular pathophysiology. ADAMs and ADAMTS activity is naturally controlled by endogenous inhibitors such as tissue inhibitors of metalloproteinases (TIMPs), and their activity can also be suppressed by synthetic small molecule inhibitors. ADAMs and ADAMTS can serve as important diagnostic biomarkers and potential therapeutic targets for cardiovascular disorders. Natural and synthetic inhibitors of ADAMs and ADAMTS could be potential therapeutic tools for the management of cardiovascular diseases.
Keywords: cytokines, metalloproteases, sheddase, vascular smooth muscle, zinc
Graphical Abstract
1. Introduction
Cardiovascular remodeling and degradation of extracellular matrix (ECM) proteins play an important role in the regulation of cardiovascular function, and can also be involved in pathological processes such as inflammation, oxidative stress, tumor metastasis and cardiovascular disease [1, 2]. Matrix metalloproteinases (MMPs) are major regulators of ECM protein degradation and tissue remodeling [3, 4]. In addition to MMPs, two families of enzymes termed a disintegrin and metalloproteinase (ADAM) and a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) have been implicated in the regulation of cardiovascular function through modulation of ECM, growth factors and cytokines. ADAMs, ADAMTS and snake venom metalloproteinase (SVM) belong to the adamalysin protein family that has a close relationship with other metalloenzymes such as MMPs and membrane-type MMPs (MT-MMPs). ADAMs and ADAMTS are very similar in their structural domains and protein sequence sharing highly conservative structure and protein homology. Both ADAM and ADAMTS members have a pro-domain, and a metalloproteinase, disintegrin and cysteine-rich domain. The metalloproteinase domain of ADAMs regulates shedding of membrane-bound growth factors, cytokines and receptors with the help of the disintegrin domain which specifically binds to integrins. The main differences are that ADAMs are largely membrane-bound proteins that are fixed on the cell membrane by a transmembrane domain, although secreted forms of ADAMs generated by alternative splicing or cleavage have also been identified in the circulation and peripheral tissues. In comparison, ADAMTS members are secreted enzymes that lack the transmembrane domain and cytoplasmic tail, but have extra ancillary thrombospondin motifs.
Most ADAMs such as ADAM 1, 2, 3, 6, 7, 18, 20, 21, 22, 28, 29, 30, 32 and 33 play a role in the reproductive system and embryo development. However, ADAM-8, 9, 10, 11, 12, 15, 17 and 19, along with ADAMTS-1, 4, 5, 16 have a broader distribution in human tissues and a wide connection with various biological processes and cardiovascular functions. Additionally, some ADAMs and ADAMTS have been suggested as potential factors in pathological conditions such as oncogenesis, autoimmune diseases, inflammation and cardiovascular diseases [5].
In this review, we will use research data published in PubMed, Web of Science and other scientific databases, as well as some information from our previous reports to provide an overview of the biochemical and biological properties of ADAM and ADAMTS family and their role in cardiovascular pathophysiology. We will discuss the ADAM and ADAMTS family structure, tissue distribution, activation process, substrates, and biological functions. The relationship between ADAMs and ADAMTS and other related families of proteases such as MMPs and SVM will also be briefly described [6]. We will describe the potential role of ADAMs and ADAMTS family members in pathological processes and cardiovascular disease. We will also describe some of the endogenous and synthetic inhibitors of ADAMs and ADAMTS and their potential usefulness in biological assays of these enzymes. We will conclude the review with insights on how ADAMs and ADAMTS can be used as potential biomarkers and therapeutic targets for the detection and management of cardiovascular disease.
2. Historical Overview of ADAM and ADAMTS Family
The discovery and classification of the ADAM family involved a large amount of important research work. ADAM-1 was first identified in 1987 as a new fertilization protein PH-30 [7]. In 1992, ADAMs were demonstrated as a new family with a close relationship to the snake venom disintegrins family, also known as snake venom metalloproteinases (SVM). In 1995, ADAM-1 was suggested to belong to the metalloproteinase/disintegrin/cysteine-rich (MDC) family and was confirmed to have a role in the fertilization process. ADAM-17 was recognized in 1997 as a TNFα converting enzyme (TACE), and subsequently ADAMs became known as sheddases [8]. Because of the large interest in ADAM-17, and its wide functions and pathological roles described in numerous publications, its crystal structure was determined in 1998. To further elucidate the role of ADAMs in biological processes and physiological functions, ADAM-17 and ADAM-10 knock-out mice were developed in in 1998 and 2002, respectively. In 2006, the crystal structure of Vascular Adhesion Protein-1 (VAP1) was revealed and helped to decipher the ADAMs’ MDC domain architecture and their unique activity sites [8]. The discovery of the C-shape scaffold, was a landmark study in deciphering the crystal structure of ADAMs and how they function [9]. In 2010 and the following years, more ADAMs knock-out mice were developed to verify their biological roles, and further research suggested potential role of ADAMs in multiple pathological conditions such as inflammation, oncogenesis and cardiovascular disease. ADAMTS family was first identified as an inflammation factor in 1997 [10]. The role of ADAMTS-2, 3, and 14 as procollagen N-endopeptidases was demonstrated in 2001 [11, 12]. In the following years, the role of different ADAMTS in various diseases such as cancer, osteoarthritis and cardiovascular disease has been illuminated.
3. ADAM Family Structure
ADAMs, formerly known as metalloproteinase/disintegrin/cysteine-rich (MDC) proteins, are a subfamily of the Metzincins superfamily of metalloproteases. ADAMs, ADAMTS, and snake venom metalloproteases (SVMs) are Zn2+-dependent catalytic enzymes (Fig. 1). They comprise the adamalysin subfamily of the metallopeptidases family of proteins.
Fig. 1.
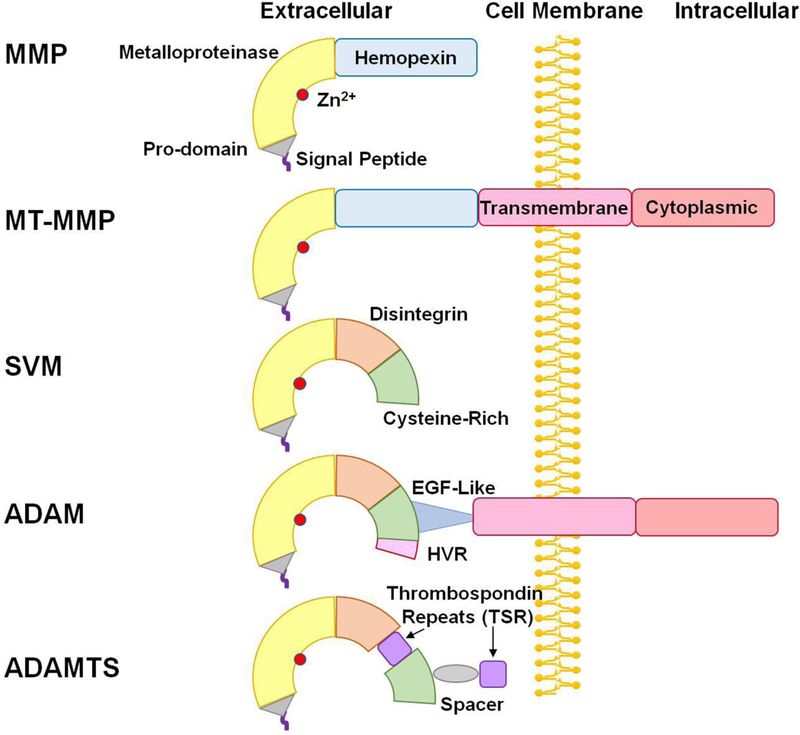
Structure of ADAM and ADAMTS. In addition to the signal peptide, pro-domain, and metalloproteinase domain in matrix metalloproteinases (MMP) and membrane-type MMPs (MT-MMP), snake venom metalloproteinases (SVM) have a disintegrin and cysteine-rich domain. Compared with SVM, ADAMs have a highly variable region (HVR) in the cysteine-rich domain, and an additional epidermal growth factor (EGF)-like region, transmembrane domain and cytoplasmic tail. ADAMTS members are similar to ADAMs, but lack the EGF-like region, transmembrane domain and cytoplasmic tail, and instead have thrombospondin repeats (TSR) and spacer.
ADAMs have various forms in different species including ciona intestinalis, mice, rats and humans [1] (Fig. 2). Currently, 37 ADAMs have been identified in rats, 34 ADAMs have been discovered in mice, and 22 ADAMs have been found in the human genome, among which 13 proteins are proteolytically active [13]. There are wide similarities in domain organization, spatial structures and protein sequences between ADAMs, ADAMTS and SVMs [14]. The common feature of the adamalysin subfamily is a pro-domain, a metalloproteinase, disintegrin, and cysteine-rich domain with some differences in protein sequence and biological function [15]:
Fig. 2.
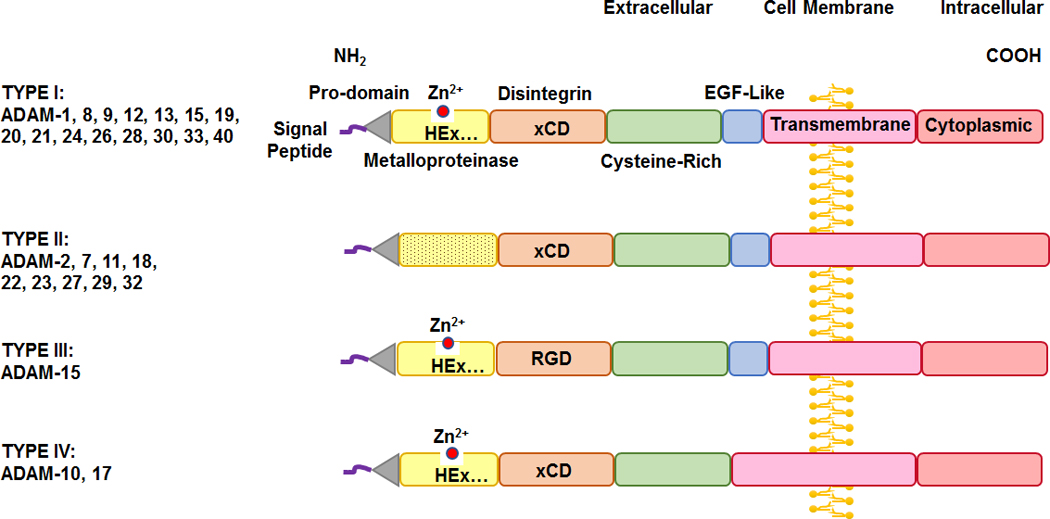
Sequence difference among different types of the ADAM family. ADAM members have four types. In Type I ADAMs the metalloprotease M-domain has the characteristic catalytic Zn2+ binding signature (HExGHxxGxxHD), and the disintegrin D-domain is based on xCD sequence. In Type II ADAMs the M-domain does not have the characteristic catalytic-Zn2+ binding signature and is more variable compared to Type I. In Type III ADAMs the M-domain contains the characteristic catalytic-Zn2+ binding signature, but similar to SVM, the D-domain is based on RGD sequence rather than the xCD sequence in most ADAMs. In Type IV ADAMs the D-domain is based on xCD sequence, but they lack the EGF-like region found in Type I, II, and III.
3.1. Signal peptide and pro-domain
ADAMs family of proteins usually have an N-terminus signal peptide which directs the enzyme to carry out its function. In most ADAMs, the signal peptide is the initial part of the molecule followed by the pro-domain. The pro-domain serves to correct protein folding, stabilize the protein, and maintain the enzyme function and catalytic latency via a “cysteine switch” mechanism, which involves blocking the catalytic zinc (Zn2+) activation site by the conserved cysteine residue in the pro-domain [16]. The cysteine switch in the pro-domain should not be confounded with the distinct cysteine-rich domain in the ADAM molecule [17].
3.2. C-shape arm
The C-shape arm is a characteristic structure in the ADAMs family, and is connected to the transmembrane domain via the EGF-like region [9]. The C-shape arm comprises the Metalloproteinase domain (M-domain), Disintegrin domain (D-domain), Cysteine-rich domain (C-domain) and the Highly Variable Region (HVR) of the C-domain. The C-shape arm is crucial for ADAM structure and function as it is responsible for target recognition, protein interaction, and proteolytic activity.
3.3. Metalloproteinase domain
Similar to other metalloenzymes such as MMPs and MT-MMPs, ADAMs have a metalloprotease domain in its active region. The metalloproteinase domain is the main functional region that proteolytically interacts with various ligands, receptors and ion channels on their own cell surface or on their neighboring cells’ membrane. Generally, the metalloproteases domain is characterized by its catalytic Zn2+ binding signature (HExGHxxGxxHD), where H denotes histidine, E glutamic acid, G glycine, D aspartic acid, and x means variable amino acid (aa). The catalytic domain assists ADAMs to serve as sheddases and mediators of cell signaling and thereby determine cell fate (proliferation or apoptosis), and contribute to cell differentiation, tissue organization and physiological function [18, 19].
3.4. Disintegrin domain
Disintegrins are small proteins that were first isolated from snake venom. Disintegrins are recognized by their common Arg-Gly-Asp (RGD) sequence, which could inhibit platelet aggregation via integrin binding. The disintegrin domain is another important functional region in the C-shape arm of ADAMs, and typically consists of 14 amino acid. The term “disintegrin” was initially assigned to this domain because it is highly similar to RGD-containing proteins in snake venom, which are highly conservative among different members of the Metzincins superfamily, and are able to adhere to integrins, thus suppressing platelet aggregation and inducing severe hemorrhage in snake bite victims [20]. However, in most ADAMs the disintegrin domain is not exactly the same as in SVMs. Instead of the highly conserved RGD recognition sequence in SVMs, the binding sequence is replaced by an ECD or xCD sequence in most ADAMs, Only ADAM-15 still conserve the RGD sequence, and is therefore considered unusual among ADAMs family members. Thus, to be more accurate, ECD or xCD sequence in ADAMs should be referred to as “disintegrin-like” domains rather than “disintegrin” domain [21]. ADAMs family present their disintegrin domain on the cell surface, which explains why integrins are common receptors for ADAMs [22].
3.5. Cysteine-rich domain
The Cysteine-rich domain (C-domain) is strongly-related to the cell adhesive and fusogenic potential. Along with cell surface syndecan, the C-domain in ADAM-12 regulates cellular adhesive interactions and in turn modulates the spreading of cells (especially inflammation cells) and their tissue distribution by mediating integrin-dependent cell spreading [23]. Compared to other ADAMs, ADAM-10 and ADAM-17 have a unique sequence and structure in the C-domain, which may partly account for their wide role in various diseases [9]. The C-domain of ADAM-13 is closely related to cell fixation, and can bind to the basement membrane proteins laminin and fibronectin [24]. Also, the highly variable region (HVR) of the C-domain is a potential protein-protein adhesive interface [9]. We should emphasize that the cysteine-rich (C-domain) is one component of the C-shape arm, and is completely different from the cysteine switch which comprises cysteine residues that are unique and exquisite structure in the ADAM pro-domain which maintains the enzyme latency by regulating the Zn2+ binding site [13, 16].
3.6. EGF-like Region, Transmembrane Domain and Cytoplasmic Tail
The epidermal growth factor (EGF)-like region, transmembrane domain and cytoplasmic tail are also important components of the ADAM molecule. These regions connect the extracellular and intracellular components of ADAMs together, and also help to fix the C-shape arm to the cell membrane and transmit the extracellular signal into the cell, and thereby regulate mRNA expression and protein phosphorylation. The cytoplasmic tail of ADAMs also has some intrinsic signaling activity and is a crucial part in regulating proteolysis [25].
3.7. Structural Differences among ADAMs Family Members
Although ADAMs share common components in the signal peptide, pro-domain, C-shape arm (which comprises the metalloproteinase M-domain, disintegrin D-domain, and cysteine-rich C-domain with the highly variable region HVR), and the EGF-like region, transmembrane domain and cytoplasmic tail, there are some differences between various ADAMs members (Fig. 2).
ADAMs members can be divided into four types. Type I is the most common and classic type among all ADAMs, its members have the common domain structure, and it includes ADAM-1, 8, 9, 12, 13, 15, 19, 20, 21, 24, 26, 28, 30, 33, 40. In type I ADAMs, the M-domain have the characterized catalytic Zn2+ binding signature (HExGHxxGxxHD), the D-domain is based on xCD sequence, and they also include the EGF-like region in their structure.
Type II is another common pattern among ADAMs members, and includes ADAM-2, 7, 11, 18, 22, 23, 27, 29, 32. In type II ADAMs, the metalloproteinase domain does not have the characteristic catalytic-Zn2+ binding signature (HExGHxxGxxHD), and is more variable than type I.
Type III is unique for ADAM-15, where the M-domain still contains the characteristic catalytic-Zn2+ binding signature (HExGHxxGxxHD). However, similar to SVM, its disintegrin domain is based on RGD sequence rather than the xCD sequence in most ADAMs members.
Type IV is unique for ADAM-10 and 17, where the disintegrin domain is based on xCD sequence. However, they lack the EGF-like region found in type I, II, and III. The high similarity in the structure of ADAM-10 and 17 may explain the similarities in some of their functions.
3.8. Unique ADAMTS Family Structure
There are 19 ADAMTS genes in mammalian genomes denoted from 1 to 20, except ADAMTS-11 which was assigned to a gene previously identified as ADAMTS-5. Therefore, there are totally 19 ADAMTS proteins in humans, the structure of which is in large part similar to that of ADAMs. Like ADAMs, ADAMTS have the common components including the signal peptide and pro-domain, metalloproteinase M-domain, disintegrin D-domain and cysteine-rich C-domain. The most obvious difference is that ADAMTS lack the EGF-like region, transmembrane domain and cytoplasmic tail, and instead, these domains are replaced by thrombospondin repeats and spacer (Fig. 3). This makes ADAMTS soluble in the circulation, where they can conduct their function in remote tissues.
Fig. 3.
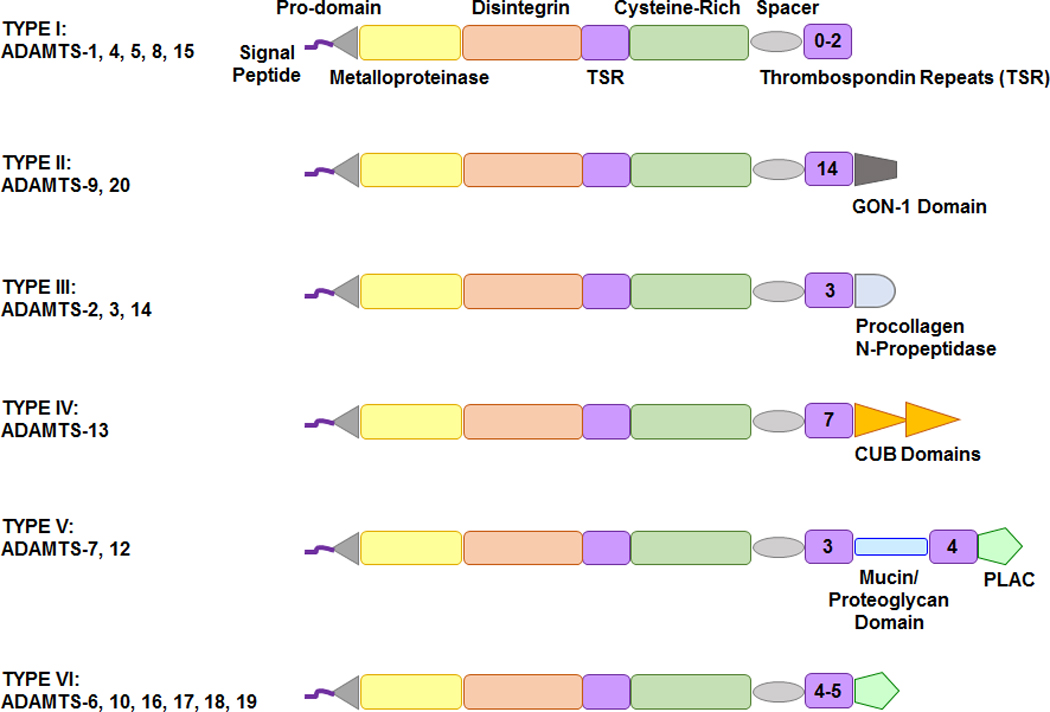
Sequence difference among different types of ADAMTS. ADAMTS members are generally classified into six types. Type I includes several members of ADAMTS and its structure comprises the basic pro-domain, metalloproteinase, disintegrin, cysteine-rich domain, thrombospondin repeats (TSR) and spacer. Type II ADAMTS has a unique GON-1 domain, type III has a procollagen N propeptidase, Type IV has a complement C1r/C1s, Uegf, Bmp1 (CUB) domain, Type V has a protease and lacunin (PLAC) structure and mucin/proteoglycan domain, and Type VI has only PLAC domain. The number inside the final TSR indicates the number of TSR repeats.
The ADAMTS unique ancillary thrombospondin-like repeats (TSR) are critical part for the enzymes’ interaction with ECM components, regulation of enzyme activity, and substrate selection and recognition. ADAMTS members have a 50 aa thrombospondin-like repeat (TSR), which is highly similar to thrombospondins 1, 2, hence the name ADAMTS [26]. After thrombospondin-like repeat (TSR), is the cysteine-rich domain, which usually consists of more than 100 aa residues. This is followed by the cysteine-free spacer region, which consists of 103 to 160 aa. The spacer region is then followed by 0 to 14 TSR modules, which in some ADAMTS are connected to GON-1 domain (present in the ADAMTS GON-1 identified in C. elegans), CUB (complement C1r/C1s, Uegf, Bmp1) domain, or PLAC (protease and lacunin) domain (Fig. 3).
ADAMTS members are generally divided into six types. Type I has the basic structure of ADAMTS, and includes several members of ADAMTS. ADAMTS type I structure comprises the pro-domain, metalloproteinase, disintegrin, cysteine-rich domain, followed by thrombospondin repeats and spacer. Type II ADAMTS has a unique GON-1, type III has a procollagen N-propeptidase, Type IV has a complement C1r/C1s, Uegf, Bmp1 (CUB) domain, type V has a protease and lacunin (PLAC) structure and mucin/proteoglycan domain, and type VI has only PLAC domain.
4. Sources and Tissue Distribution of ADAM and ADAMTS Family
ADAM and ADAMTS members are produced by different cells and have wide tissue distribution (Table 1, 2). Of note, most ADAMs, including ADAM-1, 2, 3, 4, 5, 6, 7, 20, 21, 24, 25, 26, 27, 28, 29, 30, 32 and 34 are involved in the reproduction process and therefore are often localized in the reproductive system and organs of generation such as the testis and epididymis [27]. On the other hand, ADAM-8, 9, 10, 12, 15, 17 and 19 are involved in cardiovascular development, and the abnormal expression or aberrant function of these ADAMs may lead to cardiovascular disease.
Table 1.
Human ADAM Family Members, Gene Locus, Molecular Weight, Tissue Distribution, and Substrates.
| ADAM | Other name | Gene Locus | MW (kDa) | Tissue Distribution | Substrate/Target | Biological or Pathological Role | Ref |
|---|---|---|---|---|---|---|---|
| 1 | Fertilin α, PH-30 α. | 12q24.12 | 84 | Nonfunctional pseudogene, testis | TBD | Sperm form complex, sperm-egg interaction. | [27] |
| 2 | Fertilin β, PH-30 β, Cancer/testis antigen 15 | 8p11.22 | 100 | Testis, epididymis | TBD | Sperm form complex, sperm-egg interaction. | [27] |
| 3 | Cyritestin, tMDC I | 8p11.22 | 80 | Testis, epididymis | TBD | Sperm form complex | [27] |
| 6 | tMDC IV | 14q32.33 | 60 | Testis | TBD | Sperm form complex | [27] |
| 7 | EAP-1, Sperm maturation-related glycoprotein GP-83 | 8p21.2 | 84 | Reproductive system, epididymis, testis | TBD | Transmembrane protein. | [27] |
| 8 | Cell surface antigen MS2, CD156a | 10q26.3 | 90 | Human neutrophils | CHL-1, PSGL-1, CD23, CD-30-ligand, ADAM8 pro-domain, APP, L-selectin, MBP | Osteoclast differentiation, rheumatoid arthritis, neutrophil rolling, trans-endothelial extravasation, neurodegeneration | [62–64] |
| 9 | MDC9, Meltrin γ Myeloma cell metalloprotease | 8p11.22 | 91 | Muscle cell, brain | Pro-HB-EGF, APP, fibronectin, gelatin, Angiotensin-I converting enzyme (ACE), laminin, insulin-B chain, collagen XVII, delta-like ligand 1, EGF, FGFR2IIIb, insulin-like growth factor binding protein-5 | Myogenesis, formation of myotubes; participate in endocardial cushion development (overlap with ADAM-19) | [168] |
| 10 | Kuzbanian protein homologue, CD156c | 15q21.3 | 84 | Embryonic fibroblast, endothelial cells, cardiomyocytes | Pro-HB-EGF, pro-TNFα, Notch, ephrin-α2, APP, cellular prion precursor, gelatin, DDR-1, β-cellulin, CD23, CD30, CD44, prion protein, collagen IV and XVII, fractalkine, delta-like ligand 1, desmoglein, E-cadherin, N-cadherin, VE-cadherin, EGF, ephrin A2 and A5, ERBB2, Fas-L, IL-6R, Klotho, LAG-3, L1, MICA, Pcdh-γ | Increased in dilated cardiomyopathy, atrial fibrillation. Deficiency causes embryonic lethality | [67, 68] |
| 11 | MDC | 17q21.31 | 83 | Nervous system, brain | TBD | TBD | [69] |
| 12 | Meltrin α | 10q26.2 | 100 | Placenta | IGFBP-3 & 5, pro-HB-EGF, EGF, betacellulin, Delta-like1, p-LAP, fibronectin, gelatin, collagen IV | VSMC proliferation, hypertrophic cardiomyopathy. Inhibition prevents cardiac hypertrophy, Down syndrome | [73, 74, 169] |
| 15 | Metargidin, MDC-15 | 1q21.3 | 93 | Cardiomyocyte, endothelial cell. | collage IV, gelatin, amphiregulin, CD23, E-cadherin, HB-EGF, ADAM10 | Inflammation. Contributes to pathological retina neovascularization. Increased in dilated cardiomyopathy, and in atrial fibrillation. |
[81, 170] |
| 17 | TACE, CD156b, Snake venom-like protease | 2p25.1 | 110 | Embryonic fibroblast, cardiomyocyte, smooth muscle | ECM, collagen, pro-TNF-α, pro-TGF-α, proHB-EGF, pro-amphiregulin, TRANCE, pro-neuregulin-a-2C, Notch, Fas ligand, fractalkine, L-selectin, collagen XVII, TNF receptor I and II, interleukin-1 receptor II, interleukin-6 receptor, Erb-B4/HER4, macrophage colony-stimulating factor receptor I, nerve growth factor receptor (TrkA), growth hormone receptor MUC1, APP, cellular prion precursor. | Cardiomyocyte-specific knockout exacerbates post-MI left ventricle dilation and dysfunction due to reduced angiogenesis. ADAM-17 deficient mice die perinatally due to multiple defects including thickened and mis-shaped semilunar and atrioventricular valves, and ventricular septal defect. |
[101, 102] |
| 18 | tMDC III, ADAM-27 | 8p11.22 | 79 | Testis, sperm surface protein | TBD | Highly expressed in conotruncus and endocardial cushion. | [27] |
| 19 | Meltrin β, MADDAM, Metalloprotease and disintegrin dendritic antigen marker | 5q33.3 | 105 | Peripheral nervous system, skeletal muscle, bone, heart | Pro-HB-EGF, ErbB ligands, NGR-β1 and β4, neuregulin, TNF-α, ADAM-19 | ADAM19−/− mice show defects in ventricular septum formation, immature valves, neuronal defects, and die soon after birth. | [105, 171] |
| 20 | NG | 14q24.2 | 83 | Testis specific | TBD | Embryo development | (21) |
| 21 | ADAM-31 | 14q24.2 | 85 | Testis specific | TBD | Present in mature sperm | (21, 22) |
| 22 | MDC2 | 7q21.12 | 100 | Reproductive system, brain | TBD | Expressed in both testicular somatic and germ cell | (21, 23) |
| 28 | MDC-L, ADAM-23 | 8p21.2 | 87 | Reproductive system, brain | TBD | Present in mature sperm | (21) |
| 29 | Cancer/testis antigen 73 | 4q34.1 | 127 | Testis | TBD | Present in mature sperm | (21, 25) |
| 30 | NG | 1p12 | 55 | Testis | TBD | Embryo development | [27] |
| 32 | NG | 8p11.22 | 88 | Testis | TBD | Embryo development | [27] |
| 33 | NG | 20p13 | 87 | Reproductive system | TBD | Embryo development asthma | [27] |
IGFBP-3, insulin growth factor binding protein-3; MW, molecular weight; TBD, to be determined
Table 2.
Human ADAMTS Family Members, Gene Locus, Molecular Weight, Tissue Distribution, and Substrates.
| ADAMTS | Gene Locus | MW (kDa) |
Tissue Distribution | Substrate/Target | Biological or Pathological Role | Ref |
|---|---|---|---|---|---|---|
| 1 | 21q21 | 105 | Heart, bronchial epithelial cells, fetal lung, liver, aorta, smooth muscle, colon, uterus, kidney, adrenal gland, adipocytes, placenta, uterus, ovary, prostate, bladder, spinal cord, ciliary ganglion, olfactory bulb, breast stromal fibroblasts and myoepithelial cells | Aggrecan, versican, syndecan 4, TFPI-2, semaphorin 3C, nidogen-1, −2, desmocollin-3, dystroglycan, mac-2, gelatin (denatured collagen type I), amphiregulin, TGF-α, heparin-binding EGF, VEGF | Cancer (both pro- and antitumorigenic/metastatic), antiangiogenic | [172–174] |
| 2 | 5q35 | 135 | Breast stromal fibroblasts adipocytes, heart, lung, liver, kidney, bladder, aorta, smooth muscle, skeletal muscle, tendon, bone, skin, retina, superior cervical ganglion, uterus, placenta | Fibrillar procollagens types I, II, III and V N-propeptide | Ehlers-Danlos syndrome type VIIc, dermatosparaxis (in sheep and cattle) | [41, 172, 175] |
| 3 | 4q21 | 140–150 | CD105+ endothelial cells, CD34+ cells, heart, lung, pineal gland, cartilage, bone, skeletal muscle, tendon, breast myoepithelial cells, placenta, brain | Fibrillar procollagen type II N-propeptide, biglycan, pro-VEGF-C, reelin | Lymphangiogenesis, placental angiogenesis, brain functions | [11, 41, 107, 129, 172, 176, 177] |
| 4 | 1q23 | 90 | Ovary, adrenal cortex, ciliary ganglion, trigeminal ganglion, brain, spinal cord, retina, pancreas (islets), heart, fetal lung, bladder, uterus, skeletal muscle, breast myoepithelial cells, synovial fluid | Aggrecan, versican, neurocan, reelin, biglycan, brevican, matrilin-3, α2-macroglobulin, oligomeric matrix protein (COMP) | Arthritis | [44, 172, 178] |
| 5 (11) | 21q21 | 73 | Adipocytes, bladder, uterus, placenta, breast myoepithelial cells | Aggrecan, versican, reelin, biglycan, matrilin-4, brevican, α2-macroglobulin | Osteoarthritis, cancer (anti-tumorigenic, anti-angiogenic) | [44, 172, 179, 180] |
| 6 | 5q12 | 116 | Superior cervical ganglion, trigeminal ganglion, appendix, heart, breast myoepithelial cells | TBD | TBD | [172, 180] |
| 7 | 15q24 | 182 | Trigeminal ganglion, adrenal cortex, kidney, liver, pancreas, heart, smooth muscle, skeletal muscle, intervertebral disc, breast stromal fibroblasts | Cartilage oligomeric matrix protein (COMP) | Coronary artery disease (smooth muscle cell migration) | [172, 180, 181] |
| 8 | 11q25 | 80 | Skeletal muscle, heart, lung, liver, superior cervical ganglion, adrenal cortex, breast stromal fibroblasts and luminal epithelial cells, placenta, brain | Aggrecan | Anti-angiogenic | [172, 174] |
| 9 | 3p14 | 217 | Heart, lung, kidney, pancreas, colon, ovary, skeletal muscle, dorsal root ganglion, capillary endothelial cells, breast myoepithelial cells | Aggrecan, versican | Cancer | [172, 182–184] |
| 10 | 19p13 | 121 | Heart, lung, liver, pancreas, kidney, brain, placenta, CD8+ T-cells, brain, uterus, breast stromal fibroblasts | Fibrillin-1 | Weill-Marchesani syndrome | [172, 185, 186] |
| 12 | 5q35 | 178 | Liver, bone marrow, intervertebral disc, atrioventricular node, smooth muscle, breast stromal fibroblasts and myoepithelial cells | oligomeric matrix protein (COMP) | Cell adhesion, cancer | [172, 187] |
| 13 | 9q34 | 190 | Heart, lung, liver, pancreas. kidney, brain, placenta, CD71+ early erythroid cells, endothelial cells, thyroid, breast myoepithelial cells | von Willebrand factor | Deficiency leads to thrombotic thrombocytopenic purpura | [43, 110, 172, 188–191] |
| 14 | 10q21 | 134 | Fibroblasts, lung, liver, prostate, retina, placenta, thalamus, bone marrow, fetal thyroid, adipocytes, cerebellum, bone, skin, breast myoepithelial and luminal epithelial cells | Fibrillar procollagen type I N-propeptide (pNα1 and pNα2 chains) | Linked to multiple sclerosis, osteoarthritis in women | [41, 112–114, 172, 190] |
| 15 | 11q25 | 103 | Colon, brain, heart, uterus, musculoskeletal system, breast myoepithelial cells | Aggrecan, versican | Cancer (anti-tumorigenic/metastatic, anti-angiogenic) | [172, 192] |
| 16 | 5p15 | 136 | Brain, ovary, breast myoepithelial cells, aorta | TBD | Hypertension | [172, 190] |
| 17 | 15q24 | 121 | Ovary, breast myoepithelial cells | TBD | Weill-Marchesani-like syndrome | [172, 190] |
| 18 | 16q23 | 135 | Endothelium, prostate, brain, ciliary ganglion, heart, skin, breast myoepithelial cells | TBD | Microcornea, myopic chorioretinal atrophy and telecanthus |
[43, 44, 142, 172, 190] |
| 19 | 5q31 | 134 | Dorsal root ganglion, breast myoepithelial cells | TBD | TBD | [172] |
| 20 | 12q12 | 215 | Heart, lung, pancreas, prostate, testis, ovary, placenta, brain, appendix, liver, skeletal muscle, pituitary, trigeminal ganglion, breast myoepithelial cells | Versican | Colorectal cancer | [172, 183, 193] |
MW, molecular weight of full-length protein; TBD, to be determined
5. ADAMs and ADAMTS Activation
ADAM family can be activated by cytokines, proteinases, protein kinase C activators, G-protein coupled receptor (GPCR) agonists and Ca2+ ionophores. Compared to MMPs, whose activation occurs in ECM after the pro-domain is removed [2], ADAMs activation occurs in intracellular region where their pro-domain is removed by pro-protein convertases in the Golgi system during transit process. Pro-protein convertase is the most important proteinase during ADAMs activation, as it is responsible for cleaving the Rx(R/K)R motif of the pro-domain. The pro-protein convertase then adds Zn2+ coordination to the metalloprotease domain so that ADAM can be catalytically activated and perform its physiological function and sheddase activity [28].
There are some exceptions in ADAMs activation, and the activation process and the cellular location of active proteinase are markedly different among various ADAMs. For example, ADAM-8 and ADAM-28 are activated in ECM. Also, whether cleavage of the pro-domain is required for the biological activity of ADAM members with an inactive metalloproteinase domain remains an important question. Some ADAMs such as ADAM-12 are stored as already active proteins in the cell, and are transferred to the cell membrane to perform their function when the cell is simulated by cytokines [29]. Therefore, the pro-domain removing procedure is not necessary for ADAM-12 activation, and ADAM-12 can still conserve its function without proteolytic processing which is crucial for other ADAMs [30]. Although, the pro-domain and metalloprotease domain are crucial components for most ADAMs, and the pro-domain acts as an activation switch through its cysteine residue (cysteine switch), some ADAMs auto-catalytically remove the pro-domain in order to complete their transformation from inactive pro-protein to active protein, and proceed to the following activation process [31].
The intracellular cytoplasmic tail and protein phosphorylation may also contribute to ADAMs activation, signal transmit and interaction with other proteins and receptors. However, the role of cytoplasmic tail has been debated, and some reports suggest that ADAM-17 and ADAM-19 can still act as sheddases without a cytoplasmic tail, and rely mainly on their transmembrane domain [32]. For example, although phosphorylated Ser819 was suggested as an important reaction site during activation of ADAM-17, mutation of this site did not suppress ADAM-17 activity and cleavage of its substrates transforming growth factor-α (TGFα), TNFα or TNFα receptors. These observations suggested that the cytoplasmic tail is not responsible for ADAM-17 sheddase activity [32], and instead may have other unique functions. For instance, the cytoplasmic tail of ADAM-17 is strongly correlated with G-protein coupled receptor (GPCR) mediated activation of epidermal growth factor receptor (EGFR) signaling [19]. During these activation processes, Src causes stimulation of phosphatidylinositol 3-kinase (PI3K) and subsequent activation of phosphoinositide-dependent kinase-1 (PDK1), which directly phosphorylates ADAM-17 [33]. Also, agonist-induced phosphorylation of Thr735 of ADAM-17 enhances cleavage of the TrkA neurotrophin receptor in tumor cells and cardiomyocytes. Thr735 phosphorylation also accounts for the transfer of ADAM-17 from the endoplasmic reticulum to the cell surface [34].
The activation process of ADAMTS is basically similar to that of ADAMs. In this respect, ADAMTS activation involves cleavage of the N-terminal pro-domain by proprotein convertases such as furin and furin-like enzymes [35]. These serine proteases efficiently cleave after a consensus sequence Arg-Xaa-(Arg/Lys)-Arg, and the cleavage likely occurs in the trans-Golgi or at the cell surface [36, 37]. Similar to other metalloproteases, the activity of ADAMTS requires a neutral to slightly basic pH and the presence of Zn2+ and Ca2+ ions [38]. However, there are some exception as to the importance and the site of cleavage for ADAMTS activation. ADAMTS-13 has an unusually short propeptide, and ADAMTS-9 and ADAMTS-13 zymogens appear to be active despite retention of the propeptide [39, 40]. Also, for some ADAMTS, cleavage could occur in their C-terminal domain. Studies on ADAMTS-2 have shown that an autocatalytic cleavage occurring within the C-terminal end, possibly in the procollagen N-propetidase domain, increases its activity [35, 41]. A similar autocatalytic activation process has been reported for ADAMTS-1, 4, 8, 9 and 12 [42], suggesting that this autocatalytic activation and regulation could be a common feature of most ADAMTS proteases [41]. C-terminal cleavage of ADAMTS could also have crucial impacts on their bio-disponibility, substrate recognition and activity.
6. ADAMs and ADAMTS Substrates
ADAMs and ADAMTS have multiple substrates in different tissues and organs (Table 1, 2). Similar to MMPs and SVMs, which have a metalloprotease in their crystal structure, ADAMs were initially presumed to cleave ECM components and substrates. ADAM family of enzymes was then recognized to be active proteases. The C-shape arm in ADAMs, comprising the metalloproteinase M-domain, disintegrin D-domain, and the cysteine-rich C-domain with the highly variable region (HVR), is crucial for ADAM function and substrate recognition, proteolytic processing and degradation. HVR regulates cell-cell and cell-matrix interactions in proteolytically inactive ADAMs, and guide ADAMs to recognize specific substrates and mediators. After recognition by HVR, the metalloproteinase M-domain can conduct its cleavage function as a sheddase or proteolytic enzyme (Fig. 4).
Fig. 4.
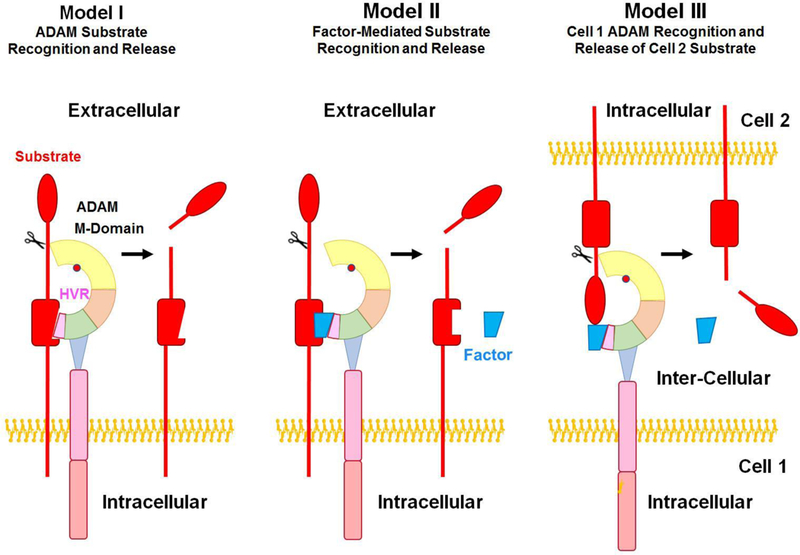
ADAM-substrate interaction. There are three models of ADAM-substrate interaction and ectodomain shedding. In Model I, the highly variable region (HVR) of the cysteine-rich domain directly recognizes specific region of membrane-anchored substrate and the metalloproteinase M-domain accounts for proteolytic activity and substrate shedding and release. In Model II, the substrate recognition requires a specific factor to combine with the substrate so that HVR can recognize and interact with the substrate. Model III is an inter-cellular interaction model, where ADAM on cell 1 surface, with the help of a factor, can shed the substrate from cell 2 surface.
‘Sheddases’ refers to the enzymes which regulate ‘Ectodomain shedding’. Shedding is a proteolytic cleavage process of the extracellular domain of membrane-bound molecules. The shedding could have one of two aspects: 1) converting the extracellular domain into a soluble protein or cytokine that enters into the circulation to perform its whole-body effect; 2) inhibiting or suppressing the activation of a specific receptor and thus down-regulating its signal pathway and function. In addition to their extracellular sheddases activity, ADAMs also possess the ability to regulate intracellular functions and various signaling pathways (Table 1).
There are three models of ADAM-mediated ectodomain shedding. Model I and II involve an extracellular recognition process, whereas Model III is an inter-cellular process (Fig. 4). In Model I, the highly variable region (HVR) of ADAM directly recognizes specific region of membrane-anchored substrate and the M-domain accounts for proteolytic activity. In Model II, the substrate recognition requires a specific factor to combine with the substrate so that HVR can recognize and interact with the substrate. Model III is an inter-cellular interaction model, where with the help of a specific factor, ADAM on one cell surface can shed the cytokine from receptor on another cell surface.
In comparison with ADAMS, ADAMTS are secreted proteases that are primarily associated with proteolytic events in ECM. Most ADAMTS substrates are ECM components (Table 2) [43, 44].
7. ADAMs and ADAMTS Inhibitors
Basically, there are two ways to inhibit ADAMs activity, one way is turn off the cysteine switch, and the other way is to generate more pro-domains to inactivate the active ADAMs. Similar to MMPs, ADAMs activity is suppressed by tissue inhibitor of metalloproteinases (TIMPs) in normal physiological conditions. TIMPs are endogenous four-member family that block the functions of MMPs [2]. While TIMPs have a wide-range inhibitory effect on most of the MMP family, its inhibitory effect on ADAMs is more specific. For instance, TIMP-1 and TIMP-3 can suppress the catalytic activity of ADAM-10 in in vitro conditions, while ADAM-8, 9 and 19 are not inhibited by any TIMP [45, 46]. TIMP-3 inhibits ADAM-17 activity by forming chemical bonds with ADAM-17 dimer on the cell membrane [46]. ADAM-33, which is closely related to the reproduction system, can be inhibited by TIMP-3 and TIMP-4, but is not affected by TIMP-1 [47].
Although TIMPs could inhibit both MMPs and ADAMs, the underlying mechanisms are different for the two families of metalloproteinases. The N-terminal domain of TIMPs is the main domain that account for inhibiting MMPs activity, while the N-terminus of TIMP-1 or TIMP-3 almost has no effect on ADAM-10. Also, the N-terminus of TIMP-2 possesses a potent effect on MMPs, but does not exhibit measurable effects on ADAM-17 activity. Additionally, while TIMP-3 has a solid inhibitory effect on the isolated catalytic domain of ADAM-17, when the other domains are added or some parts of the domains such as cysteine-rich domain are modified, the TIMP-3 inhibitory effect is markedly decreased [18]. Of note, the ‘cysteine-switch’ is another potent ADAMs inhibitory mechanism, whereby the cysteine residues in the pro-domain of some ADAMs have a selective inhibitory effect on themselves, as has been shown for ADAM-10 and ADAM-17 [48, 49].
In addition to endogenous inhibitors, several synthetic small molecule ADAM inhibitors have been developed (Table 3). Most synthetic inhibitors target the catalytic Zn2+, and therefore are not very specific and often have a broad inhibitory spectrum towards both ADAMs and MMPs. Small molecule drugs such as Zn2+ chelators 1,10-phenanthroline and hydroxamate exhibit a potent inhibitory effect on ADAMs activity [50]. Other small molecule synthetic inhibitors have shown relative specificity to ADAMs. For instance, small molecule inhibitor INCB3619 have a potent inhibitory effect on ADAM-10 and ADAM-17, with less potent effect on ADAM-8, 9, and 33 [51]. TAPI-1, TAPI-2, GI254023 and Batimastat (BB-94) are also potent inhibitors of ADAM-17 (TACE), and therefore block the shedding of cytokine receptors effectively, but could also affect MMPs. ,Other inhibitors such as CGS27023, GW280264, and GI254023 exhibit specific inhibitory effect on ADAM-9, 10 and 17 [52]. Recent evidence also suggests that glycosylation of a substrate such as TNFα, upregulated ADAM-8 and 17 activities, but inhibited ADAM-10 activity [53]. Based on this mechanism, more novel drugs which target ADAM-17 can be developed, and could be useful in cardiovascular diseases and cancer treatment.
Table 3.
Endogenous and synthetic inhibitors of ADAMs and ADAMTS.
| Inhibitor | Target | Reference |
|---|---|---|
|
Endogenous: TIMP-1 |
ADAM-10, 17 | [194] |
| TIMP-2 | ADAM-17, 33 | [195] |
| ADAMTS-1 | ||
| TIMP-3 | ADAM-10, 12, 17, 28, 33 | [41, 55, 194, |
| ADAMTS-1, 2, 4, 5 | 196] | |
| TIMP-4 | ADAM-17, 28, 33 | [197] |
| α−2 macroglobulin | ADAMTS-2, 4, 5 | [41, 55, 198] |
| Papilin | ADAMTS-2 | [60] |
|
Synthetic: Batimastat (BB-94) |
ADAM-17, MMPs | [197] |
| Calcium pentosan polysulfate | ADAMTS-7, 12 | [199] |
| CGS27023 | ADAM-9 | [200] |
| Cis-1(S)2(R)-amino-2-indanol-based compounds | ADAMTS-4, 5 | [201] |
| GI254023 | ADAM-10 | [16] |
| GI254023X | ADAM-10, MMP9 | [16] |
| Granulin-epithelin precursor | ADAMTS-7, 12 | [172] |
| GW280264 | ADAM-10, 17 | [16] |
| INCB3619 | ADAM-8, 9, 10, 17, 33 | [16] |
| KP457 | ADAM-10, ADAM 17 | [16] |
| TAPI-1 | ADAM-17/TACE | [202] |
| TAPI-2 | TACE, ADAMs, MMPs | [202] |
Similar to ADAMs, ADAMTS also show restricted susceptibility to endogenous and synthetic inhibitors [54]. As for other proteinases, α2-macroglobulin can physically entraps ADAMTS-2 after cleavage within its bait region [41]. TIMP-3 is an effective inhibitor of ADAMTS-2 and ADAMTS-4 [55, 56]. The function of ADAMTS-1 can be suppressed by TIMP-2 and 3, but is not affected by TIMP-1 and 4 [57]. While TIMP-3 can effectively inhibit ADAMTS-4 and 5, aggrecan may promote this inhibitory effect by reacting with TSR and spacer regions of ADAMTS-4 [58]. Also, heparan sulfate glycosaminoglycans (GAGs) enhance TIMP-3 activity and therefore modulate its inhibitory effect on ADAMTS [59]. Papilin inhibits ADAMTS-2 by binding both the enzyme and its substrate [60]. Other selective small molecule inhibitors such as cis-1(S)2(R)-amino-2-indanol-based compounds are effective inhibitors of ADAMTS-4 and 5. Calcium pentosan polysulfate or granulin-epithelin precursor can block substrate interactions by binding to the ancillary domains of ADAMTS-7 and 12 [61].
8. ADAMs Functions
ADAMs have multiple functions and roles under physiological condition. There are 22 ADAMs in human [62], 14 of these ADAMs including ADAM-1, 2, 3, 6, 7, 18, 20, 21, 22, 28, 29, 30, 32 and 33 are restricted to embryo development, and are mainly expressed and conduct their function in the reproductive system such as testis, epididymis and uterus. Other ADAMs including ADAM-8, 9, 10, 11, 12, 15, 17 and 19 have a broader expression and distribution in human tissues, and wide connection to different pathological conditions. These ADAMs also interact with substrates in the cardiovascular system and regulate cardiovascular physiological condition and may be involved in the pathogenesis of cardiovascular disease, and therefore will be discussed in more details.
ADAM-8 is related to multiple pathological conditions and plays a wide role in rheumatoid arthritis, asthma and inflammation. ADAM-8 also regulates the ovulation process, osteoclast differentiation, neutrophil rolling, and trans-endothelial extravasation. ADAM-8 is expressed in human breast cancer, hematopoietic cells, and human neutrophils, where it is usually stored in granules and transported to the cell surface when it is stimulated. ADAM-8 can also mediate L-selectin, which acts as a “homing receptor” for lymphocytes, and its shedding by ADAM-8 plays an important role in regulation of neutrophil rolling and trans-endothelial extravasation. To further investigate the role of ADAM-8, an inactive mutant of ADAM-8 mouse was developed and showed suppressed reaction to autoimmune arthritis. Although ADAM-8−/− mice develop and grow normally without a disease phenotype [63], ADAM-8 could contribute to cardiac inflammation as its levels are increased in the peri-infarct myocardium following myocardial infarction, which is often associated with infiltration of inflammatory cells [64].
ADAM-9 has a broad expression and is catalytically active. It is involved in heparin-binding EGF-like growth factor (HB-EGF) ectodomain shedding, and may protect against the development of Alzheimer’s disease by reducing the processing of Amyloid Precursor Protein (APP). ADAM-9 was initially found to regulate the fusion of muscle cells development and to have a regulatory role in myogenesis and formation of myotubes and myocardium development. However, when ADAM-9−/− mice were later developed, no major defects in muscle formation, HBA-EGF shedding function or APP α-secretase products formation were observed (73), suggesting that the function of ADAM-9 can be compensated by other ADAMs under physiological conditions.
ADAM-10 is mainly involved in embryonic development, ECM degradation, ectodomain shedding of cell surface proteins, and signal regulation in distant cells. ADAM-10 is one of the hub regulatory factors during the Notch and Eph/ephrin pathways processing. By inducing Notch cleavage and controlling subsequent “regulated intramembrane proteolysis” (RIP), ADAMs regulate the generation of cleaved intracellular domains which conduct its function by transferring to the nucleus and regulating gene transcription and mRNA expression. ADAM-10 regulation of Notch signaling is an important step in the cardiovascular system embryonic development [65, 66]. Both ADAM-10 and Notch signaling are of great importance for cardiovascular development [67, 68]. Because of the crucial role of ADAM-10 in various physiological process including RIP, Notch/Delta signaling and APP processing, ADAM-10 knock-out mice do not survive during early embryonic development. ADAM-10−/− mice usually show multiple defects in the central nervous system, cardiovascular system and somites, which closely resemble those seen in mice deficient in Notch/Delta signaling [65]. ADAM-10 mice die of severe cardiovascular defects after embryonic day 10, which is very similar to Notch1 deficient mice [66].
ADAM-11 is mainly distributed in the nervous system especially in the hippocampus and cerebellum, with little expression in other tissues such as the heart, kidney, eyes and brown fat. ADAM-11 may serve as an adhesion component for synapses formation in the heart, kidney and eyes. ADAM-11−/− mice have a normal development phenotype, but suffer from learning deficiency [69].
ADAM-12 was initially thought to mainly take part in myogenesis during embryonic development [70]. ADAM-12 has two splice-variants, ADAM-12L and ADAM-12S, both have proteolytic activity and affect distant cell physiological targets by controlling the shedding of various membrane-bound ectodomain substrates from the cell surface. ADAM-12L can mediate cardiomyocyte hypertrophy by controlling the shedding of HB-EGF [71, 72], and Notch signal pathway [73], and thereby regulates the disease progression [73, 74]. ADAM-12−/− mice usually develop as adults without specific signs or functional deficiencies, although some reports found 30% mortality in ADAM-12−/− pups within the first week [75], and therefore there is no agreement regarding the physiological necessity of ADAM-12.
ADAM-15 cleaves E-cadherin, thus compromising epithelial and endothelial connections and reducing cell connections and tissue stability, and in turn inducing migration and infiltration of inflammatory cells in the cardiovascular system [76, 77]. By activation of Src kinase, phosphorylation of ERK1/2 and cleaving E-cadherin, ADAM-15 decreases the stability of endothelial cells thus promoting endothelial cell permeability, neutrophil trans-endothelial migration and inflammatory cell infiltration [78]. By inducing inflammation, ADAM-15 could contribute to cardiovascular diseases such as atherosclerotic lesions, myocarditis and myocardial infarction. ADAM-15 is also related to vessel formation, and has been shown to induce the expression of vascular endothelial growth factor (VEGF), VEGFR1 and VEGFR2, which are closely related to angiogenesis [79]. ADAM-15 could also participate in cancer metastasis by increasing the migration and invasiveness of tumor cells [80]. ADAM-15−/− mice have been developed, and while no overt phenotype is found, tumor cell migration and invasion are reduced in these mice [81].
ADAM-17 was first and simultaneously described by two research groups in 1994 as the enzyme that releases membrane-bound TNFα precursor to a soluble form. Because of this sheddase function of membrane-bound TNFα, ADAM-17 was first termed TNFα converting enzyme (TACE) [82–84], but was later classified as a member of ADAM family. ADAM-17 has very similar sequence and crystal structure to ADAM-10, suggesting similarity in function. ADAM-17 has wide functions during embryonic development including fertilization, neurogenesis, and angiogenesis. Because of its wide involvement in development, ADAM-17−/− embryos die perinatally [85], supporting its essential role in embryonic development [86]. ADAM-17 plays a regulatory role on TNFα processing and function. TNFα is over-expressed and its plasma levels are elevated in cardiovascular disease. Experimental studies in animal models have suggested that inhibiting the TNFα pathway could have therapeutic benefits in cardiovascular disease [87, 88]; however, anti-TNFα treatment did not show much improvement in patients with late-stage heart failure [89, 90]. In contrast with the perinatal death in ADAM-17 deficient mice, TNFα-deficient mice develop normally and are viable and fertile, indicating that ADAM-17 has other important functions and is not limited to TNFα processing [86]. Of note, TNFα processing is not completely inhibited by the lack of ADAM-17 in ADAM-17 knockout cells, and other proteins including ADAM-9 and 10 [91, 92], and MT4-MMP (MMP17) [93] may also be involved in the shedding of TNFα. Interestingly, ADAM-17 could have a dual role in modulating TNFα. On one hand, ADAM-17 helps release soluble TNFα into the circulation to activate its receptor. On the other hand, ADAM-17 can also cleave TNFα receptors, especially TNFα-RI (p55) and TNFα-RII (p57), and the shedding of the TNFα receptors could reduce the sensitivity to TNFα and ultimately lead to a negative effect even if large amount of the TNFα ligand is released into the circulation [32, 94]. Some studies suggest that ADAM-17 mediated cleavage of TNFα receptor accounts for the primary effect [95, 96]. ADAM-17 also regulates the shedding of other ectodomain membrane-anchored proteins including the adhesion molecules selectins, vascular cell adhesion molecule-1 (VCAM-1), and intercellular adhesion molecule-1 (ICAM-1), the cytokines interleukin-1 receptor-2 (IL1-R2) and IL6-R, and the growth factors TGFα, HB-EGF, VEGF, neuregulins and VEGFR2 [18, 97]. Activation of EGFR, through the shedding of membrane-anchored pro-TGFα [85] and pro-HB-EGF [98], is among the critical functions of ADAM-17 [99–101], when compared to other candidate HB-EGF sheddases such as ADAM-9, 12 and 15 [18, 102]. ADAM-17 knock-out mice embryos’ heart valves are aberrant, thickened and mis-shaped, similar to the mice lacking HB-EGF, demonstrating wide pathogenic role of ADAM-17 [99–101, 103]. Thus, ADAM-17 could regulate various physiological functions including inflammation, growth, angiogenesis and neural function through regulation of multiple substrates, and could play a role in cardiovascular physiological and pathological changes.
ADAM-19 is mainly distributed in the peripheral nervous system, skeletal muscle and the heart [70, 104]. ADAM-19 is responsible for endocardial cushion development and controls the epithelial-to-mesenchymal transformation of embryonic endocardial epithelial cells which generates most of the cushion mesenchymes that constitute the main components of the cardiac septa and valves. To verify its physiological role, ADAM-19−/− mice have been developed and show severe deficiency in ventricular septum development and other system development failure, and ultimately die soon after birth [105].
9. ADAMTS Functions
ADAMTS are widely distributed in different tissues and organs, and could play crucial embryologic or physiological roles in humans. The important role of ADAMTS in mammalian physiology has been supported by strong phenotypes compromising morphogenesis, mobility or reproduction in several ADAMTS gene knockout mice. Mutations in ADAMTS-2, 3, 10, 13, 17 and 20 have been associated with Mendelian disorders or birth defects [43]. The procollagen N-proteinases ADAMTS-2, 3 and 14 play an important role in procollagen processing and collagen fibrils formation, maturation and assembly [41]. ADAMTS-2 deficient mice have been developed and phenotypically characterized. Although ADAMTS-2 heterozygous male and female mice display a normal phenotype at birth, they show progressively fragile skin by two weeks of age, and adult males display abnormal seminiferous tubules and lack of spermatozoids production and become sterile [106]. ADAMTS-3 is important for lymphangiogenesis, and ADAMTS-3 deficient mouse embryos do not survive past 15 days of gestation due to severe lymphedema resulting from lack of lymphangiogenesis [107]. ADAMTS-5 has been implicated in the pathogenesis of osteoarthritis via degradation of its substrate aggrecan, a major structural proteoglycan in cartilage [43]. Mutations in ADAMTS-10 and 17 have been associated with Weill–Marchesani syndrome, characterized by short stature, acromelic dysplasia and disproportionate distal limb shortening [43, 108, 109]. ADAMTS-13 cleaves ultra-large vWF multimers, which are prothrombogenic and favor platelet adhesion, into an optimal size required for normal hemostasis [110]. Therefore, ADAMTS-13 may function as a hemostatic agent [111]. Genetic association studies have linked ADAMTS-14 to the predisposition to multiple sclerosis and knee osteoarthritis in women by unclear mechanisms [41, 112–114]. ADAMTS-20 variant has been associated with cleft lip and palate [115].
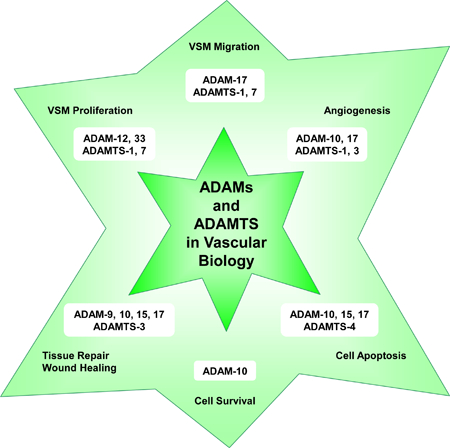
10. Role of ADAMs and ADAMTS in Vascular Biology
ADAM and ADAMTS family members play an important role in numerous vascular biological functions including VSM cell proliferation and migration, angiogenesis, vascular cell apoptosis and survival, and tissue repair and wound healing (Fig. 5).
Fig. 5.
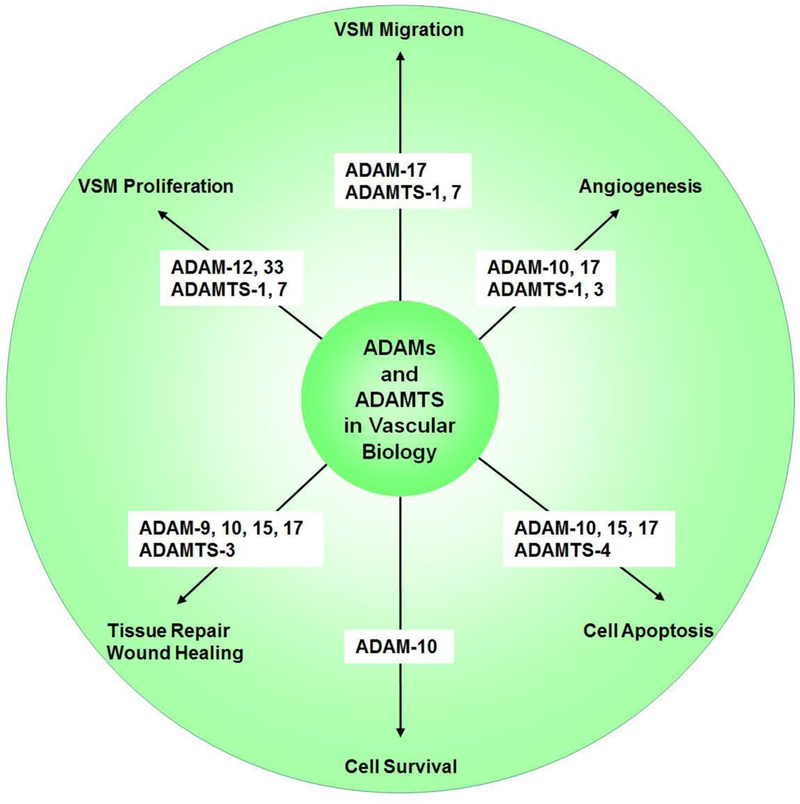
Role of ADAMs and ADAMTS in vascular biology.
ADAM-12 and 33 are crucial factors in airway smooth muscle and VSM proliferation, and are therefore closely related to bronchial asthma [116] and carotid artery lesions [117]. ADAM-17 is strongly associated with hypertrophy and hyperplasia of VSM cells [118]. ADAMTS-7 promotes VSM proliferation both in vivo and in vitro [119].
ADAMTS-1 appears to promote VSM proliferation and migration, as MiR-365b-3p and miR-362–3p inhibit VSM proliferation and migration and attenuates atherosclerosis by targeting ADAMTS-1 [120, 121]. ADAMTS-7 mediates VSM cell migration and neointima formation in balloon-injured rat arteries [122]. Also, ADAMTS-7 knock-out mice show reduced risk of atherosclerosis likely due to loss of the effect of ADAMTS-7 on VSM migration [123]. ADAMTS-7 accumulates in VSMCs in coronary and carotid atherosclerotic plaques, and genome-wide association studies have suggested an association between variation at ADAMTS-7 locus and susceptibility to coronary artery disease. In a population-based study cohort, an inverse association was observed between atherosclerosis prevalence and rs3825807, a nonsynonymous single nucleotide polymorphism SNP (A to G) leading to a Ser-to-Pro substitution in ADAMTS-7 pro-domain. Additional mechanistic studies of this rs3825807 variant associated with protection from atherosclerosis and coronary artery disease showed reduced ADAMTS-7 maturation and pro-domain cleavage and function, a decrease in the substrate thrombospondin-5 cleavage, and reduced VSMC migration [124].
During angiogenesis, ADAMs are activated by VEGF, and in turn induce Notch ectodomain shedding. Although ADAM-10 and ADAM-17 have been implicated in Notch-signaling, they appear to have different roles in angiogenesis. In mouse retinas, ADAM-10 inhibition induced vascular sprouting, whereas inhibition of both ADAM-10 and ADAM-17 produced the opposite phenotype. Besides Notch, ADAM-17 appeared to regulate other cytokines and signaling pathways during angiogenesis, and these pathways were not affected by ADAM-10. ADAM-17 inhibition induced the expression of a naturally occurring inhibitor of angiogenesis thrombospondin 1 (TSP1), whereas ADAM-10 inhibition did not [125]. These observations have suggested that ADAM-10 and ADAM-17 have opposite effects on sprouting angiogenesis that may be unrelated to Notch signaling and may involve differentially expressed anti-angiogenic proteins such as TSP1 [125]. ADAMTS-1 has been associated with angiogenesis in gastric cancer [126] and metastasis [127], and lung cancer [128]. ADAMTS-3 could cleave and activate the substrate pro-VEGF-C into active VEGF-C and in turn play a role in embryonic lymphangiogenesis and the regulation of placental angiogenesis [107]. ADAMTS-3 also participates in triggering VEGFR-3 signaling, a cornerstone for the differentiation and function of lymphatic endothelial cells [129], and loss of ADAMTS-3 activity has been associated with Hennekam lymphangiectasia-lymphedema syndrome [130]. On the other hand, recombinant ADAMTS-2 reduces proliferation of endothelial cells, decreases branching of capillary-like structures formed by endothelial cells, and inhibits vessels formation in embryoid bodies [131]. These ADAMTS-2 antiangiogenic properties may not require catalytically active enzyme, and may involve interaction of one of its C-terminal domains with nucleolin, a nuclear protein and potential receptor [41, 131]. Similarly, ADAMTS-12 shows antiangiogenic effects independent of its enzymatic activity as a mutated inactive form of the enzyme is similarly efficient in inhibiting endothelial cell sprouting in the aortic ring assay when compared to the control wild-type form, and these ADAMTS-12 antiangiogenic properties could protect the host from tumor progression [132]. ADAMTS-15 also inhibits angiogenesis and liver cancer metastasis by modulating cell-ECM interactions [133].
ADAM-10, 15 and 17 are involved in vascular cell apoptosis. ADAM-10 regulates the cleavage of N-cadherin to increase apoptosis in human atherosclerotic lesions and promote the risk of vulnerable plaque [134]. Recombinant human ADAM-15 induces vascular apoptosis and inhibits tumor formation and angiogenesis [135]. ADAM-17 can induce vascular apoptosis and mediate the invasion and migration of liver cancer stem cells, and suppression of ADAM17 could be a promising target to enhance cancer therapy [136, 137]. Also, deletion of Cdc42 increases the activity of ADAM-17 and enhances VEGFR-2 shedding and reduces vascular endothelial cell survival and angiogenesis, and inhibition of ADAM-17 can partially reverse Cdc42 deletion-induced EC apoptosis [137]. Also, ADAMTS-4 induces VSMC apoptosis, degrades versican, and promotes inflammatory cell infiltration of blood vessels [138]. Conversely, some studies suggest that ADAM-10 may be associated with vascular cell survival as ionizing radiation reduces ADAM-10 expression and induces neurovascular cell senescence and death [139].
ADAM and ADAMTS show different expression in wound healing tissues and normal tissues, suggesting that they play a role in tissue repair and wound healing processes. ADAM-9, 10, 15 and 17 are highly expressed in wound healing tissues [140]. ADAMTS-3, also shows differential expression in wound healing tissues [140].
11. Role of ADAMs and ADAMTS in Cardiovascular Pathology
ADAMs and ADAMTS have been associated with pathological changes associated with cardiovascular diseases such as atherosclerosis, hypertension, aneurysm, coronary artery disease, myocardial infarction and heart failure (Fig. 6).
Fig. 6.
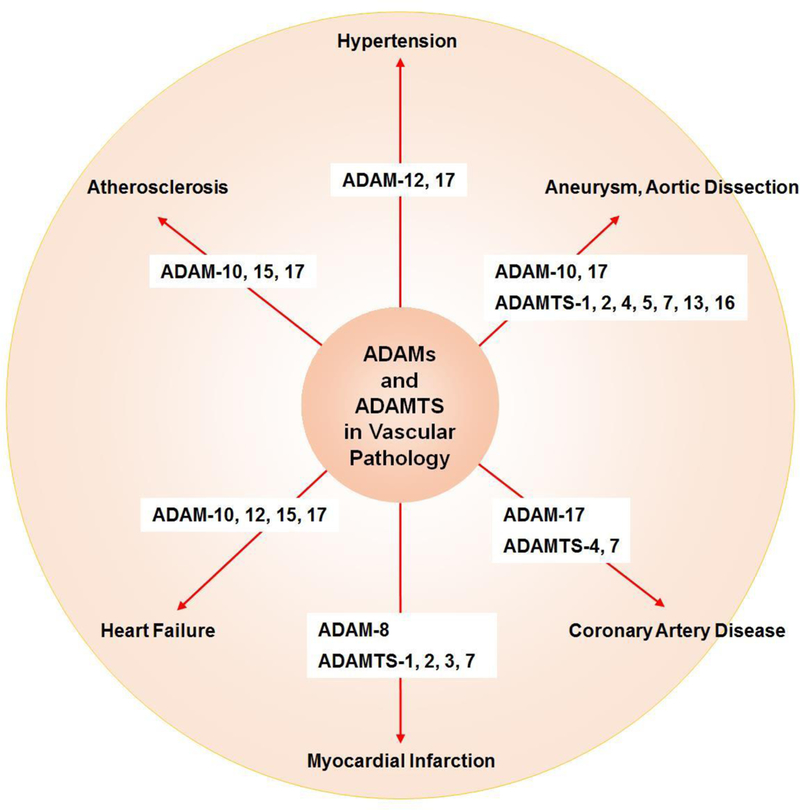
Role of ADAMs and ADAMTS in cardiovascular pathology.
Atherosclerosis is a chronic inflammatory disease characterized by aberrant accumulation of lipids and inflammatory cytokines in blood vessels, and ADAM-10, 15, 17 could promote the atherosclerotic process through inflammatory cell recruitment and differentiation. ADAM-10, 15, 17 control macrophage phenotype and function by cleaving TNFα and TNFα-Rs. ADAM-17 also decomposes the scavenger receptor CD163 and in turn modulates macrophage polarization and M2 macrophage activity. ADAM-10 and 17 are also involved in the shedding of VEGF-R2 and Tie-2. Also, angiogenesis plays a role in atherosclerotic lesion growth and plaque stability, and ADAM-10, 17 could regulate the atherosclerotic process through modulating neovessel formation [141].
ADAMs and ADAMTS are also associated with hypertension. Loss and mutation of ADAM-17 suppress angiotensin II-induced hypertension and end-organ damage [118]. On the other hand, ADAMTS-18 knock-out mice show visceral adiposity and metabolic syndrome with increased risk of diabetes, cardiovascular disease and hypertension [142].
ADAM-10, ADAM-17, and ADAMTS-1, 4, 5, 7 and 16 have been associated with aortic aneurysm formation. Clinical studies have suggested that ADAM-10 and ADAM-17 positive microvesicles are linked to increased risk of abdominal aortic aneurysm in smokers [143]. ADAM-17 has also been linked to the formation of abdominal aortic aneurysm induced by angiotensin II [144]. ADAMTS-1, 5, and 16 are increased in patients with acute aortic dissection and thoracic aortic aneurysm [145, 146]. ADAMTS-4 has been linked to sporadic aortic aneurysm and dissection in mice. ADAMTS-4 has been shown to decompose and degrade poly ADP-ribose polymerase-1, which is a key factor in DNA repair and cell survival, leading to smooth muscle cell apoptosis, vascular damage and aortic aneurysm and dissection. In support, ADAMTS-4 knock out mice show reduced challenge-induced aortic diameter enlargement, aneurysm and dissection formation, and risk of artery rupture [147]. Upregulation of ADAMTS-7 and downregulation of cartilage oligomeric matrix protein (COMP) could also be important factors underlying the pathogenesis and formation of aortic aneurysm [148].
Genetic ADAMTS-2 variant rs11750568 has been associated with cerebral aneurysm and pediatric stroke [149, 150]. Also, genetic ADAMTS-13 variants rs2301612 and rs2285489: have been associated with the risk of cerebral aneurysm, While ADAMTS-12 variant rs1364044: and ADAMTS-13 variants rs739469 and rs4962153 have been associated with a protective effect in cerebral aneurysm pathogenesis [149].
ADAMs and ADAMTS may play a role in coronary artery diseases, ADAM-17 related substrates have been linked to the recursion of cardiovascular events in patients suffering from atherosclerosis [151]. ADAM-17 may also regulate angiogenesis and myocardial cell repair post-myocardial infarction in patients with coronary artery disease [141, 152]. ADAMTS-4 is increased in the serum of patients who suffer from coronary artery disease and acute coronary syndromes [153], and statin therapy reduces the serum levels of ADAMTS-4 and ameliorates the symptom of coronary artery disease [154]. ADAMTS-7 promotes atherosclerosis and has been associated with coronary artery disease [43]. Plasma ADAMTS-7 is elevated in patients with coronary artery disease [155]. Genome-wide association studies (GWAS) and single nucleotide polymorphism (SNP) maps have led to a Ser214Pro substitution in the ADAMTS-7 propeptide that could be associated with coronary artery disease [156]. The Pro variant likely impairs ADAMTS-7 propeptide excision by furin, which is required for its proteolytic activity. Therefore, the Pro variant may have lower activity and individuals with the Pro/Pro ADAMTS-7 protein could have reduced protease activity compared to those with Ser/Ser variants [124]. ADAMTS-7 may also inhibit re-endothelialization and promote neointima formation in wire-injured carotid arteries of mice, as ADAMTS-7 deficiency in ADAMTS-7−/− mice promoted vascular re-endothelialization and ameliorated neointima formation post-injury. ADAMTS-7 may be a potential treatment target for post-injury vascular intimal hyperplasia [157].
ADAMs and ADAMTS could play a role in myocardial infarction. In a rat model of myocardial infarction induced by ligation of the left anterior coronary artery, the expression of ADAM-8 was increased in myocardial infarction rats compared with controls. Immunohistochemistry in the myocardial infarction grafts revealed that ADAM-8 was localized to the vicinity of the area of the developing infarction as well as in intramyocardial arteries remote to the infarction area, suggesting global myocardial impact of myocardial infarction [64]. On the other hand, ADAM-17, through regulating angiogenesis may play a role in post-myocardial infarction repair. Cardiomyocyte-specific ADAM-17 knockdown mice are more vulnerable to myocardial infarction following ligation of the left anterior descending coronary artery [152]. ADAM-17 knockdown mice have unfavorable survival, higher rates of cardiac rupture, more severe ventricular dilation and decreased ejection fraction compared with control wild-type group. ADAM-17 knockdown mice also have suppressed VEGFR2 expression likely through modulation of nuclear factor-κB activation and DNA binding [152]. ADAMTS-1 usually have a medium expression in normal tissues, but is markedly increased in pathological conditions associated with inflammation and hypoxia [158]. ADAMTS-2 and 3 are more abundant in the culprit plaques from patients presenting with acute myocardial infarction versus stable angina, and they colocalize with CD31 and CD68 suggesting their expression by endothelial cells and macrophages [159]. Plasma ADAMTS-7 levels have also been associated with ventricular remodeling in patients with acute myocardial infarction [160]. Other studies have shown that ADAMTS-13 markedly decreases in patients with ST-elevation myocardial infarction (STEMI) and intramyocardial hemorrhage, and that intracoronary administration of recombinant ADAMTS-13 does not decrease infarct size or intramyocardial hemorrhage in a porcine model of myocardial ischemia-reperfusion [161]. Myocardial infarction may also affect some ADAM members, it can induce increased remote ADAM-8 expression of rat hearts after cardiac arrest in a very early period [64].
ADAMs and ADAMTS could be involved in heart failure. Excessive expression of ADAM-10, 12, 15 and 17 cause cardiac hypertrophy and dilated cardiomyopathy, whereas the aberrant expression of ADAM-10 and 15 induce atrial fibrillation [14, 162]. ADAM-12 is differentially expressed in the heart during cardiac hypertrophy. In a mouse model of heart failure induced by aorta-to-vena cava arteriovenous shunt, the cardiac levels of ADAM-12 and tissue fibrosis were increased [163]. Other data suggest that hydrogen sulfide (H2S) donors could ameliorate heart failure by decreasing ADAM-12 [163–165]. ADAMTS-2 has been implicated in the pathogenesis of cardiac hypertrophy and cardiomyopathy [166]. ADAMTS-2 participates in the cleavage of the N-propeptide from types I, II, III, and V procollagens, thus allowing the formation and assembly of collagen fibrils. ADAMTS-2 knockout mice exhibit severe cardiac hypertrophy and heart failure likely because in the absence of ADAMTS-2 collagen fibrils are abnormal and do not provide the required mechanical strength to counterbalance the pressure overload, and overexpression of ADAMTS-2 in mice reverses this damaging phenotype [167].
12. Perspective
ADAMs are metalloproteases related to MMPs with characteristic disintegrin and transmembrane domains. ADAMTS members lack the transmembrane domain and instead have thrombospondin motifs. ADAMs play a role in the shedding of various membrane-bound proteins, and can regulate multiple cellular functions and physiological conditions by regulating various cytokines and their receptions. ADAMs are widely related to different disease processes, especially cardiovascular diseases. ADAM-10, and 17 are promising biomarkers and therapeutic targets in cardiovascular diseases. Like ADAMs, ADAMTS have important functions and roles in cardiovascular biology and disease. TIMPs are endogenous inhibitors of ADAMs and ADAMTS. Synthetic ADAM and ADAMTS inhibitors have been developed with varying specificity and selectivity. Further research into the potential substrates and function of ADAMs and ADAMTS should help develop specific modulators with potential usefulness in the management of cardiovascular disease.
Acknowledgements
This work was supported by grants from National Heart, Lung, and Blood Institute (HL-65998, HL-111775). Dr. S. Zhong was a visiting scholar from The First Hospital of Jilin University, Changchun, Jilin, P.R. China.
List of Abbreviations:
- aa
amino acid
- ADAM
A Disintegrin and Metalloproteinase
- ADAMTS
A Disintegrin and Metalloproteinase with Thrombospondin Motifs
- APP
amyloid precursor protein
- C-domain
cysteine-rich domain
- COMP
cartilage oligomeric matrix protein
- CUB
C1r/C1s Uegf Bmp1
- D-domain
disintegrin domain
- ECM
extracellular matrix
- EGF
epidermal growth factor
- EGFR
EGF Receptor
- GPCR
G-protein coupled receptor
- HB-EGF
heparin-binding EGF-like growth factor
- HVR
highly variable region
- M-domain
metalloproteinase domain
- MDC
metalloproteinase/disintegrin/cysteine-rich
- MMP
matrix metalloproteinase
- MT-MMP
membrane-type MMP
- PLAC
protease and lacunin
- RGD
Arg-Gly-Asp
- RIP
regulated intramembrane proteolysis
- SVM
snake venom metalloproteinase
- TNFα
tumor necrosis factor-α
- TACE
TNFα-converting enzyme
- TIMP
tissue inhibitor of metalloproteinases
- TSR
thrombospondin repeat
- VAP1
vascular adhesion protein-1
- VCAM-1
vascular cell adhesion molecule-1
- VEGF
vascular endothelial growth factor
- VEGFR
VEGF receptor
- VSM
vascular smooth muscle
- VSMC
VSM cell
- VWF
von Willebrand factor
Footnotes
Conflict of Interest
None
References
- [1].Fan D, Creemers EE, Kassiri Z. Matrix as an interstitial transport system. Circulation research 2014;114:889–902. [DOI] [PubMed] [Google Scholar]
- [2].Takawale A, Sakamuri SS, Kassiri Z. Extracellular matrix communication and turnover in cardiac physiology and pathology. Comprehensive Physiology 2015;5:687–719. [DOI] [PubMed] [Google Scholar]
- [3].Cui N, Hu M, Khalil RA. Biochemical and Biological Attributes of Matrix Metalloproteinases. Prog Mol Biol Transl Sci 2017;147:1–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wang X, Khalil RA. Matrix Metalloproteinases, Vascular Remodeling, and Vascular Disease. Adv Pharmacol 2018;81:241–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes & development 2003;17:7–30. [DOI] [PubMed] [Google Scholar]
- [6].Raffetto JD, Khalil RA. Matrix metalloproteinases and their inhibitors in vascular remodeling and vascular disease. Biochemical pharmacology 2008;75:346–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wong GE, Zhu X, Prater CE, Oh E, Evans JP. Analysis of fertilin alpha (ADAM1)-mediated sperm-egg cell adhesion during fertilization and identification of an adhesion-mediating sequence in the disintegrin-like domain. The Journal of biological chemistry 2001;276:24937–45. [DOI] [PubMed] [Google Scholar]
- [8].Giebeler N, Zigrino P. A Disintegrin and Metalloprotease (ADAM): Historical Overview of Their Functions. Toxins 2016;8:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Takeda S, Igarashi T, Mori H, Araki S. Crystal structures of VAP1 reveal ADAMs’ MDC domain architecture and its unique C-shaped scaffold. The EMBO journal 2006;25:2388–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kuno K, Kanada N, Nakashima E, Fujiki F, Ichimura F, Matsushima K. Molecular cloning of a gene encoding a new type of metalloproteinase-disintegrin family protein with thrombospondin motifs as an inflammation associated gene. The Journal of biological chemistry 1997;272:556–62. [DOI] [PubMed] [Google Scholar]
- [11].Fernandes RJ, Hirohata S, Engle JM, Colige A, Cohn DH, Eyre DR, et al. Procollagen II amino propeptide processing by ADAMTS-3. Insights on dermatosparaxis. The Journal of biological chemistry 2001;276:31502–9. [DOI] [PubMed] [Google Scholar]
- [12].Bolz H, Ramirez A, von Brederlow B, Kubisch C. Characterization of ADAMTS14, a novel member of the ADAMTS metalloproteinase family. Biochim Biophys Acta 2001;1522:221–5. [DOI] [PubMed] [Google Scholar]
- [13].Bode W, Gomis-Ruth FX, Stockler W. Astacins, serralysins, snake venom and matrix metalloproteinases exhibit identical zinc-binding environments (HEXXHXXGXXH and Met-turn) and topologies and should be grouped into a common family, the ‘metzincins’. FEBS letters 1993;331:134–40. [DOI] [PubMed] [Google Scholar]
- [14].Fedak PW, Moravec CS, McCarthy PM, Altamentova SM, Wong AP, Skrtic M, et al. Altered expression of disintegrin metalloproteinases and their inhibitor in human dilated cardiomyopathy. Circulation 2006;113:238–45. [DOI] [PubMed] [Google Scholar]
- [15].Takeda S. ADAM and ADAMTS Family Proteins and Snake Venom Metalloproteinases: A Structural Overview. Toxins 2016;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhang P, Shen M, Fernandez-Patron C, Kassiri Z. ADAMs family and relatives in cardiovascular physiology and pathology. Journal of molecular and cellular cardiology 2016;93:186–99. [DOI] [PubMed] [Google Scholar]
- [17].Van Wart HE, Birkedal-Hansen H. The cysteine switch: a principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family. Proceedings of the National Academy of Sciences of the United States of America 1990;87:5578–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Blobel CP. ADAMs: key components in EGFR signalling and development. Nature reviews Molecular cell biology 2005;6:32–43. [DOI] [PubMed] [Google Scholar]
- [19].Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Molecular aspects of medicine 2008;29:258–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gould RJ, Polokoff MA, Friedman PA, Huang TF, Holt JC, Cook JJ, et al. Disintegrins: a family of integrin inhibitory proteins from viper venoms. Proceedings of the Society for Experimental Biology and Medicine Society for Experimental Biology and Medicine (New York, NY) 1990;195:168–71. [DOI] [PubMed] [Google Scholar]
- [21].Blobel CP. Metalloprotease-disintegrins: links to cell adhesion and cleavage of TNF alpha and Notch. Cell 1997;90:589–92. [DOI] [PubMed] [Google Scholar]
- [22].Blobel CP, White JM. Structure, function and evolutionary relationship of proteins containing a disintegrin domain. Current opinion in cell biology 1992;4:760–5. [DOI] [PubMed] [Google Scholar]
- [23].Iba K, Albrechtsen R, Gilpin B, Frohlich C, Loechel F, Zolkiewska A, et al. The cysteine-rich domain of human ADAM 12 supports cell adhesion through syndecans and triggers signaling events that lead to beta1 integrin-dependent cell spreading. The Journal of cell biology 2000;149:1143–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Smith KM, Gaultier A, Cousin H, Alfandari D, White JM, DeSimone DW. The cysteine-rich domain regulates ADAM protease function in vivo. The Journal of cell biology 2002;159:893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Stone AL, Kroeger M, Sang QX. Structure-function analysis of the ADAM family of disintegrin-like and metalloproteinase-containing proteins (review). Journal of protein chemistry 1999;18:447–65. [DOI] [PubMed] [Google Scholar]
- [26].Adams JC, Lawler J. The thrombospondins. Cold Spring Harbor perspectives in biology 2011;3:a009712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Cho C. Testicular and epididymal ADAMs: expression and function during fertilization. Nature reviews Urology 2012;9:550–60. [DOI] [PubMed] [Google Scholar]
- [28].Anders A, Gilbert S, Garten W, Postina R, Fahrenholz F. Regulation of the alpha-secretase ADAM10 by its prodomain and proprotein convertases. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 2001;15:1837–9. [DOI] [PubMed] [Google Scholar]
- [29].Sundberg C, Thodeti CK, Kveiborg M, Larsson C, Parker P, Albrechtsen R, et al. Regulation of ADAM12 cell-surface expression by protein kinase C epsilon. The Journal of biological chemistry 2004;279:51601–11. [DOI] [PubMed] [Google Scholar]
- [30].Cao Y, Kang Q, Zhao Z, Zolkiewska A. Intracellular processing of metalloprotease disintegrin ADAM12. The Journal of biological chemistry 2002;277:26403–11. [DOI] [PubMed] [Google Scholar]
- [31].Schlomann U, Wildeboer D, Webster A, Antropova O, Zeuschner D, Knight CG, et al. The metalloprotease disintegrin ADAM8. Processing by autocatalysis is required for proteolytic activity and cell adhesion. The Journal of biological chemistry 2002;277:48210–9. [DOI] [PubMed] [Google Scholar]
- [32].Reddy P, Slack JL, Davis R, Cerretti DP, Kozlosky CJ, Blanton RA, et al. Functional analysis of the domain structure of tumor necrosis factor-alpha converting enzyme. The Journal of biological chemistry 2000;275:14608–14. [DOI] [PubMed] [Google Scholar]
- [33].Zhang Q, Thomas SM, Lui VW, Xi S, Siegfried JM, Fan H, et al. Phosphorylation of TNF-alpha converting enzyme by gastrin-releasing peptide induces amphiregulin release and EGF receptor activation. Proceedings of the National Academy of Sciences of the United States of America 2006;103:6901–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Soond SM, Everson B, Riches DW, Murphy G. ERK-mediated phosphorylation of Thr735 in TNFalpha-converting enzyme and its potential role in TACE protein trafficking. Journal of cell science 2005;118:2371–80. [DOI] [PubMed] [Google Scholar]
- [35].Colige A, Ruggiero F, Vandenberghe I, Dubail J, Kesteloot F, Van Beeumen J, et al. Domains and maturation processes that regulate the activity of ADAMTS-2, a metalloproteinase cleaving the aminopropeptide of fibrillar procollagens types I-III and V. The Journal of biological chemistry 2005;280:34397–408. [DOI] [PubMed] [Google Scholar]
- [36].Somerville RP, Longpre JM, Apel ED, Lewis RM, Wang LW, Sanes JR, et al. ADAMTS7B, the full-length product of the ADAMTS7 gene, is a chondroitin sulfate proteoglycan containing a mucin domain. The Journal of biological chemistry 2004;279:35159–75. [DOI] [PubMed] [Google Scholar]
- [37].Koo BH, Apte SS. Cell-surface processing of the metalloprotease pro-ADAMTS9 is influenced by the chaperone GRP94/gp96. The Journal of biological chemistry 2010;285:197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Colige A, Beschin A, Samyn B, Goebels Y, Van Beeumen J, Nusgens BV, et al. Characterization and partial amino acid sequencing of a 107-kDa procollagen I N-proteinase purified by affinity chromatography on immobilized type XIV collagen. The Journal of biological chemistry 1995;270:16724–30. [DOI] [PubMed] [Google Scholar]
- [39].Koo BH, Longpre JM, Somerville RP, Alexander JP, Leduc R, Apte SS. Regulation of ADAMTS9 secretion and enzymatic activity by its propeptide. The Journal of biological chemistry 2007;282:16146–54. [DOI] [PubMed] [Google Scholar]
- [40].Majerus EM, Zheng X, Tuley EA, Sadler JE. Cleavage of the ADAMTS13 propeptide is not required for protease activity. The Journal of biological chemistry 2003;278:46643–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bekhouche M, Colige A. The procollagen N-proteinases ADAMTS2, 3 and 14 in pathophysiology. Matrix Biol 2015;44-46:46–53. [DOI] [PubMed] [Google Scholar]
- [42].Porter S, Clark IM, Kevorkian L, Edwards DR. The ADAMTS metalloproteinases. Biochem J 2005;386:15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Mead TJ, Apte SS. ADAMTS proteins in human disorders. Matrix Biol 2018;71-72:225–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Dubail J, Apte SS. Insights on ADAMTS proteases and ADAMTS-like proteins from mammalian genetics. Matrix Biol 2015;44-46:24–37. [DOI] [PubMed] [Google Scholar]
- [45].Amour A, Knight CG, English WR, Webster A, Slocombe PM, Knauper V, et al. The enzymatic activity of ADAM8 and ADAM9 is not regulated by TIMPs. FEBS letters 2002;524:154–8. [DOI] [PubMed] [Google Scholar]
- [46].Chesneau V, Becherer JD, Zheng Y, Erdjument-Bromage H, Tempst P, Blobel CP. Catalytic properties of ADAM19. The Journal of biological chemistry 2003;278:22331–40. [DOI] [PubMed] [Google Scholar]
- [47].Zou J, Zhu F, Liu J, Wang W, Zhang R, Garlisi CG, et al. Catalytic activity of human ADAM33. The Journal of biological chemistry 2004;279:9818–30. [DOI] [PubMed] [Google Scholar]
- [48].Lemjabbar H, Basbaum C. Platelet-activating factor receptor and ADAM10 mediate responses to Staphylococcus aureus in epithelial cells. Nature medicine 2002;8:41–6. [DOI] [PubMed] [Google Scholar]
- [49].Ohtsu H, Dempsey PJ, Eguchi S. ADAMs as mediators of EGF receptor transactivation by G protein-coupled receptors. American journal of physiology Cell physiology 2006;291:C1–10. [DOI] [PubMed] [Google Scholar]
- [50].Glassey B, Civetta A. Positive selection at reproductive ADAM genes with potential intercellular binding activity. Molecular biology and evolution 2004;21:851–9. [DOI] [PubMed] [Google Scholar]
- [51].Fridman JS, Caulder E, Hansbury M, Liu X, Yang G, Wang Q, et al. Selective inhibition of ADAM metalloproteases as a novel approach for modulating ErbB pathways in cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2007;13:1892–902. [DOI] [PubMed] [Google Scholar]
- [52].Ludwig A, Hundhausen C, Lambert MH, Broadway N, Andrews RC, Bickett DM, et al. Metalloproteinase inhibitors for the disintegrin-like metalloproteinases ADAM10 and ADAM17 that differentially block constitutive and phorbol ester-inducible shedding of cell surface molecules. Combinatorial chemistry & high throughput screening 2005;8:161–71. [DOI] [PubMed] [Google Scholar]
- [53].Minond D, Cudic M, Bionda N, Giulianotti M, Maida L, Houghten RA, et al. Discovery of novel inhibitors of a disintegrin and metalloprotease 17 (ADAM17) using glycosylated and non-glycosylated substrates. The Journal of biological chemistry 2012;287:36473–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Murphy G. Tissue inhibitors of metalloproteinases. Genome biology 2011;12:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wang WM, Ge G, Lim NH, Nagase H, Greenspan DS. TIMP-3 inhibits the procollagen N-proteinase ADAMTS-2. Biochem J 2006;398:515–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hashimoto G, Aoki T, Nakamura H, Tanzawa K, Okada Y. Inhibition of ADAMTS4 (aggrecanase-1) by tissue inhibitors of metalloproteinases (TIMP-1, 2, 3 and 4). FEBS letters 2001;494:192–5. [DOI] [PubMed] [Google Scholar]
- [57].Rodriguez-Manzaneque JC, Westling J, Thai SN, Luque A, Knauper V, Murphy G, et al. ADAMTS1 cleaves aggrecan at multiple sites and is differentially inhibited by metalloproteinase inhibitors. Biochemical and biophysical research communications 2002;293:501–8. [DOI] [PubMed] [Google Scholar]
- [58].Troeberg L, Fushimi K, Scilabra SD, Nakamura H, Dive V, Thogersen IB, et al. The C-terminal domains of ADAMTS-4 and ADAMTS-5 promote association with N-TIMP-3. Matrix biology : journal of the International Society for Matrix Biology 2009;28:463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Troeberg L, Lazenbatt C, Anower EKMF, Freeman C, Federov O, Habuchi H, et al. Sulfated glycosaminoglycans control the extracellular trafficking and the activity of the metalloprotease inhibitor TIMP-3. Chemistry & biology 2014;21:1300–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kramerova IA, Kawaguchi N, Fessler LI, Nelson RE, Chen Y, Kramerov AA, et al. Papilin in development; a pericellular protein with a homology to the ADAMTS metalloproteinases. Development 2000;127:5475–85. [DOI] [PubMed] [Google Scholar]
- [61].Nusgens BV, Verellen-Dumoulin C, Hermanns-Le T, De Paepe A, Nuytinck L, Pierard GE, et al. Evidence for a relationship between Ehlers-Danlos type VII C in humans and bovine dermatosparaxis. Nature genetics 1992;1:214–7. [DOI] [PubMed] [Google Scholar]
- [62].Klein T, Bischoff R. Active metalloproteases of the A Disintegrin and Metalloprotease (ADAM) family: biological function and structure. Journal of proteome research 2011;10:17–33. [DOI] [PubMed] [Google Scholar]
- [63].Kelly K, Hutchinson G, Nebenius-Oosthuizen D, Smith AJ, Bartsch JW, Horiuchi K, et al. Metalloprotease-disintegrin ADAM8: expression analysis and targeted deletion in mice. Developmental dynamics : an official publication of the American Association of Anatomists 2005;232:221–31. [DOI] [PubMed] [Google Scholar]
- [64].Vuohelainen V, Raitoharju E, Levula M, Lehtimaki T, Pelto-Huikko M, Honkanen T, et al. Myocardial infarction induces early increased remote ADAM8 expression of rat hearts after cardiac arrest. Scandinavian journal of clinical and laboratory investigation 2011;71:553–62. [DOI] [PubMed] [Google Scholar]
- [65].Cong L, Jia J. Promoter polymorphisms which regulate ADAM9 transcription are protective against sporadic Alzheimer’s disease. Neurobiology of aging 2011;32:54–62. [DOI] [PubMed] [Google Scholar]
- [66].Howard L, Glynn P. Membrane-associated metalloproteinase recognized by characteristic cleavage of myelin basic protein: assay and isolation. Methods in enzymology 1995;248:388–95. [DOI] [PubMed] [Google Scholar]
- [67].Hartmann D, de Strooper B, Serneels L, Craessaerts K, Herreman A, Annaert W, et al. The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Human molecular genetics 2002;11:2615–24. [DOI] [PubMed] [Google Scholar]
- [68].Zhang C, Tian L, Chi C, Wu X, Yang X, Han M, et al. Adam10 is essential for early embryonic cardiovascular development. Developmental dynamics : an official publication of the American Association of Anatomists 2010;239:2594–602. [DOI] [PubMed] [Google Scholar]
- [69].Takahashi E, Sagane K, Oki T, Yamazaki K, Nagasu T, Kuromitsu J. Deficits in spatial learning and motor coordination in ADAM11-deficient mice. BMC neuroscience 2006;7:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Yagami-Hiromasa T, Sato T, Kurisaki T, Kamijo K, Nabeshima Y, Fujisawa-Sehara A. A metalloprotease-disintegrin participating in myoblast fusion. Nature 1995;377:652–6. [DOI] [PubMed] [Google Scholar]
- [71].Jorissen E, Prox J, Bernreuther C, Weber S, Schwanbeck R, Serneels L, et al. The disintegrin/metalloproteinase ADAM10 is essential for the establishment of the brain cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience 2010;30:4833–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Karkkainen I, Rybnikova E, Pelto-Huikko M, Huovila AP. Metalloprotease-disintegrin (ADAM) genes are widely and differentially expressed in the adult CNS. Molecular and cellular neurosciences 2000;15:547–60. [DOI] [PubMed] [Google Scholar]
- [73].Asakura M, Kitakaze M, Takashima S, Liao Y, Ishikura F, Yoshinaka T, et al. Cardiac hypertrophy is inhibited by antagonism of ADAM12 processing of HB-EGF: metalloproteinase inhibitors as a new therapy. Nature medicine 2002;8:35–40. [DOI] [PubMed] [Google Scholar]
- [74].Kurisaki T, Masuda A, Sudo K, Sakagami J, Higashiyama S, Matsuda Y, et al. Phenotypic analysis of Meltrin alpha (ADAM12)-deficient mice: involvement of Meltrin alpha in adipogenesis and myogenesis. Molecular and cellular biology 2003;23:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Dyczynska E, Sun D, Yi H, Sehara-Fujisawa A, Blobel CP, Zolkiewska A. Proteolytic processing of delta-like 1 by ADAM proteases. The Journal of biological chemistry 2007;282:436–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Najy AJ, Day KC, Day ML. The ectodomain shedding of E-cadherin by ADAM15 supports ErbB receptor activation. The Journal of biological chemistry 2008;283:18393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Sun C, Wu MH, Lee ES, Yuan SY. A disintegrin and metalloproteinase 15 contributes to atherosclerosis by mediating endothelial barrier dysfunction via Src family kinase activity. Arteriosclerosis, thrombosis, and vascular biology 2012;32:2444–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Sun C, Wu MH, Guo M, Day ML, Lee ES, Yuan SY. ADAM15 regulates endothelial permeability and neutrophil migration via Src/ERK1/2 signalling. Cardiovascular research 2010;87:348–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Xie B, Shen J, Dong A, Swaim M, Hackett SF, Wyder L, et al. An Adam15 amplification loop promotes vascular endothelial growth factor-induced ocular neovascularization. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 2008;22:2775–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Hou Y, Chu M, Du FF, Lei JY, Chen Y, Zhu RY, et al. Recombinant disintegrin domain of ADAM15 inhibits the proliferation and migration of Bel-7402 cells. Biochemical and biophysical research communications 2013;435:640–5. [DOI] [PubMed] [Google Scholar]
- [81].Horiuchi K, Weskamp G, Lum L, Hammes HP, Cai H, Brodie TA, et al. Potential role for ADAM15 in pathological neovascularization in mice. Molecular and cellular biology 2003;23:5614–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature 1997;385:729–33. [DOI] [PubMed] [Google Scholar]
- [83].Moss ML, Jin SL, Milla ME, Bickett DM, Burkhart W, Carter HL, et al. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-alpha. Nature 1997;385:733–6. [DOI] [PubMed] [Google Scholar]
- [84].Niu A, Wang B, Li YP. TNFalpha shedding in mechanically stressed cardiomyocytes is mediated by Src activation of TACE. Journal of cellular biochemistry 2015;116:559–65. [DOI] [PubMed] [Google Scholar]
- [85].Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg SW, Lee DC, et al. An essential role for ectodomain shedding in mammalian development. Science (New York, NY) 1998;282:1281–4. [DOI] [PubMed] [Google Scholar]
- [86].Canault M, Certel K, Schatzberg D, Wagner DD, Hynes RO. The lack of ADAM17 activity during embryonic development causes hemorrhage and impairs vessel formation. PloS one 2010;5:e13433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Sato T, Suzuki H, Shibata M, Kusuyama T, Omori Y, Soda T, et al. Tumor-necrosis-factor-alpha-gene-deficient mice have improved cardiac function through reduction of intercellular adhesion molecule-1 in myocardial infarction. Circulation journal : official journal of the Japanese Circulation Society 2006;70:1635–42. [DOI] [PubMed] [Google Scholar]
- [88].Sun M, Chen M, Dawood F, Zurawska U, Li JY, Parker T, et al. Tumor necrosis factor-alpha mediates cardiac remodeling and ventricular dysfunction after pressure overload state. Circulation 2007;115:1398–407. [DOI] [PubMed] [Google Scholar]
- [89].Coletta AP, Clark AL, Banarjee P, Cleland JG. Clinical trials update: RENEWAL (RENAISSANCE and RECOVER) and ATTACH. European journal of heart failure 2002;4:559–61. [DOI] [PubMed] [Google Scholar]
- [90].Anker SD, Coats AJ. How to RECOVER from RENAISSANCE? The significance of the results of RECOVER, RENAISSANCE, RENEWAL and ATTACH. International journal of cardiology 2002;86:123–30. [DOI] [PubMed] [Google Scholar]
- [91].Lunn CA, Fan X, Dalie B, Miller K, Zavodny PJ, Narula SK, et al. Purification of ADAM 10 from bovine spleen as a TNFalpha convertase. FEBS letters 1997;400:333–5. [DOI] [PubMed] [Google Scholar]
- [92].Rosendahl MS, Ko SC, Long DL, Brewer MT, Rosenzweig B, Hedl E, et al. Identification and characterization of a pro-tumor necrosis factor-alpha-processing enzyme from the ADAM family of zinc metalloproteases. The Journal of biological chemistry 1997;272:24588–93. [DOI] [PubMed] [Google Scholar]
- [93].English WR, Puente XS, Freije JM, Knauper V, Amour A, Merryweather A, et al. Membrane type 4 matrix metalloproteinase (MMP17) has tumor necrosis factor-alpha convertase activity but does not activate pro-MMP2. The Journal of biological chemistry 2000;275:14046–55. [DOI] [PubMed] [Google Scholar]
- [94].Weskamp G, Schlondorff J, Lum L, Becherer JD, Kim TW, Saftig P, et al. Evidence for a critical role of the tumor necrosis factor alpha convertase (TACE) in ectodomain shedding of the p75 neurotrophin receptor (p75NTR). The Journal of biological chemistry 2004;279:4241–9. [DOI] [PubMed] [Google Scholar]
- [95].McClurg UL, Danjo K, King HO, Scott GB, Robinson PA, Crabtree JE. Epithelial cell ADAM17 activation by Helicobacter pylori: role of ADAM17 C-terminus and Threonine-735 phosphorylation. Microbes and infection 2015;17:205–14. [DOI] [PubMed] [Google Scholar]
- [96].Zeinieh M, Salehi A, Rajkumar V, Barker PA. p75NTR-dependent Rac1 activation requires receptor cleavage and activation of an NRAGE and NEDD9 signaling cascade. Journal of cell science 2015;128:447–59. [DOI] [PubMed] [Google Scholar]
- [97].Sommer A, Duppe M, Baumecker L, Kordowski F, Buch J, Chico JF, et al. Extracellular sphingomyelinase activity impairs TNF-alpha-induced endothelial cell death via ADAM17 activation and TNF receptor 1 shedding. Oncotarget 2017;8:72584–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Higashiyama S, Nanba D. ADAM-mediated ectodomain shedding of HB-EGF in receptor cross-talk. Biochimica et biophysica acta 2005;1751:110–7. [DOI] [PubMed] [Google Scholar]
- [99].Sahul ZH, Mukherjee R, Song J, McAteer J, Stroud RE, Dione DP, et al. Targeted imaging of the spatial and temporal variation of matrix metalloproteinase activity in a porcine model of postinfarct remodeling: relationship to myocardial dysfunction. Circulation Cardiovascular imaging 2011;4:381–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Lee DC, Sunnarborg SW, Hinkle CL, Myers TJ, Stevenson MY, Russell WE, et al. TACE/ADAM17 processing of EGFR ligands indicates a role as a physiological convertase. Annals of the New York Academy of Sciences 2003;995:22–38. [DOI] [PubMed] [Google Scholar]
- [101].Jackson LF, Qiu TH, Sunnarborg SW, Chang A, Zhang C, Patterson C, et al. Defective valvulogenesis in HB-EGF and TACE-null mice is associated with aberrant BMP signaling. The EMBO journal 2003;22:2704–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Sahin U, Weskamp G, Kelly K, Zhou HM, Higashiyama S, Peschon J, et al. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. The Journal of cell biology 2004;164:769–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Shi W, Chen H, Sun J, Buckley S, Zhao J, Anderson KD, et al. TACE is required for fetal murine cardiac development and modeling. Developmental biology 2003;261:371–80. [DOI] [PubMed] [Google Scholar]
- [104].Kurisaki T, Masuda A, Osumi N, Nabeshima Y, Fujisawa-Sehara A. Spatially- and temporally-restricted expression of meltrin alpha (ADAM12) and beta (ADAM19) in mouse embryo. Mechanisms of development 1998;73:211–5. [DOI] [PubMed] [Google Scholar]
- [105].Kurohara K, Komatsu K, Kurisaki T, Masuda A, Irie N, Asano M, et al. Essential roles of Meltrin beta (ADAM19) in heart development. Developmental biology 2004;267:14–28. [DOI] [PubMed] [Google Scholar]
- [106].Li SW, Arita M, Fertala A, Bao Y, Kopen GC, Langsjo TK, et al. Transgenic mice with inactive alleles for procollagen N-proteinase (ADAMTS-2) develop fragile skin and male sterility. Biochem J 2001;355:271–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Janssen L, Dupont L, Bekhouche M, Noel A, Leduc C, Voz M, et al. ADAMTS3 activity is mandatory for embryonic lymphangiogenesis and regulates placental angiogenesis. Angiogenesis 2016;19:53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Le Goff C, Cormier-Daire V. Genetic and molecular aspects of acromelic dysplasia. Pediatr Endocrinol Rev 2009;6:418–23. [PubMed] [Google Scholar]
- [109].Hubmacher D, Apte SS. ADAMTS proteins as modulators of microfibril formation and function. Matrix Biol 2015;47:34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Zheng XL. ADAMTS13 and von Willebrand factor in thrombotic thrombocytopenic purpura. Annual review of medicine 2015;66:211–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Zheng X, Majerus EM, Sadler JE. ADAMTS13 and TTP. Curr Opin Hematol 2002;9:389–94. [DOI] [PubMed] [Google Scholar]
- [112].Goertsches R, Comabella M, Navarro A, Perkal H, Montalban X. Genetic association between polymorphisms in the ADAMTS14 gene and multiple sclerosis. J Neuroimmunol 2005;164:140–7. [DOI] [PubMed] [Google Scholar]
- [113].Rodriguez-Lopez J, Pombo-Suarez M, Loughlin J, Tsezou A, Blanco FJ, Meulenbelt I, et al. Association of a nsSNP in ADAMTS14 to some osteoarthritis phenotypes. Osteoarthritis Cartilage 2009;17:321–7. [DOI] [PubMed] [Google Scholar]
- [114].Poonpet T, Honsawek S, Tammachote N, Kanitnate S, Tammachote R. ADAMTS14 gene polymorphism associated with knee osteoarthritis in Thai women. Genet Mol Res 2013;12:5301–9. [DOI] [PubMed] [Google Scholar]
- [115].Wolf ZT, Brand HA, Shaffer JR, Leslie EJ, Arzi B, Willet CE, et al. Genome-wide association studies in dogs and humans identify ADAMTS20 as a risk variant for cleft lip and palate. PLoS Genet 2015;11:e1005059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Kim SH, Pei QM, Jiang P, Yang M, Qian XJ, Liu JB. Effect of active vitamin D3 on VEGF-induced ADAM33 expression and proliferation in human airway smooth muscle cells: implications for asthma treatment. Respiratory research 2017;18:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Pelisek J, Pongratz J, Deutsch L, Reeps C, Stadlbauer T, Eckstein HH. Expression and cellular localization of metalloproteases ADAMs in high graded carotid artery lesions. Scandinavian journal of clinical and laboratory investigation 2012;72:648–56. [DOI] [PubMed] [Google Scholar]
- [118].Shen M, Morton J, Davidge ST, Kassiri Z. Loss of smooth muscle cell disintegrin and metalloproteinase 17 transiently suppresses angiotensin II-induced hypertension and end-organ damage. Journal of molecular and cellular cardiology 2017;103:11–21. [DOI] [PubMed] [Google Scholar]
- [119].Zhang L, Yu F, Wang L, Zheng J, Du Y, Huang Y, et al. ADAMTS-7 promotes vascular smooth muscle cells proliferation in vitro and in vivo. Science China Life sciences 2015;58:674–81. [DOI] [PubMed] [Google Scholar]
- [120].Li M, Liu Q, Lei J, Wang X, Chen X, Ding Y. MiR-362–3p inhibits the proliferation and migration of vascular smooth muscle cells in atherosclerosis by targeting ADAMTS1. Biochemical and biophysical research communications 2017;493:270–6. [DOI] [PubMed] [Google Scholar]
- [121].Qu Y, Zhang N. miR-365b-3p inhibits the cell proliferation and migration of human coronary artery smooth muscle cells by directly targeting ADAMTS1 in coronary atherosclerosis. Experimental and therapeutic medicine 2018;16:4239–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Wang L, Zheng J, Bai X, Liu B, Liu CJ, Xu Q, et al. ADAMTS-7 mediates vascular smooth muscle cell migration and neointima formation in balloon-injured rat arteries. Circulation research 2009;104:688–98. [DOI] [PubMed] [Google Scholar]
- [123].Bauer RC, Tohyama J, Cui J, Cheng L, Yang J, Zhang X, et al. Knockout of Adamts7, a novel coronary artery disease locus in humans, reduces atherosclerosis in mice. Circulation 2015;131:1202–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Pu X, Xiao Q, Kiechl S, Chan K, Ng FL, Gor S, et al. ADAMTS7 cleavage and vascular smooth muscle cell migration is affected by a coronary-artery-disease-associated variant. American journal of human genetics 2013;92:366–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Caolo V, Swennen G, Chalaris A, Wagenaar A, Verbruggen S, Rose-John S, et al. ADAM10 and ADAM17 have opposite roles during sprouting angiogenesis. Angiogenesis 2015;18:13–22. [DOI] [PubMed] [Google Scholar]
- [126].Kilic MO, Aynekin B, Kara A, Icen D, Demircan K. Differentially regulated ADAMTS1, 8, and 18 in gastric adenocarcinoma. Bratislavske lekarske listy 2017;118:71–6. [DOI] [PubMed] [Google Scholar]
- [127].Chen J, Zhi Y, Chang X, Zhang S, Dai D. Expression of ADAMTS1 and its correlation with angiogenesis in primary gastric cancer and lymph node metastasis. Digestive diseases and sciences 2013;58:405–13. [DOI] [PubMed] [Google Scholar]
- [128].Tang JH, Zhang HM, Zhang ZH, Zhang XL. Effect of tetramethylpyrazine combined with cisplatin on VEGF, KLF4 and ADAMTS1 in Lewis lung cancer mice. Asian Pacific journal of tropical medicine 2017;10:813–8. [DOI] [PubMed] [Google Scholar]
- [129].Jeltsch M, Jha SK, Tvorogov D, Anisimov A, Leppanen VM, Holopainen T, et al. CCBE1 enhances lymphangiogenesis via A disintegrin and metalloprotease with thrombospondin motifs-3-mediated vascular endothelial growth factor-C activation. Circulation 2014;129:1962–71. [DOI] [PubMed] [Google Scholar]
- [130].Brouillard P, Dupont L, Helaers R, Coulie R, Tiller GE, Peeden J, et al. Loss of ADAMTS3 activity causes Hennekam lymphangiectasia-lymphedema syndrome 3. Hum Mol Genet 2017;26:4095–104. [DOI] [PubMed] [Google Scholar]
- [131].Dubail J, Kesteloot F, Deroanne C, Motte P, Lambert V, Rakic JM, et al. ADAMTS-2 functions as anti-angiogenic and anti-tumoral molecule independently of its catalytic activity. Cell Mol Life Sci 2010;67:4213–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].El Hour M, Moncada-Pazos A, Blacher S, Masset A, Cal S, Berndt S, et al. Higher sensitivity of Adamts12-deficient mice to tumor growth and angiogenesis. Oncogene 2010;29:3025–32. [DOI] [PubMed] [Google Scholar]
- [133].Kelwick R, Wagstaff L, Decock J, Roghi C, Cooley LS, Robinson SD, et al. Metalloproteinase-dependent and -independent processes contribute to inhibition of breast cancer cell migration, angiogenesis and liver metastasis by a disintegrin and metalloproteinase with thrombospondin motifs-15. International journal of cancer 2015;136:E14–26. [DOI] [PubMed] [Google Scholar]
- [134].Schulz B, Pruessmeyer J, Maretzky T, Ludwig A, Blobel CP, Saftig P, et al. ADAM10 regulates endothelial permeability and T-Cell transmigration by proteolysis of vascular endothelial cadherin. Circulation research 2008;102:1192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Hou Y, Chu M, Cai Y, Lei J, Chen Y, Zhu R, et al. Antitumor and anti-angiogenic activity of the recombinant human disintegrin domain of A disintegrin and metalloproteinase 15. Molecular medicine reports 2015;12:2360–6. [DOI] [PubMed] [Google Scholar]
- [136].Wang R, Li Y, Tsung A, Huang H, Du Q, Yang M, et al. iNOS promotes CD24(+)CD133(+) liver cancer stem cell phenotype through a TACE/ADAM17-dependent Notch signaling pathway. Proc Natl Acad Sci U S A 2018;115:E10127–E36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Jin Y, Liu Y, Lin Q, Li J, Druso JE, Antonyak MA, et al. Deletion of Cdc42 enhances ADAM17-mediated vascular endothelial growth factor receptor 2 shedding and impairs vascular endothelial cell survival and vasculogenesis. Molecular and cellular biology 2013;33:4181–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Ren P, Hughes M, Krishnamoorthy S, Zou S, Zhang L, Wu D, et al. Critical Role of ADAMTS-4 in the Development of Sporadic Aortic Aneurysm and Dissection in Mice. Sci Rep 2017;7:12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].McRobb LS, McKay MJ, Gamble JR, Grace M, Moutrie V, Santos ED, et al. Ionizing radiation reduces ADAM10 expression in brain microvascular endothelial cells undergoing stress-induced senescence. Aging 2017;9:1248–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Hodgkinson LM, Wang L, Duncan G, Edwards DR, Wormstone IM. ADAM and ADAMTS gene expression in native and wound healing human lens epithelial cells. Molecular vision 2010;16:2765–76. [PMC free article] [PubMed] [Google Scholar]
- [141].van der Vorst EP, Keijbeck AA, de Winther MP, Donners MM. A disintegrin and metalloproteases: molecular scissors in angiogenesis, inflammation and atherosclerosis. Atherosclerosis 2012;224:302–8. [DOI] [PubMed] [Google Scholar]
- [142].Zhu R, Cheng M, Lu T, Yang N, Ye S, Pan YH, et al. A Disintegrin and Metalloproteinase with Thrombospondin Motifs 18 Deficiency Leads to Visceral Adiposity and Associated Metabolic Syndrome in Mice. The American journal of pathology 2018;188:461–73. [DOI] [PubMed] [Google Scholar]
- [143].Folkesson M, Li C, Frebelius S, Swedenborg J, Wagsater D, Williams KJ, et al. Proteolytically active ADAM10 and ADAM17 carried on membrane microvesicles in human abdominal aortic aneurysms. Thrombosis and haemostasis 2015;114:1165–74. [DOI] [PubMed] [Google Scholar]
- [144].Kawai T, Takayanagi T, Forrester SJ, Preston KJ, Obama T, Tsuji T, et al. Vascular ADAM17 (a Disintegrin and Metalloproteinase Domain 17) Is Required for Angiotensin II/beta-Aminopropionitrile-Induced Abdominal Aortic Aneurysm. Hypertension (Dallas, Tex : 1979) 2017;70:959–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Gao Y, Wu W, Yu C, Zhong F, Li G, Kong W, et al. A disintegrin and metalloproteinase with thrombospondin motif 1 (ADAMTS1) expression increases in acute aortic dissection. Science China Life sciences 2016;59:59–67. [DOI] [PubMed] [Google Scholar]
- [146].Gunes MF, Akpinar MB, Comertoglu I, Akyol S, Demircelik B, Gurel OM, et al. The Investigation of a Disintegrin and Metalloproteinase with ThromboSpondin Motifs (ADAMTS) 1, 5 and 16 in Thoracic Aortic Aneurysms and Dissections. Clinical laboratory 2016;62:425–33. [PubMed] [Google Scholar]
- [147].Ren P, Hughes M, Krishnamoorthy S, Zou S, Zhang L, Wu D, et al. Critical Role of ADAMTS-4 in the Development of Sporadic Aortic Aneurysm and Dissection in Mice 2017;7:12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Qin W, Cao Y, Li L, Chen W, Chen X. Upregulation of ADAMTS7 and downregulation of COMP are associated with aortic aneurysm. Molecular medicine reports 2017;16:5459–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Arning A, Jeibmann A, Kohnemann S, Brokinkel B, Ewelt C, Berger K, et al. ADAMTS genes and the risk of cerebral aneurysm. J Neurosurg 2016;125:269–74. [DOI] [PubMed] [Google Scholar]
- [150].Arning A, Hiersche M, Witten A, Kurlemann G, Kurnik K, Manner D, et al. A genome-wide association study identifies a gene network of ADAMTS genes in the predisposition to pediatric stroke. Blood 2012;120:5231–6. [DOI] [PubMed] [Google Scholar]
- [151].Rizza S, Copetti M, Cardellini M, Menghini R, Pecchioli C, Luzi A, et al. A score including ADAM17 substrates correlates to recurring cardiovascular event in subjects with atherosclerosis. Atherosclerosis 2015;239:459–64. [DOI] [PubMed] [Google Scholar]
- [152].Fan D, Takawale A, Shen M, Wang W, Wang X, Basu R, et al. Cardiomyocyte A Disintegrin And Metalloproteinase 17 (ADAM17) Is Essential in Post-Myocardial Infarction Repair by Regulating Angiogenesis. Circulation Heart failure 2015;8:970–9. [DOI] [PubMed] [Google Scholar]
- [153].Zha Y, Chen Y, Xu F, Li T, Zhao C, Cui L. ADAMTS4 level in patients with stable coronary artery disease and acute coronary syndromes. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie 2010;64:160–4. [DOI] [PubMed] [Google Scholar]
- [154].Chen L, Yang L, Zha Y, Cui L. Association of serum a disintegrin and metalloproteinase with thrombospodin motif 4 levels with the presence and severity of coronary artery disease. Coronary artery disease 2011;22:570–6. [DOI] [PubMed] [Google Scholar]
- [155].Yu J, Zhou B, Yu H, Han J, Cui M, Zhang F, et al. Association between plasma ADAMTS-7 levels and severity of disease in patients with stable obstructive coronary artery disease. Medicine (Baltimore) 2016;95:e5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Reilly MP, Li M, He J, Ferguson JF, Stylianou IM, Mehta NN, et al. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet 2011;377:383–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Kessler T, Zhang L, Liu Z, Yin X, Huang Y, Wang Y, et al. ADAMTS-7 inhibits re-endothelialization of injured arteries and promotes vascular remodeling through cleavage of thrombospondin-1. Circulation 2015;131:1191–201. [DOI] [PubMed] [Google Scholar]
- [158].Hirohata S, Inagaki J, Ohtsuki T. Diverse Functions of a Disintegrin and Metalloproteinase with Thrombospondin Motif-1. Yakugaku zasshi : Journal of the Pharmaceutical Society of Japan 2017;137:811–4. [DOI] [PubMed] [Google Scholar]
- [159].Lee CW, Hwang I, Park CS, Lee H, Park DW, Kang SJ, et al. Expression of ADAMTS-2, −3, −13, and −14 in culprit coronary lesions in patients with acute myocardial infarction or stable angina. J Thromb Thrombolysis 2012;33:362–70. [DOI] [PubMed] [Google Scholar]
- [160].Wu W, Zhou Y, Li Y, Li J, Ke Y, Wang Y, et al. Association between plasma ADAMTS-7 levels and ventricular remodeling in patients with acute myocardial infarction. Eur J Med Res 2015;20:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Eerenberg ES, Teunissen PF, van den Born BJ, Meijers JC, Hollander MR, Jansen M, et al. The role of ADAMTS13 in acute myocardial infarction: cause or consequence? Cardiovascular research 2016;111:194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Arndt M, Lendeckel U, Rocken C, Nepple K, Wolke C, Spiess A, et al. Altered expression of ADAMs (A Disintegrin And Metalloproteinase) in fibrillating human atria. Circulation 2002;105:720–5. [DOI] [PubMed] [Google Scholar]
- [163].Mishra PK, Tyagi N, Sen U, Givvimani S, Tyagi SC. H2S ameliorates oxidative and proteolytic stresses and protects the heart against adverse remodeling in chronic heart failure. American journal of physiology Heart and circulatory physiology 2010;298:H451–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Donnarumma E, Trivedi RK, Lefer DJ. Protective Actions of H2S in Acute Myocardial Infarction and Heart Failure. Comprehensive Physiology 2017;7:583–602. [DOI] [PubMed] [Google Scholar]
- [165].Salloum FN. Hydrogen sulfide and cardioprotection--Mechanistic insights and clinical translatability. Pharmacology & therapeutics 2015;152:11–7. [DOI] [PubMed] [Google Scholar]
- [166].Rau CD, Romay MC, Tuteryan M, Wang JJ, Santolini M, Ren S, et al. Systems Genetics Approach Identifies Gene Pathways and Adamts2 as Drivers of Isoproterenol-Induced Cardiac Hypertrophy and Cardiomyopathy in Mice. Cell Syst 2017;4:121–8 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Wang X, Chen W, Zhang J, Khan A, Li L, Huang F, et al. Critical Role of ADAMTS2 (A Disintegrin and Metalloproteinase With Thrombospondin Motifs 2) in Cardiac Hypertrophy Induced by Pressure Overload. Hypertension (Dallas, Tex : 1979) 2017;69:1060–9. [DOI] [PubMed] [Google Scholar]
- [168].Horiuchi K, Zhou HM, Kelly K, Manova K, Blobel CP. Evaluation of the contributions of ADAMs 9, 12, 15, 17, and 19 to heart development and ectodomain shedding of neuregulins beta1 and beta2. Developmental biology 2005;283:459–71. [DOI] [PubMed] [Google Scholar]
- [169].Smiljanic K, Dobutovic B, Obradovic M, Nikolic D, Marche P, Isenovic ER. Involvement of the ADAM 12 in thrombin-induced rat’s VSMCs proliferation. Current medicinal chemistry 2011;18:3382–6. [DOI] [PubMed] [Google Scholar]
- [170].Charrier-Hisamuddin L, Laboisse CL, Merlin D. ADAM-15: a metalloprotease that mediates inflammation. FASEB J 2008;22:641–53. [DOI] [PubMed] [Google Scholar]
- [171].Zhou HM, Weskamp G, Chesneau V, Sahin U, Vortkamp A, Horiuchi K, et al. Essential role for ADAM19 in cardiovascular morphogenesis. Molecular and cellular biology 2004;24:96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Kelwick R, Desanlis I, Wheeler GN, Edwards DR. The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin motifs) family. Genome biology 2015;16:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Toba H, de Castro Bras LE, Baicu CF, Zile MR, Lindsey ML. Increased ADAMTS1 mediates SPARC-dependent collagen deposition in the aging myocardium 2016;310:E1027–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Vazquez F, Hastings G, Ortega MA, Lane TF, Oikemus S, Lombardo M, et al. METH-1, a human ortholog of ADAMTS-1, and METH-2 are members of a new family of proteins with angio-inhibitory activity. The Journal of biological chemistry 1999;274:23349–57. [DOI] [PubMed] [Google Scholar]
- [175].Colige A, Li SW, Sieron AL, Nusgens BV, Prockop DJ, Lapiere CM. cDNA cloning and expression of bovine procollagen I N-proteinase: a new member of the superfamily of zinc-metalloproteinases with binding sites for cells and other matrix components. Proc Natl Acad Sci U S A 1997;94:2374–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Ogino H, Hisanaga A, Kohno T, Kondo Y, Okumura K, Kamei T, et al. Secreted Metalloproteinase ADAMTS-3 Inactivates Reelin. J Neurosci 2017;37:3181–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Jha SK, Rauniyar K, Karpanen T, Leppanen VM, Brouillard P, Vikkula M, et al. Efficient activation of the lymphangiogenic growth factor VEGF-C requires the C-terminal domain of VEGF-C and the N-terminal domain of CCBE1. Sci Rep 2017;7:4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Boerboom D, Lafond JF, Zheng X, Lapointe E, Mittaz L, Boyer A, et al. Partially redundant functions of Adamts1 and Adamts4 in the perinatal development of the renal medulla. Developmental dynamics : an official publication of the American Association of Anatomists 2011;240:1806–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Abbaszade I, Liu RQ, Yang F, Rosenfeld SA, Ross OH, Link JR, et al. Cloning and characterization of ADAMTS11, an aggrecanase from the ADAMTS family. The Journal of biological chemistry 1999;274:23443–50. [DOI] [PubMed] [Google Scholar]
- [180].Hurskainen TL, Hirohata S, Seldin MF, Apte SS. ADAM-TS5, ADAM-TS6, and ADAM-TS7, novel members of a new family of zinc metalloproteases. General features and genomic distribution of the ADAM-TS family. The Journal of biological chemistry 1999;274:25555–63. [DOI] [PubMed] [Google Scholar]
- [181].Hanby HA, Zheng XL. Biochemistry and physiological functions of ADAMTS7 metalloprotease. Adv Biochem 2013;1. [DOI] [PMC free article] [PubMed]
- [182].Desanlis I, Felstead HL, Edwards DR, Wheeler GN. ADAMTS9, a member of the ADAMTS family, in Xenopus development. Gene expression patterns : GEP 2018;29:72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Somerville RP, Longpre JM, Jungers KA, Engle JM, Ross M, Evanko S, et al. Characterization of ADAMTS-9 and ADAMTS-20 as a distinct ADAMTS subfamily related to Caenorhabditis elegans GON-1. The Journal of biological chemistry 2003;278:9503–13. [DOI] [PubMed] [Google Scholar]
- [184].Koo BH, Coe DM, Dixon LJ, Somerville RP, Nelson CM, Wang LW, et al. ADAMTS9 is a cell-autonomously acting, anti-angiogenic metalloprotease expressed by microvascular endothelial cells. Am J Pathol 2010;176:1494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Somerville RP, Jungers KA, Apte SS. Discovery and characterization of a novel, widely expressed metalloprotease, ADAMTS10, and its proteolytic activation. The Journal of biological chemistry 2004;279:51208–17. [DOI] [PubMed] [Google Scholar]
- [186].Kutz WE, Wang LW, Bader HL, Majors AK, Iwata K, Traboulsi EI, et al. ADAMTS10 protein interacts with fibrillin-1 and promotes its deposition in extracellular matrix of cultured fibroblasts. The Journal of biological chemistry 2011;286:17156–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Bai XH, Wang DW, Luan Y, Yu XP, Liu CJ. Regulation of chondrocyte differentiation by ADAMTS-12 metalloproteinase depends on its enzymatic activity. Cellular and molecular life sciences : CMLS 2009;66:667–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Moake JL. von Willebrand factor, ADAMTS-13, and thrombotic thrombocytopenic purpura. Semin Hematol 2004;41:4–14. [DOI] [PubMed] [Google Scholar]
- [189].Lopez JA, Dong JF. Cleavage of von Willebrand factor by ADAMTS-13 on endothelial cells. Semin Hematol 2004;41:15–23. [DOI] [PubMed] [Google Scholar]
- [190].Cal S, Obaya AJ, Llamazares M, Garabaya C, Quesada V, Lopez-Otin C. Cloning, expression analysis, and structural characterization of seven novel human ADAMTSs, a family of metalloproteinases with disintegrin and thrombospondin-1 domains. Gene 2002;283:49–62. [DOI] [PubMed] [Google Scholar]
- [191].Zheng X, Chung D, Takayama TK, Majerus EM, Sadler JE, Fujikawa K. Structure of von Willebrand factor-cleaving protease (ADAMTS13), a metalloprotease involved in thrombotic thrombocytopenic purpura. The Journal of biological chemistry 2001;276:41059–63. [DOI] [PubMed] [Google Scholar]
- [192].Dancevic CM, Fraser FW, Smith AD, Stupka N, Ward AC, McCulloch DR. Biosynthesis and expression of a disintegrin-like and metalloproteinase domain with thrombospondin-1 repeats-15: a novel versican-cleaving proteoglycanase. The Journal of biological chemistry 2013;288:37267–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Llamazares M, Cal S, Quesada V, Lopez-Otin C. Identification and characterization of ADAMTS-20 defines a novel subfamily of metalloproteinases-disintegrins with multiple thrombospondin-1 repeats and a unique GON domain. The Journal of biological chemistry 2003;278:13382–9. [DOI] [PubMed] [Google Scholar]
- [194].Rapti M, Atkinson SJ, Lee MH, Trim A, Moss M, Murphy G. The isolated N-terminal domains of TIMP-1 and TIMP-3 are insufficient for ADAM10 inhibition. The Biochemical journal 2008;411:433–9. [DOI] [PubMed] [Google Scholar]
- [195].Kveiborg M, Jacobsen J, Lee MH, Nagase H, Wewer UM, Murphy G. Selective inhibition of ADAM12 catalytic activity through engineering of tissue inhibitor of metalloproteinase 2 (TIMP-2). The Biochemical journal 2010;430:79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Kashiwagi M, Tortorella M, Nagase H, Brew K. TIMP-3 is a potent inhibitor of aggrecanase 1 (ADAM-TS4) and aggrecanase 2 (ADAM-TS5). The Journal of biological chemistry 2001;276:12501–4. [DOI] [PubMed] [Google Scholar]
- [197].Heijink IH, Brandenburg SM, Noordhoek JA, Slebos DJ, Postma DS, van Oosterhout AJ. Role of aberrant metalloproteinase activity in the pro-inflammatory phenotype of bronchial epithelium in COPD. Respiratory research 2011;12:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Tortorella MD, Arner EC, Hills R, Easton A, Korte-Sarfaty J, Fok K, et al. Alpha2-macroglobulin is a novel substrate for ADAMTS-4 and ADAMTS-5 and represents an endogenous inhibitor of these enzymes. The Journal of biological chemistry 2004;279:17554–61. [DOI] [PubMed] [Google Scholar]
- [199].Takizawa M, Yatabe T, Okada A, Chijiiwa M, Mochizuki S, Ghosh P, et al. Calcium pentosan polysulfate directly inhibits enzymatic activity of ADAMTS4 (aggrecanase-1) in osteoarthritic chondrocytes. FEBS letters 2008;582:2945–9. [DOI] [PubMed] [Google Scholar]
- [200].Qian LW, Mizumoto K, Urashima T, Nagai E, Maehara N, Sato N, et al. Radiation-induced increase in invasive potential of human pancreatic cancer cells and its blockade by a matrix metalloproteinase inhibitor, CGS27023. Clinical cancer research : an official journal of the American Association for Cancer Research 2002;8:1223–7. [PubMed] [Google Scholar]
- [201].Tortorella MD, Tomasselli AG, Mathis KJ, Schnute ME, Woodard SS, Munie G, et al. Structural and inhibition analysis reveals the mechanism of selectivity of a series of aggrecanase inhibitors. The Journal of biological chemistry 2009;284:24185–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Moss ML, Rasmussen FH. Fluorescent substrates for the proteinases ADAM17, ADAM10, ADAM8, and ADAM12 useful for high-throughput inhibitor screening. Analytical biochemistry 2007;366:144–8. [DOI] [PubMed] [Google Scholar]


