Abstract
Background
Citrullinemia type I (CTLN1) is a rare autosomal recessive disorder of the urea cycle caused by a deficiency in the argininosuccinate synthetase (ASS1) enzyme due to mutations in the ASS1 gene. Only a few Chinese patients with CTLN1 have been reported, and ASS1 gene mutations have been identified sporadically in China.
Case presentation
A Chinese family with one member affected with mild CTLN1 was enrolled. Targeted exome sequencing was performed on the proband, and Sanger sequencing was used to validate the detected mutation. We also reviewed the genetic and clinical characteristics of CTLN1 in Chinese patients that have been published to date. Newborn screening showed remarkably increased concentrations of citrulline with elevated ratios of citrulline/arginine and citrulline/phenylalanine, and the patient presented with a speech delay at age three. The urinary organic acid profiles were normal. A novel homozygous splicing variant c.773 + 4A > C in the ASS1 gene was identified in the proband, and it was predicted to affect splicing by in silico analysis. To date, only nine Chinese patients with CTLN1 have been reported, with a total of 15 ASS1 mutations identified and no high frequency or hot spot mutations found; the mutation spectrum of Chinese patients with CTLN1 was heterogeneous.
Conclusions
We described a mild Chinese CTLN1 case with a novel homozygous splicing variant c.773 + 4A > C and reviewed previous genotypes and phenotypes in Chinese patients with CTLN1. Thus, our findings contribute to understanding the molecular genetic background and clinical phenotype of CTLN1 in this population.
Electronic supplementary material
The online version of this article (10.1186/s12881-019-0836-5) contains supplementary material, which is available to authorized users.
Keywords: Citrullinemia type I, ASS1, Novel variant, Mutation spectrum
Background
Citrullinemia type I (CTLN1, MIM# 215700) is a rare autosomal recessive disorder of the urea cycle caused by a deficiency of the argininosuccinate synthetase (ASS, EC 6.3.4.5) enzyme due to mutations in the ASS1 gene [1]. CTLN1 encompasses a spectrum of varying clinical phenotypes. Patients that present with fatal neonatal hyperammonemia are said to have classical citrullinemia, patients with late onset and/or mild symptoms are said to have mild citrullinemia, and a considerable number of asymptomatic individuals detected by expanded newborn screening (NBS) have only a biochemical phenotype [2, 3].
Biochemically, CTLN1 with elevated citrulline concentrations can be detected by NBS. Nonetheless, increased levels of citrulline can also be found in other inherited metabolic disorders such as citrullinemia type II, argininosuccinate lyase deficiency, and pyruvate carboxylase deficiency [4, 5]. Therefore, definitive diagnosis of CTLN1 mainly relies on an ASS enzyme assay and identification of ASS1 gene mutations. However, determination of enzyme activity in liver tissue requires an invasive procedure, and direct measurement of ASS activity or indirect measurement using a C14 incorporation assay in fibroblasts have not yet been evaluated in patients with mild CTLN1 [6]. Therefore, molecular genetic testing is paramount, not only for clinical diagnosis but also for future prenatal testing and family member screening [7].
The ASS1 gene is located on chromosome 9q34.1 and contains 16 exons, with the translation start codon in exon 3, encoding 412 amino acids [2, 8]. To date, At least 137 mutations that cause CTLN1 have been reported in the ASS1 gene [9]. However, only a few Chinese patients with CTLN1 have been reported, and ASS1 gene mutations have been identified only sporadically in China [10–17]. In this study, we present biochemical, clinical, and genetic characteristics of a new Chinese patient with CTLN1. In addition, we reviewed previous genotypes and phenotypes of Chinese patients with CTLN1, to help better understand the genetic background of this disease in the Chinese population.
Case presentation
Case report
This study was approved by the Ethical Committee of Quanzhou Maternity and Children’s Hospital. Written informed consent was obtained from the parents of the patient, who agreed to join this study, with the intent of using the resulting medical data for scientific research and publication. The proband was born at a gestational age of 38 weeks and 1 day via caesarean section; his weight at birth was 3700 g. He was the third born child of consanguineous parents of Chinese descent. There was no significant family history of inherited metabolic diseases. NBS via ACQUITY TQD tandem mass spectrometry (MS/MS) (Waters, Milford, MA, USA) analysis on dried blood spots (DBS) was performed on the proband after birth. The initial NBS results showed an elevated citrulline concentration with increased ratios of citrulline/arginine and citrulline/phenylalanine. The hypercitrullinemia and increased citrulline/phenylalanine ratios persisted, and the concentrations of citrulline fluctuated between 71.82–120.99 μmol/L during follow up, while the citrulline/arginine ratios were persistent within the reference range (Table 1). Subsequently, urinary organic acid analysis by gas chromatography-mass spectrometry (7890B/5977A, Agilent Technologies, Santa Clara, CA, USA) and auxiliary biochemical tests were carried out. Increased levels of orotic acid were not observed in urinary organic acid analysis. The blood ammonia levels were slightly elevated at 1 month and 11 months, which may have been transitional, and returned to the normal reference range later. The patient exhibited normal growth and development during follow up, but a speech delay was noted at 3 years of age.
Table 1.
Detection results of MS/MS and biochemical testing in the patient
| Testing time | MS/MS analysis in dried blood spots | Biochemical testing | ||||||
|---|---|---|---|---|---|---|---|---|
| Citrulline (5.5–30 μmol/L) | Citrulline/Arginine (0.35–15) | Citrulline/Phenylalanine (0.12–0.83) | Arginine (1–50 μmol/L) | Blood ammonia (10–47 μmol/L) | Total bilirubin (5.1–19 μmol/L) | Direct bilirubin (0–6.8 μmol/L) | AFP (ng/ml) | |
| 2015.9.21a | 90.05 | 46.18 | 1.17 | 1.95 | ||||
| 2015.10.9 | 89.87 | 6.58 | 2.26 | 13.65 | ||||
| 2015.10.19 | 88.14 | 4.72 | 2.82 | 18.69 | 70 | 142 | 15.6 | 6508.14 |
| 2016.8.26 | 118.24 | 3.9 | 2.56 | 30.3 | 49 | 4.2 | 0.9 | 2.37 |
| 2016.10.10 | 87.66 | 4.8 | 1.9 | 18.25 | 33 | |||
| 2016.11.8 | 81.25 | 3.52 | 2.25 | 23.05 | ||||
| 2017.8.4 | 71.82 | 5.19 | 1.37 | 13.85 | 29 | 6.2 | 1.6 | <1.3 |
| 2018.9.7 | 120.99 | 3.4 | 1.45 | 35.63 | 45 | 3.2 | 2.1 | |
aNewborn screening results
Genetic analysis
Genomic DNA was extracted from whole blood of the proband and his family members using Qiagen Blood DNA mini kit (Qiagen®, Hilden, Germany). The DNA of the proband was used for NGS. Targeted enrichment of target region sequences was performed by multiple probe hybridization using metabolic abnormality of common amino acids capture oligo, which was designed by Genuine Diagnostics (Zhejiang, China), and included 40 genes (ABCD4, ACSF3, ARG1, AMT, ASS1, ASL, BCAT1, BCAT2, BCKDHA, BCKDHB, CBS, CPS1, CTH, D2HGDH, ETHE1, GCH1, GCSH, GLDC, GPHN, GRHPR, HGD, HPD, L2HGDH, LMBRD1, MOCS1, MOCS2, MTR, OAT, OGDH, OPA3, PAH, PCBD1, PRODH, PTS, QDPR, SERAC1, SLC25A13, SLC25A15, SPR, and SUOX). The sequencing libraries were quantified using the Illumina DNA Standads and Primer Premix Kit (Kapa Biosystems, Boston, MA, USA), and then massively parallel sequenced using the Illumina HiSeq 2500 platform (Illumina, San Diego, CA, USA). After sequencing and filtering out low-quality reads, high-quality reads were compared to the human genome reference sequence (GRCh37.p13, hg19). The quality control data are listed in Additional file 1: Table S1. Variants were called using the GATK software. Next, candidate variants were further confirmed by Sanger sequencing. Sequencing was performed on ABI 3500xL (Applied Biosystems, Foster City, CA, USA), and the results were analysed using DNASTAR software. The primers used for polymerase chain reaction (PCR) and Sanger sequencing are listed in Additional file 2: Table S2.
The identified variants were annotated to public databases, such as the Human Gene Mutation Database (HGMD, http://www.hgmd.cf.ac.uk/ac/index.php), ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/), the Leiden Open Variation Database (LOVD, http://www.lovd.nl/3.0/home), dbSNP (https://www.ncbi.nlm.nih.gov/projects/SNP/), the 1000 Genome Project (http://www.1000genomes.org/), and the ExAC consortium (http://exac.broadinstitute.org/). In addition, the variant was further assessed for possible pathogenicity using HSF (http://www.umd.be/HSF3/), MutationTaster (http://www.mutationtaster.org/), and regSNP-intron (http://clark.compbio.iupui.edu/regsnp_intron_web/). To exclude any polymorphisms, 100 healthy controls underwent Sanger sequencing of the ASS1 exon 11. Pathogenicity analysis of the variant was performed to comply with the American College of Medical Genetics and Genomics (ACMG) guidelines [18].
Mutation analysis
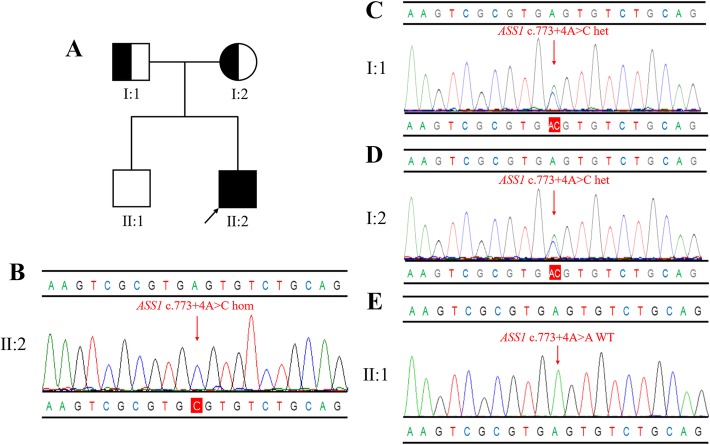
We identified a homozygous ASS1 gene variant c.773 + 4A > C in the proband, which was inherited from the parents. In addition, no ASS1 mutations were detected in the proband’s older brother via family genetic screening (Fig. 1). The c.773 + 4A > C variant is located in intron 11 of the ASS1 gene, with the 5th nucleotide changed from adenine to cytosine. This variant has not been previously reported in the literature and was not detected in 100 healthy controls. It was found only in a heterozygous state in the East Asian population with an allele frequency of 1.387e-03 in the ExAC database, and the allele frequency in the overall population was 9.892e-05, indicating that individuals in the population with this heterozygous variation were just carriers. It was absent from the dbSNP and 1000 Genomes databases, and was also absent from disease databases such as HGMD, ClinVar, and LOVD. Furthermore, in silico analysis by HSF, MutationTaster, and regSNP-intron all suggested that the variant most likely affects splicing (Table 2). According to the ACMG guidelines, the c.773 + 4A > C variant was classified as variant of uncertain clinical significance.
Fig. 1.
a: Pedigree of the family. The filled black symbols represent the affected members and the arrow denotes the proband. b-e: Sequence analysis of the ASS1 gene independently identified the c.773 + 4A > C variant in the proband (b), his father (c), his mother (d), and his old brother (e)
Table 2.
The effect of c.773 + 4A > C on protein function by in silico analysis
| Software | Score | Predicted signal |
|---|---|---|
| HSFa | Site broken (−42.85) | Alteration of the WT donor site, most probably affect splicing |
| MutationTasterb | 1 | Disease causing |
| regSNP-intronc | 0.822174865 | Disease causing |
aHSF: www.umd.be/HSF/. The score less than 0 is site broken
bMutationTaster: www.mutationtaster.org. The score is between 0 and 1, it is more likely to be disease causing with the score closer to 1
cregSNP-intron: http://clark.compbio.iupui.edu/regsnp_intron_web/. The score is between 0 and 1, it is more likely to be disease causing with the score closer to 1
In a review of Chinese CTLN1 cases published to date, we found that only ten Chinese patients with CTLN1 received genetic testing. The mutations in the ASS1 gene from our study and from previously reported Chinese patients are summarized in Table 3. In total, 15 ASS1 mutations have been identified in Chinese patients to date. Among them, 11 are missense mutations, three are splice mutations, and one is a deletion mutation. 50% of the mutant alleles are clustered in exons 7, 13, and 14. Four mutations (p.Arg127Gln, p.Arg265Cys, p.Gly324Ser, and p.Gly390Arg) proved to be disease-causing. Four patients had homozygous or compound heterozygous splicing/frameshift mutations, resulting in neonatal onsets and with poor outcomes, and thus were classified as having neonatal CTLN1. Three patients developed symptoms after the neonatal period with moderate outcomes and were classified as having late-onset CTLN1. The other three patients without obvious clinical symptoms and with good prognosis were classified as having a mild form of CTLN1.
Table 3.
Clinical presentations, biochemical, and genetic investigations of ten Chinese patients with citrullinemia type 1
| Patient no. | Gender | Age of onset | Clinical presentation | Citrulline levels (μmol/L)a | Blood ammonia (μmol/L)b | Mutaion 1 | Mutaion 2 | Outcome | Ref. | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Location | c.DNAc | Protein | Location | c.DNA | Protien | ||||||||
| 1 | Male | 3 y | Mild form | 90.05 | 70 | Intron 11 | c.773 + 4A > C | Intron 11 | c.773 + 4A > C | Well | This study | ||
| 2 | n.a. | 2 d | Neonatal form | 487.69 | 286 | Exon 6 | c.380G > A | p.Arg127Gln | Exon 6 | c.380G > A | p.Arg127Gln | Died | [12] |
| 3 | Male | n.p. | Mild form | 961.42 | 91 | Intron 4 | c.174 + 1G > A | Exon 7 | c.422 T > C | p.Val141Gly | Well | [13] | |
| 4 | Female | 4 d | Neonatal form | 1085.41 | 231 | Intron 11 | c.773 + 1G > A | Exon 12 | c.793C > T | p.Arg265Cys | Moderate | [13] | |
| 5 | Female | 3 m | Late-onset form | n.a. | 311 | Exon 7 | c.431C > G | p.Pro144Arg | Exon 14 | c.1087C > T | p.Arg363Trp | Moderate | [17] |
| 6 | Male | n.p. | Mild form | 111.21 | 17 | Exon 3 | c.53C > T | p.Ser18Leu | Exon 15 | c.1168G > A | p.Gly390Arg | Well | [16] |
| 7 | Female | 1 y, 3 m | Late-onset form | 928.77 | 160 | Exon 13 | c.847G > A | p.Glu283Lys | Exon 14 | c.1009 T > C | p.Cys337Arg | Moderate | [10] |
| 8 | Female | 1 y, 5 m | Late-onset form | 653 | 126 | Exon 5 | c.236C > T | p.Ser79Phe | Exon 7 | c.431C > G | p.Pro144Arg | n.a. | [14] |
| 9 | Female | 2 d | Neonatal form | 1577.7 | 670 | Exon 13 | c.951delT | p.F317LfsX375 | Exon 14 | c.1087C > T | p.Arg363Trp | Died | [11] |
| 10 | Male | 2 d | Neonatal form | 2513.5 | n.a. | Exon 13 | c.970G > A | p.Gly324Ser | Exon 13 | c.970G > A | p.Gly324Ser | n.a. | [15] |
aReference range: 5.5–30 μmol/L; b Reference range: 10–47 μmol/L; c novel mutations are in bold character
n.p. not present, n.a. not available, d day, y year
Discussion and conclusions
In this study, we described a Chinese family with one child having a mild form of CTLN1. The patient had an elevated citrulline level, which was detected by MS-based NBS. No abnormalities were found in urinary organic acid analysis. The patient had normal growth and development during follow up, and the main clinical manifestation was a speech delay. A homozygous ASS1 gene variant c.773 + 4A > C was identified in the patient. This variant has not been previously reported in the literature, and was predicted through bioinformatics analysis to cause a broken WT donor site and to affect splicing. Furthermore, it is likely to truncate the monomers, impairing the synthetase-binding domain. Similar variants leading to protein truncation have been reported previously [13, 19, 20]. Therefore, we believe that the variant c.773 + 4A > C is associated with the pathogenesis of CTLN1. However, further functional studies are needed to validate the pathogenicity of this variant.
After combining our findings with those from previously reported Chinese patients with CTLN1, we performed mutation spectrum analysis. The results showed that the mutation spectrum of Chinese patients with CTLN1 was heterogeneous, with no high frequency or hot spot mutations. In comparison, many mutations have been documented at high frequencies in various other populations and the mutation spectra differ among different ethnic groups. p.Gly390Arg is by far the most common mutation and is widely distributed around the world [9]. It has been proposed that a CpG dinucleotide in the coding region could be the cause of recurring mutations in this region [21], and therefore the recurrent nature of this variant could be explained by its location in a CpG dinucleotide. For instance, the allelic frequencies of p.Gly390Arg in Indian and Turkish patients are 42.7 and 50%, respectively [22, 23]. Likewise, 17.3% of German patients carry at least one p.Gly390Arg allele. p.Gly390Arg is also regarded as a recurrent mutation in a limited geographic area of Argentina [24]. Similarly, the p.Val263Met variant seems to be common in the Pacific Island population [25]. The most frequent mutations in Korean patients are c.421-2A > G, p.Gly324Ser, and c.1128-6_1188dup67, and in Japanese patients the predominant mutations are c.421-2A > G, p. Arg265His, and p.Arg304Trp [26–30]. However, it is surprising that the particularly frequent mutation c.421-2A > G, reported in Korean and Japanese patients, has not yet been detected in Chinese patients.
To date, some mutations have been elucidated with clear genotype-phenotype correlations [9], while most Chinese patients were in a compound heterozygous state, rendering it more difficult to investigate the relationship between genotype and phenotype. Though p.Arg127Gln was proven to be inactive in previous studies, in the neonatal CTLN1 group, a patient (no.2) in our study homozygous for p.Arg127Gln died shortly after birth, confirming previous reports and highlighting the severity of this mutation [31]. Previous enzyme studies revealed that both p.Arg265Cys and p.Gly324Ser yielded < 2% of ASS wild-type activity, and both are known to be associated with a severe phenotype [32]. Supporting these findings, a patient (no.4) who was compound heterozygous for p.Arg265Cys with a splicing mutation presented with early onset neonatal citrullinmia; of note is that an older sibling in this family progressed to severe encephalopathy and died 4 days after birth. A patient (no.10) homozygous for p.Gly324Ser presented with acute hyperammonemia and encephalopathy, again confirming previous studies. p.Arg363Trp was reported to be associated with neonatal CTLN1; consistent with this, a patient (no.9) with p.Arg363Trp in combination with a frameshift mutation died shortly after birth [9]. Regarding mild/late-onset form CTLN1, a patient (no.1) homozygous for c.773 + 4A > C presented no clinical symptoms until 3 years of age, indicating that this variant may be related to mild symptoms. The remaining patients are all compound heterozygotes, and it is likely that the mutations p.Ser18Leu, p.Val141Gly, p.Pro144Arg, and p.Cys337Arg may allow for some residual ASS function, because the second allelic mutations in these patients are known to drastically impair ASS activity [9, 13, 32].
In summary, we described one mild Chinese CTLN1 case with a novel splicing variant c.773 + 4A > C. We also reviewed previous genotypes and phenotypes of Chinese patients with CTLN1, hereby adding to our understanding of the molecular genetic background and clinical phenotype of CTLN1 in this population. The mutation spectrum of Chinese patients with CTLN1 was heterogeneous. More functional research is needed to elucidate the genotype-phenotype correlation in patients with CTLN1.
Additional files
Table S1. Summary of targeted gene sequencing data in the proband. (DOCX 13 kb)
Table S2. Primers used for PCR and Sanger sequencing of exon 11 of ASS1. (DOCX 14 kb)
Acknowledgements
We thank all the participants for their co-operation.
Abbreviations
- ACMG
American College of Medical Genetics Association of Clinical Genetics
- CTN1
Citrullinemia type I
- MAF
Minor allele frequency
- MS/MS
Tandem mass spectrometry
- NBS
Newborn screening
- NGS
Next-generation sequencing
- PCR
Polymerase chain reaction
Authors’ contributions
YL designed the study, performed experimental work, wrote the paper, and conducted the literature review. HG and SZ helped with data collection and interpretation of the data. BL, LZ, and MJ carried out the genetic tests, mutation analysis, and paper editing. TZ and WL followed the patients and collected the clinical data. QF conceived the study design. All authors read and approved the final manuscript.
Funding
This study was supported by the Quanzhou Municipal Science and Technology Plan Project (Grant No. 2018Z160 and 2018N085S). The role of the funding body was to sponsor the two-generation pedigree analysis, the NGS, and the other tests we conducted for this case.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
This study was approved by the ethics committee of The Maternal and Children’s Hospital of Quanzhou. The family have signed written informed consent and agree for themselves and their children to take part in this study, and to the use of the relevant data and information for scientific research.
Consent for publication
We confirm that this family has signed written informed consent for publication of their own and their children’s genetic data, clinical details, and/or any accompanying images.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yiming Lin, Email: 9537237@qq.com.
Hongzhi Gao, Email: hongzhi_gao@hotmail.com.
Bin Lu, Email: lubin@biosan.cn.
Shuang Zhou, Email: 27293988@qq.com.
Tianwen Zheng, Email: zhengtianwen@sina.com.
Weihua Lin, Email: linweihua@sina.com.
Lin Zhu, Email: zhulin@biosan.cn.
Mengyi Jiang, Phone: +086 571 26205108, Email: jiangmengyi@biosan.cn.
Qingliu Fu, Phone: +086 595 22192325, Email: wrightlym@sina.com.
References
- 1.Beaudet AL, O'Brien WE, Bock HG, Freytag SO, Su TS. The human argininosuccinate synthetase locus and citrullinemia. Adv Hum Genet. 1986;15:161–196. doi: 10.1007/978-1-4615-8356-1_3. [DOI] [PubMed] [Google Scholar]
- 2.Haberle J, Pauli S, Linnebank M, Kleijer WJ, Bakker HD, Wanders RJ, Harms E, Koch HG. Structure of the human argininosuccinate synthetase gene and an improved system for molecular diagnostics in patients with classical and mild citrullinemia. Hum Genet. 2002;110(4):327–333. doi: 10.1007/s00439-002-0686-6. [DOI] [PubMed] [Google Scholar]
- 3.Haberle J, Pauli S, Schmidt E, Schulze-Eilfing B, Berning C, Koch HG. Mild citrullinemia in Caucasians is an allelic variant of argininosuccinate synthetase deficiency (citrullinemia type 1) Mol Genet Metab. 2003;80(3):302–306. doi: 10.1016/j.ymgme.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Haberle J, Boddaert N, Burlina A, Chakrapani A, Dixon M, Huemer M, Karall D, Martinelli D, Crespo PS, Santer R, et al. Suggested guidelines for the diagnosis and management of urea cycle disorders. Orphanet J Rare Dis. 2012;7:32. doi: 10.1186/1750-1172-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sander J, Janzen N, Sander S, Steuerwald U, Das AM, Scholl S, Trefz FK, Koch HG, Haberle J, Korall H, et al. Neonatal screening for citrullinaemia. Eur J Pediatr. 2003;162(6):417–420. doi: 10.1007/s00431-003-1177-z. [DOI] [PubMed] [Google Scholar]
- 6.Engel K, Hohne W, Haberle J. Mutations and polymorphisms in the human argininosuccinate synthetase (ASS1) gene. Hum Mutat. 2009;30(3):300–307. doi: 10.1002/humu.20847. [DOI] [PubMed] [Google Scholar]
- 7.Miller MJ, Soler-Alfonso CR, Grund JE, Fang P, Sun Q, Elsea SH, Sutton VR. Improved standards for prenatal diagnosis of citrullinemia. Mol Genet Metab. 2014;112(3):205–209. doi: 10.1016/j.ymgme.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Freytag SO, Beaudet AL, Bock HG, O'Brien WE. Molecular structure of the human argininosuccinate synthetase gene: occurrence of alternative mRNA splicing. Mol Cell Biol. 1984;4(10):1978–1984. doi: 10.1128/MCB.4.10.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diez-Fernandez C, Rufenacht V, Haberle J. Mutations in the human Argininosuccinate Synthetase (ASS1) gene, impact on patients, common changes, and structural considerations. Hum Mutat. 2017;38(5):471–484. doi: 10.1002/humu.23184. [DOI] [PubMed] [Google Scholar]
- 10.Wu TF, Liu YP, Li XY, Wang Q, Song JQ, Yang YL. Prenatal diagnosis of citrullinemia type 1: a Chinese family with a novel mutation of the ASS1 gene. Brain Dev. 2014;36(3):264–267. doi: 10.1016/j.braindev.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 11.Xie B, Chen R, Wang J, Luo J, Li W, Chen S. ASS1 gene mutation in a neonate with citrullinemia type I. Chinese J Pediatr. 2014;52(10):788–791. [PubMed] [Google Scholar]
- 12.Sun J, Shen Y, Yan C, Gong X. [identification of a homozygous ASS1 mutation in a child with citrullinemia type with high-melting curve method]. Chinese. J Med Genet. 2018;35(3):429–433. doi: 10.3760/cma.j.issn.1003-9406.2018.03.028. [DOI] [PubMed] [Google Scholar]
- 13.Kimani JK, Wei T, Chol K, Li Y, Yu P, Ye S, Huang X, Qi M. Functional analysis of novel splicing and missense mutations identified in the ASS1 gene in classical citrullinemia patients. Clin Chim Acta. 2015;438:323–329. doi: 10.1016/j.cca.2014.08.028. [DOI] [PubMed] [Google Scholar]
- 14.Wen P, Chen Z, Wang G, Liu X, Chen L, Chen S, Wan L, Cui D, Shang Y, Li C. [genetic analysis of ASS1, ASL and SLC25A13 in citrullinemia patients]. Chinese. J Med Genet. 2014;31(3):268–271. doi: 10.3760/cma.j.issn.1003-9406.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Hu P, Zhou XY, Ma DY, Sun Y, Zhang XJ, Han SP, Yu ZB, Jiang T, Chen YL, Xu Z. [ASS1 mutation leading to citrullinemia I in a Chinese Han family]. Chinese. J Med Genet. 2011;28(6):630–633. doi: 10.3760/cma.j.issn.1003-9406.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 16.Sun Y, Ma DY, Yang GJ, Wang YY, Yang B, Jiang T. [clinical features and ASSl gene mutation analysis of a case of citrullinemia]. Chinese. J Med Genet. 2015;32(5):745–747. [Google Scholar]
- 17.Cheng AL, Han LS, Feng Y, Jin B. [the magnetic resonance imaging features and clinical manifestations of citrullinemia]. Journal of. Clin Pediatr. 2015;33(5):466–469. [Google Scholar]
- 18.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laine CM, Chung BD, Susic M, Prescott T, Semler O, Fiskerstrand T, D'Eufemia P, Castori M, Pekkinen M, Sochett E, et al. Novel mutations affecting LRP5 splicing in patients with osteoporosis-pseudoglioma syndrome (OPPG) Eur J Hum Genet. 2011;19(8):875–881. doi: 10.1038/ejhg.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohtani R, Inazu A, Noji Y, Wakasugi T, Miwa K, Tada H, Kawashiri MA, Noguchi T, Nohara A, Kobayashi J, et al. Novel mutations of cholesteryl ester transfer protein (CETP) gene in Japanese hyperalphalipoproteinemic subjects. Clin Chim Acta. 2012;413(5–6):537–543. doi: 10.1016/j.cca.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 21.Morgan GE, Figueiredo MS, Winship PR, Baker R, Bolton-Maggs PH, Brownlee GG. The high frequency of the -6G-->a factor IX promoter mutation is the result both of a founder effect and recurrent mutation at a CpG dinucleotide. Br J Haematol. 1995;89(3):672–674. doi: 10.1111/j.1365-2141.1995.tb08388.x. [DOI] [PubMed] [Google Scholar]
- 22.Kose E, Unal O, Bulbul S, Gunduz M, Haberle J, Arslan N. Identification of three novel mutations in fourteen patients with citrullinemia type 1. Clin Biochem. 2017;50(12):686–689. doi: 10.1016/j.clinbiochem.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 23.Bijarnia-Mahay S, Haberle J, Jalan AB, Puri RD, Kohli S, Kudalkar K, Rufenacht V, Gupta D, Maurya D, Verma J, et al. Urea cycle disorders in India: clinical course, biochemical and genetic investigations, and prenatal testing. Orphanet J Rare Dis. 2018;13(1):174. doi: 10.1186/s13023-018-0908-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larovere LE, Angaroni CJ, Antonozzi SL, Bezard MB, Shimohama M, de Kremer RD. Citrullinemia type I, classical variant. Identification of ASS-p~G390R (c.1168G>a) mutation in families of a limited geographic area of Argentina: a possible population cluster. Clin Biochem. 2009;42(10–11):1166–1168. doi: 10.1016/j.clinbiochem.2009.03.024. [DOI] [PubMed] [Google Scholar]
- 25.Glamuzina E, Marquis-Nicholson R, Knoll D, Love DR, Wilson C. Citrullinaemia type I: a common mutation in the Pacific Island population. J Paediatr Child Health. 2011;47(5):262–265. doi: 10.1111/j.1440-1754.2010.01948.x. [DOI] [PubMed] [Google Scholar]
- 26.Woo HI, Ki CS, Lee SY, Kim JW, Song J, Jin DK, Park WS, Lee DH, Lee YW, Park HD. Mutation spectrum of the ASS1 gene in Korean patients with citrullinemia type I. Clin Biochem. 2013;46(3):209–213. doi: 10.1016/j.clinbiochem.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Lee BH, Kim YM, Heo SH, Kim GH, Choi IH, Lee BS, Kim EA, Kim KS, Jhang WK, Park SJ, et al. High prevalence of neonatal presentation in Korean patients with citrullinemia type 1, and their shared mutations. Mol Genet Metab. 2013;108(1):18–24. doi: 10.1016/j.ymgme.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 28.Gao HZ, Kobayashi K, Tabata A, Tsuge H, Iijima M, Yasuda T, Kalkanoglu HS, Dursun A, Tokatli A, Coskun T, et al. Identification of 16 novel mutations in the argininosuccinate synthetase gene and genotype-phenotype correlation in 38 classical citrullinemia patients. Hum Mutat. 2003;22(1):24–34. doi: 10.1002/humu.10230. [DOI] [PubMed] [Google Scholar]
- 29.Hong KM, Shin CH, Choi YB, Song WK, Lee SD, Rhee KI, Jang P, Pak GS, Kim JK, Paik MK, et al. Mutation analysis of Korean patients with citrullinemia. Mol Cells. 2000;10(4):465–468. [PubMed] [Google Scholar]
- 30.Kobayashi K, Kakinoki H, Fukushige T, Shaheen N, Terazono H, Saheki T. Nature and frequency of mutations in the argininosuccinate synthetase gene that cause classical citrullinemia. Hum Genet. 1995;96(4):454–463. doi: 10.1007/BF00191806. [DOI] [PubMed] [Google Scholar]
- 31.Diez-Fernandez C, Wellauer O, Gemperle C, Rufenacht V, Fingerhut R, Haberle J. Kinetic mutations in argininosuccinate synthetase deficiency: characterisation and in vitro correction by substrate supplementation. J Med Genet. 2016;53(10):710–719. doi: 10.1136/jmedgenet-2016-103937. [DOI] [PubMed] [Google Scholar]
- 32.Berning C, Bieger I, Pauli S, Vermeulen T, Vogl T, Rummel T, Hohne W, Koch HG, Rolinski B, Gempel K, et al. Investigation of citrullinemia type I variants by in vitro expression studies. Hum Mutat. 2008;29(10):1222–1227. doi: 10.1002/humu.20784. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Summary of targeted gene sequencing data in the proband. (DOCX 13 kb)
Table S2. Primers used for PCR and Sanger sequencing of exon 11 of ASS1. (DOCX 14 kb)
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author upon reasonable request.