Abstract
The use of zebrafish in whole organism phenotypic assays has become a valuable strategy throughout the drug discovery process. Zebrafish assays can be used not only to screen libraries of compounds at the earliest stages but also to evaluate advanced leads for their effects on specific biological pathways or for toxicity. However, when confronted with inactivity of a compound in a zebrafish assay, there are little data that can be used to judge if the compound is truly biologically inert or inactive due to a lack of permeability into the model organism. While medicinal chemistry principles suggest parameters that are predictive of human oral bioavailability, cellular permeability, and even bacterial permeability, there have been no such parameters developed for zebrafish absorption. To address this question, we compiled a set of 700 compounds reported in the literature to be active in zebrafish assays, evaluated their properties, and compared them to properties derived from a set of historical drugs and a set of recently approved oral drugs. While some properties overlap, the averages and 10th and 90th percentiles of molecular weight, octanol–water partition coefficient (logP), H-bond counts, and polar surface area for zebrafish-active compounds are statistically different from those of known drugs. This analysis should be useful to scientists interpreting structure–activity relationships based on data from zebrafish assays and help to inform the translation from fish to mammals.
Keywords: Zebrafish, physicochemical properties, drug-like properties, permeability, Lipinski Rule-of-Five, phenotypic assays
Whole organism phenotypic screening has become an important complement to more traditional target-based and cell-based assays throughout the drug discovery process.1 Benefits include the ability to evaluate multiple parameters, such as biological effects, permeability, and toxicity, in a single assay. Zebrafish (Danio rerio), in particular, have been widely used in this regard. Their small size, transparent embryos, ease and low cost of breeding, rapid development, evolutionary similarity to humans, and well understood genetics make zebrafish a powerful model organism for screening of small molecules, and for evaluating advanced leads for effects in a biological pathway2−5 or for toxicity.6 Indeed, zebrafish have been used as models for a variety of disease states including kidney disease,7 cancer,8 and central nervous system disorders,9 among others.
In most cases, small molecules are dispensed in the fish water, offering continuous exposure of the zebrafish to drug substance.10 Small molecules can also be directly injected into the fish to ensure exposure, but this approach is not common, particularly in a screening paradigm.4 When dispensed in tank water, compound absorption occurs either through permeation of the skin layer, transport through gills, or, when using embryos, via absorption through the yolk sac. After treatment, effects are monitored and/or quantified. When no effects are observed, conclusions are either (1) the compound was absorbed and had no effect on the biological system or the organism (inactive) or (2) the compound was not absorbed by the fish, and therefore, conclusions about its effects on the biological target or the organism cannot be made. The ability to make this distinction can have important consequences; for example, when zebrafish assays are used to assess safety, and lack of permeability could be incorrectly interpreted as lack of toxicity.
There is an abundance of literature on which physical properties of small molecules predict human oral bioavailability. Lipinski and co-workers correlated preferred values for molecular weight (MW ≤ 500), lipophilicity as measured by octanol–water partition coefficients (logP ≤ 5), and the numbers of hydrogen bond donors (HBD ≤ 5) and acceptors (HBA ≤ 10) to oral bioavailability (Lipinski Rules, Rule of Five11). Veber et al. suggested that total polar surface area (tPSA ≤ 140 Å) of the molecule and the number of rotatable bonds (≤10) could predict oral bioavailability.12 Others have described characteristics of molecules that are important for cell permeability13 and even bacterial permeability.14 However, there is little that correlates physical properties such as molecular weight, lipophilicity, and solubility to zebrafish absorption. In this absence, the assumption is usually made that the properties that are predictive of human/mammalian absorption will translate to fish; moreover, since the ultimate goal is to treat mammals, not fish, designing for human oral bioavailability is appropriate. However, significant physiological differences between fish and mammals raise questions about the validity of this assumption, and the absence of data may lead to incorrect conclusions about whether a compound is truly biologically inert or did not permeate the fish, thereby complicating any structure–activity relationship (SAR) analysis or potentially leading to false conclusions about the toxicity of small molecules.
To address this knowledge gap, we assembled structures of compounds reported in the literature to exhibit biological activity in zebrafish assays and made the assumption that biological activity necessitated permeability/absorption into this model organism (whether it be passive or active). We then calculated and predicted a series of properties and property descriptors and asked (1) what is the range of drug-like properties [e.g., molecular weight (MW), logP, polar surface area (PSA), number of rotatable bonds, H-bond donors (HBD), H-bond acceptors (HBA), solubility, and permeability] exhibited by compounds that are absorbed by zebrafish?; (2) do the general characteristics of zebrafish-active compounds differ from the standard physicochemical property guidelines for oral bioavailability used by medicinal chemists or from those of known drugs?; and (3) can we develop a set of guidelines that can be used to interpret zebrafish assay data? This report details our analysis.
To compile a set of zebrafish-permeable compounds, we used keywords such as “zebrafish assay” and “zebrafish screen” to search the Scifinder and Pubmed databases for publications reporting data on small molecules’ activity in zebrafish assays. We also directly searched medicinal chemistry and chemical biology journals such as ACS Chemical Biology, ACS Medicinal Chemistry Letters, Bioorganic & Medicinal Chemistry, Bioorganic & Medicinal Chemistry Letters, European Journal of Medicinal Chemistry, Journal of Medicinal Chemistry, and MedChemComm. In total, 158 unique publications were identified, and the structures of active compounds were extracted from them. To supplement the literature data set, we reviewed Pubchem for data reported in zebrafish assays. We eliminated assays designated as “screening” as they typically report high false positive rates. Thirty-one compounds labeled as “active” in four different confirmatory or secondary assays (AIDs 691, 1137, 504373, 652284) were added to the data set. While it is likely that some publications were not captured, we believe the number and diversity of compounds identified (vide infra) provide a representative data set. Figure 1 outlines the data compilation and curation process.
Figure 1.
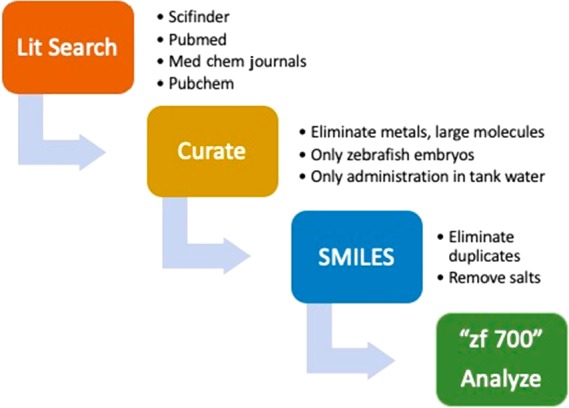
Process for data accumulation and curation.
Structures of compounds, administered in tank water, that were reported to be active in a zebrafish embryo assay, regardless of potency, degree of efficacy, or time of exposure were collected. We eliminated structures that contained metals or were large peptides or proteins, and those that bypassed the absorption process by administration via injection or other direct method. Structures were prepared as SMILES strings (simplified molecular-input line-entry system); salt counterions and any duplicates were removed. Applying these criteria, 700 unique small molecules (“zf 700”) were identified (see SI).
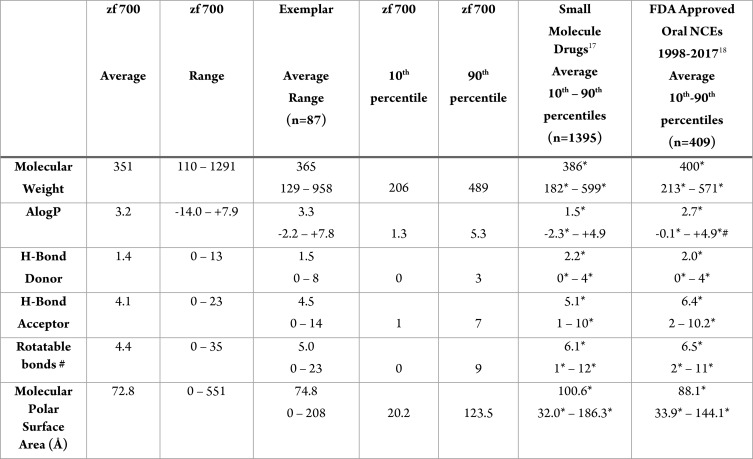
We calculated MW, logP (as AlogP), HBD, HBA, number of rotatable bonds, and molecular PSA.15Table 1 shows the range and averages for these properties for the zf 700. Not unexpectedly, the averages all fall within the typical Lipinski/Veber guidelines: MW = 351; AlogP = 3.2; HBD = 1.4; HBA = 4.1; rotatable bonds = 4.4; and tPSA = 72.8 Å. Notable, however, is the wide range of values that appear to be compatible with zebrafish absorption, particularly molecular weight (110 to 1291 Da), AlogP (−14.0 to +7.9), and tPSA (0 to 551 Å). Other properties evaluated, including HBD, HBA, and rotatable bonds, also reveal a very wide range associated with zebrafish active compounds. While some of the extreme values are represented only sparsely, this data suggests that compounds with properties far outside what is typically considered drug-like can, in some cases, be permeable to zebrafish.
Table 1. Calculated Properties of Zebrafish Active, Small Molecule Drugs, and Recently Approved Oral Drugsa.
Small molecule drugs derived from DrugBank;17 1998–2017 values adapted from Table 5 in ref (18); p values for means based on one-way ANOVA using GraphPad Prism7 software and posthoc analysis using Tukey’s multiple comparison test; p values for 10th and 90th percentile values are based on nonparametric Wilcoxon signed-rank test and are relative to zf 700; p < 0.05 considered significant, indicated with*; #AlogP value for 1998–2017 drug set based on Stardrop clogP.
To ensure that disproportionate representation of multiple close structural analogs in the zf 700 set were not skewing the averages, we used a computational algorithm (“Affinity Propagation Clustering”16) that clustered the 700 based on structural similarity. This algorithm identified 87 exemplars (see SI), representative of the structural diversity of the entire set, for which we calculated the same values for comparison with the full zf 700 set. An independent evaluation confirmed the diversity of this Exemplar 87 set (SI). As seen in Table 1, even though the ranges are narrower, the averages calculated for the 87 exemplars is, in most cases, very similar to those generated from the full set of 700. This analysis provides evidence that the presence of large numbers of closely related compounds in the zf 700 set did not bias the calculated averages.
We calculated the 10th and 90th percentiles of the values in Table 1 for the zf 700 set to interrogate the boundaries that can be applied to predict zebrafish absorption. For comparison, we also completed a similar analysis of a set of historical small molecule drugs (approved and experimental, regardless of route of administration) derived from DrugBank (n = 1395)17 and a set of >400 oral drugs approved by the FDA in a recent 20-year (1998–2017) span.18 This data set (Table 1) highlights the differences between the characteristics of the majority of zebrafish-permeable compounds and characteristics of approved drugs, and by extension the properties medicinal chemists typically strive for when developing drug-like and/or orally available molecules. Inclusion of the 1998–2017 set allows for specific comparison with orally administered drugs, and also takes into account some of the considerable debate in the literature on which properties reflect “drug-likeness.”18
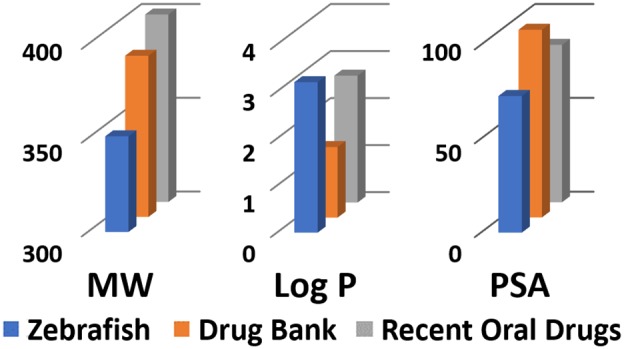
The majority of zebrafish-permeable compounds fall within the molecular weight range of 200–500 Da, with an average MW of 351. These numbers are shifted to lower values compared to both drug groups, and by ∼50 Da compared to the recent oral drug set. This shift is most apparent at the higher end of the molecular weight range with the 90th percentile of the zf 700 set (489 Da) being ∼100 Da lower than the two drug sets (599 and 571 Da), but the 10th percentiles being very close. The molecular weight differences reached statistical significance for all values.
Also divergent are the AlogP values. The zf 700 average (3.2) is significantly higher than the two drug groups (1.5 and 2.7 for the small molecule and recent oral drug sets, respectively), tending toward greater lipophilicity. The 90th percentile for the zf 700 is also greater than the same values for the historical and recent oral drugs (5.3 vs 4.9 for both drug sets); however, for the 90th percentiles, only the difference between the zebrafish and recent oral drug set was significant. This preference for higher lipophilicity could be attributed to zebrafish-active molecules being predominantly absorbed through the yolk sac, which contains high concentrations of lipids.
In accordance with the higher values for AlogP, the HBD and HBA counts for the zf 700 are also shifted toward more lipophilic properties. The 90th percentiles for HBD and HBA are 3 and 7, respectively, with averages of 1.4 (HBD) and 4.1 (HBA) vs the more typical limits (90th percentile) for HBD of 4 and HBA of 10 for both the small molecule and recent oral drugs. The averages for the two drug sets (HBD, 2 for both; HBA, 5.1 and 6.4 for DrugBank and recent oral drugs, respectively) were significantly higher than the zf 700 averages. Consistent with the trend toward more lipophilic characteristics, the average PSA of the zebrafish active set was 72.8 Å, and its 90th percentile equaled 123 Å; both significantly lower than the two drug categories. The rotatable bond count average (4.4) is also lower in the zf 700 set compared to the drug sets (6.1 and 6.5), as are the 10th and 90th percentile values.
Overall, zebrafish-absorbed molecules tend to be more lipophilic than known drugs, and in a most cases, their physicochemical properties fall within a narrower range of values. Based on this analysis, in particular the 90th percentiles (which were loosely applied to derive the Lipinski rules), we suggest that compounds most likely to be absorbed by zebrafish will have the following characteristics:
MW ≤ 500
clogP ≤ 5.3
HBD ≤ 3
HBA ≤ 7
tPSA ≤ 124 Å
rotatable bonds ≤ 9
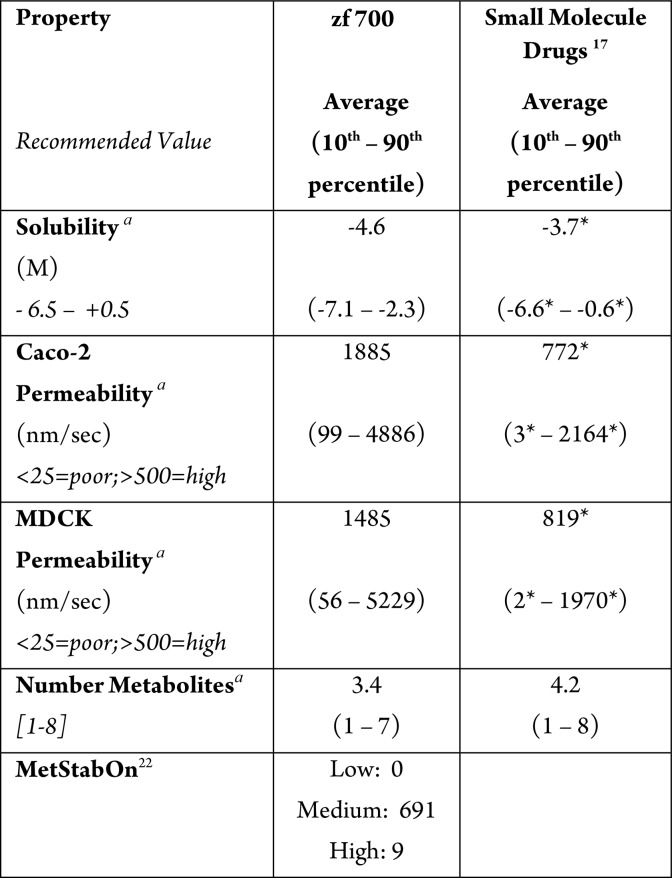
To develop a further understanding of the characteristics of zebrafish-permeable compounds, we predicted19 additional properties such as solubility, various cellular permeability rates, and number of predicted metabolites. Table 2 shows those results and a comparison to the DrugBank drug set.17 Of note, the same analysis with the exemplar set was, again, consistent with the full set of 700 compounds (data not shown). Solubility trended toward lower values (log S average = −4.6 M) compared to typical goals of log S > −4.2 M20 and the drug set (log S = −3.7 M), perhaps confirming the shift toward greater lipophilicity. This result was somewhat unexpected given that the zebrafish actives were administered in tank water, which requires aqueous solubility at relatively high concentrations (typically ≥10 μM) with low concentrations (typically <0.5% or less) of cosolvents (e.g., DMSO, EtOH, MeOH) present.10
Table 2. Predicted Properties of zf 700 and Small Molecule Drugs.
Values predicted for 684/700 compounds; p values for means based on one-way ANOVA using GraphPad Prism7 software and posthoc analysis using Tukey’s multiple comparison test; 10th and 90th percentiles calculated using R. p values for 10th and 90th percentile values are based on nonparametric Wilcoxon signed-rank test and are relative to zf 700; p < 0.05 considered significant, indicated with *.
Averages for cellular permeability predictions (Caco-2 and MDCK cell lines) were far above the >500 nm/s value that is generally characterized as “excellent”, as were the averages calculated for the small molecule drug set. Of interest, however, the 10th percentiles for both cell lines for the zf 700 set were also well above the “poor” boundary of <25 nm/s, with values equal to 99 and 56 nm/s, respectively. This metric is at variance from the historic drug set, where the 10th percentiles for both Caco-2 (3 nm/s) and MDCK (2 nm/s) permeability fell into the “poor” descriptor. This suggestion that adequate compound permeability is more important for fish embryo absorption than mammalian cell absorption is not unexpected and could be due to a number of factors including differences between mammalian and fish membrane composition or in the activity of ABC efflux transporters.21 However, this analysis suggests that predicted permeability in either Caco-2 or MDCK cell lines may be useful in evaluating the potential of compounds to be absorbed into zebrafish, and that activity is unlikely to be observed with compounds predicted to be poorly permeable (i.e., <25 nm/s).
Finally, we considered whether zebrafish-actives would be distinguished from other sets by differences in predicted metabolism. Since the zebrafish yolk is highly metabolically active22 and assays are often run over the course of days, one could hypothesize that activity in zebrafish assays requires metabolic stability. The prediction of numbers of metabolites indicated that the zf 700 set (average = 3.4; 10th percentile = 1; 90th percentile = 7) fell within the recommended value of 1–8 and was only marginally different from values calculated for small molecule drugs (Table 2).
To evaluate this hypothesis further, we used the open source program MetStabOn23 to predict human metabolic stability. Of the 700 zebrafish-active compounds, 0 were predicted to exhibit high metabolic stability, and 691 were predicted to have medium metabolic stability; low metabolic stability was predicted for only nine compounds. Based on this analysis, one can conclude that zf-active compounds, as a whole, are not exceptionally stable compared to typical drugs.
We believe this analysis should be useful when interpreting data from small molecule testing in zebrafish assays; however, there are some limitations to the data set. First, practical limitations in zebrafish testing/assay capacity and/or incorporation of zebrafish assays late in the drug discovery process may have resulted in compounds in the zf 700 data set being prechosen for good/excellent drug-like properties, rather than for a more diverse sampling. For example, when used for screening, libraries that very often contain known drugs10 or molecules that are more drug-like than lead-like are frequently used. In other instances, our literature search identified advanced compounds, already optimized for good drug-like properties, that were evaluated in zebrafish assays for specific effects, such as toxicity. In this way, compounds in the zf 700 data set may not represent the broadly diverse scaffolds that are common in more traditional HTS libraries. However, if a preselection of compounds had influenced the data, we would have expected no or minimal difference between the zf 700 set and the known drug sets, rather than the significant differences observed.
Second, our criteria for inclusion was binary (active/inactive) and considered neither potency nor degree of efficacy. While most compounds in the zf 700 set exhibited activity at concentrations of 10–20 μM, some were active at concentrations as high as 100 μM. Therefore, future studies should focus on how potencies correlate with properties. Third, due to a lack of data, we do not understand what precludes absorption into zebrafish and can only infer the properties that zebrafish active molecules possess.
Despite these caveats, the reported analyses should be of value to medicinal chemists who rely on SAR data from zebrafish assays to guide drug discovery efforts and translation into mammals. Importantly, our results provide guidelines to aid in determining which compounds are likely to be absorbed by zebrafish and, as a result, can subsequently be used to rationalize when an “inactive” report in a zebrafish assay should be attributed to a lack of biological activity versus a lack of permeability. The observation that zebrafish-actives display a different range of the properties typically considered drug-like should help focus medicinal chemistry design that relies on zebrafish assay data and also ensure that optimization of activity in fish can be translated to humans.
Acknowledgments
The authors thank Matthew C. Kostman for help with literature searching, Zoe Vaughn (University of Pittsburgh) for help with bibliographies, and Jay Kostman for review of the manuscript.
Glossary
ABBREVIATIONS
- AlogP
atom based logP prediction
- HBA
hydrogen bond donor
- HBA
hydrogen bond acceptor
- MDCK
Madin–Darby Canine Kidney cells
- QPlogS
predicted aqueous solubility
- QPPCaco
predicted apparent Caco-2 permeability
- QPPMDCK
predicted apparent MDCK permeability
- SMILES
simplified molecular-input line-entry system
- PSA
polar surface area
- zf
zebrafish
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.9b00063.
Methods (PDF)
Zf 700: identity, SMILES, references, calculated properties, QikProp predictive properties, statistical analysis (XLSX)
Eighty-seven exemplars: identity, SMILES, calculated properties, statistical analysis (XLSX)
DrugBank small molecules: identity, SMILES, calculated properties QikProp predictive properties (XLSX)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
This work was funded in whole or in part with funds from the US Department of Defense, Grant #W81XWH-17-1-0610 and by NIH Grant R01 CA216815.
The authors declare no competing financial interest.
Supplementary Material
References
- Cho Y. S.; Kwon H. J. Identification and validation of bioactive small molecule target through phenotypic screening. Bioorg. Med. Chem. 2012, 20, 1922–1928. 10.1016/j.bmc.2011.11.021. [DOI] [PubMed] [Google Scholar]
- Taylor K. L.; Grant N. J.; Temperley N. D.; Patton E. E. Small molecule screening in zebrafish: an in vivo approach to identifying new chemical tools and drug leads. Cell Commun. Signaling 2010, 8, 11. 10.1186/1478-811X-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miscevic F.; Rotstein O.; Wen X. Y. Advances in zebrafish high content and high throughput technologies. Comb. Chem. High Throughput Screening 2012, 15, 515–21. 10.2174/138620712801619140. [DOI] [PubMed] [Google Scholar]
- Rennekamp A. J.; Peterson R. T. 15 years of zebrafish chemial screening. Curr. Opin. Chem. Biol. 2015, 24, 58–70. 10.1016/j.cbpa.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoganantharjah P.; Gibert Y. The use of the zebrafish model to aid in drug discovery and target validation. Curr. Top. Med. Chem. 2017, 17, 2041–2055. 10.2174/1568026617666170130112109. [DOI] [PubMed] [Google Scholar]
- Rubinstein A. L. Zebrafish assays for drug toxicity screening. Expert Opin. Drug Metab. Toxicol. 2006, 2, 231–240. 10.1517/17425255.2.2.231. [DOI] [PubMed] [Google Scholar]
- Cirio M. C.; de Caestecker M. P.; Hukriede N. A. Zebrafish models of kidney damage and repair. Curr. Pathobiol. Rep. 2015, 3, 163–170. 10.1007/s40139-015-0080-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriuso J.; Nagaraju R.; Hurlstone A. Zebrafish: a new companion for translational research in oncology. Clin. Cancer Res. 2015, 21, 969–975. 10.1158/1078-0432.CCR-14-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart A. M.; Braubach O.; Spitsbergen J.; Gerlai R.; Kalueff A. V. Zebrafish models for translational neuroscience resaerch: from tank to bedside. Trends Neurosci. 2014, 37, 264–278. 10.1016/j.tins.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H.; Lin S.. Chemical screening with zebrafish embryos. In Drug Design and Discovery. Methods in Molecular Biology (Methods and Protocols); Satyanarayanajois S., Ed.; Humana Press, 2011; pp 193–205. [DOI] [PubMed] [Google Scholar]
- Lipinski C. A.; Lombardo F.; Dominy B. W.; Feeney P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 2001, 46, 3–26. 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Veber D. F.; Johnson S. R.; Cheng H. Y.; Smith B. R.; Ward K. W.; Kopple K. D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–23. 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- Yang N. J.; Hinner M. J. Getting across the cell membrane: an overview for small molecules, peptides and proteins. Methods Mol. Biol. 2015, 1266, 29–53. 10.1007/978-1-4939-2272-7_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M. F.; Drown B. S.; Riley P. A.; Garcia A.; Shirai T.; Svec R. L.; Hergenrother P. J. Predictive rules for compound accumulation yield a broad-spectrum antibiotic. Nature 2017, 545, 299–304. 10.1038/nature22308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassault Systèmes. BIOVIA Discovery Studio; Dassault Systèmes: San Diego, 2018. [Google Scholar]
- Athanasiadis E.; Cournia Z.; Spyrou G. ChemBioServer: a web-based pipeline for filtering, clustering and visualization of chemical compounds used in drug discovery. Bioinformatics 2012, 28, 3002–3003. http://chembioserver.vi-seem.eu 10.1093/bioinformatics/bts551. [DOI] [PubMed] [Google Scholar]
- Wishart D. S.; Feunang Y. D.; Guo A. C.; Lo E. J.; Marcu A.; Grant J. R.; Sajed T.; Johnson D.; Li C.; Sayeeda Z.; Assempour N.; Iynkkaran I.; Liu Y.; Maciewjewski A.; Gale N.; Wilson A.; Chin L.; Cummings R.; Le D.; Pon A.; Knox C.; Wilson M. DrugBank5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz M. D. Two decades under the influence of the rule of five and the changing properties of approved oral drugs. J. Med. Chem. 2019, 62, 1701. 10.1021/acs.jmedchem.8b00686. [DOI] [PubMed] [Google Scholar]
- Schrödinger Release 2016-1: Maestro; Schrödinger, LLC: New York, 2018. [Google Scholar]
- Di L.; Fish P. V.; Mano T. Bridging solubility between drug discovery and development. Drug Discovery Today 2012, 17, 486–495. 10.1016/j.drudis.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Fischer S.; Klüver N.; Burkhardt-Medicke K.; Pietsch M.; Schmidt A.-M.; Wellner P.; Schirmer K.; Luckenbach T. Abcb4 acts as a multixenobiotic transporter and active barrier against chemical uptake in zebrafish (Danio rerio) embryos. BMC Biol. 2013, 11, 69. 10.1186/1741-7007-11-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraher D.; Sanigorski A.; Mellett N. A.; Meikle P. J.; Sinclair A. J.; Gibert Y. Zebrafish embryonic lipidomic analysis reveals that the yolk cell is metabolically active in processing lipid. Cell Rep. 2016, 14, 1317–1329. 10.1016/j.celrep.2016.01.016. [DOI] [PubMed] [Google Scholar]
- Podlewska S.; Kafel R. MetStabOn- online platform for metabolic stability prediction. Int. J. Mol. Sci. 2018, 4, 1040–1057. 10.3390/ijms19041040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.