Abstract
Background
Cultivated rice (Oryza sativa L.) is one of the staple food for over half of the world’s population. Thus, improvement of cultivated rice is important for the development of the world. It has been shown that abundant elite genes exist in rice landraces in previous studies.
Results
A genome-wide association study (GWAS) performed with EMMAX for 12 agronomic traits measured in both Guangzhou and Hangzhou was carried out using 150 accessions of Ting’s core collection selected based on 48 phenotypic traits from 2262 accessions of Ting’s collection, the GWAS included more than 3.8 million SNPs. Within Ting’s core collection, which has a simple population structure, low relatedness, and rapid linkage disequilibrium (LD) decay, we found 32 peaks located closely to previously cloned genes such as Hd1, SD1, Ghd7, GW8, and GL7 or mapped QTL, and these loci might be natural variations in the cloned genes or QTL which influence potentially agronomic traits. Furthermore, we also detected 32 regions where new genes might be located, and some peaks of these new candidate genes such as the signal on chromosome 11 for heading days were even higher than that of Hd1. Detailed annotation of these significant loci were shown in this study. Moreover, according to the estimated LD decay distance of 100 to 350 kb on the 12 chromosomes in this study, we found 13 identical significant regions in the two locations.
Conclusions
This research provided important information for further mining these elite genes within Ting’s core collection and using them for rice breeding.
Electronic supplementary material
The online version of this article (10.1186/s12870-019-1842-7) contains supplementary material, which is available to authorized users.
Keywords: Ting’s core collection, Rice (Oryza sativa L.), Agronomic traits, Genome-wide association study, Elite genes
Background
Cultivated rice (Oryza sativa L.) is one of the staple foods for over half of the world’s population. Uncovering the genetic basis of natural variations in important agronomic traits in rice landraces is indispensable for ensuring the world’s food supply.
In general, linkage mapping is a conventional method for gene mining in rice. However, association mapping based on linkage disequilibrium (LD) has been widely used in rice studies since it was firstly reported in maize [1, 2]. Association mapping could overcome the limitations (i.e., limited alleles, high cost and poor mapping resolution) of linkage mapping [3] and enable researchers to use modern genetic technologies for exploiting natural genetic diversity and identifying elite genes in the genome [4]. Furthermore, many candidate genes or loci have been identified in rice through genome-wide association study (GWASs) of agronomic traits [5–10], abiotic stress tolerance [11–13] and metabolites [14, 15].
A population with diverse landraces or cultivars which could be used in crops GWASs is supposed to be a permanent resource and be rephenotyped for many traits [2]. Sampling populations (e.g., core collections and mini core collections) created from rice landraces might be a suitable choice for rice GWASs [16]. Rice landraces are easier to be utilized for breeding than wild rice because they have greater genetic diversity than elite cultivars and represent an intermediate stage of domestication history between wild rice and cultivars [17]. As early as 1920–1964, Ying Ting collected more than 7128 rice landraces from all over China and from some of the other main rice-cultivating countries. This collection is one of the earliest collections of rice germplasm resources in China and was named Ting’s collection [18]. Moreover, a rice core collection called Ting’s core collection and consisting of 150 accessions selected based on 48 phenotypic traits has been constructed from 2262 accessions of Ting’s collection [18]. In Ting’s core collection, the average polymorphism information content (PIC) is 0.48, and the average genetic diversity is 0.54 [19]. Furthermore, Ting’s core collection has been used in association mapping of 12 agronomic traits [20] and aluminum tolerance [21] with 274 SSR markers. However, no association mapping with higher resolution has been performed for agronomic traits within Ting’s core collection.
In the present study, a GWAS of 12 rice agronomic traits was carried out using Ting’s core collection of rice landraces with more than 3.8 million high-quality 3.8 million SNPs by whole-genome re-sequencing. Regions identified by the GWAS were compared with those identified as QTL and candidate genes in previous studies. This information will be very useful for rice breeders to improve elite cultivars.
Results
Comparison between Ting’s core collection and other populations used in GWASs
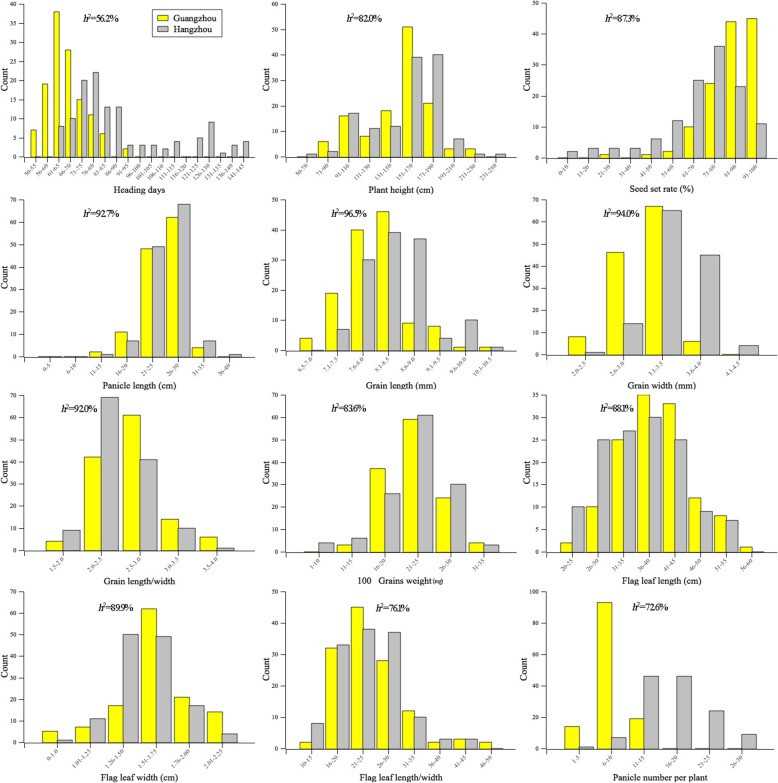
Ting’s core collection consists of 150 rice landraces that were collected from 20 different provinces of China and from North Korea, Japan, the Philippines, Brazil, Sulawesi, Java, Oceania, and Vietnam (Additional file 2: Table S1). The number of varieties in Ting’s core collection is lower than that in a population of Chinese rice landraces [5], a global collection [9] and a mini core collection of japonica rice [8], however, the phenotypic diversity in several agronomic traits in Ting’s core collection are comparable to those in above mentioned collections or even higher for some agronomic traits (Fig. 1).
Fig. 1.
Frequency distribution of agronomic traits in Ting’s core collection
Genome re-sequencing and SNP identification
Whole-genome re-sequencing of Ting’s core collection was performed, resulting in a total of 522.4 Gb of clean data with an average sequencing depth of 7.3× and an average coverage of 82.9% of the reference genome (Additional file 2: Table S2). The distribution of SNP positions along each chromosome are shown in Additional file 1: Figure S1. A total of 3,808,730 SNPs and 391,756 InDels with a minor allele frequency > 0.05 were generated, and 386,562 SNPs were found in the CDS region (Additional file 2: Table S3).
Phenotypic variation
A wide range of phenotypic variation in the 12 agronomic traits was revealed in Ting’s core collection both in Guangzhou and Hangzhou (Fig. 1). Plant height, grain length, grain width, grain length/width, 100 grains weight, flag leaf length, flag leaf width and flag leaf length/width showed similar distributions in the two locations, while heading days, seed set rate, panicle length and panicle number per plant had different distributions in the two locations. The broad-sense heritability ranged from 56.2% (Heading days) to 96.5% (Grain length) for these traits (Fig. 1).
Population structure and LD estimation in Ting’s core collection
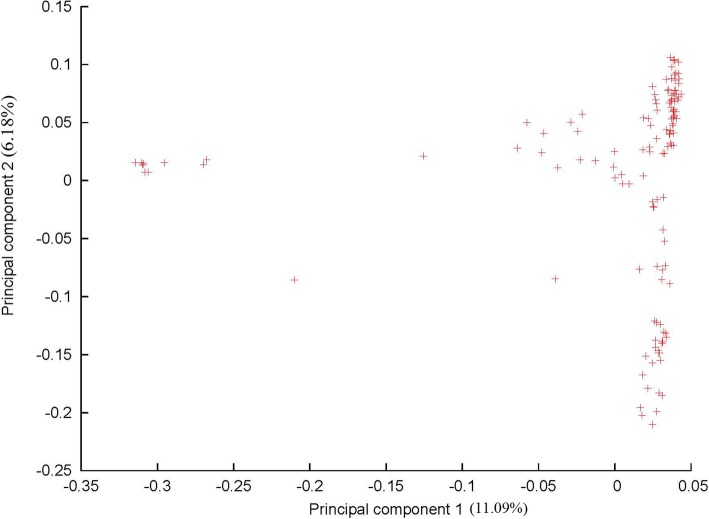
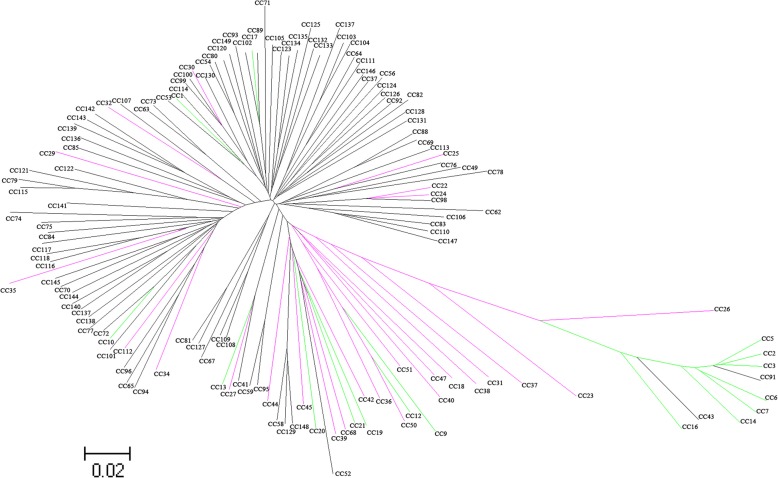
We performed PCA to identify the population structure of Ting’s core collection with all SNPs data, and we observed two subpopulations in Ting’s core collection (Fig. 2). The discrimination obtained via a NJ tree based on the SNP data was not identical to that based on Cheng’s index method (Additional file 2: Table S1) [19] and showed fairly consistent results with that from the PCA (Fig. 3). Moreover, the LD dropped to the half of its maximum value at a distance of 100~350 kb on the 12 chromosomes, which is agreement with previous measurements [5, 9, 22, 23] (Additional file 1: Figure S2).
Fig. 2.
Principal component analysis on 3.8 million SNPs of Ting’s core collection. PC 1 and PC 2 refer to the first and second principal components, respectively. The numbers in parentheses refer to the proportion of variance explained by the corresponding axes. Symbols represent each variety in Ting’s core collection
Fig. 3.
Unrooted neighbor-joining trees of 150 rice varieties in Ting’s core collection. Root with different colors represent the subpopulation identified in our previous study in which population structure was estimated by using 274 SSR markers (Zhang et al., 2011), i.e. Black, green and purple represent indica, japonica and mixed, respectively
Relative kinship among varieties in Ting’s core collection and the effect of controlling type I error using EMMAX
In Ting’s core collection, most kinship estimates between varieties were zero, and none of the kinship values were larger than 0.5, indicating that these varieties were unrelated (Additional file 1: Figure S3).
Observed versus expected P values for each signal were graphed for estimating the effect of controlling for type I errors. As deviations from expected values demonstrate that the statistical analysis may cause spurious associations [24]. Our result indicated that the false positives were unlikely for all traits except grain length/width for the EMMAX method used in this study (Additional file 1: Figure S4).
GWAS of 12 agronomic traits
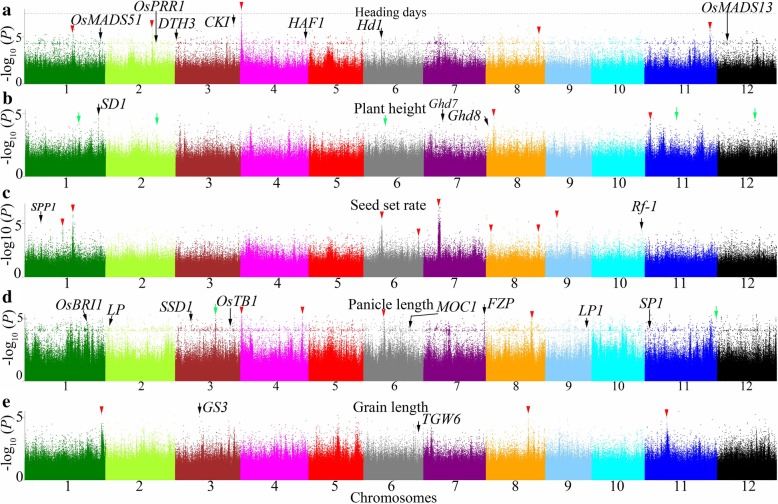
A total of 3,808,730 SNPs were included in a GWAS of 12 agronomic traits using the EMMAX method. Only one association signal’s -log10(P) value was higher than 6.58 (this value was the significant threshold in this study, please see methods section)—a signal for heading days (Fig. 4a). Thus, we used -log10(mBF) = 4.97 as the significance threshold for different traits in our study. A total of 1308 and 4272 significant loci were identified for the 12 agronomic traits in Guangzhou and Hangzhou, respectively (Table 1). The top-ranking candidate gene-based association signals for each trait are shown in Additional file 3: Table S4.
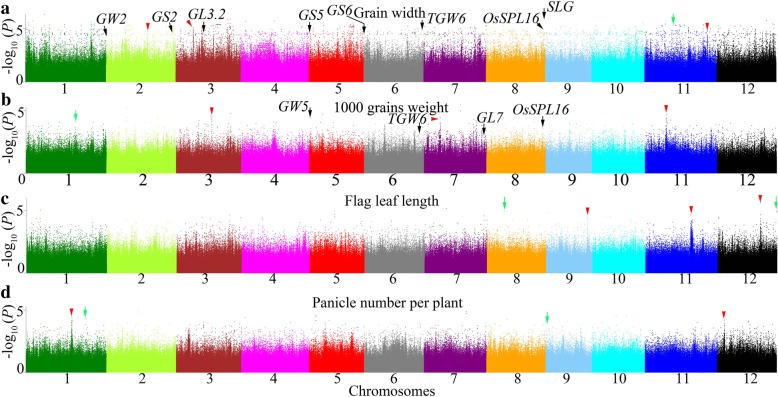
Fig. 4.
Manhattan plots of EMMAX for 5 agronomic traits in genome-wide association studies. Negative log10(P) values from a genome-wide scan are plotted against position on each of 12 chromosomes. a Manhattan plots of EMMAX for heading days. Red horizontal dashed line indicates the genome-wide significant threshold; b Manhattan plots of EMMAX for plant height; c Manhattan plots of EMMAX for seed set rate; d Manhattan plots of EMMAX for panicle length; e Manhattan plots of EMMAX for grain length. Black, red and green arrow represent the loci close to previous genes, new loci and identical in Guangzhou and Hangzhou, respectively
Table 1.
Summary of association mapping results for 12 agronomic traits using EMMAX
| Trait | Number of significant loci | Identical genome region (IRGSP 1.0) | ||
|---|---|---|---|---|
| Guangzhou | Hangzhou | Position (Guangzhou) | Position (Hangzhou) | |
| Heading days | 18 | 174 | – | – |
| Plant height | 3 | 2 | – | – |
| Seed set rate | 943 | 78 | Chr05_25567352 | Chr05_25408291 |
| Chr06_8915912~Chr06_9551431 | Chr06_9230285 | |||
| Chr07_8779751 | Chr07_8438294~Chr07_8467097 | |||
| Panicle length | 1 | 129 | – | – |
| Grain length | 5 | 7 | – | – |
| Grain width | 2 | 54 | – | – |
| Grain length/width | 0 | 3772 | – | – |
| 100 grains weight | 0 | 1 | – | – |
| Flag leaf length | 3 | 3 | – | – |
| Flag leaf width | 9 | 31 | – | – |
| Flag leaf length/width | 323 | 19 | Chr10_12103594~Chr10_12266458 | Chr10_12442627 |
| Chr12_8018851~Chr12_8206256 | Chr12_8234635 | |||
| Panicle number per plant | 1 | 2 | – | – |
| Total | 1308 | 4272 | – | – |
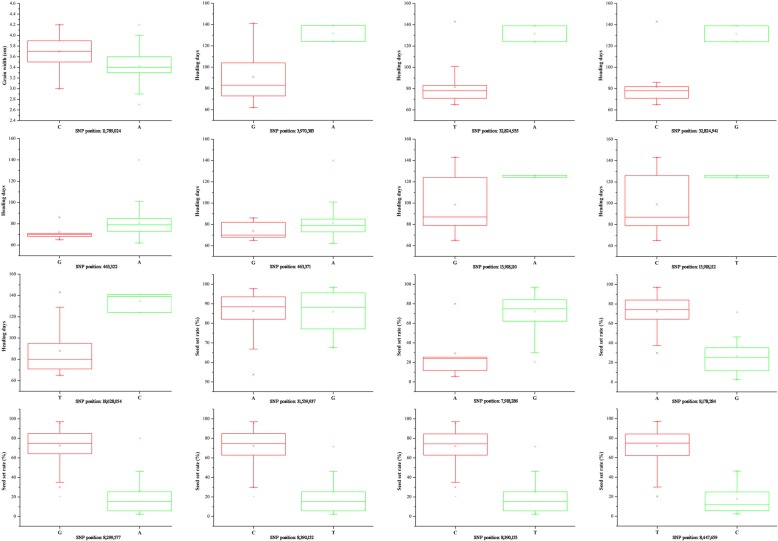
Furthermore, Si et al. (2016) indicated that they considered analyzing the 11 predicted genes within the 260-kb interval centered on the index SNP from the GWAS given the estimated LD decay rate of about 100 to 200 kb [25]. Thus, we analyzed whether some of the significant detections for each trait were identical in the two locations according to the estimated distance of LD decay of 100 to 350 kb on the 12 chromosomes (Additional file 1: Figure S2). Three significant regions (located on chromosomes 5, 6 and 7) for seed set rate were detected both in Guangzhou and Hangzhou. Moreover, two significant regions for flag leaf length/width were detected (located on chromosomes 10 and 12) in both locations (Figs. 4b, d, 5a, b, c, d and Table 1). Moreover, we chose the top 16 most significant signals (P value < 1 × 10− 6) for in-depth analysis (Tables 2 and 3). The significant association signals with smaller P values and higher consecutive peaks for each trait are summarized in Table 3, Figs. 4 and 5, these signals might be located in candidate genes/regions. In addition, a detailed distribution of these new gene-based association signals is included in Additional file 4: Table S5 To confirm the effect of different alleles at the top 16 significant SNPs in the present study, we performed allelic analysis to these SNPs. Accessions in Ting’s core collection carrying different alleles for most of the 16 SNPs showed distinct discrepancies of phenotypes (Fig. 6).
Fig. 5.
Manhattan plots of EMMAX for 4 agronomic traits in genome-wide association studies. Negative log10(P) values from a genome-wide scan are plotted against position on each of 12 chromosomes. a Manhattan plots of EMMAX for grain width; b Manhattan plots of EMMAX for 100 grains weight; c Manhattan plots of EMMAX for flag leaf length; d Manhattan plots of EMMAX for panicle number per plant. Black, red and green arrow represent the loci close to previous genes, new loci and identical in Guangzhou and Hangzhou, respectively
Table 2.
Information of new genome-wide significant association signals using EMMAX
| Trait | Chromosome | SNP position (IRGSP 1.0) |
|---|---|---|
| Heading days | 1 | 23,170,046~23,178,871 |
| 2 | 23,561,650~23,647,315 | |
| 4 | 501,174~599,922 | |
| 8 | 24,711,213~24,788,877 | |
| 11 | 25,420,422~25,527,993 | |
| Plant height | 8 | 4,006,947~4,099,049 |
| 11 | 2,700,225~2,766,157 | |
| Seed set rate | 1 | 18,520,148~18,579,781 |
| 1 | 22,939,166~22,978,931 | |
| 6 | 8,983,019~9,267,052 | |
| 6 | 27,910,112~27,985,250 | |
| 7 | 7,750,765~9,020,981 | |
| 8 | 2,701,887~2,793,377 | |
| 8 | 23,506,423~23,515,115 | |
| 9 | 5,476,906~5,579,147 | |
| Panicle length | 4 | 431,949~574,020 |
| 4 | 30,763,510~31,085,620 | |
| 6 | 10,410,902~10,540,457 | |
| 8 | 19,760,055~19,773,993 | |
| Grain length | 1 | 40,952,595~41,000,011 |
| 8 | 18,807,278~18,877,159 | |
| 11 | 10,147,830~10,203,596 | |
| Grain width | 3 | 11,852,705~11,908,957 |
| 11 | 24,258,724~24,290,332 | |
| 100 grains weight | 3 | 20,308,101~20,374,967 |
| 7 | 8,265,240~8,330,871 | |
| 11 | 9,821,720~9,835,992 | |
| Flag leaf length | 9 | 19,484,679~19,503,424 |
| 11 | 17,945,459~17,974,773 | |
| 12 | 17,992,241~18,042,367 | |
| Panicle number per plant | 1 | 22,182,883~22,220,448 |
| 12 | 4,146,868~4,199,211 |
Table 3.
Top highest genome-wide significant association signals of agronomic traits using EMMAX
| Trait | Chr. | Position (IRGSP 1.0) | Reference allele | Alternative allele | Alternative allele frequency | -log10(P) | R2 (%)a | Candidate/known geneb |
|---|---|---|---|---|---|---|---|---|
| Grain width | 1 | 11,789,024 | C | A | 0.77 | 6.29 | 9.41 | |
| Heading days | 2 | 3,970,385 | G | A | 0.05 | 6.20 | 11.54 | |
| 3 | 32,824,935 | T | A | 0.05 | 6.21 | 8.57 | CK1 [29] | |
| 3 | 32,824,941 | C | G | 0.05 | 6.08 | 10.75 | CK1 [29] | |
| 4 | 463,322 | G | A | 0.95 | 7.86 | 12.86 | ||
| 4 | 463,371 | G | A | 0.94 | 7.41 | 10.97 | ||
| 8 | 15,918,110 | G | A | 0.05 | 6.21 | 7.42 | ||
| 8 | 15,918,112 | C | T | 0.05 | 6.10 | 4.38 | ||
| 9 | 18,628,054 | T | C | 0.13 | 6.04 | 6.87 | ||
| Seed set rate | 4 | 31,539,937 | A | G | 0.18 | 7.35 | 9.45 | LOC_Os04g52940.1 |
| 7 | 7,918,286 | G | A | 0.07 | 6.16 | 5.21 | ||
| 7 | 8,178,284 | A | G | 0.12 | 6.47 | 3.15 | ||
| 7 | 8,299,577 | G | A | 0.07 | 6.33 | 14.26 | ||
| 7 | 8,390,152 | C | T | 0.07 | 6.08 | 5.27 | ||
| 7 | 8,390,155 | C | T | 0.07 | 6.09 | 8.25 | ||
| 7 | 8,447,659 | T | C | 0.09 | 6.45 | 9.64 | LOC_Os07g14800.1 |
Note: aR2 represents the genetic variants explained by the significant SNPs. bGene ID of MSU rice genome annotation project (http://rice.plantbiology.msu.edu/)
Fig. 6.
The box plots showing phenotypic distribution for Ting’s core collection carrying the different alleles at the top 16 significant SNPs in Table 3. The middle line indicates the median, the box indicates the range of the 25th to 75th percentiles of the total data, the whiskers indicate the inter-quartile range and the outer dots are outliers
In our study, we also identified some genes that were reported in previous studies according to the estimated distance of LD decay of 100 to 350 kb on the 12 chromosomes. We think a SNP is close to a cloned gene when it locates in 350 kb from the cloned gene. For heading days, significant association signals close to OsMADS51 on chromosome 1, OsPRR1 [26] on chromosome 2, DTH3 [27] on chromosome 3, CKI [28] on chromosome 3, HAF1 [29] on chromosome 4, Hd1 [30] on chromosome 6 and OsMADS13 [31] on chromosome 12 were detected (Fig. 4a and Table 4). For plant height, significant association signals close to SD1 [32], Ghd7 [33] and Ghd8 [34] were identified (Fig. 4b and Table 4). For seed set rate, signals close to SPP1 [35] and Rf-1 [36] were found (Fig. 4c and Table 4). For panicle length, significant association signals close to OsBRI1 [37], LP [38], SSD1 [39], FZP [40], LP1 [41] and SP1 [42] were found (Fig. 4d and Table 4). For grain length, significant association signals close to GS3 [43] and TGW6 [44] were detected (Fig. 4e and Table 4). For grain width, significant association signals close to GW2 [45], GS2 [46], GL3.2 [47], GS5 [48], GS6 [49], TGW6 [44], OsSPL16-GW8 [50] and SLG [51] were detected (Fig. 5a and Table 4). For 100 grains weight, significant association signals close to GW5 [52], TGW6 [43], GL7 [53] and OsSPL16 [50] were identified (Fig. 5b and Table 4).
Table 4.
Top highest genome-wide significant association signals of agronomic traits using EMMAX
| Trait | Significant loci | -log 10 (P) | genes | Genes position |
|---|---|---|---|---|
| Heading days | Chr1_40,209,752 | 4.56 | OsMADS51 | 40,344,329~40,364,584 |
| Chr2_24,800,400 | 4.42 | OSPRR1 | 24,569,294~24,572,560 | |
| Chr3_1,342,491 | 4.59 | DTH3 | 1,269,856~1,271,783 | |
| Chr3_32,824,935 | 6.21 | CKI | 32,999,502~33,006,898 | |
| Chr4_32,895,071 | 4.69 | HAF1 | 33,022,716~33,028,387 | |
| Chr6_9,156,215 | 4.78 | Hd1 | 9,336,359~9,338,643 | |
| Chr12_5,600,578 | 4.55 | OsMADS13 | 5,586,131~5,590,285 | |
| Plant height | Chr1_38,483,533 | 4.85 | SD1 | 38,382,382~38,385,504 |
| Chr7_9,235,801 | 4.38 | Ghd7 | 9,152,402~9,155,185 | |
| Chr8_4,056,392 | 4.70 | Ghd8 | 4,333,717~4,335,434 | |
| Seed set rate | Chr1_6,369,510 | 5.15 | SPP1 | 6,528,797~6,630,463 |
| Chr10_18,962,735 | 4.41 | Rf-1 | 18,935,690~18,942,573 | |
| Panicle length | Chr1_29,750,254 | 4.39 | OsBRI1 | 29,927,543~29,931,487 |
| Chr2_9,035,894 | 4.31 | LP | 9,042,076~9,046,141 | |
| Chr3_10,779,794 | 4.69 | SSD1 | 10,684,315~10,688,955 | |
| Chr7_28,613,922 | 5.20 | FZP | 28,299,591~28,301,089 | |
| Chr9_16,891,286 | 4.12 | LP1 | 17,182,867~17,188,378 | |
| Chr11_7,007,154 | 4.23 | SP1 | 7,193,230~7,198,552 | |
| Grain length | Chr3_16,876,884 | 4.12 | GS3 | 16,729,501~16,735,109 |
| Chr6_25,249,340 | 4.46 | TGW6 | 25,093,242~25,094,294 | |
| Grain width | Chr2_8,073,466 | 4.28 | GW2 | 8,114,961~8,121,925 |
| Chr2_28,875,239 | 4.33 | GS2 | 28,863,173~28,866,997 | |
| Chr3_17,360,192 | 4.45 | GL3.2 | 17,340,415~17,342,284 | |
| Chr5_3,576,630 | 4.45 | GS5 | 3,439,259~3,443,769 | |
| Chr6_1,281,784 | 5.07 | GS6 | 1,465,499~1,468,600 | |
| Chr6_25,355,332 | 4.62 | TGW6 | 25,093,242~25,094,294 | |
| Chr8_26,162,707 | 4.61 | OsSPL16 | 26,501,167~26,506,218 | |
| Chr8_28,114,414 | 4.44 | SLG | 28,162,970~28,165,431 | |
| 100 grains weight | Chr5_5,539,341 | 3.73 | GW5 | 5,365,122~5,366,701 |
| Chr6_25,216,303 | 4.01 | TGW6 | 25,093,242~25,094,294 | |
| Chr7_24,377,379 | 3,69 | GL7 | 24,664,168~24,669,324 | |
| Chr8_26,475,471 | 3.78 | OsSPL16 | 26,501,167~26,506,218 |
Discussion
The abundant genetic variation in Ting’s core collection makes it an important reservoir of genetic diversity and potential source of beneficial alleles for rice breeding (Fig. 1). It is very difficult to mine and utilize the exotic genes in all the rice accessions (i.e., 775,000) in the world [54] by either linkage mapping or association mapping. The maximum population size used for GWAS was 1495 rice accessions in a previous study [10]. One of the methods of utilizing a large set of germplasm in a GWAS is to construct a core collection [16]. A rice core collection consisting of 150 accessions selected based on 48 morphological traits from 2262 accessions of Ting’s collection has been constructed and used in rice association mapping with low resolution [19, 20]. Therefore, we performed a GWAS by whole-genome re-sequencing for getting higher resolution within Ting’s core collection.
Although the population size of Ting’s core collection is smaller than that of three other populations [5, 8, 9], the phenotypic diversity of several agronomic traits was comparable to that of these populations or even higher for some agronomic traits. Moreover, more than 3.8 million SNPs in Ting’s core collection were developed. The ratio of SNPs to population size in Ting’s core collection is higher than that in previous studies in which the ratio were approximately 3.6 million SNPs to 517 rice landraces [5], 0.04 million SNPs to 413 diverse landraces and cultivars [9], 4.1 million SNPs to 950 worldwide varieties [6], 1.6 million SNPs to 1495 elite hybrid varieties [10] and 0.04 million SNPs to 176 japonica varieties [8]. Furthermore, a simpler population structure (Figs. 2 and 3), more rapid LD decay (Additional file 1: Figure S2) and more distant relatedness (Additional file 1: Figure S3) among accessions were found in Ting’s core collection than in other collections. The above mentioned information illuminates and supports the fact that Ting’s core collection is suitable for GWASs.
Population structure in the present study was not identical to that in our previous study [55, 56]. This discrepancy might be due to molecular markers density used in two studies. In our previous study, 274 SSR markers were included to detect the population structure while about 3.8 million SNPs were used in present study.
A total of 3,808,730 SNPs from 150 varieties were used for the GWAS (Additional file 2: Table S3). A mixed model was performed using EMMAX software [55, 56]. EMMAX not only can correct for a wide range of sample structures by explicitly accounting for pairwise relatedness between individuals, using high-density markers to model the phenotype distribution. But also can reduce computational time [55, 56]. The value obtained from a rough Bonferroni correction of P = 1/n, where n is the total number of markers used in the GWAS, is widely applied as the threshold P value for significance [5–8, 10]. The threshold P value for significance in our study was P ≤ 2.63 × 10− 7, corresponding to -log10(P) = 6.58. However, only one peak, i.e., one on chromosome 4 for heading days was higher than this threshold value in Fig. 4a. Hence, we chose a lower -log10(mBF) value as the significance threshold for different traits in our study (Table 1) because there will be no significant locus according to the theoretical threshold P value. We speculated that this result might due to population size in our study. However, Ting’s core collection is suitable for GWASs because the peaks located in well-known genes such as SD1, GS2, GS3, GS5, GL7, GW8 and TGW6 were also much lower than the theoretical threshold value (Figs. 4 and 5).
In our study, some significant association signals were identified through a GWAS of Ting’s core collection. First, loci significantly associated with agronomic traits were uncovered close to cloned genes such as Hd1, SD1, Ghd7, GW8, and GL7 (Figs. 4, 5 and Table 4) that were reported in previous studies. Moreover, some of these loci were located by coincidence in these genes, and they might be natural variations of these genes, which could be functional (Table 2 and Additional file 3: Table S4). Second, Si et al. [25] indicated that some significant loci within the distance of LD decay might be identical to each other. However, there were no identical significant loci in the two locations overall (Table 1), but some identical significant regions were discovered in the two locations when the estimated distance of LD decay of 100 to 350 kb was considered in Ting’s core collection (Table 1, Figs. 4 and 5). Third, some new significant association signals that might be candidate genes were detected in our study (Figs. 4, 5 and Additional file 4: Table S5). Some peaks of these candidate genes such as the peak on chromosome 4 for heading days (Fig. 4a) were even higher than the threshold value. Further, the peak on chromosome 11 for heading days (Fig. 4a) was higher than that of some famous genes such as Hd1. It would be valuable to test the functions of these candidate genes because some loci or regions were also detected by previous studies. For instance, the region on chromosome 8 for plant height, the region at position 23,300,000 on chromosome 1 for heading days and the region at position 21,650,000 on chromosome 2 were found to be significantly associated with related traits in the study of Zhao et al. [9].
Conclusions
In this study, Ting’s core collection showed abundant genetic variation for agronomic traits and was proved to be a suitable natural population that could be comparable to other populations used in previous GWASs. Moreover, according to this study, core collections constructed from large natural populations of other plants might be good choices for GWASs. Furthermore, some natural variations in cloned genes were founded in this study, and these variations could be used for functional analysis of these genes. In addition, new candidate genes identified in this study could be very useful for rice improvement. In sum, this study provided important information for further mining these elite genes within Ting’s core collection and using them for rice breeding.
Methods
Plant material
Ting’s core collection with 150 accessions of rice landraces [18], was used in this study. The information for these accessions is shown in Additional file 2: Table S1.
Phenotyping
In total, 12 agronomic traits of Ting’s core collection were measured in two locations. The methods of measuring these 12 agronomic traits were identical to those described in detail in our previous study [20].
A randomized complete block design with three replications was used in two locations. First, Ting’s core collection was cultivated at the farm of South China Agricultural University, Guangzhou (23°16′’ N, 113°8′ E), during the late season (July–November) in 2009. The design and methods of this research in Guangzhou were described in detail in our previous study [20]. Second, Ting’s core collection was cultivated at the farm of China National Rice Research Institute, Hangzhou (30°3′ N, 120°2′ E), during the late season (May–October) in 2016. A randomized complete block design with three replications, as in Guangzhou, was used during this season in Hangzhou. The space between rows and between plants was set to 26 and 20 cm, respectively. Twenty-four plants of each variety were grown in four rows with 6 plants per row. For each block, the five plants in the middle position of the second and third row of each variety were selected to prevent edge effects. The broad-sense heritability (H2) was calculated as , where is the genetic variance, is the environmental variance.
DNA isolation and genome sequencing
Total genomic DNA was extracted using a modified SDS method. Then, each landrace’s DNA was sheared randomly into ~ 500-bp fragments by Covaris, and the DNA fragments were loaded on 2% agarose gels. Fragments of ~ 500 bp were recovered and purified, and adapters were then added to each fragment. After making libraries for the clusters, they were loaded into an Illumina HiSeq™ 4000 for 2× 150-bp paired-end sequencing at 6~7-fold genome coverage.
The 150-bp paired-end reads were mapped onto the rice reference genome (IRGSP 1.0) using bwamem with the –M option in BWA software [57]. The mapped reads were realigned by using RealignerTargetCreator and IndelRealigner in GATK [58]. UnifiedGenotyper in GATK was used with the −glm BOTH option to label SNPs and indels. After removing nucleotide variants with a missing rate ≥ 0.25 and a minor allele frequency > 0.05, a total of 3,808,730 SNPs and 391,756 indels were generated.
Population genetic analyses
Principal component analysis (PCA), construction of a neighbor-joining (NJ) tree, determination of LD decay level and kinship analysis among landraces were performed based on SNPs. The population structure of the 150 varieties was estimated with PCA by using the software EIGENSTRAT [59]. PHYLIP version 3.695 software (http://evolution.genetics.washington.edu/phylip/getme-new1.html) was used to construct the NJ tree on the basis of similarity measures. The software MEGA V5.2 was used to observe the NJ tree [60]. The LD in Ting’s core collection was evaluated using squared Pearson’s correlation coefficients (r2) calculated with the −r2 command in the software PLINK [61]. A Q matrix was obtained from the membership probability of each variety using ADMIXTURE Version 1.22 software [62]. The Q matrix was used for further association mapping. The Loiselle algorithm was chosen to construct a kinship matrix (K) with the software SPAGeDi [63]. Moreover, all negative kinship values were set to zero.
GWAS
A total of 3,808,730 SNPs from 150 varieties were used for GWAS. A mixed model was performed using EMMAX software [56]. P ≤ 2.63 × 10− 7 (P = 1/n, n = total number of markers used [7], which is a rough Bonferroni correction, corresponding to -log10(P) = 6.58). However, no significant loci were detected based on this threshold, hence, we calculated another significance threshold, i.e., a minimum Bayes factor (mBF), based on the P value threshold for significance. The mBF was calculated using the following formula: mBF = −e*P*ln(P) [64]. Thus, the significance threshold in this study was -log10(P) = 4.97.
Additional files
Figure S1. SNP distribution along position in each chromosome. Figure S2. Genome-wide average LD decay estimated in Ting’s core collection on 12 chromosomes. Figure S3. Distribution of pairwise relative 1 kinship values based on 3.8 million SNPs in Ting’s core collection. The height of blue bar represents the percentage of varieties in different range of kinships. Figure S4. Plots of observed versus expected P-values using EMMAX for 12 agronomic traits. Red symbol represents expected P-values, and Blue symbol represents observed P-values. (DOCX 1084 kb)
Table S1. Accessions, variety names, origin and germplasm types of 150 rice varieties in Ting’s core collection. Table S2. Re-sequencing average read depth and coverage in Ting’s core collection. Table S3. Summary of categorized SNPs and InDels. (DOC 246 kb)
Table S4. List of all P-value ranked genes in the gene-based association analysis of heading days/plant height/seed set rate/panicle length/grain length/100 grains weight/flag leaf length/flag leaf width/panicle number per plant. (XLSX 169 kb)
Table S5. List of new loci in association analysis of heading days/plant height/seed set rate/panicle length/grain length/grain width/100 grains weight/ flag leaf length/panicle number per plant. (XLSX 51 kb)
Acknowledgments
We are grateful to Dr. Jinquan Li from Max Planck Institute for Plant Breeding Research for his advice and assistance and Dr. Xiangdong Liu from South China Agricultural University for supplying Ting’s core collection. We would like to thank the anonymous reviewers for valuable suggestions and American Journal Experts (https://www.aje.com) for English language editing.
Abbreviations
- CDS
Coding sequence
- DNA
Deoxyribonucleic acid
- EMMAX
Efficient mixed model association eXpedited
- IRGSP
International Rice Genome Sequencing Project
- QTL
Quantitative trait locus
- SDS
Sodium dodecyl sulfate
- SNPs
Single Nucleotide Polymorphisms
- SSR
Simple Sequence Repeats
Authors’ contributions
Conceived and designed the experiments: PZ and HT. Performed the experiments: PZ and KZ. Analyzed the data: PZ, KZ, ZZ and HT. Contributed reagents/materials/analysis tools: PZ and HT. Wrote the paper: PZ, KZ and HT. All authors read and approved the final manuscript
Funding
This work was supported by three funds of the National Natural Science Foundation of China (31701401, 31872862 and 31601287), a fund from Zhejiang Province Public Welfare Technology Application Research Project (LGN19C130005), a fund from the State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources (SKLCUSA-b201713), as well as a fund from Shanghai Agrobiological Gene Center (201503). The funding bodies did not play any role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets used during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peng Zhang and Kaizhen Zhong contributed equally to this study.
Contributor Information
Peng Zhang, Email: zhangpeng03@caas.cn.
Kaizhen Zhong, Email: zyzhang_311@163.com.
Zhengzheng Zhong, Email: zhongzhengzheng@caas.cn.
Hanhua Tong, Email: htonghz@126.com.
References
- 1.Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, Kresovich S, Goodman MM, Buckler ET. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc Natl Acad Sci U S A. 2001;98(20):11479–11484. doi: 10.1073/pnas.201394398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang X, Han B. Natural variations and genome-wide association studies in crop plants. Annu Rev Plant Biol. 2014;65:531–551. doi: 10.1146/annurev-arplant-050213-035715. [DOI] [PubMed] [Google Scholar]
- 3.Kraakman A, Niks RE, Van den Berg P, Stam P, Van Eeuwijk FA. Linkage disequilibrium mapping of yield and yield stability in modern spring barley cultivars. Genetics. 2004;168(1):435–446. doi: 10.1534/genetics.104.026831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu C, Gore M, Buckler ES, Yu J. Status and prospects of association mapping in plants. Plant Genome. 2008;1(1):5–20. doi: 10.3835/plantgenome2008.02.0089. [DOI] [Google Scholar]
- 5.Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, Li C, Zhu C, Lu T, Zhang Z, et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet. 2010;42(11):961–976. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- 6.Huang X, Zhao Y, Wei X, Li C, Wang A, Zhao Q, Li W, Guo Y, Deng L, Zhu C, et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat Genet. 2012;44(1):32–53. doi: 10.1038/ng.1018. [DOI] [PubMed] [Google Scholar]
- 7.Yang W, Guo Z, Huang C, Duan L, Chen G, Jiang N, Fang W, Feng H, Xie W, Lian X, et al. Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat Commun. 2014;5:5087. doi: 10.1038/ncomms6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yano K, Yamamoto E, Aya K, Takeuchi H, Lo P, Hu L, Yamasaki M, Yoshida S, Kitano H, Hirano K, et al. Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat Genet. 2016;48(8):927. doi: 10.1038/ng.3596. [DOI] [PubMed] [Google Scholar]
- 9.Zhao K, Tung C, Eizenga GC, Wright MH, Ali ML, Price AH, Norton GJ, Islam MR, Reynolds A, Mezey J, et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat Commun. 2011;(9):467. [DOI] [PMC free article] [PubMed]
- 10.Huang X, Yang S, Gong J, Zhao Y, Feng Q, Gong H, Li W, Zhan Q, Cheng B, Xia J, et al. Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nat Commun. 2015;6:6258. doi: 10.1038/ncomms7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Famoso AN, Zhao K, Clark RT, Tung C, Wright MH, Bustamante C, Kochian LV, McCouch SR. Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. PLoS Genet. 2011;7:e10022218. doi: 10.1371/journal.pgen.1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ueda Y, Frimpong F, Qi Y, Matthus E, Wu L, Hoeller S, Kraska T, Frei M. Genetic dissection of ozone tolerance in rice (Oryza sativa L.) by a genome-wide association study. J Exp Bot. 2015;66(1):293–306. doi: 10.1093/jxb/eru419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Norton GJ, Douglas A, Lahner B, Yakubova E, Guerinot ML, Pinson SRM, Tarpley L, Eizenga GC, McGrath SP, Zhao F, et al. Genome wide association mapping of grain arsenic, copper, molybdenum and zinc in rice (Oryza sativa L.) grown at four international field sites. PLoS One. 2014;9:e896852. doi: 10.1371/journal.pone.0089685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen W, Gao Y, Xie W, Gong L, Lu K, Wang W, Li Y, Liu X, Zhang H, Dong H, et al. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat Genet. 2014;46(7):714–721. doi: 10.1038/ng.3007. [DOI] [PubMed] [Google Scholar]
- 15.Matsuda F, Nakabayashi R, Yang Z, Okazaki Y, Yonemaru J, Ebana K, Yano M, Saito K. Metabolome-genome-wide association study dissects genetic architecture for generating natural variation in rice secondary metabolism. Plant J. 2015;81(1):13–23. doi: 10.1111/tpj.12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang P, Zhong K, Shahid MQ, Tong H. Association analysis in rice: from application to utilization. Front Plant Sci. 2016;7:1202. doi: 10.3389/fpls.2016.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Londo JP, Chiang YC, Hung KH, Chiang TY, Schaal BA. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc Natl Acad Sci U S A. 2006;103(25):9578–9583. doi: 10.1073/pnas.0603152103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Lu Y, Li J, Xu H, Shahid MQ. Strategies on sample size determination and qualitative and quantitative traits integration to construct core collection of rice (Oryza sativa) Rice Sci. 2011;18(1):46–55. doi: 10.1016/S1672-6308(11)60007-3. [DOI] [Google Scholar]
- 19.Zhang P, Li J, Li X, Liu X, Zhao X, Lu Y. Population structure and genetic diversity in a rice core collection (Oryza sativa L.) investigated with SSR markers. PLoS One. 2011;6:e2756512. doi: 10.1371/annotation/4056d03c-20ed-4eca-9568-3e9400e2312e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang P, Liu X, Tong H, Lu Y, Li J. Association mapping for important agronomic traits in core collection of rice (Oryza sativa L.) with SSR markers. PLoS One. 2014;9:e11150810. doi: 10.1371/journal.pone.0111508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang P, Zhong K, Tong H, Shahid MQ, Li J. Association mapping for aluminum tolerance in a core collection of rice landraces. Front Plant Sci. 2016;7:1415. doi: 10.3389/fpls.2016.01415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mather KA, Caicedo AL, Polato NR, Olsen KM, McCouch S, Purugganan MD. The extent of inkage disequilibrium in rice (Oryza sativa L.) Genetics. 2007;177(4):2223–2232. doi: 10.1534/genetics.107.079616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNally KL, Childs KL, Bohnert R, Davidson RM, Zhao K, Ulat VJ, Zeller G, Clark RM, Hoen DR, Bureau TE, et al. Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proc Natl Acad Sci U S A. 2009;106(30):12273–12278. doi: 10.1073/pnas.0900992106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X, Yan J, Shah T, Warburton ML, Li Q, Li L, Gao Y, Chai Y, Fu Z, Zhou Y, et al. Genetic analysis and characterization of a new maize association mapping panel for quantitative trait loci dissection. Theor Appl Genet. 2010;121(3):417–431. doi: 10.1007/s00122-010-1320-y. [DOI] [PubMed] [Google Scholar]
- 25.Si L, Chen J, Huang X, Gong H, Luo J, Hou Q, Zhou T, Lu T, Zhu J, Shangguan Y, et al. OsSPL13 controls grain size in cultivated rice. Nat Genet. 2016;48(4):447–456. doi: 10.1038/ng.3518. [DOI] [PubMed] [Google Scholar]
- 26.Kim SL, Lee S, Kim HJ, Nam HG, An G. OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol. 2008;147(1):438. doi: 10.1104/pp.104.900258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murakami M, Ashikari M, Miura K, Yamashino T, Mizuno T. The evolutionarily conserved OsPRR quintet: Rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol. 2003;44(11):1229–1236. doi: 10.1093/pcp/pcg135. [DOI] [PubMed] [Google Scholar]
- 28.Bian X, Liu X, Zhao Z, Jiang L, Gao H, Zhang Y, Zheng M, Chen L, Liu S, Zhai H, et al. Heading date gene, dth3 controlled late flowering in O. Glaberrima Steud. By down-regulating Ehd1. Plant Cell Rep. 2011;30(12):2243–2254. doi: 10.1007/s00299-011-1129-4. [DOI] [PubMed] [Google Scholar]
- 29.Dai C, Xue H. Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J. 2010;29(11):1916–1927. doi: 10.1038/emboj.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y, Fu D, Zhu C, He Y, Zhang H, Liu T, Li X, Wu C. The RING-finger ubiquitin ligase HAF1 mediates heading date 1 degradation during photoperiodic flowering in rice. Plant Cell. 2015;27(9):2455–2468. doi: 10.1105/tpc.15.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the arabidopsis flowering time gene CONSTANS. Plant Cell. 2000;12(12):2473–2483. doi: 10.1105/tpc.12.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Y, Liang W, Yin C, Yang X, Ping B, Li A, Jia R, Chen M, Luo Z, Cai Q, et al. Interactions of OsMADS1 with floral homeotic genes in rice flower development. Mol Plant. 2015;8(9):1366–1384. doi: 10.1016/j.molp.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 33.Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, et al. Green revolution: a mutant gibberellin-synthesis gene in rice. Nature. 2002;416(6882):701–702. doi: 10.1038/416701a. [DOI] [PubMed] [Google Scholar]
- 34.Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40(6):761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- 35.Yan W, Wang P, Chen H, Zhou H, Li Q, Wang C, Ding Z, Zhang Y, Yu S, Xing Y, et al. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol Plant. 2011;4(2):319–330. doi: 10.1093/mp/ssq070. [DOI] [PubMed] [Google Scholar]
- 36.Liu T, Mao D, Zhang S, Xu C, Xing Y. Fine mapping SPP1, a QTL controlling the number of spikelets per panicle, to a BAC clone in rice (Oryza sativa) Theor Appl Genet. 2009;118(8):1509–1517. doi: 10.1007/s00122-009-0999-0. [DOI] [PubMed] [Google Scholar]
- 37.Akagi H, Nakamura A, Yokozeki-Misono Y, Inagaki A, Takahashi H, Mori K, Fujimura T. Positional cloning of the rice Rf-1 gene, a restorer of BT-type cytoplasmic male sterility that encodes a mitochondria-targeting PPR protein. Theor Appl Genet. 2004;108(8):1449–1457. doi: 10.1007/s00122-004-1591-2. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura A, Fujioka S, Sunohara H, Kamiya N, Hong Z, Inukai Y, Miura K, Takatsuto S, Yoshida S, Ueguchi-Tanaka M, et al. The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiol. 2006;140(2):580–590. doi: 10.1104/pp.105.072330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li M, Tang D, Wang K, Wu X, Lu L, Yu H, Gu M, Yan C, Cheng Z. Mutations 1 in the F-box gene LARGER PANICLE improve the panicle architecture and enhance the grain yield in rice. Plant Biotechnol J. 2011;9(9):1002–1013. doi: 10.1111/j.1467-7652.2011.00610.x. [DOI] [PubMed] [Google Scholar]
- 40.Asano K, Miyao A, Hirochika H, Kitano H, Matsuoka M, Ashikari M. SSD1, which encodes plant-specific novel protein, controls plant elongation by regulating cell division in rice. Proc Jpn Acad Ser B Phys Biol Sci. 2010;86(3):265–273. doi: 10.2183/pjab.86.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komatsu M, Chujo A, Nagato Y, Shimamoto K, Kyozuka J. FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development. 2003;130(16):3841–3850. doi: 10.1242/dev.00564. [DOI] [PubMed] [Google Scholar]
- 42.Liu E, Liu Y, Wu G, Zeng S, Thi TGT, Liang L, Liang Y, Dong Z, She D, Wang H, et al. Identification of a candidate gene for panicle length in rice (Oryza sativa L.) via association and linkage analysis. Front Plant Sci. 2016;7:596. doi: 10.3389/fpls.2016.00596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S, Qian Q, Fu Z, Zeng D, Meng X, Kyozuka J, Maekawa M, Zhu X, Zhang J, Li J, et al. Short panicle1 encodes a putative PTR family transporter and determines rice panicle size. Plant J. 2009;58(4):592–605. doi: 10.1111/j.1365-313X.2009.03799.x. [DOI] [PubMed] [Google Scholar]
- 44.Fan C, Xing Y, Mao H, Lu T, Han B, Xu C, Li X, Zhang Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet. 2006;112(6):1164–1171. doi: 10.1007/s00122-006-0218-1. [DOI] [PubMed] [Google Scholar]
- 45.Ishimaru K, Hirotsu N, Madoka Y, Murakami N, Hara N, Onodera H, Kashiwagi T, Ujiie K, Shimizu B, Onishi A, et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat Genet. 2013;45(6):707. doi: 10.1038/ng.2612. [DOI] [PubMed] [Google Scholar]
- 46.Song X, Huang W, Shi M, Zhu M, Lin H. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet. 2007;39(5):623–630. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- 47.Hu J, Wang Y, Fang Y, Zeng L, Xu J, Yu H, Shi Z, Pan J, Zhang D, Kang S, et al. A rare allele of GS2 enhances grain size and grain yield in rice. Mol Plant. 2015;8(10):1455–1465. doi: 10.1016/j.molp.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 48.Xu F, Fang J, Ou S, Gao S, Zhang F, Du L, Xiao Y, Wang H, Sun X, Chu J, et al. Variations in CYP78A13 coding region influence grain size and yield in rice. Plant Cell Environ. 2015;38(4):800–811. doi: 10.1111/pce.12452. [DOI] [PubMed] [Google Scholar]
- 49.Li Y, Fan C, Xing Y, Jiang Y, Luo L, Sun L, Shao D, Xu C, Li X, Xiao J, et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet. 2011;43(12):1266–1269. doi: 10.1038/ng.977. [DOI] [PubMed] [Google Scholar]
- 50.Sun L, Li X, Fu Y, Zhu Z, Tan L, Liu F, Sun X, Sun X, Sun C. GS6, a member of the GRAS gene family, negatively regulates grain size in rice. J Integr Plant Biol. 2013;55(10):938–949. doi: 10.1111/jipb.12062. [DOI] [PubMed] [Google Scholar]
- 51.Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X, Zeng R, Zhu H, Dong G, Qian Q, et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet. 2012;44(8):950. doi: 10.1038/ng.2327. [DOI] [PubMed] [Google Scholar]
- 52.Feng Z, Wu C, Wang C, Roh J, Zhang L, Chen J, Zhang S, Zhang H, Yang C, Hu J, et al. SLG controls grain size and leaf angle by modulating brassinosteroid homeostasis in rice. J Exp Bot. 2016;67(14):4241–4253. doi: 10.1093/jxb/erw204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu J, Chen J, Zheng X, Wu F, Lin Q, Heng Y, Tian P, Cheng Z, Yu X, Zhou K, et al. GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat Plants. 2017;3:17043. doi: 10.1038/nplants.2017.43. [DOI] [PubMed] [Google Scholar]
- 54.Wang Y, Xiong G, Hu J, Jiang L, Yu H, Xu J, Fang Y, Zeng L, Xu E, Xu J, et al. Copy numbe variation at the GL7 locus contributes to grain size diversity in rice. Nat Genet. 2015;47(8):944. doi: 10.1038/ng.3346. [DOI] [PubMed] [Google Scholar]
- 55.FAO . The second report on the state of the world's plant genetic 1 resources for food and agriculture. Commission on genetic resources for food and agriculture. 2010. [Google Scholar]
- 56.Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42(4):348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H, Durbin R. Fast and accurate long-read alignment with burrows-wheeler transform. Bioinformatics. 2010;26:589. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, Del Angel G, Rivas MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 60.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 61.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hardy Olivier J., Vekemans Xavier. spagedi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes. 2002;2(4):618–620. doi: 10.1046/j.1471-8286.2002.00305.x. [DOI] [Google Scholar]
- 64.Goodman SN. Of p-values and Bayes: a modest proposal. Epidemiology. 2001;12:295–297. doi: 10.1097/00001648-200105000-00006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. SNP distribution along position in each chromosome. Figure S2. Genome-wide average LD decay estimated in Ting’s core collection on 12 chromosomes. Figure S3. Distribution of pairwise relative 1 kinship values based on 3.8 million SNPs in Ting’s core collection. The height of blue bar represents the percentage of varieties in different range of kinships. Figure S4. Plots of observed versus expected P-values using EMMAX for 12 agronomic traits. Red symbol represents expected P-values, and Blue symbol represents observed P-values. (DOCX 1084 kb)
Table S1. Accessions, variety names, origin and germplasm types of 150 rice varieties in Ting’s core collection. Table S2. Re-sequencing average read depth and coverage in Ting’s core collection. Table S3. Summary of categorized SNPs and InDels. (DOC 246 kb)
Table S4. List of all P-value ranked genes in the gene-based association analysis of heading days/plant height/seed set rate/panicle length/grain length/100 grains weight/flag leaf length/flag leaf width/panicle number per plant. (XLSX 169 kb)
Table S5. List of new loci in association analysis of heading days/plant height/seed set rate/panicle length/grain length/grain width/100 grains weight/ flag leaf length/panicle number per plant. (XLSX 51 kb)
Data Availability Statement
The datasets used during the current study are available from the corresponding author on reasonable request.