Abstract
The diagnosis and prognostication of glioblastoma (GBM) remain to be solely dependent on histopathological findings and few molecular markers, despite the clinical heterogeneity in this entity. To address this issue, we investigated the prognostic impact of copy number alterations (CNAs) using two population-based IDH-wild-type GBM cohorts: an original Japanese cohort and a dataset from The Cancer Genome Atlas (TCGA). The molecular disproportions between these cohorts were dissected in light of cohort differences in GBM. The Japanese cohort was collected from cases registered in Kansai Molecular Diagnosis Network for CNS tumors (KNBTG). The somatic landscape around CNAs was analyzed for 212 KNBTG cases and 359 TCGA cases. Next, the clinical impacts of CNA profiles were investigated for 140 KNBTG cases and 152 TCGA cases treated by standard adjuvant therapy using temozolomide-based chemoradiation. The comparative profiling indicated unequal distribution of specific CNAs such as EGFR, CDKN2A, and PTEN among the two cohorts. Especially, the triple overlap CNAs in these loci (triple CNA) were much higher in frequency in TCGA (70.5%) than KNBTG (24.3%), and its prognostic impact was independently validated in both cohorts. The KNBTG cohort significantly showed better prognosis than the TCGA cohort (median overall survival 19.3 vs 15.6 months). This survival difference between the two cohorts completely resolved after subclassifying all cases according to the triple CNA status. The prognostic significance of triple CNA was identified in IDH-wild-type GBM. Distribution difference in prognostic CNA profiles potentially could cause survival differences across cohorts in clinical studies.
Electronic supplementary material
The online version of this article (10.1186/s40478-019-0749-8) contains supplementary material, which is available to authorized users.
Keywords: Glioblastoma, Copy number alteration, EGFR, CDKN2A, PTEN
Introduction
The recent comprehensive molecular analysis of glioblastoma (GBM), including The Cancer Genome Atlas (TCGA) projects, has revealed the tumor genetic landscape and various functional relations between genes and pathways in tumorigenesis [4]. Several molecular alterations by somatic mutation, genomic rearrangement, and copy number alteration (CNA) have been shown to be closely involved in GBM. One of the major interests with regards to these genetic changes is towards the availability of prognostic and/or diagnostic stratification or potential targetability of such genetic changes in the development of novel therapeutic agents.
The most important molecular change in glioma is IDH1/2 mutations [24, 29], which define biological characteristics in glioma through the change of global DNA methylation and histone modification. These mutations have been well validated to be favorable prognostic factor in GBM patients in several studies [22]. In the 2016 WHO Classification of Tumors of the Central Nervous System (CNS WHO), IDH-wild-type GBM came to be regarded as a more delineated entity: the most common and malignant astrocytic glioma [19]. O6-methylguanine-DNA-methyltransferase (MGMT) methylation status is also regarded as a prognostic/predictive marker [13, 26], while it has little value as a diagnostic marker and does not define any subtypes of GBMs with distinct molecular features [17].
The CNA in GBM is another significant somatic alteration which often shows a distinctive landscape with synchronous genomic gains and/or losses. Long before discovering IDH1/2 mutation, a chromosomal aberration in GBM was initially detected by gain of EGFR [18], which was subsequently followed by various CNAs. CNAs such as gain in chromosome 7 and loss of chromosomes 9 and 10 were regarded as hallmarks especially for IDH-wild-type GBM [10], and have become increasingly recognized to play a key role in GBM oncogenic pathways, according to CNA profiles by transcriptome subtypes [27]. Some CNAs, such as loss of chromosome 10 [23] and NFKBIA deletion [2], were also reported to be solely associated with survival in GBM patients. Nevertheless, since their correlation with survival has not been fully validated in the subsequent studies, no consensus has been reached as to which CNA has a universal prognostic value beyond the WHO grading of GBM.
It remains a problem that the diagnosis and prognostication of GBM largely depend on histopathological findings and few molecular features in IDH1/2 and MGMT [28], despite the heterogenous clinical courses of IDH-wild-type GBM and even in the subsets with the same status of MGMT methylation. For achieving accurate prognostic stratification in GBM, we investigated molecular distribution of CNAs and its relationship to clinical outcome using two independent population-based cohorts of GBM including an original Japanese cohort and a large dataset from TCGA. When targeting two different population from east Asia and TCGA, another concern is the geographical diversity in clinical and molecular profiles of GBMs. For instance, the lower EGFR amplification rates of GBM patients from Asia were recently reported during a screening for two randomized GBM trials with depatux-m: INTELLANCE1 and INTELLANCE2 (10.1093/annonc/mdx366.002). Therefore, a comparative study between two cohorts should deserve special consideration to help in identifying the cohort disproportion in GBM practice.
Here, we examined the survival impact of CNAs in Japanese and TCGA GBM cohorts. The differences of molecular profiles and clinical outcome were carefully investigated between these two cohorts. This investigation conclusively showed that a combination of EGFR, CDKN2A, and PTEN CNA status had a prognostic impact in GBM patients, and that the differences in the frequencies of these molecular profiles resulted in the different survival between the two cohorts.
Materials and methods
Study design
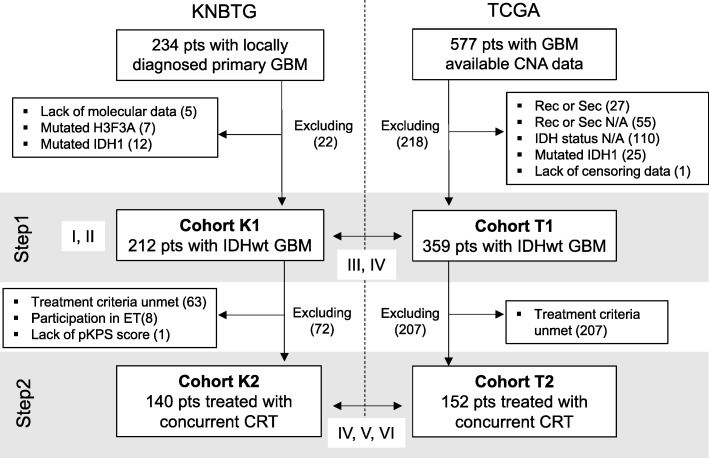
The analyses of this retrospective study included two steps. The detail of this study design and flowchart of patient selection are provided below and are summarized in Fig. 1. Two cohorts were collected as described below: the Japanese and TCGA cohorts. Briefly, Step1 aimed to investigate the somatic landscape including CNAs in the primary IDH wild-type GBM and compared the somatic landscapes of the two cohorts with each other. In Step2, we investigated the clinical impact of CNA profiles in primary IDH wild-type GBM cases treated with chemoradiotherapy with TMZ after initial surgery using the Japanese and TCGA cohorts [26]. The Japanese cohorts collected for Step1 and 2 analyses were Cohort K1 and K2, respectively. Similarly, TCGA cohorts subjected to each step were Cohort T1 and T2, respectively.
Fig. 1.
Study design and patient selection. This study consisted of two steps (Step1 and 2). In Step1, 212 primary GBM cases in KNBTG enrolled as Cohort K1. From TCGA dataset, 359 cases conclusively diagnosed with primary IDH-wild-type GBM were selected as Cohort T1. In Step2, 140 patients from Cohort K1 and 152 patients from Cohort T1 were further extracted as Cohort K2 and T2, who were concurrently treated with TMZ and RT. Targeting for each cohort or step, the analyses were conducted in the Roman numerical order as follows: I, somatic genetic landscape of GBM; II, interaction among each genetic alteration; III, comparison of frequency of genetic alterations among cohorts; IV, survival difference among cohorts; V, exploration of prognostic biomarkers; VI, survival analysis between cohorts by adjustment with common prognostic biomarkers. Abbreviations: CRT, chemoradiation therapy; ET, experimental treatment; N/A, not available; pKPS, preoperative KPS; pts., patients; Rec or Sec, recurrent or secondary GBM; wt, wild type
A Japanese GBM cohort of Kansai molecular diagnosis network for CNS tumors (KNBTG)
Japanese cohort was collected from the cases registered in Kansai Molecular Diagnosis Network for CNS tumors (KNBTG): a consortium where neuro-oncologists, neurosurgeons, pediatricians, pathologists, and basic scientists are conducting cooperative researches for malignant brain tumor [25]. The researchers are affiliated to university hospitals, regional medical centers, and research institutes mainly in the Kansai area of Western Japan. This network routinely collects glioma cases registered from the participating institutions. Tumor samples and clinical information of patients treated at the affiliated institutions are collected and registered in this data bank after obtaining written informed consent. The detailed clinical information including preoperative Karnofsky Performance status (KPS), extent of resection, and adjuvant therapy are routinely obtained from each institution. Since this is a population-based study in Japan, subjects are exclusively composed of East Asians and under the universal health care insurance system of Japan.
The initial screening criteria from enrolled gliomas in KNBTG was as follows: local diagnosis of primary GBM and availability of genomic DNA for molecular analysis of the naïve specimens. Primary GBM was clinically regarded as the GBM arising de novo, with no known lower-grade precursor lesion. The inclusion criteria for Cohort K1 was as follows: central review of a histopathological diagnosis of IDH-wild-type GBM based on the 2016 CNS WHO, molecular data available for analysis, and absence of IDH1/2 and H3F3A mutation, and 1p/19q codeletion. Out of 234 primary GBM initially collected from seven institutions participating in this study, 212 cases were enrolled as Cohort K1.
To analyze the prognostic impact of CNAs, cases in which patients underwent radiotherapy (RT) of 50–65 Gy and concurrent TMZ were further selected from Cohort K1. Those treated with either RT alone, TMZ alone, or short-course RT plus TMZ were excluded from any survival analysis. Cases ineligible for treatment criteria and lacking in KPS score were excluded. In addition, those participating in clinical studies for testing experimental treatment add-on to RT plus TMZ (n = 8) were excluded. Finally, 140 cases were available for Step2 analysis as Cohort K2.
Central pathology review of KNBTG cohort
All molecular-pathological diagnosis of IDH-wild-type GBM eligible for this investigation had reached an agreement with an experienced neuropathologist (Y.K.) and was according to the 2016 WHO classification for central nervous system tumors.
Molecular analysis of KNBTG cohort
All tissue specimens were obtained at the time of the initial treatment before chemoradiation between December 2006 and November 2017. Tumor genomic DNA was extracted with NucleoSpin Tissue kit (Macherey-Nagel, Inc., Bethlehem, PA) or DNeasy Blood & Tissue Kit (Qiagen, Tokyo, Japan), according to the manufacturer’s protocol. The same batch of DNA samples was used for each technique as follows.
Mutational status of IDH1/2, TERT promoter, H3F3A, HIST1H3B, and TP53 were determined using the Sanger technique. The details of Sanger sequencing have been previously reported [1] and is additionally provided in the supplementary information (SI). The methylation status of the MGMT promoter was assessed using quantitative methylation specific PCR (qMSP) following the bisulfite modification of tumor genomic DNA. The details of the qMSP protocol for testing MGMT methylation are described in SI.
To assess CNAs in GBM, we performed Multiplex Ligation-dependent Probe Amplification (MLPA) using the SALSA MLPA KIT P105 (version D2) and P088 (version C2), in accordance with the manufacturer’s protocol (MRC Holland, Amsterdam, Netherland) [15]. The P105 kit is designed to detect CNAs typical in gliomas, and includes probes against PDGFRA, EGFR, CDKN2A, PTEN, TP53, CDK4, MDM2, and NFKBIA genes. P088 kit was designed to assess mainly 1p/19q codeletion, and we used this kit only for the IDH-mutated-GBM to exclude oligodendroglial tumors. Based on previous publications, the CNA category was classified by the following thresholds: homozygous deletion (x ≤ 0.4), hemizygous deletion (0.4 < x ≤ 0.7), gain (1.3 ≤ x < 2.0), amplification (x ≥ 2.0) [16]. Any other details of the MLPA protocol and its data analysis to assess CNAs are described in SI.
The cancer genome atlas (TCGA)
An independent and extensive molecular data and clinical information including survival data and treatment of GBM in TCGA (n = 577) were collected from cBioPortal for Cancer Genomics (https://cbioportal.org) [6, 11] and the supplemental data of the previous publication by TCGA [3, 5]. Based on the racial distribution available from cBioPortal (n = 550), whites accounted for a much higher percentage (88.5%, n = 487) than blacks (9.1%, n = 50) and Asians (2.4%, n = 13).
Out of 577 cases initially collected, 359 cases were enrolled as Cohort T1, who were conclusively diagnosed with primary IDH-wild-type GBM. Recurrent, secondary, or IDH-mutant-GBMs along with those that lacked any such information were differentially excluded from this step. For the purpose of survival analysis in Step1, only one case was additionally excluded due to a lack of censoring data.
From Cohort T1, 152 cases receiving TMZ chemoradiation as initial treatment were selected for external validation (Cohort T2) [3]. Those initially treated with either RT alone, TMZ alone, or alkylating chemotherapy other than TMZ, along with those where any such information was unavailable, were excluded. Thus, the inclusion criteria of the cohort from TCGA were similar to that for the Japanese cases in each step.
Molecular and clinical data acquisition from a larger dataset in TCGA
All data regarding CNA in each patient from TCGA were downloaded from the online resource provided in cBioPortal for Cancer Genomics on July 15th, 2018. To identify CNA profiles, we employed putative copy number calls generated by the GISTIC or RAE algorithms in the portal, such as “-2”, “-1”, “0”, “1”, and “2” [6, 11]. These acquired calls were converted to foregoing categories as follows: − 2 to homozygous deletion, − 1 to hemizygous deletion, 0 to neutral, 1 to gain, and 2 to amplification. The mutation status in TP53 was also downloaded from the portal. Further molecular data about MGMT methylation and various clinical information were available from the supplemental data of the previous publication by TCGA [3, 5]. IDH mutational status could be obtained from both data sources and obtainable results were fully cross-validated with each other. There were few data regarding TERT promoter mutation in any of the data sources.
Statistical analysis
All statistical analyses were performed using a JMP Pro version 13 software (SAS Institute, Cary, NC, USA). The difference was considered significant if the p-value was < 0.05.
Patients were subdivided into two groups on the basis of age (≥ 65 or < 65 years), preoperative KPS (≤ 70 or 80–100%), and extent of resection (< 90% or 90–100%) for the purposes of statistical analysis.
Pair associations of molecular variables were evaluated using a two-tailed Fisher’s exact test and computing the log odds ratio (LOR). Concurrent with significant correlation (p < 0.05, Fisher’s exact), LOR below − 2.0 and above 2.0 were respectively defined as association toward mutual exclusivity and co-occurrence. LOR from − 2.0 to − 1.5 and 1.5 to 2.0 were defined to have a tendency for these associations.
For survival analysis, overall survival (OS) was defined as the interval between the initial operative day and the date of either death or the last follow-up date on which the patient was known to be alive, with a cut-off date of 28 February 2018 for the KNBTG cohort. Patients who were still alive at the last follow-up were considered as a censored event. The survival data were analyzed using the log-rank test and Cox regression analyses. In Cox analysis, the hazard ratio (HR) and p-value were computed. Variables with a significant p-value in univariate analysis were subsequently used to build a multivariate Cox model. Stepwise procedure was used for constructing multivariate Cox regression model for survival. After excluding non-significant factor one by one in the multivariate analysis, the remaining variables (p < 0.05) were considered to be independent predictors of survival. Extent of resection and TERT promoter mutation status were not considered in Cox regression model for the TCGA cohort because of a lack of information for all or the majority of patients in this dataset.
Results
Genetic characteristics of cohort K1 (step 1)
TERT promoter mutation, MGMT promoter methylation, and TP53 mutation were observed in 125 cases (59.0%), 98 cases (46.2%), and 80 cases (37.7%), respectively. The major CNA, frequently observed in Cohort K1, were CDKN2A deletion (62.3%), EGFR amp/gain (56.1%), and PTEN deletion (44.3%), as shown in Table 1. Association or tendency toward co-occurrence was observed among TERT promoter mutation, EGFR gain/amp, and PTEN deletion with each other. TERT promoter mutation also had a tendency of mutual exclusivity with PDGFRA amplification (LOR − 1.97, p<0.001). Association toward mutual exclusivity was observed in the pairs of PDGFRA amplification/ EGFR amplification, CDKN2A deletion/CDK4 amplification, and TP53 mutation/MDM2 amp/gain. MGMT methylation status was not significantly associated with any genetic alterations. These mutually exclusive or co-occurring sets are shown in Additional file 1: Figure S1. In the case of pair association, the details for computed LORs and p-values from the Fisher’s exact test are provided in Additional file 2: Table S3 and S4.
Table 1.
Comparison of molecular and clinical characteristics of patients with IDH-wild-type GBM
| Step 1 | Step 2 | |||||
|---|---|---|---|---|---|---|
| Cohort K1 (n = 212) | Cohort T1 (n = 359) | Cohort K2 (n = 140) | Cohort T2 (n = 152) | |||
| Clinical status | Age (years) at diagnosis | Median (range) | 67.0 (18–93) | 61.0 (18–89) | 64.0 (18–82) | 59.0 (18–86) |
| elderly (≥65) | 125 (59.0%) | 138 (38.4%) | 67 (47.9%) | 41 (27.0%) | ||
| Gender | Male | 114 (53.8%) | 221 (61.6%) | 77 (55.0%) | 93 (61.2%) | |
| Female | 98 (46.2%) | 138 (38.4%) | 63 (45.0%) | 59 (38.8%) | ||
| Preoperative KPS (%) | 80–100 | 99 (47.4%) | 195 (71.2%) | 79 (56.4%) | 100 (79.4%) | |
| 0–70 | 110 (52.6%) | 79 (28.8%) | 61 (43.6%) | 26 (20.6%) | ||
| N/A | 3 | 85 | 0 | 26 | ||
| Extent of resection | ≥ 90% | 104 (49.8%) | – | 68 (48.6%) | – | |
| N/A | 3 | – | 0 | – | ||
| TERT promoter | Mut | 125 (59.0%) | – | 81 (57.9%) | – | |
| N/A | 0 | – | 0 | – | ||
| MGMT promoter | Met | 98 (46.2%) | 110 (41.5%) | 59 (42.1%) | 56 (44.8%) | |
| N/A | 0 | 94 | 0 | 27 | ||
| TP53 | Mut | 80 (37.7%) | 60 (26.7%) | 53 (37.9%) | 22 (24.4%) | |
| N/A | 0 | 134 | 0 | 62 | ||
| CNA | PDGFRA | Amp | 33 (15.6%) | 53 (14.8%) | 20 (14.3%) | 19 (12.5%) |
| Gain | 10 (4.7%) | 20 (5.6%) | 8 (5.7%) | 11 (7.2%) | ||
| EGFR | Amp | 54 (25.5%) | 181 (50.4%) | 39 (27.9%) | 76 (50.0%) | |
| Gain | 65 (30.7%) | 159 (44.3%) | 39 (27.9%) | 67 (44.1%) | ||
| CDKN2A | Homo | 79 (37.3%) | 210 (58.5%) | 58 (41.4%) | 87 (57.2%) | |
| Hemi | 53 (25.0%) | 61(17.0%) | 30 (21.4%) | 23 (15.1%) | ||
| PTEN | Homo | 4 (1.9%) | 41 (11.4%) | 3 (2.1%) | 14 (9.2%) | |
| Hemi | 90 (42.5%) | 300 (83.6%) | 58 (41.4%) | 132 (86.8%) | ||
| CDK4 | Amp | 23 (10.8%) | 57 (15.9%) | 13 (9.3%) | 19 (12.5%) | |
| Gain | 7 (3.3%) | 33 (9.2%) | 7 (5.0%) | 17 (11.2%) | ||
| MDM2 | Amp | 18 (8.5%) | 33 (9.2%) | 10 (7.1%) | 12 (7.9%) | |
| Gain | 3 (1.4%) | 28 (7.8%) | 3 (2.1%) | 15 (9.9%) | ||
| NFKBIA | Homo | 1 (0.5%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | |
| Hemi | 46 (21.7%) | 109 (30.4%) | 33 (23.6%) | 45 (29.6%) | ||
| TP53 | Homo | 8 (3.8%) | 6 (1.7%) | 5 (3.6%) | 3 (2.0%) | |
| Hemi | 78 (36.8%) | 56 (15.6%) | 50 (35.7%) | 22 (14.5%) | ||
| Mut/Del | 122 (57.5%) | 96 (38.6%) | 80 (57.1%) | 39 (38.6%) | ||
| Triple CNA | Triple | 51 (24.1%) | 253 (70.5%) | 34 (24.3%) | 103 (67.8%) | |
| Non triple | 161 (75.9%) | 106 (29.5%) | 106 (75.7%) | 49 (32.2%) | ||
Abbreviations: Amp Amplification, Del homozygous and/or hemizygous deletion, Hemi hemizygous deletion, Homo homozygous deletion, Met methylated, Mut mutated, Mut/Del Mut or Del, N/A not available
Frequencies of several genetic abnormalities differed across the cohorts KNBTG and TCGA (Step 1)
The molecular characteristics of each cohort are shown in Table 1. The details for each case from Cohort K1 and T1 are provided in Additional file 2: Table S1 and S2, respectively.
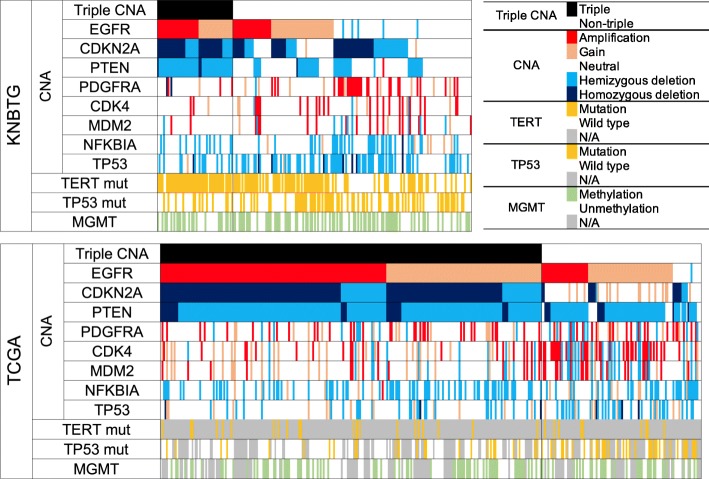
The frequencies of genetic abnormalities, especially in EGFR, CDKN2A, and PTEN, in Cohort K1 were lower than that in Cohort T1, with the difference in excess of 20%. The triple overlapping presence of CNAs in EGFR, CDKN2A, and PTEN (termed triple CNA) especially showed the apparent distinction between Cohort K1 (24.1%) and T1 (70.5%). The frequencies of TP53 mutation and deletion were higher in Cohort K1 (57.5%) than in T1 (38.6%). The distribution of MGMT methylation status between the two cohorts was roughly equivalent (46.2% versus 41.5%). The patient molecular status of each cohort is shown in Fig. 2.
Fig. 2.
Genetic distribution in IDH-wild-type GBM among the two cohorts. The diagram shows the landscape of the molecular characteristics of IDH-wild-type GBM from KNBTG (upper figure) and TCGA (lower figure), which are sorted by CNAs in EGFR, CDKN2A, and PTEN. The triple overlap CNAs in EGFR, PTEN, and CDKN2A (triple CNA) proved to be approximately three-fold higher in frequency in TCGA. Abbreviations: Alteration, mutation and/or copy number alteration; mut, mutation; N/A, not available
Comparative survival analysis revealed favorable outcome in the Japanese cohort (K1) (Step1)
Cohort K1 showed significantly longer survival (median OS, 16.7 months) than Cohort T1(median OS, 12.9 months) (p < 0.001, log-rank test) (Additional file 1: Figure S2). We compared the clinical characteristics of K1 and T1 to identify the factors that caused the survival difference. As shown in Table 1, the proportion of elderly patients (≥ 65) in Cohort K1 (59.0%) was higher than that in T1 (38.4%). Although KPS status in TCGA was not fully obtained, patients with low preoperative KPS (≤ 70%) were more common in Cohort K1 (52.6%) versus T1 (28.8%). Unfavorable prognostic factors were more frequent in the Japanese cohort, contrary to our first hypothesis that the Japanese cohort harbor clinical advantages for survival. Therefore, we formulated another hypothesis that the differences in genetic backgrounds including CNAs may cause the discrepancies of clinical outcomes.
Exploration of potential prognostic biomarkers based on CNAs (Step 2)
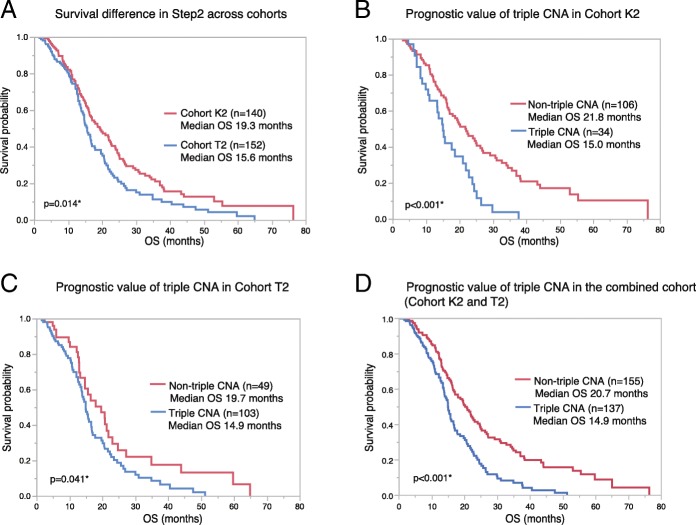
To evaluate the impact of molecular profiles on the survival difference across the cohorts, we collected the cases with homogenous treatment background from KNBTG and TCGA (Cohort K2 and T2, respectively). Similar to that in Step1, OS of Cohort T2 (median OS, 15.6 months) was significantly shorter than that of Cohort K2 (median OS, 19.3 months) (p = 0.014) (Fig. 3a). Thus, population from KNBTG showed longer survival than that from TCGA, regardless of postoperative TMZ chemoradiation. When conducting comparative profiling of these cohorts (Table 1), patients with advanced age and lower KPS were similarly more frequent in Cohort K2. We concluded that these cohorts (K2 and T2) were suitable for investigating the impact of molecular profiles on the survival difference.
Fig. 3.
Survival difference in Step2 across cohorts and prognostic value of triple CNA. a. Kaplan–Meier estimates of OS in Step2 for Cohort K2 (n = 140) and T2(n = 152) are shown. OS of Cohort T2 showed significantly shorter survival than that of Cohort K2 (p = 0.014) as well as in Step1, even after the adjustment of postoperative treatment background. b-c. Kaplan–Meier estimates of OS according to triple CNA are shown on each cohort: Cohort K2 (n = 140) (b) and Cohort T2 (n = 152) (c). Cases with triple CNA significantly showed a worse prognosis than that without triple CNA both in Cohort K2 (p < 0.001, Log-rank test) and Cohort T2 (p = 0.041). Median OS of cases with triple CNA were roughly 15 months that was comparable in the two cohorts. d. In the combined cohort of 292 patients (both Cohort K2 and T2), triple CNA was also a negative prognostic indicator (p < 0.001, log-rank test). Abbreviations: OS, overall survival; triple CNA, the triple overlap of copy number alterations in EGFR, PTEN and CDKN2A
The prognostic impact of each clinical or molecular factors were examined in Cohort K2 (Table 2, Additional file 2: Table S5). In univariate Cox regression analysis, advanced age (≥ 65) at diagnosis, preoperative KPS ≤ 70%, extent of resection < 90%, MGMT unmethylation, and TERT promoter mutation were associated with unfavorable prognosis. Focusing on CNAs, PTEN deletion and NFKBIA deletion were associated with shorter survival, while EGFR amp/gain, CDKN2A deletion, and TP53 deletion were not associated with OS. Note that the cases with triple CNA significantly showed a worse prognosis (HR 2.134, p = 0.002). Median OS was significantly different between cases in Cohort K2 with triple CNA (15.0 months) and without triple CNA (21.8 months) (p < 0.001, log-rank test). Kaplan–Meier estimates according to triple CNA is shown in Fig. 3b. Next, we proceeded to multivariate analysis using a model including the possible explanatory variables in univariate analysis (age at diagnosis, preoperative KPS, extent of resection, MGMT, and probable CNA profiles). After excluding non-significant factors by a stepwise procedure, triple CNA was found to be an independent prognostic factor along with age, extent of resection, NFKBIA, and MGMT in Cohort K2 (Table 2).
Table 2.
Cox proportional hazards models in Step 2
| Univariate | Multivariate | ||||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI for HR | p-value | HR | 95% CI for HR | p-value | ||
| Cohort K2 (n = 140) | |||||||
| Agea | 1.027 | 1.009–1.047 | 0.002† | 1.035 | 1.016–1.056 | < 0.001† | |
| KPS | ≤70% | 1.611 | 1.082–2.391 | 0.019† | excluded by factor selection with step-wise method | ||
| ≥80% | Ref | – | – | ||||
| Extent of resection | < 90% | 1.555 | 1.051–2.310 | 0.027† | 1.860 | 1.246–2.790 | 0.002† |
| ≥90% | Ref | – | – | Ref | |||
| MGMT promoter | Un-Met | 2.037 | 1.351–3.123 | < 0.001† | 2.447 | 1.601–3.808 | < 0.001† |
| Met | Ref | – | – | Ref | |||
| PTEN | Del | 1.552 | 1.041–2.314 | 0.031† | excluded by factor selection with step-wise method | ||
| Neutral | Ref | – | – | ||||
| NFKBIA | Del | 1.613 | 1.016–2.490 | 0.043† | 1.886 | 1.181–2.930 | 0.009† |
| Neutral | Ref | – | – | Ref | |||
| Triple CNA | Triple | 2.134 | 1.348–3.303 | 0.002† | 2.361 | 1.475–3.702 | < 0.001† |
| Non-triple | Ref | – | – | Ref | |||
| Cohort T2 (n = 152) | |||||||
| Agea | 1.023 | 1.005–1.041 | 0.009† | excluded by factor selection with step-wise method | |||
| MGMT | Un-Met | 2.316 | 1.419–3.855 | < 0.001† | 2.320 | 1.422–3.860 | < 0.001† |
| Met | Ref | Ref | |||||
| NFKBIA | Del | 1.767 | 1.118–2.735 | 0.016† | excluded by factor selection with step-wise method | ||
| Neutral | Ref | ||||||
| Triple CNA | Triple | 1.576 | 1.027–2.489 | 0.037† | 1.736 | 1.053–2.980 | 0.030† |
| Non-triple | Ref | Ref | |||||
Abbreviations: CI confidence interval, Del homozygous and/or hemizygous deletion, Met methylated, Ref Reference: aHR is for each 1 year increase. †Statistically significant (p < 0.05)
In univariate Cox regression analysis for Cohort T2, age ≥ 65 at diagnosis, MGMT unmethylation, NFKBIA deletion, and triple CNA were unfavorable prognostic factors, similar to the results of the analysis using Cohort K2 (Table 2, Additional file 2: Table S5). Median OS varied widely between cases in Cohort T2 with triple CNA (14.9 months) and without triple CNA (19.7 months) (p = 0.041, log-rank test). Kaplan–Meier estimates according to triple CNA is shown in Fig. 3c. In a multivariate Cox model incorporating the significant factors in univariate analysis (age, MGMT, NFKBIA, and triple CNA), MGMT methylation and triple CNA remained as independent prognostic factors (Table 2). Thus, the clinical significance of MGMT methylation and triple CNA was validated in both Cohorts K2 and T2. The details regarding univariate analysis for each molecular variable is shown in Additional file 2: Table S5. In the combined cohort of 292 patients (both Cohort K2 and T2), triple CNA was also the negative prognostic indicator (p < 0.001, log-rank test) (Fig. 3d).
Adjustment for common prognostic biomarkers in survival analysis
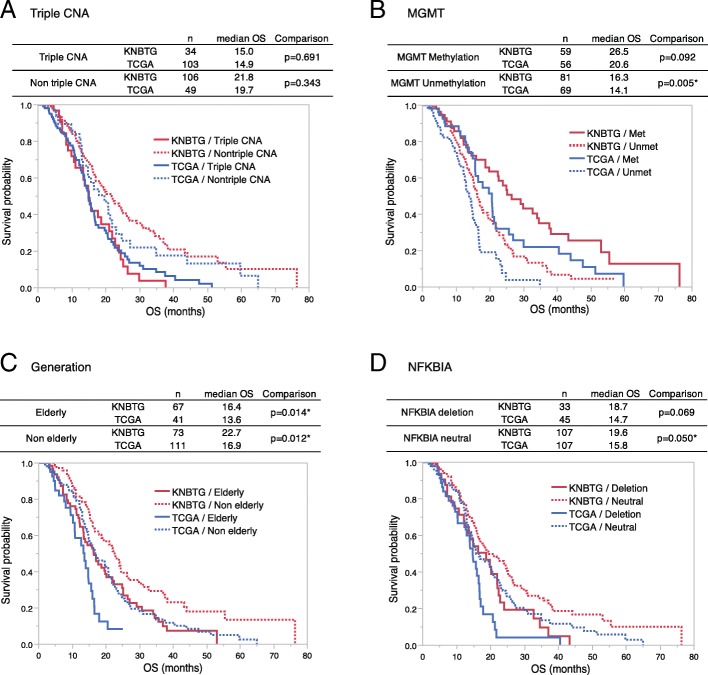
Here, all cases in Step2 (n = 292) were subdivided into subgroups with or without the common prognostic markers in univariate analysis (Age, MGMT, NFKBIA, and triple CNA) to apply their adjustment, and Kaplan-Meier curve comparison for cohorts with log-rank test was shown in Fig. 4 (four curves for each biomarker). Remarkably, the statistical discrepancies of OS between KNBTG and TCGA completely resolved after adjusting by the triple CNA status (Fig. 4a). However, even after adjustment by other prognostic factors including MGMT methylation status, generation, and NFKBIA status, survival differences remained significant in at least one of the subgroups representing presence or absence of these factors. (Fig. 4b-d) Accordingly, triple CNA was validated as a universal prognostic factor responsible for the survival difference between KNBTG and TCGA.
Fig. 4.
Kaplan-Meier analysis of overall survival between KNBTG and TCGA after stratification by the common prognostic biomarkers. Population in Step2 (both Cohort K2 and T2) were subdivided into two subgroups either present or absent of triple CNA, MGMT methylation, elderly, or NFKBIA deletion, respectively. OS comparison between KNBTG and TCGA for each subgroup was performed with log-rank test and Kaplan–Meier plot are shown. The status of biomarkers is recognizable by solid or dotted line. Red curves indicate KNBTG, while blue ones indicate TCGA. a. The solid curves (triple CNA) are almost overlapped. There is also crossover between dotted curves (non-triple CNA). The statistical discrepancies of OS between KNBTG and TCGA resolved completely both in triple CNA subgroup (p = 0.691, Log-rank test) and non-triple CNA subgroup (p = 0.343) as shown in a table above. b-d. Even after being stratified by other prognostic factors (MGMT, generation, and NFKBIA), survival differences remained significant in at least one of the subgroups (either present or absent of these factors) as shown in each table above. Abbreviations: KNBTG, Kansai Molecular Diagnosis Network for CNS tumors; Met, methylated; OS, overall survival; TCGA, The Cancer Genome Atlas; triple CNA, the triple overlap of copy number alterations in EGFR, PTEN and CDKN2A; Unmet, unmethylated
Discussion
In this study on IDH-wild-type GBM, we conducted comparative profiling for CNA status and clinical outcome targeting two large cohorts: KNBTG and TCGA. Firstly, CNA frequencies of EGFR, PTEN, and CDKN2A turned out to exhibit extreme variation between these cohorts. Secondly, the triple overlap in these three loci (triple CNA) proved to be approximately three-fold higher in frequency in TCGA (70.5%) than KNBTG (24.1%). The prognostic impact of this finding was validated in both cohorts, which consisted entirely of GBM patients homogenously treated by standard chemoradiation. Although KNBTG cohort significantly showed the better prognosis than TCGA, its survival difference completely resolved after subclassifying all cases by triple CNA status, in contrast to other common prognostic molecular markers. Consequently, the discrepancies in the distribution of triple CNA in two cohorts could provide a full account of the cohort disparities of survival duration.
The prognostic impact of CNAs is still controversial; however, some of them have the potential to be related with clinical outcomes in GBM. The impact of NFKBIA deletion on survival was observed in both KNBTG and TCGA cohorts in the present study, and this result was in line with a previous report [2]. Even though the CNAs in EGFR, CDKN2A, and PTEN affected patients’ clinical outcome insufficiently by itself, these markers proved to commonly show a prognostic impact when combined. In another attempt to provide prognostic assessment based on CNA molecular subtype, the classification defined by chromosome 1 gain, 19 gain, and CDK4/MDM2 co-amplification was recently proposed through an analysis on the dataset from TCGA and German Glioma Network [9]. The prognostic discrimination according to this classification was incompletely validated in IDH-wild-type GBM; however, their findings supported the impact of CNAs on survival. These clinical impacts of CNA profiles immediately raise a question of whether any molecular marker could make “authentic” GBMs further distinguishable from preexisting entities diagnosed by histopathological findings and IDH1/2 status. Our findings support the need to introduce molecular markers in the diagnostic criteria of GBM and re-evaluate the definition of GBMs.
The underlying reason for the unequal distribution of CNAs between Japan and TCGA cohorts is poorly understood; however, there are two presumptive causes. The first hypothesis is the interobserver variances in the histopathological diagnosis of GBMs. In practice, the diagnosis of GBM in our cohort was solely based on the universal guidelines in WHO classification. However, even the universal criteria may pose some limitations, and, hence, molecular marker seems to be of practical value in GBM diagnostics. The second hypothesis is the geographical variances in the frequencies of CNAs. The regional difference has been widely recognized in other malignant tumors such as lung adenocarcinoma [20, 30], breast cancer [14], and endometrial cancer [12]. A well-known example is the high prevalence of EGFR mutations in pulmonary adenocarcinoma in Asia. Despite few data concerning GBM, the low prevalence of TP53 mutations in whites and homozygous deletion of CDKN2 gene in Japanese were reported [7, 21], along with the regional disproportion of EGFR amplification in GBM. These findings are roughly compatible with our results in that the most striking disproportion of genetic aberrations is mainly in the loci of EGFR, CDKN2A, PTEN, and TP53. Our study may provide an additional evidence for the interregional molecular disproportion of GBM. Further study is needed to elucidate regional variances and its clinical value.
This is an illustrative cohort study showing that different clinical outcomes across cohorts could be attributed to the uneven distributions of prognostic genetic backgrounds. We formed two independent cohorts consisting of population subjected to the same treatment, and successfully validated the common prognostic value of triple CNA. Notably, the survival discrepancy between the cohorts resolved after subclassifying these two cohorts by triple CNA. In population-based research, treatment options are submitted to physicians in local institutions and depend on the socioeconomical status of patients. Therefore, population-based cohort is reflective of the general population, but is likely to be a heterogeneously treated population, which makes it difficult to elucidate the clinical value of molecular markers. Our results indicate that some underlying selection biases remain even after adjusting the treatment backgrounds, and that one of those biases is the differences in prognostic CNA profiles. Along with our results, Cimino et al. showed an inherent bias that enrolled patients for the prospective cohort in clinical trial are liable to bear favorable molecular markers for survival, using molecular profiles by their original CNA subclassification [8]: the prospective cohort in clinical trials thus, less likely mirrors the general population. Therefore, the potential biases in genetic background including CNA profiles should be taken into consideration while interpreting clinical trials or biomarker studies.
This study harbors the limitations as a retrospective analysis susceptible to selection biases; the initial inclusion criteria for molecular data availability probably reflected one of the selection biases. Instead, the cohorts in this study could be regarded as reflective of the general population with homogeneous treatment background. The other limitation is the different molecular biological techniques utilized in the two cohorts. We evaluated CNAs by MLPA, which is a reliable and cost-effective method, but lower throughput than the comprehensive high-throughput array of the TCGA cohort. The resultant CNA profiling in our cohort seems dependable, because interactive CNA relationships such as co-occurrence and mutual exclusivity showed mostly equivalent outcomes to previous reports (Additional file 1: Figure S1). It is, however, difficult to discriminate by MLPA technique whether the CNA involves entire chromosomal arm or focal locus; low-copy EGFR amplification and 7p gain is hard to be distinguished. Nonetheless, the putative copy number calls downloaded from cBioPortal was virtually equivalent to that generated by MLPA because focal gene copy number was equally estimated rather than the entire chromosomal aberrations. Our analyses incorporated hemizygous and homozygous deletion of CDKN2A as one of the molecular markers. The biological significance of hemizygous deletion is not relevant in the molecular pathway while CDKN2A homozygous deletion is regarded as one of the representative causes for disruptions of Rb1 pathway. When incorporating only homozygous deletion of CDKN2A for the definition of triple CNA, the prognostic impact of this combination is retained in the Japan cohort but not in the TCGA cohort (data not shown). Further investigation using homogeneous molecular techniques in independent larger cohorts is needed to validate our findings and select the optimal combination of CNAs. However, we believe that this study adequately addressed the clinical importance and geographical distribution of CNAs in GBMs.
Conclusions
We conclude that specific CNA profiles harbor significant impacts on the survival of GBM patients, and that the distribution of CNAs potentially affects clinical outcomes across cohorts. We believe that the GBM classification according to copy number profiles would be a prognostic indicator and provide new insight into the interpretation and comparison of interregional clinical trials.
Additional files
Figure S1. Combinations (or pairs) of genetic alterations showing co-occurrence or mutual exclusivity in KNBTG. Figure S2. Kaplan-Meier analysis of overall survival in Step1 between KNBTG and TCGA. (PPTX 281 kb)
Table S1. Dataset of patients with glioblastoma from Kansai Molecular Diagnosis Network for CNS tumors (n = 234). Table S2. Dataset of patients with glioblastoma from The Cancer Genome Atlas (n = 577). Table S3. Log odds ratio in pair association analysis for genetic aberrations. Table S4. P-value from two-tailed Fisher’s exact test in pair association analysis for genetic aberrations. Table S5. Univariate analysis for each molecular variable using Cox proportional hazards models in Step2. Table S6. Sequences of primers used for amplification and Sanger sequencing. Table S7. Sequences of primers used for quantitative methylation specific PCR. (ZIP 256 kb)
Acknowledgements
The authors thank Ms. Ai Takada at the Institute for Clinical Research, Osaka National Hospital, National Hospital Organization, and Ms. Rieko Kugo at the Department of Neurosurgery, Osaka University Graduate School of Medicine for their generous assistance. We gratefully acknowledge all the investigators participating in the TCGA analysis of GBMs and for providing accessibility to the molecular and clinical data. Finally, we are grateful to all the patients who participated in this study.
Abbreviations
- CNA
copy number alteration
- CNS WHO
WHO Classification of Tumors of the Central Nervous System
- GBM
Glioblastoma
- HR
hazard ratio
- KNBTG
Kansai Molecular Diagnosis Network for CNS tumors
- KPS
Karnofsky Performance status
- LOR
log odds ratio
- MGMT
O6-methylguanine-DNA-methyltransferase
- MLPA
Multiplex Ligation-dependent Probe Amplification
- OS
overall survival
- qMSP
quantitative methylation specific PCR
- RT
radiotherapy
- TCGA
The Cancer Genome Atlas
Authors’ contributions
Study design: HA, MK, KM, YK Data collection: EM, TS, DK, MK, YK, MM, NK, YF, YO, MN, KN, TU, NT, JF, KF, DS Data analysis: TU, HA Interpretation: HA, KM, HK, YK Manuscript writing: TU, HA, YK. All authors read and approved the final manuscript.
Funding
This investigation was supported by JSPS KAKENHI 17 K16646 (T.U.) and 16 K20006 (H.A.), and the Research award of Osaka Cancer Society (H.A.).
Availability of data and materials
The anonymized datasets analyzed in the present study are provided in supplementary information.
Ethics approval and consent to participate
This study was carried out in accordance with the principles of the Helsinki Declaration, and approval was obtained from the internal ethical review boards of Osaka University Graduate School of Medicine (approval number: 13244), Osaka National Hospital (No. 713), and all collaborative institutes. Written informed consent was obtained from all patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Toru Umehara, Email: t-umehara@nsurg.med.osaka-u.ac.jp.
Hideyuki Arita, Phone: +81-6-6879-3652, Email: h-arita@nsurg.med.osaka-u.ac.jp.
Ema Yoshioka, Email: e.hirai@onh.go.jp.
Tomoko Shofuda, Email: tshofuda@onh.go.jp.
Daisuke Kanematsu, Email: kanematu@onh.go.jp.
Manabu Kinoshita, Email: mail@manabukinoshita.com.
Yoshinori Kodama, Email: kodamayo@cosmos.ocn.ne.jp.
Masayuki Mano, Email: mano-ma@onh.go.jp.
Naoki Kagawa, Email: nkagawa@nsurg.med.osaka-u.ac.jp.
Yasunori Fujimoto, Email: yasufujimoto@gmail.com.
Yoshiko Okita, Email: yokita4246@gmail.com.
Masahiro Nonaka, Email: nonakam@hirakata.kmu.ac.jp.
Kosuke Nakajo, Email: kousuke19841984@yahoo.co.jp.
Takehiro Uda, Email: uda@med.osaka-cu.ac.jp.
Naohiro Tsuyuguchi, Email: ntsuyuguchi@gmail.com.
Junya Fukai, Email: junfukai@wakayama-med.ac.jp.
Koji Fujita, Email: kfujita@wakayama-med.ac.jp.
Daisuke Sakamoto, Email: d.sakamoto.ns@gmail.com.
Kanji Mori, Email: kanji_m@mac.com.
Haruhiko Kishima, Email: hkishima@nsurg.med.osaka-u.ac.jp.
Yonehiro Kanemura, Email: yonehirok@gmail.com.
References
- 1.Arita H, Yamasaki K, Matsushita Y, Nakamura T, Shimokawa A, Takami H, et al. A combination of TERT promoter mutation and MGMT methylation status predicts clinically relevant subgroups of newly diagnosed glioblastomas. Acta Neuropathol Commun. 2016;4:79. doi: 10.1186/s40478-016-0351-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bredel M, Scholtens DM, Yadav AK, Alvarez AA, Renfrow JJ, Chandler JP, et al. NFKBIA deletion in glioblastomas. N Engl J Med. 2011;364:627–637. doi: 10.1056/NEJMoa1006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research N Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.Cd-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen P, Aldape K, Wiencke JK, Kelsey KT, Miike R, Davis RL, et al. Ethnicity delineates different genetic pathways in malignant glioma. Cancer Res. 2001;61:3949–3954. [PubMed] [Google Scholar]
- 8.Cimino Patrick J, McFerrin Lisa, Wirsching Hans-Georg, Arora Sonali, Bolouri Hamid, Rabadan Raul, Weller Michael, Holland Eric C. Copy number profiling across glioblastoma populations has implications for clinical trial design. Neuro-Oncology. 2018;20(10):1368–1373. doi: 10.1093/neuonc/noy108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cimino PJ, Zager M, McFerrin L, Wirsching HG, Bolouri H, Hentschel B, et al. Multidimensional scaling of diffuse gliomas: application to the 2016 World Health Organization classification system with prognostically relevant molecular subtype discovery. Acta Neuropathol Commun. 2017;5:39. doi: 10.1186/s40478-017-0443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen A, Sato M, Aldape K, Mason CC, Alfaro-Munoz K, Heathcock L, et al. DNA copy number analysis of grade II-III and grade IV gliomas reveals differences in molecular ontogeny including chromothripsis associated with IDH mutation status. Acta Neuropathol Commun. 2015;3:34. doi: 10.1186/s40478-015-0213-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guttery DS, Blighe K, Polymeros K, Symonds RP, Macip S, Moss EL. Racial differences in endometrial cancer molecular portraits in the Cancer genome atlas. Oncotarget. 2018;9:17093–17103. doi: 10.18632/oncotarget.24907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 14.Huo D, Hu H, Rhie SK, Gamazon ER, Cherniack AD, Liu J, et al. Comparison of breast Cancer molecular features and survival by African and European ancestry in the Cancer genome atlas. JAMA Oncol. 2017;3:1654–1662. doi: 10.1001/jamaoncol.2017.0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeuken J, Cornelissen S, Boots-Sprenger S, Gijsen S, Wesseling P. Multiplex ligation-dependent probe amplification: a diagnostic tool for simultaneous identification of different genetic markers in glial tumors. J Mol Diagn. 2006;8:433–443. doi: 10.2353/jmoldx.2006.060012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeuken J, Sijben A, Alenda C, Rijntjes J, Dekkers M, Boots-Sprenger S et al (2009) Robust detection of EGFR copy number changes and EGFR variant III: technical aspects and relevance for glioma diagnostics. Brain Pathol 19:661–671. https://doi.org/10.1111/j.1750–3639.2009.00320.x [DOI] [PMC free article] [PubMed]
- 17.Kessler Tobias, Sahm Felix, Sadik Ahmed, Stichel Damian, Hertenstein Anne, Reifenberger Guido, Zacher Angela, Sabel Michael, Tabatabai Ghazaleh, Steinbach Joachim, Sure Ulrich, Krex Dietmar, Grosu Anca-L, Bewerunge-Hudler Melanie, Jones David, Pfister Stefan M, Weller Michael, Opitz Christiane, Bendszus Martin, von Deimling Andreas, Platten Michael, Wick Wolfgang. Molecular differences in IDH wildtype glioblastoma according to MGMT promoter methylation. Neuro-Oncology. 2017;20(3):367–379. doi: 10.1093/neuonc/nox160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Libermann TA, Nusbaum HR, Razon N, Kris R, Lax I, Soreq H, et al. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature. 1985;313:144–147. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- 19.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 20.Ma BB, Hui EP, Mok TS. Population-based differences in treatment outcome following anticancer drug therapies. Lancet Oncol. 2010;11:75–84. doi: 10.1016/s1470-2045(09)70160-3. [DOI] [PubMed] [Google Scholar]
- 21.Mochizuki S, Iwadate Y, Namba H, Yoshida Y, Yamaura A, Sakiyama S, et al. Homozygous deletion of the p16/MTS-1/CDKN2 gene in malignant gliomas is infrequent among Japanese patients. Int J Oncol. 1999;15:983–989. doi: 10.3892/ijo.15.5.983. [DOI] [PubMed] [Google Scholar]
- 22.Nobusawa S, Watanabe T, Kleihues P, Ohgaki H. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res. 2009;15:6002–6007. doi: 10.1158/1078-0432.Ccr-09-0715. [DOI] [PubMed] [Google Scholar]
- 23.Ohgaki H, Dessen P, Jourde B, Horstmann S, Nishikawa T, Di Patre PL, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004;64:6892–6899. doi: 10.1158/0008-5472.Can-04-1337. [DOI] [PubMed] [Google Scholar]
- 24.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasaki Takahiro, Fukai Junya, Kodama Yoshinori, Hirose Takanori, Okita Yoshiko, Moriuchi Shusuke, Nonaka Masahiro, Tsuyuguchi Naohiro, Terakawa Yuzo, Uda Takehiro, Tomogane Yusuke, Kinoshita Manabu, Nishida Namiko, Izumoto Shuichi, Nakajima Yoshikazu, Arita Hideyuki, Ishibashi Kenichi, Shofuda Tomoko, Kanematsu Daisuke, Yoshioka Ema, Mano Masayuki, Fujita Koji, Uematsu Yuji, Nakao Naoyuki, Mori Kanji, Kanemura Yonehiro. Characteristics and outcomes of elderly patients with diffuse gliomas: a multi-institutional cohort study by Kansai Molecular Diagnosis Network for CNS Tumors. Journal of Neuro-Oncology. 2018;140(2):329–339. doi: 10.1007/s11060-018-2957-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 27.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weller M, Felsberg J, Hartmann C, Berger H, Steinbach JP, Schramm J, et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German glioma network. J Clin Oncol. 2009;27:5743–5750. doi: 10.1200/jco.2009.23.0805. [DOI] [PubMed] [Google Scholar]
- 29.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang W, Edwards A, Flemington EK, Zhang K. Racial disparities in patient survival and tumor mutation burden, and the association between tumor mutation burden and cancer incidence rate. Sci Rep. 2017;7:13639. doi: 10.1038/s41598-017-13091-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Combinations (or pairs) of genetic alterations showing co-occurrence or mutual exclusivity in KNBTG. Figure S2. Kaplan-Meier analysis of overall survival in Step1 between KNBTG and TCGA. (PPTX 281 kb)
Table S1. Dataset of patients with glioblastoma from Kansai Molecular Diagnosis Network for CNS tumors (n = 234). Table S2. Dataset of patients with glioblastoma from The Cancer Genome Atlas (n = 577). Table S3. Log odds ratio in pair association analysis for genetic aberrations. Table S4. P-value from two-tailed Fisher’s exact test in pair association analysis for genetic aberrations. Table S5. Univariate analysis for each molecular variable using Cox proportional hazards models in Step2. Table S6. Sequences of primers used for amplification and Sanger sequencing. Table S7. Sequences of primers used for quantitative methylation specific PCR. (ZIP 256 kb)
Data Availability Statement
The anonymized datasets analyzed in the present study are provided in supplementary information.