Abstract
Rationale: The contributions of diverse cell populations in the human lung to pulmonary fibrosis pathogenesis are poorly understood. Single-cell RNA sequencing can reveal changes within individual cell populations during pulmonary fibrosis that are important for disease pathogenesis.
Objectives: To determine whether single-cell RNA sequencing can reveal disease-related heterogeneity within alveolar macrophages, epithelial cells, or other cell types in lung tissue from subjects with pulmonary fibrosis compared with control subjects.
Methods: We performed single-cell RNA sequencing on lung tissue obtained from eight transplant donors and eight recipients with pulmonary fibrosis and on one bronchoscopic cryobiospy sample from a patient with idiopathic pulmonary fibrosis. We validated these data using in situ RNA hybridization, immunohistochemistry, and bulk RNA-sequencing on flow-sorted cells from 22 additional subjects.
Measurements and Main Results: We identified a distinct, novel population of profibrotic alveolar macrophages exclusively in patients with fibrosis. Within epithelial cells, the expression of genes involved in Wnt secretion and response was restricted to nonoverlapping cells. We identified rare cell populations including airway stem cells and senescent cells emerging during pulmonary fibrosis. We developed a web-based tool to explore these data.
Conclusions: We generated a single-cell atlas of pulmonary fibrosis. Using this atlas, we demonstrated heterogeneity within alveolar macrophages and epithelial cells from subjects with pulmonary fibrosis. These results support the feasibility of discovery-based approaches using next-generation sequencing technologies to identify signaling pathways for targeting in the development of personalized therapies for patients with pulmonary fibrosis.
Keywords: pulmonary fibrosis, RNA sequencing, alveolar macrophages, alveolar type II cells
At a Glance Commentary
Scientific Knowledge on the Subject
The contributions of diverse cell populations in the human lung to pulmonary fibrosis pathogenesis are poorly understood. Single-cell RNA sequencing can reveal changes within individual cell populations during pulmonary fibrosis that are important for disease pathogenesis.
What This Study Adds to the Field
We generated a single-cell atlas of pulmonary fibrosis. Using this atlas, we demonstrated heterogeneity within alveolar macrophages and epithelial cells from subjects with pulmonary fibrosis.
Pulmonary fibrosis is defined as the progressive replacement of alveolar tissue with fibrotic scar that threatens alveolar gas exchange and reduces lung compliance (1). The resulting increased work of breathing and hypoxemia lead to progressive respiratory failure and eventual death. Although the cause of pulmonary fibrosis is often not identified (idiopathic pulmonary fibrosis [IPF]), in some patients pulmonary fibrosis can be attributed to connective tissue disease, environmental exposures (hypersensitivity pneumonitis), occupational exposures (silicosis, asbestosis), or drugs. Diagnostic approaches to distinguish between these causes of pulmonary fibrosis, including surgical lung biopsy, are imprecise, and there are few laboratory features that predict responsiveness to therapy (2, 3). The clinical and economic burden of pulmonary fibrosis is sizeable: the worldwide incidence of IPF is increasing and attributable medical costs for patients with IPF in the United States alone have been estimated at nearly 2 billion dollars (4, 5). Despite decades of research, only a handful of therapies are available. Although these therapies slow disease progression, the response is variable, and lung transplantation represents the only option for temporary cure. Accordingly, there has been a call to leverage cutting-edge genomic techniques and systems biology approaches to provide molecular insights into pulmonary fibrosis that can be used to develop personalized approaches to diagnosis and therapy (6, 7).
Although every cell in the body has an identical genome, the quantity of mRNA molecules encoded by individual genes, collectively referred to as the transcriptome, differs widely as a function of cell type. Indeed, investigators have used next-generation sequencing technology (RNA-Seq) to develop an organ-level transcriptomic map of the human body (8). The lung is a complex tissue comprised of more than 40 cell populations (9). Because each of these populations has a distinct transcriptome, previous investigations of the “lung transcriptome” during fibrosis using whole-lung tissue were limited by changes in the cellular composition of the lung during disease, which could mask relevant changes in individual cell populations (10–13). Transcriptomic analysis of flow cytometry–sorted cell populations from the lung provides an alternative, but requires a priori assumptions about cell surface markers whose expression may change during disease. The advent of single-cell RNA-Seq allows reliable identification of even closely related cell populations (14). Single-cell RNA-Seq methods also allow for the identification of known or novel cell populations for which there are no reliable surface markers, and provide the opportunity to assess heterogeneity of gene expression in individual lung cell populations during health and disease (15).
Methods
Here, we used single-cell RNA-Seq to analyze lung tissue from patients with pulmonary fibrosis and lung tissue from transplant donors, which we used as a normal comparison. We compared these data with bulk RNA-Seq data from whole-lung tissue and flow cytometry–sorted alveolar macrophages and alveolar type II cells generated from a separate cohort. Combined with in situ RNA hybridization, these data provide a molecular atlas of disease pathobiology. We observed emergence of a distinct, novel population of macrophages exclusively in patients with fibrosis that demonstrated enhanced expression of profibrotic genes. Within epithelial cells, we observed that the expression of genes involved in Wnt secretion and response was restricted to nonoverlapping cells. We identified rare cell populations including airway stem cells and senescent cells emerging during pulmonary fibrosis in the single-cell RNA-Seq data. We performed analysis of a cryobiopsy specimen from a patient with early disease, supporting the clinical application of single-cell RNA-Seq to develop personalized approaches to therapy. Some of the results of these studies have been previously reported in the form of a preprint (https://doi.org/10.1101/296608) and conference abstracts (16, 17). The dataset is available at nupulmonary.org/resources/.
Results
Study Population
Single-cell RNA-Seq was performed on eight donor lung biopsies and eight lung explants from patients with pulmonary fibrosis attributed to IPF (four patients), systemic sclerosis (two patients), polymyositis (one patient), and chronic hypersensitivity pneumonitis (one patient). All samples were obtained at the time of transplantation. Separately, we performed single-cell RNA-Seq using one bronchoscopic cryobiopsy sample from a patient subsequently diagnosed with IPF. Bulk RNA-Seq was performed on samples of lung biopsy tissue obtained from 14 donors before transplantation and eight lung explants from transplant recipients with pulmonary fibrosis. The median age of patients with pulmonary fibrosis was 56.0 years (interquartile range, 41.5–70.5 yr). Eight (47.0%) were male and six (35.3%) were former smokers. Characteristics of patients with pulmonary fibrosis are reported in Table 1, and representative histology from these lungs is provided in Figure E1A in the online supplement. Clinical characteristics of donors are reported in Table 2, and representative histology from donor lung samples adjacent to the region used for single-cell RNA-Seq analysis is provided in Figure E1B.
Table 1.
Characteristics of Patients with Pulmonary Fibrosis
| Age (yr) | Sex | Diagnosis | History of Smoking | Supplemental Oxygen Use before Transplant | Sample ID |
|---|---|---|---|---|---|
| 66* | M | IPF | Yes | Yes | IPF 1 |
| 60* | M | IPF | No | Yes | IPF 2 |
| 68* | M | IPF | Yes | Yes | IPF 3 |
| 72* | F | IPF | No | Yes | IPF 4 |
| 55* | F | Hypersensitivity pneumonitis | No | Yes | HP |
| 39* | F | Systemic sclerosis–associated ILD | No | Yes | SSc-ILD 1 |
| 53* | F | Systemic sclerosis–associated ILD | No | Yes | SSc-ILD 2 |
| 37* | F | Myositis-associated ILD | No | Yes | PM-ILD |
| 71† | F | IPF | Yes | No | Cryobiopsy |
| 42‡ | M | Systemic sclerosis–associated ILD | No | Yes | NA |
| 65‡ | M | IPF | No | Yes | NA |
| 52‡ | M | Systemic sclerosis–associated ILD | Yes | Yes | NA |
| 52‡ | F | Systemic sclerosis–associated ILD | No | Yes | NA |
| 55‡ | F | Myositis-associated ILD | No | Yes | NA |
| 56‡ | F | Hypersensitivity pneumonitis | No | Yes | NA |
| 60‡ | M | Myositis-associated ILD | Yes | Yes | NA |
| 67‡ | M | IPF | Yes | No | NA |
Definition of abbreviations: HP = hypersensitivity pneumonitis; ILD = interstitial lung disease; IPF = idiopathic pulmonary fibrosis; NA = not applicable; PM = polymyositis; SSc = systemic sclerosis.
Representative histopathology with pathologist interpretation is found in Figure E1A.
Transplant recipients with pulmonary fibrosis included in single-cell RNA-Seq analysis.
Patient with IPF undergoing cryobiopsy included in single-cell RNA-Seq analysis.
Transplant recipients with pulmonary fibrosis included in bulk RNA-Seq analysis.
Table 2.
Characteristics of Lung Transplant Donors
| Age (yr) | Sex | Race | Smoking Status | Cause of Death | Sample ID |
|---|---|---|---|---|---|
| 41 | M | White | Never | Stroke | NA |
| 57 | M | White | Active | Intracranial hemorrhage after blunt trauma | NA |
| 64 | M | African American | Never | Intracranial hemorrhage | NA |
| 49 | F | White | Never | Intracranial hemorrhage | NA |
| 50 | M | African American | Never | Intracranial hemorrhage | NA |
| 19 | M | African American | Never | Stroke after gunshot wound | NA |
| 54 | M | White | Never | Intracranial hemorrhage after blunt trauma | NA |
| 43 | M | Hispanic | Never | Anoxic brain injury after opiate overdose | NA |
| 21 | M | White | Never | Intracranial hemorrhage after blunt trauma | NA |
| 43 | F | White | Former | Anoxic brain injury | NA |
| 50 | M | Hispanic | Active | Intracranial hemorrhage after blunt trauma | NA |
| 51 | M | African American | Active | Stroke | NA |
| 26 | F | African American | Active | Anoxic brain injury after opiate overdose | NA |
| 40 | M | Hispanic | Active | Intracranial hemorrhage after blunt trauma | NA |
| 63* | F | African American | Never | Stroke | Donor 1 |
| 55* | M | Asian | Former | Intracranial hemorrhage | Donor 2 |
| 29* | F | African American | Never | Anoxic brain injury | Donor 3 |
| 57* | F | African American | Never | Anoxic brain injury | Donor 4 |
| 49* | F | White | Active | Intracranial hemorrhage | Donor 5 |
| 22* | F | African American | Never | Anoxic brain injury after seizure | Donor 6 |
| 47* | F | White | Active | Intracranial hemorrhage | Donor 7 |
| 21* | M | African American | Never | Head trauma from gunshot wound | Donor 8 |
Definition of abbreviation: NA = not applicable.
Representative histopathology is found in Figure E1B.
Used for single-cell RNA-Seq analysis.
Single-Cell RNA-Seq Identifies Multiple Cell Populations in the Human Lung
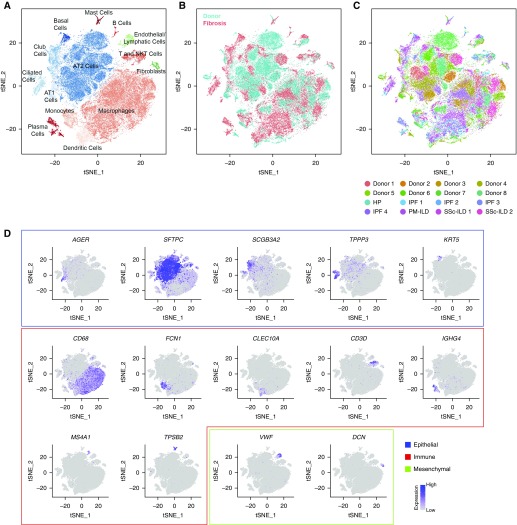
In total, 76,070 cells were used for integrated single-cell RNA-Seq analysis. We performed a similar analysis on 13,822 cells from two normal mouse lungs. We assigned cell types to each cluster based on the expression of established markers from the LungMAP and ImmGen databases (Figures 1A–1D; see Figures E2A–E2D and Tables E1 and E2) (interactive web tool is available at nupulmonary.org/resources/) (18, 19). In the human lung, we identified alveolar type II cells; alveolar type I cells; ciliated, club, and basal airway epithelial cells; alveolar macrophages; dendritic cells; T cells and natural killer T cells; plasma cells and B cells; fibroblasts; and endothelial and lymphatic cells (Figure 1A; see Table E1). Each cluster included cells from donors and patients with pulmonary fibrosis (Figure 1B). In the mouse, we were able to identify all cell types seen in the human lung and several rare and difficult to isolate cell populations, including additional endothelial and lymphatic cell populations; megakaryocytes; innate lymphoid cells; and mesothelial cells (see Figure E2B and Table E2). Each cluster included cells from each individual mouse (see Figure E2D). Expression of cell cycle genes was similar between donor and fibrotic lungs within the 14 clusters (see Figures E3A and E3B).
Figure 1.
Integrated single-cell RNA-Seq analysis of patients with pulmonary fibrosis identifies diverse lung cell populations. Single-cell RNA-Seq was performed on single-cell suspensions generated from eight lung biopsies from transplant donors and eight lung explants from transplant recipients with pulmonary fibrosis. All 16 samples were analyzed using canonical correlation analysis within the Seurat R package. Cells were clustered using a graph-based shared nearest neighbor clustering approach and visualized using a t-distributed Stochastic Neighbor Embedding (tSNE) plot. (A) Cellular populations identified. (B) Cells on the tSNE plot of all 16 samples were colored as originating either from a donor or from a patient with pulmonary fibrosis. (C) Each population included cells from donors and patients with pulmonary fibrosis. (D) Canonical cell markers were used to label clusters by cell identity as represented in the tSNE plot. Cell types were classified as epithelial, immune, or mesenchymal as indicated in the legend. AT1 = alveolar type I; HP = hypersensitivity pneumonitis; ILD = interstitial lung disease; IPF = idiopathic pulmonary fibrosis; NK = natural killer; PM = polymyositis; SSc = systemic sclerosis.
Single-Cell RNA-Seq Analysis Reveals Fibrosis-associated Transcriptomic Changes in Key Lung Cell Populations
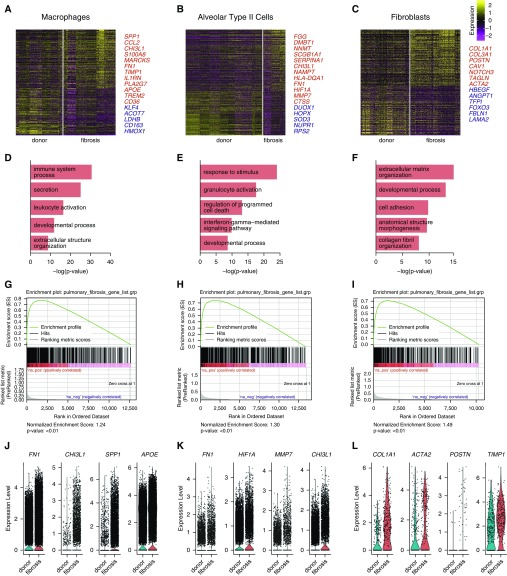
We estimated differential gene expression between cells from normal and fibrotic lungs in three key cell populations: macrophages, alveolar type II cells, and fibroblasts (Figures 2A–2C; see Table E3). Gene Ontology enrichment analysis of the differentially expressed genes identified cell type–specific processes relevant to pulmonary fibrosis (Figures 2D–2F) (20–23). We performed Gene Set Enrichment Analysis to determine whether integrated single-cell RNA-Seq data can recapitulate gene expression signatures associated with pulmonary fibrosis in the literature using the Comparative Toxicogenomics Database Pulmonary Fibrosis Gene Set (24–26). All three cell populations showed significant enrichment for genes in this dataset with normalized enrichment scores of 1.24, 1.30, and 1.49 for macrophages, alveolar type II cells, and fibroblasts, respectively (false discovery rate q value < 0.01) (Figures 2G–2I). Violin plots for representative differentially expressed genes from each of the populations are shown (Figures 2J–2L). It should be noted that for each of the three cell populations examined, differentially expressed genes included some that are not normally expressed by individual cell populations (e.g., surfactant proteins genes or immunoglobulin genes; see Table E3). This likely resulted from contaminating ambient RNA released during tissue processing, a known problem in single-cell RNA-Seq experiments (27). Computational methods to address this problem are being developed, but are not yet validated (28).
Figure 2.
Differential expression analysis of single-cell RNA-Seq data from normal and fibrotic lungs identifies genes characteristic of pulmonary fibrosis. (A–C) Differential expression analysis was performed comparing cells from normal and fibrotic lungs within macrophages, alveolar type II cells, and fibroblasts (Wilcoxon rank sum test as implemented within Seurat toolkit). Heatmaps are shown representing the upregulated and downregulated genes in macrophages, alveolar type II cells, and fibroblasts, highlighting genes involved in fibrosis. The full table of genes is found in Table E3. (D–F) Functional enrichment analysis with GO Biological Processes was performed using GOrilla with the significantly upregulated genes in cells from fibrotic compared with normal lungs. Representative significantly enriched GO processes are shown for macrophages, alveolar type II cells, and fibroblasts. (G–I) Gene Set Enrichment Analysis was performed using the Comparative Toxicogenomics Database Pulmonary Fibrosis Gene Set with genes ranked by log difference in average expression between fibrotic and normal lungs. Enrichment plots together with normalized enrichment scores and false discovery rate q values are shown for macrophages, alveolar type II cells, and fibroblasts. (J–L) Violin plots of expression for select genes significantly upregulated in patients with fibrotic compared with normal lungs.
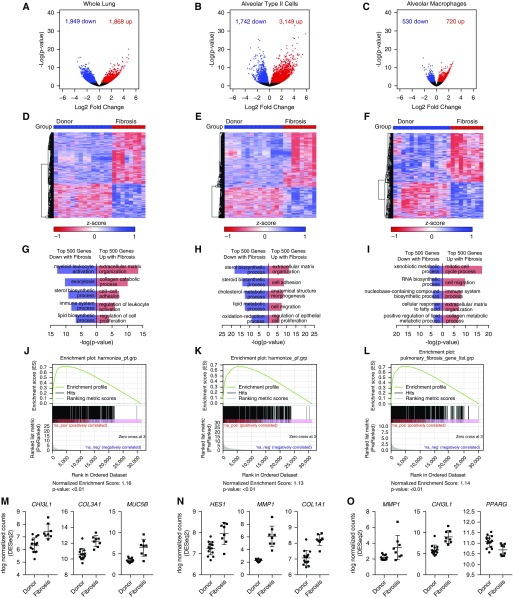
We compared the single-cell RNA-Seq analysis with bulk RNA-Seq analysis of flow cytometry–sorted alveolar macrophages, alveolar type II cells, and whole-lung tissue from 14 donor lung biopsies compared with explants from eight transplant recipients with pulmonary fibrosis (Tables 1 and 2). Estimation of differential gene expression identified 3,817 genes in whole-lung tissue, 4,891 in alveolar type II cells, and 1,238 in alveolar macrophages (Figures 3A–3C; see Table E4). Hierarchical clustering of samples and estimation of differentially expressed genes between fibrotic and normal lungs was performed (Figures 3D–3F). Consistent with the single-cell RNA-Seq data, whole-lung tissue, alveolar macrophages and alveolar type II cells from patients with pulmonary fibrosis were significantly enriched for Gene Ontology processes relevant to pulmonary fibrosis and for genes in the curated Comparative Toxicogenomics Database Pulmonary Fibrosis Gene Set by Gene Set Enrichment Analysis (Figures 3G–3O). We then asked if single-cell RNA-Seq and bulk RNA-Seq identify similar changes in alveolar macrophages and alveolar type II cells. After filtering genes that demonstrated little change between donor and fibrosis (absolute log fold change < 0.05), we found significant positive correlation between donor and fibrosis log fold change in the single-cell and bulk datasets in alveolar type II cells and alveolar macrophages (Pearson product-moment correlation, r = 0.43, P < 2.2 × 10−16 and r = 0.59, P < 2.2 × 10−16, respectively) (see Figures E4A and E4B).
Figure 3.
Bulk RNA-Seq of whole-lung tissue and flow cytometry–sorted alveolar type II cells and alveolar macrophages from normal and fibrotic lungs validates the single-cell RNA-Seq analysis. (A–C) Bulk RNA-Seq was performed on whole-lung tissue and flow cytometry–sorted alveolar type II cells and alveolar macrophages from 14 normal and 8 fibrotic lungs. Estimation of differential gene expression using DESeq2 was performed comparing fibrotic with normal lungs. Volcano plots are shown for whole lung, alveolar type II cells, and alveolar macrophages, respectively. (D–F) Hierarchical clustering heatmaps of significant differentially expressed genes were generated using GENE-E. (G–I) Functional enrichment analysis with GO Biological Processes was performed using GOrilla with the top 500 genes upregulated and downregulated in fibrotic compared with donor lungs. Representative GO processes are shown. (J–L) Gene Set Enrichment Analysis was performed using the Comparative Toxicogenomics Database Pulmonary Fibrosis Gene Set with genes ranked by −log(P value). Enrichment plots together with normalized enrichment scores and false discovery rate q values are shown for whole lung, alveolar type II cells, and alveolar macrophages. (M–O) Expression of selected significant differentially expressed genes previously described to be important in pulmonary fibrosis.
Localization of Known Pulmonary Fibrosis-associated Signaling Pathways to Specific Cell Populations
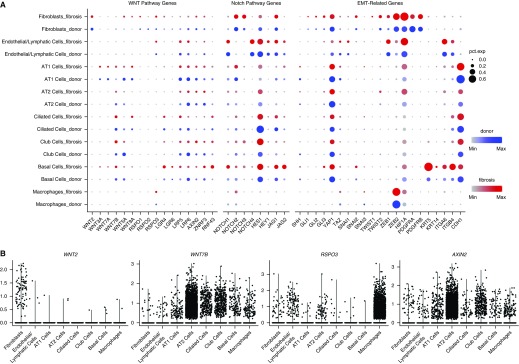
The pathobiology of pulmonary fibrosis in animal models involves the aberrant activation of developmental pathways in the lung, including Notch, Wnt/β-catenin, and signaling pathways associated with epithelial to mesenchymal transition (29). Our single-cell RNA-Seq data allowed us to localize the expression of genes associated with these pathways to specific cellular populations within the diseased lung (Figures 4A and 4B). We saw upregulation of Notch ligands and Notch target gene expression in alveolar type II cells and club cells, with downregulation of Notch target gene expression in endothelial cells. We detected low-level expression of several Wnt ligands in epithelial cell populations and fibroblasts in the normal and fibrotic lungs. Our findings were similar in the murine lung, although they differed from the observations in a recently published smaller murine single-cell RNA-Seq dataset (see Figure E4C) (30). The expression of the Wnt target gene AXIN2 was detected in a minority of epithelial cells in normal lungs and showed only modest increases during fibrosis. Several genes related to epithelial to mesenchymal transition were upregulated in fibroblasts during fibrosis. R-spondins are structurally distinct from Wnts but work synergistically to sustain Wnt/β-catenin signaling (31). As expected, we found expression of R-spondins in fibroblasts and endothelial cells, but we were surprised to observe it in macrophages. None of the populations expressing R-spondins expressed Wnt ligands or the Wnt target, AXIN2.
Figure 4.
Single-cell RNA-Seq analysis reveals distinct contributions of individual cell populations to pathways implicated in the pathogenesis of pulmonary fibrosis. (A) Expression of selected Wnt pathway, Notch pathway, and epithelial-to-mesenchymal transition–related genes is shown in fibroblasts, endothelial cells, alveolar type I cells, alveolar type II cells, ciliated cells, club cells, basal cells, and macrophages, separated by donor (blue) or pulmonary fibrosis (red) origin. Dot size corresponds to the percentage of cells in the cluster expressing a gene, and dot color corresponds to the average expression level for the gene in the cluster. (B) Violin plots of WNT2, WNT7B, AXIN2, and RSPO3 expression are shown, suggesting the presence of distinct Wnt-expressor and Wnt-responder cell populations in the human lung. AT 1/2 = alveolar type I/II; EMT = epithelial to mesenchymal transition.
These findings are consistent with the emerging concept that Wnt signaling, particularly in progenitor populations, is optimized through defined epithelial/mesenchymal pairings that comprise a niche (30–32). Accordingly, we examined our dataset for cells responsible for maintaining this progenitor niche. LGR5 and LGR6 have been recently reported to mark a mesenchymal population in the lung necessary for maintenance of a progenitor niche (31, 33). Although LGR5 was not detected in the normal or fibrotic human lung or in the naive adult mouse lung, possibly because of underrepresentation of mesenchymal cells, LGR4 and LGR6 were detected in a small subset of epithelial cells and fibroblast/mesenchymal cells (Figure 4A; see Figure E4C). In mice, a subset of lung fibroblasts expressing Pdgfra, Porcn, Wls, Wnt5a, and other Wnts was reported to form a niche responsible for maintenance of a rare population of Axin2-positive alveolar type II cells that differentiate into type I cells during injury (30). These genes were detectable in fibroblasts from human and murine lungs (Figure 4A; see Figure E4C).
Single-Cell RNA-Seq Reveals Novel Subpopulations of Alveolar Macrophages and Alveolar Epithelial Cells during Pulmonary Fibrosis
Single-cell RNA-Seq data are uniquely suited to look for heterogeneity within cell populations that might emerge during disease. Because integrative canonical correlation analysis tends to mask heterogeneity within individual populations, we performed clustering of cells from each of the individual subjects and assigned cell type identities to each cluster based on the expression of established markers in the curated publicly available LungMAP and ImmGen databases (see Figure E5 and Table E5) (18, 19). Survey of the individual subjects revealed several distinct cell populations that were not observed in the canonical correlation analysis. In total, we identified 22 distinct cell types by individual annotation of each subject, compared with the 14 cell populations identified by the integrated analysis. Many of the populations not observed using the integrated analysis were represented only in a minority of patients or in a single patient and were combined with other cell populations in the integrated analysis. In a patient with IPF (IPF 2), we were able to resolve FOXP3- and CD4-expressing regulatory T cells and ITGAE- and CD8-expressing lung-resident T-cell subsets (see Figure E6A). In the patient with hypersensitivity pneumonitis, we identified two small populations of dendritic cells that did not form distinct clusters in other subjects (see Figure E6B). The first cluster (30 cells) expressed CLEC4C, TCL1A, IRF8, and TLR7, markers associated with plasmacytoid dendritic cells. The second cluster (31 cells) expressed CLEC9A, a typical marker of a subset of conventional dendritic cells (DC1). These populations were classified as macrophages and dendritic cells, respectively, in the integrated analysis. The distinction between classical and nonclassical monocytes, which was apparent in the subjects IPF 1, HP, Donor 6, and SSc-ILD 1, was not identified in the integrated analysis (see Figure E6C). Having accurately identified all specific cellular populations, we then performed combined analysis of the alveolar macrophages and epithelial cells.
Alveolar Macrophages
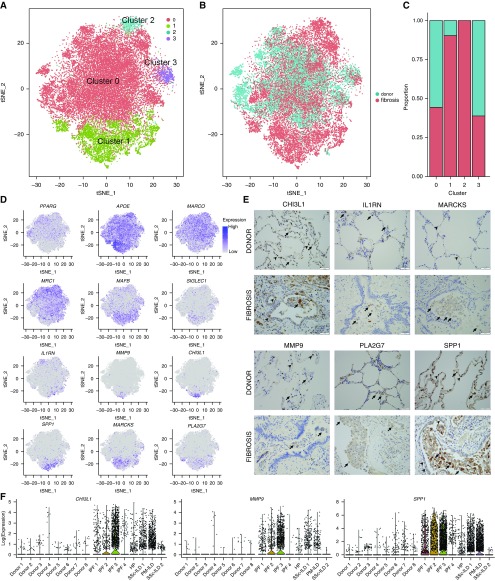
We have reported that in a mouse model of bleomycin-induced pulmonary fibrosis alveolar macrophages are comprised of two ontologically distinct populations: tissue-resident alveolar macrophages and monocyte-derived alveolar macrophages (34). In that system, we showed that selective genetic deletion of monocyte-derived alveolar macrophages while leaving tissue-resident alveolar macrophages intact resulted in improved fibrosis, a finding that has been subsequently confirmed by an independent group (35). These studies predict the presence of homeostatic tissue-resident alveolar macrophages in the normal lung, and a combination of tissue-resident alveolar macrophages and profibrotic, monocyte-derived alveolar macrophages in the fibrotic lung. We performed individual annotation of our eight normal and eight fibrotic lungs and then combined all clusters of cells identified as macrophages and repeated clustering. In this analysis, we identified four macrophage clusters (Figure 5A; see Table E6). Cluster 0 contained almost all of the alveolar macrophages from the donor lungs, and some alveolar macrophages from patients with pulmonary fibrosis, irrespective of the diagnosis (Figures 5B and 5C). In contrast, clusters 1 and 2 originated largely from the lungs of patients with fibrosis (Figure 5C). Consistent with the hypothesis that cells in cluster 0 represent homeostatic tissue-resident alveolar macrophages, they expressed higher levels of PPARG, MRC1, and MARCO, and lower levels of APOE and MAFB, which we and others have reported rise and fall, respectively, as monocytes differentiate into alveolar macrophages (Figure 5D) (34, 36). Functional enrichment analysis with GO Biological Processes demonstrated that cluster 1 was enriched for processes associated with fibrosis including “exocytosis,” “secretion,” “regulation of cell migration,” and “extracellular matrix organization.” In contrast, enriched processes in cluster 0 were suggestive of homeostatic functions of macrophages including “immune system processes,” “response to lipid,” and “response to organic/inorganic substance.” A full list of GO processes for all four clusters is provided in Table E7. Although our study includes only a small number of patients, we did not see differences in clustering in patients with IPF compared with other causes of pulmonary fibrosis, possibly because of the limitations of current computational approaches (see Figures E7A and E7B).
Figure 5.
Distinct populations of alveolar macrophages emerge during pulmonary fibrosis. (A) Cells identified as macrophages by individual annotation of single-cell RNA-Seq data from eight normal and eight fibrotic lungs were combined and then clustered, revealing four clusters. (B and C) Relative contributions of alveolar macrophages from normal and fibrotic lungs to each cluster as shown by t-distributed Stochastic Neighbor Embedding plot and by bar plots. (D) Feature plots demonstrating differential expression of selected alveolar macrophage maturation genes (PPARG, APOE, MARCO, MRC1, MAFB, and SIGLEC1), and genes associated with fibrosis (IL1RN, MMP9, CHI3L1, SPP1, MARCKS, and PLA2G7). (E) Immunohistochemistry on lung sections from the same patients confirms heterogeneity in fibrotic gene expression within alveolar macrophages. Arrows indicate positive staining in alveolar macrophages; arrowheads indicate positive staining in alveolar epithelium for CHI3L1 and SPP1, endothelium for MARCKS, and neutrophils for MMP9. Scale bars, 50 μm. (F) Violin plots representing heterogeneity in expression of CHI3L1, MMP9, and SPP1 in macrophages from donor and fibrotic lungs. HP = hypersensitivity pneumonitis; ILD = interstitial lung disease; IPF = idiopathic pulmonary fibrosis; PM = polymyositis; SSc = systemic sclerosis; tSNE = t-distributed Stochastic Neighbor Embedding.
To validate the heterogeneity of alveolar macrophages in pulmonary fibrosis that we observed in the single-cell RNA-Seq data, we performed immunohistochemistry. We selected CHI3L1, MARCKS, IL1RN, PLA2G7, MMP9, and SPP1 because they 1) were differentially expressed in alveolar macrophages between fibrotic compared with normal lungs in both bulk and single-cell RNA-Seq, 2) are associated with pulmonary fibrosis in the Comparative Toxicogenomics Database Pulmonary Fibrosis Gene Set, and 3) have antibodies validated in the Human Protein Atlas (37). Consistent with the single-cell RNA-Seq data, we found that alveolar macrophages from normal lungs did not stain for these markers, and only a subpopulation of cells in any given patient with pulmonary fibrosis expressed profibrotic proteins (Figure 5E).
Epithelial Cells
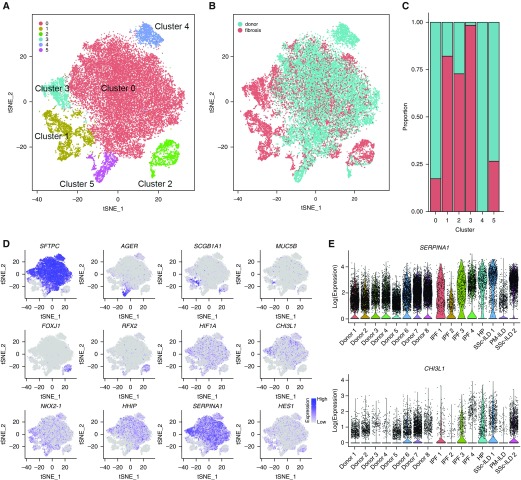
To capture potentially novel epithelial cell populations in patients with fibrosis, we performed combined analysis of all cells identified as belonging to any epithelial population from our individual annotation of the eight normal and eight fibrotic lungs. Epithelial cells contained clusters representing the major known lung epithelial cell types: ciliated epithelial cells (FOXJ1), club cells (SCGB1A1), alveolar type I cells (AGER), and alveolar type II cells (SFTPC and LAMP3). Ciliated cells and alveolar type I cells from fibrotic lungs clustered with cells from normal lungs. In contrast, cells expressing canonical alveolar type II cell markers (SFTPC, LAMP3) formed three clusters (Figures 6A–6D, only SFTPC is shown for alveolar type II cells; see Table E8). Similar to alveolar macrophages, cluster 0 contained cells from normal and fibrotic lungs, whereas cluster 3 almost exclusively contained cells from fibrotic lungs, and was characterized by increased expression of genes previously reported to be associated with pulmonary fibrosis and regulation of immune response (DMBT1, SERPINA1, and CHI3L1). The expression of these genes was variable among the patients with pulmonary fibrosis (Figure 6E).
Figure 6.
Distinct populations of alveolar epithelial cells emerge during fibrosis. (A) Six clusters were identified after epithelial cells from each of eight normal and eight fibrotic lungs were combined and clustered. (B and C) Relative contributions of epithelial cells from normal and fibrotic lungs to each cluster as shown by t-distributed Stochastic Neighbor Embedding plot and by bar plots. (D) Feature plots demonstrating differential expression of selected epithelial marker genes: SFTPC (alveolar type II cells), AGER (alveolar type I cells), SCGB1A1 (club cells), FOXJ1, and RFX2 (ciliated airway epithelial cells). Also shown are genes implicated in pulmonary fibrosis (HIF1A, CHI3L1, NKX2-1, HHIP, FASN, and HES1). (E) Violin plots representing heterogeneity in expression of SERPINA1 and CHI3L1 in epithelial cells from normal and fibrotic lungs. For definition of abbreviations, see Figure 5.
In Situ RNA Hybridization with Amplification Confirms Coexistence of Two Distinct Populations of Alveolar Macrophages in the Same Anatomic Niche in Patients with Pulmonary Fibrosis
Our finding of increased heterogeneity within alveolar macrophages in fibrotic lungs is potentially clinically relevant. Accordingly, we looked for heterogeneity within alveolar macrophages in single-cell RNA-Seq data from each of our eight normal and fibrotic lungs (see Figure E5). We observed heterogeneity within macrophages in most of the fibrotic lungs, with distinct clusters of cells enriched for profibrotic genes (e.g., CHI3L1 or SPP1) (see Figure E7D). In contrast, macrophages from seven of the normal lungs were homogenous and were characterized by expression of homeostatic genes, including PPARG and FABP4. In the remaining normal lung (Donor 6), we observed a distinct subpopulation of macrophages, characterized by expression of CCL3/CCL4 (encoding MIP1a, a chemokine produced by macrophages in response to bacterial endotoxin and controlling recruitment of neutrophils) (see Figure E7E). This lung also had a distinct population of cells with a gene expression profile suggesting that they were recruited monocyte-derived inflammatory macrophages. This lung was scored as abnormal in our histopathology assessment (see Figure E1B, Donor 6), and a review of premortem laboratory studies showed the corresponding donor’s respiratory cultures were positive for Klebsiella pneumoniae.
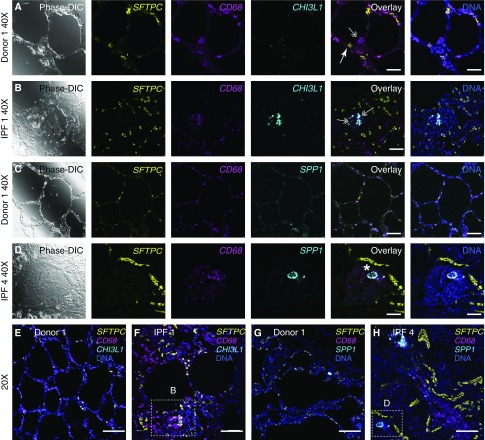
To determine whether single-cell RNA-Seq accurately localized profibrotic gene expression to specific cell populations, we performed a highly sensitive modified in situ RNA hybridization procedure (RNAscope) on samples of the same tissues that we used for single-cell RNA-Seq analysis. We focused on SPP1 and CHI3L1 for two reasons. First, these genes have been causally implicated in the development of organ fibrosis in animal models (38, 39). Second, single-cell RNA-Seq data suggested that SPP1 expression was increased specifically in alveolar macrophages during fibrosis, whereas CHI3L1 was increased in both alveolar macrophages and alveolar type II cells. Using selective marker genes (CD68 for alveolar macrophages and SFTPC for alveolar type II cells), we were able to confirm the predicted emergence of CHI3L1- and SPP1-positive alveolar macrophages and increased expression of CHI3L1 in alveolar type 2 cells in patients with pulmonary fibrosis (Figures 7A–7H; see Figures E8A–E8D). This analysis confirms the prediction from single-cell RNA-Seq data that two distinct populations of alveolar macrophages coexist in the same anatomic niche in the fibrotic lung.
Figure 7.
In situ RNA hybridization with amplification confirms the emergence of distinct populations of alveolar macrophages in patients with pulmonary fibrosis. (A–D) Lung sections from Donor 1 (A and C), IPF 1 (B), and IPF 4 (D) were hybridized with indicated target probes. High-magnification (×40) images for differential interference contrast phase are shown on the left, followed by single-channel images for SFTPC (yellow), CD68 (magenta), and either CHI3L1 or SPP1 (cyan), with overlay images (with and without nuclei staining, Hoechst, blue) shown on the right. (A and B) CHI3L1-positive and CHI3L1-negative alveolar macrophages coexist in the same niche in patients with pulmonary fibrosis. Arrows show alveolar type II cells (double positive for SFTPC and CHI3L1), double arrows show alveolar macrophages (CD68-positive) negative or positive for CHI3L1 in the donor and fibrotic lung, respectively. (C and D) SPP1-positive and SPP1-negative alveolar macrophages coexist in the same niche in patients with pulmonary fibrosis. Note that donor alveolar macrophages lack expression of SPP1 (C), whereas alveolar macrophages from the fibrotic lung exhibit heterogeneity of SPP1 expression (D); double-positive CD68 and SPP1 are indicated with asterisk. Scale bars, 50 μm. (E–H) Low-magnification (×20) overlay images from the same subjects. Boxes indicate areas shown on B and D, respectively. Scale bars, 50 μm. See Figures E8A–E8D for corresponding single-channel panels. DIC = differential interference contrast; IPF = idiopathic pulmonary fibrosis.
Wnt Secretion and Response Are Restricted to Distinct Nonoverlapping Epithelial Cells
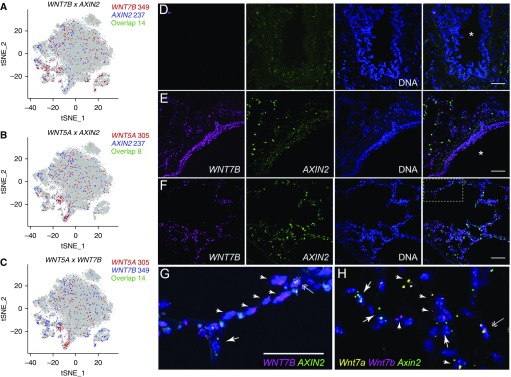
We used in situ RNA hybridization in combination with single-cell RNA-Seq data to explore the expression of ligands and target genes of the Wnt/β-catenin pathway in the human lung during homeostasis and fibrosis. Likely because of increased sampling, we were able to identify higher rates of AXIN2 expression in alveolar type II cells in our single-cell RNA-Seq data in both mice and humans than were recently reported (see Figures E4C and E8E) (30). We found low levels of expression of most Wnt ligands in epithelial cells from humans, but WNT5A and WNT7B were by far the most abundantly expressed (see Figure E8E). Among queried Wnt ligands, we found nonoverlapping patterns of expression between cell populations that express Wnt ligands and the canonical Wnt target, AXIN2 (Figures 8A and 8B). Furthermore, although many cells within the lung expressed Wnt ligands, including alveolar type II cells, individual cells to a large extent expressed a single Wnt ligand (Figure 8C). We confirmed these findings using in situ RNA hybridization with amplification in human tissues. Consistent with the single-cell RNA-Seq data, we observed that airway epithelia largely expressed WNT7B, but we did not detect expression of AXIN2. In the alveolar epithelium, we confirmed increased expression of WNT7B relative to WNT7A, and found that cells expressing either WNT7A or WNT7B were generally distinct from those expressing AXIN2 (Figures 8D–8G). This distinction was not absolute, because we were able to identify a small number of alveolar epithelial cells expressing both WNT7B and AXIN2 (Figure 8G).
Figure 8.
Wnt secretion and response are restricted to distinct nonoverlapping epithelial cells. (A–C) Expression of WNT7B, WNT5A, and AXIN2 in epithelial cells from normal and fibrotic lungs by single-cell RNA-Seq. Total number of single-positive or double-positive cells is indicated. (D and E) Fluorescence RNA in situ hybridization with amplification reveals Wnt-expresser and Wnt-responder cells in the proximal airways of human lung. Images were obtained from human donor lung sections incubated without (D) or with designated target probes, WNT7B (magenta) and AXIN2 (green) (E–G). Nuclei are stained with Hoechst (blue) and overlay images are shown on the far right. Note WNT7B is abundantly detected in small airway epithelial cells (E), consistent with its prominent detection in club cells by single-cell RNA-Seq. AXIN2 is not similarly enriched in club cells and is only specifically detected in alveolar regions (F) under higher magnification (G is a higher-magnification image of the boxed region in F). (H) High magnification of mouse lung alveoli subjected to fluorescence RNA in situ hybridization with amplification with probes for Wnt7a (yellow), Wnt7b (magenta), and Axin2 (green). Arrows indicate cells that are only positive for Axin2; arrowheads indicate specified Wnts. Double arrows indicate cells that may express both Wnts and Axin2, although quantification suggests cells that highly express Wnts and Axin2 are rare (Figure E9D). Asterisks indicate airway lumen. Scale bars, 50 μm. tSNE = t-distributed Stochastic Neighbor Embedding.
The observation that Wnt secretion and response are largely restricted to distinct nonoverlapping cells in the human lung was confirmed by single-cell RNA-Seq and in situ RNA hybridization with amplification analysis of mouse lung. Specifically, murine alveolar type II cells expressed Wnt3a, Wnt7b, Wnt4, and Wnt9a, whereas alveolar type I cells expressed Wnt3a, Wnt7a, Wnt9a, and Wnt10b (see Figure E4C). As in the human lung, we mostly detected only one Wnt-ligand at a time (see Figure E9A). We also confirmed that Wnt-ligand and Axin2 expression in mouse alveolar type II cells seems to be mutually exclusive by both single-cell RNA-Seq (see Figure E9B) and in situ RNA hybridization analysis (Figures 8H; see Figures E9C and E9D).
Single-Cell RNA-Seq Identifies Rare Cell Populations in Human Lung
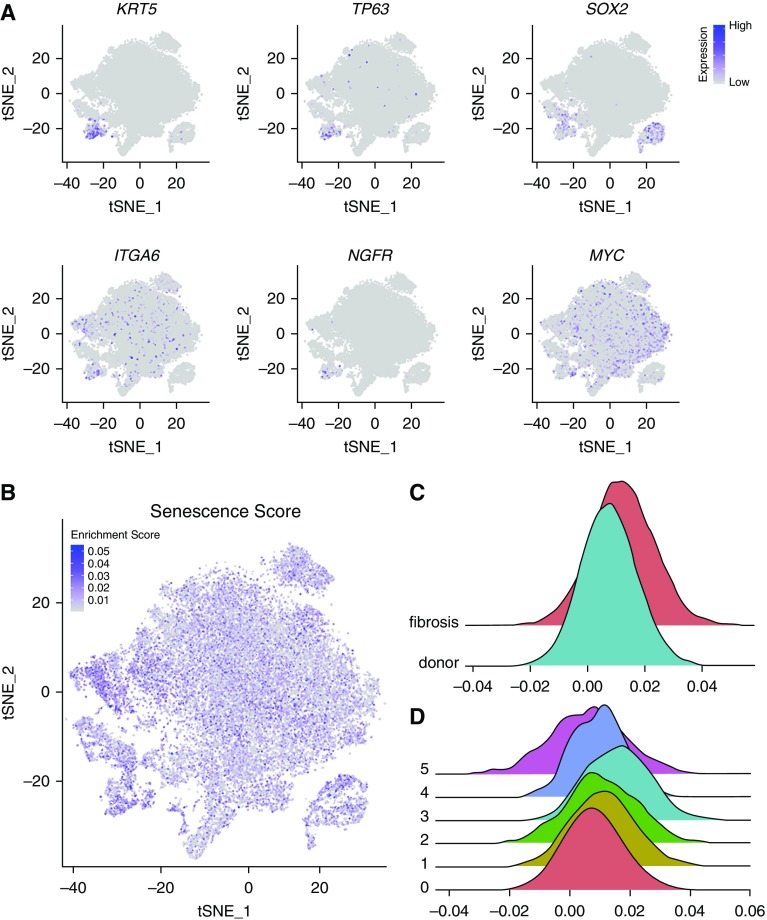
To determine whether single-cell RNA-Seq can identify rare cell populations in the lung, we queried our data using marker genes associated with stem cell populations or senescent cell populations, both of which have been implicated in the pathobiology of pulmonary fibrosis. Investigators identified a population of basal cells in the airway that expand after tracheal injury (40). Using single-cell RNA-Seq, we identified a cluster of cells expressing many of the genes distinctly associated with these cells including KRT5, TP63, and SOX2 with variable expression of ITGA6, NGF, and MYC (Figure 9A). Two recent reports identified a subpopulation of alveolar type II cells expressing high levels of Axin2/AXIN2 (2–20% of mouse and 29% of human alveolar type II cells), and demonstrated these cells serve as a facultative progenitor cell population that regenerates the alveoli after injury (30, 41). Although Axin2/AXIN2-positive alveolar type II cells express all of the established markers of this cell type (30, 41), a study by Zacharias and colleagues (41) identified 875 and 2,773 genes that were differentially expressed in the AXIN2/Axin2-positive progenitor cell population relative to AXIN2/Axin2-negative alveolar type II cells in the human and mouse lung, respectively. Accordingly, we asked whether single-cell RNA-Seq can detect a population of cells resembling Axin2-positive facultative epithelial progenitors in the normal mouse alveolar type II cells. However, unbiased clustering of the 1,821 alveolar type II cells we sampled did not identify clusters enriched for expression of Axin2 or Tm4sf1 (see Figures E10A–E10C and Table E9). To provide additional power to this guided analysis, we used the 500 differentially expressed genes that were reported to be upregulated in Axin2-positive progenitor cells as a gene module for the AddModuleScore function implemented in Seurat package to highlight cells enriched for these genes. We did not find subclusters of alveolar type II cells enriched for the genes associated with Axin2-positive progenitors in the previous studies (see Figures E10D and E10E). We then searched for this subpopulation within human alveolar type II cells from each of the eight normal lungs and performed clustering, which did not reveal additional clusters enriched for these genes (see Figure E10F). These data suggest that single-cell RNA-Seq can readily identify transcriptional differences between dedicated progenitor populations (e.g., basal cells), and that alveolar type II cells in normal mouse and human lungs represent a relatively homogenous population.
Figure 9.
Identification of rare cell populations among lung epithelial cells using single-cell RNA-Seq. (A) Expression of markers associated with airway basal stem cells. (B) Feature plot showing cells enriched for senescence-associated genes using a senescence score for each cell. (C) Histograms showing the distribution of senescence scores in all epithelial cells from eight fibrotic compared with eight normal lungs (P = 0.0001; Student’s t test; n = 8 per group). (D) Histograms showing the distribution of senescence scores by epithelial cell cluster (see Figure 6A for cluster details). tSNE = t-distributed Stochastic Neighbor Embedding.
Terminally differentiated cells in culture undergo a relatively constant number of population doublings before undergoing replicative senescence. The development of senescence is associated with characteristic changes in gene expression that include the induction of p16, p21, and p53, which slow the cell cycle and promote resistance to apoptosis. Senescence also results in the production and secretion of a distinct set of proteins collectively referred to as senescence-associated secretory proteins (42). These include insulin-like growth factor-binding proteins, interleukins, transforming growth factor type-β, and plasminogen activator inhibitor-1. A growing body of evidence supports a link between aging, cellular senescence in the lung and the susceptibility to lung disease. For example, investigators have reported increased markers of senescence in the lungs of patients with IPF, and have found that myofibroblasts obtained from the lungs of patients with IPF demonstrate a senescence-associated resistance to apoptosis (43–45). However, even in pulmonary fibrosis, the percentage of cells in the lung that are positive for senescence markers is very low, precluding their identification using bulk RNA or protein analysis. We did not observe a distinct cluster of epithelial cells expressing canonical genes associated with senescence (CDKN2A, GLB1, SERPINE1, and IL6; see Figure E10G). Therefore, we used the expression of 1,311 replicative senescence-associated genes to generate a senescence score for any individual cell and applied it to epithelial cells in our dataset (Figure 9B, see Figure 6 for a description of the clusters) (46). This score was significantly higher in fibrotic compared with normal lungs (Figure 9C; P = 0.0001). Additionally, this senescence score was highest in the population of alveolar type II cells we identified as originating almost exclusively from fibrotic lungs (cluster 3, Figure 9D). These results suggest that single-cell RNA-Seq offers promise for the quantification of cells expressing senescence-associated genes in the lung.
Single-Cell RNA-Seq of Lung Biopsy Tissue from a Living Patient
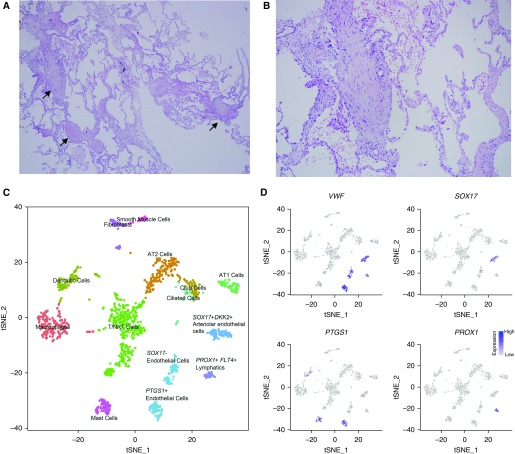
We next examined the feasibility of applying these technologies to a cryobiopsy specimen obtained ex vivo from an explanted lung specimen from a patient with systemic sclerosis–associated interstitial lung disease using flow cytometry. We were able to resolve alveolar macrophages and alveolar epithelial cells, both with good viability (see Figure E11A). Accordingly, we obtained a cryobiopsy specimen from a patient at the time of video-assisted thoracoscopic biopsy. The patient was subsequently diagnosed with IPF based on histopathologic examination of the biopsy (Figures 10A and 10B). We identified 1,516 cells corresponding to 13 different cell populations (Figure 10C). Among the detected cells, we were able to resolve several populations of endothelial cells (characterized by expression of VWF) including a distinct population of cells characterized by expression of PROX1, MMRN1, TBX1, and RELN, suggesting that these cells represent lymphatic progenitors, which were underrepresented in samples obtained at the time of transplant (Figure 10D, only PROX1 shown; see Table E5). We performed clustering of the 192 alveolar macrophages and 364 epithelial cells identified in the cryobiopsy specimen together with those from the eight normal and eight fibrotic lungs. We found that most cryobiopsy alveolar macrophages were in a cluster containing cells from both normal and fibrotic lungs, whereas only a minority were in a cluster comprised almost exclusively of cells from fibrotic lungs (see Figures E11B–E11D). We found that the cryobiopsy epithelial cells seemed to be distributed more among clusters primarily containing cells from fibrotic lungs than clusters primarily containing cells from donor lungs (see Figures E11E–E11G). These results support the feasibility of single-cell RNA-Seq analysis applied to lung tissue from patients with early disease.
Figure 10.
Single-cell RNA-Seq of a cryobiopsy from a living patient with idiopathic pulmonary fibrosis. (A and B) Representative histology (hematoxylin and eosin) showing fibroblastic foci (arrows), ×40 and ×100 magnification, respectively. (C) tSNE plot clustering of 1,516 cells into 13 distinct cellular types. (D) Subsets of endothelial cells were identified by expression of VWF, SOX17, and PTGS1; lymphatics were identified by expression of PROX1. AT 1/2 = alveolar type I/II; NK = natural killer; tSNE = t-distributed Stochastic Neighbor Embedding.
Discussion
We compared single-cell RNA-Seq data from freshly isolated lung tissue obtained from lung transplant donors and patients with pulmonary fibrosis with bulk RNA-Seq data we generated from flow cytometry–sorted alveolar macrophages and alveolar type II cells. Our analysis validates the utility of single-cell RNA-Seq analysis for localizing the expression of profibrotic genes to specific lung cell populations in patients with pulmonary fibrosis. In findings validated with in situ RNA hybridization and immunohistochemistry, single-cell RNA-Seq analysis revealed transcriptionally distinct populations of alveolar macrophages expressing profibrotic genes in patients with pulmonary fibrosis that were predicted from mouse models (34, 35). Single-cell RNA-Seq combined with in situ RNA hybridization provided novel biologic insights into the multicellular and spatially restricted nature of Wnt signaling niches in the normal and fibrotic lung. Cells expressing Wnt ligands were largely distinct from those expressing the Wnt target gene AXIN2/Axin2. We identified some previously described rare cell populations in normal and fibrotic lungs through analysis of single-cell RNA-Seq. We found that the application of single-cell RNA-Seq to lung tissue obtained bronchoscopically via cryobiopsy was feasible. Taken together, our findings suggest that the application of single-cell RNA-Seq analysis to alveolar macrophages or lung tissue obtained during routine care, can be used to develop a molecular approach to improve the diagnosis and therapeutic monitoring of patients with pulmonary fibrosis.
Analysis of biologic processes implicated in pulmonary fibrosis through the prism of single-cell RNA-Seq data demonstrates the unique power of this approach in understanding disease pathogenesis. Specifically, in manually querying this single-cell data set, we noticed nonoverlapping patterns of expression of Wnt ligands and AXIN2/Axin2-expressing cells in both the human and mouse lung. Because of the sparse nature and shallow sequencing depth of single-cell RNA-Seq data, we validated this finding using a highly sensitive in situ RNA hybridization technique. These findings extend our understanding of the complexity of the multicellular signaling niches that sustain Wnt/β-catenin signaling in the normal and fibrotic lung. We were similarly able to use in situ RNA hybridization and immunohistochemistry to validate the presence of heterogeneity within alveolar macrophages from the same patient during fibrosis. Collectively, these data suggest the combination of high-throughput single-cell RNA-Seq with sensitive spatial transcriptomic methods will allow us to functionally and spatially reconstruct multicellular signaling niches during pulmonary fibrosis (47).
In a recent report, single-cell RNA-seq was used to distinguish subpopulations of goblet and tufts cells in the mouse airways, and to identify a rare population of ionocytes (48). Accordingly, we tested the ability of single-cell RNA-Seq to detect rare cell populations implicated in the development of pulmonary fibrosis. We were able to identify markers of some mesenchymal cell populations suggested to play a role in Wnt/β-catenin signaling during lung regeneration (31, 49). However, we did not observe any wide-scale upregulation of Wnts in samples from patients with pulmonary fibrosis, nor expansion of Wnt/Axin2-double positive alveolar type II cells as observed in the hyperoxia model of murine lung injury (30). We identified a likely population of TP63- and KRT5-expressing airway progenitor cells (basal cells) in the normal and fibrotic lung, and we largely confirmed the observation that Axin2-positive alveolar type II cells are a minor subset of the murine alveolar type II cell population (30, 41). Nevertheless, we were unable to identify distinct transcriptional signatures in AXIN2-expressing compared with nonexpressing cells. Similarly, although an expansion of senescent cells during pulmonary fibrosis has been suggested to play a role in disease pathogenesis, our single-cell RNA-Seq data did not identify a transcriptionally distinct cluster of cells expressing senescence markers, nor were we able to identify senescent cells from a handful of canonical markers. However, we were able to identify enriched expression of senescence-associated genes in epithelial cells from patients with pulmonary fibrosis compared with donor lungs. These findings must be interpreted with caution because transcriptional changes defining these cell types may lay below the resolution of single-cell RNA-seq technology (50).
Transcriptomic analysis of whole-lung tissue is heavily influenced by changes in the cellular composition of the tissue that mask changes in gene expression within cell populations (11–13, 51, 52). Combining bulk RNA-Seq applied to flow cytometry–sorted cell populations and single-cell RNA-Seq avoided this problem. Each of these approaches has advantages and limitations. Single-cell RNA-Seq reliably assigned profibrotic gene expression to distinct cell populations in the lung and revealed heterogeneity in gene expression within cell populations. For example, unbiased analysis of single-cell RNA-Seq data identified a distinct population of alveolar macrophages during fibrosis that was predicted from genetic lineage tracing studies during fibrosis in mice (34). Single-cell RNA-Seq analysis was limited, however, by the relatively shallow depth of sequencing and by contamination from ambient RNA liberated during tissue digestion, both of which reduce the reliability of differential gene expression analysis (28). Bulk RNA-Seq of flow cytometry–sorted cells detects more genes at significantly reduced cost. However, this approach cannot identify heterogeneity in gene expression within a given cell population, and inherently assumes a relatively constant level of surface marker expression during health and disease. Few markers are explicitly validated for identification of cellular populations during disease states, and this problem is exacerbated when antibodies are used for which the gene encoding the antigen is not known (53).
The falling costs of transcriptomic profiling make it a promising clinical tool to identify biomarkers that predict disease severity or response to therapy. Ideally, the cellular material used for transcriptomic profiling should be safe and easy to collect so that it can be readily sampled before and after the initiation of therapy. Although peripheral blood fulfils these criteria, transcriptomic analyses of peripheral blood mononuclear cells have not generated markers of disease outcome with sufficient predictive accuracy to be adopted into clinical practice (54). To determine the feasibility of transcriptomic profiling in tissue samples obtained during routine patient care, we performed single-cell RNA-Seq analysis on tissue obtained from a bronchoscopic cryobiopsy (55). Alternatively, single-cell analysis of alveolar macrophages is attractive from a clinical perspective, because large numbers of cells can be safely and repeatedly sampled using bronchoscopic alveolar lavage, even in patients who are ill. Our analysis of alveolar macrophages suggests that single-cell RNA-Seq identifies a distinct population of alveolar macrophages with higher expression of profibrotic genes, and accurately reveals differences in fibrotic gene expression in macrophages from different patients. Single-cell analyses that combine sequencing with imaging of lung biopsies or alveolar macrophages from a larger number of patients might therefore identify patient-specific markers that could inform a personalized approach to therapy.
A particular strength of our study was the availability of small 1- to 2-cm-sized biopsies from the donor lung. Although these samples still had limitations (such as overrepresentation of the distal lung parenchyma and corresponding paucity of large airways), they were likely as close to normal as could be pragmatically obtained. Because pulmonary fibrosis is a temporally and spatially heterogenous disease, some investigators suggest the use of “normal” regions from the fibrotic lung as controls. We abandoned this approach because we were unable to identify lung regions free of disease in the explanted lungs. Furthermore, this approach is problematic, because investigators have recently observed changes in gene expression in tissue distant from areas of severe fibrosis (56). Similar observations have been made in lung tissue distant from radiographically localized lung cancers (57).
There are also limitations to our study that highlight the need for further work to optimize and expand single-cell RNA-Seq datasets for pulmonary fibrosis and other diseases. First, although our single-cell RNA-Seq dataset is the largest in the literature to date, it includes a relatively small number of donors and patients with pulmonary fibrosis. Even in this small cohort, however, we were able to identify many of the same genes that we detected in flow cytometry–sorted cell populations from an independent cohort of patients, and many of the genes identified in the literature. Second, the lung is a complex tissue, consisting of at least 40 cell types tightly embedded into extracellular matrix, and both the cellular composition and matrix characteristics change during disease (9). Isolation of intact cell populations faithfully representing the composition of the lung tissue via current enzymatic digestion protocols is therefore not feasible. Alternative approaches, such as single-nucleus RNA-Seq, will likely be necessary to address this limitation (58, 59). Third, computational approaches for analyzing single-cell RNA-Seq datasets, unbiased identification of the cell types, and integrative analysis are in their infancy. As such, they are heavily biased by the experimentalists’ familiarity with the biology of the systems being studied, and are not yet adapted to distinguish and correct for batch effect or ambient RNA because of sample processing from true biologic effect associated with inter-subject variability or disease conditions. The analyses and annotations of cell types presented here are not exhaustive, and will likely be improved in the future, particularly with the help of computational tools developed by research consortia, such as Human Cell Atlas (60). Our findings suggest that the generation of larger datasets from normal and diseased samples, coupled with spatial information from RNA in situ hybridization and immunohistochemistry will drive improvements in these tools, and expand the current atlas of the cellular types and states.
In conclusion, single-cell RNA sequencing of normal and fibrotic lung tissue reveals shared and distinct patterns of fibrotic gene expression in individual cell populations and emphasizes the importance of intracellular communication in disease pathobiology. Single-cell RNA-Seq analysis offers promise for the discovery of multicellular pathways important for disease pathogenesis, and for the generation or exclusion of hypotheses regarding the function of distinct cell populations that emerge during disease. Single-cell RNA-Seq can be performed on lung samples from patients with early disease. These technologies may therefore be used clinically to identify patients most likely to benefit from targeted therapy, and to monitor their response over the course of disease.
Supplementary Material
Acknowledgments
Acknowledgment
Next-generation sequencing on the Illumina HiSeq 4000 was performed by the NUSeq Core Facility, which is supported by the Northwestern University Center for Genetic Medicine, Feinberg School of Medicine, and Shared and Core Facilities of the University’s Office for Research. Northwestern University Flow Cytometry Facility, Center for Advanced Microscopy, and Pathology Core Facility are supported by NCI Cancer Center Support Grant P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center. This research was supported in part through the computational resources and staff contributions provided by the Genomics Computing Cluster (Genomic Nodes on Quest), which is jointly supported by the Feinberg School of Medicine, the Center for Genetic Medicine, and Feinberg’s Department of Biochemistry and Molecular Genetics, the Office of the Provost, the Office for Research, and Northwestern Information Technology. The Genomics Computing Cluster is part of Quest, Northwestern University’s high-performance computing facility, with the purpose to advance research in genomics. This publication is part of the Human Cell Atlas (www.humancellatlas.org/publications).
Footnotes
P.A.R. is supported by Northwestern University’s Lung Sciences Training Program 5T32HL076139-13 and 1F32HL136111-01A1. J.M.W. is supported by Northwestern University’s Lung Sciences Training Program 5T32HL076139-14 and Dixon Translational Research Grant. D.R.W. is supported by the Northwestern Memorial Foundation Dixon Award, the Arthritis National Research Foundation, the American Lung Association, the Scleroderma Foundation, and ATS Foundation/Mallinckrodt Pharmaceuticals Inc. Research Fellowship. S.H. is supported by Northwestern University’s Lung Sciences Training Program 5T32HL076139. C.A.B. is funded by HL119995. R.B.H. is supported by AR066579 and ATS Foundation Unrestricted Grant. R.D.G. is supported by HL125910. G.M.M. is supported by NIH grants ES015024, ES025644, and ES0236718. A.B. is supported by NIH grant HL125940 and matching funds from Thoracic Surgery Foundation, research grant from Society of University Surgeons, and John H. Gibbon Jr. Research Scholarship from American Association of Thoracic Surgery. M.J. is supported by The Veterans Administration grant BX000201. B.D.S. is supported by HL128867 and the Parker B. Francis Research Opportunity Award. J.I.S. is supported by NIH grants AG049665, HL048129, HL071643, and HL085534. K.M.R. is supported by NIH grants HL128194, HL071643, and AG049665. H.P. is supported by NIH grants AR064546, HL134375, AG049665, and UH2AR067687 and the United States-Israel Binational Science Foundation (2013247), the Rheumatology Research Foundation (Agmt 05/06/14), Mabel Greene Myers Professor of Medicine, and generous donations to the Rheumatology Precision Medicine Fund. C.J.G. and A.P.L. are supported by NIH grant HL143800. A.P.L. is supported by NIH HL127245, Scleroderma Foundation, and Respiratory Health Association grants. G.R.S.B. is supported by NIH grants ES013995, HL071643, AG049665, the Veterans Administration Grant BX000201, and Department of Defense grant PR141319. A.V.M. is supported by NIH grants HL135124 and AI135964 and Department of Defense grant PR141319. This work was supported by the Office of the Assistant Secretary of Defense for Health Affairs, through the Peer Reviewed Medical Research Program under Award W81XWH-15-1-0215 (G.R.S.B. and A.V.M.). Opinions, interpretations, conclusions, and recommendations are those of the author and are not necessarily endorsed by the Department of Defense. Flow cytometry cell sorting was performed on a BD FACSAria SORP system, purchased through the support of NIH 1S10OD011996-01. Multiphoton microscopy was performed on a Nikon A1R multiphoton microscope, acquired through the support of NIH 1S10OD010398-01.
Author Contributions: P.A.R. contributed to conceptualization, methodology, software, validation, formal analysis, investigation, data curation, writing, visualization, and supervision. J.M.W. contributed to methodology, formal analysis, investigation, data curation, writing, and visualization. N.J. contributed to methodology, formal analysis, investigation, visualization, and writing. K.R.A. contributed to methodology, software, formal analysis, data curation, and visualization. A.C.M.-P. contributed to methodology, validation, formal analysis, investigation, data curation, and writing. S.C. contributed to methodology, investigation, formal analysis, and data curation. R.F. and M.A. contributed to methodology, formal analysis, and data curation. C.-IC. contributed to methodology, validation, and investigation. Z.R. contributed to methodology, software, validation, formal analysis, and visualization. R.V. contributed to methodology, software, validation, and visualization. H.A.-V. contributed to investigation, resources, data analysis, data curation, and project administration. K.N. and M.C. contributed to methodology, investigation, and data curation. D.R.W. contributed to methodology, software, validation, formal analysis, data curation, writing, and visualization. S.H. contributed to methodology, investigation, and data curation. F.J.G.-G., S.S., T.T.N., and S.A.M. contributed to methodology and investigation. S.W. and K.J.N.W. contributed to methodology, validation, formal analysis, and investigation. A.S.F. contributed to methodology, validation, formal analysis, investigation, and vizualization. C.L.H., R.D.G., C.A.B., R.B., R.B.H., A.C.A., C.T.G., J.D., and A.P.L. contributed to methodology, validation, formal analysis, investigation, and data curation. M.J. contributed to conceptualization, data curation, writing, and supervision. B.D.S. contributed to methodology, validation, formal analysis, writing, and visualization. K.M.R. contributed to conceptualization, resources, writing, supervision, and project administration. C.J.G. contributed to conceptualization, methodology, validation, formal analysis, investigation, resources, writing, supervision, visualization, and project administration. A.V.Y. contributed to methodology, investigation, and formal analysis. V.K.M. contributed to methodology, validation, investigation, and data curation. M.H. contributed to resources, data curation, writing, and supervision. A.I.S. contributed to conceptualization, methodology, validation, formal analysis, investigation, resources, and data curation. G.M.M. contributed to conceptualization, methodology, validation, formal analysis, investigation, resources, data curation, writing, visualization, supervision, and project administration. L.A.N.A. contributed to conceptualization, software, writing, and supervision. A.S. contributed to methodology and resources. H.P., G.R.S.B., and A.V.M. contributed to conceptualization, methodology, validation, formal analysis, investigation, resources, data curation, writing, visualization, supervision, project administration, and funding acquisition. J.I.S. contributed to conceptualization, resources, data curation, writing, supervision, and project administration. A.B. and S.M.B. contributed to resources, data curation, writing, supervision, and project administration.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201712-2410OC on December 15, 2018
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Lederer DJ, Martinez FJ. Idiopathic pulmonary fibrosis. N Engl J Med. 2018;378:1811–1823. doi: 10.1056/NEJMra1705751. [DOI] [PubMed] [Google Scholar]
- 2.Richeldi L. Idiopathic pulmonary fibrosis: current challenges and future perspectives. Eur Respir Rev. 2013;22:103–105. doi: 10.1183/09059180.00001413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez FJ, Chisholm A, Collard HR, Flaherty KR, Myers J, Raghu G, et al. The diagnosis of idiopathic pulmonary fibrosis: current and future approaches. Lancet Respir Med. 2017;5:61–71. doi: 10.1016/S2213-2600(16)30325-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hutchinson J, Fogarty A, Hubbard R, McKeever T. Global incidence and mortality of idiopathic pulmonary fibrosis: a systematic review. Eur Respir J. 2015;46:795–806. doi: 10.1183/09031936.00185114. [DOI] [PubMed] [Google Scholar]
- 5.Collard HR, Chen SY, Yeh WS, Li Q, Lee YC, Wang A, et al. Health care utilization and costs of idiopathic pulmonary fibrosis in US Medicare beneficiaries aged 65 years and older. Ann Am Thorac Soc. 2015;12:981–987. doi: 10.1513/AnnalsATS.201412-553OC. [DOI] [PubMed] [Google Scholar]
- 6.White ES, Borok Z, Brown KK, Eickelberg O, Guenther A, Jenkins RG, et al. American Thoracic Society Respiratory Cell, Molecular Biology Assembly Working Group on Pulmonary Fibrosis. An American Thoracic Society official research statement: future directions in lung fibrosis research. Am J Respir Crit Care Med. 2016;193:792–800. doi: 10.1164/rccm.201602-0254ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolters PJ, Blackwell TS, Eickelberg O, Loyd JE, Kaminski N, Jenkins G, et al. Time for a change: is idiopathic pulmonary fibrosis still idiopathic and only fibrotic? Lancet Respir Med. 2018;6:154–160. doi: 10.1016/S2213-2600(18)30007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.GTEx Consortium. Human genomics: the Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franks TJ, Colby TV, Travis WD, Tuder RM, Reynolds HY, Brody AR, et al. Resident cellular components of the human lung: current knowledge and goals for research on cell phenotyping and function. Proc Am Thorac Soc. 2008;5:763–766. doi: 10.1513/pats.200803-025HR. [DOI] [PubMed] [Google Scholar]
- 10.Bauer Y, Tedrow J, de Bernard S, Birker-Robaczewska M, Gibson KF, Guardela BJ, et al. A novel genomic signature with translational significance for human idiopathic pulmonary fibrosis. Am J Respir Cell Mol Biol. 2015;52:217–231. doi: 10.1165/rcmb.2013-0310OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim SY, Diggans J, Pankratz D, Huang J, Pagan M, Sindy N, et al. Classification of usual interstitial pneumonia in patients with interstitial lung disease: assessment of a machine learning approach using high-dimensional transcriptional data. Lancet Respir Med. 2015;3:473–482. doi: 10.1016/S2213-2600(15)00140-X. [DOI] [PubMed] [Google Scholar]
- 12.Kusko RL, Brothers JF, II, Tedrow J, Pandit K, Huleihel L, Perdomo C, et al. Integrated genomics reveals convergent transcriptomic networks underlying chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2016;194:948–960. doi: 10.1164/rccm.201510-2026OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selman M, Pardo A, Barrera L, Estrada A, Watson SR, Wilson K, et al. Gene expression profiles distinguish idiopathic pulmonary fibrosis from hypersensitivity pneumonitis. Am J Respir Crit Care Med. 2006;173:188–198. doi: 10.1164/rccm.200504-644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paul F, Arkin Y, Giladi A, Jaitin DA, Kenigsberg E, Keren-Shaul H, et al. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell. 2015;163:1663–1677. doi: 10.1016/j.cell.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 15.Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356:eaah4573. doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reyfman PA, Walter JM, Joshi NR, Anekalla KR, Quattie-Pimentel ACM, Chiu S, et al. Integrated single-cell transcriptomic analysis of human lung reveals distinct patterns of intercellular heterogeneity in idiopathic pulmonary fibrosis [abstract] Am J Respir Crit Care Med. 2018;197:A2286. [Google Scholar]
- 17.Walter JM, Reyfman PA, Joshi N, Anekalla KR, Quattie-Pimentel ACM, Chiu S, et al. Single-cell RNA sequencing of a transbronchial cyrobiopsy identifies unique cell populations in a patient with pulmonary fibrosis [abstract] Am J Respir Crit Care Med. 2018;197:A4352. [Google Scholar]
- 18.Ardini-Poleske ME, Clark RF, Ansong C, Carson JP, Corley RA, Deutsch GH, et al. LungMAP: the molecular atlas of lung development program. Am J Physiol Lung Cell Mol Physiol. 2017;313:L733–L740. doi: 10.1152/ajplung.00139.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heng TS, Painter MW Immunological Genome Project Consortium. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 20.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The Gene Ontology Consortium. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017;45:D331–D338. doi: 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eden E, Lipson D, Yogev S, Yakhini Z. Discovering motifs in ranked lists of DNA sequences. PLoS Comput Biol. 2007;3:e39. doi: 10.1371/journal.pcbi.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rouillard AD, Gundersen GW, Fernandez NF, Wang Z, Monteiro CD, McDermott MG, et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) 2016;2016:baw100. doi: 10.1093/database/baw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis AP, Grondin CJ, Johnson RJ, Sciaky D, King BL, McMorran R, et al. The Comparative Toxicogenomics Database: update 2017. Nucleic Acids Res. 2017;45:D972–D978. doi: 10.1093/nar/gkw838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young MD, Behjati S. SoupX removes ambient RNA contamination from droplet based single cell RNA sequencing data [preprint]. bioRxiv; 2018 [accessed 2018 Nov 10]; Available from: https://www.biorxiv.org/content/10.1101/303727v1. [Google Scholar]
- 29.Wynn TA. Integrating mechanisms of pulmonary fibrosis. J Exp Med. 2011;208:1339–1350. doi: 10.1084/jem.20110551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nabhan AN, Brownfield DG, Harbury PB, Krasnow MA, Desai TJ. Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science. 2018;359:1118–1123. doi: 10.1126/science.aam6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan KS, Janda CY, Chang J, Zheng GXY, Larkin KA, Luca VC, et al. Non-equivalence of Wnt and R-spondin ligands during Lgr5+ intestinal stem-cell self-renewal. Nature. 2017;545:238–242. doi: 10.1038/nature22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zepp JA, Zacharias WJ, Frank DB, Cavanaugh CA, Zhou S, Morley MP, et al. Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell. 2017;170:1134–1148. doi: 10.1016/j.cell.2017.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 34.Misharin AV, Morales-Nebreda L, Reyfman PA, Cuda CM, Walter JM, McQuattie-Pimentel AC, et al. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J Exp Med. 2017;214:2387–2404. doi: 10.1084/jem.20162152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCubbrey AL, Barthel L, Mohning MP, Redente EF, Mould KJ, Thomas SM, et al. Deletion of c-FLIP from CD11b(hi) macrophages prevents development of bleomycin-induced lung fibrosis. Am J Respir Cell Mol Biol. 2018;58:66–78. doi: 10.1165/rcmb.2017-0154OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics: tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 38.Zhou Y, Peng H, Sun H, Peng X, Tang C, Gan Y, et al. Chitinase 3-like 1 suppresses injury and promotes fibroproliferative responses in mammalian lung fibrosis. Sci Transl Med. 2014;6:240ra276. doi: 10.1126/scitranslmed.3007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu M, Schneider DJ, Mayes MD, Assassi S, Arnett FC, Tan FK, et al. Osteopontin in systemic sclerosis and its role in dermal fibrosis. J Invest Dermatol. 2012;132:1605–1614. doi: 10.1038/jid.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci USA. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zacharias WJ, Frank DB, Zepp JA, Morley MP, Alkhaleel FA, Kong J, et al. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature. 2018;555:251–255. doi: 10.1038/nature25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campisi J. Cellular senescence and lung function during aging: yin and yang. Ann Am Thorac Soc. 2016;13:S402–S406. doi: 10.1513/AnnalsATS.201609-703AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romero Y, Bueno M, Ramirez R, Álvarez D, Sembrat JC, Goncharova EA, et al. mTORC1 activation decreases autophagy in aging and idiopathic pulmonary fibrosis and contributes to apoptosis resistance in IPF fibroblasts. Aging Cell. 2016;15:1103–1112. doi: 10.1111/acel.12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yanai H, Shteinberg A, Porat Z, Budovsky A, Braiman A, Ziesche R, et al. Cellular senescence-like features of lung fibroblasts derived from idiopathic pulmonary fibrosis patients. Aging (Albany NY) 2015;7:664–672. doi: 10.18632/aging.100807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hecker L, Logsdon NJ, Kurundkar D, Kurundkar A, Bernard K, Hock T, et al. Reversal of persistent fibrosis in aging by targeting Nox4-Nrf2 redox imbalance. Sci Transl Med. 2014;6:231ra247. doi: 10.1126/scitranslmed.3008182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hernandez-Segura A, de Jong TV, Melov S, Guryev V, Campisi J, Demaria M. Unmasking transcriptional heterogeneity in senescent cells. Curr Biol. 2017;27:2652–2660. doi: 10.1016/j.cub.2017.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagendran M, Riordan DP, Harbury PB, Desai TJ. Automated cell-type classification in intact tissues by single-cell molecular profiling. Elife. 2018;7:e30510. doi: 10.7554/eLife.30510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, Birket SE, et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature. 2018;560:319–324. doi: 10.1038/s41586-018-0393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zepp JA, Zacharias WJ, Frank DB, Cavanaugh CA, Zhou S, Morley MP, et al. Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell. 2017;170:1134–1148. doi: 10.1016/j.cell.2017.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torre E, Dueck H, Shaffer S, Gospocic J, Gupte R, Bonasio R, et al. Rare cell detection by single-cell RNA sequencing as guided by single-molecule RNA FISH. Cell Syst. 2018;6:171–179. doi: 10.1016/j.cels.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Konishi K, Gibson KF, Lindell KO, Richards TJ, Zhang Y, Dhir R, et al. Gene expression profiles of acute exacerbations of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2009;180:167–175. doi: 10.1164/rccm.200810-1596OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pardo A, Gibson K, Cisneros J, Richards TJ, Yang Y, Becerril C, et al. Up-regulation and profibrotic role of osteopontin in human idiopathic pulmonary fibrosis. PLoS Med. 2005;2:e251. doi: 10.1371/journal.pmed.0020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gonzalez RF, Allen L, Gonzales L, Ballard PL, Dobbs LG. HTII-280, a biomarker specific to the apical plasma membrane of human lung alveolar type II cells. J Histochem Cytochem. 2010;58:891–901. doi: 10.1369/jhc.2010.956433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lam AP, Herazo-Maya JD, Sennello JA, Flozak AS, Russell S, Mutlu GM, et al. Wnt coreceptor Lrp5 is a driver of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2014;190:185–195. doi: 10.1164/rccm.201401-0079OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poletti V, Ravaglia C, Tomassetti S. Transbronchial cryobiopsy in diffuse parenchymal lung diseases. Curr Opin Pulm Med. 2016;22:289–296. doi: 10.1097/MCP.0000000000000272. [DOI] [PubMed] [Google Scholar]
- 56.Luzina IG, Salcedo MV, Rojas-Pena ML, Wyman AE, Galvin JR, Sachdeva A, et al. Transcriptomic evidence of immune activation in macroscopically normal-appearing and scarred lung tissues in idiopathic pulmonary fibrosis. Cell Immunol. 2018;325:1–13. doi: 10.1016/j.cellimm.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silvestri GA, Vachani A, Whitney D, Elashoff M, Porta Smith K, Ferguson JS, et al. A bronchial genomic classifier for the diagnostic evaluation of lung cancer. N Engl J Med. 2015;373:243–251. doi: 10.1056/NEJMoa1504601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Habib N, Li Y, Heidenreich M, Swiech L, Avraham-Davidi I, Trombetta JJ, et al. Div-Seq: single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science. 2016;353:925–928. doi: 10.1126/science.aad7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods. 2017;14:955–958. doi: 10.1038/nmeth.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, et al. The human cell atlas. Elife. 2017;6:e27041. doi: 10.7554/eLife.27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.