Abstract
Skeletal muscle is one of the major organs responsible for body movements and metabolism making up approximately 40% of the total body mass. During aging, skeletal muscle exhibits a degenerative age-associated decline in mass and function termed sarcopenia. This age-associated dysfunction of skeletal muscle is a major criterion of morbidity, mortality, and overall declines of quality of life in the elderly people. Therefore, researchers have focused on identifying modulators of muscle aging process including messenger RNAs, proteins, and recently small noncoding RNAs such as microRNAs (miRNAs). In particular, miRNAs have been demonstrated to play a critical role in skeletal muscle development and homeostasis. Recent studies revealed that miRNAs were also involved in muscle aging processes and the rejuvenation of aged muscle by regulating important molecules and pathways of aging including insulin-like growth factors, nicotine-adenine dinucleotide (+)-dependent protein deacetylase sirtuin-1, telomerase reverse transcriptase, and transforming growth factor-β signaling pathway. Over the years, miRNAs have emerged as promising candidates for biomarkers of sarcopenia and targets for interventions to slow muscle aging. Here, we comprehensively review the current knowledge on the role of miRNAs in skeletal muscle aging and highlight their potential as biomarkers or therapeutic targets for skeletal muscle health.
Keywords: MicroRNA, Skeletal muscle, Sarcopenia, Muscle aging, Muscle stem cells
As we age, many organs show degenerative phenotypes with morphological abnormalities and functional declines. Skeletal muscle also undergoes an inevitable degeneration such as loss of muscle mass, strength, and function during aging (1). The age-related muscle degeneration is known as sarcopenia, which can be triggered by several factors such as chronic inflammation, oxidative stress, motor neuron disorder, and abnormalities of muscle stem cells (2). Recent studies reported that sarcopenia might affect up to 50% people aged 80 years and older (3). Muscle mass started to decrease by about 1% every year from 30 years of age, and this tendency of muscle loss accelerates after the age of 70 years (4). Sarcopenia can cause not only physical disabilities but also several age-related diseases such as cardiovascular disorders, diabetes, and hypertension (5). Patients having sarcopenia are approximately four times more likely to develop cardiovascular diseases (6). Recent studies also showed that abnormalities of aged muscle stem cells are one of the major underlying causes of sarcopenia. These cells exhibit loss of their myogenic and self-renewal capacity with age, resulting in reduced regenerative potential and senescence (7).
MicroRNAs (miRNAs), which are small noncoding RNAs, have been demonstrated to play important roles in multiple biological processes including cell proliferation and metabolic homeostasis (8). miRNAs are initially transcribed as primary miRNA transcripts (pri-miRNAs) that are then cleaved by Drosha, an RNase III enzyme, into approximately 70 nt precursor miRNAs (pre-miRNAs). Pre-miRNAs are exported from nucleus via the nuclear transport protein Exportin 5. In the cytoplasm, pre-miRNAs are further cleaved by the enzyme Dicer resulting in miRNA duplexes, which harbor the mature miRNA strand. Mature miRNAs are then loaded into an RNA-induced silencing complex containing Argonaute. Mature miRNAs primarily regulate their targets by binding to complementary sites within the 3′ UTR region of protein coding genes, leading to translational repression or messenger RNA deadenylation and decay. Each individual miRNA can bind to several hundreds of target genes, hence miRNAs act as powerful posttranscriptional regulators of gene expression.
Recently, miRNAs have been also identified as the critical regulators of skeletal muscle proliferation, differentiation, and apoptosis at the cellular level (9) as well as in vivo in skeletal muscle atrophy/hypertrophy and disuse models (10). Furthermore, human studies examining miRNA expression in elderly individuals have demonstrated that miRNAs may play a role in the age-related changes of skeletal muscle (11). In this review, we aim to provide the current knowledge on the role of miRNA in muscle aging from the discovery of age-related miRNAs in skeletal muscle to the role of miRNAs in regulating development and homeostasis of muscle fibers and stem cells. In addition, we highlight the potential of miRNAs as biomarkers or therapeutic targets of muscle aging.
Discovery of Age-associated miRNAs in Skeletal Muscle
Increasing evidence has shown that miRNAs are differentially expressed in skeletal muscle with age (Table 1). Hamrick et al. (12) have profiled miRNAs in quadriceps muscle of young (aged 12 months, n = 24) and old (aged 24 months, n = 24) mice using TaqMan miRNA array. It was found that a total of 57 miRNAs were significantly changed in expression in quadriceps muscle tissues of aged mice compared with young mice. Among them, 36 miRNAs were significantly decreased whereas 21 miRNAs were significantly increased in aged muscle compared to young muscle. In this study, the age-related upregulation of miR-206, miR-7, miR-542, miR-468, and miR-698 and the age-related downregulation of miR-181a, miR-434, miR-382, miR-455, miR-124a, and miR-221 were validated by quantitative reverse transcription polymerase chain reaction (qRT-PCR) (12). Recently, Kim et al. (13) also reported genome-wide miRNA profiles in gastrocnemius muscle from young (aged 6 months, n = 6) and old (aged 24 months, n = 6) mice using next-generation sequencing. In this study, 34 miRNAs were found to be differentially expressed with age, among which miR-34a-5p, miR-146a-5p, miR-92b-3p, miR-155-5p, and miR-203-3p were validated to be upregulated whereas miR-337-3p*, miR-434-3p, miR-434-5p*, miR-136-5p, and miR-148a-3p were validated to be downregulated with age by qRT-PCR. Interestingly, approximately 50% of the downregulated miRNAs are located as a cluster in the Dlk1-Dio3 imprinted genomic region on mouse distal chromosome 12 although whether these miRNAs in the cluster are involved in muscle function needs to be further investigated. In rhesus monkeys, miRNAs were profiled in skeletal muscle tissues from young (aged 6 years, n = 4) and old (aged 26.8 years, n = 4) animals using next-generation sequencing (4). The authors found 35 differentially expressed miRNAs in old rhesus monkeys compared to young rhesus monkeys. Interestingly, the majority of miRNAs including miR-451, miR-144, miR-18a, and miR-15a were upregulated, whereas only five miRNAs such as miR-181a and miR-181b were downregulated in old monkeys compared to young monkeys. In humans, miRNA profiles of muscle tissues from young (31 ± 2 years, n = 19) and old (73 ± 3 years, n = 17) men using miRNA array were reported (11). It was found that 18 miRNAs were differentially expressed in old, adult skeletal muscle, among which eight miRNAs (let-7a, let-7b, let-7e, and let-7f, and miR-25, miR-98, miR-195, and miR-1268) were upregulated and 10 miRNAs (miR-22, miR-24, miR-27a, miR-27b, miR-30d, miR-133a, miR-133b, miR-223, miR-378, and miR-378*) were downregulated in skeletal muscle tissues of old adults compared to those of young adults. Particularly, let-7b and let-7e were validated by qRT-PCR.
Table 1.
miRNAs validated from profiling studies on skeletal muscle aging
| Validated miRNAs | Tools and models | References | |
|---|---|---|---|
| Upregulated | Downregulated | ||
| miR-206 miR-7 miR-542 miR-468 miR-698 |
miR-181a miR-434 miR-382 miR-455 miR-124a miR-221 |
TaqMan miRNA array Mouse quadriceps muscle Young (aged 12 months) and old (aged 24 months) N = 24 |
(12) |
| miR-34a-5p miR-146a-5p miR-92b-3p miR-155-5p miR-203-3p |
miR-127-3p miR-337-3p miR-434-3p miR-434-5p miR-136-5p miR-148a-3p |
Next generation sequencing Mouse gastrocnemius muscle Young (aged 6 months) and old (aged 24 months) N = 6 |
(13) |
| miR-451 miR-144 miR-18a miR-15a miR-15b |
miR-181a miR-1323 miR-653 miR-489 |
Next generation sequencing Rhesus monkeys skeletal muscle Young (aged 6 years) and old (aged 26.8 years) N = 4 |
(4) |
| let-7b let-7e |
— | miRNA array Human skeletal muscle Young (aged 31 years) and old (aged 73 years) N = 19 and 17 |
(11) |
miRNAs Regulating Myogenesis of Muscle Stem Cells Through Aging-related Pathways
One of the most obvious physical manifestations of aging can be linked to altered stem cell function. With age, the number of muscle stem cells or progenitor cells gradually decreases and their myogenic capability declines. These phenotypic changes of satellite cells are critical causal factors of sarcopenia (14). Several studies have demonstrated that both extrinsic and intrinsic factors could affect cellular homeostasis of satellite cells. Numerous studies using a parabiosis mouse model revealed that circulating factors in young serum could reverse aged phenotypes of old skeletal muscles (15). On the other hand, studies focusing on intrinsic factors in aged satellite cells demonstrated that p38 inhibitors promoted their myogenic capabilities, resulting in enhanced muscle regeneration of old skeletal muscle (16).
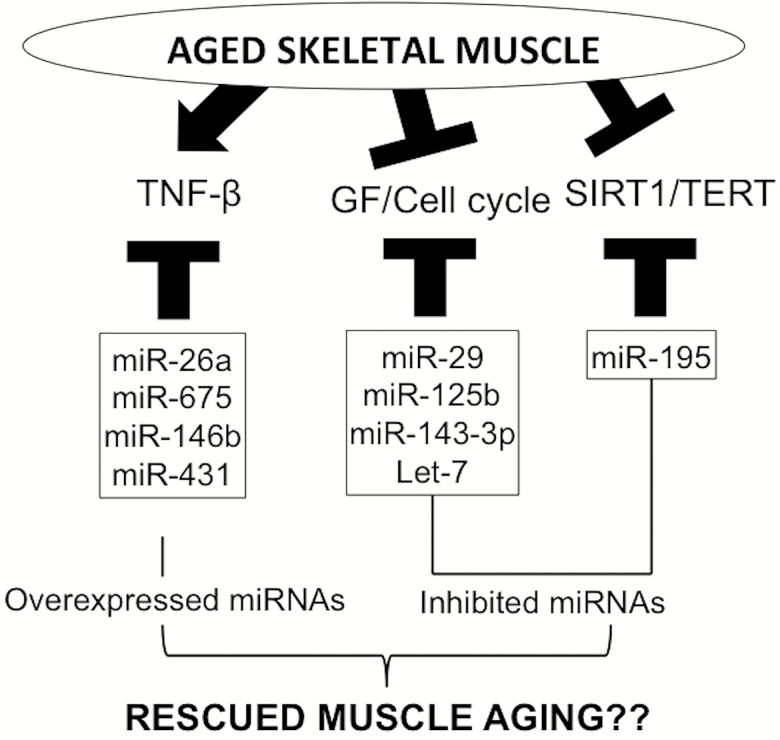
Recently, there has been increasing evidence for the role of miRNAs in muscle stem cells maintenance. Using satellite cell-specific Dicer knockout mice, Cheung et al. (17) revealed that ablation of miRNAs in muscle stem cells caused an impaired maintenance of quiescence state and induced cell death. In addition, skeletal muscle-specific knockout of Dicer causes a decrease in muscle mass and abnormal muscle formation in mice, resulting in perinatal death (18). Similarly, knockdown of Dicer gene in fully differentiated myotubes results in an atrophic phenotype, suggesting that miRNAs have an important role in muscle function (19). Furthermore, miRNAs have been reported to play a role in myogenesis by modulating the conserved genes and pathways of aging, including nicotine-adenine dinucleotide (+)-dependent protein deacetylase sirtuin-1 (SIRT1), telomerase reverse transcriptase (TERT) as well as transforming growth factor-β (TGF-β) and insulin-like growth factors (IGFs) (Figure 1).
Figure 1.
miRNAs regulating the conserved pathways of aging in skeletal muscle. During aging, TNF-β signaling pathway is excessively activated in skeletal muscle. miR-26a, miR-675, miR-146b, and miR-431 inhibit the TNF-β signaling pathway and improve the regeneration of aged skeletal muscle. In contrast, growth factors such as insulin-like growth factor-1 (IGF-1) and cell cycle regulators such as CDK6 are downregulated during the aging process of skeletal muscle. miR-29, miR-125b, miR-143-3p, and let-7 inhibit myogenesis by targeting those factors. Two well-known aging modulators, Sirt1 and Tert, are also downregulated with age and they are targeted by miR-195. These miRNAs that target the aging pathways in muscle tissues could be explored as therapeutic targets against muscle aging or aging-related muscle diseases by manipulating their expression.
TGF-β signaling has been well-known to inhibit myogenesis by downregulating the transcriptional activity of myogenic regulatory factors (20,21). Moreover, several studies have shown that TGF-β signaling delays muscle differentiation and inhibits the regeneration of aged muscle (22–25). Aged muscle tissues produce high levels of TGF-β, which in turn activates TGF-β receptors in resident muscle stem cells. Activated TGF-β receptors phosphorylate SMAD2 and SMAD3 and phosphorylated SMAD2/3 form a complex with SMAD4. These heterotrimeric complexes translocate into the nucleus and turn on the target genes that interfere with the regenerative capacity of muscle stem cells. Recently, miRNAs have been reported to be involved in this signaling pathway and thus regulate muscle differentiation and regeneration (Figure 1). Among these, miR-26a was reported to be upregulated during muscle differentiation (26). Overexpression of miR-26a promoted myogenesis, whereas inhibiting miR-26a using antisense miR-26a RNA delayed myogenesis and increased cellular proliferation, in C2C12 cells. In this study, Smad1 and Smad4, downstream effectors of TGF-β signaling, were shown to be downregulated by overexpression of miR-26a. Another study from this group has shown that miR-675, encoded in exon 1 of H19 long noncoding RNA, regulated a Smad family (27). miR-675 promoted muscle differentiation and regeneration by repression of Smad1 and Smad5, two transcription factors in the bone morphogenetic protein pathway, and repression of Cdc6, the DNA replication initiation factor. Khanna et al. (28) have reported that expression of miR-146b was induced during muscle regeneration after muscle injury. Inhibition of miR-146b prevented downregulation of Smad4, Notch1, and Hmga2 during muscle differentiation. The authors suggested that miR-146b might be a positive regulator of myogenic differentiation (28). A recent study on comparative analysis of miRNA expression profiles in young and old myoblasts has identified miR-431 as a markedly reduced miRNA in expression in aged myoblasts (29). Direct injection of miR-431 into old skeletal muscle tissues greatly improved muscle regeneration and reduced Smad4 levels. They also found that miR-431 directly interacted with the 3′ UTR of Smad4 messenger RNA, which encodes one of the downstream effectors of TGF-β signaling. The authors suggest that miR-431 plays a crucial role in maintaining the myogenic ability of aged skeletal muscle.
The satellite cell proliferation has known to be reduced with age (30) and there are studies showing that some miRNAs are involved in this process by regulating growth factors and cell cycle regulators (Figure 1). Hu et al. have found upregulation of miR-29 in muscle of aged rodents (31). Overexpression of miR-29 in muscle progenitor cells resulted in impaired proliferation with increasing SA-βgal expression, a maker of cellular senescence. miR-29 directly interacted with the 3′ UTRs of p85a and Igf-1 leading to the suppression of their translation. Direct injection of miR-29 into muscles of young mice also inhibited proliferation and induced senescence-associated proteins, showing phenotypes of aged muscle. Ge et al. (32) have shown that miR-125b regulated myogenesis by targeting Igf-2, which is an embryonic regulator of myogenesis and an autocrine factor that initiates myoblast differentiation in vitro. In this study, the expression of miR-125b was gradually decreased during myogenesis and overexpression of miR-125b suppressed myogenesis in vitro by negatively regulating Igf-2. A recent study revealed that overexpression of miR-143-3p negatively regulated myogenic differentiation of myoblasts by targeting Igfbp5 (33). The authors suggested that dysregulation of miR-143-3p might be an important cue for their aged phenotypes. Drummond et al. (34) have found that the let-7 family miRNAs, especially let-7b and let-7e, were upregulated and inhibited cellular proliferation or regeneration in aged human skeletal muscle. Using bioinformatics analysis, they identified predicted targets of let-7 involved in cell cycle regulation. Among them, cell cycle regulators (CDK6, CDC25A, and CDC34) and a myogenic transcription factor (PAX7) were negatively correlated with let-7 expression, suggesting that they are potential targets of let-7 mediating cellular outcomes in aged human skeletal muscle (34).
More recently, Kondo et al. (35) have reported that miR-195 was upregulated, whereas SIRT1 and telomerase reverse transcriptase were downregulated, in myoblasts isolated from 24-month old mice. They found that miR-195 inhibited Sirt1 expression level by directly binding on Sirt1 3′ UTR. Inhibition of miR-195 expression improved cellular reprograming in old myoblasts with restoring the expression of rejuvenation genes such as Sirt1 and Tert. The authors demonstrated that induced pluripotent stem cells from old myoblasts transduced with miR-195 inhibitor did not affect pluripotency, suggesting that the blockade of miR-195 is needed to efficiently generate induced pluripotent stem cell from old donor subjects for transplantation of induced pluripotent stem cells. Taken together, the miRNAs that target the conserved pathways of aging and thereby regulate myogenic capabilities of muscle stem cells could be potential therapeutic targets for delaying muscle aging (Figure 1).
The Role of miRNAs in Aging Human Skeletal Muscle
Emerging studies of human skeletal muscle have shown the role of aging-associated miRNAs. Zacharewicz et al. (36) have identified 26 miRNAs regulated by age, exercise, or a combination of both factors in young and old individuals before and 2 hours after exercise. They found that among those, nine miRNAs including miR-99/100 family of miRNAs were predicted to regulate genes involved in the Akt-mTOR signaling pathway, which is associated with age-related muscle wasting. Recently, Russell et al. (37) have reported that significantly deregulated striated muscle activator of Rho signaling (STARS) protein in old individuals could be regulated by miR-628-5p, which was regulated by age and exercise through the binding to the STARS 3′ UTR.
There are some studies to investigate the potential role of modulation in miRNA expression on age-associated declines in skeletal muscle anabolism in human (38). Drummond et al. (34) have reported that aging can differentially regulate human skeletal muscle miRNA expression at rest and after an anabolic stimulus of resistance exercise (RE) and essential amino acids. Particularly, they pointed out miR-1 was reduced only in the young men, whereas there was no change in older men even after exercise and ingestion of essential amino acid. Recently, Rivas et al. (39) have reported that dysregulation of miRNAs could contribute to reduced muscle plasticity with aging. They found 21 miRNAs were altered by RE in young individuals, whereas there was no change in old individuals. Particularly, miR-126, one of miRNAs not altered by RE in old individuals targeted the expression of regulators of skeletal muscle growth and activation of insulin growth factor-1 (IGF-1), suggesting that miRNAs have a role in the adaptation of muscle to anabolic stimulation.
miRNAs as Biomarkers of Muscle Function
Recently, circulating miRNAs have been recognized as potential noninvasive biomarkers due to their stability in body fluids including blood. Because a number of miRNAs are expressed in skeletal muscles, determining their circulating levels may be useful for the estimation of muscle function. Although the origin and function of circulating miRNAs are still not fully understood, recent studies have shown the correlation in the expression between circulating miRNAs and muscle-related miRNAs in muscle function by exercise training. Uhlemann et al. (40) have measured the plasma concentration of miR-133 using qRT-PCR from healthy individuals performing one of different exercise forms including maximal symptom-limited exercise, bicycling for 4 hours, running a marathon, and RE. Plasma miR-133 levels were increased during recovery after eccentric resistance training and marathon running but were unchanged following a maximal exercise test or prolonged bicycling, and this was correlated with creatine phosphokinase activity, a marker of muscle damage, suggesting that miR-133 level could be an indicator of the muscle damage (40). Another human study observed that circulating levels of muscle specific miRNAs, miR-1, miR-133, miR-133b, and miR-208b, were increased between 2 hour and 6 hour after muscle damaging eccentric exercise compared to concentric exercise, suggesting that these miRNAs could potentially be alternative biomarkers of muscle damage (41). Aoi et al. (42) have measured muscle-enriched miRNAs in human serum after acute and chronic aerobic exercise and found that the circulating level of miR-486 was decreased by both acute and chronic exercise. This reduction was negatively correlated with the maximum oxygen uptake (VO2max) for each subject, suggesting that miR-486 may be associated with metabolic changes during exercise and adaptation induced by training (42). One recent study conducted by Margolis et al. (38,43) has shown that age-associated circulating miRNAs could be predictive of anabolic resistance. They found that the expression levels of miR-19b-3p, miR-206, and miR-486 were altered with age and associated with body composition and metabolic health. Furthermore, in this study, they applied Ingenuity Pathway Analysis and found circulating miRNA expression was predictive of acute adaptations to RE, where circulating miRNA expression was associated with altered phosphorylation status of upstream and downstream targets of mTORC1.
The circulating levels of several muscle-enriched miRNAs are also altered in muscle disorders (44). Serum levels of several muscle-enriched miRNAs such as miR-1, miR-133, and miR-206 are elevated in Duchenne muscular dystrophy in humans and animals (45). In addition to these miRNAs, other candidate circulating miRNAs, such as miR-378 and miR-31, have been shown to associate with dystrophy in a recent human study (46). Furthermore, circulating levels of a muscle-enriched miRNA, miR-499, were directly associated with strength and quadriceps type I fiber proportion in patients with chronic obstructive pulmonary disease (47). Levels of muscle-specific miRNAs (miR-1, miR-133a, miR-133b, and miR-206) were also higher in the serum of patients with rhabdomyosarcoma tumors than that of control subjects (44). In muscular atrophy, miR-23a was decreased and secreted into the extracellular space after being taken up by exosomes (48).
Taken together, these studies provide a strong basis for the development of circulating miRNAs as biomarkers for muscle aging. However, there is no doubt that further studies are necessary to determine the origin and function of circulating miRNAs before being used in the clinic to screen for muscle health and function in older adults. Recently, one clue has been reported about the origin of circulating miRNAs and their role on other nonadipose tissue (49). Thomou et al. have shown that ADicerKO mice, which have an adipose-tissue-specific knockout of the miRNA-processing enzyme Dicer, exhibited a substantial decrease in levels of circulating exosomal miRNAs whereas transplantation of adipose tissue into ADicerKO mice restores the level of circulating miRNAs. They also found that the serum exosomal transfer of mice expressing human-specific miRNA in the brown adipose tissue to other mice could regulate the gene expression in the liver tissue, suggesting that circulating miRNAs have a role to affect the gene expression of distant tissues like adipokines.
Potential miRNA Therapeutics Against Muscle Aging and Muscle Diseases
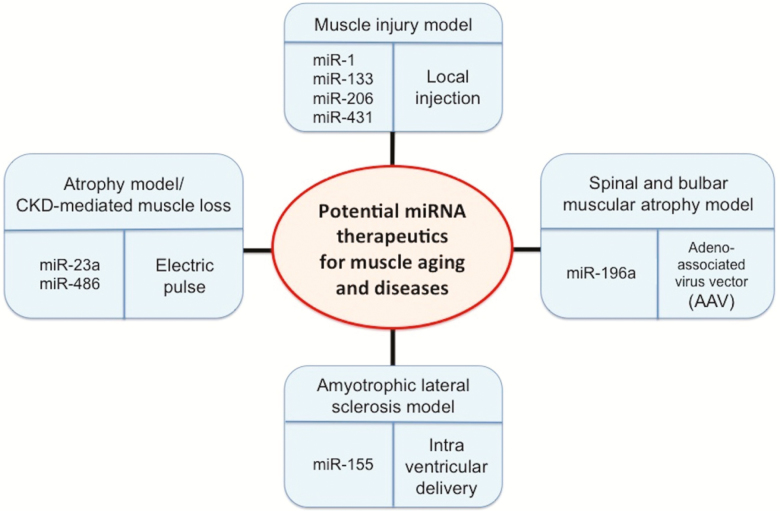
Up to date, pharmacological inhibition of myostatin has been successfully attempted in mouse models of chronic kidney disease with abnormal muscle protein metabolism and progressive muscle atrophy, and similar experiments leading to human therapy are currently under way (50). Many of the non-myomiR therapeutic intervention studies were performed on the mdx mice model as a surrogate for Duchenne muscular dystrophy and G93A-SOD1 mice model as the standard model for a familial amyotrophic lateral sclerosis, but these studies failed to translate to useful therapeutic intervention in humans (51). Given the diverse etiology of these atrophies, additional approaches are needed. Several recent studies indicate that myomiR therapy could be useful for the treatment of neuromuscular disorders (52). We reviewed recent studies that apply myomiRs to prevent muscle aging or disease phenotypes using different miRNA delivery systems (Figure 2).
Figure 2.
miRNAs as potential therapeutic targets against skeletal muscle aging and disorders. Many studies using mouse models of disease have shown that manipulation of some miRNAs such as miR-431, miR-23a, miR-196a, and miR-155 reverses aging phenotypes in muscle or protects against muscle disorders (29,53,54,56,57), suggesting that miRNAs could be potential therapeutic targets against muscle aging. In general, miRNAs that are altered in expression during muscle aging and disorders are candidates for manipulation by miRNA mimics or inhibitors to counteract the effects of aging or disorders. The candidate miRNAs can then be delivered into muscle by multiple approaches including local injection, electric pulse, intra ventricular delivery, and adeno-associated virus vector followed by studying the functional consequences in target gene expression, cellular, and organismal outcomes.
Lee et al. (29) have identified miR-431 as a downregulated miRNA in skeletal muscle during aging in mice. When they overexpressed, miR-431 improved the myogenic capacity of old myoblasts. Interestingly, they found that ectopic miR-431 injection directly to the muscle with cardiotoxin injury showed enhanced muscle regeneration and reduced SMAD4 levels, which were increased during muscle aging (29). It was suggested that age-associated miR-431 might have a role in maintaining the myogenic ability of skeletal muscle with age. Koval et al. (53) have developed oligonucleotide-based miRNA inhibitors (antisense miRNAs) that inhibit miRNAs throughout the central nervous system and in the periphery. In this study using the superoxide dismutase 1 (SOD1)G93A rodents, an amyotrophic lateral sclerosis model, the authors found a significantly extended survival by 10 days and reduced disease duration by 15 days compared to control after treating with antisense miR-155 (53). Xu et al. have reported that chronic kidney disease-induced loss of skeletal muscle was inhibited when they electroporated miR-486 into tibialis anterior muscles of chronic kidney disease mouse model (54). It was demonstrated that miR-486 protected against muscle protein degradation by targeting FoxO1 and PTEN. Nakasa et al. (55) induced the skeletal muscle injury in rat models and injected double-stranded miR-1, miR-133, and miR-206 in the surrounding area. At 1 week after injury, an injection of miRNAs was found to enhance muscle regeneration morphologically and physiologically, and to prevent fibrosis effectively compared to control with inducing expression of myogenic markers, MyoD1, myogenin and Pax7 (55). Wada et al. (56) have reported that miR-23a suppressed the translation of MAFbx/atrogin-1 and MuRF1, which are known to be prominently induced during muscle atrophy and mediate atrophy-associated protein degradation. When overexpressed using expressing vector in tibialis anterior muscle of dexamethasone-induced muscle atrophy mouse model, miR-23a counteracted muscle atrophy by inhibiting MAFbx/atrogin-1 and MuRF1 in a 3′ UTR-dependent manner (56). Furthermore, miR-23a transgenic mice had resistance against glucocorticoid-induced skeletal muscle atrophy. Miyazaki et al. (57) have reported that miR-196a expression using the adeno-associated virus vector-mediated delivery in a mouse model of spinal and bulbar muscular atrophy ameliorated spinal and bulbar muscular atrophy phenotypes by inducing the enhanced decay of the androgen receptor messenger RNA, a known causal factor of spinal and bulbar muscular atrophy, by targeting CUGBP, Elav-like family member 2 (CELF2) (57). Taken together, therapeutic potential of miRNAs is rapidly emerging. To realize this potential for clinical therapies in human, considerable hurdles remain including the efficient vehicles for the targeted delivery and increased the duration of action of miRNAs.
Summary
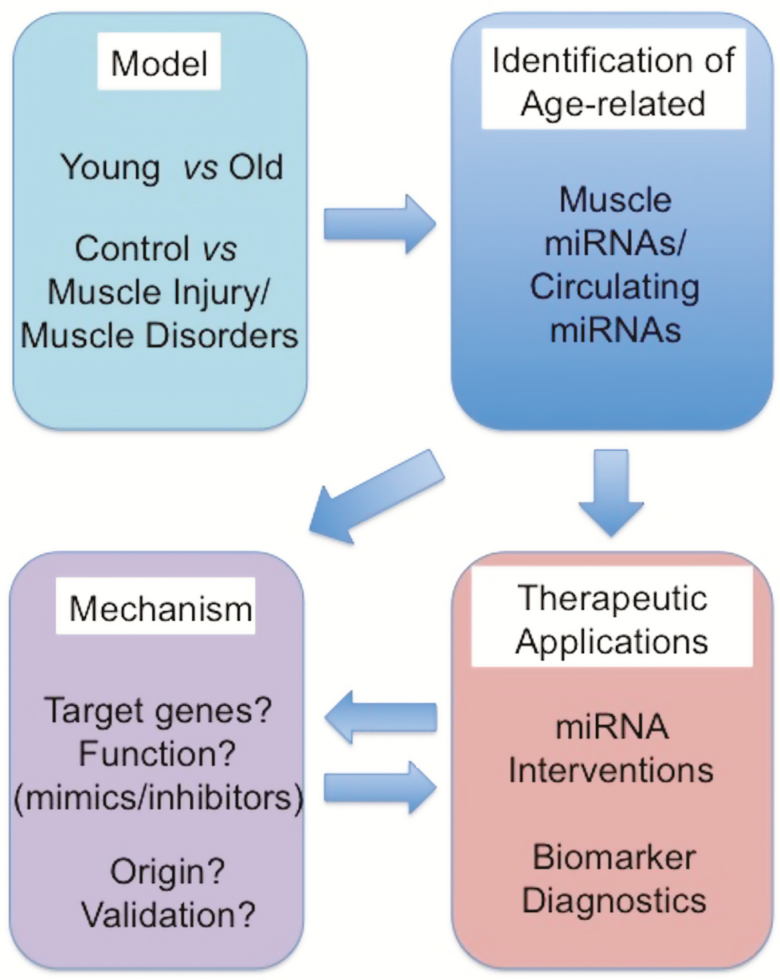
Skeletal muscle has been one of the major target tissues in aging and aging-related disease. Muscle dysfunction is one of the central aging phenotypes among elderly people significantly affecting their quality of life and health status. Many studies have been conducted to identify intrinsic and extrinsic factors of muscle aging and targets for interventions against the age-related muscle dysfunction. miRNAs have been recognized as a promising candidate. Their expression profiles are altered during aging, functionally contributing to the aging-related loss of muscle mass. In addition, miRNAs modulate the important processes of muscle cell biology including proliferation, differentiation, and stem cell renewal and maintenance by negatively regulating the expression of their target genes, many of which are involved in the conserved aging pathways. miRNAs have great potential as noninvasive biomarkers of skeletal muscle dysfunction. Furthermore, recent studies have shown that the manipulation of age-related miRNAs can restore or reverse muscle aging or disease phenotypes. To realize the full potential of miRNA as robust biomarkers for or therapeutic targets against muscle aging and aging-related muscle disorders, a large-scale validation studies have to be performed together with carefully designed functional studies using both in in vitro and in vivo systems (Figure 3).
Figure 3.
Strategy to identify age-associated muscle miRNAs and develop them as antiaging therapeutics. Age-associated muscle miRNAs are identified from aged subjects or models of muscle injury or muscle atrophy by profiling them using high-throughput technologies. From serum or plasma samples of these models, the circulating miRNAs related with aging are also identified. One identified, further functional studies are needed to elucidate the molecular mechanism underlying age- or disease-associated muscle miRNAs, which include identification of their target genes and pathways, and uncovering their biological function in in vivo and in vitro using miRNA mimics/inhibitors. Age- and disease-related circulating miRNAs can be developed as noninvasive biomarkers for diagnostics of muscle dysfunction by identifying their origin and validating in large population. Ultimately, age- or disease-related muscle miRNAs could be targeted for miRNA intervention studies against muscle aging and diseases.
Funding
This work was supported by grants from the Bio and Medical Technology Development Program (2013M3A9B6076413 to K.-S.K.) of the National Research Foundation, which is funded by the Ministry of Science, ICT and Future Planning, the Korea Research Institute of Bioscience and Biotechnology Research Initiative Program, and the National Institutes of Health grants (AG017242, GM104459, and CA180126 to Y.S.).
Conflict of interest statement
The authors have no conflict of interest to declare.
References
- 1. Nilwik R, Snijders T, Leenders M, et al. The decline in skeletal muscle mass with aging is mainly attributed to a reduction in type II muscle fiber size. Exp Gerontol. 2013;48:492–498. doi: 10.1016/j.exger.2013.02.012 [DOI] [PubMed] [Google Scholar]
- 2. Narici MV, Maffulli N. Sarcopenia: characteristics, mechanisms and functional significance. Br Med Bull. 2010;95:139–159. doi: 10.1093/bmb/ldq008 [DOI] [PubMed] [Google Scholar]
- 3. Morley JE, Anker SD, von Haehling S. Prevalence, incidence, and clinical impact of sarcopenia: facts, numbers, and epidemiology-update 2014. J Cachexia Sarcopenia Muscle. 2014;5:253–259. doi: 10.1007/s13539-014-0161-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mercken EM, Majounie E, Ding J, et al. Age-associated miRNA alterations in skeletal muscle from rhesus monkeys reversed by caloric restriction. Aging (Albany NY). 2013;5:692–703. doi: 10.18632/aging.100598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Morley JE. Sarcopenia: diagnosis and treatment. J Nutr Health Aging. 2008;12:452–456. doi: 10.1007/BF02982705 [DOI] [PubMed] [Google Scholar]
- 6. Brown JC, Harhay MO, Harhay MN. Sarcopenia and mortality among a population-based sample of community-dwelling older adults. J Cachexia Sarcopenia Muscle. 2016;7:290–298. doi: 10.1002/jcsm.12073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cosgrove BD, Gilbert PM, Porpiglia E, et al. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nat Med. 2014;20:255–264. doi: 10.1038/nm.3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jung HJ, Suh Y. Circulating miRNAs in ageing and ageing-related diseases. J Genet Genomics. 2014;41:465–472. doi: 10.1016/j.jgg.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang XH. MicroRNA in myogenesis and muscle atrophy. Curr Opin Clin Nutr Metab Care. 2013;16:258–266. doi: 10.1097/MCO.0b013e32835f81b9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jung HJ, Lee KP, Milholland B, et al. Comprehensive miRNA profiling of skeletal muscle and serum in induced and normal mouse muscle atrophy during aging. J Gerontol A Biol Sci Med Sci. 2017;72:1483–1491. doi: 10.1093/gerona/glx025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Drummond MJ, McCarthy JJ, Sinha M, et al. Aging and microRNA expression in human skeletal muscle: a microarray and bioinformatics analysis. Physiol Genomics. 2011;43:595–603. doi: 10.1152/physiolgenomics.00148.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hamrick MW, Herberg S, Arounleut P, et al. The adipokine leptin increases skeletal muscle mass and significantly alters skeletal muscle miRNA expression profile in aged mice. Biochem Biophys Res Commun. 2010;400:379–383. doi: 10.1016/j.bbrc.2010.08.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim JY, Park YK, Lee KP, et al. Genome-wide profiling of the microRNA-mRNA regulatory network in skeletal muscle with aging. Aging. 2014;6:524–544. doi: 10.18632/aging.100677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Collins CA, Zammit PS, Ruiz AP, Morgan JE, Partridge TA. A population of myogenic stem cells that survives skeletal muscle aging. Stem Cells. 2007;25:885–894. doi: 10.1634/stemcells.2006-0372 [DOI] [PubMed] [Google Scholar]
- 15. Conboy IM, Rando TA. Heterochronic parabiosis for the study of the effects of aging on stem cells and their niches. Cell Cycle. 2012;11:2260–2267. doi: 10.4161/cc.20437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perdiguero E, Ruiz-Bonilla V, Gresh L, et al. Genetic analysis of p38 MAP kinases in myogenesis: fundamental role of p38alpha in abrogating myoblast proliferation. EMBO J. 2007;26:1245–1256. doi: 10.1038/sj.emboj.7601587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheung TH, Quach NL, Charville GW, et al. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature. 2012;482:524–528. doi: 10.1038/nature10834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. O’Rourke JR, Georges SA, Seay HR, et al. Essential role for Dicer during skeletal muscle development. Dev Biol. 2007;311:359–368. doi: 10.1016/j.ydbio.2007.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hitachi K, Nakatani M, Tsuchida K. Myostatin signaling regulates Akt activity via the regulation of miR-486 expression. Int J Biochem Cell Biol. 2014;47:93–103. doi: 10.1016/j.biocel.2013.12.003 [DOI] [PubMed] [Google Scholar]
- 20. Liu D, Black BL, Derynck R. TGF-beta inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3. Genes Dev. 2001;15:2950–2966. doi: 10.1101/gad.925901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martin JF, Li L, Olson EN. Repression of myogenin function by TGF-beta 1 is targeted at the basic helix-loop-helix motif and is independent of E2A products. J Biol Chem. 1992;267:10956–10960. [PubMed] [Google Scholar]
- 22. Carlson ME, Conboy MJ, Hsu M, et al. Relative roles of TGF-beta1 and Wnt in the systemic regulation and aging of satellite cell responses. Aging Cell. 2009;8:676–689. doi: 10.1111/j.1474-9726.2009.00517.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carlson ME, Hsu M, Conboy IM. Imbalance between pSmad3 and Notch induces CDK inhibitors in old muscle stem cells. Nature. 2008;454:528–532. doi: 10.1038/nature07034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jang YC, Sinha M, Cerletti M, Dall’Osso C, Wagers AJ. Skeletal muscle stem cells: effects of aging and metabolism on muscle regenerative function. Cold Spring Harb Symp Quant Biol. 2011;76:101–111. doi: 10.1101/sqb.2011.76.010652 [DOI] [PubMed] [Google Scholar]
- 25. Gopinath SD, Rando TA. Stem cell review series: aging of the skeletal muscle stem cell niche. Aging Cell. 2008;7:590–598. doi: 10.1111/j.1474-9726.2008.00399.x [DOI] [PubMed] [Google Scholar]
- 26. Dey BK, Gagan J, Yan Z, Dutta A. miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes Dev. 2012;26:2180–2191. doi: 10.1101/gad.198085.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28:491–501. doi: 10.1101/gad.234419.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Khanna N, Ge Y, Chen J. MicroRNA-146b promotes myogenic differentiation and modulates multiple gene targets in muscle cells. PLoS One. 2014;9:e100657. doi: 10.1371/journal.pone.0100657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee KP, Shin YJ, Panda AC, et al. miR-431 promotes differentiation and regeneration of old skeletal muscle by targeting Smad4. Genes Dev. 2015;29:1605–1617. doi: 10.1101/gad.263574.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barber L, Scicchitano BM, Musaro A. Molecular and cellular mechanisms of muscle aging and sarcopenia and effects of electrical stimulation in seniors. Eur J Transl Myol. 2015;25:231–236. doi: 10.4081/ejtm.2015.5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hu Z, Klein JD, Mitch WE, Zhang L, Martinez I, Wang XH. MicroRNA-29 induces cellular senescence in aging muscle through multiple signaling pathways. Aging (Albany NY). 2014;6:160–175. doi: 10.18632/aging.100643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ge Y, Sun Y, Chen J. IGF-II is regulated by microRNA-125b in skeletal myogenesis. J Cell Biol. 2011;192:69–81. doi: 10.1083/jcb.201007165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Soriano-Arroquia A, McCormick R, Molloy AP, McArdle A, Goljanek-Whysall K. Age-related changes in miR-143-3p:Igfbp5 interactions affect muscle regeneration. Aging Cell. 2016;15:361–369. doi: 10.1111/acel.12442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Drummond MJ, McCarthy JJ, Fry CS, Esser KA, Rasmussen BB. Aging differentially affects human skeletal muscle microRNA expression at rest and after an anabolic stimulus of resistance exercise and essential amino acids. Am J Physiol Endocrinol Metab. 2008;295:E1333–E1340. doi: 10.1152/ajpendo.90562.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kondo H, Kim HW, Wang L, et al. Blockade of senescence-associated microRNA-195 in aged skeletal muscle cells facilitates reprogramming to produce induced pluripotent stem cells. Aging Cell. 2016;15:56–66. doi: 10.1111/acel.12411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zacharewicz E, Della Gatta P, Reynolds J, et al. Identification of microRNAs linked to regulators of muscle protein synthesis and regeneration in young and old skeletal muscle. PLoS One. 2014;9:e114009. doi: 10.1371/journal.pone.0114009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Russell AP, Wallace MA, Kalanon M, et al. Striated muscle activator of Rho signalling (STARS) is reduced in ageing human skeletal muscle and targeted by miR-628-5p. Acta Physiol (Oxf). 2017;220:263–274. doi: 10.1111/apha.12819 [DOI] [PubMed] [Google Scholar]
- 38. Margolis LM, Rivas DA. Potential role of microRNA in the anabolic capacity of skeletal muscle with aging. Exerc Sport Sci Rev. 2018;46:86–91. doi: 10.1249/JES.0000000000000147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rivas DA, Lessard SJ, Rice NP, et al. Diminished skeletal muscle microRNA expression with aging is associated with attenuated muscle plasticity and inhibition of IGF-1 signaling. FASEB J. 2014;28:4133–4147. doi: 10.1096/fj.14-254490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Uhlemann M, Möbius-Winkler S, Fikenzer S, et al. Circulating microRNA-126 increases after different forms of endurance exercise in healthy adults. Eur J Prev Cardiol. 2014;21:484–491. doi: 10.1177/2047487312467902 [DOI] [PubMed] [Google Scholar]
- 41. Banzet S, Chennaoui M, Girard O, et al. Changes in circulating microRNAs levels with exercise modality. J Appl Physiol (1985). 2013;115:1237–1244. doi: 10.1152/japplphysiol.00075.2013 [DOI] [PubMed] [Google Scholar]
- 42. Aoi W, Ichikawa H, Mune K, et al. Muscle-enriched microRNA miR-486 decreases in circulation in response to exercise in young men. Front Physiol. 2013;4:80. doi: 10.3389/fphys.2013.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Margolis LM, Lessard SJ, Ezzyat Y, Fielding RA, Rivas DA. Circulating MicroRNA are predictive of aging and acute adaptive response to resistance exercise in men. J Gerontol A Biol Sci Med Sci. 2017;72:1319–1326. doi: 10.1093/gerona/glw243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Miyachi M, Tsuchiya K, Yoshida H, et al. Circulating muscle-specific microRNA, miR-206, as a potential diagnostic marker for rhabdomyosarcoma. Biochem Biophys Res Commun. 2010;400:89–93. doi: 10.1016/j.bbrc.2010.08.015 [DOI] [PubMed] [Google Scholar]
- 45. Cacchiarelli D, Legnini I, Martone J, et al. miRNAs as serum biomarkers for Duchenne muscular dystrophy. EMBO Mol Med. 2011;3:258–265. doi: 10.1002/emmm.201100133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vignier N, Amor F, Fogel P, et al. Distinctive serum miRNA profile in mouse models of striated muscular pathologies. PLoS One. 2013;8:e55281. doi: 10.1371/journal.pone.0055281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Donaldson A, Natanek SA, Lewis A, et al. Increased skeletal muscle-specific microRNA in the blood of patients with COPD. Thorax. 2013;68:1140–1149. doi: 10.1136/thoraxjnl-2012-203129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hudson MB, Woodworth-Hobbs ME, Zheng B, et al. miR-23a is decreased during muscle atrophy by a mechanism that includes calcineurin signaling and exosome-mediated export. Am J Physiol Cell Physiol. 2014;306:C551–C558. doi: 10.1152/ajpcell.00266.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thomou T, Mori MA, Dreyfuss JM, et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017;542:450–455. doi: 10.1038/nature21365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang L, Rajan V, Lin E, et al. Pharmacological inhibition of myostatin suppresses systemic inflammation and muscle atrophy in mice with chronic kidney disease. FASEB J. 2011;25:1653–1663. doi: 10.1096/fj.10-176917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Scott S, Kranz JE, Cole J, et al. Design, power, and interpretation of studies in the standard murine model of ALS. Amyotroph Lateral Scler. 2008;9:4–15. doi: 10.1080/17482960701856300 [DOI] [PubMed] [Google Scholar]
- 52. Alexander MS, Kunkel LM. Skeletal muscle microRNAs: their diagnostic and therapeutic potential in human muscle diseases. J Neuromuscul Dis. 2015;2:1–11. doi: 10.3233/JND-140058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Koval ED, Shaner C, Zhang P, et al. Method for widespread microRNA-155 inhibition prolongs survival in ALS-model mice. Hum Mol Genet. 2013;22:4127–4135. doi: 10.1093/hmg/ddt261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xu J, Li R, Workeneh B, Dong Y, Wang X, Hu Z. Transcription factor FoxO1, the dominant mediator of muscle wasting in chronic kidney disease, is inhibited by microRNA-486. Kidney Int. 2012;82:401–411. doi: 10.1038/ki.2012.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nakasa T, Ishikawa M, Shi M, Shibuya H, Adachi N, Ochi M. Acceleration of muscle regeneration by local injection of muscle-specific microRNAs in rat skeletal muscle injury model. J Cell Mol Med. 2010;14:2495–2505. doi: 10.1111/j.1582-4934.2009.00898.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wada S, Kato Y, Okutsu M, et al. Translational suppression of atrophic regulators by microRNA-23a integrates resistance to skeletal muscle atrophy. J Biol Chem. 2011;286:38456–38465. doi: 10.1074/jbc.M111.271270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Miyazaki Y, Adachi H, Katsuno M, et al. Viral delivery of miR-196a ameliorates the SBMA phenotype via the silencing of CELF2. Nat Med. 2012;18:1136–1141. doi: 10.1038/nm.2791 [DOI] [PubMed] [Google Scholar]