Growth factor independence 1B (GFI1B) coordinates assembly of transcriptional repressor complexes comprised of corepressors and histone-modifying enzymes to control gene expression programs governing lineage allocation in hematopoiesis. Enforced expression of GFI1B in K562 erythroleukemia cells favors erythroid over megakaryocytic differentiation, providing a platform to define molecular determinants of binary fate decisions triggered by GFI1B.
KEYWORDS: BHC complex, Erythropoiesis, GFI1B, GSE1, HMG20A, HMG20B, hematopoiesis, LSD1, SNAG domain, transcriptional repression
ABSTRACT
Growth factor independence 1B (GFI1B) coordinates assembly of transcriptional repressor complexes comprised of corepressors and histone-modifying enzymes to control gene expression programs governing lineage allocation in hematopoiesis. Enforced expression of GFI1B in K562 erythroleukemia cells favors erythroid over megakaryocytic differentiation, providing a platform to define molecular determinants of binary fate decisions triggered by GFI1B. We deployed proteome-wide proximity labeling to identify factors whose inclusion in GFI1B complexes depends upon GFI1B’s obligate effector, lysine-specific demethylase 1 (LSD1). We show that GFI1B preferentially recruits core and putative elements of the BRAF-histone deacetylase (HDAC) (BHC) chromatin-remodeling complex (LSD1, RCOR1, HMG20A, HMG20B, HDAC1, HDAC2, PHF21A, GSE1, ZMYM2, and ZNF217) in an LSD1-dependent manner to control acquisition of erythroid traits by K562 cells. Among these elements, depletion of both HMG20A and HMG20B or of GSE1 blocks GFI1B-mediated erythroid differentiation, phenocopying impaired differentiation brought on by LSD1 depletion or disruption of GFI1B-LSD1 binding. These findings demonstrate the central role of the GFI1B-LSD1 interaction as a determinant of BHC complex recruitment to enable cell fate decisions driven by GFI1B.
INTRODUCTION
Transcriptional regulation is critical for cell growth, homeostasis, and fate determination in multicellular organisms. In hematopoiesis, cell surface receptors, signaling networks, transcription factors, and their effectors cooperate to influence chromatin state and gene expression, directing hematopoietic progenitors toward specific fates and, once these are achieved, ensuring proper function for each cell type. Failure to properly control chromatin structural dynamics governing stage-specific gene expression programs can cause qualitative and quantitative hematopoietic disorders, including malignancies. Failures can arise from altered expression of transcription factors or from malfunction or misdirection of their partners (1–3). Thus, knowing the composition of protein complexes that regulate gene expression by cell fate-specifying transcription factors is essential to understanding normal and malignant hematopoiesis and may have therapeutic relevance (4).
Growth factor independence (GFI) family transcriptional repressors GFI1 and GFI1B are master regulators of developmental hematopoiesis (5, 6). GFI1 controls hematopoietic stem cell self-renewal, B- and T-lymphopoiesis, and the granulocyte-versus-monocyte fate decision during late stages of myeloid differentiation (7–9). Notably, Gfi1-null mice are viable but are characterized by severe neutropenia with compensatory monocytosis and sensorineural hearing loss (10–12). Altered GFI1 expression is found in multiple hematologic malignancies, most notably acute myeloid leukemia (AML) and T-cell acute lymphoblastic leukemia (T-ALL) (13–17). Moreover, loss-of-function mutations in GFI1 cause severe congenital neutropenia (SCN) type 2 and nonimmune chronic idiopathic neutropenia (CIN) of adults, both of which predispose to AML (18, 19). GFI1B is required for definitive erythropoiesis and megakaryopoiesis and, like GFI1, also helps to maintain hematopoietic stem cell quiescence (20–23). In mice, Gfi1b nullizygosity is lethal at embryonic day 15 (E15) due to erythropoietic failure (24). Mutations in GFI1B are associated with both qualitative and quantitative disorders of platelets, and elevated GFI1B expression is found in AML (25–28). However, the role of GFI1B in AML is uncertain. Despite GFI1B often being increased in AML patients (29–32), a recent study suggests that low GFI1B expression correlates with poor patient outcome and that in mice, Gfi1b depletion accelerates progression to AML from myelodysplasia driven by a NUP98-HOXD13 fusion protein (33). Collectively, these observations suggest widespread contributions by GFI family proteins to lineage allocation during normal hematopoiesis and both oncogenic and oncosuppressor roles in hematologic malignancies. A clear understanding of GFI1/1B contributions to hematopoiesis and malignancy requires a comprehensive accounting of their protein partners and their dependency relationships during cell fate decision making.
GFI family proteins feature a shared structural organization (5, 34). Both GFI1 and GFI1B are comprised of a Snail/Slug/Gfi1 (SNAG) domain and a highly conserved concatemer of six C2H2-type zinc fingers (ZnFs) at their N and C termini, respectively. Linker regions with limited conservation separate the SNAG and ZnF domains. Sequence-specific DNA binding by both GFI1 and GFI1B is coordinated by ZnFs 3, 4, and 5 (32, 35–37), while ZnFs 1, 2, and 6 enable protein-protein interactions with an incompletely defined subset of transcriptional partners (38–41). Linker regions provide a platform for protein binding and posttranslational modifications that regulate innate functions of GFI proteins (40, 42). GFI1 and GFI1B SNAG domains differ only by a conservative S→T substitution at amino acid 14; they are otherwise identical. Moreover, the SNAG domain is near invariant among orthologs from human to zebrafish, underscoring the critical role of the SNAG domain and its binding partners. Prominent among these is lysine-specific demethylase 1 (LSD1) (KDM1A, AOF2, BHC110, NPAO, and KIAA0601), which is required for transcriptional repression by GFI proteins (10, 43). The SNAG domain is necessary and sufficient for LSD1 binding by GFI1 and GFI1B (44). SNAG domain deletion, alanine substitution at proline 2 (P2A), or leucine substitution at lysine 8 (K8L) of the SNAG domain abolishes LSD1 binding to GFI family proteins and profoundly impairs their activity in multiple functional assays (43, 45–47).
LSD1 is a flavin-dependent monoamine oxidase that demethylates mono- and dimethylated histone 3 lysines 4 and 9 (H3K4me1/2 and H3K9me1/2) to impact chromatin structure and gene expression (48, 49). LSD1 also demethylates nonhistone targets, including p53, STAT3, estrogen receptor α (ERα), MYPT1, hypoxia-inducible factor 1α (HIF1α), DNMT1, E2F, and SOX2, suggesting a broad influence over cellular homeostasis (50–57). LSD1 operates within multiple assemblies that regulate transcription, including nucleosome remodeling and deacetylase (NuRD), C-terminal binding protein (CtBP), mixed-lineage leukemia (MLL) coactivator, and BRAF-histone deacetylase (BHC)/CoREST complexes (58–62). Its inclusion in distinct multiprotein assemblies suggests both enzymatic and structural contributions to transcriptional control, yet affiliations between LSD1-containing complexes and the DNA binding proteins that recruit them are poorly understood.
We leveraged GFI1B-driven erythroid differentiation in K562 cells, GFI1B fusion proteins with a promiscuous biotin ligase (BirA*) (63, 64), and unbiased mass spectral quantitation to define proximity relationships between GFI1B and partners that depend upon GFI1B-LSD1 binding. We show that GFI1B-LSD1 binding is necessary but not sufficient for erythroid differentiation in K562 cells and that the BHC complex is recruited by GFI1B through LSD1 to coordinate GFI1B-mediated erythroid differentiation. Our findings highlight the need to consider protein assemblies as modules comprised of factors acting together to accomplish tasks associated with cell fate acquisition.
RESULTS
LSD1 depletion impairs GFI1B-mediated erythroid differentiation.
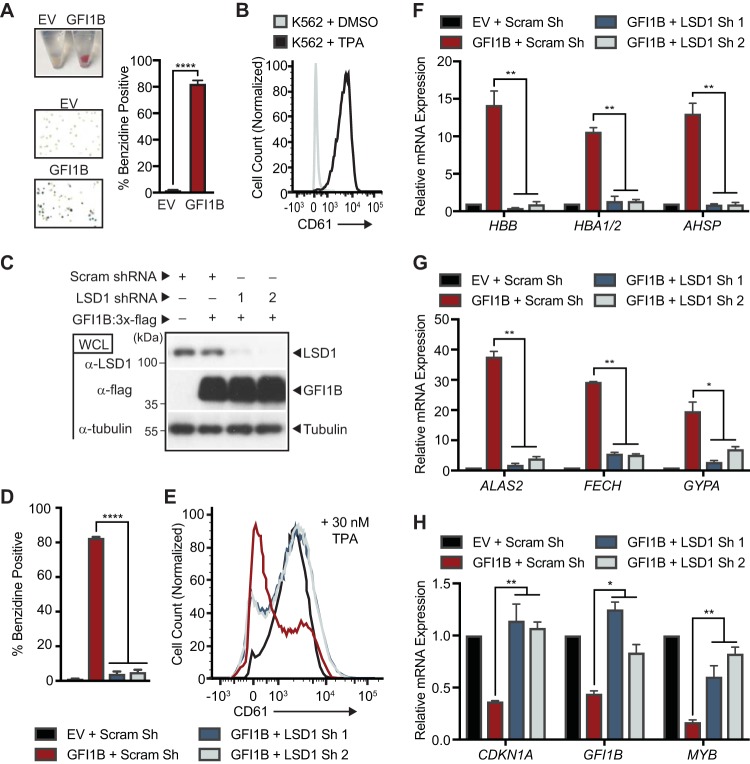
The human erythroleukemia cell line K562 displays erythroid differentiation with enforced GFI1B expression (65), as reflected by hemoglobinization and benzidine-positive staining (Fig. 1A). Likewise, K562 cells adopt megakaryocytic features, including CD61 expression, with tetradecanoyl-1-phorbol-13-acetate (TPA) treatment (Fig. 1B). These dual fates are mutually exclusive. While not completely faithful to normal hematopoietic development, K562 cell differentiation offers a platform to discern GFI1B dependency relationships that are relevant to lineage allocation. Notably, deletion of the GFI1B SNAG domain or placement of an epitope tag at the GFI1B N terminus abolishes GFI1B-driven erythroid differentiation, suggesting primacy of the SNAG domain for this phenotype (65–67). Because the SNAG domain is necessary and sufficient for LSD1 binding, we surmised that disrupting or blocking access to the SNAG domain would impair LSD1 recruitment and, by extension, that LSD1 function would be required for GFI1B-driven erythroid differentiation. To confirm this, we depleted K562 cells of LSD1 and tested the ability of GFI1B to induce erythroid differentiation and of TPA to trigger cell surface expression of CD61. Immunoblots show depletion of LSD1 by two distinct small hairpin RNA (shRNA) constructs relative to a scramble control sequence and equivalent expression of GFI1B (Fig. 1C). LSD1 depletion almost completely eliminates the formation of benzidine-positive cells in response to enforced expression of GFI1B compared to controls, yet it had limited impact on CD61 expression at the cell surface following TPA treatment (Fig. 1D and E). LSD1 depletion also blocked expression of RNAs for alpha (HBA1/2)- and beta (HBB)-globins and alpha-hemoglobin-stabilizing protein (AHSP), as well as the RNAs for 5-aminolevulinic acid synthase 2 (ALAS2) and ferrochelatase (FECH), which are rate-limiting enzymes required for heme biosynthesis. Expression of the RNA for the erythroid fate marker glycophorin A (GYPA) was also significantly impaired (Fig. 1F and G). To confirm contributions to GFI1B-mediated transcriptional repression, we examined mRNA levels for GFI1B direct targets CDKN1A, GFI1B, and MYB in the context of LSD1 depletion. Repression of each gene was reversed by LSD1 depletion (Fig. 1H). Together these data show that LSD1 enables transcriptional repression by GFI1B to trigger an erythroid fate-specifying program in K562 cells and that LSD1 is dispensable for TPA-driven megakaryocytic differentiation.
FIG 1.
LSD1 depletion impedes GFI1B-mediated erythroid differentiation. (A) GFI1B overexpression promotes K562 cell hemoglobinization. Cells were infected with empty vector (EV) or virus expressing GFI1B. GFI1B-expressing cells become red and are benzidine positive. Benzidine-positive cells are quantified as mean ± standard deviation (SD) from four biological replicates. (B) TPA triggers cell surface expression of CD61 in K562 cells. CD61 expression was quantified in vehicle- versus TPA-treated viable cells by flow cytometry. (C) LSD1 depletion and GFI1B overexpression in K562 cells. Small hairpin RNAs targeting LSD1 (LSD1 shRNA) or content-matched scramble control (Scram shRNA) were inducibly transcribed in K562 cells, followed by infection at day +3 with GFI1B:3×-Flag-expressing virus or EV. LSD1 and GFI1B were quantified by Western blotting of whole-cell lysates (WCLs). Tubulin served as a loading control. (D) LSD1 depletion blocks GFI1B-mediated K562 cell hemoglobinization. (E) LSD1 depletion enables TPA-induced CD61 expression in the context of GFI1B overexpression. (F and G) LSD1 depletion impairs expression of erythroid fate genes in response to GFI1B overexpression. Expression of the alpha- and beta-globin (HBA1/2 and HBB), alpha-globin-stabilizing protein (AHSP), aminolevulinic acid synthase 2 (ALAS2), ferrochelatase (FECH), and glycophorin A (GYPA) genes is shown. (H) LSD1 depletion restores expression of GFI1B-repressed target genes. Expression of the CDKN1A, GFI1B, and MYB genes is shown. Gene expression was quantified by RT-qPCR normalized to GUS. Results are expressed as mean ± 2 SD from two independent experiments performed in triplicate. *, P < 0.05; **, P < 0.005; ****, P < 0.00005.
GFI1B-LSD1 binding is required for GFI1B-mediated erythroid differentiation.
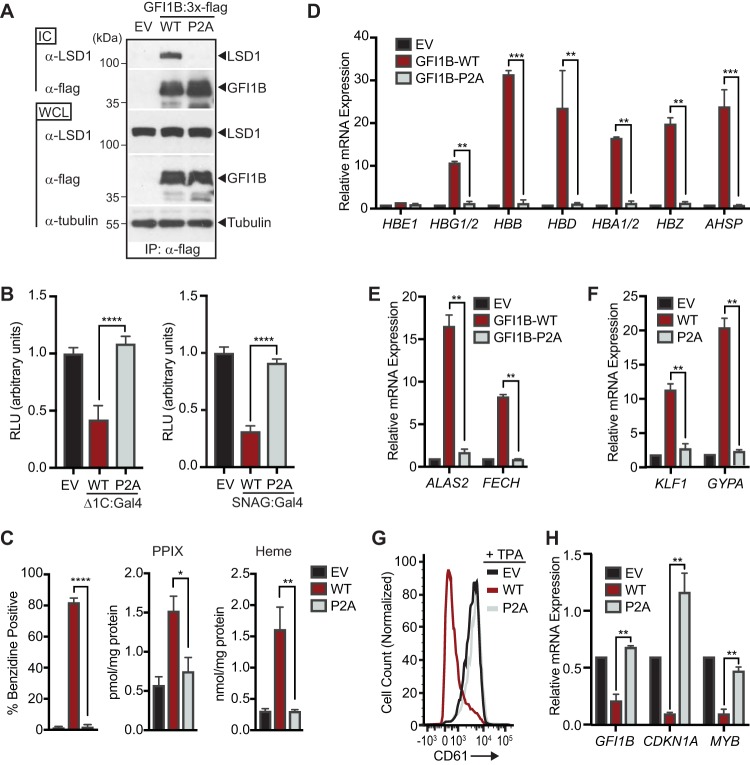
LSD1 cooperates with an extensive roster of DNA binding proteins to influence gene expression (68) and therefore could influence this binary fate decision through partnerships with factors other than GFI1B. To test whether an interaction between GFI1B and LSD1 is needed for GFI1B-mediated erythroid differentiation, we compared K562 cells with enforced expression of wild-type GFI1B (GFI1B-WT) or its P2A derivative (GFI1B-P2A). The P2A substitution abolishes GFI1B-LSD1 binding, as well as transcriptional repression by fusion proteins between Gal4 and either GFI1B or the SNAG domain in isolation (Fig. 2A and B). While GFI1B-WT triggers an erythroid differentiation program in K562 cells, as reflected by hemoglobinization and erythroid-specific gene expression, GFI1B-P2A fails to do so (Fig. 2C to F). GFI1B-WT expression also blocks TPA-induced megakaryocytic differentiation in K562 cells, yet GFI1B-P2A displays no such antagonism (Fig. 2G). Additionally, failure by GFI1B-P2A either to trigger erythroid differentiation or to block CD61 expression in response to TPA correlates with impaired transcriptional repression of direct GFI1B targets (Fig. 2H). These findings indicate that LSD1 recruitment by GFI1B is necessary to direct the erythroid fate-specifying program in K562 cells and to block acquisition of the alternative megakaryocytic fate induced by TPA.
FIG 2.
LSD1 binding is required for GFI1B-regulated cell fate decisions in K562 cells. (A) P2A substitution in GFI1B impairs GFI1B-LSD1 binding. K562 cells were transduced with constructs expressing wild-type GFI1B:3×-Flag, its P2A variant, or empty vector (EV) as shown. GFI1B forms were immunopurified with anti-Flag (M2) antibody and protein G-Sepharose. LSD1 was quantified in immune complexes (ICs) by Western blotting. Equal expression (LSD1 and GFI1B) and precipitation (GFI1B) were evaluated by Western blotting of whole-cell lysates (WCLs) and ICs, respectively. Tubulin was used to confirm equal gel loading. (B) P2A substitution abolishes transcriptional repression by GFI1B. 293-T-Rex-5×Gal-luciferase cells were transfected with WT or P2A variants of GFI1B-Δ1C:Gal4 (left) or SNAG:Gal4 (right) fusion proteins. Firefly luciferase was measured and normalized to constitutively expressed, cotransfected Renilla luciferase. Reporter activity is expressed as mean ± 2 SD from two experiments performed in triplicate. (C) P2A substitution impairs GFI1B-mediated hemoglobinization. Benzidine staining was quantified as for Fig. 1A. Protoporphyrin IX (PPIX) and heme were quantified by UPLC in K562 cells transduced with EV, GFI1B-WT, or GFI1B-P2A. (D to F) P2A substitution impairs expression of erythroid fate genes in K562 cells. Expression of anti-globin cluster genes AHSP, ALAS2, FECH, GYPA, and Kruppel-like factor 1 gene (KLF1) are shown. (G) P2A substitution abolishes GFI1B-mediated suppression of TPA-induced CD61 expression. Cell surface expression of CD61 in TPA-treated cells expressing GFI1B-WT or its P2A variant was determined by flow cytometry. (H) P2A substitution reverses repression of GFI1B target genes GFI1B, CDKN1A, and MYB. Results expressed as mean ± 2 SD. *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ****, P < 0.00005.
GFI1B partners identified by proximity-dependent biotinylation.
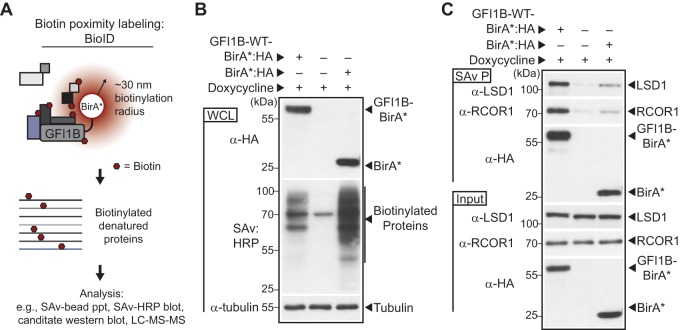
GFI1B-LSD1 binding is required for GFI1B-mediated transcriptional control and fate-specifying functions in hematopoiesis (43, 45). However, LSD1 is a constituent of multiple transcriptional regulatory complexes. Which among these governs GFI1B function to direct erythroid differentiation is unknown. To more comprehensively catalog GFI1B partners, we engineered a chimeric protein, GFI1B-WT-BirA*:HA, comprised of the complete GFI1B primary structure fused in frame with a promiscuous variant of the biotin ligase BirA (BirA*) from Aquifex aeolicus (63, 64, 69) and a C-terminal hemagglutinin (HA) epitope tag under the control of a doxycycline (Doxy)-inducible promoter. Proteins with a proximity relationship to BirA*:HA or GFI1B-WT-BirA*:HA are biotinylated spontaneously and can be further characterized (Fig. 3A). GFI1B-WT-BirA*:HA or BirA*:HA was inducibly expressed in K562 cells, and proteome-wide biotinylation was compared to that of K562 cells transduced with vector only. GFI1B-WT-BirA*:HA and BirA*:HA were equivalently expressed and produced a broad array of biotinylated protein substrates as reflected by horseradish peroxidase (HRP)-conjugated streptavidin (SAv)-mediated detection in whole-cell extracts (Fig. 3B). Both qualitative and quantitative differences in the pattern of biotinylated proteins are apparent, suggesting preferential labeling of targets by BirA*:HA when anchored to GFI1B. In the vector control, the 72-kDa band likely represents propionyl coenzyme A (propionyl-CoA) carboxylase, which is naturally biotinylated and expressed in K562 cells (70). To further validate the proximity labeling technique, we confirmed enrichment for established GFI1B-associated proteins, LSD1 and RCOR1, among biotinylated proteins from cells expressing GFI1B-WT-BirA*:HA relative to those expressing nontargeted BirA*:HA. LSD1 binds directly to the GFI1/1B SNAG domain, while RCOR1 engages GFI1B through LSD1 (43). LSD1 and RCOR1 were identified readily among biotinylated proteins from GFI1B-WT-BirA*:HA-expressing cells collected on SAv-Sepharose beads but only minimally so from cells expressing BirA*:HA or empty vector (EV) (Fig. 3C). Serving as an internal control for equal target biotinylation potential, GFI1B-WT-BirA*:HA and BirA*:HA were comparably modified with biotin, purified on SAv-Sepharose, and detected by anti-HA Western blotting (Fig. 3C). These findings indicate preferential biotinylation of factors known to have a proximity relationship with GFI1B. Moreover, they confirm detection of primary (intrinsic), secondary (direct binding), and tertiary (indirect binding) labeling events and validate this platform for identifying GFI1B partners proteome-wide.
FIG 3.
Proximity labeling of GFI1B binding proteins with GFIB-BirA*. (A) Schematic representation of the strategy for biotin modification of GFI1B partners. GFI1B-WT is fused to the promiscuous biotin ligase with an HA epitope tag (BirA*:HA) to form GFI1B-WT-BirA*:HA. GFI1B partners are spontaneously biotinylated due to their proximity to BirA*:HA anchored to GFI1B in cis. Biotinylated products are captured on streptavidin (SAv)-conjugated beads for downstream analysis via candidate-based or screening approaches. (B) BirA*:HA and GFI1B-WT-BirA*:HA fusion biotinylate diverse targets in K562 cells. Cells were transduced with constructs inducibly expressing GFI1B-WT-BirA*:HA, BirA*:HA, or EV, treated with doxycycline, and harvested. GFI1B-WT-BirA*:HA and BirA*:HA expression was confirmed by anti-HA Western blotting and biotinylated proteins detected by SAv:HRP. Tubulin served as a loading control. (C) GFI1B-interacting proteins LSD1 and RCOR1 are enriched among proteins biotinylated by GFI1B-WT-BirA*:HA compared to BirA*:HA or EV. Biotin-modified proteins were collected on SAv-Sepharose. LSD1, RCOR1, and the BirA*:HA fusion proteins themselves were quantified among purified proteins (SAv P) by Western blotting. Equal expression was confirmed by Western blotting in whole-cell lysates (input).
A “proximitome” of GFI1B partners defined by a quantitative mass spectral screen.
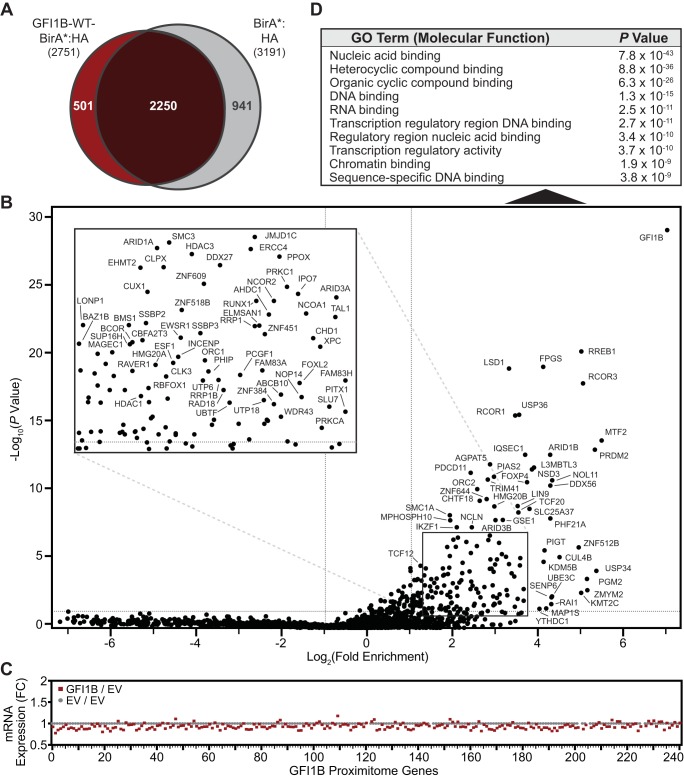
We coined the term “proximitome” to represent the subset of all cellular proteins having a proximity relationship to a factor of interest determined from unbiased, proteome-wide proximity labeling and quantitative mass spectral identification of biotinylated proteins purified on SAv-Sepharose beads. Included proximity partners reflect the spectrum of proteins found to be enriched 2-fold or more with expression of a factor:BirA*:HA fusion protein relative to BirA*:HA alone and with a P value from triplicate analysis of 0.05 or less. To comprehensively catalog GFI1B proximity partners, biotinylated proteins were purified from whole-cell extracts of K562 cells inducibly expressing BirA*:HA or GFI1B-WT-BirA*:HA for 48 h. This allowed cells time to reconfigure their complement of proteins following induced expression of GFI1B-WT-BirA*:HA or BirA*:HA alone. Purified proteins were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) to unequivocally establish their identities and extrapolate their abundances from sum read intensities (see Data Set S1 in the supplemental material). Proteins identified in GFI1B-WT-BirA*:HA- or BirA*:HA-expressing K562 cells are compared in a Venn diagram in Fig. 4A. The diagram reveals strong concordance between proteins identified in each data set, suggesting comparable access to potential partners. The overlap sector represents 82% of labeled proteins in GFI1B-WT-BirA*:HA-expressing cells. A subset of 501 proteins was found exclusively in the GFI1B-WT-BirA*:HA-expressing cells, while 941 proteins were detected uniquely in cells expressing BirA*:HA alone. To establish candidate GFI1B proximity partners, we calculated fold enrichment (x axis) for each protein and plotted these ratios relative to P values (y axis) in a volcano plot with logarithmic axes. We identified 241 statistically significant outliers (hits), which are labeled on the plot and inset (Fig. 4B; see Table S1 in the supplemental material). As expected, with GFI1B covalently linked to BirA*:HA it displays the greatest fold enrichment (141-fold) and most significant P value (5.05 × 10−29) in the GFI1B-WT-BirA*:HA proximitome. Moreover, several known GFI1B binding proteins, including LSD1, RCOR1, HDAC1, PIAS2, and CBFA2T3, were also readily detected, supporting the validity of the approach to defining GFI1B proximity partners.
FIG 4.
GFIB-WT-BirA*:HA defines a GFI1B-WT proximitome. (A) Venn diagram showing proteins identified from mass spectral screening of biotin-modified proteins from K562 cells expressing GFI1B-WT-BirA*:HA or BirA*:HA. Common and unique proteins are indicated. (B) GFI1B-WT-BirA*:HA identifies members of a GFI1B-WT proximitome. Factors enriched among biotinylated and SAv-Sepharose-purified proteins modified by GFI1-WT-BirA*:HA (test) compared to BirA*:HA (control) are shown. Fold enrichment is plotted logarithmically as a ratio of average sum read intensities between test and control samples against their P values calculated from their 20 closest-ranked neighbors in the data set as described in Materials and Methods. Individual proteins are labeled with their respective gene names. The inset shows an expanded view of the data contained in the boxed area. (C) Fold change in mRNA levels for the 241 hits identified in the GFI1B-WT proximitome. mRNAs for each hit are listed in rank order from highest to lowest to match their order in the GFI1B-WT proximitome, also from highest to lowest. (D) Gene ontology (GO) analysis for the 241 hits identified in the GFI1B-WT proximitome. The top 10 functional assignment are shown with their corresponding P values.
Notably, no phenotypic changes were seen in cells at 48 h when proximitomes were determined, but those expressing GFI1B-WT-BirA*:HA went on to acquire erythroid features at 7 days postinduction, while those expressing BirA*:HA only did not (data not shown). To address the possibility that proteins were misidentified as hits due to increased expression of their corresponding genes, we performed RNA sequencing (RNA-seq) in K562 cells expressing GFI1B-WT or vector alone. Among the 241 proteins identified as hits in the GFI1B-WT-BirA*:HA proximitome, only 31 showed increased mRNA levels and none rose to 1.5-fold (the maximum fold change observed was 1.175 for L3MBTL3). Other hits showed either no change or modestly decreased mRNA expression (Fig. 4C). While it is conceivable that increased protein levels arising from reduced turnover could lead to spurious identification of a hit due to more frequent but stochastic biotinylation, these RNA-seq data strongly suggest that hits reflect true proximity partnerships with GFI1B.
GFI1B proximity partners are members of complexes that control gene expression.
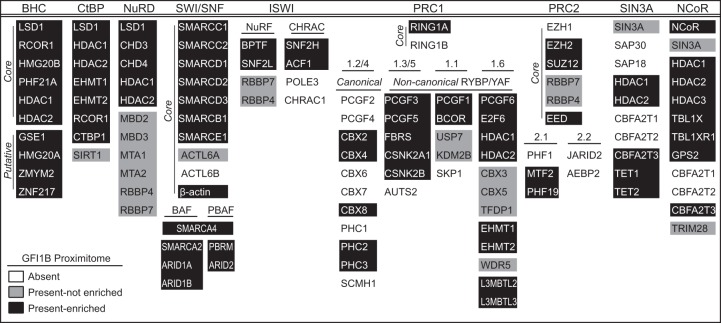
GFI1B is best known as a transcriptional repressor, and its contributions to multiple functional assays can be traced to this role (20, 71–76). Indeed, the top 25 functional groupings in gene ontology (GO) analysis of the 241 GFI1B-WT proximity partners were related to gene expression and chromatin structure. The top 10 are listed in Fig. 4D. Therefore, we focused first on partnerships between GFI1B and determinants of chromatin structure and transcriptional control. In the GFI1B-WT-BirA*:HA proximitome, we identified elements of multiple complexes highly relevant to regulated gene expression (Fig. 5). Prominent among those proteins present and enriched in the GFI1B-WT-BirA*:HA proximitome are members of the BHC, CtBP, and SWI/SNF complexes, from which we discovered 10/10 (core plus putative), 7/8, and 14/16 complex members, respectively. Similarly, we observed each component of the NuRD complex in the GFI1B-WT-BirA*:HA proximitome, yet only a subset of these were found to be enriched relative to the BirA*:HA control. Components from several other complexes (NuRF, CHRAC, canonical and noncanonical PRC1, PRC2, SIN3A, and NCoR) were found, and for each, some elements were enriched, suggesting that they also contribute to GFI1B-mediated transcriptional control and lineage allocation.
FIG 5.
Components of transcriptional repression and chromatin-remodeling complexes identified by GFI1B-WT proximity labeling. A roster of transcriptionally relevant complexes and their component members are shown. Proteins are listed according to HUGO gene nomenclature. With respect to the GFI1B-WT proximitome (GFI1B-WT-BirA*:HA versus BirA*:HA), proteins present and enriched are shown on a black background. Those present but not significantly enriched (P < 0.05) are shown on a gray background. Those not present (average sum read intensity below that for EV) are shown on a white background. Where appropriate, complexes are subdivided into core and putative members and/or with canonical versus noncanonical designations.
Identification of LSD1-dependent GFI1B binding partners.
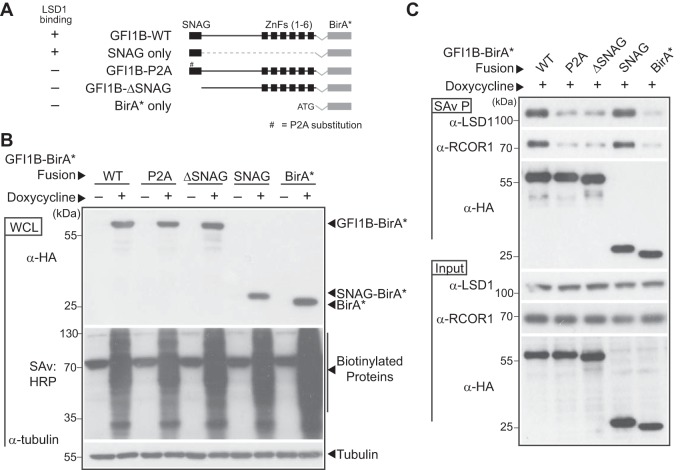
LSD1 binding is necessary for GFI1B function (10, 43). However, discovering elements from functionally distinct transcriptional regulatory complexes within the GFI1B proximitome suggests an important interplay between these elements and LSD1. Moreover, at least three of these complexes (BHC, NuRD, and CtBP) contain LSD1 as a core component (58, 60, 77). Given the importance of LSD1 for GFI1B function, we hypothesized that one or more transcriptional regulatory complexes represented in the GFI1B proximitome would be recruited in an LSD1-dependent manner and, if its components were depleted, would impair GFI1B function even when GFI1B-LSD1 binding potential was preserved. To address this hypothesis, we engineered expression constructs encoding fusion proteins comprised of BirA*:HA and either LSD1 binding-competent forms of GFI1B (GFI1B-WT-BirA*:HA and SNAG-BirA*:HA) or GFI1B variants with impaired LSD1 binding (GFI1B-P2A-BirA*:HA and GFI1B-ΔSNAG-BirA*:HA). A BirA*:HA construct served as an additional negative control (Fig. 6A). Fusion proteins were inducibly expressed in K562 cells and their biotinylating ability compared in whole-cell lysates by SDS-PAGE, transblotting, and detection with HRP-conjugated SAv (Fig. 6B). Equivalent expression and robust, proteome-wide biotinylation were observed for each construct upon induction with doxycycline. We then tested the ability of each fusion protein to establish proximity relationships with the known SNAG domain binding proteins LSD1 and RCOR1 (43, 45). As expected, GFI1B-WT-BirA*:HA and SNAG-BirA*:HA display clear enrichment for biotinylated LSD1 and RCOR1 in SAv-Sepharose pulldowns, while the LSD1 nonbinding variants (P2A and ΔSNAG) show biotinylation comparable to the background seen for the BirA*:HA negative control (Fig. 6C). Equally important, we observed comparable biotinylation proteome-wide for BirA*:HA and each BirA*:HA fusion protein, suggesting an equal capacity for spontaneous biotinylation and mitigating concerns that the fusion partner might adversely impact BirA*:HA activity. Collectively, these results support the use of GFI1B variants to unambiguously define the LSD1-dependent elements of GFI1B complexes governing transcriptional control and cell fate specification.
FIG 6.
LSD1-nonbinding variants of GFI1B-BirA*:HA fail to biotinylate LSD1 and RCOR1. (A) Graphical representation of GFI1B-BirA*:HA fusion proteins employed in experiments. Each fusion protein is comprised of the GFI1B regions shown in frame with the BirA* expression cassette and a C-terminal HA epitope tag to ensure equivalent expression of all forms. The BirA*:HA construct lacks GFI1B structures and instead begins with an ATG. Wild-type (WT) GFI1B is shown. The SNAG and zinc finger (ZnF) domains (1 to 6) of GFI1B are shown as black boxes. A thin line represents the GFI1B linker and connections between ZnFs. The proline-to-alanine substitution (P2A) in the GFI1B SNAG domain is indicated. ΔSNAG represents deletion of the SNAG domain but preservation of the remaining GFI1B primary structure. (B) Expression of GFI1B-BirA*:HA forms and proteome-wide biotin modification in K562 cells. K562 cells were transduced with lentivirus to inducibly express the GFI1B-BirA*:HA forms shown and stable isolates selected as polyclonal populations. Doxycycline-inducible expression of each fusion protein was confirmed by Western blotting against the common HA epitope tag and biotinylating activity confirmed proteome-wide by SAv:HRP detection of transblotted total cellular protein. Tubulin served as a loading control. (C) LSD1 and RCOR1 are enriched among biotinylated proteins generated by BirA*:HA fusions competent for LSD1 binding. Biotin-modified proteins from K562 cells transduced with the BirA*:HA fusion proteins shown were purified from whole-cell extracts (SAv P), fractionated by SDS-PAGE, and subjected to Western blotting with anti-LSD1, anti-RCOR1, and anti-HA antibodies. Equivalent expression of each protein in whole-cell lysates (input) was confirmed by Western blotting with these same antibodies.
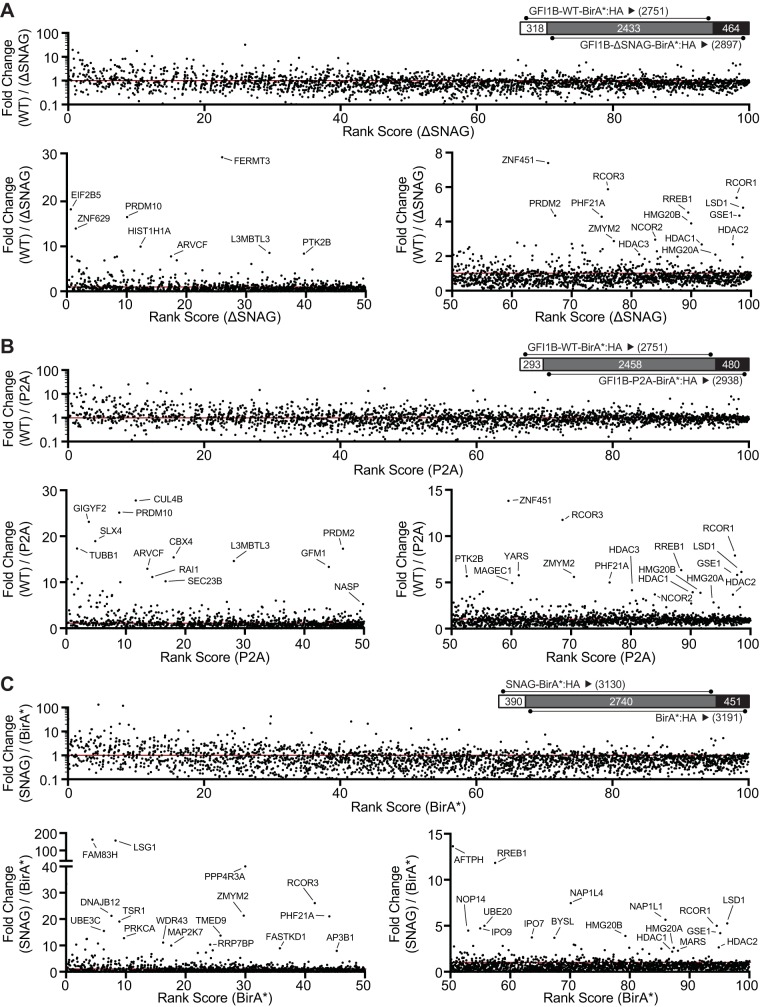
In pursuit of this goal, we deployed these tools for quantitative proximitome analyses to compare partnerships between GFI1B forms which are either capable of (GFI1B-WT-BirA*:HA and SNAG-BirA*:HA) or deficient in (GFI1B-P2A-BirA*:HA, GFI1B-ΔSNAG-BirA*:HA, and BirA*:HA) LSD1 binding. Compared to LSD1 nonbinding variants GFI1B-ΔSNAG-BirA*:HA (Fig. 7A) and GFI1B-P2A-BirA*:HA (Fig. 7B), we observed enrichment for nine of the 10 core (LSD1, RCOR1, PHF21A, HMG20B, HDAC1, and HDAC2) and putative (GSE1, HMG20A, and ZMYM2) components of the BHC complex in the GFI1B-WT-BirA*:HA proximitome. These results indicate that BHC complex recruitment is lost with impaired GFI1B-LSD1 binding. To focus specifically on the SNAG-LSD1 interaction as a determinant of BHC complex recruitment, we determined enrichment for BHC complex components in the SNAG-BirA*:HA proximitome relative to BirA*:HA only. Again, we found enrichment for these nine BHC complex components in the SNAG-BirA*:HA proximitome (Fig. 7C).
FIG 7.
GFI1B recruits the BHC complex in an LSD1-dependent manner via the SNAG domain. K562 cells stably and inducibly expressing the BirA*:HA fusion proteins shown in Fig. 6A were deployed to define GFI1B proximity partners whose recruitment is LSD1 dependent. Fusion protein expression was induced by doxycycline addition and biotinylation permitted in situ. Biotin-modified proteins were captured on SAv-Sepharose beads and analyzed by LC-MS/MS as described in Materials and Methods. Dot plots compare the fold change in average sum read intensity among fusion proteins with intact SNAG domains and capable of LSD1 binding (GFI1B-WT-BirA*:HA and SNAG-BirA*:HA) to those with disrupted or absent SNAG domains and deficient in LSD1 binding (GFI1B-P2A-BirA*:HA, GFI1B-ΔSNAG-BirA*:HA, and BirA*:HA). For each comparison (GFI1B-WT-BirA*:HA versus GFI1B-ΔSNAG-BirA*:HA [A], GFI1B-WT-BirA* versus GFI1B-P2A-BirA*:HA [B], and SNAG-BirA*:HA versus BirA*:HA [C]), average sum read intensities were ranked according to the LSD1-nonbinding sample and plotted on the x axis. The fold change in average sum read intensities, represented by a ratio between the LSD1 binding and LSD1 nonbinding samples in each comparison, is plotted on the y axis. Each comparison (A to C) is represented by three panels. On top in each is the entire data set for that comparison, along with a Venn diagram comparing common and unique proteins for each data set, and below are two panels dividing the percentile rank score from 0 to 50 and from 50 to 100 with expanded y axis limits to facilitate visualization of proteins enriched in the data set for the LSD1 binding form. Outlying proteins are labeled with their gene names. A line indicates equal read intensities, and thus no enrichment, for the LSD1 binding over the LSD1 nonbinding sample.
LSD1, HDAC1, and HDAC2 are shared among the BHC, CtBP, and NuRD complexes, while RCOR1 is found in both the BHC and CtBP complexes. To confirm the LSD1 dependence of BHC complex recruitment specifically, we surveyed the top 100 enriched proteins in each binary proximitome comparison (GFI1B-WT-BirA*:HA versus GFI1B-ΔSNAG-BirA*:HA, GFI1B-WT-BirA*:HA versus GFI1B-P2A-BirA*:HA, and SNAG-BirA*:HA versus BirA*:HA) for the unique components of BHC, CtBP, and NuRD complexes (Tables 1 and 2; see Table S2 in the supplemental material). While unique components of the BHC complex were found in each binary proximitome comparison performed, none of those unique to the CtBP or NuRD complexes were observed. These findings indicate that among LSD1-containing complexes detected in the GFI1B proximitome, only the BHC complex displays LSD1 dependence.
TABLE 1.
Components of the transcriptional regulatory complexes
| Complex | No. of components |
|
|---|---|---|
| Total | Unique | |
| BHC | 10 | 6 |
| CtBP | 8 | 4 |
| NuRD | 11 | 8 |
TABLE 2.
Comparison of GFI1B LSD1-dependent proximitomes
| Complex | No. of components present in top 100 enriched proteins |
|||||
|---|---|---|---|---|---|---|
| WT vs ΔSNAG |
WT vs P2A |
SNAG vs BirA* |
||||
| Total | Unique | Total | Unique | Total | Unique | |
| BHC | 7 | 4 | 8 | 5 | 5 | 3 |
| CtBP | 2 | 0 | 3 | 0 | 2 | 0 |
| NuRD | 2 | 0 | 2 | 0 | 1 | 0 |
GFI1B-mediated transcriptional repression and cell fate require the BHC complex.
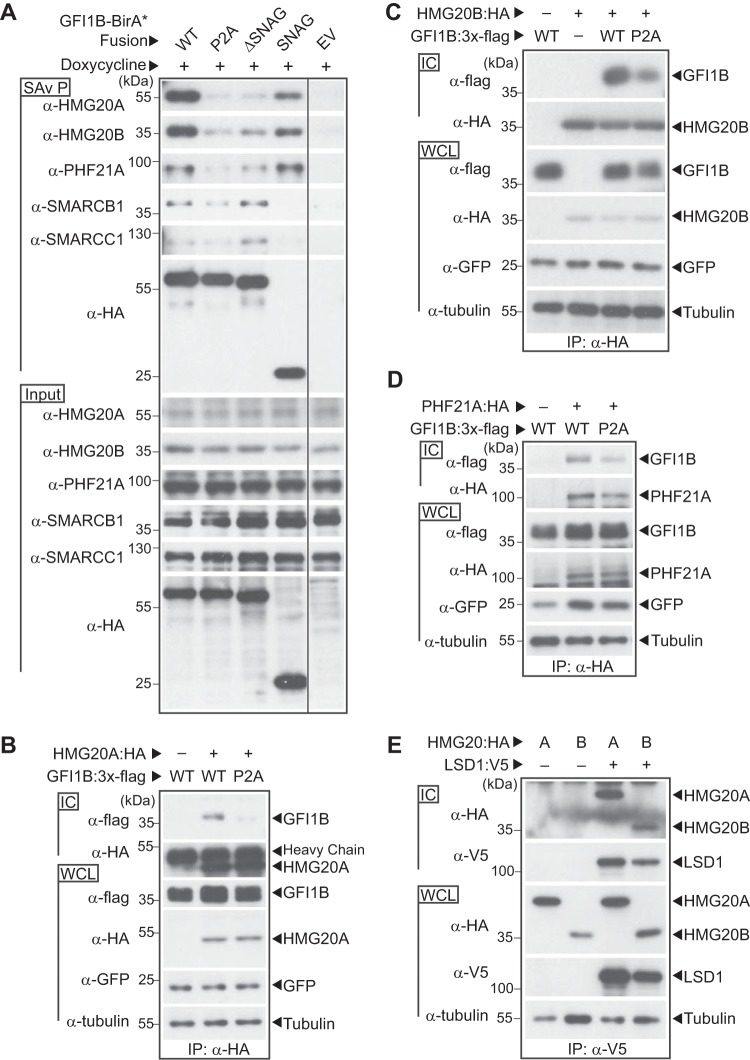
To confirm the findings from mass spectral screens, we first assessed associations between GFI1B and candidate BHC complex components, specifically testing their dependence upon GFI1B-LSD1 binding. We focused on unique elements of the BHC complex. GFI1B-WT-BirA*:HA, GFI1B-P2A-BirA*:HA, GFI1B-ΔSNAG-BirA*:HA, and SNAG-BirA*:HA were inducibly expressed in K562 cells and biotinylated proteins purified on SAv-Sepharose beads. HMG20A, HMG20B, and PHF21A were then quantified among purified proteins by Western blotting. Each was enriched among proteins purified from cells expressing BirA*:HA fusions with GFI1B variants capable of LSD1 binding relative to nonbinding controls (Fig. 8A). Notably, no such enrichment was observed for SMARCB1 or SMARCC1, both members of the SWI/SNF complex that showed enrichment but no LSD1 dependence in GFI1B proximitome analyses; nor did SMARCB1 or SMARCC1 show a proximity relationship when SNAG-BirA*:HA was employed, confirming a distinct and LSD1-independent means of associating with GFI1B. Using Flag-tagged wild-type and P2A forms of GFI1B, we extended the findings from proximity labeling studies to traditional coprecipitation assays. For HMG20A, HMG20B, and PHF21A, binding to GFI1B-P2A was significantly impaired relative to binding to wild-type GFI1B, confirming dependence upon LSD1 recruitment (Fig. 8B to D). This observation is further supported by coprecipitation assays showing interactions with LSD1 for both HMG20A and HMG20B (Fig. 8E).
FIG 8.
BHC complex components HMG20A, HMG20B, and PHF21A bind GFI1B in an LSD1-dependent manner. (A) BHC complex components are preferentially enriched in proximity labeling assays with fusion proteins competent for LSD1 binding. Biotin-modified proteins were purified from K562 cells inducibly expressing the BirA*:HA fusion proteins shown or an empty vector control (EV). HMG20A, HMG20B, PHF21A, SMARCB1, and SMARCC1, as well as the BirA*:HA fusion proteins, were quantified in SAv-Sepharose pulldowns (SAv P) by Western blotting. Equivalent input for each protein was confirmed by Western blotting of whole-cell lysates (input). (B to D) GFI1B binding to HMG20A, HMG20B, and PHF21A requires GFI1B-LSD1 binding. COS7L cells were transfected with HA-tagged HMG20A (B), HMG20B (C), or PHF21A (D) and wild-type (WT) or P2A variants of GFI1B:3×-Flag. HMG20A/B and PHF21A proteins and their binding partners were purified in anti-HA immune complexes (ICs). Coprecipitating GFI1B forms were detected in immune complexes by Western blotting using anti-Flag antibody. Equivalent expression and precipitation were evaluated in whole-cell lysates (WCLs) and immune complexes (ICs), respectively. GFP served as a transfection control. (E) HMG20A and HMG20B bind LSD1. COS7L cells were transfected with HA-tagged HMG20A or HMG20B along with a V5-tagged LSD1. LSD1 and its binding partners were collected in anti-V5 ICs. Coprecipitating HMG20A and HMG20B were detected in immune complexes by Western blotting using anti-HA antibody. Equivalent expression and precipitation were evaluated in the WCLs and ICs, respectively.
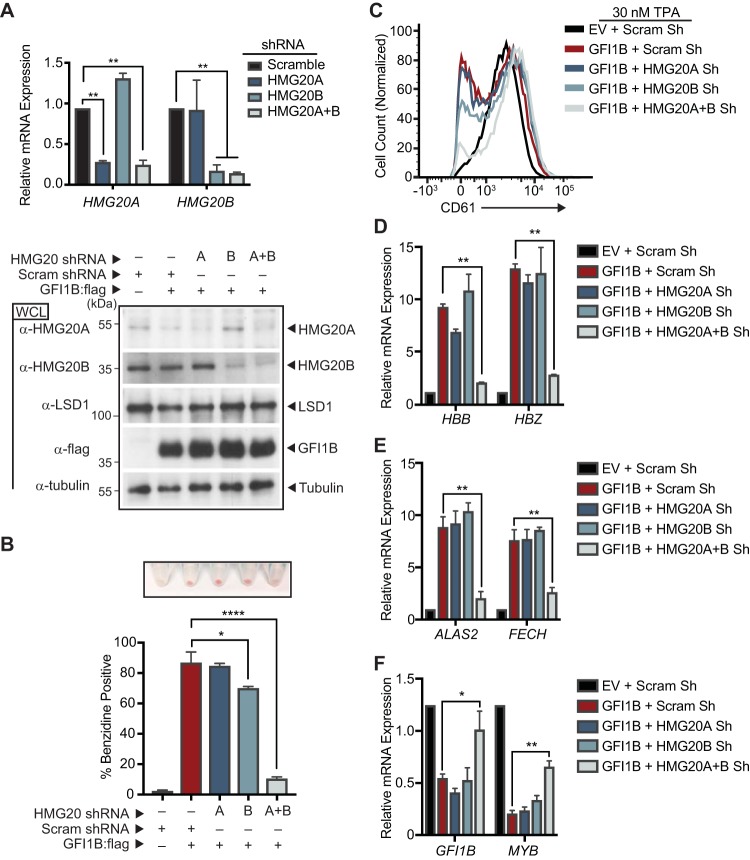
LSD1 is required for both GFI1B function and BHC complex recruitment, but whether LSD1 exerts its impact through BHC complex recruitment is not known. To address this question, we targeted unique BHC complex components for depletion in K562 cells and then tested GFI1B’s ability to direct changes in cell fate and gene expression (Fig. 9). HMG20A and HMG20B are known to bind structured elements in DNA in a sequence-nonspecific manner (78–80). They have an oppositional role in neuronal differentiation via modulation of REST-responsive genes, but limited information exists regarding their contributions to hematopoietic differentiation (77, 78, 81). HMG20A and HMG20B were depleted alone or together using inducible shRNA constructs (Fig. 9A), and then effects of enforced GFI1B expression on K562 cell differentiation and gene expression were assessed. While neither HMG20A nor HMG20B depletion alone had a notable impact on K562 cell hemoglobinization, simultaneous depletion of both factors prevented this GFI1B-driven phenotype (Fig. 9B). Likewise, impaired TPA-induced megakaryocytic differentiation brought on by enforced GFI1B expression was not significantly affected by depletion of either HMG20A or HMG20B alone, yet their concurrent depletion enabled accumulation of CD61 on the K562 cell surface despite GFI1B overexpression (Fig. 9C). Erythroid differentiation blockade brought on by concurrent depletion of HMG20A and HMG20B correlated with reduced expression of genes required for heme and globin chain synthesis and with loss of repression of GFI1B target genes (Fig. 9D to F). Notably, LSD1 expression is unaffected by HMG20A or HMG20B depletion (Fig. 9A). Thus, HMG20A and HMG20B make functional contributions to LSD1-dependent outcomes driven by GFI1B. Their concurrent absence impairs GFI1B-driven erythroid differentiation in K562 cells and enables alternate, TPA-driven megakaryocytic differentiation despite enforced GFI1B expression that would normally block it. Moreover, unlike the oppositional relationship between HMG20A and HMG20B observed in REST-regulated neuronal differentiation, these findings intimate functional redundancy between them in GFI1B-mediated transcription and its control of cellular identity.
FIG 9.
HMG20A and HMG20B are required for GFI1B-mediated cell fate changes in K562 cells. (A) HMG20A and HMG20B depletion in K562 cells. RNA (top) and protein (bottom) were collected from K562 cells transduced with shRNA targeting HMG20A, HMG20B, HMG20A, and HMG20B (HMG20A+B) or a content-matched scrambled control. HMG20A and HMG20B expression levels were evaluated by RT-qPCR and normalized to GUS. HMG20A, HMG20B, LSD1, and GFI1B:3×-Flag levels were determined by Western blotting. Tubulin served as a loading control. (B to F) HMG20A/B depletion abolishes GFI1B-dependent cell fate determination in K562 cells. K562 cells were transduced with inducible shRNAs targeting HMG20A, HMG20B, HMG20A, and HMG20B or a scrambled control. The shRNAs were induced with doxycycline, cells were infected with GFI1B:3×-Flag-expressing retrovirus, and erythroid differentiation was allowed to proceed. (B) Benzidine staining was quantified by counting positive cells in bright-field microscopy, while hemoglobinized cell pellets are shown above each condition shown in the bar graph. The experiment was performed in triplicate. (C) GFI1B-dependent inhibition of TPA-induced CD61 expression requires HMG20A/B. K562 cells overexpressing GFI1B and depleted of HMG20A, HMG20B, or HMG20A and -B were treated with TPA and cell surface expression of CD61 quantified by flow cytometry. The histograms presented are representative of three independent experiments. (D to F) mRNAs for globin genes HBB and HBZ (D), heme biosynthesis genes ALAS2 and FECH (E), and GFI1B-repressed target genes GFI1B and MYB (F) were quantified by RT-qPCR normalized to the GUS reference gene. Results are expressed as mean ± 2 SD. Statistical significance was determined by the two-sided unpaired t test: *, P < 0.05; **, P < 0.005; ****, P < 0.00005.
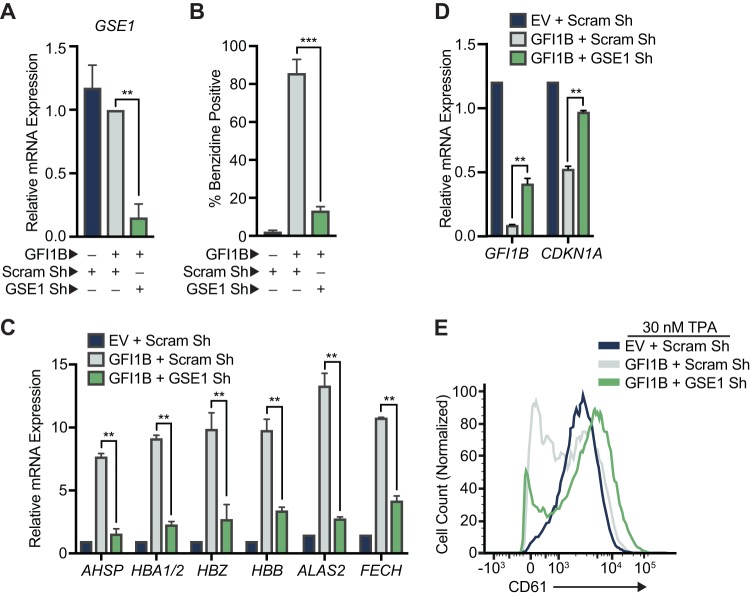
To extend observations made with HMG20A and HMG20B, we tested the effects of GSE1 depletion on GFI1B functions in transcriptional repression and erythroid differentiation in K562 cells. GSE1 is a proline-rich coiled-coil domain protein implicated in pathogenesis of breast and gastric cancers (82, 83). It is widely expressed in human cells, but its role in cell fate decisions in hematopoiesis has not been studied. Like for HMG20 proteins, reduced GSE1 expression blocks erythroid differentiation and gene expression changes in K562 cells which depend upon the GFI1B-LSD1 binding relationship (Fig. 10). Together with the results from LSD1, HMG20A, and HMG20B depletion, these data highlight that LSD1 is necessary but not sufficient for GFI1B-driven outcomes and showcase a critical role for the BHC complex as an LSD1-dependent determinant of GFI1B-mediated transcriptional control and cell fate decisions.
FIG 10.
GSE1 is required for GFI1B-mediated cell fate changes in K562 cells. (A) GSE1 depletion in K562 cells. RNA was collected from K562 cells transduced with shRNA targeting GSE1 or a content-matched scrambled control. GSE1 expression levels were evaluated by RT-qPCR and normalized to GUS. (B to D) GSE1 depletion abolishes GFI1B-dependent cell fate determination in K562 cells. K562 cells were transduced with inducible shRNAs targeting GSE1 or a scrambled control. The shRNAs were induced with doxycycline, cells were infected with GFI1B:3×-Flag-expressing retrovirus, and erythroid differentiation was allowed to proceed. (B) Benzidine staining was quantified by counting positive cells in bright-field microscopy. (C and D) mRNAs for globin and related genes AHSP, HBA1/2, HBZ, and HBB and heme biosynthesis genes ALAS2 and FECH (C) and GFI1B-repressed target genes GFI1B and CDKN1A (D) were quantified by RT-qPCR and normalized to the GUS reference gene. Results are expressed as mean ± 2 SD. (E) GFI1B-dependent inhibition of TPA-induced CD61 expression requires GSE1. K562 cells overexpressing GFI1B and depleted of GSE1 were treated with TPA, and cell surface expression of CD61 was quantified by flow cytometry compared to control cells. The histograms presented are representative of three independent experiments. Statistical significance was determined by the two-sided unpaired t test: **, P < 0.005; ****, P < 0.00005.
DISCUSSION
The control of gene expression involves DNA binding proteins that recognize discrete sequence elements in regulatory regions of genes, directing site-selective assembly of multiprotein complexes that modulate chromatin structure and, ultimately, access to coding regions by the basal transcription machinery (84, 85). By deploying individual elements in a modular fashion, cells ensure flexibility and economy as they orchestrate transcriptional programs to control the specification of cell fate. Proper function of these complexes is layered upon a spatial and temporal order of events that combine to create a flow of information needed for a specific outcome among the myriad possibilities. Yet, this strategy necessarily creates dependency relationships between factors cooperating to accomplish functions required for an outcome to be realized. Malfunction of just one complex component or disordered operation of a complex in space or time may impair functional outputs and thus mask the underlying molecular complexities required to achieve them.
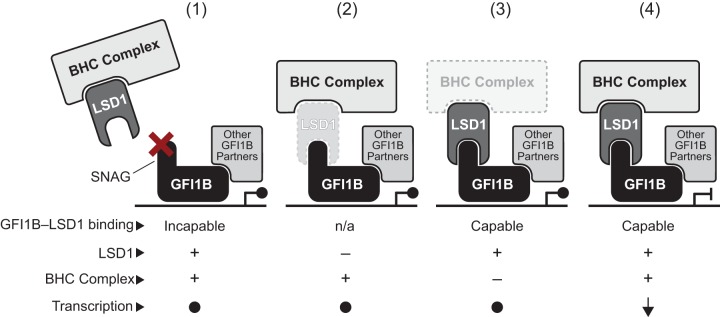
The relationship between GFI family proteins and LSD1 exemplifies this theme. Both GFI1 and GFI1B require LSD1 binding for transcriptional repression, and point mutations in their SNAG domains completely abolish activity in multiple functional assays (43, 45, 46, 86). It is not, however, immediately apparent that other factors might enable LSD1’s central role, and identifying these collaborators is technically challenging. We deployed an unbiased, proteome-wide, quantitative proximity labeling strategy (referred to as proximitome analysis) based upon a promiscuous variant of BirA (BirA*) to prospectively and systematically label with biotin factors that dwell in the vicinity of GFI1B and then to determine functionally relevant partners which are recruited in a manner requiring LSD1, its obligate effector. In so doing, we show that while LSD1 recruitment is critical to GFI1B-driven erythroid differentiation, it does not operate alone as its singular depletion in K562 cells may suggest. Rather, LSD1 is responsible for selective recruitment of the BHC complex, implying that its contributions to GFI1B-mediated outcomes may reflect both its intrinsic demethylase activity and its role as a scaffolding component for this multiprotein complex. A model summarizing the collaborative relationship between LSD1, BHC complex components, and other partners in GFI1/1B-mediated transcriptional repression is shown in Fig. 11.
FIG 11.
A working model for contributions of the BHC complex to GF1B-mediated repression. LSD1 serves as a bridge between GFI1B and other components of the BHC complex, including HDAC1/2, HMG20A, HMG20B, PHF21A, RCOR1, ZMYM2, ZNF217, and GSE1. Therefore, BHC complex recruitment is rendered LSD1 dependent. A functional GFI1B transcriptional repression complex requires coincident recruitment of these factors, along with other partners brought to the promoter in an LSD1-independent manner by GFI1B or DNA binding proteins with which it collaborates. Dysfunction resulting from an impaired, SNAG-dependent GFI1B-LSD1 recruitment mechanism (1), LSD1 depletion or an intrinsic change that disrupts SNAG binding (2), or ineffective formation or operation of BHC complex components (3) could each poison the GFI1B-LSD1 functional axis to create a permissive state for misexpression of GFI1B-regulated genes. Only when each element is present and functioning properly (4) is the appropriate repressive outcome achieved. trans regulation by each component toward the others, or toward partners that engage GFI1B in an LSD1-independent manner, may also make important contributions to enabling and fine-tuning target gene expression.
LSD1 displays in vitro demethylase activity toward peptides containing mono- and dimethylated lysines (K), best exemplified by demethylation of H3K4me1/me2, and toward purified histones (48, 87). However, in a chromatinized context, methylated histones are poor substrates for LSD1 (88). Efficient demethylation of chromatinized histones by LSD1 requires concurrent RCOR1 binding, and this activity can be further stimulated by an interaction between RCOR1 and SUMO2/3 (88, 89). Likewise, LSD1’s demethylase activity is favored by HDAC1-mediated deacetylation of LSD1-K374ac (90). Observations such as these support the notion that LSD1’s demethylase activity may hinge upon its inclusion in multiprotein complexes to enable its proper posttranslational modifications and allosteric modulation. Additionally, by serving as a scaffold, LSD1 may enable regulation of other components of the BHC complex or perhaps discrete regulatory modules recruited to operationalize GFI1B activity as a transcriptional repressor. Absent these cooperative relationships, LSD1 may fail to execute its obligate effector role for GFI1B despite the means of its recruitment by GFI1B being intact. Investigating these possibilities presents a daunting challenge, but it is one that can now be tackled using the situational capabilities of proximity labeling approaches. Moreover, improvements in these technologies are emerging at a rapid pace. For example, a variant of BirA optimized for substrate processivity, known as TurboID, enables significantly shorter labeling times and thereby a refined definition of proximity partners for a factor of interest (91). Likewise, proximity labeling strategies using ascorbate peroxidase (APEX2), whose biotinylating activity is dependent upon hydrogen peroxide and limited by its half-life, can be deployed to assess rapid alterations in proximity relationships (92). When combined with first-generation BirA* (BioID and BioID2), these strategies can illuminate changes in protein complexes arising from rapid regulatory events, track the choreography of protein complex formation and change, and even collect an historical record of relationships experienced by a given protein in its lifetime.
A view of LSD1 that accounts for both enzymatic and structural contributions is consistent not only with an absolute requirement for its presence but also with its dependence upon partnerships to enable repressor and cell fate-specifying activities of GFI1B. To establish evidence supporting this view, we focused on BHC complex components HMG20A and HMG20B as LSD1 partners. This focus was stimulated by studies in Schizosaccharomyces pombe concerning the role of Lsd1/2 in replication fork pausing associated with mating type class switching (93). Lsd1/2 is required for growth and viability in S. pombe. Notably, S. pombe strains rendered catalytically inactive at lsd1/lsd2 are viable and do not phenocopy lsd1/lsd2 deletion, suggesting that they have a nonenzymatic role. Moreover, Lsd1 contains a C-terminal HMG domain that is lacking in mammalian LSD1, and HMG domain mutations impair mating type class switching in S. pombe through defective imprinting at the mat1 locus brought on by dysfunctional replication fork pausing (48, 93). The importance of the Lsd1 HMG domain to a phenotype dependent upon structural change in DNA, the absence of the HMG domain in mammalian LSD1, the suggestion of a nonenzymatic role for Lsd1, and the inclusion of HMG20 proteins in the GFI1B proximitome made HMG20A and HMG20B attractive candidates in GFI1B-LSD1-dependent outputs in K562 cells. Our finding that concurrent HMG20A/B depletion blocks LSD1-dependent functions of GFI1B suggests that their inclusion in the BHC complex is essential to enable LSD1 actions. Similar observations made by depleting GSE1 reinforce this notion. Notably, HMG20A and HMG20B are ubiquitously expressed (94). They contain N-terminal HMG boxes which bind DNA in a sequence-nonspecific manner and C-terminal coiled-coil domains required for dimerization and incorporation into multiprotein complexes (81, 95). In neural development, HMG20A and HMG20B display an oppositional relationship at REST-regulated genes (77, 81, 96). Yet, our data suggest that these proteins may also compensate for deficiencies in one another and that perhaps the relative abundance of each may fine-tune transcriptional activity of the BHC complex at sites to which it has been recruited. Thus, with site-selective DNA binding proteins (e.g., GFI1B), epigenetic modifiers (e.g., LSD1 and HDAC1/2), their cofactors (e.g., RCOR1), and readers of epigenetic state (e.g., PHF21A) as partners, a network of interdependent proteins may cooperate to direct the quality, quantity, and timing of transcriptional programs governing the specification of cell fate. Our findings provide a framework for exploring the interdependent and situational relationships among these classes of proteins.
MATERIALS AND METHODS
Cell culture and viral transduction.
COS7L cells, HEK293T cells (American Type Culture Collection), MEL cells, and 293-T-Rex-5xGal-luciferase cells (generously provided by Raphael Margueron) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). K562 cells were maintained in Roswell Park Memorial Institute (RPMI) medium supplemented with 10% FBS and 25 μM HEPES. All cell culture media were supplemented with 2 μM l-glutamine, 50 U/ml penicillin, and 50 μg/ml streptomycin. All cell media and additives were obtained from Thermo Fisher. All cell lines were verified using the Geneprint 10 system (Promega). For viral production, HEK293T cells were transfected with a viral vector containing the transgene along with the respective lentiviral (psPAX2 and pMD2G) or retroviral (gag/pol and vesicular stomatitis virus G [VSVG]) packaging plasmids using polyethylenimine (PEI) (3 μg PEI to 1 μg DNA). Viral supernatant was collected at 48 and 72 h posttransfection and clarified by filtration (Corning, 45 μm). K562 cells were transduced with viral particles in the presence of 8 μg/ml Polybrene for 16 h. Stable cells were obtained by selection with 5 μg/ml puromycin (Puro) for 4 days and with 400 μg/ml G418/neomycin (Neo) or hygromycin (Hygro) for 10 days. For LSD1 and HMG20B small hairpin RNA (shRNA) experiments, K562 cells were transduced with Tet-pLKO-neo (a gift from Dmitri Wiederschain; Addgene plasmid 21916) containing either LSD1- or HMG20B-targeted shRNA or a scrambled control and selected in neomycin to establish stable isolates. For HMG20A and GSE1 shRNA experiments, K562 cells were transduced with EZ-Tet-pLKO-Hygro (a gift from Cindy Miranti; Addgene plasmid 85972) containing either HMG20A- or GSE1-targeted shRNA or a scrambled control and selected in hygromycin. To obtain double HMG20A and -B shRNA knockdown, K562 cells were first transduced with either HMG20B or scrambled shRNA vector, selected with neomycin, and subsequently transduced with either HMG20A or scrambled shRNA vector and selected with hygromycin. shRNA expression was achieved by treating transduced cells with 500 ng/ml doxycycline every 24 h. shRNA sequences are listed in Table S3 in the supplemental material.
RNA extraction and gene expression analysis.
Total RNA was prepared using RNeasy (Qiagen) as per the manufacturer’s instructions, including on-column DNase I digestion to avoid potential genomic DNA (gDNA) contamination. The cDNA was generated from 100 to 300 ng of total RNA using iScript Reverse Transcription Supermix for real-time quantitative PCR (RT-qPCR) (Bio-Rad). cDNA was diluted 2× with water, and 1 μl of cDNA was used as a template for RT-qPCR. Quantitation was performed by RT-qPCR with the CFX96 real-time system (Bio-Rad), using 5 μl SsoAdvanced Universal SYBR green Supermix (Bio-Rad) and 330 nM concentrations of the indicated forward and reverse primers in a 10-μl reaction mixture. Relative mRNA expression was determined by the ΔΔCT method and normalized to a GUS reference gene. Each experiment was performed in triplicate. Statistical significance was determined by two-sided unpaired t tests using GraphPad Prism 7.0. Sequences of all oligonucleotides used are listed in Table S3 in the supplemental material. For RNA sequencing, stable K562 Tet-On Advanced cells were infected with pLVX-Tight-Puro empty vector or pLVX-Tight-Puro-GFI1B:3×-Flag and were selected in puromycin for 3 days. Expression of GFI1B:3×-Flag or EV was achieved by treating 1 × 106 cells with 100 ng/ml doxycycline for 48 h. To confirm GFI1B expression, half of the sample was used to collect whole-cell lysates, and protein levels were evaluated by Western blotting. The other half of the sample was used to collect total RNA as described above. Each experimental condition was performed in biological triplicates. RNA quality was assessed by the Agilent RNA ScreenTape assay. The Illumina TruSeq Stranded RNA kit with Ribo-Zero Gold was used for sequencing library preparation. An Illumina HiSeq 50 Cycle single-read sequencing version 4 sequencing protocol was performed, and reads were aligned to the hg38 human genome assembly. Fold changes in expression of the 241 GFI1B proximitome genes were calculated by averaging the triplicate read count of each gene and dividing it by the average read count of the empty vector. Read counts were normalized to ATP5F1A, GAPDH, GUSB, PPIA, and YWHA2. A fold change of greater than 1.5 was deemed significant.
Constructs and subcloning.
GFI1B (NM_008114.3), PHF21A (NM_138755.2), HMG20A (NM_025812.3), and HMG20B (NM_010440.3) were amplified by PCR (Platinum SuperFi DNA polymerase; Invitrogen) from cDNA obtained from MEL cells and subcloned into pcDNA3.1(−) (Invitrogen) with either a 3×-Flag, HA, or V5 tag on the 3′ end of the gene. The GFI1B-P2A mutation was made by site-directed mutagenesis. Retroviral GFI1B:3×-Flag vectors were produced by subcloning GFI1B into the pMSCV (murine stem cell virus)-IRES-Puro vector using EcoRI. GFI1B fusion proteins to Gal4 were generated by PCR amplification of the GFI1B SNAG domain (amino acids 1 to 20) or GFI1B-Δ1C (amino acids 1 to 244 only, eliminating ZnFs 4, 5, and 6, which remove intrinsic DNA binding via the GFI1B sequence) and inserted into pcDNA3.1 N terminally and in frame with the Gal4 DNA binding domain (amino acids 1 to 147) by using EcoRI/XbaI. GFI1B fusion proteins with BirA*:HA were made by subcloning GFI1B-WT, -P2A, or -ΔSNAG (amino acids 21 to 330) or the SNAG domain (amino acids 1 to 20) into EcoRI/BamHI sites in frame to BirA*:HA in the MCS-BioID2-HA vector, a gift from Kyle Roux (Addgene plasmid 74224). Subsequently, GFI1B-BirA*:HA forms were expressed from PCR-amplified sequences subcloned into pLVX-Tight-Puro (Clontech) with NotI/MluI for doxycycline-inducible expression with the Tet-On Advanced (Clontech) system. The BirA*:HA-only vector was created by inserting a start codon and an EcoRI site into AgeI/BamH1 (5′ of BirA*:HA) in pcDNA3.1 and subsequently put into pLVX-Tight-Puro by using EcoRI. Primers used for cloning are listed in Table S3 in the supplemental material. The LSD1 construct was a generous gift from Sunil Sharma. Complete open reading frame sequences for all constructs were confirmed by automated dideoxy sequencing in the University of Utah DNA sequencing HSC Core Facility.
Reagents and antibodies.
Mouse monoclonal anti-Flag (M2) and antitubulin (clone B-5-1-2) were obtained from Sigma. Rabbit polyclonal anti-HA (ab9110) and anti-HMG20B (ab167415) were obtained from Abcam. Rabbit polyclonal anti-LSD1 (C69G12) and anti-RCOR1 (D6I2U) were obtained from Cell Signaling Technology. Mouse monoclonal anti-HMG20A (D-5) and anti-green fluorescent protein (anti-GFP) (B-2) were obtained from Santa Cruz Biotechnology. Rabbit polyclonal anti-SMARCB1 (A301-087A) and anti-PHF21A (A303-603A) were obtained from Bethyl Laboratories, and anti-SMARCC1 (17722-1-AP) was obtained from Proteintech. Mouse monoclonal anti-V5 (46-0705) was obtained from Invitrogen. Horseradish peroxidase (HRP)-conjugated anti-mouse (715-035-150) and anti-rabbit (711-035-152) antibodies were obtained from Jackson Immunoresearch. HRP-conjugated streptavidin (RPN1231) (SAv) was obtained from GE Healthcare UK Limited. Allophycocyanin (APC)-conjugated anti-human CD61 (VI-PI.2) and 4′,6-diamidino-2-phenylindole, dilactate (DAPI) (422801), were obtained from BioLegend. Restriction endonucleases and ligases were obtained from New England Biosciences. Puromycin (Puro), hygromycin (Hygro), and G418/Geneticin (Neo) were obtained from Invitrogen. Protein G-Sepharose 4 Fast Flow and SAv-Sepharose beads were obtained from GE Healthcare Bio-Sciences AB. Biotin was obtained from TCI Chemicals, and doxycycline (Doxy) was obtained from Fisher Scientific. Benzidine dihydrochloride, 12-O-tetradecanoylphorbol-13-acetate (TPA), and Polybrene were obtained Sigma. Polyethylenimine (PEI) (25-kDa linear form) was obtained from Polysciences (23966-2). All other materials were of reagent grade.
Transient transfection and coimmunoprecipitation.
Transient transfections in COS7L cells were performed using expression plasmids in the indicated combinations and Lipofectamine 2000 per the manufacturer’s instructions. Monolayers were washed twice with phosphate-buffered saline (PBS), scraped into ice-cold lysis buffer (50 mM Tris, 150 mM NaCl, 10% glycerol, 1 mM EDTA, 1 mM dithiothreitol [DTT], 1% Triton X-100, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 μg/ml aprotinin [pH 8.0]), disrupted by sonication (10 1-s pulses at 5 W using a Virtis Virsonic sonicator), rocked for 30 min at 4°C, and clarified by centrifugation. As indicated in the figures, antibodies were added to clarified extracts and left for 90 min, and then the immune complexes (ICs) were captured by Protein G-Sepharose beads washed in lysis buffer. Immune complexes were washed 4 times with lysis buffer and boiled in SDS-PAGE sample buffer supplemented with DTT.
Immunoblotting.
After separation by SDS-PAGE, cleared lysates or immune complexes were examined by immunoblotting following standard methods. Briefly, separated protein products were transferred to nitrocellulose filters (0.45 μm; Genesee Scientific) using 25 mM Tris–192 mM glycine (pH 8.35) for 1 h at 4°C. Nitrocellulose filters were blocked with PBS-T (1× PBS, 0.05% Tween 20) supplemented with 0.25% gelatin and 0.2% NaN3 (GBB) and incubated with primary antibodies overnight at 4°C as indicated in the figures. Filters were then washed successively with PBS-T and incubated with species-appropriate HRP-conjugated antibody for 1 h at room temperature. After PBS-T washes, proteins were visualized by chemiluminescence detection (ECL) (30 mM Tris [pH 8.5], 375 μM Luminol, 67.5 μM p-coumaric acid, 0.02% H2O2) using autoradiography film (Genesee Scientific).
Luciferase reporter assays.
Luciferase assays were performed using the dual-luciferase reporter assay system (Promega) as per the manufacturer’s instructions. Briefly, 3.25 × 105 293-T-Rex-5×Gal-luciferase cells were plated per well in 6-well plates. Transient transfection of the respective GFI1B:Gal4 expression constructs was performed with Lipofectamine 2000 as per the manufacturer’s instructions. Reporter activity was measured in cell lysates collected at 48 h posttransfection using a Modulus microplate reader (Turner BioSystems). Firefly luciferase activity was normalized to a constitutively expressed, cotransfected Renilla luciferase control. Statistical significance was determined by two-sided unpaired t tests using GraphPad Prism 7.0.
Chemical differentiation and phenotypic assays.
K562 cells (4 × 105) were treated with 30 nM TPA. Cells were harvested postinduction, washed, and incubated with anti-CD61 antibody diluted 1:100 in Hanks buffered saline solution (HBSS) (Gibco) supplemented with 0.5% bovine serum albumin (BSA) (RPI Corp). The fluorescence signal from DAPI-negative cells was detected by flow cytometry (Fortessa; BD Biosciences) and analyzed using FlowJo analysis software (TreeStar). Heme/porphyrin levels were analyzed by ultraperformance liquid chromatography (UPLC) with an Acquity UPLC BEH C18 column (1.7 μm; 2.1 by 100 mm) at the Iron and Heme Core Facility of the University of Utah. Benzidine staining was performed by collecting 1 × 106 K562 cells, washing with PBS, and staining with 0.2% benzidine dihydrochloride in 0.5% acetic acid for 15 min at room temperature. Benzidine-positive cells were quantified with a hemocytometer using bright-field microscopy. Experiments were all performed in triplicate with 200 cells or more counted from randomly selected fields for each condition. Statistical significance was determined by two-sided unpaired t tests using GraphPad Prism 7.0.
SAv affinity capture of biotin-modified proteins.
To induce GFI1B-WT-BirA*:HA or its derivatives in K562 cells, 6 × 107 cells were treated with 1 μg/ml doxycycline for 48 h, and 30 μM biotin was added 16 h before harvesting. Cells were collected, washed twice with ice-cold PBS, lysed with ice-cold lysis buffer (50 mM Tris [pH 7.5], 500 mM NaCl, 0.5 mM EDTA, 1% Triton X-100), sonicated, and cleared by centrifugation. Biotinylated complexes were captured by streptavidin (Sav)-Sepharose high-performance beads and rocked at 4°C overnight. Beads were washed 5 times with lysis buffer supplemented with SDS, boiled in DTT-containing sample buffer, and fractionated by SDS-PAGE (10% T/2.67% C, 4% stack). The gel was stained with colloidal Coomassie blue (Invitrogen). Lanes were divided into two equal sections for downstream LC-MS/MS analysis.
Protein identification by MS.
Mass spectrometry (MS) was performed by the Taplin Mass Spectrometry Facility, Cell Biology Department, Harvard Medical School. Briefly, samples were reduced using 1 mM DTT in 50 mM ammonium bicarbonate for 30 min at 60°C and cooled, and 5 mM iodoacetamide in 50 mM ammonium bicarbonate was added for 15 min and left in the dark at ambient temperature. DTT (5 mM) was added to quench iodoacetamide derivatization, and then 5 ng/μl of sequencing-grade trypsin (Promega) was added and proteins digested overnight at 37°C. Samples were then desalted and reconstituted in 5 to 10 μl of high-pressure liquid chromatography (HPLC) solvent A (2.5% acetonitrile, 0.1% formic acid). A nanoscale reverse-phase HPLC capillary column was created by packing 2.6-μm C18 spherical silica beads into a fused silica capillary (100-μm inner diameter, ∼30-cm length) with a flame-drawn tip. After column equilibration, each sample was loaded via a Famos autosampler (LC Packings) onto the column. A linear gradient was formed to elute peptides with increasing concentrations of solvent B (97.5% acetonitrile, 0.1% formic acid). Eluted peptides were subjected to electrospray ionization coupled to an LTQ Orbitrap Velos Pro ion trap mass spectrometer (Thermo Fisher Scientific) to produce tandem mass spectra whose b and y ion series patterns were compared to theoretical digests of the proteome in Sequest (Thermo Fisher Scientific) to establish protein identity. Data were filtered to a 1 to 2% peptide false-discovery rate.
Statistical analysis of MS data.
All assays were performed in triplicate (biological replicates) and read intensities averaged for each sample. To establish the GFI1B proximitome (assessment of GFI1B-WT-BirA*:HA versus BirA*:HA only), sum read intensities among detected proteins were ranked from 0 to 100%. A total of 3,191 proteins were detected in the BirA*:HA-only sample and utilized to establish a background intensity ranking. A total of 2,751 unique proteins were identified in the GFI1B-WT-BirA*:HA proximitome. An intensity fold change for each ranked protein was determined by dividing the GFI1B-WT-BirA*:HA average read intensity by the BirA*:HA-only average read intensity. This calculated fold change in intensity was compared to the average intensity rank and was observed to follow a strong diminution of variance with increasing values, as expected. To identify proteins with aberrantly increased detection, z-scores were calculated for each of the 2,751 proteins as the deviance in rank from the localized (across the background intensity axis) mean rank compared to the localized standard deviation of the ranks after the manner of Eubank (97). The corresponding P value from a t distribution of a 20-protein smoothing selection was determined and utilized to rank prioritize proteins showcasing the most consistent and increased abundance compared to expectation. Proteins with a P value of <0.05 were considered hits. This approach was deemed appropriate, as the background intensity of pertinent proteins varied and detecting relative abundance compared to background was paramount. A volcano plot was created from the –log10 of the P value and the log2 of the intensity fold change. Read intensities detected in GFI1B-WT-BirA*:HA but not in the BirA*:HA-only sample (501 proteins) were analyzed separately, considering only the top 50 to 100 percentiles of ranked GFI1B-WT-BirA*:HA proximity partners as hits (a total of 35 proteins). Likewise, assessment of GFI1B-WT-BirA*:HA versus GFI1B-ΔSNAG-BirA*:HA, GFI1B-WT-BirA*:HA versus GFI1B-P2A-BirA*:HA, and SNAG-BirA*:HA versus BirA*:HA only was done by establishing a background percentile ranking of GFI1B-ΔSNAG-BirA*:HA (2,897 proteins), GFI1B-P2A-BirA*:HA (2,938 proteins), and BirA*:HA (3,191 proteins). Proteins were then plotted according to background rank score and fold change. Statistical analysis was performed as described above for GFI1B-WT-BirA*:HA versus BirA*:HA using nonparametric regression and spline smoothing (98). Gene ontology (GO) analysis was performed on the 241 GFI1B proximitome by g:Profiler (99). SAS version 9.4 and Microsoft Excel were used for statistical calculations and data comparisons.
Supplementary Material
ACKNOWLEDGMENTS
We thank Katharine Ullman, Jason Gertz, Trudy Oliver, Charles Murtaugh, Rodney Stewart, and Mahesh Chandrasekharan for their helpful insights and suggestions during preparation of the manuscript. We acknowledge the following University of Utah Core facilities: DNA sequencing and cell line authentication were performed at the DNA Sequencing Core Facility, oligonucleotides were synthesized by the DNA/Peptide Facility of the Health Sciences Center (HSC), flow cytometry was performed by the Flow Cytometry Facility, and heme/porphyrin analysis was performed at the Heme & Iron Facility. Mass spectrometry was performed at the Taplin Mass Spectrometry Facility, Cell Biology Department, Harvard Medical School, with special thanks going to Ross Tomaino.
This work was supported by National Institutes of Health grants R01CA201235 (M.E.E.) and T32 DK007115 (D.M. and M.J.C.) and by grants from the American Cancer Society, Alex’s Lemonade Stand Foundation, and the Hyundai Hope on Wheels Foundation. We acknowledge support for C.C.M. from the Intermountain Healthcare and Primary Children's Hospital Foundations. The Flow Cytometry Facility is supported by the National Cancer Institute (grant 5P30CA042014-24), and the Heme & Iron Facility is supported in part by a grant from the NIH National Institute of Diabetes and Digestive and Kidney Diseases (U54DK110858).
The supporters had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We declare no conflicts of interest associated with this article.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00020-19.
REFERENCES
- 1.Feinberg AP, Koldobskiy MA, Göndör A. 2016. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet 17:284–299. doi: 10.1038/nrg.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avgustinova A, Benitah SA. 2016. Epigenetic control of adult stem cell function. Nat Rev Mol Cell Biol 17:643–658. doi: 10.1038/nrm.2016.76. [DOI] [PubMed] [Google Scholar]
- 3.Ntziachristos P, Abdel-Wahab O, Aifantis I. 2016. Emerging concepts of epigenetic dysregulation in hematological malignancies. Nat Immunol 17:1016–1024. doi: 10.1038/ni.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egger G, Liang G, Aparicio A, Jones PA. 2004. Epigenetics in human disease and prospects for epigenetic therapy. Nature 429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 5.van der Meer LT, Jansen JH, van der Reijden BA. 2010. Gfi1 and Gfi1b: key regulators of hematopoiesis. Leukemia 24:1834–1843. doi: 10.1038/leu.2010.195. [DOI] [PubMed] [Google Scholar]
- 6.Lancrin C, Mazan M, Stefanska M, Patel R, Lichtinger M, Costa G, Vargel O, Wilson NK, Möröy T, Bonifer C, Göttgens B, Kouskoff V, Lacaud G. 2012. GFI1 and GFI1B control the loss of endothelial identity of hemogenic endothelium during hematopoietic commitment. Blood 120:314–322. doi: 10.1182/blood-2011-10-386094. [DOI] [PubMed] [Google Scholar]
- 7.Zeng H, Yücel R, Kosan C, Klein-Hitpass L, Möröy T. 2004. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J 23:4116–4125. doi: 10.1038/sj.emboj.7600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hock H, Hamblen MJ, Rooke HM, Schindler JW, Saleque S, Fujiwara Y, Orkin SH. 2004. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature 431:1002–1007. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- 9.Yücel R, Karsunky H, Klein-Hitpass L, Möröy T. 2003. The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit+ T cell progenitors and CD4/CD8 lineage decision in the thymus. J Exp Med 197:831–844. doi: 10.1084/jem.20021417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiolka K, Hertzano R, Vassen L, Zeng H, Hermesh O, Avraham KB, Dührsen U, Möröy T. 2006. Gfi1 and Gfi1b act equivalently in haematopoiesis, but have distinct, non-overlapping functions in inner ear development. EMBO Rep 7:326–333. doi: 10.1038/sj.embor.7400618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karsunky H, Zeng H, Schmidt T, Zevnik B, Kluge R, Schmid KW, Dührsen U, Möröy T. 2002. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat Genet 30:295–300. doi: 10.1038/ng831. [DOI] [PubMed] [Google Scholar]
- 12.Hock H, Hamblen MJ, Rooke HM, Traver D, Bronson RT, Cameron S, Orkin SH. 2003. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity 18:109–120. doi: 10.1016/S1074-7613(02)00501-0. [DOI] [PubMed] [Google Scholar]
- 13.Zörnig M, Schmidt T, Karsunky H, Grzeschiczek A, Moroy T. 1996. Zinc finger protein GFI-1 cooperates with myc and pim-1 in T-cell lymphomagenesis by reducing the requirements for IL-2. Oncogene 12:1789–1801. [PubMed] [Google Scholar]
- 14.Khandanpour C, Thiede C, Valk PJM, Sharif-Askari E, Nuckel H, Lohmann D, Horsthemke B, Siffert W, Neubauer A, Grzeschik K-H, Bloomfield CD, Marcucci G, Maharry K, Slovak ML, van der Reijden BA, Jansen JH, Schackert HK, Afshar K, Schnittger S, Peeters JK, Kroschinsky F, Ehninger G, Lowenberg B, Duhrsen U, Moroy T. 2010. A variant allele of growth factor independence 1 (GFI1) is associated with acute myeloid leukemia. Blood 115:2462–2472. doi: 10.1182/blood-2009-08-239822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khandanpour C. 2017. Growth factor independence 1 (Gfi1) regulates the AML supporting function of mesenchymal stromal cells. Exp Hematol 53:S90. doi: 10.1016/j.exphem.2017.06.209. [DOI] [Google Scholar]
- 16.Möröy T. 2014. The zinc finger protein Gfi1 maintains development and progression of lymphoid leukemia by blocking the activation of the tumor suppressor p53. Exp Hematol 42:S7. doi: 10.1016/j.exphem.2014.07.024. [DOI] [Google Scholar]
- 17.Volpe G, Walton DS, Grainger DE, Ward C, Cauchy P, Blakemore D, Coleman DJL, Cockerill PN, Garcia P, Frampton J. 2017. Prognostic significance of high GFI1 expression in AML of normal karyotype and its association with a FLT3-ITD signature. Sci Rep 7:11148. doi: 10.1038/s41598-017-11718-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia J, Bolyard AA, Rodger E, Stein S, Aprikyan AA, Dale DC, Link DC. 2009. Prevalence of mutations in ELANE, GFI1, HAX1, SBDS, WAS and G6PC3 in patients with severe congenital neutropenia. Br J Haematol 147:535–542. doi: 10.1111/j.1365-2141.2009.07888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Person RE, Li F-Q, Duan Z, Benson KF, Wechsler J, Papadaki HA, Eliopoulos G, Kaufman C, Bertolone SJ, Nakamoto B, Papayannopoulou T, Grimes HL, Horwitz M. 2003. Mutations in proto-oncogene GFI1 cause human neutropenia and target ELA2. Nat Genet 34:308–312. doi: 10.1038/ng1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Randrianarison-Huetz V, Laurent B, Bardet V, Blobe GC, Huetz F, Duménil D. 2010. Gfi-1B controls human erythroid and megakaryocytic differentiation by regulating TGF- signaling at the bipotent erythro-megakaryocytic progenitor stage. Blood 115:2784–2795. doi: 10.1182/blood-2009-09-241752. [DOI] [PubMed] [Google Scholar]
- 21.Vassen L, Beauchemin H, Lemsaddek W, Krongold J, Trudel M, Möröy T. 2014. Growth factor independence 1b (gfi1b) is important for the maturation of erythroid cells and the regulation of embryonic globin expression. PLoS One 9:e96636. doi: 10.1371/journal.pone.0096636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh D, Upadhyay G, Sengupta A, Biplob MA, Chakyayil S, George T, Saleque S. 2016. Cooperative stimulation of megakaryocytic differentiation by Gfi1b gene targets Kindlin3 and Talin1. PLoS One 11:e0164506. doi: 10.1371/journal.pone.0164506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beauchemin H, Shooshtarizadeh P, Vadnais C, Vassen L, Pastore YD, Möröy T. 2017. Gfi1b controls integrin signaling-dependent cytoskeleton dynamics and organization in megakaryocytes. Haematologica 102:484–497. doi: 10.3324/haematol.2016.150375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saleque S, Cameron S, Orkin SH. 2002. The zinc-finger proto-oncogene Gfi-1b is essential for development of the erythroid and megakaryocytic lineages. Genes Dev 16:301–306. doi: 10.1101/gad.959102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevenson WS, Morel-Kopp M-C, Chen Q, Liang HP, Bromhead CJ, Wright S, Turakulov R, Ng AP, Roberts AW, Bahlo M, Ward CM. 2013. GFI1B mutation causes a bleeding disorder with abnormal platelet function. J Thromb Haemost 11:2039–2047. doi: 10.1111/jth.12368. [DOI] [PubMed] [Google Scholar]
- 26.Monteferrario D, Bolar NA, Marneth AE, Hebeda KM, Bergevoet SM, Veenstra H, Laros-van Gorkom BAP, MacKenzie MA, Khandanpour C, Botezatu L, Fransen E, Van Camp G, Duijnhouwer AL, Salemink S, Willemsen B, Huls G, Preijers F, Van Heerde W, Jansen JH, Kempers MJE, Loeys BL, Van Laer L, Van der Reijden BA. 2014. A dominant-negative GFI1B mutation in the gray platelet syndrome. N Engl J Med 370:245–253. doi: 10.1056/NEJMoa1308130. [DOI] [PubMed] [Google Scholar]
- 27.Uchiyama Y, Ogawa Y, Kunishima S, Shiina M, Nakashima M, Yanagisawa K, Yokohama A, Imagawa E, Miyatake S, Mizuguchi T, Takata A, Miyake N, Ogata K, Handa H, Matsumoto N. 2018. A novel GFI1B mutation at the first zinc finger domain causes congenital macrothrombocytopenia. Br J Haematol 181:843–847. doi: 10.1111/bjh.14710. [DOI] [PubMed] [Google Scholar]
- 28.Kitamura K, Okuno Y, Yoshida K, Sanada M, Shiraishi Y, Muramatsu H, Kobayashi R, Furukawa K, Miyano S, Kojima S, Ogawa S, Kunishima S. 2016. Functional characterization of a novel GFI1B mutation causing congenital macrothrombocytopenia. J Thromb Haemost 14:1462–1469. doi: 10.1111/jth.13350. [DOI] [PubMed] [Google Scholar]
- 29.Vassen L, Khandanpour C, Ebeling P, Van der Reijden BA, Jansen JH, Mahlmann S, Dührsen U, Möröy T. 2009. Growth factor independent 1b (Gfi1b) and a new splice variant of Gfi1b are highly expressed in patients with acute and chronic leukemia. Int J Hematol 89:422–430. doi: 10.1007/s12185-009-0286-5. [DOI] [PubMed] [Google Scholar]
- 30.Elmaagacli AH, Koldehoff M, Zakrzewski JL, Steckel NK, Ottinger H, Beelen DW. 2007. Growth factor-independent 1B gene (GFI1B) is overexpressed in erythropoietic and megakaryocytic malignancies and increases their proliferation rate. Br J Haematol 136:212–219. doi: 10.1111/j.1365-2141.2006.06407.x. [DOI] [PubMed] [Google Scholar]
- 31.Hernández A, Villegas A, Anguita E. 2010. Human promoter mutations unveil Oct-1 and GATA-1 opposite action on Gfi1b regulation. Ann Hematol 89:759–765. doi: 10.1007/s00277-009-0900-x. [DOI] [PubMed] [Google Scholar]
- 32.Tong B, Grimes HL, Yang TY, Bear SE, Qin Z, Du K, El-Deiry WS, Tsichlis PN. 1998. The Gfi-1B proto-oncoprotein represses p21WAF1 and inhibits myeloid cell differentiation. Mol Cell Biol 18:2462–2473. doi: 10.1128/MCB.18.5.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thivakaran A, Botezatu L, Hönes JM, Schütte J, Vassen L, Al-Matary YS, Patnana P, Zeller A, Heuser M, Thol F, Gabdoulline R, Olberding N, Frank D, Suslo M, Köster R, Lennartz K, Görgens A, Giebel B, Opalka B, Dührsen U, Khandanpour C. 2018. Gfi1b: a key player in the genesis and maintenance of acute myeloid leukemia and myelodysplastic syndrome. Haematologica 103:614–625. doi: 10.3324/haematol.2017.167288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Möröy T, Vassen L, Wilkes B, Khandanpour C. 2015. From cytopenia to leukemia: the role of Gfi1 and Gfi1b in blood formation. Blood 126:2561–2569. doi: 10.1182/blood-2015-06-655043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zweidler-McKay PA, Grimes HL, Flubacher MM, Tsichlis PN. 1996. Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol Cell Biol 16:4024–4034. doi: 10.1128/MCB.16.8.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee S, Doddapaneni K, Hogue A, McGhee L, Meyers S, Wu Z. 2010. Solution structure of Gfi-1 zinc domain bound to consensus DNA. J Mol Biol 397:1055–1066. doi: 10.1016/j.jmb.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 37.Anguita E, Villegas A, Iborra F, Hernández A. 2010. GFI1B controls its own expression binding to multiple sites. Haematologica 95:36–46. doi: 10.3324/haematol.2009.012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakazawa Y, Suzuki M, Manabe N, Yamada T, Kihara-Negishi F, Sakurai T, Tenen DG, Iwama A, Mochizuki M, Oikawa T. 2007. Cooperative interaction between ETS1 and GFI1 transcription factors in the repression of Bax gene expression. Oncogene 26:3541–3550. doi: 10.1038/sj.onc.1210140. [DOI] [PubMed] [Google Scholar]
- 39.Huang D-Y, Kuo Y-Y, Chang Z-F. 2005. GATA-1 mediates auto-regulation of Gfi-1B transcription in K562 cells. Nucleic Acids Res 33:5331–5342. doi: 10.1093/nar/gki838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marteijn JAF, van der Meer LT, van Emst L, van Reijmersdal S, Wissink W, de Witte T, Jansen JH, Van der Reijden BA. 2007. Gfi1 ubiquitination and proteasomal degradation is inhibited by the ubiquitin ligase Triad1. Blood 110:3128–3135. doi: 10.1182/blood-2006-11-058602. [DOI] [PubMed] [Google Scholar]
- 41.Dahl R, Iyer SR, Owens KS, Cuylear DD, Simon MC. 2007. The transcriptional repressor GFI-1 antagonizes PU.1 activity through protein-protein interaction. J Biol Chem 282:6473–6483. doi: 10.1074/jbc.M607613200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andrade D, Velinder M, Singer J, Maese L, Bareyan D, Nguyen H, Chandrasekharan MB, Lucente H, McClellan D, Jones D, Sharma S, Liu F, Engel ME. 2016. SUMOylation regulates growth factor independence 1 in transcriptional control and hematopoiesis. Mol Cell Biol 36:1438–1450. doi: 10.1128/MCB.01001-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saleque S, Kim J, Rooke HM, Orkin SH. 2007. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol Cell 27:562–572. doi: 10.1016/j.molcel.2007.06.039. [DOI] [PubMed] [Google Scholar]
- 44.Lin Y, Wu Y, Li J, Dong C, Ye X, Chi Y-I, Evers BM, Zhou BP. 2010. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J 29:1803–1816. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laurent B, Randrianarison-Huetz V, Frisan E, Andrieu-Soler C, Soler E, Fontenay M, Dusanter-Fourt I, Duménil D. 2012. A short Gfi-1B isoform controls erythroid differentiation by recruiting the LSD1-CoREST complex through the dimethylation of its SNAG domain. J Cell Sci 125:993–1002. doi: 10.1242/jcs.095877. [DOI] [PubMed] [Google Scholar]
- 46.Velinder M, Singer J, Bareyan D, Meznarich J, Tracy CM, Fulcher JM, McClellan D, Lucente H, Franklin S, Sharma S, Engel ME. 2017. GFI1 functions in transcriptional control and cell fate determination require SNAG domain methylation to recruit LSD1. Biochem J 474:2951. doi: 10.1042/BCJ-2016-0558_COR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Upadhyay G, Chowdhury AH, Vaidyanathan B, Kim D, Saleque S. 2014. Antagonistic actions of Rcor proteins regulate LSD1 activity and cellular differentiation. Proc Natl Acad Sci U S A 111:8071–8076. doi: 10.1073/pnas.1404292111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 49.Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters A, Günther T, Buettner R, Schüle R. 2005. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 50.Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, Berger SL. 2007. p53 is regulated by the lysine demethylase LSD1. Nature 449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 51.Yang J, Huang J, Dasgupta M, Sears N, Miyagi M, Wang B, Chance MR, Chen X, Du Y, Wang Y, An L, Wang Q, Lu T, Zhang X, Wang Z, Stark GR. 2010. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci U S A 107:21499–21504. doi: 10.1073/pnas.1016147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang X, Tanaka K, Yan J, Li J, Peng D, Jiang Y, Yang Z, Barton MC, Wen H, Shi X. 2013. Regulation of estrogen receptor α by histone methyltransferase SMYD2-mediated protein methylation. Proc Natl Acad Sci U S A 110:17284–17289. doi: 10.1073/pnas.1307959110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cho H-S, Suzuki T, Dohmae N, Hayami S, Unoki M, Yoshimatsu M, Toyokawa G, Takawa M, Chen T, Kurash JK, Field HI, Ponder BAJ, Nakamura Y, Hamamoto R. 2011. Demethylation of RB regulator MYPT1 by histone demethylase LSD1 promotes cell cycle progression in cancer cells. Cancer Res 71:655–660. doi: 10.1158/0008-5472.CAN-10-2446. [DOI] [PubMed] [Google Scholar]
- 54.Lee J-Y, Park J-H, Choi H-J, Won H-Y, Joo H-S, Shin D-H, Park MK, Han B, Kim KP, Lee TJ, Croce CM, Kong G. 2017. LSD1 demethylates HIF1α to inhibit hydroxylation and ubiquitin-mediated degradation in tumor angiogenesis. Oncogene 36:5512–5521. doi: 10.1038/onc.2017.158. [DOI] [PubMed] [Google Scholar]
- 55.Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, Gaudet F, Li E, Chen T. 2009. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet 41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 56.Kontaki H, Talianidis I. 2010. Lysine methylation regulates E2F1-induced cell death. Mol Cell 39:152–160. doi: 10.1016/j.molcel.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 57.Zhang C, Hoang N, Leng F, Saxena L, Lee L, Alejo S, Qi D, Khal A, Sun H, Lu F, Zhang H. 2018. LSD1 demethylase and the methyl-binding protein PHF20L1 prevent SET7 methyltransferase-dependent proteolysis of the stem-cell protein SOX2. J Biol Chem 293:3663–3674. doi: 10.1074/jbc.RA117.000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, Liang J, Sun L, Yang X, Shi L, Li R, Li Y, Zhang Y, Li Q, Yi X, Shang Y. 2009. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell 138:660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 59.Yang Y, Huang W, Qiu R, Liu R, Zeng Y, Gao J, Zheng Y, Hou Y, Wang S, Yu W, Leng S, Feng D, Wang Y. 2018. LSD1 coordinates with the SIN3A/HDAC complex and maintains sensitivity to chemotherapy in breast cancer. J Mol Cell Biol 10:285–301. doi: 10.1093/jmcb/mjy021. [DOI] [PubMed] [Google Scholar]
- 60.Ray SK, Li HJ, Metzger E, Schüle R, Leiter AB. 2014. CtBP and associated LSD1 are required for transcriptional activation by NeuroD1 in gastrointestinal endocrine cells. Mol Cell Biol 34:2308–2317. doi: 10.1128/MCB.01600-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsai M-C, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shi Y-J, Matson C, Lan F, Iwase S, Baba T, Shi Y. 2005. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell 19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 63.Roux KJ, Kim DI, Raida M, Burke B. 2012. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim DI, Jensen SC, Noble KA, Kc B, Roux KH, Motamedchaboki K, Roux KJ. 2016. An improved smaller biotin ligase for BioID proximity labeling. Mol Biol Cell 27:1188–1196. doi: 10.1091/mbc.E15-12-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Garçon L, Lacout C, Svinartchouk F, Le Couédic J-P, Villeval J-L, Vainchenker W, Duménil D. 2005. Gfi-1B plays a critical role in terminal differentiation of normal and transformed erythroid progenitor cells. Blood 105:1448–1455. doi: 10.1182/blood-2003-11-4068. [DOI] [PubMed] [Google Scholar]
- 66.Lam LT, Ronchini C, Norton J, Capobianco AJ, Bresnick EH. 2000. Suppression of erythroid but not megakaryocytic differentiation of human K562 erythroleukemic cells by notch-1. J Biol Chem 275:19676–19684. doi: 10.1074/jbc.M002866200. [DOI] [PubMed] [Google Scholar]
- 67.Meng Y-S, Khoury H, Dick JE, Minden MD. 2005. Oncogenic potential of the transcription factor LYL1 in acute myeloblastic leukemia. Leukemia 19:1941–1947. doi: 10.1038/sj.leu.2403836. [DOI] [PubMed] [Google Scholar]
- 68.Maiques-Diaz A, Somervaille TC. 2016. LSD1: biologic roles and therapeutic targeting. Epigenomics 8:1103–1116. doi: 10.2217/epi-2016-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim DI, Roux KJ. 2016. Filling the void: proximity-based labeling of proteins in living cells. Trends Cell Biol 26:804–817. doi: 10.1016/j.tcb.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leon-Del-Rio A, Gravel RA. 1994. Sequence requirements for the biotinylation of carboxyl-terminal fragments of human propionyl-CoA carboxylase alpha subunit expressed in Escherichia coli. J Biol Chem 269:22964–22968. [PubMed] [Google Scholar]
- 71.Vassen L, Fiolka K, Mahlmann S, Möröy T. 2005. Direct transcriptional repression of the genes encoding the zinc-finger proteins Gfi1b and Gfi1 by Gfi1b. Nucleic Acids Res 33:987–998. doi: 10.1093/nar/gki243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rodriguez P, Bonte E, Krijgsveld J, Kolodziej KE, Guyot B, Heck AJR, Vyas P, de Boer E, Grosveld F, Strouboulis J. 2005. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J 24:2354–2366. doi: 10.1038/sj.emboj.7600702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuo Y-Y, Chang Z-F. 2007. GATA-1 and Gfi-1B interplay to regulate Bcl-xL transcription. Mol Cell Biol 27:4261–4272. doi: 10.1128/MCB.02212-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jegalian AG, Wu H. 2002. Regulation of Socs gene expression by the proto-oncoprotein GFI-1B: two routes for STAT5 target gene induction by erythropoietin. J Biol Chem 277:2345–2352. doi: 10.1074/jbc.M105575200. [DOI] [PubMed] [Google Scholar]
- 75.Chowdhury AH, Ramroop JR, Upadhyay G, Sengupta A, Andrzejczyk A, Saleque S. 2013. Differential transcriptional regulation of meis1 by Gfi1b and its co-factors LSD1 and CoREST. PLoS One 8:e53666. doi: 10.1371/journal.pone.0053666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vassen L, Fiolka K, Möröy T. 2006. Gfi1b alters histone methylation at target gene promoters and sites of gamma-satellite containing heterochromatin. EMBO J 25:2409–2419. doi: 10.1038/sj.emboj.7601124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ceballos-Chávez M, Rivero S, García-Gutiérrez P, Rodríguez-Paredes M, García-Domínguez M, Bhattacharya S, Reyes JC. 2012. Control of neuronal differentiation by sumoylation of BRAF35, a subunit of the LSD1-CoREST histone demethylase complex. Proc Natl Acad Sci U S A 109:8085–8090. doi: 10.1073/pnas.1121522109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Esteghamat F, van Dijk TB, Braun H, Dekker S, van der Linden R, Hou J, Fanis P, Demmers J, van IJcken W, Ozgür Z, Horos R, Pourfarzad F, Lindern von M, Philipsen S. 2011. The DNA binding factor Hmg20b is a repressor of erythroid differentiation. Haematologica 96:1252–1260. doi: 10.3324/haematol.2011.045211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hock R, Furusawa T, Ueda T, Bustin M. 2007. HMG chromosomal proteins in development and disease. Trends Cell Biol 17:72–79. doi: 10.1016/j.tcb.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Malarkey CS, Churchill M. 2012. The high mobility group box: the ultimate utility player of a cell. Trends Biochem Sci 37:553–562. doi: 10.1016/j.tibs.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wynder C, Hakimi M-A, Epstein JA, Shilatifard A, Shiekhattar R. 2005. Recruitment of MLL by HMG-domain protein iBRAF promotes neural differentiation. Nat Cell Biol 7:1113–1117. doi: 10.1038/ncb1312. [DOI] [PubMed] [Google Scholar]
- 82.Chai P, Tian J, Zhao D, Zhang H, Cui J, Ding K, Liu B. 2016. GSE1 negative regulation by miR-489-5p promotes breast cancer cell proliferation and invasion. Biochem Biophys Res Commun 471:123–128. doi: 10.1016/j.bbrc.2016.01.168. [DOI] [PubMed] [Google Scholar]
- 83.Ding K, Tan S, Huang X, Wang X, Li X, Fan R, Zhu Y, Lobie PE, Wang W, Wu Z. 2018. GSE1 predicts poor survival outcome in gastric cancer patients by SLC7A5 enhancement of tumor growth and metastasis. J Biol Chem 293:3949–3964. doi: 10.1074/jbc.RA117.001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, Weirauch MT. 2018. The human transcription factors. Cell 172:650–665. doi: 10.1016/j.cell.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 85.Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu Rev Biochem 78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 86.Grimes HL, Chan TO, Zweidler-McKay PA, Tong B, Tsichlis PN. 1996. The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol Cell Biol 16:6263–6272. doi: 10.1128/MCB.16.11.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen Y, Yang Y, Wang F, Wan K, Yamane K, Zhang Y, Lei M. 2006. Crystal structure of human histone lysine-specific demethylase 1 (LSD1). Proc Natl Acad Sci U S A 103:13956–13961. doi: 10.1073/pnas.0606381103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang M, Gocke CB, Luo X, Borek D, Tomchick DR, Machius M, Otwinowski Z, Yu H. 2006. Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol Cell 23:377–387. doi: 10.1016/j.molcel.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 89.Ouyang J, Shi Y, Valin A, Xuan Y, Gill G. 2009. Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex. Mol Cell 34:145–154. doi: 10.1016/j.molcel.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nalawansha DA, Pflum M. 2017. LSD1 substrate binding and gene expression are affected by HDAC1-mediated deacetylation. ACS Chem Biol 12:254–264. doi: 10.1021/acschembio.6b00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Branon TC, Bosch JA, Sanchez AD, Udeshi ND, Svinkina T, Carr SA, Feldman JL, Perrimon N, Ting AY. 2018. Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol 36:880–887. doi: 10.1038/nbt.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lam SS, Martell JD, Kamer KJ, Deerinck TJ, Ellisman MH, Mootha VK, Ting AY. 2015. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat Methods 12:51–54. doi: 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Holmes A, Roseaulin L, Schurra C, Waxin H, Lambert S, Zaratiegui M, Martienssen RA, Arcangioli B. 2012. Lsd1 and lsd2 control programmed replication fork pauses and imprinting in fission yeast. Cell Rep 2:1513–1520. doi: 10.1016/j.celrep.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sumoy L, Carim L, Escarceller M, Nadal M, Gratacòs M, Pujana MA, Estivill X, Peral B. 2000. HMG20A and HMG20B map to human chromosomes 15q24 and 19p13.3 and constitute a distinct class of HMG-box genes with ubiquitous expression. Cytogenet Cell Genet 88:62–67. doi: 10.1159/000015486. [DOI] [PubMed] [Google Scholar]
- 95.Rivero S, Ceballos-Chávez M, Bhattacharya SS, Reyes JC. 2015. HMG20A is required for SNAI1-mediated epithelial to mesenchymal transition. Oncogene 34:5264–5276. doi: 10.1038/onc.2014.446. [DOI] [PubMed] [Google Scholar]
- 96.Hakimi M-A, Bochar DA, Chenoweth J, Lane WS, Mandel G, Shiekhattar R. 2002. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc Natl Acad Sci U S A 99:7420–7425. doi: 10.1073/pnas.112008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Eubank RL. 1999. Nonparametric regression and spline smoothing, 2nd ed CRC Press, Boca Raton, FL. [Google Scholar]
- 98.Ke C, Wang Y. 2004. Smoothing spline nonlinear nonparametric regression models. J Am Stat Assoc 99:1166–1175. doi: 10.1198/016214504000000755. [DOI] [Google Scholar]
- 99.Reimand J, Arak T, Adler P, Kolberg L, Reisberg S, Peterson H, Vilo J. 2016. g:Profiler—a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res 44:W83–W89. doi: 10.1093/nar/gkw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.