Abstract
The host-microbiome supraorganism appears to have coevolved and the unperturbed microbial component of the dyad renders host health sustainable. This coevolution has likely shaped evolving phenotypes in all life forms on this predominantly microbial planet. The microbiota seems to exert effects on the next generation from gestation, via maternal microbiota and immune responses. The microbiota ecosystems develop, restricted to their epithelial niches by the host immune system, concomitantly with the host chronological development, providing early modulation of physiological host development and functions for nutrition, immunity and resistance to pathogens at all ages. Here, we review the role of the microbiome in human development, including evolutionary considerations, and the maternal/fetal relationships, contributions to nutrition and growth. We also discuss what constitutes a healthy microbiota, how antimicrobial modern practices are impacting the human microbiota, the associations between microbiota perturbations, host responses and diseases rocketing in urban societies and potential for future restoration.
Keywords: human microbiome, development, evolution, perturbations, restoration
Evolution of the microbiota
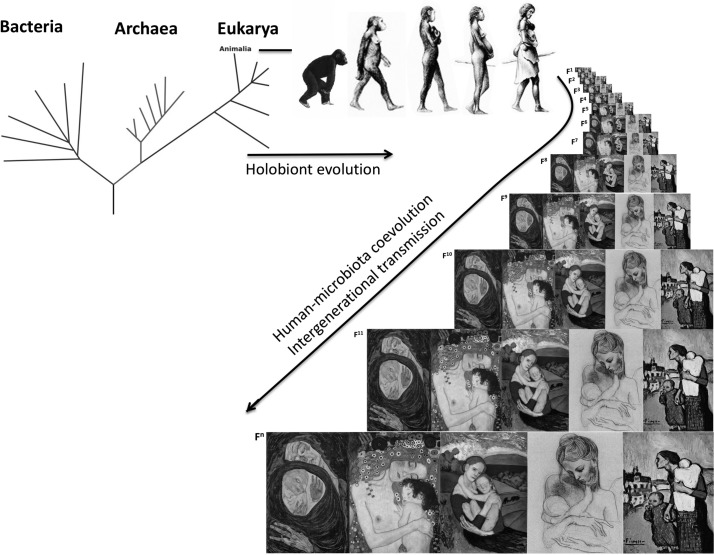
Bacteria arose about 3.8 billion years ago,1 and the eukaryotic lineage, which includes humans, arose after the oxygenation of earth’s atmosphere 2.2–2.4 billion years ago.2 Together with archaea, protists and fungi, bacteria remained free-living single cells although some became host-associated. Thus, an animal holobiont (the animal host and its evolved microbial communities)3 spans the phylogenetic tree: the animal host, plus its associated microbiota such as bacteria, archaea, fungi, protists, helminths and viruses (figure 1). The collective genome content of microbiota or the microbial metagenome was coined the microbiome,4 although microbiome and microbiota are currently used interchangeably.
Figure 1.
Evolution of the holobiont and vertical transmission through human generations.
By coevolving with the host, the microbiome has shaped phenotypes in our ancestral lineages. The congruence of the phylogenetic trees of intestinal bacterial microbiota and primates5 demonstrates host-microbiota coevolution and implies within-species transmission of microbes across generations. Through the process of natural selection, mutations lead to evolutionary adaptations to environmental conditions and increased fitness in these environments. Human environments have changed dramatically during human evolution, and dietary changes and exposures to famine have been major selective pressures. While there is evidence of adaptive survival traits to starvation on the human genome,6 human microbiome adaptations that offer energy-sparing traits for the human host remain unknown. Abrupt changes in environmental conditions can lead to mal-adaptations (adaptations that were beneficial when first took place, but not anymore under new environmental conditions). Today, modernisation and urbanisation pose exactly this challenge to human health.
Together with their microbionts (microbiota members), hosts evolved an immune system, which prevents microbial colonisation in the topological interior of the body. Host immune systems evolved complex mechanisms to identify and destroy invading microbes, whether they are microbionts or primary pathogens that cross into forbidden territories. Immune molecules evolved more than 500 million years ago, in choanoflagellates, unicellular progenitors of metazoans,7 and there is growing evidence that the innate immune system—antimicrobial peptides and repertoire of pattern recognition receptors—evolved in response to the need for controlling the epithelium-colonising microbiota.8
The human immune system restricts microbiota to their natural niches in the body ‘exterior’ and invaginations: epithelia that cover the body (such as skin and mucosa) and the gut, which, strictly speaking, is a hollow tube that traverses the body with the influx of external materials (diet). Thus, the microbiota occupies the interface between our bodies and the exterior, and interactions with the environment (including diet, sun-light, bathing, cosmetics) cross this interface. The microbiota is at the same time self and non-self: it is part of our biology, but consists of fast-evolving entities that respond rapidly on physiological, ecological and evolutionary timescales to external perturbations in ways that affect our phenotypes (figure 1). The gut microbiota have been shown to impact diverse physiological processes ranging from adiposity/obesity, to energy metabolism, blood pressure control, glucose homeostasis, clotting risks or even behaviour. In each case, there are mechanistic ties between gut microbes, metabolites they generate and host receptors and phenotypic responses. Evolutionary considerations are crucial to understanding the nature of microbial-host interactions, perturbations and health consequences and will ultimately need to be understood and exploited in order to prevent and treat ‘modern’ diseases.
Development and the microbiota: from fertilisation to birth
In some insects, bacteria colonise egg capsules during mating, and the individual is colonised before hatching.9 In mammals, fertilisation occurs in an immune-protected organ, the uterus. However, immune protection means lack of colonisation, but not necessarily sterility at all times. Indeed, it seems possible that some bacterial cells of the uterine cervix10 may enter with the sperm during fertilisation and reach the egg at the time of fertilisation, implantation or early embryonic development. Regardless, immunity appears to preclude the establishment of a microbial community in immune-protected organs. Uterus, placenta, fetus as well as blood appear void of a microbiota, although they may contain bacterial DNA or even some isolated live bacteria. There is a current controversy about whether the presence of bacterial DNA contradicts the notion of sterility, but the presence of circulating bacterial DNA, such in the blood11 or placenta,12 or even sporadic presence of an alive intruder bacteria does not demonstrate a living blood microbiota and does not challenge the current paradigm of sterility in immune-protected organs. There might be a transitory ‘mini-sepsis’ when live cells enter the blood after injuries, microabrasions or mucosal ‘leaking’13—including transient bacteraemia due to tooth-brushing14—, but in healthy individuals, the intruders are cleared by phagocytic cells rather than colonising and assembling microbial communities. Fetal development is an important period for the reproduction of placental species, and gestational infection and inflammation reduce fecundity and increase the risk of spontaneous preterm birth15 16 The concept of sterile fetal development remains, and little is known about mechanisms and functions of transplacental trafficking of free nucleic acids.
The maternal microbiota may exert an indirect effect on the fetus via maternal factors such as maternal immune responses or microbial metabolites that cross the placenta17–19 or more indirectly via factors that may mediate epigenetic programming in the fetus, such as diet,20 stress21 or neuroendocrine exposure,22–24 which also affect the maternal microbiota. The gut25 and vaginal26 maternal microbiota change with gestation, and whether or not these changes have adaptive value for the mother or baby is still unknown. It has been suggested that they allow the fetus to derive energy from the mother’s blood, more efficiently,25 or that butyrate-producing bacteria may sustain gut epithelial functions and promote immune tolerance in the mother.27
Labour and birth represent the first major exposure to a complex microbiota and is the primordial mechanism for intergenerational microbiota transfer in mammals. Ancestral vertebrates (birds, reptiles, finned fishes) and, exceptionally among mammals, the Monotremes, lay eggs through a single canal—the cloaca—shared for excretion and reproduction. Placental mammals evolved separate canals for reproduction (vagina), excretion of faeces (anus) and urine (urethra), and the birth canal is always adjacent to the rectum (but not the urethra), providing an efficient mechanism for intergenerational transmission of both vaginal and gut microbes. Rupture of the chorioamniotic membrane allows exposure of the baby to the maternal vaginal and perineal faecal microbes. Indeed, prolonged labour poses a risk of infection by opportunistic microbionts.28 Infants are naturally born with their skin and mouth covered by maternal inocula and have swallowed these microbes,29 30 supported by the observation of both DNA31 and live bacteria31 in the meconium. Thus, we inherit the primordial microbiota from our mothers, grandmothers and further on the matrilineal line, with microbial vertical transmission extending back to earlier ancestors32 (box 1). Whether the primordial inoculum contains most microbes that will be nurtured by the child, and which maternal strains colonise which parts of the baby’s body and their functions, the paternal and sibling contribution along with the infant’s microbial diversity33 and the extent to which modern practices reduce intergenerational transmission, are still not completely understood. C-section as intrapartum antibiotics during vaginal delivery alter bacterial colonisation in the neonates.34
Box 1. Highlights of the holobiont evolution.
Evolution of all complex life forms has occurred in associations with bacteria, the first forms of life on earth.
The human body carries representatives of all branches of the tree of life (Animalia-Homo sapiens, and protozoa, fungi, archaea, bacteria conforming the microbiota).
The microbiota has been transferred throughout generations of humans, with the matrilineal line transferring the primordial birth microbiota.
The vertical human transmission has led to conservation of a phylogenetic signal in human microbiota communities.
Postnatal development of the microbiota
By definition, placental mammals develop in a placenta, are born through the maternal vagina and drink maternal milk during the initial developmental window during which remarkable changes occur. Animals can develop without microbiota, as shown by the existence of germ-free mice, rats, chickens and pigs, but they have abnormal phenotypes and the microbiota is believed to be required for normal development. Pioneer neonatal bacteria prime the development of the microbiota, immune, metabolic, hormonal and nervous systems in the neonate.35 36 Under natural conditions, the neonate and the microbiota develop in an orchestrated fashion under the nutritional, immunological, hormonal and prebiotic effect of maternal milk—a single food of complex biological formulation.37 Bacteria acquired during labour include lactic acid bacteria that digest lactose, and others that use substrates that are indigestible for the babies (indigestible milk glycans known as human milk oligosaccharides, or HMOs),38 39 with polymorphisms, such as in fucose transferase gene FUT2, associated with selective effects of HMOs on the infant microbiota composition,40 which in turn can affect the susceptibility to immune diseases later in life.41 Milk also includes urea and oxalate, two end-products of human metabolism. Why would nature include such indigestible molecules in the diet of young mammals? There are beneficial microbes that can use these molecules such as carbon, nitrogen or other energy sources. The types of glycans found in breast milk can shape the infant gut microbiota and the microbial composition of breast milk, specifically of Bifidobacterium species.42 The degree of development of the sensory and motor capabilities of the brain in neonates during strict lactation is remarkable. Ultimately, understanding this period, the functions of milk glycans and other molecules, and the microbes they select, will be critical to understand human development.
Other ways in which the microbiome has been related to development include the synthesis of vitamins during postnatal development. There is poor vitamin K diffusion through the placental barrier,43 thus neonates are born with low vitamin K levels.44 Later, intestinal bacteria will provide K2 or menaquinone, and older children will consume it from vegetables in the form of phylloquinone. Vitamin K is necessary to synthesise functional forms of coagulation factors II, VII, IX, and X in the liver.45 Often clinicians consider that neonates are vitamin K-deficient, but again, from the evolutionary perspective, we need to ask why has this trait been selected during our evolution. Similarly, it is thought that babies that are exclusively breastfed may become deficient in vitamin B12 due to the lack of solid foods rich in this vitamin.46 Does it hold adaptive value or is it a maladaptation? The answer to this question is important, since clinical interpretations lead to public health measures that affect millions of infants, such as the recommended vitamin K boost to all neonates.47 We need to understand our biology first, before we define pathologic scenarios and intervene. Much research is needed to identify adaptations that we must respect, and then define the maladaptations that need to be addressed.
The microbiota development trajectory from birth follows dynamic changes. Immediately after birth, there seems to be a decrease in gut alpha diversity48 49 probably reflecting the selective pressure of the substrate constraints of milk, and by 1 week of age, the gut microbiota is already very similar to that in a month-old baby.50 Infants develop during the first 6 months under the selective pressure of milk shaping the gut microbial communities, whose metabolites promote peripheral regulatory T-cell generation.51 Bacteria given to germ-free mice induce germinal centres (lymphoid cells) to produce IgA+ B cells.52 Bacterial molecules also induce mucosa-associated lymphoid tissue of the intestine, via Toll-like receptors, and shape the intestinal Th-cell mediated immunity.53 Thus, antigen-driven priming/activation, polarisation and expansion of naïve T cells yield Th1 and/or Th17 effector cells,54 which enter the systemic circulation and home to the gut to help destroy the invading pathogens.55 GF animals consistently exhibit impaired development of Peyer’s patches,56 have reduced numbers of T-helper (Th)1 and Th17 cells, with the intestinal T-cell immune response primarily controlled by Th2 cells.57 Importantly, the imbalances in Th-cell responses in GF mice can be reversed by restoring the microbiota.52 Th17/Tregs are involved in tolerance of and is induced by microbionts such as H. pylori and commensal Clostridia-related bacteria.58 59 Perturbations that reduce transmission and early colonisation of human bacteria lead to reduced numbers of Th17 cells in the small intestine.52 Environmental variables may also affect the microbiota, such as number of siblings (babies with siblings have increased gut Bifidobacterium catenulatum) and sex (girls having higher gut B. fragilis and Lactobacillus spp. than boys).60 Finally, evidence suggests that longer duration of breastfeeding is associated with decrease in risk of overweight.61
In the large intestine and colon, bacteria can either colonise the epithelial mucosa, digesta particles or live free in suspension in the liquid phase. Particles and liquid colonisation is dictated in part by transit time in the intestine. After strict lactation ends, dentition begins, and the GI system of the baby has matured to handle dietary solids that reach the postabsorptive sites and bring new substrates. These solids change the conditions in the hindgut, selecting for bacterial populations with relevant metabolic activities and the microbial diversity of the intestine increases steadily until at least age 3 years.62 This increase in diversity may reflect the increased chemical diversity of a complex diet and the gut and immune maturation of the host.
The new solid diet has components refractory to proximal intestinal digestion, such as some starches and cell wall polysaccharides of plant origin, which are fermented by bacteria in the large intestine. The products, short chain fatty acids, have modulatory roles in host metabolism and immunity. Butyrate has beneficial effects, being energy source for colonocytes, maintaining epithelial integrity in the gut,63 supporting Treg differentiation and driving anti-inflammatory responses,51 as shown with bacterial butyrate producers, such as Faecalibacterium prausnitzii,,64 or by direct supplementation of butyrate to mice.65 Propionate also potentiates de novo Treg-cell generation in the periphery.51 Butyrate and acetate regulate satiety,66 67 with acetate being obesogenic.67 High Bacteroides and low acetogens and methanogens have been associated with reduced weight gain.68
The convergence in metabolic products produced by the repertoire of microbes in the gut ecosystem is an example of functional redundancy. Functional redundancy is a recognised trait of the microbiota in human adults,69 leading to high interindividual variability and, importantly, increasing resilience of the ecosystem. Redundancy probably increases with age, at least during the first 3 years of life, when diversity is gained,62 but this phenomenon is still poorly understood. We are depleting our ancestral microbiome diversity and its functional repertoire, and this results in compromising redundancy, with detrimental effects on the resilience that governs complex metabolic interactions.
Modern disruptors of the microbiota and modern diseases
Industrial urban societies have dramatically changed human lifestyle in relation to traditional societies, and the world is becoming increasingly urban. The changes are complex, including housing, urban plan, human density, home architecture, technologic isolation of houses from the environment, ventilation, diet, clothing, exercise, personal care products and medicines. Understanding what changes with urbanisation requires a multidisciplinary approach and is important because sudden environmental changes may lead to maladaptations. Urbanisation is indeed associated with increased risks of immune and metabolic diseases, including obesity, T1D, behavioural disorders, IBD and asthma, all of which have been increasing in recent decades,70–73 and with reduced gut microbiota diversity.62 74 Although human genetics affects host development as well as the structure of the microbiota,75 the effect of environmental factors on the microbiota is known to be substantial. Practices that significantly impair transmission and colonisation of bacteria early in life are abundant in modern societies, and we have learnt from ecological studies that compounded impacts or removal of high competitive populations reduces alpha diversity, while removal of more cooperative bacteria (or of redundant bacteria) has a smaller effect on diversity. Selective pressures that are shaping microbiome characteristics within high-income countries may include prenatal and postnatal antibiotics exposure, dietary antimicrobials, toothpaste, soaps and perhaps even consumption of chlorinated water. The direction of changes of the microbial ecosystem after perturbations depends on the ecosystem and does not always imply reduction of diversity. For example, perturbations in the microbiota of the vagina and of the stomach, which are naturally of low alpha diversity, increase richness and evenness.
There are connections between increased disease risks and microbiota. Obesity risk has been epidemiologically associated with C-section birthing and early antibiotic exposure.73 Evidence of obesity causation has been shown in mice,76 with population-scale studies leading to identification of bacteria that when transplanted to mice had physiological effect on body mass.77
Early life microbiota functions are likely to be key in understanding the aetiology of chronic immune diseases of urban societies and where potential for their prevention resides. In the gut, reduced microbiota diversity is consistent with reduction in resilience—the capacity to bounce back after perturbations—and in resistance—to pathogens. In clean urban settings, where sanitation and vaccines diminish colonisation by pathogens, reduction in resistance might not be as important as it was as in ancestral human societies exposed to more infectious challenges). However, with the compounded effects of perturbations exerted by the antimicrobial practices of modern life, loss of resilience might be important. Moreover, much research is needed to understand the role of gut microbiota in vaccine and immunisation efficacy, the timing of vaccines provided to infants and children and the impact of not just antibiotics, but other drugs78 on microbial community structure.
Gestational use of antibiotics affects microbiota colonisation in the infants.79 In the USA, about one in two women is prescribed an antibiotic during pregnancy or at term, and beta-lactams, vancomycin, nitrofurantoin, metronidazole, clindamycin and fosfomycin are generally considered safe. However, antibiotics given 4–5 days before birth in mice changed proportions in gut bacteria in the litters and affected lung lymphoid cell development.80 Lack of maternal microbiota exposure at birth, as happens in the C-section born, leads to alterations in the microbiota of babies30 49 and in fatty acid and bile acid metabolism.60 Formula also alters the baby microbiota81 and the modern practice by working mothers, of bottle feeding breast milk -rather than directly breastfeeding- may also have effects. This practice involves refrigerating or freezing - thawing and reheating breast milk, and reduces maternal-baby contact. How this might affect microbial transmission has not been studied. Thus, the compounded effect of prenatal, perinatal and postnatal antibiotics, C-section birth, formula feeding, reduced skin to skin and mouth-breast contact between babies and mothers, extensive bathing of the neonate and other hospital interventions and a built environment isolated from the natural environment, might all count and be cumulative in their effects.
More information is needed to understand the functions of the early microbiota and its relation with later health conditions. Cohort studies are currently ongoing; ClinicalTrials.org shows 17 longitudinal clinical trials with interventions and 18 without interventions, being performed in infants (table 1).
Table 1.
Clinical studies on the development and restoration of the infant microbiome
| Intervention/Follow-up up without intervention | ClinicalTrials.gov Identifier | Participant enrolment |
| Antibiotics at birth | NCT02477423 | 80 |
| Antibiotics in preterm | NCT02784821 | 420 |
| At-birth restoration with vaginal fluids | NCT03298334 | 800 |
| Diet supplementation—eggs | NCT03385252 | 662 |
| Diet supplementation—sweet potato vs pear | NCT03229863 | 120 |
| Diet supplementation—Human milk fortifier | NCT03214822 | 30 |
| Diet supplementation-maternal—Omega-3 | NCT03297801 | 109 |
| Diet supplementation—protein sources | NCT02142647 | 40 |
| Human milk donor | NCT02573779 | 125 |
| Probiotic/prebiotic supplemented formula | NCT03320837 | 108 |
| Probiotics | NCT03388112 | 30 |
| Probiotics B. breve | NCT03219931 | 320 |
| Probiotics in preterm | NCT02197468 | 60 |
| Probiotics in preterm | NCT02695784 | 40 |
| Rotavirus vaccine | NCT03031743 | 88 |
| Rotavirus vaccine | NCT02220439 | 76 |
| Skin-to-skin contact | NCT03181269 | 88 |
| Follow-up without intervention | NCT03001167, NCT02121938, NCT03365583, NCT03236194, NCT01789268, NCT02778750, NCT02526004, NCT02843087, NCT03229967, NCT01661491, NCT03235635, NCT03296631, NCT03396198, NCT02836119, NCT03335202, NCT02061306, NCT03213275, NCT03373721 | Variable |
The interventions include important issues such as at birth exposure to vaginal fluids and skin-to-skin contact, dietary supplementation with probiotics, proteins, carbohydrates, fortified milk, antibiotics in preterm babies. Outcomes include microbiota development, bacteriophage populations during development, infant growth, urinary metabolites, immune profiles, incidence of infections, the development of infantile colic, celiac disease and bronchopulmonary dysplasia as well as sleep behaviour and neurodevelopment. Causation studies are difficult in humans, and normally involve longitudinal randomised clinical trials, which are expensive, and in the USA typically require an Investigational New Drug (IND) approval from the Food and Drug Administration (FDA), which adds costs, complexity and time to the studies. Certainly, more studies are needed to determine reproducibility, safety and benefits of early restorations to C-section-born babies, particularly in the context of randomised control studies addressing the risk of asthma, atopy and other relevant clinical endpoints.
Medicine is one of the great human creations, and its life-saving capability has driven the substantial increase in human lifespans. Medical interventions such as vaccines, antibiotics and surgery have contributed dramatically to improve life expectancy. For example, antibiotics treat major killers including diarrheal disease and pneumonia and C-sections save both infants and mothers, with formula nourishing and supplementing children that cannot be breastfed. However, these interventions come with costs that have been underestimated, with the consequent overuse and abuse. Such costs are only justified when the intervention is needed. Changes in practice will only arrest the current trend, and restoration efforts will be needed to decrease the intervention costs. Identifying the ‘when’ and ‘what’, the timing of interventions and the breadth of influence of specific microbial species and strains, is essential if we are to intervene effectively. Restoration efforts ought to be carefully considered, weighing risks and potential benefits. For example, restoration of the neonatal microbiota of C-section born neonates with maternal vaginal fluids82 has raised concerns of infection risks to neonates posed to by vaginal exposure83 84 (see also ACOG note in https://www.acog.org/Clinical-Guidance-and-Publications/Committee-Opinions/Committee-on-Obstetric-Practice/Vaginal-Seeding), and although this exposure is natural and has been conserved over the millions of years of mammalian evolution, only solid scientific demonstration of health benefits will pave the road for the practice to become standard clinical practice. Restoration approaches are also promising to counteract the associations between altered microbiology in successfully treated childhood cancers and the consequent persistent increased risk of inflammatory diseases.85
Creating a synthetic human milk
Research on prebiotic and probiotic functions of human maternal milk could lead to the design of synbiotic formulas that respect the developmental biology of the child and drive a healthy infant gut, although frankly, it will take years to produce a biologically appropriate synthetic human milk that includes the changing circadian and developmental levels of glycans (HMOs), hormones, cells and antibodies. We are far from being there, but the initial efforts have started with the Danish Biotech Glycom adding N- Acetyl-D-Neuraminic acid to formula.86 Understanding the coevolution of milk glycans, the immune system and gut bacteria in infancy across mammals may provide a translational model for modulation of the gut microbiota.
Research on prebiotic and probiotic functions of human maternal milk could provide an important information base to design synbiotic formulas that respect the developmental biology of the child, to drive a healthy infant gut. We are far from being there, but the initial efforts have started with the Danish Biotech Glycom adding N- Acetyl-D-Neuraminic acid to formula,86 but of course milk is far more than that and contains glycans (HMOs), hormones and antibodies. A recent oral synbiotic preparation of a Lactobacillus plantarum and fructooligosaccharide resulted in a reduction of neonatal sepsis in rural Indian newborns.87 Understanding the coevolution of milk glycans, the immune system and gut bacteria in infancy across mammals may be critical in improving human health in infants and provides a translational model for modulation of the gut microbiota.
Restorations of mothers to hand out the next generation microbiota
The idea of freezing healthy stools and using them to restore after antibiotic treatment has not been implemented but seems ecologically plausible. Babies could have their predisease microbiota restored, and adults too, particularly women, who play a particularly important role in transmitting the human microbiota to the next generation. There is a need for services provided by companies, who allow families to regularly store the growing infant microbiota, for this purpose; being a self-transplant, it is not clear what will be required from regulatory agencies such as the FDA.
What is a healthy microbiome?
Individuals differ enormously in the taxonomic content of their microbiota, and even the same person over time can appear dramatically different from their own prior representation. Functional redundancy makes the characterisation of the healthy microbiome extremely complex, because different taxonomic profiles can lead to ecosystems with similar behaviour. It is also unclear whether ‘normal’ in a human population implies healthy, because the health optimum might be context-dependent both at a population and at an individual level—is the average microbiome of lean 20-year-olds, half of whom will become chronically ill 70-year-olds, really healthy? Studies of healthy children in 10 locales in Asia showed substantial variation in the composition of the gut microbiota, yet there was a clear North-South pattern in terms of predominant taxa, likely related to different levels of socioeconomic modernisation and market integration.88 Thus, we do not yet know what are the key features of healthy microbiomes, beyond the descriptive composition that characterises body sites: Staphylococcus, Streptococcus, Actinomyces, Veillonella, Fusobacterium, Porphyromonas or Treponema species in the oral cavity89, with shared lifestyle, environment and genetic factors playing a role;90 Acinetobacter and Aeribacillus in the ocular surface, Pseudomonas on the lid margin and conjunctiva;91 Actinobacteria (Corynebacteriaceae and Propionibacteriaceae and Firmicutes—mostly Staphylococcaceae, Bacteroidetes and Proteobacteria in the skin,92 lipophilic organisms such as Propionibacterium spp. and the fungus Malassezia spp. in areas of higher density of sebaceous glands (face or back),93 94 Firmicutes and Bacteroidetes, including Bacteroides, Prevotella, Ruminococcus, Bifidobacterium, Streptococcus, Enterobacteriaceae, Enterococcus, Lactobacillus, the Verrucomicrobia Akkermansia and the archaeal Methanobrevibacter smithii in the mucosal surfaces of the gastrointestinal tract,95–100 and Lactobacillus spp. in the women’s genital tract.101–105 The ubiquity of the ‘core’ dominant metabolisms69 contrasts with the variability of niche-specific low abundant functions, many of which remain uncharacterised. A possible approach to the complexity of the human microbiome variability and disease risks is to obtain longitudinal data from multiple cohorts in global studies from which subjects developing any diseases throughout their lifespan are excluded, and only the healthy subjects (lacking a disease phenotype) are considered. In ecosystems like the gut, the extent of diversity is one proxy for health. Immigrants from developing countries lose diversity across human generations, as they develop westernised lifestyles and diseases.106
In children, we urgently need prospective studies that assess how well the microbiome matures across a population of healthy individuals, just as we normalise the maturation of height and weight in children and then compare those with disease states—essentially a growth curve for the developing microbiome. Just as for these physical attributes, knowledge of normal development allows abnormalities to be detected. Studies in humans using such approaches now indicate that it is possible to recognise the effects of disease states, for example, malnutrition107 and also the effects of perturbations, such as C-section or antibiotic exposures. Physicians have begun to use concepts of maturation in pathological states, such as in recipients of bone marrow transplantation or after faecal transplant to treat C. difficile infection to predict who might have a more successful outcome.
Focusing on functions, rather than taxa, may be important in addressing some research and clinical questions but may not be applicable to others, because each strain delivers a combination of functions, under multiple selection pressures and thus it is difficult to determine which components of the ecosystem can be manipulated without unintended consequences. Understanding the dynamics and effects of microbiome changes may be analogous to predicting the weather. We can see some general outlines that help us with the 3-day forecast, but as we attempt to forecast further out, the complexity of the system overwhelms the available tools. Because this is a young field, as knowledge grows and tools become more refined, our ability to classify and predict will correspondingly grow.
Future perspectives
The human holobiont is progressively being understood, as the collective microbiome and host functions are better characterised in health and disease, and as we assess both correlation and causal relationships. Efforts to standardise specimen preparation108 and analytical protocols and to increase the availability of the growing body of data109–111 are increasing. These technical efforts as well as robust clinical studies will improve characterisation of the variation in the global human microbiomes, functions of redundancy, trajectories of development, effect of lifestyles, immigration,106 disease biomarkers, all of which will establish the basis to understand the progression from health to disease and to efficiently discover new preventive interventions and therapies.
Footnotes
Contributors: MGD-B coordinated efforts in the review and all authors wrote the manuscript.
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: MGD-B, MJB and RK are members of the Scientific Advisory Board of CommenSe. RK reports personal fees from Biota, CommenSe, Prometheus, Metagenics and Genentech, outside the submitted work. MJB reports advisory board positions for Dupont Nutrition, Procter & Gamble, Seed, Ubiome, outside the submitted work. This publication does not endorse any commercial products.
Patient consent: Not required.
Provenance and peer review: Not commissioned; internally peer reviewed.
References
- 1. Mojzsis SJ, Arrhenius G, McKeegan KD, et al. Evidence for life on Earth before 3,800 million years ago. Nature 1996;384:55–9. 10.1038/384055a0 [DOI] [PubMed] [Google Scholar]
- 2. Kappler A, Pasquero C, Konhauser KO, et al. Deposition of banded iron formations by anoxygenic phototrophic Fe(II)-oxidizing bacteria. Geology 2005;33:865–8. 10.1130/G21658.1 [DOI] [Google Scholar]
- 3. Rosenberg E, Zilber-Rosenberg I. Microbiotas are transmitted between holobiont generations The hologenome concept: human, animal and plant microbiota. Cham: Springer, 2013. [Google Scholar]
- 4. Lederberg J, McCray AT. “Ome Sweet Omics”—a genealogical treasury of words. Scientist 2001;15. [Google Scholar]
- 5. Ochman H, Worobey M, Kuo CH, et al. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol 2010;8:e1000546 10.1371/journal.pbio.1000546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hancock AM, Witonsky DB, Ehler E, et al. Colloquium paper: human adaptations to diet, subsistence, and ecoregion are due to subtle shifts in allele frequency. Proc Natl Acad Sci U S A 2010;107(Supplement_2):8924–30. 10.1073/pnas.0914625107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. King N. The unicellular ancestry of animal development. Dev Cell 2004;7:313–25. 10.1016/j.devcel.2004.08.010 [DOI] [PubMed] [Google Scholar]
- 8. Bosch TC. Rethinking the role of immunity: lessons from Hydra. Trends Immunol 2014;35:495–502. 10.1016/j.it.2014.07.008 [DOI] [PubMed] [Google Scholar]
- 9. Davidson SK, Stahl DA. Transmission of nephridial bacteria of the earthworm Eisenia fetida. Appl Environ Microbiol 2006;72:769–75. 10.1128/AEM.72.1.769-775.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chiu CY, Chan YL, Tsai YS, et al. Airway microbial diversity is inversely associated with mite-sensitized rhinitis and asthma in early childhood. Sci Rep 2017;7:1820 10.1038/s41598-017-02067-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Damgaard C, Magnussen K, Enevold C, et al. Viable bacteria associated with red blood cells and plasma in freshly drawn blood donations. PLoS One 2015;10:e0120826 10.1371/journal.pone.0120826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aagaard K, Ma J, Antony KM, et al. The Placenta Harbors a Unique Microbiome. Sci Transl Med 2014;6:ra265 10.1126/scitranslmed.3008599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lopetuso LR, Scaldaferri F, Bruno G, et al. The therapeutic management of gut barrier leaking: the emerging role for mucosal barrier protectors. Eur Rev Med Pharmacol Sci 2015;19:1068–76. [PubMed] [Google Scholar]
- 14. Tomás I, Diz P, Tobías A, et al. Periodontal health status and bacteraemia from daily oral activities: systematic review/meta-analysis. J Clin Periodontol 2012;39:213–28. 10.1111/j.1600-051X.2011.01784.x [DOI] [PubMed] [Google Scholar]
- 15. Dixon CL, Richardson L, Sheller-Miller S, et al. A distinct mechanism of senescence activation in amnion epithelial cells by infection, inflammation, and oxidative stress. Am J Reprod Immunol 2018;79 10.1111/aji.12790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tan SL, Scammell G, Houang E. The midcycle cervical microbial flora as studied by the weighed-swab method, and its possible correlation with results of sperm cervical mucus penetration tests. Fertil Steril 1987;47:941–6. 10.1016/S0015-0282(16)59227-9 [DOI] [PubMed] [Google Scholar]
- 17. Gomez de Agüero M, Ganal-Vonarburg SC, Fuhrer T, et al. The maternal microbiota drives early postnatal innate immune development. Science 2016;351:1296–302. 10.1126/science.aad2571 [DOI] [PubMed] [Google Scholar]
- 18. Romano-Keeler J, Weitkamp JH. Maternal influences on fetal microbial colonization and immune development. Pediatr Res 2015;77(1-2):189–95. 10.1038/pr.2014.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Enders AC, Blankenship TN. Comparative placental structure. Adv Drug Deliv Rev 1999;38:3–15. 10.1016/S0169-409X(99)00003-4 [DOI] [PubMed] [Google Scholar]
- 20. Hoffman DJ, Reynolds RM, Hardy DB. Developmental origins of health and disease: current knowledge and potential mechanisms. Nutr Rev 2017;75:951–70. 10.1093/nutrit/nux053 [DOI] [PubMed] [Google Scholar]
- 21. Weaver IC, Korgan AC, Lee K, et al. Stress and the emerging roles of chromatin remodeling in signal integration and stable transmission of reversible phenotypes. Front Behav Neurosci 2017;11:41 10.3389/fnbeh.2017.00041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Solomon O, Yousefi P, Huen K, et al. Prenatal phthalate exposure and altered patterns of DNA methylation in cord blood. Environ Mol Mutagen 2017;58:398–410. 10.1002/em.22095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu X, Zhao B, Su Y, et al. Association of prenatal organochlorine pesticide-dichlorodiphenyltrichloroethane exposure with fetal genome-wide DNA methylation. Life Sci 2018;200:81–6. 10.1016/j.lfs.2018.03.030 [DOI] [PubMed] [Google Scholar]
- 24. Montrose L, Padmanabhan V, Goodrich JM, et al. Maternal levels of endocrine disrupting chemicals in the first trimester of pregnancy are associated with infant cord blood DNA methylation. Epigenetics 2018;13:301–9. 10.1080/15592294.2018.1448680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koren O, Goodrich JK, Cullender TC, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 2012;150:470–80. 10.1016/j.cell.2012.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aagaard K, Riehle K, Ma J, et al. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS One 2012;7:e36466 10.1371/journal.pone.0036466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Blaser MJ, Dominguez-Bello MG. The Human Microbiome before Birth. Cell Host Microbe 2016;20:558–60. 10.1016/j.chom.2016.10.014 [DOI] [PubMed] [Google Scholar]
- 28. Bahn SA, Jacobson J, Petersen F. Maternal and neonatal outcome following prolonged labor induction. Obstet Gynecol 1998;92:403–7. [DOI] [PubMed] [Google Scholar]
- 29. Hansen R, Scott KP, Khan S, et al. First-pass meconium samples from healthy term vaginally-delivered neonates: an analysis of the microbiota. PLoS One 2015;10:e0133320 10.1371/journal.pone.0133320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A 2010;107:11971–5. 10.1073/pnas.1002601107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nagpal R, Tsuji H, Takahashi T, et al. Sensitive quantitative analysis of the meconium bacterial microbiota in healthy term infants born vaginally or by cesarean section. Front Microbiol 2016;7:1997 10.3389/fmicb.2016.01997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moeller AH, Li Y, Mpoudi Ngole E, et al. Rapid changes in the gut microbiome during human evolution. Proc Natl Acad Sci U S A 2014;111:16431–5. 10.1073/pnas.1419136111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Song SJ, Lauber C, Costello EK, et al. Cohabiting family members share microbiota with one another and with their dogs. Elife 2013;2:e00458 10.7554/eLife.00458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stearns JC, Simioni J, Gunn E, et al. Intrapartum antibiotics for GBS prophylaxis alter colonization patterns in the early infant gut microbiome of low risk infants. Sci Rep 2017;7:16527 10.1038/s41598-017-16606-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith CL, Dickinson P, Forster T, et al. Identification of a human neonatal immune-metabolic network associated with bacterial infection. Nat Commun 2014;5:4649 10.1038/ncomms5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Diaz Heijtz R, Wang S, Anuar F, et al. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A 2011;108:3047–52. 10.1073/pnas.1010529108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pacheco AR, Barile D, Underwood MA, et al. The impact of the milk glycobiome on the neonate gut microbiota. Annu Rev Anim Biosci 2015;3:419–45. 10.1146/annurev-animal-022114-111112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zivkovic AM, German JB, Lebrilla CB, et al. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc Natl Acad Sci U S A 2011;108(Suppl_1):4653–8. 10.1073/pnas.1000083107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hamilton MK, Ronveaux CC, Rust BM, et al. Prebiotic milk oligosaccharides prevent development of obese phenotype, impairment of gut permeability, and microbial dysbiosis in high fat-fed mice. Am J Physiol Gastrointest Liver Physiol 2017;312:G474–87. 10.1152/ajpgi.00427.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lewis ZT, Totten SM, Smilowitz JT, et al. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome 2015;3:13 10.1186/s40168-015-0071-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Seppo AE, Autran CA, Bode L, et al. Human milk oligosaccharides and development of cow’s milk allergy in infants. J Allergy Clin Immunol 2017;139:708–11. 10.1016/j.jaci.2016.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Garrido D, Ruiz-Moyano S, Jimenez-Espinoza R, et al. Utilization of galactooligosaccharides by Bifidobacterium longum subsp. infantis isolates. Food Microbiol 2013;33:262–70. 10.1016/j.fm.2012.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lippi G. Vitamin K in neonates: facts and myths. Blood Transfus 2011;9:4–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Van Winckel M, De Bruyne R, Van De Velde S, et al. Vitamin K, an update for the paediatrician. Eur J Pediatr 2009;168:127–34. 10.1007/s00431-008-0856-1 [DOI] [PubMed] [Google Scholar]
- 45. Shearer MJ. Vitamin K metabolism and nutriture. Blood Rev 1992;6:92–104. 10.1016/0268-960X(92)90011-E [DOI] [PubMed] [Google Scholar]
- 46. Roumeliotis N, Dix D, Lipson A. Vitamin B(12) deficiency in infants secondary to maternal causes. CMAJ 2012;184:1593–8. 10.1503/cmaj.112170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. American Academy of Pediatrics VKAHTF. Controversies concerning vitamin K and the newborn. Pediatrics 1993;91:1001–3. [PubMed] [Google Scholar]
- 48. Pantoja-Feliciano IG, Clemente JC, Costello EK, et al. Biphasic assembly of the murine intestinal microbiota during early development. Isme J 2013;7:1112–5. 10.1038/ismej.2013.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bokulich NA, Chung J, Battaglia T, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med 2016;8:343ra82 10.1126/scitranslmed.aad7121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hill CJ, Lynch DB, Murphy K, et al. Evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET Cohort. Microbiome 2017;5:4 10.1186/s40168-016-0213-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013;504:451–5. 10.1038/nature12726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kamada N, Núñez G. Regulation of the immune system by the resident intestinal bacteria. Gastroenterology 2014;146:1477–88. 10.1053/j.gastro.2014.01.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bouskra D, Brézillon C, Bérard M, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 2008;456:507–10. 10.1038/nature07450 [DOI] [PubMed] [Google Scholar]
- 54. Agace WW. T-cell recruitment to the intestinal mucosa. Trends Immunol 2008;29:514–22. 10.1016/j.it.2008.08.003 [DOI] [PubMed] [Google Scholar]
- 55. Koboziev I, Karlsson F, Grisham MB. Gut-associated lymphoid tissue, T cell trafficking, and chronic intestinal inflammation. Ann N Y Acad Sci 2010;1207(Suppl 3):E86–93. 10.1111/j.1749-6632.2010.05711.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Durkin HG, Bazin H, Waksman BH. Origin and fate of IgE-bearing lymphocytes. I. Peyer’s patches as differentiation site of cells. Simultaneously bearing IgA and IgE. J Exp Med 1981;154:640–8. 10.1084/jem.154.3.640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mazmanian SK, Liu CH, Tzianabos AO, et al. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005;122:107–18. 10.1016/j.cell.2005.05.007 [DOI] [PubMed] [Google Scholar]
- 58. Luzza F, Parrello T, Monteleone G, et al. Up-regulation of IL-17 is associated with bioactive IL-8 expression in Helicobacter pylori-infected human gastric mucosa. J Immunol 2000;165:5332–7. 10.4049/jimmunol.165.9.5332 [DOI] [PubMed] [Google Scholar]
- 59. Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009;139:485–98. 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Martin R, Makino H, Cetinyurek Yavuz A, et al. Early-life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS One 2016;11:e0158498 10.1371/journal.pone.0158498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Harder T, Bergmann R, Kallischnigg G, et al. Duration of breastfeeding and risk of overweight: a meta-analysis. Am J Epidemiol 2005;162:397–403. 10.1093/aje/kwi222 [DOI] [PubMed] [Google Scholar]
- 62. Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature 2012;486:222–7. 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Scott KP, Gratz SW, Sheridan PO, et al. The influence of diet on the gut microbiota. Pharmacol Res 2013;69:52–60. 10.1016/j.phrs.2012.10.020 [DOI] [PubMed] [Google Scholar]
- 64. Zhang M, Zhou Q, Dorfman RG, et al. Butyrate inhibits interleukin-17 and generates Tregs to ameliorate colorectal colitis in rats. BMC Gastroenterol 2016;16:84 10.1186/s12876-016-0500-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jiminez JA, Uwiera TC, Abbott DW, et al. Butyrate Supplementation at High Concentrations Alters Enteric Bacterial Communities and Reduces Intestinal Inflammation in Mice Infected with Citrobacter rodentium . mSphere 2017;2 10.1128/mSphere.00243-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cani PD, Lecourt E, Dewulf EM, et al. Gut microbiota fermentation of prebiotics increases satietogenic and incretin gut peptide production with consequences for appetite sensation and glucose response after a meal. Am J Clin Nutr 2009;90:1236–43. 10.3945/ajcn.2009.28095 [DOI] [PubMed] [Google Scholar]
- 67. Perry RJ, Peng L, Barry NA, et al. Acetate mediates a microbiome-brain-β-cell axis to promote metabolic syndrome. Nature 2016;534:213–7. 10.1038/nature18309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006;444:1027–31. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- 69. Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207–14. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bach J-F. Infections and autoimmune diseases. J Autoimmun 2005;25(Suppl):74–80. 10.1016/j.jaut.2005.09.024 [DOI] [PubMed] [Google Scholar]
- 71. Cox AJ, West NP, Cripps AW. Obesity, inflammation, and the gut microbiota. Lancet Diabetes Endocrinol 2015;3 10.1016/S2213-8587(14)70134-2 [DOI] [PubMed] [Google Scholar]
- 72. Semenkovich CF, Danska J, Darsow T, et al. American Diabetes Association and JDRF Research Symposium: Diabetes and the Microbiome. Diabetes 2015;64:3967–77. 10.2337/db15-0597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Blustein J, Liu J. Time to consider the risks of caesarean delivery for long term child health. BMJ 2015;350:h2410 10.1136/bmj.h2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Clemente JC, Pehrsson EC, Blaser MJ, et al. The microbiome of uncontacted Amerindians. Sci Adv 2015;1:e1500183 10.1126/sciadv.1500183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Goodrich JK, Waters JL, Poole AC, et al. Human genetics shape the gut microbiome. Cell 2014;159:789–99. 10.1016/j.cell.2014.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Martinez KA, Devlin JC, Lacher CR, et al. Increased weight gain by C-section: Functional significance of the primordial microbiome. Sci Adv 2017;3:eaao1874 10.1126/sciadv.aao1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cox LM, Blaser MJ. Pathways in microbe-induced obesity. Cell Metab 2013;17:883–94. 10.1016/j.cmet.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Maier L, Pruteanu M, Kuhn M, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 2018;555:623–8. 10.1038/nature25979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Schwiertz A, Gruhl B, Löbnitz M, et al. Development of the intestinal bacterial composition in hospitalized preterm infants in comparison with breast-fed, full-term infants. Pediatr Res 2003;54:393–9. 10.1203/01.PDR.0000078274.74607.7A [DOI] [PubMed] [Google Scholar]
- 80. Deshmukh HS, Liu Y, Menkiti OR, et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med 2014;20:524–30. 10.1038/nm.3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Pannaraj PS, Li F, Cerini C, et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr 2017;171:647–54. 10.1001/jamapediatrics.2017.0378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat Med 2016;22:250–3. 10.1038/nm.4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Cunnington AJ, Sim K, Deierl A, et al. "Vaginal seeding" of infants born by caesarean section. BMJ 2016;352:i227 10.1136/bmj.i227 [DOI] [PubMed] [Google Scholar]
- 84. Clemente JC, Dominguez-Bello MG. Safety of vaginal microbial transfer in infants delivered by caesarean, and expected health outcomes. BMJ 2016;352:i1707 10.1136/bmj.i1707 [DOI] [PubMed] [Google Scholar]
- 85. Chua LL, Rajasuriar R, Azanan MS, et al. Reduced microbial diversity in adult survivors of childhood acute lymphoblastic leukemia and microbial associations with increased immune activation. Microbiome 2017;5:35 10.1186/s40168-017-0250-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Röhrig CH, Choi SS, Baldwin N. The nutritional role of free sialic acid, a human milk monosaccharide, and its application as a functional food ingredient. Crit Rev Food Sci Nutr 2017;57:1017–38. 10.1080/10408398.2015.1040113 [DOI] [PubMed] [Google Scholar]
- 87. Panigrahi P, Parida S, Nanda NC, et al. A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature 2017;548:407–12. 10.1038/nature23480 [DOI] [PubMed] [Google Scholar]
- 88. Nakayama J, Watanabe K, Jiang J, et al. Diversity in gut bacterial community of school-age children in Asia. Sci Rep 2015;5:8397 10.1038/srep08397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jenkinson HF, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol 2005;13:589–95. 10.1016/j.tim.2005.09.006 [DOI] [PubMed] [Google Scholar]
- 90. Kort R, Caspers M, van de Graaf A, et al. Shaping the oral microbiota through intimate kissing. Microbiome 2014;2:41 10.1186/2049-2618-2-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ozkan J, Willcox M, Wemheuer B, et al. Biogeography of the human ocular microbiota. Ocul Surf 2018. 10.1016/j.jtos.2018.11.005 [DOI] [PubMed] [Google Scholar]
- 92. Roth RR, James WD. Microbial ecology of the skin. Annu Rev Microbiol 1988;42:441–64. 10.1146/annurev.mi.42.100188.002301 [DOI] [PubMed] [Google Scholar]
- 93. Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol 2011;9:244–53. 10.1038/nrmicro2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Gao Z, Perez-Perez GI, Chen Y, et al. Quantitation of major human cutaneous bacterial and fungal populations. J Clin Microbiol 2010;48:3575–81. 10.1128/JCM.00597-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Naito Y, Uchiyama K, Takagi T. A next-generation beneficial microbe: Akkermansia muciniphila . J Clin Biochem Nutr 2018;63:33–5. 10.3164/jcbn.18-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Bäckhed F, Ley RE, Sonnenburg JL, et al. Host-bacterial mutualism in the human intestine. Science 2005;307:1915–20. 10.1126/science.1104816 [DOI] [PubMed] [Google Scholar]
- 97. Bjursell MK, Martens EC, Gordon JI. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J Biol Chem 2006;281:36269–79. 10.1074/jbc.M606509200 [DOI] [PubMed] [Google Scholar]
- 98. Avershina E, Lundgård K, Sekelja M, et al. Transition from infant- to adult-like gut microbiota. Environ Microbiol 2016;18:2226–36. 10.1111/1462-2920.13248 [DOI] [PubMed] [Google Scholar]
- 99. Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature 2009;457:480–4. 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol 2009;587:4153–8. 10.1113/jphysiol.2009.174136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Blum HE. The human microbiome. Adv Med Sci 2017;62:414–20. 10.1016/j.advms.2017.04.005 [DOI] [PubMed] [Google Scholar]
- 102. Rampersaud R, Randis TM, Ratner AJ. Microbiota of the upper and lower genital tract. Semin Fetal Neonatal Med 2012;17:51–7. 10.1016/j.siny.2011.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Linhares IM, Summers PR, Larsen B, et al. Contemporary perspectives on vaginal pH and lactobacilli. Am J Obstet Gynecol 2011;204:120.e1–5. 10.1016/j.ajog.2010.07.010 [DOI] [PubMed] [Google Scholar]
- 104. Ravel J, Gajer P, Abdo Z, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 2011;108(Suppl 1):4680–7. 10.1073/pnas.1002611107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Forney LJ, Foster JA, Ledger W. The vaginal flora of healthy women is not always dominated by Lactobacillus species. J Infect Dis 2006;194:1468–9. author reply 1469-1470 10.1086/508497 [DOI] [PubMed] [Google Scholar]
- 106. Vangay P, Johnson AJ, Ward TL, et al. US Immigration Westernizes the Human Gut Microbiome. Cell 2018;175:962–72. 10.1016/j.cell.2018.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Subramanian S, Huq S, Yatsunenko T, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 2014;510:417–21. 10.1038/nature13421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Costea PI, Zeller G, Sunagawa S, et al. Towards standards for human fecal sample processing in metagenomic studies. Nat Biotechnol 2017;35:1069–76. 10.1038/nbt.3960 [DOI] [PubMed] [Google Scholar]
- 109. Gonzalez A, Navas-Molina JA, Kosciolek T, et al. Qiita: rapid, web-enabled microbiome meta-analysis. Nat Methods 2018;15:796–8. 10.1038/s41592-018-0141-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Allali I, Arnold JW, Roach J, et al. A comparison of sequencing platforms and bioinformatics pipelines for compositional analysis of the gut microbiome. BMC Microbiol 2017;17:194 10.1186/s12866-017-1101-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Dhariwal A, Chong J, Habib S, et al. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res 2017;45:W180–8. 10.1093/nar/gkx295 [DOI] [PMC free article] [PubMed] [Google Scholar]