Abstract
Objective
We aimed to evaluate the performance of the newly developed deep learning Radiomics of elastography (DLRE) for assessing liver fibrosis stages. DLRE adopts the radiomic strategy for quantitative analysis of the heterogeneity in two-dimensional shear wave elastography (2D-SWE) images.
Design
A prospective multicentre study was conducted to assess its accuracy in patients with chronic hepatitis B, in comparison with 2D-SWE, aspartate transaminase-to-platelet ratio index and fibrosis index based on four factors, by using liver biopsy as the reference standard. Its accuracy and robustness were also investigated by applying different number of acquisitions and different training cohorts, respectively. Data of 654 potentially eligible patients were prospectively enrolled from 12 hospitals, and finally 398 patients with 1990 images were included. Analysis of receiver operating characteristic (ROC) curves was performed to calculate the optimal area under the ROC curve (AUC) for cirrhosis (F4), advanced fibrosis (≥F3) and significance fibrosis (≥F2).
Results
AUCs of DLRE were 0.97 for F4 (95% CI 0.94 to 0.99), 0.98 for ≥F3 (95% CI 0.96 to 1.00) and 0.85 (95% CI 0.81 to 0.89) for ≥F2, which were significantly better than other methods except 2D-SWE in ≥F2. Its diagnostic accuracy improved as more images (especially ≥3 images) were acquired from each individual. No significant variation of the performance was found if different training cohorts were applied.
Conclusion
DLRE shows the best overall performance in predicting liver fibrosis stages compared with 2D-SWE and biomarkers. It is valuable and practical for the non-invasive accurate diagnosis of liver fibrosis stages in HBV-infected patients.
Trial registration number
NCT02313649; Post-results.
Keywords: hepatitis B, cirrhosis, ultrasonography
Significance of this study.
What is already known on this subject?
There are more than 93 million chronic hepatitis B (CHB) carriers in China, and accurate assessment of liver fibrosis is essential for patients with CHB.
Liver stiffness measurement (LSM) by two-dimensional shear wave elastography (2D-SWE) is widely applied, but different studies showed great variability of cut-off values for staging liver fibrosis.
Radiomics for quantitative analysis of medical images has been proven to be a powerful tool, but its application in 2D-SWE images for classifying liver fibrosis stages in patients with CHB has not been systematically studied.
What are the new findings?
The deep learning Radiomics of elastography (DLRE) showed similar diagnostic efficacy with the liver biopsy for assessing cirrhosis (area under the receiver operating characteristic curve (AUC) 0.97) and advanced fibrosis (AUC 0.98), which were significantly better than LSM in 2D-SWE and biomarkers.
The diagnostic accuracy of DLRE improved as acquiring more 2D-SWE images from each patient.
There was no significant variation of the DLRE performance if enough 2D-SWE images were applied to train it, no matter training images came from which hospitals.
How might it impact on clinical practice in the foreseeable future?
As a non-invasive tool, DLRE in 2D-SWE may achieve a better overall diagnostic accuracy than LSM in 2D-SWE for assessing liver fibrosis stages in patients with CHB.
Introduction
HBV infection is a serious problem in China, causing more than one-third of the world’s HBV-infected people (approximately 93 million) residing in this country.1 Liver fibrosis is a progressive condition in chronic hepatitis B (CHB), and the accurate assessment of fibrosis is essential for prognosis, surveillance and management of patients with CHB.2
Liver biopsy is considered the reference standard for hepatic fibrosis staging. However, it is invasive and limited by sample errors, interobserver variability and various potential complications.3 Biomarkers, such as aspartate transaminase-to-platelet ratio index (APRI) and fibrosis index based on four factors (FIB-4), are also used to assess liver fibrosis, but their diagnostic performance remains controversial in HBV-infected patients.4 Recently, liver stiffness measurement (LSM) based on non-invasive ultrasonic imaging technologies is strongly recommended by many guidelines because of its effectivity and feasibility in the liver fibrosis evaluation.2 5
Two-dimensional (2D) shear wave elastography (SWE) is a new LSM technology with many advantages. Compared with transient elastography (TE), its application is not limited by ascites.6 It integrates B-mode imaging and colour-coded tissue stiffness map in real time, so that non-target structure and artefacts can be effectively avoided for acquiring more reliable LSM.7 Furthermore, it also can be used to detect focal liver lesions or assess liver morphological and blood flow changes.8 Therefore, 2D-SWE has been widely applied for the surveillance of HBV-infected patients in more than 400 Chinese hospitals in recent years. Several studies demonstrated that the diagnostic performance of 2D-SWE was comparable or even better than that of TE or point SWE in assessing liver fibrosis.9 10 However, despite these advantages, LSM of 2D-SWE is still affected by many factors. Important criteria for defining the optimal region of interest (ROI) of LSM, distinguishing reliable and unreliable measurements and controlling the overall image quality are still ambiguous in guidelines. As a consequence, the cut-off of 2D-SWE values for identifying cirrhosis in HBV-infected patients showed great variability ranging from 10.1 to 11.7 kPa in several studies.11 to 14 Therefore, the conventional strategy of using 2D-SWE values alone is likely to be insufficient for accurate assessment of liver fibrosis stages.
In contrast, an emerging technology named Radiomics can provide automated quantification of large amounts of image features (termed radiographic phenotypes) from medical images, which has the potential to uncover disease characteristics that fail to be appreciated by naked eyes.15 Radiomics has been proven to be useful in clinical oncology, where CT and/or MR images were acquired for analysis.16 17 We hypothesised that a distinctive radiomic technique might be able to use more valuable information from 2D-SWE images rather than just rely on the 2D-SWE value alone, and thus may provide better liver fibrosis staging accuracy.
There are only a few studies that applied radiomic methods on ultrasound images for chronic liver disease (CLD) diagnosis.18–21 They all successfully demonstrated the feasibility and potential benefits of using Radiomics for quantitative analysis of ultrasound images. However, there were some inherent limitations among these studies, such as lack of liver biopsy as reference, lack of a thorough comparison between proposed radiomic techniques and other well-established methods, not a prospective multicentre study focused on HBV-infected patients, or used engineered features (hard-coded features) for quantitative analysis, which is suitable for relatively smaller sample size. Different from these studies, our study sought to investigate the diagnostic performance of a deep learning method, named convolutional neural network (CNN),22 in 2D-SWE images for liver fibrosis staging in multicentre patients with HBV infection. Deep learning radiomic methods can learn features included in neural nets’ hidden layers automatically from imaging data, and thus they do not need object segmentation and hard-coded feature extraction, but their application requires a relatively large amount of imaging data.23
Here, we successfully enrolled 398 patients from 12 hospitals in China, with 1990 2D-SWE images, which we believe were suitable for the application of the deep learning radiomic method. To the best of our knowledge, this is the first prospective multicentre study that applied the deep learning radiomic method on 2D-SWE images for staging liver fibrosis in patients with CHB. Furthermore, in this study, histology obtained from liver biopsy was used as reference, as well as 2D-SWE and biomarkers were employed for the comparison with this new quantitative diagnostic strategy, named deep learning Radiomics of elastography (DLRE).
Patients and methods
Design and overview
This was a multicentre, prospective study. A new diagnostic approach named DLRE was used to assess liver fibrosis stages. Liver histology was used as the reference standard, and DLRE was compared with 2D-SWE, APRI and FIB-4. From January 2015 to January 2016, patients with CHB who provided informed consent to participate in this study were enrolled from 12 Chinese hospitals in different regions. This multicentre study was approved by the ethics committee of the principal investigator’s hospital and is registered at ClinicalTrials.gov (NCT02313649).
Patient enrolments
The inclusion criteria were as follows: (1) HBsAg positive more than 6 months; (2) older than 18 years; and (3) liver fibrosis stage scheduled for liver biopsy assessment. The exclusion criteria were as follows: (1) companied with other liver disease, including alcoholic CLD, haemochromatosis, autoimmune hepatitis, or intrahepatic biliary tract disease; (2) coinfection with HIV or any other viral hepatitis; (3) previous liver transplantation; (4) antiviral treatment in the previous 6 months; (5) unqualified histological samples (length was smaller than 15 mm, or the portal tract number was less than 6; (6) missing important serological results; and (7) unsuccessful 2D-SWE measurements. The demographic and clinical data of the patients (gender, age, height, weight and body mass index (BMI)) were recorded.
Two-dimensional shear wave elastography
Measurements of the 2D-SWE value were obtained by using the Aixplorer US imaging system (SuperSonic Imagine, SSI, France). The protocol of performing 2D-SWE was described in our previous studies,24 which is also recommended by the latest EFSUMB guidelines.6 B-mode ultrasound scan was first performed, and then 2D-SWE was performed in a well-visualised area that was free of large vessels. The size of the 2D-SWE ROI was 4 cm×3 cm, and it was located 1–2 cm under the liver capsule. A 2 cm diameter circular Q-Box ROI was placed in the 2D-SWE image, and the mean, maximum, minimum and SD of the elasticity within it were automatically calculated and displayed (figure 1A). Five independent 2D-SWE values and corresponding five 2D-SWE images were obtained from each patient, and the median value was used for statistical analysis. To be emphasised, strict quality controls were taken throughout the entire procedure. Operators who have performed more than 300 abdominal ultrasound scans or more than 50 supervised 2D-SWE examinations were enrolled in this multicentre study, and they were all strictly trained for the 2D-SWE measurement using the uniform procedure.24 Measurements were considered as failed or unqualified when little or no signal was obtained in the 2D-SWE ROI for every acquisition. Two 2D-SWE operators with more than 1-year 2D-SWE and 10 years of ultrasound operating experience were employed as quality controllers for reviewing all 2D-SWE images and excluding unqualified acquisitions.
Figure 1.
Illustration of the two-dimensional shear wave elastography (2D-SWE) measurement and the deep learning Radiomics of elastography (DLRE) flow chart. (A) The top shows the 2D-SWE region of interest (ROI) (pseudocolour area), Q-Box (white circle area within 2D-SWE ROI) and DLRE ROI (red square area). The obtained 2D-SWE values are displayed on the right yellow box. The bottom is the corresponding B-mode ultrasound image. (B) An input layer (DLRE ROI) is analysed by using four convolution-pooling procedures (C1-P1 to C4-P4), and then last pooled maps are fully connected with 32 neural nodes to calculate its probability for classification. The neural nodes and other parameters of the convolutional neural network (CNN) model were automatically optimised by using all 2D-SWE images in the training cohort.
Liver biopsy
Liver biopsy was performed in the right lobe of a liver by using a 16 or 18 G needle (Bard Magnum, GA, USA) within 1 week of the 2D-SWE scan. All the biopsy specimens were transported to one centre and examined by two liver pathologists. Each of them had more than 6 years of work experience, and they were both blind to 2D-SWE and clinical results. Unqualified samples including length less than 15 mm and portal tracts less than 6 were strictly excluded. Histological staging of fibrosis was based on METAVIR scoring system, and the grades of ≥F2, ≥F3 and F4 indicated the significant fibrosis, advanced fibrosis and cirrhosis, respectively.25
Serological examinations
Serological examinations were performed within 1 week of 2D-SWE. The platelet count, fasting blood glucose, aspartate aminotransferase (AST), alanine aminotransferase (ALT), gamma-glutamyl transpeptidase, total bilirubin, direct bilirubin, indirect bilirubin, albumin and prothrombin activity levels were recorded. Two biomarker models were employed and calculated as: APRI=[(AST/upper limit of normal AST)×100]/platelet count (109/L) and FIB-4=[age (year)]×[AST (U/L)]/[platelet count (109/L)]×[ALT (U/L)1⁄2].26 27
Deep learning Radiomics of elastography
For using Radiomics, the enrolled patients were randomly divided into the training cohort and validation (or testing) cohort. One is for training the radiomic model to optimise its parameters, the other is to validate the performance of the generated model. In the training cohort, to reduce the potential bias caused by the unbalanced data for binary classification, a strategy called data augmentation was applied before the training procedure.28 2D-SWE images in the training cohort were augmented through a number of random transformations, which increased the training data pool and decreased the overfitting of the generated radiomic model.
In this study, DLRE adopted the CNN method, one of the deep learning radiomic techniques, for the automatic analysis of 2D-SWE images. The three major operations of CNN are the convolution, activation and pooling, and the entire process can be divided into two steps, the forward computation and the back propagation.29 Finally, online supplementary figure 1 defines the termination of the process in building the CNN model. The detailed introduction and the mathematical descriptions of these operations and steps are demonstrated in the online supplementary materials.
gutjnl-2018-316204supp001.pdf (765.6KB, pdf)
For applying DLRE, a square DLRE ROI containing the entire 2D-SWE ROI with the size of 250×250 pixels was manually selected as the input layer (figure 1B), and then the CNN model was triggered. Four hidden layers (convolutional layers) were designed in CNN, and each followed with a max pooling layer to combine the neuron clusters at the prior layer into a single neuron in the next layer. The first hidden layer contained 16 feature maps, and each of the rest three contained 32 feature maps, which were obtained by applying 16 or 32 convolution filters (3×3 pixels) to the prior layer. The pooling size was 2×2 pixels. At the end, a fully connected layer with 32 nodes was applied to connect every neuron in the fourth pooling layer, so that the binary classification can be calculated in the output layer in the form of probabilities (figure 1B). The DLRE model generated by using the training cohort of this study is available at http://www.casmi.science/index.php/s/WZrE61nXlrZupi9. Some 2D-SWE images of four patients can also be downloaded as examples for testing the DLRE model.
Assessing the overall diagnostic accuracy of DLRE
Two-thirds of the enrolled patients were randomly selected, and their corresponding 2D-SWE images and histological results were used as the training cohort of DLRE. Images were sent to the input layer of the CNN model directly, so that the low-level to high-level features included in neural nets’ hidden layers were automatically extracted. DLRE then learnt these features to fine-tune its parameters and finally established its classification model for liver fibrosis staging. The 2D-SWE images and histological results of the other one-third patients were used as the validation cohort to evaluate the diagnostic accuracy of DLRE. All five images acquired from each patient were employed in this assessment. The diagnostic accuracy of DLRE was compared with 2D-SWE and biomarkers. After that, all enrolled patients with CHB were further divided into subgroups regarding to their ALT, BMI and inflammation levels. Then, the diagnostic performances of DLRE and 2D-SWE were compared in different subgroups for each fibrosis stage (online supplementary materials).
Assessing the diagnostic accuracy versus the number of acquisitions
DLRE was trained by one, three and five 2D-SWE images of each patient in the training cohort, respectively, and then the corresponding three DLRE models were used to assess liver fibrosis stages in the validation cohort. As for using 2D-SWE values, one, three and five measurements of each individual were also separately employed for liver fibrosis classification. For each staging strategy, the diagnostic accuracy of using three images/measurements was compared with that of using one and five images/measurements, respectively (intrastrategy comparison). Moreover, for using the same number of images/measurements, the diagnostic accuracies of these two strategies were also compared in each classification of liver fibrosis stages (interstrategy comparison).
Assessing the diagnostic robustness of DLRE
There were 12 Chinese hospitals (coded as A–L) participating in this study. Three different training cohorts were composed of patients enrolled from different combinations of hospitals, whereas patients in the rest hospitals consisted the three corresponding validation cohorts. These combinations were all random, but we still kept about two-thirds of the enrolled patients for training, and the rest of patients for validation in all three cases. Then, the diagnostic robustness of DLRE for liver fibrosis staging was evaluated through these different arrangements. Five 2D-SWE images of each patient were all employed in this experiment.
Statistical analysis
Descriptive statistics were summarised as mean±SD or median and IQR. Comparisons between groups were made with the Student’s t test or Mann-Whitney U test, when appropriate, for quantitative variables and with the Χ2test or Fisher’s test for qualitative variables. Area under the receiver operating characteristic (ROC) curve (AUC) was used to estimate the probability of the correct prediction of liver fibrosis stages. Differences between various AUCs were compared by using a Delong test. Sensitivity, specificity, positive and negative predictive values, and positive and negative diagnostic likelihood ratios were calculated. All statistical tests were two sided, and p values less than 0.05 indicated statistical significance. The statistical analyses were performed using SPSS software for Windows, V.20.0 (SPSS) and MedCalc software (V.11.2; 2011 MedCalc Software bvba, Mariakerke, Belgium).
Results
Baseline characters
Between January 2015 and January 2016, up to 654 potentially eligible patients form 12 Chinese hospitals were prospectively enrolled in this study. Among them, 256 patients were excluded because of the combination with other diseases, antiviral treatment, as well as unqualified histological, serological and/or 2D-SWE results. Thus, 398 patients with 1990 2D-SWE images were finally enrolled for analysis (figure 2). The mean liver biopsy length of all patients is 17.7 mm.
Figure 2.
The results of the multicentre patient enrolments. In total, 398 out of 654 patients from 12 Chinese hospitals were enrolled in this study. 2D-SWE, two-dimensional shear wave elastography.
After randomisation of these patients, 266 patients with 1330 images were assigned to the training cohort, and the other 132 patients with 660 images composed the validation cohort. Their characteristics are summarised in table 1. Between the training and validation cohorts, there were neither significant differences in all baseline characters (p>0.05), nor the distribution of patients among all fibrosis stages (p>0.05).
Table 1.
Baseline characters of patients
| Variables | All patients | Training cohort | Validation cohort | P values |
| Number of patients (%) | 398 | 266 (66.8%) | 132 (33.2%) | – |
| Age (years) | 38.6±12.1 | 38.8±12.1 | 38.1±12.1 | 0.997 |
| Gender (male) | 265 (66.6%) | 175 (65.8%) | 90 (68.2%) | 0.634 |
| BMI (kg/m2) | 22.8±3.26 | 22.8±3.3 | 22.8±3.2 | 0.951 |
| FBG (g/L) | 5.2±1.2 | 5.2±1.1 | 5.1±1.4 | 0.860 |
| PLT (109/L) | 177.8±64.0 | 175.0±61.2 | 183.4±69.1 | 0.218 |
| AST (IU/L) | 43.9±46.5 | 43.5±42.2 | 44.6±54.5 | 0.832 |
| ALT (IU/L) | 54.9±53.9 | 54.9±57.5 | 54.9±46.0 | 0.997 |
| GGT (IU/L) | 47.5±62.1 | 49.2±64.6 | 43.9±56.7 | 0.428 |
| TB (µmol/L) | 15.9±8.6 | 16.0±9.2 | 15.7±7.3 | 0.724 |
| DB (µmol/L) | 5.3±5.2 | 5.6±5.9 | 4.7±2.9 | 0.113 |
| IB (µmol/L) | 10.9±5.8 | 10.8±6.1 | 11.0±5.2 | 0.727 |
| ALP (IU/L) | 83.8±33.5 | 82.1±32,5 | 87.5±35.2 | 0.131 |
| ALB (g/L) | 44.8±26.5 | 43.4±8.5 | 47.6±44.4 | 0.134 |
| PT (%) | 90.7±13.1 | 90.9±13.4 | 90.1±12.3 | 0.557 |
| HBV status | ||||
| HBeAg (+/−) | 150/248 | 96/170 | 54/78 | 0.428 |
| HBeAg (+/−) CI | 66/143 | 45/97 | 21/46 | 0.913 |
| Fibrosis stages | ||||
| F0-1 | 65 (16.3%) | 43 (16.1%) | 22 (16.7%) | 0.993 |
| F2 | 109 (27.4%) | 72 (27.1%) | 37 (28.0%) | 0.944 |
| F3 | 126 (31.7%) | 85 (32.0%) | 41 (31.1%) | 0.946 |
| F4 | 98 (24.6%) | 66 (24.8%) | 32 (24.2%) | 0.994 |
| Inflammation grades | ||||
| A0 | 1 (0.3%) | 1 (0.4%) | 0 (0%) | – |
| A1 | 137 (34.4%) | 84 (31.6%) | 53 (40.2%) | 0.132 |
| A2 | 148 (37.2%) | 106 (39.8%) | 42 (31.8%) | 0.149 |
| A3 | 112 (28.1%) | 75 (28.2%) | 37 (28.0%) | 0.939 |
Qualitative variables are in n (%), and quantitative variables are in mean±SD, when appropriate.
HBV status was categorised according to 2017 European Association for the Study of the Liver (EASL) guideline.
P values were calculated between the training and validation cohorts.
ALB, albumin; ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BMI, body mass index; DB, direct bilirubin; FBG, fasting blood glucose; GGT, gamma-glutamyl transpeptidase; HBeAg, hepatitis Be antigen; HBeAg (+/−) CI, hepatitis Be antigen (+/−) chronic infection; IB, indirect bilirubin; PLT, platelet count; PT, prothrombin activity; TB, total bilirubin.
Overall diagnostic accuracy of DLRE in comparison with 2D-SWE, APRI and FIB-4
In the training cohort, DLRE demonstrated the highest diagnostic accuracy compared with all other methods for classifying of F4, ≥F3 and ≥F2 (figure 3A–C), and differences of AUCs were all statistically significant (p<0.001, table 2). AUCs of DLRE reached startling 1.00, 0.99 and 0.99 for three stratifications, respectively, which were 0.13, 0.18 and 0.25 higher than these of 2D-SWE who offered the second highest AUCs. The sensitivity and specificity analyses also demonstrated that DLRE was universally better than 2D-SWE and biomarkers (table 2).
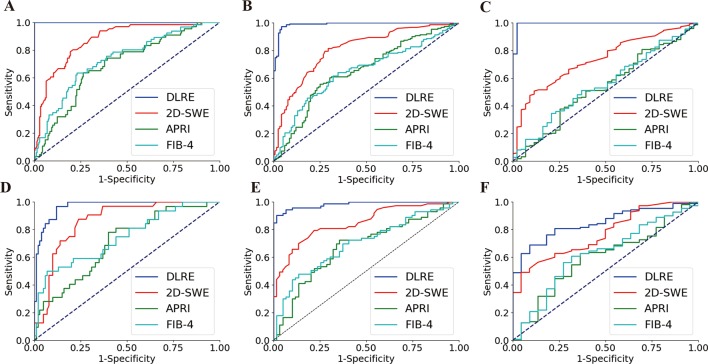
Figure 3.
Comparison of ROC curves between DLRE, 2D-SWE and biomarkers for the assessment of liver fibrosis stages in training and validation cohorts, respectively. (A, D) F0-F3 versus F4 (F4) in training and validation cohorts. (B, E) F0-F2 versus F3-F4 (≥F3) in training and validation cohorts. (C, F) F0-F1 versus F2-F4 (≥F2) in training and validation cohorts. 2D-SWE, two-dimensional shear wave elastography; APRI, aspartate transaminase-to-platelet ratio index; DLRE, deep learning Radiomics of elastography; FIB-4, fibrosis index based on four factors.
Table 2.
Diagnostic performance of DLRE, 2D-SWE, APRI and FIB-4 for the assessment of liver fibrosis stages in training and validation cohorts
| n (P) | AUC | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | LR+ | LR− | ||
| Cirrhosis (F4) | |||||||||
| DLRE | T | 266 (24.8%) | 1.00 (0.99 to 1.00) |
100.0 (98.0 to 100.0) |
100.0 (98.0 to 100.0) |
100.0 (99.0 to 100.0) |
100.0 (99.0 to 100.0) |
– | 0.0 (0.0 to 0.0) |
| V | 132 (24.2%) | 0.97 (0.94 to 0.99) |
96.9 (94.7 to 99.1) |
88.0 (82.8 to 92.2) |
72.1 (64.4 to 79.8) |
99.9 (98.4 to 100.0) |
8.1 (7.9 to 8.2) |
0.0 (0.0 to 0.1) |
|
| 2D-SWE | T | 266 (24.8%) | 0.87*** (0.83 to 0.91) |
80.3 (68.7 to 89.1) |
79.0 (72.7 to 84.4) |
55.8 (45.2 to 66.0) |
92.4 (87.4 to 95.9) |
3.8 (3.3 to 4.4) |
0.3 (0.1 to 0.4) |
| V | 132 (24.2%) | 0.86** (0.79 to 0.92) |
87.5 (71.0 to 96.5) |
76.0 (66.4 to 84.0) |
53.8 (39.5 to 67.8) |
95.0 (87.7 to 98.6) |
3.7 (3.1 to 4.3) |
0.2 (0.1 to 0.4) |
|
| APRI | T | 266 (24.8%) | 0.69*** (0.63 to 0.75) |
63.6 (50.9 to 75.1) |
73.5 (66.8 to 79.5) |
44.2 (34.0 to 54.8) |
86.0 (79.8 to 90.8) |
2.4 (2.0 to 2.9) |
0.5 (0.3 to 0.7) |
| V | 132 (24.2%) | 0.70*** (0.62 to 0.78) |
78.1 (60.0 to 90.7) |
60.0 (49.7 to 69.7) |
38.5 (26.7 to 51.4) |
89.6 (79.7 to 95.7) |
2.0 (1.5 to 2.5) |
0.4 (0.2 to 0.7) |
|
| FIB-4 | T | 266 (24.8%) | 0.73*** (0.68 to 0.78) |
63.6 (50.9 to 75.1) |
77.0 (70.5 to 82.6) |
47.7 (37.0 to 58.6) |
86.5 (80.6 to 91.2) |
2.8 (2.3 to 3.4) |
0.5 (0.3 to 0.7) |
| V | 132 (24.2%) | 0.75*** (0.66 to 0.82) |
50.0 (31.9 to 68.1) |
93.0 (86.1 to 97.1) |
69.6 (46.5 to 87.1) |
85.3 (77.3 to 91.4) |
7.1 (5.0 to 10.1) |
0.5 (0.2 to 1.2) |
|
| Advanced fibrosis (≥F3) | |||||||||
| DLRE | T | 266 (56.8%) | 0.99 (0.97 to 1.00) |
97.4 (93.2 to 99.1) |
95.7 (91.4 to 98.6) |
95.6 (89.4 to 98.2) |
71.9 (63.5 to 80.1) |
22.4 (20.1 to 24.5) |
0.0 (0.0 to 0.0) |
| V | 132 (55.3%) | 0.98 (0.96 to 1.00) |
90.4 (84.2 to 94.6) |
98.3 (95.3 to 99.3) |
98.5 (93.7 to 99.7) |
89.2 (82.4 to 94.3) |
53.3 (49.7 to 57.3) |
0.1 (0.1 to 0.1) |
|
| 2D-SWE | T | 266 (56.8%) | 0.81*** (0.75 to 0.85) |
81.5 (74.3 to 87.3) |
70.4 (61.2 to 78.6) |
78.3 (71.1 to 84.5) |
74.3 (65.0 to 82.2) |
2.8 (2.4 to 3.2) |
0.3 (0.2 to 0.4) |
| V | 132 (55.3%) | 0.85*** (0.78 to 0.90) |
79.5 (68.4 to 88.0) |
78.0 (65.3 to 87.7) |
81.7 (70.6 to 89.9) |
75.4 (62.7 to 85.5) |
3.6 (3.0 to 4.3) |
0.3 (0.1 to 0.5) |
|
| APRI | T | 266 (56.8%) | 0.65*** (0.59 to 0.71) |
55.6 (47.3 to 63.7) |
74.8 (65.8 to 82.4) |
74.3 (65.2 to 82.1) |
56.2 (48.0 to 64.2) |
2.2 (1.8 to 2.6) |
0.6 (0.4 to 0.9) |
| V | 132 (55.3%) | 0.68*** (0.59 to 0.76) |
72.6 (60.9 to 82.4) |
64.4 (50.9 to 76.4) |
71.6 (59.9 to 81.5) |
65.5 (51.8 to 77.6) |
2.0 (1.6 to 2.6) |
0.4 (0.3 to 0.7) |
|
| FIB-4 | T | 266 (56.8%) | 0.65*** (0.59 to 0.71) |
64.2 (56.0 to 71.9) |
65.2 (55.8 to 73.9) |
70.8 (62.4 to 78.3) |
58.1 (49.1 to 66.8) |
1.9 (1.5 to 2.2) |
0.6 (0.4 to 0.8) |
| V | 132 (55.3%) | 0.70*** (0.61 to 0.78) |
48.0 (36.1 to 60.0) |
86.4 (75.0 to 94.0) |
81.4 (66.4 to 91.7) |
57.3 (46.4 to 67.7) |
3.5 (2.7 to 4.6) |
0.6 (0.3 to 1.2) |
|
| Significance fibrosis (≥F2) | |||||||||
| DLRE | T | 266 (83.8%) | 0.99 (0.97 to 1.00) |
100.0 (99.0 to 100.0) |
97.7 (92.5 to 99.3) |
99.6 (97.4 to 100.0) |
100.0 (99.0 to 100.0) |
43.0 (41.8 to 44.5) |
0.0 (0.0 to 0.0) |
| V | 132 (83.3%) | 0.85 (0.81 to 0.89) |
69.1 (64.3 to 73.5) |
90.9 (83.6 to 96.7) |
97.4 (95.1 to 99.2) |
37.0 (34.5 to 39.1) |
7.6 (6.8 to 8.2) |
0.3 (0.3 to 0.4) |
|
| 2D-SWE | T | 266 (83.8%) |
0.74*** (0.68 to 0.79) |
50.7 (43.9 to 57.4) |
88.4 (74.9 to 96.1) |
95.8 (90.4 to 98.6) |
25.7 (18.8 to 33.5) |
4.4 (3.7 to 5.2) |
0.6 (0.2 to 1.3) |
| V | 132 (83.3%) |
0.77 (0.69 to 0.84) |
49.1 (39.4 to 58.8) |
95.5 (77.2 to 99.9) |
98.2 (90.2 to 100.0) |
27.3 (17.7 to 38.7) |
10.8 (8.7 to 13.3) |
0.5 (0.1 to 3.7) |
|
| APRI | T | 266 (83.8%) |
0.54*** (0.47 to 0.60) |
37.7 (31.3 to 44.4) |
74.4 (58.8 to 86.5) |
88.4 (80.2 to 94.1) |
18.7 (13.2 to 25.4) |
1.5 (1.2 to 1.9) |
0.8 (0.5 to 1.4) |
| V | 132 (83.3%) |
0.60*** (0.51 to 0.68) |
62.7 (53.0 to 71.8) |
63.6 (40.7 to 82.8) |
89.6 (80.6 to 95.4) |
25.5 (14.6 to 39.1) |
1.7 (1.2 to 2.4) |
0.6 (0.3 to 1.1) |
|
| FIB-4 | T | 266 (83.8%) | 0.56*** (0.49 to 0.62) |
51.1 (44.4 to 57.9) |
62.8 (46.7 to 77.0) |
87.7 (80.8 to 92.8) |
19.9 (13.5 to 27.6) |
1.4 (1.1 to 1.8) |
0.8 (0.5 to 1.2) |
| V | 132 (83.3%) | 0.62** (0.53 to 0.71) |
60.9 (51.1 to 70.1) |
68.2 (45.1 to 86.1) |
90.5 (81.5 to 96.1) |
25.9 (15.2 to 39.2) |
1.9 (1.4 to 2.6) |
0.6 (0.3 to 1.1) |
|
Statistical quantifications were demonstrated with 95% CI, when applicable.
AUC of DLRE was statistically compared with AUC of 2D-SWE, APRI and FIB-4, respectively, in the same fibrosis stage (*P<0.05; **P<0.01; ***P<0.001).
2D-SWE, two-dimensional shear wave elastography; APRI, aspartate transaminase-to-platelet ratio index; AUC, area under the receiver operating characteristic curve; DLRE, deep learning Radiomics of elastography; FIB-4, fibrosis index based on four factors; LR+, positive diagnostic likelihood ratio; LR−, negative diagnostic likelihood ratio; n, number of patients; NPV, negative predictive value; P, prevalence; PPV, positive predictive value; T, training cohort; V, validation cohort.
In the validation cohort, AUCs of DLRE dropped slightly for the diagnosis of F4 and ≥F3 (figure 3D,E), but they still reached 0.97 and 0.98, which were significantly higher than other methods (p<0.01 or p<0.001, table 2). However, the performance of DLRE for ≥F2 became much poorer than it was in the training cohort (figure 3F). AUC decreased from 0.99 to 0.85. It still demonstrated the highest AUC, and was significantly better than APRI (p<0.001) and FIB-4 (p<0.01), but no significant difference was found between DLRE and 2D-SWE (p>0.05, table 2).
For all 398 patients, the performances of DLRE and 2D-SWE did not show significant differences among ≥F2, ≥F3 and F4 regarding to different ALT and BMI levels (online supplementary tables 1 and 2 and figures 2 and 3). However, for F4, AUC of 2D-SWE in non-severe inflammation (A0-2) group was significantly higher than that in severe inflammation (A3) group (0.88 vs 0.69, p<0.001), whereas no significant difference was found between AUCs of DLRE in different inflammation subgroups (online supplementary table 3 and figure 4).
Diagnostic accuracy versus number of acquisitions: intrastrategy and interstrategy comparison of DLRE and 2D-SWE
For the intrastrategy comparison, when 2D-SWE separately adopted one, three and five stiffness measurements of each patient to assess liver fibrosis stages, its diagnostic accuracy showed no significant variation for classifying F4, ≥F3 and ≥F2 (table 3). Their ROC curves overlapped each other in all three fibrosis staging cases (figure 4), which indicated that the sensitivity and specificity of 2D-SWE had no obvious correlation with the number of acquisitions. This phenomenon was confirmed in both training and validation cohorts (figure 4).
Table 3.
Intrastrategy and interstrategy comparisons of DLRE and 2D-SWE for their relationship of diagnostic accuracy versus the number of image/measurement acquisitions in assessing liver fibrosis stages in training and validation cohorts
| n (P) | AUC | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | LR+ | LR− | ||
| Cirrhosis (F4) | |||||||||
| DLRE | |||||||||
| 1 image | T | 266 (24.8%) | 0.94** (0.91 to 0.97) |
95.5 (94.2 to 96.8) |
84.5 (79.6 to 89.4) |
67.0 (60.4 to 73.8) |
98.3 (96.1 to 99.6) |
6.2 (5.8 to 6.4) |
0.1 (0.0 to 0.1) |
| V | 132 (24.2%) | 0.84*** (0.81 to 0.87) |
75.0 (72.6 to 77.4) |
82.0 (77.3 to 86.7) |
57.1 (50.4 to 63.4) |
91.1 (85.4 to 97.2) |
4.2 (3.9 to 4.3) |
0.3 (0.3 to 0.4) |
|
| 3 images | T | 266 (24.8%) | 1.00 (0.98 to 1.00) |
100.0 (98.0 to 100.0) |
100.0 (98.0 to 100.0) |
100.0 (99.0 to 100.0) |
100.0 (99.0 to 100.0) |
– | 0.0 (0.0 to 0.0) |
| V | 132 (24.2%) | 0.96 (0.93 to 0.98) |
84.4 (76.9 to 91.9) |
95.0 (91.2 to 98.7) |
84.4 (78.5 to 90.1) |
95.0 (90.1 to 98.9) |
16.9 (14.9 to 18.6) |
0.2 (0.1 to 0.2) |
|
| 5 images | T | 266 (24.8%) | 1.00 (0.99 to 1.00) |
100.0 (98.0 to 100.0) |
100.0 (98.0 to 100.0) |
100.0 (99.0 to 100.0) |
100.0 (99.0 to 100.0) |
– | 0.0 (0.0 to 0.0) |
| V | 132 (24.2%) | 0.97 (0.94 to 0.99) |
96.9 (94.7 to 99.1) |
88.0 (82.8 to 92.2) |
72.1 (65.8 to 79.1) |
98.9 (94.8 to 99.7) |
8.1 (7.9 to 8.3) |
0.0 (0.0 to 0.1) |
|
| 2D-SWE | |||||||||
| 1 measurement | T | 266 (24.8%) | 0.87****** (0.83 to 0.91) |
84.9 (73.9 to 92.5) |
75.5 (68.9 to 81.3) |
53.3 (43.3 to 63.1) |
93.8 (88.9 to 97.0) |
3.5 (3.0 to 3.9) |
0.2 (0.1 to 0.4) |
| V | 132 (24.2%) | 0.86 (0.78 to 0.91) |
93.8 (79.2 to 99.2) |
74.0 (64.3 to 82.3) |
53.6 (39.7 to 67.0) |
97.4 (90.8 to 99.7) |
3.6 (3.1 to 4.2) |
0.1 (0.0 to 0.3) |
|
| 3 measurements | T | 266 (24.8%) | 0.88****** (0.83 to 0.92) |
86.4 (75.7 to 93.6) |
74.5 (67.9 to 80.4) |
52.8 (42.9 to 62.5) |
94.3 (89.5 to 97.4) |
3.4 (3.0 to 3.8) |
0.2 (0.1 to 0.4) |
| V | 132 (24.2%) | 0.85***** (0.78 to 0.91) |
87.5 (71.0 to 96.5) |
76.0 (66.4 to 84.0) |
53.8 (39.5 to 67.8) |
95.0 (87.7 to 98.6) |
3.7 (3.1 to 4.3) |
0.2 (0.1 to 0.4) |
|
| 5 measurements | T | 266 (24.8%) | 0.87****** (0.83 to 0.91) |
80.3 (68.7 to 89.1) |
79.0 (72.7 to 84.4) |
55.8 (45.2 to 66.0) |
92.4 (87.4 to 95.9) |
3.8 (3.3 to 4.4) |
0.3 (0.1 to 0.4) |
| V | 132 (24.2%) | 0.86***** (0.79 to 0.92) |
87.5 (71.0 to 96.5) |
76.0 (66.4 to 84.0) |
53.8 (39.5 to 67.8) |
95.0 (87.7 to 98.6) |
3.7 (3.1 to 4.3) |
0.2 (0.1 to 0.4) |
|
| Advanced fibrosis (≥F3) | |||||||||
| DLRE | |||||||||
| 1 image | T | 266 (83.8%) | 0.91* (0.86 to 0.95) |
86.8 (81.2 to 92.4) |
81.7 (76.4 to 87.1) |
86.2 (80.6 to 91.5) |
82.5 (77.1 to 87.5) |
4.8 (4.2 to 5.3) |
0.2 (0.1 to 0.2) |
| V | 132 (83.3%) | 0.82** (0.75 to 0.89) |
75.3 (70.3 to 80.3) |
79.7 (71.4 to 87.7) |
82.1 (77.0 to 87.6) |
72.3 (67.1 to 77.9) |
3.7 (3.5 to 3.9) |
0.3 (0.3 to 0.3) |
|
| 3 images | T | 266 (83.8%) | 0.96 (0.92 to 0.99) |
90.1 (86.8 to 93.4) |
88.7 (82.3 to 95.1) |
91.3 (83.8 to 97.3) |
81.2 (74.8 to 87.6) |
8.0 (7.7 to 8.2) |
0.1 (0.1 to 0.1) |
| V | 132 (83.3%) | 0.95 (0.91 to 0.98) |
94.5 (90.8 to 98.2) |
86.4 (81.2 to 91.6) |
89.6 (82.5 to 95.6) |
92.7 (87.5 to 96.8) |
7.0 (6.5 to 7.4) |
0.1 (0.1 to 0.1) |
|
| 5 images | T | 266 (83.8%) | 0.99 (0.97 to 1.00) |
97.4 (93.2 to 99.1) |
95.7 (91.4 to 98.6) |
95.6 (90.2 to 97.8) |
71.9 (63.8 to 78.5) |
22.4 (21.5 to 23.3) |
0.0 (0.0 to 0.0) |
| V | 132 (83.3%) | 0.98 (0.96 to 1.00) |
90.4 (84.2 to 94.6) |
98.3 (95.3 to 99.3) |
98.5 (95.8 to 99.8) |
89.2 (83.5 to 94.5) |
53.3 (51.0 to 55.3) |
0.1 (0.1 to 0.1) |
|
| 2D-SWE | |||||||||
| 1 measurement | T | 266 (83.8%) | 0.81****** (0.75 to 0.82) |
72.2 (64.3 to 79.2) |
75.7 (66.8 to 83.2) |
79.6 (71.8 to 86.0) |
67.4 (58.6 to 75.5) |
3.0 (2.6 to 3.4) |
0.4 (0.2 to 0.6) |
| V | 132 (83.3%) | 0.83 (0.75 to 0.89) |
68.5 (56.6 to 78.9) |
89.8 (79.2 to 96.2) |
89.3 (78.0 to 96.0) |
69.7 (58.1 to 79.8) |
6.7 (5.6 to 8.0) |
0.4 (0.2 to 0.8) |
|
| 3 measurements | T | 266 (83.8%) | 0.81****** (0.75 to 0.85) |
80.79 (73.6 to 86.7) |
68.7 (59.4 to 77.0) |
77.2 (69.9 to 83.5) |
73.1 (63.7 to 81.3) |
2.6 (2.2 to 3.0) |
0.3 (0.2 to 0.4) |
| V | 132 (83.3%) | 0.84***** (0.77 to 0.90) |
78.1 (66.9 to 86.9) |
79.7 (67.2 to 89.0) |
82.6 (71.5 to 90.7) |
74.6 (61.9 to 84.8) |
3.8 (3.2 to 4.6) |
0.3 (0.1 to 0.5) |
|
| 5 measurements | T | 266 (56.8%) | 0.81****** (0.75 to 0.85) |
81.5 (74.3 to 87.3) |
70.4 (61.2 to 78.6) |
78.3 (71.1 to 84.5) |
74.3 (65.0 to 82.2) |
2.8 (2.4 to 3.2) |
0.3 (0.2 to 0.4) |
| V | 132 (55.3%) | 0.85****** (0.78 to 0.90) |
79.5 (68.4 to 88.0) |
78.0 (65.3 to 87.7) |
81.7 (70.6 to 89.9) |
75.4 (62.7 to 85.5) |
3.6 (3.0 to 4.3) |
0.3 (0.1 to 0.5) |
|
| Significance fibrosis (≥F2) | |||||||||
| DLRE | |||||||||
| 1 image | T | 266 (83.8%) | 0.95 (0.93 to 0.97) |
89.7 (83.5 to 95.9) |
93.0 (87.5 to 98.5) |
98.5 (95.7 to 99.7) |
63.5 (58.4 to 68.1) |
12.9 (11.6 to 13.5) |
0.1 (0.1 to 0.1) |
| V | 132 (83.3%) | 0.74 (0.68 to 0.79) |
76.4 (71.3 to 81.5) |
72.7 (68.1 to 77.3) |
93.3 (96.7 to 98.1) |
38.1 (35.5 to 40.5) |
2.8 (2.7 to 2.9) |
0.3 (0.3 to 0.4) |
|
| 3 images | T | 266 (83.8%) | 0.97 (0.94 to 0.99) |
97.8 (92.4 to 99.9) |
95.3 (92.5 to 98.1) |
99.1 (97.2 to 99.9) |
89.1 (83.4 to 94.5) |
21.0 (19.8 to 22.2) |
0.0 (0.0 to 0.0) |
| V | 132 (83.3%) | 0.82 (0.78 to 0.86) |
67.3 (62.6 to 72.1) |
95.5 (91.8 to 99.1) |
98.7 (95.8 to 99.7) |
36.8 (34.8 to 38.4) |
14.8 (13.4 to 16.0) |
0.3 (0.3 to 0.4) |
|
| 5 images | T | 266 (83.8%) | 0.99 (0.97 to 1.00) |
100.0 (99.0 to 100.0) |
97.7 (92.5 to 99.3) |
99.5 (97.8 to 99.9) |
51.2 (48.5 to 53.9) |
43.0 (41.8 to 44.5) |
0.0 (0.0 to 0.0) |
| V | 132 (83.3%) | 0.85 (0.81 to 0.89) |
69.1 (64.3 to 73.5) |
90.9 (83.6 to 96.7) |
97.4 (96.8 to 99.7) |
37.0 (35.8 to 38.1) |
7.6 (6.9 to 8.4) |
0.3 (0.3 to 0.4) |
|
| 2D-SWE | |||||||||
| 1 measurement | T | 266 (83.8%) | 0.72****** (0.66 to 0.77) |
59.2 (52.4 to 65.7) |
79.1 (64.0 to 90.0) |
93.6 (88.2 to 97.0) |
27.2 (19.6 to 35.9) |
2.8 (2.3 to 3.4) |
0.5 (0.3 to 0.9) |
| V | 132 (83.3%) | 0.76 (0.67 to 0.83) |
60.0 (50.2 to 69.2) |
86.4 (65.1 to 97.1) |
95.7 (87.7 to 99.1) |
30.2 (19.2 to 43.0) |
4.4 (3.5 to 5.5) |
0.5 (0.2 to 1.4) |
|
| 3 measurements | T | 266 (83.8%) | 0.74****** (0.68 to 0.79) |
54.3 (47.5 to 60.9) |
83.7 (69.3 to 93.2) |
94.5 (89.0 to 97.8) |
26.1 (19.0 to 34.2) |
3.3 (2.8 to 4.0) |
0.6 (0.3 to 1.1) |
| V | 132 (83.3%) | 0.77 (0.69 to 0.84) |
51.8 (42.1 to 61.4) |
95.5 (77.2 to 99.9) |
98.3 (90.7 to 100.0) |
28.4 (18.5 to 40.1) |
11.4 (9.3 to 14.0) |
0.5 (0.1 to 3.5) |
|
| 5 measurements | T | 266 (83.8%) | 0.74****** (0.68 to 0.79) |
50.7 (43.9 to 57.4) |
88.4 (74.9 to 96.1) |
95.8 (90.4 to 98.6) |
25.7 (18.8 to 33.5) |
4.4 (3.7 to 5.2) |
0.6 (0.2 to 1.3) |
| V | 132 (83.3%) | 0.77 (0.69 to 0.84) |
49.1 (39.4 to 58.8) |
95.5 (77.2 to 99.9) |
98.2 (90.2 to 100.0) |
27.3 (17.7 to 38.7) |
10.8 (8.7 to 13.3) |
0.5 (0.1 to 3.7) |
|
Statistical quantifications were demonstrated with 95% CI, when applicable.
Intrastrategy comparison: for either DLRE or 2D-SWE, AUCs obtained by analysing three images/measurements were compared with these obtained by analysing one and five images/measurements, respectively (*P<0.05; **P<0.01; ***P<0.001).
Interstrategy comparison: for using the same number of images/measurements, AUCs obtained by DLRE were compared with these obtained by 2D-SWE in each liver fibrosis classification, respectively (****P<0.05; *****P<0.01; ******P<0.001).
2D-SWE, two-dimensional shear wave elastography; AUC, area under the receiver operating characteristic curve; DLRE, deep learning Radiomics of elastography; LR+, positive diagnostic likelihood ratio; LR−, negative diagnostic likelihood ratio; n, number of patients; NPV, negative predictive value; P, prevalence; PPV, positive predictive value; T, training cohort; V, validation cohort.
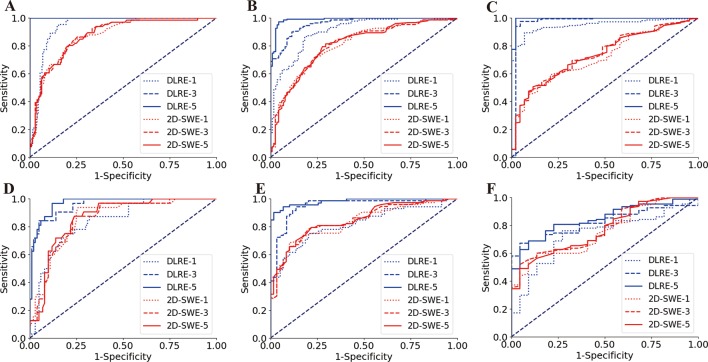
Figure 4.
Comparison of receiver operating characteristic (ROC) curves between deep learning Radiomics of elastography (DLRE) and two-dimensional shear wave elastography (2D-SWE) using different number of image acquisitions/measurements (1, 3 and 5) of each patient for the assessment of liver fibrosis stages. (A, D) F0-F3 versus F4 (F4) in training and validation cohorts. (B, E) F0-F2 versus F3-F4 (≥F3) in training and validation cohorts. (C, F) F0-F1 versus F2-F4 (≥F2) in training and validation cohorts.
However, DLRE demonstrated a very different nature. Its diagnostic accuracy increased as more 2D-SWE images of each individual were added to the training procedure (table 3). This was particularly obvious in the assessment of F4 and ≥F3 (figure 4A,B,D,E), in which significant improvements of AUC were found from using one to three images in both training (F4, AUC: 0.94 vs 1.00, p<0.01; ≥F3, AUC: 0.91 vs 0.96, p<0.05) and validation (F4, AUC: 0.84 vs 0.96, p<0.001; ≥F3, AUC: 0.82 vs 0.95, p<0.01) cohorts. Although there were no statistically significant improvements from using three to five images, AUCs still increased in all cases, unless it already reached 1.00 with three images (table 3). For ≥F2, AUCs of DLRE increased in both cohorts, when more numbers of images were employed, but these increases were not significant.
For the interstrategy comparison, DLRE showed different performances in the training (figure 4A–C) and validation (figure 4D–F) cohorts. In the training cohort, AUCs of DLRE were significantly better than those of 2D-SWE in all stratifications when using the same number of images/measurements (table 3). However, in the validation cohort, DLRE offered similar accuracy with 2D-SWE when only employing one image/measurement from each patient. If more images were adopted, DLRE outperformed 2D-SWE in the stratification of F4 and ≥F3 (all p<0.01), but it did not offer significantly higher AUC for ≥F2.
Diagnostic robustness of DLRE
Three randomly selected combinations of hospitals were employed to establish three different training cohorts (with similar number of patients), so that the DLRE model with three sets of parameters was obtained, respectively. For each fibrosis classification in either training or validation cohort, the resulted three ROC curves always overlapped each other (figure 5), and no significant differences were found (table 4). This revealed that DLRE demonstrated robust and consistent performances regardless of the training data coming from which hospitals, as long as the number of enrolled patients in different training cohorts was fairly constant.
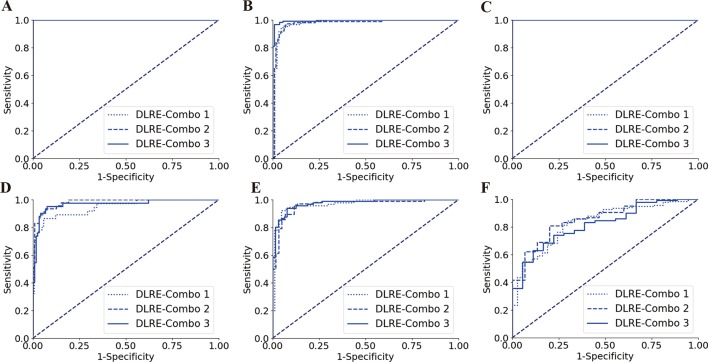
Figure 5.
Comparison of receiver operating characteristic (ROC) curves between different combinations of hospitals for training deep learning Radiomics of elastography (DLRE) in the classification of liver fibrosis stages. (A, D) F0-F3 versus F4 (F4) in training (combination of hospitals B, D, G, E, H and J) and validation cohorts. (B, E) F0-F2 versus F3-F4 (≥F3) in training (combination of hospitals A, C and K) and validation cohorts. (C, F) F0-F1 versus F2-F4 (≥F2) in training (combination of hospitals A, G and K) and validation cohorts. Note: three ROC curves completely overlap each other in (A) and (C), as they all reach the optimal profile (area under the receiver operating characteristic curve (AUC)=1).
Table 4.
Comparisons using different combinations of hospitals for training DLRE to classify liver fibrosis stages in training and validation cohorts
| n (P) | AUC | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | LR+ | LR− | ||
| Cirrhosis (F4) | |||||||||
| Combo1 | T | 221 (27.6%) | 1.00 (0.99 to 1.00) |
100.0 (99.0 to 100.0) |
100.0 (99.0 to 100.0) |
100.0 (99.0 to 100.0) |
100.0 (99.0 to 100.0) |
– | 0.0 (0.0 to 0.0) |
| V | 177 (20.9%) | 0.95 (0.92 to 0.98) |
86.5 (81.4 to 91.6) |
94.3 (90.2 to 98.4) |
80.0 (73.8 to 86.4) |
96.4 (89.7 to 98.5) |
15.1 (14.8 to 15.4) |
0.1 (0.1 to 0.2) |
|
| Combo 2 | T | 221 (27.6%) | 1.00 (0.99 to 1.0) |
100.0 (99.0 to 100.0) |
100.0 (99.0 to 100.0) |
100.0 (99.0 to 100.0) |
100.0 (99.0 to 100.0) |
– | 0.0 (0.0 to 0.0) |
| V | 177 (20.9%) | 0.98 (0.96 to 0.99) |
93.6 (88.4 to 98.8) |
93.3 (89.4 to 97.2) |
84.6 (79.5 to 89.2) |
97.4 (92.6 to 99.2) |
13.9 (12.1 to 15.7) |
0.1 (0.1 to 0.1) |
|
| Combo 3 | T | 221 (27.6%) | 1.00 (0.99 to 1.0) |
100.0 (99.0 to 100.0) |
100.0 (99.0 to 100.0) |
100.0 (99.0 to 100.0) |
100.0 (99.0 to 100.0) |
– | 0.0 (0.0 to 0.0) |
| V | 177 (20.9%) | 0.97 (0.95 to 0.99) |
95.2 (92.1 to 98.3) |
92.4 (88.2 to 96.6) |
81.6 (73.8 to 88.4) |
98.2 (94.8 to 99.7) |
12.6 (117 to 13.5) |
0.1 (0.0 to 0.1) |
|
| Advanced fibrosis (≥F3) | |||||||||
| Combo 1 | T | 221 (59.7%) | 0.98 (0.96 to 0.99) |
94.7 (91.2 to 98.2) |
95.5 (91.7 to 99.3) |
96.9 (91.5 to 98.9) |
92.4 (87.5 to 96.5) |
21.1 (20.4 to 21.8) |
0.1 (0.1 to 0.1) |
| V | 177 (52.0%) | 0.97 (0.95 to 0.99) |
92.4 (88.7 to 96.1) |
95.3 (91.9 to 98.7) |
95.5 (90.5 to 97.8) |
92.1 (85.6 to 97.8) |
19.6 (18.3 to 20.8) |
0.1 (0.1 to 0.1) |
|
| Combo 2 | T | 221 (59.7%) | 0.98 (0.97 to 0.99) |
95.8 (92.7 to 98.9) |
93.9 (90.1 to 97.7) |
94.2 (87.5 to 97.8) |
95.5 (88.5 to 97.9) |
15.6 (14.9 to 16.3) |
0.05 (0.0 to 0.1) |
| V | 177 (52.0%) | 0.97 (0.95 to 0.98) |
96.2 (92.6 to 98.7) |
88.3 (83.5 to 93.1) |
93.6 (86.9 to 96.8) |
93.0 (96.4 to 96.4) |
8.3 (8.0 to 8.5) |
0.0 (0.0 to 0.1) |
|
| Combo 3 | T | 221 (59.7%) | 1.00 (0.99 to 1.00) |
96.9 (93.8 to 99.9) |
99.1 (95.8 to 99.9) |
99.2 (97.1 to 100.0) |
96.5 (92.5 to 99.7) |
106.5 (100.2 to 112.0) |
0.0 (0.0 to 0.0) |
| V | 177 (52.0%) | 0.97 (0.96 to 0.99) |
93.8 (89.4 to 98.2) |
92.2 (87.8 to 96.7) |
94.8 (89.5 to 98.6) |
90.8 (84.5 to 96.2) |
12.0 (10.9 to 13.2) |
0.1 (0.1 to 0.1) |
|
| Significance fibrosis (≥F2) | |||||||||
| Combo 1 | T | 221 (87.3%) | 1.00 (0.99 to 1.00) |
100.0 (99.0 to 100.0) |
100.0 (99.0 to 100.0) |
100.0 (99.0 to 100.0) |
100.0 (99.0 to 100.0) |
– | 0.0 (0.0 to 0.0) |
| V | 177 (79.1%) | 0.83 (0.79 to 0.87) |
84.3 (80.1 to 88.5) |
70.3 (66.8 to 73.9) |
91.5 (85.7 to 97.2) |
54.2 (49.5 to 59.2) |
2.8 (2.6 to 3.1) |
0.2 (0.2 to 0.2) |
|
| Combo 2 | T | 221 (87.3%) | 1.00 (0.99 to 1.00) |
100.0 (99.0 to 100.0) |
100.0 (99.0 to 100.0) |
100.0 (99.0 to 100.0) |
100.0 (99.0 to 100.0) |
– | 0.0 (0.0 to 0.0) |
| V | 177 (79.1%) | 0.86 (0.81 to 0.91) |
80.8 (76.2 to 85.4) |
80.0 (74.6 to 85.4) |
97.6 (92.8 to 99.6) |
29.3 (27.1 to 31.5) |
4.0 (3.8 to 4.2) |
0.2 (0.2 to 0.3) |
|
| Combo 3 | T | 221 (87.3%) | 1.00 (0.99 to 1.00) |
100.0 (99.0 to 100.0) |
100.0 (99.0 to 100.0) |
100.0 (99.0 to 100.0) |
100.0 (99.0 to 100.0) |
– | 0.0 (0.0 to 0.0) |
| V | 177 (79.1%) | 0.82 (0.78 to 0.87) |
74.1 (69.8 to 78.4) |
77.8 (72.4 to 83.2) |
96.4 (90.5 to 98.9) |
27.5 (25.8 to 29.1) |
3.3 (3.2 to 3.5) |
0.3 (0.3 to 0.4) |
|
Statistical quantifications were demonstrated with 95% CI, when applicable.
AUCs obtained by three different combinations of hospitals were statistically compared with each other in each classification and each cohort (*P<0.05; **P<0.01; ***P<0.001).
AUC, area under the receiver operating characteristic curve; Combo, combination of hospitals for training and validation cohorts; DLRE, deep learning Radiomics of elastography; LR+, positive diagnostic likelihood ratio; LR−, negative diagnostic likelihood ratio; n, number of patients; NPV, negative predictive value; P, prevalence; PPV, positive predictive value; T, training cohort; V, validation cohort.
Discussion
In this multicentre prospective study, the diagnostic accuracy of DLRE, 2D-SWE and biomarkers in assessing liver fibrosis stages was compared against histology in patients with CHB. For assessing cirrhosis and advanced fibrosis, DLRE demonstrated significant improvements compared with 2D-SWE and biomarkers. In the training cohort, AUCs of DLRE reached 1.00 and 0.99, and in the validation cohort, they were 0.97 and 0.98, which indicated that DLRE provided similar diagnostic efficacy with the reference standard liver biopsy. 2D-SWE showed the second highest diagnostic accuracy, with AUCs of 0.87 and 0.81 in the training cohort, as well as 0.86 and 0.85 in the validation cohort. AUCs of biomarkers were all ≤0.75 in both stratifications and both cohorts. In the assessment of ≥F2, DLRE (AUC: 0.99) still performed significantly better than the other methods (AUC: ≤0.74) in the training cohort. However, its accuracy decreased in the validation cohort (AUC: 0.85), which did not show a significant difference with 2D-SWE, but was significantly better than biomarkers.
In order to investigate whether different levels of ALT, BMI and inflammation affected the performances of 2D-SWE and DLRE or not, stratification analysis in subgroups was performed. The results revealed that for F4, the inflammation grade did show significant impact on the performance of 2D-SWE, whereas its impact on that of DLRE was not significant.
These findings suggest that DLRE can be successfully used for the assessment of liver fibrosis stages in patients with CHB, and provides comparable diagnostic accuracy with current reference standard in classifying cirrhosis and advanced fibrosis. Its diagnostic accuracy was higher than 2D-SWE, and it may overcome the influence of inflammation for cirrhosis evaluation, which is likely to be a potential breakthrough in elastography diagnosis.
DLRE was completely established on analysing 2D-SWE images with the Radiomics concept. It uses exactly the same images as 2D-SWE stiffness measurement does, but it has two major advantages with respect to 2D-SWE. First, for the manual initiation, the input layer of DLRE contained the entire 2D-SWE ROI, whereas 2D-SWE performed LSM inside the Q-Box, which was only a portion of the 2D-SWE ROI. Therefore, DLRE fully used the 2D-SWE ROI (area about 10.5 cm2) instead of just using Q-Box (area about 3.1 cm2) for quantitative analysis. Second, DLRE employed the CNN method to achieve automatic feature extraction and deep learning in 2D-SWE images. Instead of solely measuring the average liver stiffness inside the Q-Box based on shear wave velocities, a large variety of features included in multiple hidden layers of 2D-SWE images, which reflected the heterogeneity of intensity and texture of these images, were quantitatively analysed to classify liver fibrosis stages. This offered a more thorough and comprehensive assessment compared with using 2D-SWE values as a single parameter for diagnosis. As a result, DLRE significantly improved the accuracy in the assessment of cirrhosis and advanced fibrosis.
In the assessment of significant fibrosis, the performance of DLRE became worse in the validation cohort, even though it was very accurate in the training. It is commonly seen that differentiating F0-F1 from F2-F4 is more challenging in many studies.7 9 30 to 32 This is because the heterogeneity of liver fibrosis is more severe in ≥F2 compared with that in ≥F3 and F4, which reduces the accuracy of all classification strategies in general, and DLRE was no exception. One possible way to overcome this challenge is to integrate multiple strategies for fibrosis classification. The current DLRE model still has tremendous room for improvements. If DLRE can be further optimised and integrated with other approaches, such as LSM by 2D-SWE and biomarkers, it might be possible to achieve a better performance in classifying ≥F2. Furthermore, only 16.3% enrolled patients were in the F0-F1 stage in this study (table 1), which was much less than portions of patients in other stages. The unbalanced data further compromised the efficacy of DLRE. This was probably because the involved 12 hospitals were all high-level teaching hospitals all over China, thus their patients were more likely to be in a severe condition. Since CNN requires larger data volume for more complicated classification, it is likely that DLRE may achieve better accuracy in assessing significant fibrosis, if the sample population of F0-F1 could be further extended in future studies.
The second finding of our study was that DLRE was highly data volume dependent. If more 2D-SWE images were acquired from each patient to train the DLRE model, it showed systematic improvements of the diagnostic accuracy in the assessment of all fibrosis stages in both training and validation cohorts. Different from DLRE, 2D-SWE did not show any significant differences with the increase of data volume, which was consistent with our previous studies.33 EFSUMB guideline suggests at least three measurements of each individual for assessing liver fibrosis using elastography.7 Coincidentally, with three acquisitions per person, our study demonstrated that DLRE significantly improved the diagnostic accuracy for assessing cirrhosis and advanced fibrosis compared with 2D-SWE. Moreover, the results also suggested five acquisitions might be even better when using DLRE.
Last but not the least, DLRE showed remarkable robustness in this multicentre study. When three randomly selected combinations of hospitals were used to build training cohorts, no significant variation was found for classifying liver fibrosis in training and validation cohorts using DLRE, and the diagnostic accuracy (table 4) matched the overall diagnostic accuracy of DLRE (table 2). These findings proved DLRE to be robust and reliable, which was valuable of clinical generalisation in China. Utilising the data acquired from limited number of hospitals to train and establish the DLRE model is likely to be sufficient in applying it for assessing liver fibrosis stages in other hospitals with a consistent accuracy.
To the best of our knowledge, this is the first prospective study that aimed to compare the diagnostic accuracy of liver fibrosis by means of deep learning Radiomics on 2D-SWE images, 2D-SWE and biomarkers in patients with CHB who underwent liver biopsy. About 2000 images obtained from 12 hospitals were enrolled here, which, we believe, is also the largest study of investigating Radiomics in diagnosing liver fibrosis stages with 2D-SWE so far. Strict quality control was applied for all image acquisition and histological analysis in every individual. Furthermore, this study only enrolled HBV-infected patients as a single-disease investigation to eliminate unnecessary interference. The final results proved that applying DLRE for the quantitative analysis of 2D-SWE images offered valuable benefits of diagnosing liver fibrosis in patients with CHB. Once the DLRE model is established, operators only need to perform a standardised selection of DLRE ROI in the daily workflow of 2D-SWE to conduct such analysis, which is extremely easy for clinical applications.
There is only one study we found that applied Radiomics for 2D-SWE analysis besides us. Gatos et al reported a multicentre study (126 patients) that adopted 35 hard-coded radiomic features extracted from 2D-SWE images to identify patients with CLD from healthy people.20 AUC reached 0.87 for the proposed machine learning method. However, their machine learning was fundamentally different from our deep learning approach, and their method was neither used to assess liver fibrosis stages, nor compared with any other diagnostic strategies.
The major limitations in our study were the limited population size, the unbalanced distribution of the patient population and the still developing DLRE method. Future studies need to involve more patients with CHB in a larger scale, as well as to achieve an equal distribution of patients in all fibrosis stages, so that the deep learning model can be better trained. The model itself also needs to be further optimised with better engineering design, as well as further developed with more comprehensive integration of other clinical data, such as serological results. All these effects can be made, so that more available data can be thoroughly analysed to enhance the overall performance of DLRE and achieve more accurate non-invasive diagnosis of liver fibrosis stages. Besides these limitations, our study did not investigate the performance of DLRE for classifying patients with CHB of different ethnic populations, for classifying patients with other aetiologies (chronic hepatitis C, non-alcoholic fatty liver disease, and so on), as well as its efficacy of using different commercial 2D-SWE systems, which are also worthy of further studies in the future.
In conclusion, this study demonstrated that DLRE was more accurate than 2D-SWE in assessing cirrhosis and advanced fibrosis, and more accurate than biomarkers in assessing all three liver fibrosis stages in patients with CHB. With more imaging acquisitions of each patient, DLRE provided increased diagnostic accuracy. With different training cohorts, DLRE also showed excellent robustness. All of these suggested a good potential of DLRE for clinical generalisation. Further studies in larger patient populations and balanced patient distribution are still needed.
Acknowledgments
We thank Doctor Rongkui Luo and Doctor Jing Zhao from Zhongshan Hospital Fudan University for their help with the pathological diagnoses.
Footnotes
KW, XL, HZ and YG contributed equally.
Contributors: Study conception: RZ, JT and PL. Data collection: KW, XL, HZ, YG, JZ, MT, CW, CL, LH, TJ, FM, YL, HA, XYX and LY. Data analysis: KW, XL, HZ and YG. Administrative support: RZ and PL. Manuscript drafting: KW, XL and HZ. All authors read and approved the final version of the manuscript.
Funding: The work is supported by the National Key Research and Development Program of China under Grant No 2017YFA0205200, the National Natural Science Foundation of China under Grant Nos 81227901, 61231004, 61671449 and 61401462, and Beijing Municipal Science and Technology Commission No Z161100002616022.
Competing interests: None declared.
Patient consent: Obtained.
Ethics approval: Chinese PLA General Hospital.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Cui Y, Jia J. Update on epidemiology of hepatitis B and C in China. J Gastroenterol Hepatol 2013;28 Suppl 1(Suppl 1):7–10. 10.1111/jgh.12220 [DOI] [PubMed] [Google Scholar]
- 2. Terrault NA, Lok ASF, McMahon BJ, et al. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology 2018;67:1560–99. 10.1002/hep.29800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bravo AA, Sheth SG, Chopra S, et al. N Engl J Med 2001;344:495–500. [DOI] [PubMed] [Google Scholar]
- 4. Kim WR, Berg T, Asselah T, et al. Evaluation of APRI and FIB-4 scoring systems for non-invasive assessment of hepatic fibrosis in chronic hepatitis B patients. J Hepatol 2016;64:773–80. 10.1016/j.jhep.2015.11.012 [DOI] [PubMed] [Google Scholar]
- 5. European Association for the Study of the Liver. Electronic address: easloffice@easloffice.eu European Association for the Study of the Liver. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol 2017;67:370–98. 10.1016/j.jhep.2017.03.021 [DOI] [PubMed] [Google Scholar]
- 6. Dietrich CF, Bamber J, Berzigotti A, et al. EFSUMB Guidelines and Recommendations on the Clinical Use of Liver Ultrasound Elastography, Update 2017 (Long Version). Ultraschall Med 2017;38:e16–e47. 10.1055/s-0043-103952 [DOI] [PubMed] [Google Scholar]
- 7. Barr RG, Ferraioli G, Palmeri ML, et al. Elastography Assessment of Liver Fibrosis: Society of Radiologists in Ultrasound Consensus Conference Statement. Radiology 2015;276:845–61. 10.1148/radiol.2015150619 [DOI] [PubMed] [Google Scholar]
- 8. Lu Q, Ling W, Lu C, et al. Hepatocellular carcinoma: stiffness value and ratio to discriminate malignant from benign focal liver lesions. Radiology 2015;275:880–8. 10.1148/radiol.14131164 [DOI] [PubMed] [Google Scholar]
- 9. Cassinotto C, Lapuyade B, Mouries A, et al. Non-invasive assessment of liver fibrosis with impulse elastography: comparison of Supersonic Shear Imaging with ARFI and FibroScan®. J Hepatol 2014;61:550–7. 10.1016/j.jhep.2014.04.044 [DOI] [PubMed] [Google Scholar]
- 10. Ferraioli G, Tinelli C, Dal Bello B, et al. Accuracy of real-time shear wave elastography for assessing liver fibrosis in chronic hepatitis C: a pilot study. Hepatology 2012;56:2125–33. 10.1002/hep.25936 [DOI] [PubMed] [Google Scholar]
- 11. Leung VY, Shen J, Wong VW, et al. Quantitative elastography of liver fibrosis and spleen stiffness in chronic hepatitis B carriers: comparison of shear-wave elastography and transient elastography with liver biopsy correlation. Radiology 2013;269:910–8. 10.1148/radiol.13130128 [DOI] [PubMed] [Google Scholar]
- 12. Zhuang Y, Ding H, Zhang Y, et al. Two-dimensional Shear-Wave Elastography Performance in the Noninvasive Evaluation of Liver Fibrosis in Patients with Chronic Hepatitis B: Comparison with Serum Fibrosis Indexes. Radiology 2017;283:873–82. 10.1148/radiol.2016160131 [DOI] [PubMed] [Google Scholar]
- 13. Zheng J, Guo H, Zeng J, et al. Two-dimensional shear-wave elastography and conventional US: the optimal evaluation of liver fibrosis and cirrhosis. Radiology 2015;275:290–300. 10.1148/radiol.14140828 [DOI] [PubMed] [Google Scholar]
- 14. Samir AE, Dhyani M, Vij A, et al. Shear-wave elastography for the estimation of liver fibrosis in chronic liver disease: determining accuracy and ideal site for measurement. Radiology 2015;274:888–96. 10.1148/radiol.14140839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016;278:563–77. 10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang YQ, Liang CH, He L, et al. Development and Validation of a Radiomics Nomogram for Preoperative Prediction of Lymph Node Metastasis in Colorectal Cancer. J Clin Oncol 2016;34:2157–64. 10.1200/JCO.2015.65.9128 [DOI] [PubMed] [Google Scholar]
- 17. Nie K, Shi L, Chen Q, et al. Rectal Cancer: Assessment of Neoadjuvant Chemoradiation Outcome based on Radiomics of Multiparametric MRI. Clin Cancer Res 2016;22:5256–64. 10.1158/1078-0432.CCR-15-2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meng D, Zhang L, Cao G, et al. Liver fibrosis classification based on transfer learning and FCNet for ultrasound images. IEEE Access 2017;5:1–10. 10.1109/ACCESS.2017.2689058 [DOI] [Google Scholar]
- 19. Chen Y, Luo Y, Huang W, et al. Machine-learning-based classification of real-time tissue elastography for hepatic fibrosis in patients with chronic hepatitis B. Comput Biol Med 2017;89:18–23. 10.1016/j.compbiomed.2017.07.012 [DOI] [PubMed] [Google Scholar]
- 20. Gatos I, Tsantis S, Spiliopoulos S, et al. A Machine-Learning Algorithm Toward Color Analysis for Chronic Liver Disease Classification, Employing Ultrasound Shear Wave Elastography. Ultrasound Med Biol 2017;43:1797–810. 10.1016/j.ultrasmedbio.2017.05.002 [DOI] [PubMed] [Google Scholar]
- 21. Liu X, Song JL, Wang SH, et al. Learning to Diagnose Cirrhosis with Liver Capsule Guided Ultrasound Image Classification. Sensors 2017;17:149 10.3390/s17010149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. LeCun Y, Bengio Y, Hinton G, et al. Nature 2015;521:436–44. [DOI] [PubMed] [Google Scholar]
- 23. Tajbakhsh N, Shin JY, Gurudu SR, et al. Convolutional Neural Networks for Medical Image Analysis: Full Training or Fine Tuning? IEEE Trans Med Imaging 2016;35:1299–312. 10.1109/TMI.2016.2535302 [DOI] [PubMed] [Google Scholar]
- 24. Zeng J, Liu GJ, Huang ZP, et al. Diagnostic accuracy of two-dimensional shear wave elastography for the non-invasive staging of hepatic fibrosis in chronic hepatitis B: a cohort study with internal validation. Eur Radiol 2014;24:2572–81. 10.1007/s00330-014-3292-9 [DOI] [PubMed] [Google Scholar]
- 25. Bedossa P, Poynard T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology 1996;24:289–93. 10.1002/hep.510240201 [DOI] [PubMed] [Google Scholar]
- 26. Wai CT, Greenson JK, Fontana RJ, et al. A simple noninvasive index can predict both significant fibrosis and cirrhosis in patients with chronic hepatitis C. Hepatology 2003;38:518–26. 10.1053/jhep.2003.50346 [DOI] [PubMed] [Google Scholar]
- 27. Sterling RK, Lissen E, Clumeck N, et al. Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatology 2006;43:1317–25. 10.1002/hep.21178 [DOI] [PubMed] [Google Scholar]
- 28. Nitesh VC, Kevin WB, Lawrence OH, et al. SMOTE: synthetic minority over-sampling technique. J Artif Intell Res 2002;16:321–57. [Google Scholar]
- 29. Shin HC, Roth HR, Gao M, et al. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans Med Imaging 2016;35:1285–98. 10.1109/TMI.2016.2528162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Afdhal NH, Bacon BR, Patel K, et al. Accuracy of fibroscan, compared with histology, in analysis of liver fibrosis in patients with hepatitis B or C: a United States multicenter study. Clin Gastroenterol Hepatol 2015;13:772–9. 10.1016/j.cgh.2014.12.014 [DOI] [PubMed] [Google Scholar]
- 31. Herrmann E, de Lédinghen V, Cassinotto C, et al. Assessment of biopsy-proven liver fibrosis by two-dimensional shear wave elastography: An individual patient data-based meta-analysis. Hepatology 2018;67:260–72. 10.1002/hep.29179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Degos F, Perez P, Roche B, et al. Diagnostic accuracy of FibroScan and comparison to liver fibrosis biomarkers in chronic viral hepatitis: a multicenter prospective study (the FIBROSTIC study). J Hepatol 2010;53:1013 to 21. 10.1016/j.jhep.2010.05.035 [DOI] [PubMed] [Google Scholar]
- 33. Huang ZP, Zhang XL, Zeng J, et al. Study of detection times for liver stiffness evaluation by shear wave elastography. World J Gastroenterol 2014;20:9578–84. 10.3748/wjg.v20.i28.9578 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2018-316204supp001.pdf (765.6KB, pdf)