Abstract
Aims
Kinesin family member 11 (Kif11) is a member of the kinesin family motor proteins, which is associated with spindle formation and tumour genesis. In this study, we investigated the relationship between Kif11 expression and clear cell renal cell carcinoma (CCRCC) development.
Methods
The relationship between Kif11 expression and CCRCC development was analysed by quantitative real-time (qRT)-PCR analyses, and tissue immunohistochemistry. The prognostic significance of Kif11 expression was explored by univariable and multivariable survival analyses of 143 included patients. Furthermore, SB743921 was used as a specific Kif11 inhibitor to treat 786-O cells with the epithelial to mesenchymal transition (EMT) process analysed by qRT-PCR, and cell survival rates analysed with Annexin V-FITC/PI staining followed by flow cytometric analyses. Disease-free survival curves of Kif11 with different cancers and the relationships between Kif11 and the von Hippel-Lindau disease tumour suppressor gene (VHL), and proliferating cell nuclear antigen (PCNA) in kidney cancer were further analysed using the GEPIA database.
Results
The levels of Kif11 mRNA were significantly higher in CCRCC tissues compared with corresponding non-cancerous tissues. The results of immunohistochemistry demonstrated that the expression of Kif11 protein was significantly associated with clinicopathologial parameters, including nuclear grade and TNM stage. The Kaplan-Meier survival curve indicated that high Kif11 expression, nuclear grade and TNM stage were independent factors to predict poor prognosis in patients with CCRCC. In addition, inhibition of Kif11 expression by SB743921 suppressed cell proliferation, migration and the EMT process with increased apoptosis rate.
Conclusions
These results combined with bioinformation analyses suggest that high Kif11 expression was associated with unfavourable prognosis in CCRCC and could be used as a potential prognostic marker in the clinical diagnosis of CCRCC.
Keywords: Kif11, Clear cell renal cell carcinoma, prognosis, EMT
Introduction
More than 63 000 Americans were diagnosed with renal cancer and nearly 14 000 died of this disease in 2017.1 Clear cell renal cell carcinoma (CCRCC), which accounts for approximately 90% of renal neoplasms and 3% of adult malignancies, is the most common pathological type of renal cancer.2 According to the National Central Cancer Registry of China, new cases of CCRCC have reached 6 68 000 and 2 34 000 patients have died of this disease. Traditional clinical treatment for renal cancer usually consisted of symptomatic treatments, such as chemotherapy, interferon therapy, interleukin therapy and nephrectomy.3 However, even following curative nephrectomies, over 20% of patients with CCRCC would experience recurrence within 5 years.4
In recent years, breakthroughs in cancer treatments have resulted from improved tumour diagnosis and treatment methods. Various methods have been used to optimise the prognostic factors of CCRCC and other cancers. Traditionally, the tumour grade, Eastern Cooperative Oncology Group performance status (ECOG PS), and tumour necrosis were determined to be classic clinicopathological prognostic factors.5 In genetics, mutation of the von Hippel-Lindau disease tumour suppressor gene (VHL) was thought to be associated with a more favourable prognosis.6 Despite remarkable advances in the diagnosis and treatment of cancer, the clinical outcomes of patients with CCRCC remain unsatisfactory. Following tumour metastasis, the prognosis for patients with CCRCC is markedly poor, although immunotherapy and targeted agents could partially improve survival rates. It is therefore crucial to optimise novel biomarkers for CCRCC to predict which patients are likely to develop disease recurrence after nephrectomy surgery.
Kinesin family member 11 (Kif11; also known as Eg5, Kinesin-5, kinesin spindle protein or KSP) is a plus-end-directed motor localised to interpolar spindle microtubules and to the spindle poles.7–10 Kif11 plays essential roles in the formation of bipolar spindle and maintenance in the early prometaphase during mitosis.11 12 Several studies have evaluated the correlation between Kif11 expression and clinicopathological characteristics in various malignancies such as prostate cancer,13 breast cancer,14 malignant pleural mesothelioma,15 pancreatic cancer,16 laryngeal squamous cell carcinoma17 and gastric cancer with intestinal mucin phenotype.18
In this study, our goal was to confirm the expression of Kif11 in CCRCC tissues and corresponding non-cancerous tissues, and to further determine whether Kif11 could be used as a prognostic marker for patients with CCRCC with specific Kif11 inhibition treatments and bioinformation analyses. The mRNA levels of Kif11 in fresh cancer and corresponding non-cancerous tissues were detected by quantitative real-time (qRT)-PCR while the location of Kif11 expression was detected by immunohistochemistry (IHC) analyses. In addition, the correlations between Kif11 protein expression levels and clinicopathological parameters of patients with CCRCC were evaluated. Furthermore, 786-O cells, one of the typical CCRCC cell lines (including ACHN, Caki-1, Caki-2, A498 and 786-O), were chosen and treated with SB743921, a Kif11-specific inhibitor. The epithelial to mesenchymal transition (EMT) process was analysed with qRT-PCR, and cell survival rates were analysed with Annexin V-FITC/PI staining followed by flow cytometric analyses according to previous studies of the CCRCC cell model.19 20 To further confirm the role of Kif11 in clinical cancer development, the disease-free survival curves of Kif11 with different cancers were further analysed using the GEPIA database.
Materials and methods
Patient specimens
Thirty pairs of fresh CCRCC tissues and corresponding non-cancerous tissues (normal adjacent kidney tissue, ~5 cm distance from the cancer tissues) were collected from the department of pathology, Affiliated Hospital of Nantong University. Simultaneously, a total of 143 paraffin-embedded CCRCC tissue samples and the matched non-cancerous tissue samples were collected from the department of pathology, Affiliated Hospital of Nantong University, between August 2009 and April 2013.
All diagnoses were validated by two pathologists in the department, according to the latest WHO criteria. None of the included patients received radiotherapy or chemotherapy before surgery. Each included patient signed a written informed consent for this study before our research started. The study protocol was approved by the ethics committee of the Affiliated Hospital of Nantong University and all experiments were performed in accordance with the university's approved guidelines.
qRT-PCR analyses
Thirty pairs of fresh CCRCC tissue samples and corresponding adjacent tissue samples were used for the qRT-PCR analyses. Total RNA was extracted from tissues with Trizol (79306, Gibco) and cDNA was synthesised with commercial kits (PrimeScript RT reagent Kit with gDNA Eraser, RR047A; TaKaRa, China). Amplifications were performed in an AB Verti96 well thermal cycler or a Thermo Pikoreal96 using commercial kits (Premix Taq, TaKaRa) and specific synthesised primers. The ubiquitously expressed β-actin gene was used as an internal control. All experiments were performed in triplicate.
The specific primers for Kif11 were as follows: forward primer 5’-GAACAATCA TTAGCAGCAGAA-3’ and reversed primer 5’-TCAGTATAGACACCA CAGTTG-3’. The β-actin gene was used as an internal control, and the specific primers for β-actin were as follows: forward TAATCTTCGCCTTAATACTT, reversed AGCCTTCATACATCTCAA. The relative Kif11 mRNA expression was calculated using the 2-ΔΔCt method.
IHC analyses
A total of 143 CCRCC tissue samples and corresponding adjacent tissue sample paraffin sections were deparaffinised with xylene and rehydrated in graded ethanol (100%–95%–85%–75%), then washed with phosphate-buffered saline solution (PBS, 0.01 M, pH 7.0). Antigen retrieval was achieved by boiling under pressure in citrate buffer (0.01 M, pH 6.0). The non-specific bindings were blocked through incubation with 5% goat blocking serum (Solarbio, SL039) in PBS solution for 30 min at 37℃. All paraffin sections were incubated with rabbit polyclonal anti-Kif11 antibody (1:200, Abcam, ab61199) and subsequently with goat anti-rabbit HRP secondary antibody (ZSGB-BIO, ZDR-5306). The IHC analyses for proliferating cell nuclear antigen (PCNA) (1:200, Abcam, ab18197) was performed at the same time under the same conditions. For a colour reaction, sections were incubated with DAB substrate chromogen solution (Solarbio, DA1010) for 8 min. Subsequently sections were counterstained with hematoxylin solution.6
Immunostained sections were scored by two independent pathologists under blinded experimental conditions according to intensity and percentage of Kif11-positive cells under light microscope (Axio imager A2, Zeiss). The intensity of Kif11-positive cells was scored as follows: 0 (negative), 1 (weak), 2 (moderate) and 3 (strong intensity). The percentage of Kif11-positive cells was scored as follows: 0 (no cytoplasm expression), 1 (1%–25% positive tumour cytoplasm), 2 (26%–50% positive tumour cytoplasm), 3 (51%–75% positive tumour cytoplasm) and 4 (76%–100% positive tumour cytoplasm). The multiplication of the intensity and percentage scores led to the final Kif11 staining score and was defined as follows: a staining score less than 6 was considered as low expression, while a staining score of 7 or more was considered as high expression, in accordance with other reported studies.21 Using these definitions, two independent pathologists, with no knowledge of the clinical/pathological parameters or the outcome for included patients in our research, reviewed the scores in each specimen. Inter-observer differences were resolved by consensus review using a double-headed microscope after independent review.
Cell cultures
Human kidney cancer cells (786-O) were cultured in RPMI 1640 medium (22400089, Invitrogen, China) containing 10% fetal bovine serum (FBS; 10099141, Invitrogen) and 100 U/mL penicilin/strapmycin antibotic (P/S, 15070063, Invitrogen) at 37°C in a cell incubator with a humidified atmosphere containing 5% CO2.
Kif11 inhibitor assay
To evaluate the essential role of Kif11 in kidney cancer development, 786-O cells were treated with 0, 1.0 or 5.0 nM of a small compound inhibitor against Kif11 (SB743921; Selleck, S2182). These cells were collected and used for the following analyses.
Wound healing assay
The wound healing assay was used to measure the migration ability of 786-O cells. After SB743921 treatments, a wound was made by scratching with a plastic pipette tip followed by washing twice with PBS solution to remove cellular debris and non-adherent cells. Then the cells were incubated with growth medium for 24 hours. Cells migrated into the wounded empty space, and photographs were taken at 0 and 24 hours after wounding.
Cell proliferation assay
CCK-8 assay was performed with the aim of estimating the proliferative ability of 786-O cells with SB743921 treatment. A 96-well plate (Corning) was used, with 5 × 103 786 O cells per well. At 0, 24, 48 and 72 hours after seeding, 10 µL CCK-8 solution (Solarbio, CA1210, China) was added to each well, followed by incubation for 1 hour at 37 °C in a humidified incubator containing 5% CO2. Finally, the optical density at 450 nm of the 96-well plate was detected with Multiskan GO (Thermo Scientific).
EMT process analyses
After treatment for 72 hours, the EMT process was analysed by qRT-PCR. Total RNA was extracted from cells with Trizol reagent (Invitrogen), according to the manufacturer's instructions. Reverse transcription was conducted with PrimeScript reagent kit with genomic DNA earser (RR047A, TaKaRa) on the basis of the manufacturer's protocols. qRT-PCR was carried out with SYBR prime script RT-PCR kits (TaKaRa) based on the manufacturer's instructions.
The levels of E-cadherin, Vimentin, ZEB-1 and Kif11 mRNA were separately calculated with the 2−ΔΔCt method, which were normalised to that of β-actin mRNA. All assays were conducted in triplicate. The primers of E-cadherin, Vimentin and ZEB-1 mRNA were as follows.
E-cadherin
Forward CCCTTCACAGCAGAACTAACA,
Reversed TTGGGTTGGGTCGTTGTA.
Vimentin
Forward AATGACCGCTTCGCCAAC,
Reversed CCGCATCTCCTCCTCGTAG.
ZEB-1
Forward CCTGTCCATATTGTGATAGAGGC,
Reversed ACCCAGACTGCGTCACATGT.
Flow cytometric analyses of cell apoptosis rate
After the SB743921 treatment for 24 hours, the cells were harvested and the early apoptosis rate was detected by FACS with Annexin V-FITC apoptosis detection kits (Cwbiotech, CW2574, China) with the manufacturer's instructions.
Comparison of Kif11 gene expression level in different cancers
GEPIA (http://gepia.cancer-pku.cn/index.html) is a newly developed interactive web server that analyses the RNA sequencing expression data of 9736 tumours and 8587 normal samples from The Cancer Genome Atlas (TCGA) and the Genotype Tissue Expression (GTEx) projects with a standard processing pipeline.21 GEPIA provides customisable functions such as tumour and normal differential expression models and Kif11 expression could be demonstrated for different cancers such as adrenocortical carcinoma (ACC), bladder urothelial carcinoma (BLCA), kidney chromophobe (KICH), kidney renal papillary cell carcinoma (KIRP), sarcoma (SARC), liver hepatocellular carcinoma (LIHC), pancreatic adenocarcinoma (PAAD), brain lower grade glioma (LGG) and uveal melanoma (UVM). In addition, the relationships between Kif11, PCNA and VHL in kidney renal clear cell carcinoma (KIRC) were further detected by the GEPIA database.
Statistical analysis
The expression levels of Kif11 mRNA in fresh CCRCC tissues and corresponding adjacent tissues were calculated by the Wilcoxon signed-rank non-parametric test. Pearson’s χ 2 was used to examine the correlations between Kif11 protein expression and clinicopathological parameters. The Kaplan-Meier and log-rank tests were performed to calculate the survival curves. Factors of prognostic significance in the univariable analyses were further analysed by the multivariable Cox regression model. For all tests, p values less than 0.05 were considered significant. All statistical work was analysed by the Statistical Package for the Social Sciences software (SPSS, IBM, version 19.0).
Results
Kif11 mRNA expression in CCRCC tissues by qRT-PCR analyses
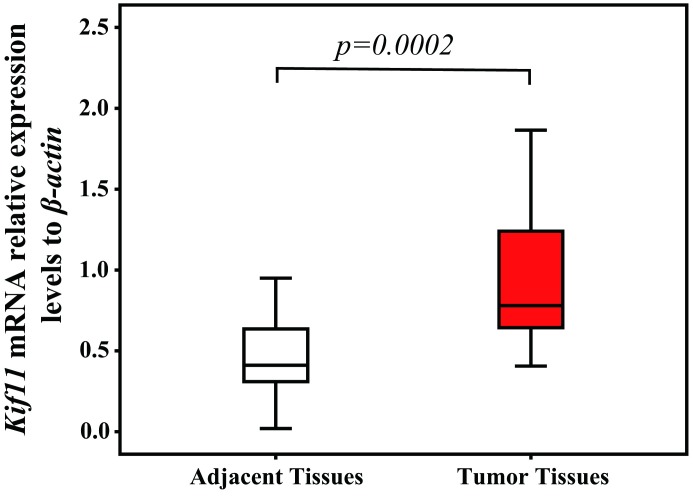
The mRNA expression levels of Kif11 were detected by qRT-PCR analyses in 30 pairs of CCRCC tissues and corresponding adjacent tissues. Compared with the transcript levels in adjacent tissues, the transcript levels of Kif11 mRNA were significantly higher in CCRCC tissues (Z=−4.391, p=0.0002, with Wilcoxon signed-rank non-parametric test) (figure 1).
Figure 1.
Quantitative real-time PCR analyses were used to detect the kinesin family member 11 (Kif11) mRNA expression levels in clear cell renal cell carcinoma (CCRCC) tissues and the corresponding adjacent tissues. The Kif11 mRNA median level in CCRCC tissues is 0.7799 (IQR 0.6227–1.2454), compared with the median level in the adjacent tissues which is 0.4111 (IQR 0.3066–0.6487) (Z=−4.391, p<0.001, with Wilcoxon signed-rank non-parametric test).
Location and expression of Kif11 protein in patients with CCRCC using IHC analyses
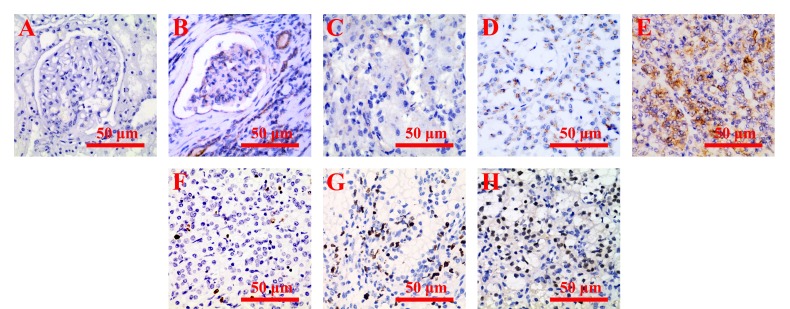
The IHC analyses were performed to determine the protein location and expression of Kif11 in CCRCC tissues and adjacent tissues. As shown in figure 2, the location of Kif11 expression was detected primarily in the cytoplasm of CCRCC tissues. According to the results of the χ 2 test with included data, a high Kif11 expression level was detected in 62.24% (89/143) of CCRCC tissues, significantly higher (p<0.001 with McNemar test) than that in 34.26% (49/143) of non-cancerous tissues. PCNA was mainly expressed in the nucleus of tumour cells in CCRCC tissues.
Figure 2.
Representative images of the kinesin family member 11 (Kif11) protein and proliferating cell nuclear antigen (PCNA) location and expression in clear cell renal cell carcinoma (CCRCC) tissues. (A) No immunohistochemistry (IHC) staining of Kif11 in the cytoplasm of normal kidney cells. (B) High IHC staining of Kif11 in the cytoplasm of normal kidney cells. (C) Low IHC staining of Kif11 in the cytoplasm of CCRCC cells. (D) Moderate IHC staining of Kif11 in the cytoplasm of CCRCC cells. (E) High IHC staining of Kif11 in the cytoplasm of CCRCC cells. (F) Low IHC staining of PCNA in the nucleus of CCRCC cells. (G) Moderate IHC staining of PCNA in the nucleus of CCRCC cells. (H) High IHC staining of PCNA in the nucleus of CCRCC cells. Bar=50 µm.
Correlations between Kif11 protein expression and clinicopathological attributes of patients with CCRCC
The correlations between Kif11 protein levels and clinicopathological attributes of patients with CCRCC were investigated and are shown in table 1. Higher expression of Kif11 was significantly associated with tumour nuclear grade (p=0.021) and TNM stage (p<0.001). Higher expression of PCNA was associated with tumour nuclear grade (p=0.039) and TNM stage (p=0.001). However, Kif11 protein and PCNA protein expression in CCRCC cells was not significantly associated with age, gender, location or tumour sizes. Kif11 and PCNA expression was positive in 65 cases and negative in 40 cases, with weak positive correlation (χ2=30.165, r=0.459, p<0.01).
Table 1.
Association of Kif11 and PCNA expression with clinical characteristics and selected biological markers of CCRCC
| Characteristic | n | Kif11 expression (%) | PCNA expression (%) | ||||||
| High | Low or none | χ2 | P value | High | Low or none | χ2 | P value | ||
| Age (years) | 0.245 | 0.621 | 0.200 | 0.655 | |||||
| ≤60 | 70 | 45 (64.29) | 25 (35.71) | 40 (57.14) | 30 (42.9) | ||||
| >60 | 73 | 44 (60.27) | 29 (39.73) | 39 (53.42) | 34 (46.6) | ||||
| Location | 0.632 | 0.426 | 0.110 | 0.740 | |||||
| Left | 76 | 45 (59.21) | 31 (40.79) | 41 (53.95) | 35 (46.1) | ||||
| Right | 67 | 44 (65.67) | 23 (34.33) | 38 (56.72) | 29 (43.3) | ||||
| Gender | 1.483 | 0.223 | 0.414 | 0.520 | |||||
| Male | 100 | 59 (59.00) | 41 (41.00) | 57 (57.00) | 43 (43.0) | ||||
| Female | 43 | 30 (69.77) | 13 (30.23) | 22 (51.16) | 21 (48.8) | ||||
| Tumour size (cm) | 2.483 | 0.115 | 1.526 | 0.217 | |||||
| ≤7 | 70 | 39 (55.71) | 31 (44.29) | 35 (50.00) | 35 (50.0) | ||||
| >7 | 73 | 50 (68.49) | 23 (31.51) | 44 (60.27) | 29 (39.7) | ||||
| Nuclear grade | 7.737 | 0.021 | 6.513 | 0.039 | |||||
| G1 | 52 | 25 (48.08) | 27 (51.92) | 22 (42.31) | 30 (57.69) | ||||
| G2 | 61 | 41 (67.21) | 20 (32.79) | 36 (59.02) | 25 (40.98) | ||||
| G3–4 | 30 | 23 (76.67) | 7 (23.33) | 21 (70.00) | 9 (30.00) | ||||
| TNM stage | 22.55 | <0.001 | 13.030 | 0.001 | |||||
| Ⅰ | 57 | 22 (38.60) | 35 (61.40) | 21 (36.84) | 36 (63.16) | ||||
| Ⅱ | 60 | 47 (78.33) | 13 (21.67) | 40 (66.67) | 20 (33.33) | ||||
| Ⅲ+Ⅳ | 26 | 20 (76.92) | 6 (23.08) | 18 (69.23) | 8 (30.77) | ||||
CCRCC, clear cell renal cell carcinoma; Kif11, kinesin family member 11; PCNA, proliferating cell nuclear antigen.
High Kif11 protein expression predicts poor prognosis in patients with CCRCC
Subsequently, the univariable analyses were conducted to evaluate the Kif11 protein expression and clinicopathological factors of patients with CCRCC. The results showed that high expression levels of Kif11 (HR=4.592, p=0.001), nuclear grade (HR=2.839, p<0.001) and TNM stage (HR=2.571, p<0.001) were significantly associated with poor overall survival of patients with CCRCC (table 2). These prognostic factors were further evaluated by multivariable Cox proportional hazards regression model analyses. With the Kaplan-Meier analysis, high Kif11 expression (HR=2.782, p=0.022), nuclear grade (HR=2.432, p<0.001) and TNM stage (HR=2.320, p=0.001) were all independent prognostic markers of poor 5-year overall survival (table 2). Patients with CCRCC with high expression of Kif11 protein had significantly shorter overall survival compared with those with low or no Kif11 expression (figure 3).
Table 2.
Univariable and multivariable analyses of prognostic factors in CCRCC for 5-year overall survival
| Univariable analyses | Multivariable analyses | |||||
| HR | P value | 95% CI | HR | P value | 95% CI | |
| Expression of Kif11 | ||||||
| High vs low or no expression | 4.592 | 0.001 | 1.937 to 10.891 | 2.782 | 0.022 | 1.162 to 6.664 |
| Age (years) | ||||||
| ≤60 vs >60 | 1.453 | 0.227 | 0.793 to 2.664 | |||
| Location | ||||||
| Left vs right | 0.720 | 0.293 | 0.391 to 1.328 | |||
| Gender | ||||||
| Male vs female | 1.114 | 0.739 | 0.589 to 2.109 | |||
| Nuclear grade | ||||||
| G1 vs G2 vs G3–4 | 2.839 | <0.001 | 1.855 to 4.346 | 2.432 | <0.001 | 1.613 to 3.665 |
| Tumour size (cm) | ||||||
| ≤7 cm vs>7 | 0.727 | 0.299 | 0.398 to 1.327 | |||
| PCNA | ||||||
| High vs low | 1.815 | 0.067 | 0.959 to 3.437 | |||
| TNM stage | ||||||
| I vs Ⅱ vs Ⅲ+Ⅳ | 2.571 | <0.001 | 1.69 to 3.90 | 2.32 | 0.001 | 1.440 to 3.737 |
CCRCC, clear cell renal cell carcinoma; Kif11, kinesin family member 11; PCNA, proliferating cell nuclear antigen.
Figure 3.
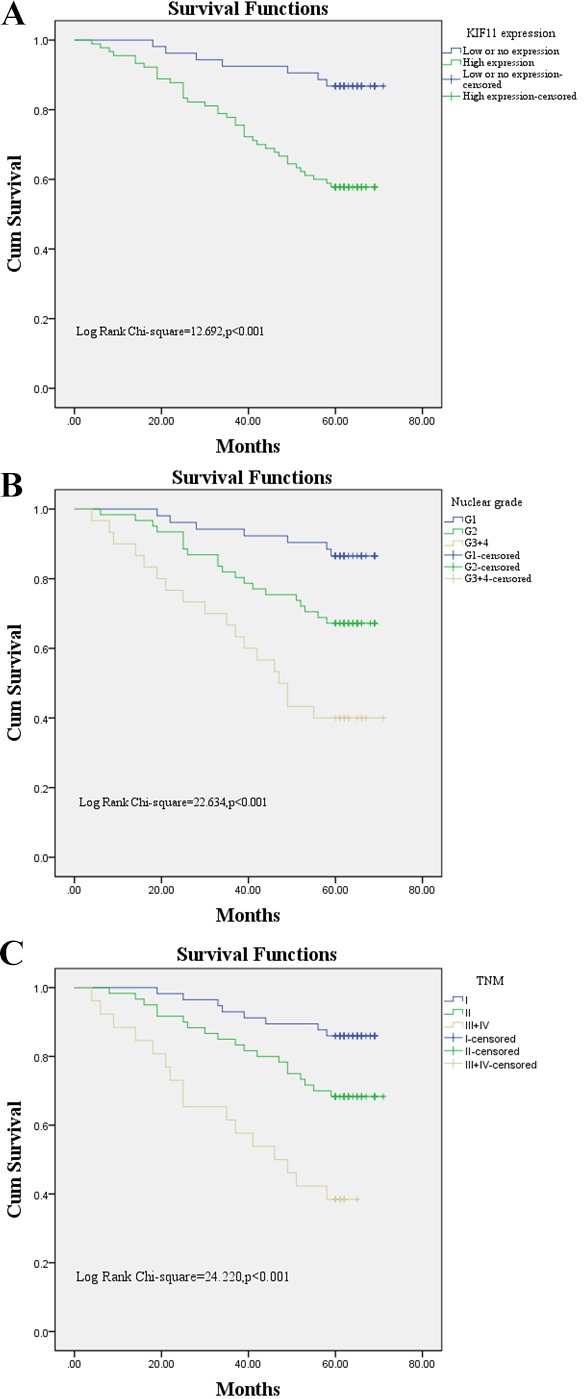
Survival analyses of patients with clear cell renal cell carcinoma (CCRCC) using the Kaplan-Meier test. (A) Overall survival rate in patients with CCRCC and high expression levels of kinesin family member 11 (Kif11) (green line) was statistically lower than that in patients with CCRCC and low or no Kif11 expression (blue line). (B) Overall survival rate in patients with CCRCC and nuclear grade G3–4 was statistically higher than that in patients with CCRCC and nuclear grade G1 and G2. (C) Overall survival rate in patients with CCRCC and TNM stage Ⅲ+Ⅳ was statistically higher than that in patients with CCRCC and TNM stageⅠ and Ⅱ.
Growth inhibition in kidney cancer cells by specific inhibition of Kif11 expression
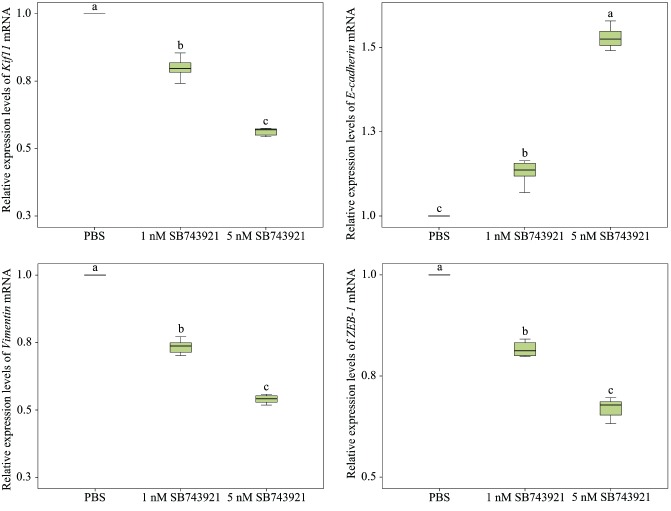
To confirm the effect of Kif11 expression on kidney cancer cells, 786-O cells were treated with SB743921. The results of CCK8 showed that inhibition of Kif11 expression with SB743921 significantly decreased the proliferation rates of 786-O cells in a dose-dependent manner (figure 4A). To elucidate the effect of Kif11 on kidney cancer cell apoptosis, we further performed FACS after SB743921 treatments. The results of Annexin V assay showed that inhibition of Kif11 expression significantly increased the early apoptosis rate in 786-O cells (figure 4B). The migration results showed that with the inhibition of Kif11, the migration abilities of 786-O cells significantly decreased (figure 4C). The EMT process was further analysed. After 72 hours of incubation with SB743921, the results of qRT-PCR showed that the EMT process significantly decreased in a dose-dependent manner with SB743921 treatments (figure 5). The results showed that the E-cadherin mRNA relative expression levels significantly increased (p<0.05) with Vimentin and ZEB-1 mRNA relative expression levels significantly decreased (p<0.05). Meanwhile, the Kif11 mRNA relative expression levels significantly decreased in a dose-dependent manner with SB743921 treatments (p<0.05).
Figure 4.
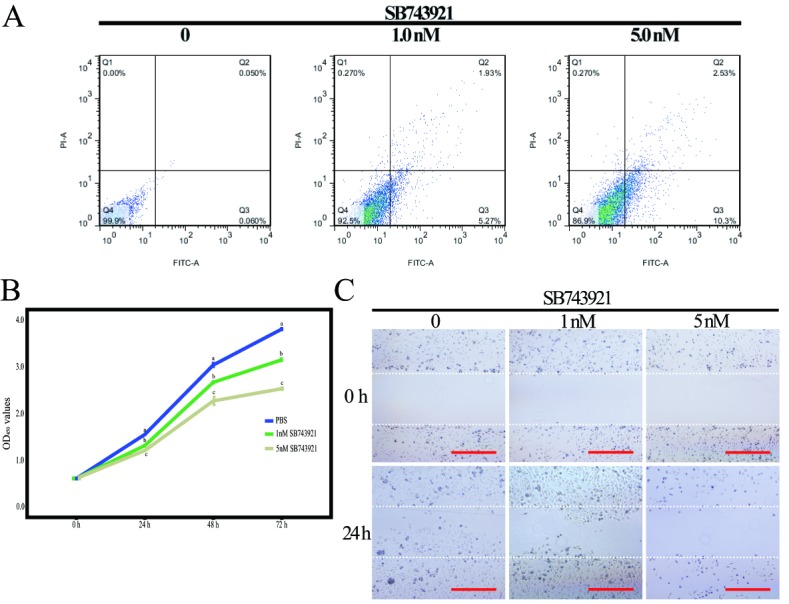
Inhibition of kinesin family member 11 (Kif11) in 786-O cells by SB743921 treatments. (A) Early apoptosis rate of 786-O cells by SB743921 treatments by FACS. (B) Effects of SB743921 on 786-O cell growth over 72 hours. Different letters indicate significant difference (p<0.05). (C) Representative micrographs of the motility of 786-O cells in the wound healing assay at 0 and 24 hours. PBS, phosphate-buffered saline.
Figure 5.
Expression of epithelial to mesenchymal transition (EMT) related genes and kinesin family member 11 (Kif11) mRNA in 786-O cells with SB743921 treatments. Different letters in each column indicate significant difference (p<0.05). PBS, phosphate-buffered saline.
Survival analyses of target gene
High expression of Kif11 confers a survival disadvantage to patients with ACC (log rank p=2.8e-06, HR (high)=5.30), patients with BLCA (log rank p=0.045, HR (high)=1.40), patients with KICH (log rank p=0.02, HR (high)=5.20), patients with KIRP (log rank p=0.00028, HR (high)=3.00), patients with SARC (log rank p=0.00057, HR (high)=1.90), patients with LIHC (log rank p=0.00024, HR (high)=1.80), patients with AAD (log rank p=0.043, HR (high)=1.60), patients with LGG (log rank p=0.0087, HR (high)=1.50) and patients with UVM (log rank p=0.014, HR (high)=3.30) (figure 6).
Figure 6.
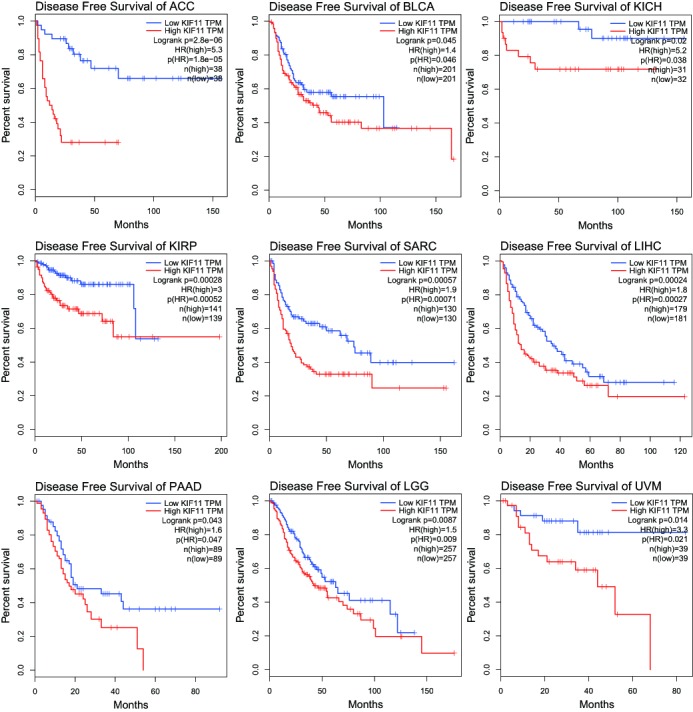
High expression of kinesin family member 11 (Kif11) was correlated with poorer prognosis in different cancers. Overall survival rates in patients with ACC, BLCA, KICH, KIRP, SARC, LIHC, LGG, PAAD and UVM with high expression levels of Kif11 protein (red line) were statistically lower than those in patients with low and no Kif11 expression (blue line). ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; LGG, brain lower grade glioma; KICH, kidney chromophobe; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma;PAAD, pancreatic adenocarcinoma; SARC, sarcoma; TPM, transcripts per kilobase of exonmodel per million mapped reads; UVM. uveal melanoma.
The relationship between Kif11, PCNA and VHL showed that in KIRC tissues, the expression of Kif11 was positively related with that of PCNA (p<0.01, R=0.74) and VHL (p<0.01, R=0.47) (figure 7).
Figure 7.
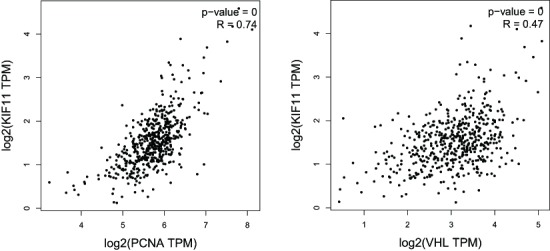
Relationship between kinesin family member 11 (Kif11), proliferating cell nuclear antigen (PCNA) and the von Hippel-Lindau disease tumour suppressor gene (VHL) in kidney renal clear cell carcinoma tissues. TPM, transcripts per kilobase of exonmodel per million mapped reads.
Discussion
To our knowledge, the management of CCRCC remains a therapeutic challenge for renal diseases because of its late presentation and resistance to chemotherapy.22 For patients with local disease, surgical resection represents the only curative treatment modality. However, more than 30% of patients who undergo resection of localised disease still present with recurrence, metastasis or experience progression.23 Consequently, significant attention has been directed to the optimisation of novel prognostic markers, which can be related to tumour recurrence and progression to predict prognosis of patients with CCRCC.
Unlike most other kinesins characterised thus far, full-length Kif11 is a homotetrameric, bipolar protein composed of four identical motor domains, with two motor domains at each end of a rod.8 Three of the Kif11 domains are assigned with the roles of kinesin function differentiation7 and the N-terminal globular motor domain performs the functions of microtubule binding and nucleotide hydrolysis. Variations in sequence in the tail, interrupted coiled-coil region, and neck-linker/cover neck24 dictate the specific motor proteins binding cargo, particular oligomerisation states, and control directionality of net motion of the motor domain, respectively.7 Pilocytic astrocytoma, diffuse astrocytoma, anaplastic astrocytoma and glioblastoma have Kif11 expression with a mean of 18.60% positive neoplastic cells, 13.48% positive neoplastic cells, 18.60% positive neoplastic cells, 43.59% positive neoplastic cells and 76.88% positive neoplastic cells, respectively.25 In Alzheimer's disease, the inhibition of Kif11 by amyloid beta (Aβ) could block neuronal function and cause neuronal dysfunction with the mechanism of reduced transport of neurotrophin and neurotransmitter receptors to the cell surface.26 Kif11 has been implicated in tumourigenesis and Kif11 inhibitors such as monastrol and ispinesib have been successfully used in clinical treatment.27 Dimethylenastron, one of the Kif11 inhibitors, effectively inhibited tumour cell proliferation and induced apoptosis of pancreatic cancer cells in nude mouse xenografts via the c-Jun NH2 kinase pathway.28–30 The loss of Kif11 activity by mutation,31 immunodepletion,32 inhibition33 or by RNA interference34 could result in spindle assembly defects, cell cycle arrest and apoptosis, which leads to cell death.8 35 Moreover, transgenic mice with overexpressed Kif11 expression exhibited high tendencies to develop different malignancies.36 The above results indicate that Kif11 may act as an oncogene during cancer development.
In this study, expression levels of Kif11 in CCRCC tissue samples and matched non-cancerous samples were detected with the observation that Kif11 mRNA and protein levels were significantly higher in CCRCC tumour tissues than those in the corresponding adjacent non-cancerous tissues, which is consistent with previous studies.37 In our study, high Kif11 protein levels were associated with poor overall survival in patients with CCRCC, which indicates that the Kif11 protein level may be used as an independent clinical marker in predicting unfavourable prognosis.
Different expression locations of Kif11 in tumours or cell lines, such as nuclear or cytoplasmic,30 have been reported. We mainly observed the cytoplasmic expression of Kif11 in CCRCC tissues, and rarely detected the nuclear expression of Kif11 in CCRCC tissues. We suggest that these differences are probably due to the differences in tumour types, antibodies and fixation fluid used.
Kinesin inhibitors have been shown to exhibit strong antitumour activities38 39 with many clinical studies of kinesin inhibitors ongoing. In our study, SB743921 was selected as a Kif11 inhibitor with the inhibition of the ATPase activities of Kif11.21 40 Our results showed that SB743921 decreased the proliferation of 786-O cells with subsequent apoptosis and inhibition of the EMT process. Targeting KIF11 with small molecule inhibitors, such as SB743921, may be a powerful approach for kidney cancer treatment.
Future studies that include larger numbers of tumour samples, different CCRCC cell lines or transgenic mice are necessary to further investigate the role of Kif11 in CCRCC tumour biology. Taken together, we analysed the Kif11 expression in CCRCC and demonstrated that Kif11 is involved in CCRCC development. The findings indicate that Kif11 may act as an oncogene during CCRCC development and Kif11 could be identified as a novel prognostic biomarker in CCRCC clinical treatment.
Take home messages.
High kinesin family member 11 (Kif11) expression, nuclear grade and TNM stage were independent factors to predict poor prognosis in patients with clear cell renal cell carcinoma (CCRCC).
High Kif11 expression was associated with unfavourable prognosis in CCRCC and could be used as a potential prognostic marker in the clinical diagnosis of CCRCC.
Inhibition of Kif11 expression in 786-O cells by SB743921 suppressed cell proliferation, migration and the epithelial to mesenchymal transition process with increased apoptosis rate.
Footnotes
QJ and YD contributed equally.
Handling editor: Cheok Soon Lee.
Contributors: Data curation, QJ, YD and YW; Formal analysis, QJ and SZ; Funding acquisition, YD; Investigation, QJ, YD, YW, SZ and GL; Methodology, YD, YW, SZ and GL; Project administration, QJ; Resources, QJ, SZ; Supervision, GL; Writing-original draft, GL; Writing-review & editing, GL.
Funding: This work was supported by the Inner Mongolia Science and Technology Research Project (2017ZD04).
Competing interests: None declared.
Patient consent for publication: Obtained.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin 2017;67:7–30. 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Siegel R, Xu J, et al. Cancer statistics, 2010. CA Cancer J Clin 2010;60:277–300. 10.3322/caac.20073 [DOI] [PubMed] [Google Scholar]
- 3. Vera-Badillo FE, Templeton AJ, Duran I, et al. Systemic therapy for non-clear cell renal cell carcinomas: a systematic review and meta-analysis. Eur Urol 2015;67:740–9. 10.1016/j.eururo.2014.05.010 [DOI] [PubMed] [Google Scholar]
- 4. Zisman A, Pantuck AJ, Wieder J, et al. Risk group assessment and clinical outcome algorithm to predict the natural history of patients with surgically resected renal cell carcinoma. J Clin Oncol 2002;20:4559–66. 10.1200/JCO.2002.05.111 [DOI] [PubMed] [Google Scholar]
- 5. Finley DS, Pantuck AJ, Belldegrun AS. Tumor biology and prognostic factors in renal cell carcinoma. Oncologist 2011;16 Suppl 2(Suppl 2):4–13. 10.1634/theoncologist.2011-S2-04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yao M, Yoshida M, Kishida T. VHL tumor suppressor gene alterations associated with good prognosis in sporadic clear-cell renal carcinoma. CancerSpectrum Knowledge Environment 2002;94:1569–75. 10.1093/jnci/94.20.1569 [DOI] [PubMed] [Google Scholar]
- 7. Wojcik EJ, Buckley RS, Richard J, et al. Kinesin-5: cross-bridging mechanism to targeted clinical therapy. Gene 2013;531:133–49. 10.1016/j.gene.2013.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kapitein LC, Peterman EJ, Kwok BH, et al. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 2005;435:114–8. 10.1038/nature03503 [DOI] [PubMed] [Google Scholar]
- 9. Valentine MT, Gilbert SP. To step or not to step? How biochemistry and mechanics influence processivity in kinesin and Eg5. Curr Opin Cell Biol 2007;19:75–81. 10.1016/j.ceb.2006.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tanenbaum ME, Macůrek L, Janssen A, et al. Kif15 cooperates with Eg5 to promote bipolar spindle assembly. Curr Biol 2009;19:1703–11. 10.1016/j.cub.2009.08.027 [DOI] [PubMed] [Google Scholar]
- 11. Chauvière M, Kress C, Kress M. Disruption of the mitotic kinesin Eg5 gene (Knsl1) results in early embryonic lethality. Biochem Biophys Res Commun 2008;372:513–9. 10.1016/j.bbrc.2008.04.177 [DOI] [PubMed] [Google Scholar]
- 12. Fernández JP, Agüero TH, Vega López GA, et al. Developmental expression and role of Kinesin Eg5 during Xenopus laevis embryogenesis. Dev Dyn 2014;243:527–40. 10.1002/dvdy.24094 [DOI] [PubMed] [Google Scholar]
- 13. Piao XM, Byun YJ, Jeong P, et al. Kinesin family member 11 mRNA expression predicts prostate cancer aggressiveness. Clin Genitourin Cancer 2017;15:450–4. 10.1016/j.clgc.2016.10.005 [DOI] [PubMed] [Google Scholar]
- 14. Jin Q, Huang F, Wang X, et al. High Eg5 expression predicts poor prognosis in breast cancer. Oncotarget 2017;8 10.18632/oncotarget.19215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kato T, Lee D, Wu L, et al. Kinesin family members KIF11 and KIF23 as potential therapeutic targets in malignant pleural mesothelioma. Int J Oncol 2016;49:448–56. 10.3892/ijo.2016.3566 [DOI] [PubMed] [Google Scholar]
- 16. Sun XD, Shi XJ, Sun XO, et al. Dimethylenastron suppresses human pancreatic cancer cell migration and invasion in vitro via allosteric inhibition of mitotic kinesin Eg5. Acta Pharmacol Sin 2011;32:1543–8. 10.1038/aps.2011.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lu M, Zhu H, Wang X, et al. The prognostic role of Eg5 expression in laryngeal squamous cell carcinoma. Pathology 2016;48:214–8. 10.1016/j.pathol.2016.02.008 [DOI] [PubMed] [Google Scholar]
- 18. Imai T, Oue N, Nishioka M, et al. Overexpression of KIF11 in gastric cancer with intestinal mucin phenotype. Pathobiology 2017;84:16–24. 10.1159/000447303 [DOI] [PubMed] [Google Scholar]
- 19. Lin H, Huang B, Wang H, et al. MTHFD2 overexpression predicts poor prognosis in renal cell carcinoma and is associated with cell proliferation and Vimentin-Modulated migration and invasion. Cell Physiol Biochem 2018;51:991–1000. 10.1159/000495402 [DOI] [PubMed] [Google Scholar]
- 20. Xue S, Li QW, Che JP, et al. Decreased expression of long non-coding RNA NBAT-1 is associated with poor prognosis in patients with clear cell renal cell carcinoma. Int J Clin Exp Pathol 2015;8:3765–74. [PMC free article] [PubMed] [Google Scholar]
- 21. Daigo K, Takano A, Thang PM, et al. Characterization of KIF11 as a novel prognostic biomarker and therapeutic target for oral cancer. Int J Oncol 2018;52:155–65. 10.3892/ijo.2017.4181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Buzaid AC, Todd MB. Therapeutic options in renal cell carcinoma. Semin Oncol 1989;16(1 Suppl 1):12–19. [PubMed] [Google Scholar]
- 23. Cohen HT, McGovern FJ. Renal-cell carcinoma. N Engl J Med 2005;353:2477–90. 10.1056/NEJMra043172 [DOI] [PubMed] [Google Scholar]
- 24. Hesse WR, Steiner M, Wohlever ML, et al. Modular aspects of kinesin force generation machinery. Biophys J 2013;104:1969–78. 10.1016/j.bpj.2013.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu L, Liu X, Mare M, et al. Overexpression of Eg5 correlates with high grade astrocytic neoplasm. J Neurooncol 2016;126:77–80. 10.1007/s11060-015-1954-3 [DOI] [PubMed] [Google Scholar]
- 26. Ari C, Borysov SI, Wu J, et al. Alzheimer amyloid beta inhibition of Eg5/kinesin 5 reduces neurotrophin and/or transmitter receptor function. Neurobiol Aging 2014;35:1839–49. 10.1016/j.neurobiolaging.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu Y, Feng YM. The role of kinesin family proteins in tumorigenesis and progression: potential biomarkers and molecular targets for cancer therapy. Cancer 2010;116:5150–60. 10.1002/cncr.25461 [DOI] [PubMed] [Google Scholar]
- 28. Liu M, Yu H, Huo L, et al. Validating the mitotic kinesin Eg5 as a therapeutic target in pancreatic cancer cells and tumor xenografts using a specific inhibitor. Biochem Pharmacol 2008;76:169–78. 10.1016/j.bcp.2008.04.018 [DOI] [PubMed] [Google Scholar]
- 29. Liu M, Aneja R, Sun X, et al. Parkin regulates Eg5 expression by Hsp70 ubiquitination-dependent inactivation of c-Jun NH2-terminal kinase. J Biol Chem 2008;283:35783–8. 10.1074/jbc.M806860200 [DOI] [PubMed] [Google Scholar]
- 30. Xing ND, Ding ST, Saito R, et al. A potent chemotherapeutic strategy in prostate cancer: S-(methoxytrityl)-L-cysteine, a novel Eg5 inhibitor. Asian J Androl 2011;13:236–41. 10.1038/aja.2010.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hagan I, Yanagida M. Novel potential mitotic motor protein encoded by the fission yeast cut7+ gene. Nature 1990;347:563–6. 10.1038/347563a0 [DOI] [PubMed] [Google Scholar]
- 32. Blangy A, Lane HA, d'Hérin P, et al. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 1995;83:1159–69. [DOI] [PubMed] [Google Scholar]
- 33. Mayer TU, Kapoor TM, Haggarty SJ, et al. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science 1999;286:971–4. 10.1126/science.286.5441.971 [DOI] [PubMed] [Google Scholar]
- 34. Weil D, Garçon L, Harper M, et al. Targeting the kinesin Eg5 to monitor siRNA transfection in mammalian cells. Biotechniques 2002;33:1244–8. 10.2144/02336st01 [DOI] [PubMed] [Google Scholar]
- 35. Duan Y, Huo D, Gao J, et al. Ubiquitin ligase RNF20/40 facilitates spindle assembly and promotes breast carcinogenesis through stabilizing motor protein Eg5. Nature Communications 2016;7:12648 10.1038/ncomms12648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Castillo A, Morse HC, Godfrey VL, et al. Overexpression of Eg5 causes genomic instability and tumor formation in mice. Cancer Res 2007;67:10138–47. 10.1158/0008-5472.CAN-07-0326 [DOI] [PubMed] [Google Scholar]
- 37. Sun D, Lu J, Ding K, et al. The expression of Eg5 predicts a poor outcome for patients with renal cell carcinoma. Med Oncol 2013;30:476 10.1007/s12032-013-0476-0 [DOI] [PubMed] [Google Scholar]
- 38. Sadakane K, Alrazi IMD, Maruta S. Highly efficient photocontrol of mitotic kinesin Eg5 ATPase activity using a novel photochromic compound composed of two azobenzene derivatives. J Biochem 2018;164:295–301. 10.1093/jb/mvy051 [DOI] [PubMed] [Google Scholar]
- 39. Milic B, Chakraborty A, Han K, et al. Kif15 nanomechanics and kinesin inhibitors, with implications for cancer chemotherapeutics. Proceedings of the National Academy of Sciences 2018;115:E4613–E4622. 10.1073/pnas.1801242115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhu L, Xiao F, Yu Y, et al. KSP inhibitor SB743921 inhibits growth and induces apoptosis of breast cancer cells by regulating p53, Bcl-2, and DTL. Anticancer Drugs 2016;27:863–72. 10.1097/CAD.0000000000000402 [DOI] [PMC free article] [PubMed] [Google Scholar]